Summary
We have begun an early phase of biomarker discovery in three clinically important types of breast cancer using a panel of human cell lines: HER2 positive, HER2 negative and hormone receptor positive and triple negative (HER2−, ER−, PR−). We identified and characterized the most abundant secreted, sloughed, or leaked proteins released into serum free media from these breast cancer cell lines using a combination of protein fractionation methods before LC-MS/MS mass spectrometry analysis. A total of 249 proteins were detected in the proximal fluid of 7 breast cancer cell lines. The expression of a selected group of high abundance and/or breast cancer specific potential biomarkers including thromobospondin 1, galectin-3 binding protein, cathepsin D, vimentin, zinc-α2-glycoprotein, CD44, and EGFR from the breast cancer cell lines and in their culture media were further validated by Western blot analysis. Interestingly, mass spectrometry identified a cathepsin D protein single-nucleotide polymorphism (SNP) by alanine to valine replacement from the MCF-7 breast cancer cell line. Comparison of each cell line media proteome displayed unique and consistent biosignatures regardless of the individual group classifications demonstrating the potential for stratification of breast cancer. Based on the cell line media proteome, predictive Tree software was able to categorize each cell line as HER2 positive, HER2 negative and hormone receptor positive and triple negative based on only two proteins, muscle fructose 1,6-bisphosphate aldolase and keratin 19. In addition, the predictive Tree software clearly identified MCF-7 cell line overexpresing the HER2 receptor with the SNP cathepsin D biomarker.
Keywords: breast cancer, biomarkers, blood assay, proteomics, mass spectrometry, single-nucleotide polymorphism (SNP), HER2, triple negative, proximal fluid
Introduction
Early detection is one of the most effective means to decrease cancer mortalities1. Protein biomarker discovery by mass spectrometry research represents a promising new approach to improve cancer detection and enable earlier treatment2. Currently mammographic screening is the gold standard in early detection of breast cancer3, 4. However, mammography frequently fails to detect tumors in women with increased density in breast tissue and those with lobular cancer5–7. In addition, routine mammography screening is unaffordable to many women even in the USA and the vast majority of the developing world8. The recent age-related controversy on the inability of mammography to detect tumors in younger women as well as a significant level of false positives in different age groups9, 10 further underlies the importance in developing alternative techniques. Our goal is to develop a more affordable and easily obtainable screening tool for the early detection and characterization of breast cancer. Plasma/serum is the most suitable clinical specimen for biomarker research because it is attainable by non-invasive means, extraction is feasible, and it is likely to contain tumor markers11. However, both patient populations and breast cancer are heterogeneous in nature12 which can complicate the discovery phase of the assay development. Furthermore, there are two major technical hurdles in identifying disease-related protein biomarkers in serum. First, protein concentrations vary by 10–11 orders of magnitude in serum with the useful biomarkers in the lower end of this spectrum. Second, the 20 most abundant serum proteins that make up more than 99% of total protein mass can obscure the finding of low abundance proteins13, 14. Both the wide concentration range and the interference of the high abundance proteins can mask detection of less abundant serum proteins. One way to circumvent these hurdles is to begin by analyzing breast cancer cell lines and their released proteins15 for comparison with matching biopsied tumor samples.
The analysis of proximal fluid from a homogenous cancer source, provides a pool of leaked, secreted and sloughed proteins that may be similar to the proteins found in the interstitial fluid of tumor tissue. Most of the clinically useful tumor markers such as prostrate specific antigen (PSA), cancer antigen 125 (CA125), carcinoembryonic antigen (CEA) and alpha-fetoprotein (AFP) are membrane proteins16, 17. These proteins may be released into the interstitial fluids thus enter the patients' blood circulation18–20. The additional advantages of studying the in vitro proximal fluid are reduced levels of human serum proteins and higher concentrations of tumor-related proteins allowing the identification by mass spectrometry.
Mass spectrometry currently is capable of identifying the proximal fluid proteome at a dynamic range of 1–4 orders of magnitude21, 22. Applying this approach to cancer proximal fluid biomarker discovery and characterization improves the likelihood of detecting new biomarkers that are present in serum at a dilution of 10-11 orders of magnitude13. When candidate biomarkers are identified and validated not only from studying the cancer cell lines and their proximal fluid, but also cancer tissue, then an effective, practical, and highly sensitive validation assay can be developed with corresponding antibodies to the new biomarkers. In addition, the identified peptides transition states from candidate biomarkers may be used for selective ion monitoring (SIM) during mass spectrometry analysis to screen small volumes of human serum samples while ignoring the high abundant serum proteins23.
Breast cancer is heterogeneous with alternate splicing leading to multiple protein expression, function and activity from a single gene. Currently, breast cancers are grouped into 3 clinical types, HER2 positive, estrogen receptor (ER) and/or progesterone (PR) positive/HER2 negative and triple negative based on the presence or absence of these three biomarkers24. The clinical classification of breast cancers determines the type of adjuvant therapy and predicts clinical outcomes of women with different types of breast cancer25. However, despite targeted treatment of these three breast cancer markers, the successful outcomes are not uniform with many recurrences after the initial treatment25–27. In addition, these three breast cancer markers are typically expressed in lower quantities making them difficult to identify in serum/plasma for screening use. Carcinoma antigen 27.29 (CA27.29, MUC1) and CEA are the only two circulation-borne and breast cancer related biomarkers used clinically, however lack of sensitivity and specificity in blood assays result in no early detection of breast cancer and although elevated levels in blood reflect recurrent/metastatic disease, normal levels may not indicate the lack of disease presence18–20, 28–30. Recently, new candidate tumor biomarkers such as keratin 18, keratin 8, EGFR, CD44, as well as others have been reported20, 31, 32. Taken together, further proteomic characterization is needed to refine subtypes of breast cancer, as well as improve early detection, and systemic treatment of breast cancer.
In this study we have begun an effort to identify the breast cancer proteome by LC-MS/MS in the culture media (proximal fluid) of seven human tumor cell lines representing the 3 major types of breast cancer defined clinically. The quantitative presence of several high abundance breast cancer related proteins, was validated in both the breast cancer whole cell lysates and the proximal fluid by antibody assays. Most of the validated proteins were selected based on their overlap with our previous reports of enriched N-linked glycoproteins present in cancer cell membrane fractions by the hydrazide method32 and were also found in hydrophobic fractions33. Interestingly, we also identified a single-nucleotide polymorphism (SNP) in cathepsin D by LC-MS/MS unique to MCF-7 and MCF-7HER2 cell lines. Further development of a sensitive blood assay and mass spectrometry based selective ion monitoring methods will be necessary to determine the significance of these candidate biomarkers in breast cancer patients.
Experimental Procedures
Cell Culture and Media collection
A panel of breast cancer cell lines including HER2 negative and estrogen receptor (ER) and progesterone receptor (PR) positive T47D and MCF-7, HER2 transfected MCF-7 (MCF-7HER2), HER 2 positive SKBR-3 and MDA-MB-453, and triple negative breast cancer (TNBC) MDA-MB-468 and MDA-MB-231 were maintained in DMEM culture medium supplemented with L-glutamine and sodium pyruvate, 10% FBS, and 1% penicillin and streptomycin. Cells were grown to 70% confluency in fifteen 10 cm petri dishes. Media was removed and cells were washed with serum free media three times each, and incubated with serum free media for 12 hours. Serum free media was collected from 5 of the 15 petri dishes, combined into 3 separate sets for each cell line. Cells were harvested by centrifugation (1,000 × g, 2 min 4 °C) and combined in the same manner as media. Both were stored at −80 °C until needed.
Serum free media
Serum free media (proximal fluid) was centrifuged to remove any cellular debris and a 1 ml aliquot of culture media was removed for Western dot blot validation. The remainder or the proximal fluid was concentrated via centricon (3000 MW cut off, Vivaspin 20, Sartorius Biolab Products), speed vacuum dried, solubilized in 40 mM Tris-HCl pH 8.3, 6 M guanidine HCl, 5 mM DTT, centrifuged (15,000 × g, 2 min, RT), and supernatants were diluted with <1 M guanidine HCl with 40 mM Tris-HCl pH 8.3. The sample was sequentially treated with iodoacetamide and trypsin (overnight, 37 °C) according to manufacturers protocol (Promega). The pH of the samples was adjusted to pH 3 with TCA, before centrifugation (15,000 × g, 2 min, RT) and the supernatants were passed through an activated, washed C18 spin column (The Nest Group, Inc.) according to manufacturers protocol.
Whole cell extract preparation
The frozen cell pellets were homogenized in ice cold 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM PMSF, 1% NP40, and protease cocktail (Roche Cat. No. 04693132001) using an ultrasonic cell disrupter (Fisher Scientific, sonic dismembrater model 100, at setting 4 for 2 × 10 seconds at 30 seconds intervals) on ice. Samples were centrifuged at 15,000 × g for 10 min at 4 °C to remove large debris. Protein concentration was determined by Bradford assay (Biorad), proteins were solubilized with Laemli buffer, and subjected to Western blot analysis.
Western blot analysis
Equal amounts of protein were loaded onto 4–12% SDS-PAGE, transferred to nitrocellulose and blotted with the antibodies indicating HER2, MUC1, ERα, thromobospondin 1, galectin-3 binding protein, cathepsin D, vimentin, zinc-α-glycoprotein, CD44, or EGFR (1:2000 dilution; Santa Cruz biotechnology, INC., Santa Cruz, CA). Dot blots were performed on the proximal fluid from an equal, quadruple count of breast cancer cells.
LC-MS/MS analysis
Dried samples were treated as described in Whelan et al.32, by dissolving in Buffer A (H2O/acetonitrile/formic acid, 98.9/1/0.1), separated by nanospray LC (Eskigent technologies, Inc. Dublin, CA), and analyzed by online tandem LTQ Orbitrap mass spectrometry (Thermo Fisher). Aliquots were injected (10 μl) onto a reverse phase column (New Objective C18, 15 cm, 75 μM diameter, 5 μm particle size equilibrated in Buffer A) and eluted (300 nL/min) with an increasing concentration of Buffer B (acetonitrile/water/formic acid, 98.9/1/0.1; min 0/5, 10/10, 112/40, 130/60, 135/90, 140/90). Eluted peptides were analyzed by MS and data-dependent MS/MS acquisition (collision-induced dissociation CID) selecting the 7 most abundant precursor ions for MS/MS with a dynamic exclusion duration of 15.0 seconds.
Data analysis
The mass spectra were searched against a IPI 3.73 human trypsin indexed database (two trypsin missed cleavages), in both the forward and backward directions (decoy database; 179, 304 proteins), with variable modifications of carboxyamidomethylation, deamidation and methionine oxidation using the Proteome Discoverer software 1.3 (Thermo Fisher) based on the SEQUEST algorithm and Mascot (Matrix Science, UK). Quantitative data analysis was performed using the Scaffold 3.43 (Proteome Software, Inc.) program. The Proteome Discoverer search results were uploaded into the scaffold software program and a filter with a 99% minimum protein ID probability (calculated probability of correct protein identification), with a minimum number of 2 unique peptides for one protein and a stringent minimum peptide ID probability of 95% was set (all data and parameters are included in supplemental data). Scaffold verifies peptide identification derived from MS/MS sequencing results using X! Tandem34 ProteinProphet computer algorithms35. Scaffold normalizes MS/MS data between samples with similar total protein amounts by averaging the spectral counts for all the samples and then multiplying the spectral counts in each sample by the average divided by the individual sample's sum. The proximal fluid from each breast cancer cell line is represented by three replicates each containing a combination of five separate experiments.
Statistical analysis
One at a time mean intensity comparisons across the 4 cancer cell groups (triple negative breast cancer, HER2+ and hormone negative, hormone positive and HER2 negative, and HER2 transected MCF7) were carried out using one way analysis of variance (ANOVA) methods. The overall F statistic and corresponding p value under this method is reported. A separate comparison is done for each of the top 229 proteins.
Visualization of the high dimensional intensity scattergram was carried out by computing the first two principle components. The principle components were computed from the subset of all proteins that had statistically significant mean intensity differences for at least one group compared to the others.
A multivariate classification tree analysis (CART-classification and regression tree) was also carried out in order to identify a subset of the up the top 229 proteins that best classified the observations into the four groups. The binary recursive partitioning algorithm is used by the tree.
Results
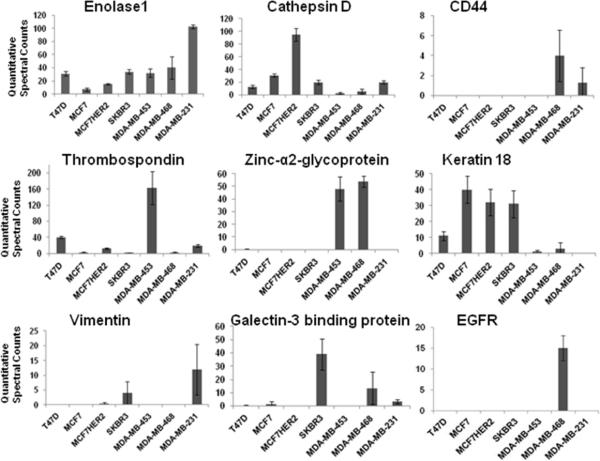
The aim of this study was to identify the most abundant proteins in the proximal fluid of growing breast cancer cells which represent the proteins sloughed off, secreted, leaked or cleaved by proteases from the cells. The breast cancer cell lines were selected instead of cancer tissues as the initial step for new biomarker discovery because they are more homogenous, can be grown to any quantity necessary, and easily manipulated experimentally. They are also readily available to other investigators to reproduce these findings and provide proximal fluid to define new cancer biomarkers. We analyzed the proximal fluids of 7 different breast cancer cell lines in triplicate from the three major categories of breast cancer, HER2 positive (SKBR-3, MDA-MB-453), HER2 negative and hormone receptor positive (T47D, MCF-7), triple negative (MDA-MB-468, MDAMB-231), and MCF-7 transfected with HER2 (MCF-7HER2). We used the MCF-7HER2 cell line to assess if it resembles its native HER2 negative parental cancer cell line or the group of HER2 positive breast cancer cell lines. The proximal fluid of each cell line was concentrated, digested and subjected to LCMS/MS mass spectrometry. All mass spectrometry data was searched using Bioworks and Mascot against the IPI human 3.73 database with a built in decoy database, then uploaded into Scaffold, to quantitatively analyze the proximal proteins of all 7 breast cancer cell lines (Table 1, top 50 proteins). While uploading data in Scaffold the data was verified by X! Tandem. A total of 249 proteins were identified in the serum free media from 7 breast cancer cell lines with a stringent scaffold filter setting with a 99% minimum protein ID probability and a minimum of 2 unique peptides for one protein (Supplemental Table 1, all 249 proteins). In addition, the averages of triplicate experiments along with the standard deviation was calculated for each identified protein (Supplemental Table 1). The standard deviation of the triplicate samples for each cell line demonstrate significant reproducibility. A significant portion of the proximal fluid proteins are localized in the extracellular region (76 proteins) and plasma membrane (54 proteins) and have diverse biological functions and processes (Supplemental Figures 2–4). However, more importantly several proteins were found to vary considerably in the serum free media of the 7 breast cancer cell lines, thrombospondin 1, galectin-3 binding protein, cathepsin D, vimentin, zinc-α2-glycoprotein (ZAG), CD44, EGFR, keratin 18 and enolase as shown in Figure 1. Seven of these proteins were selected for further validation in the whole cell lysates of each cell line and their respective proximal fluid. The criteria for selecting these proteins for validation was based on their high abundance and differential expression in the different breast cancer groups.
Table 1.
Top 50 proteins in the proximal fluid of 7 breast cancer cell lines.
| # | Identified Proteins (Top 50) | Acc. No. | M.W. | T47D | MCF-7 | MCF-7 HER2 | SKBR-3 | MDA-MB-453 | MDA-MB-468 | MDA-MB-231 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | AVG | SD | TP | Avg | SD | TP | Avg | SD | TP | AVG | SD | TP | AVG | SD | TP | AVG | SD | TP | AVG | SD | ||||
| 1 | Enolase 1 | IPI00465248 | 47 kDa | 92 | 30.7 | 3.2 | 20 | 6.7 | 2.1 | 45 | 15.0 | 1.0 | 103 | 34.3 | 3.1 | 97 | 32.3 | 6.1 | 121 | 40.3 | 16.6 | 310 | 103.3 | 3.1 |
| 2 | Actin, cytoplasmic 1 | IPI00021439 | 42 kDa | 92 | 30.7 | 6.8 | 15 | 5.0 | 5.0 | 165 | 55.0 | 3.6 | 60 | 20.0 | 4.0 | 120 | 40.0 | 8.5 | 233 | 77.7 | 18.7 | 117 | 39.0 | 7.9 |
| 3 | Thrombospondin-1 | IPI00296099 | 129 kDa | 119 | 39.7 | 2.5 | 7 | 2.3 | 1.5 | 38 | 12.7 | 2.1 | 3 | 1.0 | 1.0 | 487 | 162.3 | 41.0 | 6 | 2.0 | 2.0 | 60 | 20.0 | 3.5 |
| 4 | Keratin, type I cytoskeletal 19 | IPI00479145 | 44 kDa | 83 | 27.7 | 11.0 | 134 | 44.7 | 10.0 | 47 | 15.7 | 1.2 | 333 | 111.0 | 9.5 | 25 | 8.3 | 4.6 | 37 | 12.3 | 5.0 | 0 | 0.0 | 0.0 |
| 5 | Keratin, type II cytoskeletal 8 | IPI00554648 | 54 kDa | 97 | 32.3 | 6.0 | 128 | 42.7 | 8.4 | 100 | 33.3 | 5.1 | 288 | 96.0 | 3.0 | 10 | 3.3 | 3.2 | 23 | 7.7 | 2.5 | 3 | 1.0 | 1.7 |
| 6 | Cathepsin D | IPI00011229 | 45 kDa | 38 | 12.7 | 2.1 | 93 | 31.0 | 2.6 | 284 | 94.7 | 10.3 | 60 | 20.0 | 3.6 | 8 | 2.7 | 1.2 | 17 | 5.7 | 4.2 | 59 | 19.7 | 2.5 |
| 7 | Peptidyl-prolyl cis-trans isomerase A | IPI00419585 | 18 kDa | 89 | 29.7 | 3.1 | 29 | 9.7 | 3.8 | 49 | 16.3 | 7.6 | 45 | 15.0 | 3.6 | 106 | 35.3 | 6.7 | 175 | 58.3 | 27.4 | 77 | 25.7 | 5.5 |
| 8 | Fructose-bisphosphate aldolase A | IPI00465439 | 39 kDa | 21 | 7.0 | 3.5 | 27 | 9.0 | 2.6 | 68 | 22.7 | 1.2 | 79 | 26.3 | 8.7 | 143 | 47.7 | 4.5 | 15 | 5.0 | 7.0 | 47 | 15.7 | 1.5 |
| 9 | Clusterin | IPI00400826 | 58 kDa | 116 | 38.7 | 2.5 | 207 | 69.0 | 3.5 | 53 | 17.7 | 3.5 | 115 | 38.3 | 6.7 | 1 | 0.3 | 0.6 | 8 | 2.7 | 2.5 | 4 | 1.3 | 1.2 |
| 10 | M2 of Pyruvate kinase isozymes M1/M2 | IPI00479186 | 58 kDa | 26 | 8.7 | 1.5 | 22 | 7.3 | 3.2 | 109 | 36.3 | 4.9 | 42 | 14.0 | 6.1 | 3 | 1.0 | 1.7 | 49 | 16.3 | 19.6 | 150 | 50.0 | 12.1 |
| 11 | Insulin-like growth factor-binding protein 2 | IPI00297284 | 35 kDa | 63 | 21.0 | 1.0 | 70 | 23.3 | 6.7 | 93 | 31.0 | 1.0 | 0 | 0.0 | 0.0 | 143 | 47.7 | 1.2 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| 12 | cDNA FLI55606, Alpha-2-HS-glycoprotein | IPI00022431 | 47 kDa | 9 | 3.0 | 1.0 | 0 | 0.0 | 0.0 | 32 | 10.7 | 2.5 | 16 | 5.3 | 9.2 | 192 | 64.0 | 55.7 | 122 | 40.7 | 28.3 | 19 | 6.3 | 11.0 |
| 13 | Profilin-1 | IPI00216691 | 15 kDa | 32 | 10.7 | 3.8 | 1 | 0.3 | 0.6 | 15 | 5.0 | 1.0 | 35 | 11.7 | 4.9 | 21 | 7.0 | 2.0 | 71 | 23.7 | 16.2 | 110 | 36.7 | 12.7 |
| 14 | Glyceraldehyde-3-phosphate dehydrogenase | IPI00219018 | 36 kDa | 18 | 6.0 | 3.6 | 55 | 18.3 | 4.0 | 52 | 17.3 | 5.7 | 47 | 15.7 | 3.5 | 40 | 13.3 | 2.1 | 58 | 19.3 | 5.0 | 47 | 15.7 | 2.1 |
| 15 | Triosephosphate isomerase isoform 2 | IPI00465028 | 31 kDa | 7 | 2.3 | 0.6 | 46 | 15.3 | 4.7 | 37 | 12.3 | 1.2 | 86 | 28.7 | 6.4 | 18 | 6.0 | 2.6 | 7 | 2.3 | 2.1 | 91 | 30.3 | 13.6 |
| 16 | Cofilin-1 | IPI00012011 | 19 kDa | 20 | 6.7 | 1.5 | 46 | 15.3 | 1.5 | 63 | 21.0 | 2.6 | 31 | 10.3 | 4.7 | 53 | 17.7 | 4.0 | 24 | 8.0 | 2.6 | 48 | 16.0 | 1.0 |
| 17 | Keratin, type I cytoskeletal 18 | IPI00554788 | 48 kDa | 32 | 10.7 | 3.8 | 120 | 40.0 | 8.5 | 95 | 31.7 | 8.1 | 93 | 31.0 | 8.5 | 3 | 1.0 | 1.0 | 8 | 2.7 | 3.8 | 0 | 0.0 | 0.0 |
| 18 | Zinc-alpha-2-glycoprotein | IPI00166729 | 34 kDa | 1 | 0.3 | 0.6 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 144 | 48.0 | 9.5 | 162 | 54.0 | 4.4 | 0 | 0.0 | 0.0 |
| 19 | Agrin | IPI00374563 | 215 kDa | 129 | 43.0 | 9.2 | 62 | 20.7 | 0.6 | 24 | 8.0 | 2.6 | 1 | 0.3 | 0.6 | 7 | 2.3 | 3.2 | 17 | 5.7 | 4.0 | 3 | 1.0 | 1.0 |
| 20 | Tubulin, beta | IPI00645452 | 48 kDa | 53 | 17.7 | 6.0 | 5 | 17 | 2.9 | 21 | 7.0 | 3.0 | 17 | 5.7 | 3.2 | 21 | 7.0 | 2.6 | 85 | 28.3 | 6.0 | 41 | 13.7 | 1.5 |
| 21 | Phosphoglycerate kinase 1 | IPI00169383 | 45 kDa | 6 | 2.0 | 1.0 | 7 | 2.3 | 0.6 | 37 | 12.3 | 2.5 | 18 | 6.0 | 2.6 | 20 | 6.7 | 3.1 | 14 | 4.7 | 4.0 | 131 | 43.7 | 6.7 |
| 22 | Elongation factor 1-alpha 1 | IPI00396485 | 50 kDa | 20 | 6.7 | 1.5 | 9 | 3.0 | 1.0 | 20 | 6.7 | 2.3 | 23 | 7.7 | 2.1 | 74 | 24.7 | 7.0 | 50 | 16.7 | 22.8 | 53 | 17.7 | 2.1 |
| 23 | Glucose-6-phosphate isomerase | IPI00027497 | 63 kDa | 24 | 8.0 | 1.0 | 44 | 14.7 | 5.7 | 33 | 11.0 | 3.5 | 55 | 18.3 | 2.5 | 38 | 12.7 | 1.2 | 20 | 6.7 | 3.8 | 36 | 12.0 | 3.6 |
| 24 | Alpha-actinin-4 | IPI00013808 | 105 kDa | 16 | 5.3 | 1.5 | 24 | 8.0 | 4.0 | 39 | 13.0 | 1.7 | 27 | 9.0 | 1.0 | 7 | 2.3 | 4.0 | 39 | 13.0 | 10.8 | 67 | 22.3 | 3.1 |
| 25 | cDNA FLI60097,Tubulin alpha-ubiquitous chain | IPI00792677 | 46 kDa | 39 | 13.0 | 3.5 | 21 | 7.0 | 1.0 | 15 | 5.0 | 1.0 | 32 | 10.7 | 2.5 | 17 | 5.7 | 3.2 | 58 | 19.3 | 4.0 | 39 | 13.0 | 7.2 |
| 26 | Keratin, type II cytoskeletal 1 | IPI00220327 | 66 kDa | 23 | 7.7 | 2.5 | 5 | 1.7 | 1.5 | 38 | 12.7 | 20.2 | 53 | 17.7 | 5.0 | 39 | 13.0 | 21.7 | 0 | 0.0 | 0.0 | 54 | 18.0 | 21.4 |
| 27 | Corticosteroid-binding globulin | IPI00027482 | 45 kDa | 176 | 58.7 | 12.7 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| 28 | cDNA FLI54957, highly similar to Transketolase | IPI00643920 | 69 kDa | 15 | 5.0 | 1.0 | 39 | 13.0 | 6.1 | 4 | 1.3 | 2.3 | 93 | 31.0 | 4.6 | 38 | 12.7 | 4.0 | 7 | 2.3 | 1.5 | 15 | 5.0 | 2.0 |
| 29 | Insulin-like growth factor-binding protein 5 | IPI00029236 | 31 kDa | 68 | 22.7 | 1.5 | 1 | 0.3 | 0.6 | 0 | 0.0 | 0.0 | 4 | 1.3 | 1.2 | 112 | 37.3 | 2.9 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
| 30 | Elongation factor 2 | IPI00186290 | 95 kDa | 61 | 20.3 | 2.3 | 46 | 15.3 | 8.1 | 22 | 7.3 | 5.5 | 15 | 5.0 | 2.6 | 5 | 1.7 | 0.6 | 27 | 9.0 | 6.1 | 29 | 9.7 | 5.0 |
| 31 | Nucleoside diphosphate kinase | IPI00604590 | 33 kDa | 7 | 2.3 | 1.5 | 21 | 7.0 | 4.4 | 53 | 17.7 | 2.1 | 31 | 10.3 | 5.5 | 33 | 11.0 | 3.5 | 2 | 0.7 | 0.6 | 58 | 19.3 | 5.8 |
| 32 | L-lactate dehydrogenase A chain | IPI00217966 | 37 kDa | 18 | 6.0 | 1.7 | 8 | 2.7 | 2.5 | 3 | 1.0 | 1.0 | 40 | 13.3 | 3.2 | 0 | 0.0 | 0.0 | 13 | 4.3 | 5.1 | 115 | 38.3 | 26.7 |
| 33 | Histone H2B type 1-H | IPI00303133 | 14 kDa | 54 | 18.0 | 2.0 | 5 | 1.7 | 0.6 | 6 | 2.0 | 1.0 | 4 | 1.3 | 1.2 | 34 | 11.3 | 2.1 | 104 | 34.7 | 15.3 | 1 | 0.3 | 0.6 |
| 34 | Galectin-3-binding protein | IPI00023673 | 65 kDa | 1 | 0.3 | 0.6 | 4 | 1.3 | 2.3 | 0 | 0.0 | 0.0 | 118 | 39.3 | 11.6 | 0 | 0.0 | 0.0 | 40 | 13.3 | 12.2 | 10 | 3.3 | 1.5 |
| 35 | Isoform 1 of Sulfhydryl oxidase 1 | IPI00003590 | 83 kDa | 4 | 1.3 | 1.2 | 58 | 19.3 | 6.8 | 76 | 25.3 | 7.0 | 29 | 9.7 | 0.6 | 1 | 0.3 | 0.6 | 5 | 1.7 | 2.1 | 36 | 12.0 | 2.6 |
| 36 | Peroxiredoxin-1 | IPI00000874 | 22 kDa | 11 | 3.7 | 0.6 | 1 | 0.3 | 0.6 | 32 | 10.7 | 3.2 | 49 | 16.3 | 1.5 | 35 | 11.7 | 5.7 | 36 | 12.0 | 6.0 | 10 | 3.3 | 1.2 |
| 37 | Heat shock cognate 71 kDa protein | IPI00003865 | 71 kDa | 58 | 19.3 | 4.9 | 4 | 1.3 | 2.3 | 18 | 6.0 | 2.0 | 9 | 3.0 | 1.0 | 36 | 12.0 | 3.5 | 18 | 6.0 | 2.6 | 19 | 6.3 | 1.5 |
| 38 | Transitional endoplasmic reticulum ATPase | IPI00022774 | 89 kDa | 18 | 6.0 | 1.0 | 4 | 1.3 | 2.3 | 6 | 2.0 | 1.0 | 24 | 8.0 | 4.4 | 69 | 23.0 | 6.1 | 33 | 11.0 | 8.2 | 28 | 9.3 | 4.5 |
| 39 | Histone H4 | IPI00453473 | 11 kDa | 67 | 22.3 | 7.8 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 1 | 0.3 | 0.6 | 51 | 17.0 | 2.6 | 53 | 17.7 | 5.7 | 0 | 0.0 | 0.0 |
| 40 | Cystatin-C | IPI00032293 | 16 kDa | 24 | 8.0 | 1.0 | 22 | 7.3 | 2.1 | 97 | 32.3 | 3.8 | 2 | 0.7 | 0.6 | 18 | 6.0 | 3.6 | 0 | 0.0 | 0.0 | 4 | 1.3 | 0.6 |
| 41 | Fatty acid synthase | IPI00026781 | 273 kDa | 24 | 8.0 | 2.0 | 65 | 21.7 | 5.9 | 5 | 1.7 | 1.5 | 82 | 27.3 | 2.1 | 1 | 0.3 | 0.6 | 3 | 1.0 | 1.0 | 0 | 0.0 | 0.0 |
| 42 | Heat shock protein beta-1 | IPI00025512 | 23 kDa | 48 | 16.0 | 2.6 | 10 | 3.3 | 4.2 | 70 | 23.3 | 2.3 | 1 | 0.3 | 0.6 | 0 | 0.0 | 0.0 | 6 | 2.0 | 1.7 | 0 | 0.0 | 0.0 |
| 43 | Retinonic acid receptor responder protein 1 | IPI00410240 | 33 kDa | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 176 | 58.7 | 20.8 | 0 | 0.0 | 0.0 |
| 44 | 14-3-3 protein zeta/delta | IPI00021263 | 28 kDa | 35 | 11.7 | 1.5 | 0 | 0.0 | 0.0 | 50 | 16.7 | 2.1 | 0 | 0.0 | 0.0 | 18 | 6.0 | 1.7 | 22 | 7.3 | 8.4 | 11 | 3.7 | 2.9 |
| 45 | Sap-mu-0 of Proactivator polypeptide | IPI00012503 | 58 kDa | 21 | 7.0 | 1.0 | 46 | 15.3 | 4.2 | 0 | 0.0 | 0.0 | 49 | 16.3 | 3.5 | 0 | 0.0 | 0.0 | 10 | 3.3 | 3.5 | 22 | 7.3 | 5.5 |
| 46 | Beta-2-microglobulin | IPI00004656 | 14 kDa | 20 | 6.7 | 0.6 | 13 | 4.3 | 2.5 | 26 | 8.7 | 2.3 | 13 | 4.3 | 1.2 | 58 | 19.3 | 1.5 | 0 | 0.0 | 0.0 | 6 | 2.0 | 1.0 |
| 47 | Transgelin-2 | IPI00550363 | 22 kDa | 28 | 9.3 | 1.5 | 3 | 1.0 | 1.7 | 6 | 2.0 | 0.0 | 24 | 8.0 | 1.7 | 2 | 0.7 | 1.2 | 15 | 5.0 | 3.6 | 33 | 11.0 | 1.0 |
| 48 | Macrophage migration inhibitory factor | IPI00293276 | 12 kDa | 9 | 3.0 | 0.0 | 18 | 6.0 | 3.5 | 11 | 3.7 | 2.9 | 12 | 4.0 | 1.0 | 6 | 2.0 | 1.7 | 64 | 21.3 | 7.1 | 11 | 3.7 | 1.2 |
| 49 | Keratin, type II cytoskeletal 7 | IPI00306959 | 51 kDa | 2 | 0.7 | 0.6 | 0 | 0.0 | 0.0 | 3 | 1.0 | 1.0 | 93 | 31.0 | 2.6 | 2 | 0.7 | 1.2 | 8 | 2.7 | 4.6 | 0 | 0.0 | 0.0 |
| 50 | Trefoil factor 1 | IPI00022283 | 9 kDa | 0 | 0.0 | 0.0 | 46 | 15.3 | 11.4 | 84 | 28.0 | 3.6 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 | 0 | 0.0 | 0.0 |
Figure 1.
Quantitative comparison of proteins detected by LC-MS/MS in the proximal fluid of seven breast cancer cell lines. In triplicate, MS/MS data of proteins identified in the proximal fluid from each breast cancer cell line was quantitatively determined by spectral counting in Scaffold software. Enolase 1, cathepsin D, CD44,Thrombospondin 1, zinc-α2-glycoprotein, keratin 18, vimentin, galectin-3 binding protein, and EGFR were chosen to represent HER2 negative and hormone receptor positive cell lines T47D and MCF-7, HER2 positive SKBR-3 and MDA-MB-453, triple negative breast cancer MDA-MB-468 and MDA-MB-231, and hormone receptor positive MCF-7 transfected with HER2 (MCF-7HER2).
Cell lysates of the 7 breast cancer cell lines were subjected to Western blot analysis in triplicate (1 replicate= average of 5 combined experiments). In most cases the expression and levels of each protein in the cells as detected by Western blot (Figure 2) were consistent with the findings of mass spectrometry analysis. Western blot analysis of HER2, ERα and MUC1 were incorporated as a quality control of the assay (Figure 2). MCF-7 and MCF-7HER2 cell lines both contained ERα and MUC1 consistent with the literature, as well as thrombospondin 1 and low levels of galectin-3 binding protein. Although MCF-7 and MCF-7HER2 had high levels of cathepsin D by mass spectrometry analysis, a monoclonal antibody to cathepsin D (antigen 1–75) failed to detect this protein in the Western blot of the same cell lysates. This discrepancy between the two assays may have been due to a posttranslational modification or mutated amino acid in the first 75 amino acids that interferes with the monoclonal antibody binding site. Both HER2 positive cell lines had high levels of HER2 and thrombospondin 1. However, SKBR-3 had high levels of galectin-3 binding protein and cathepsin D, while another HER2 positive cell line MDA-MB-453 had high levels of zinc-α2-glycoprotein (ZAG) and low levels cathepsin D. Signature cancer proteins, EGFR and CD44, were both found in a triple negative cell line, MDA-MB-468, as well as ZAG, galectin-3 binding protein and cathepsin D. Although another triple negative cell line, MDA-MB-231, had higher levels of cathepsin D and galectin-3 binding protein and significant levels of vimentin, reduced CD44 and no detectable levels of ZAG, EGFR, and MUC1 were seen. Our study suggests that even within the same subtype of breast cancer, significant differences exist in each breast cancer cell line.
Figure 2.
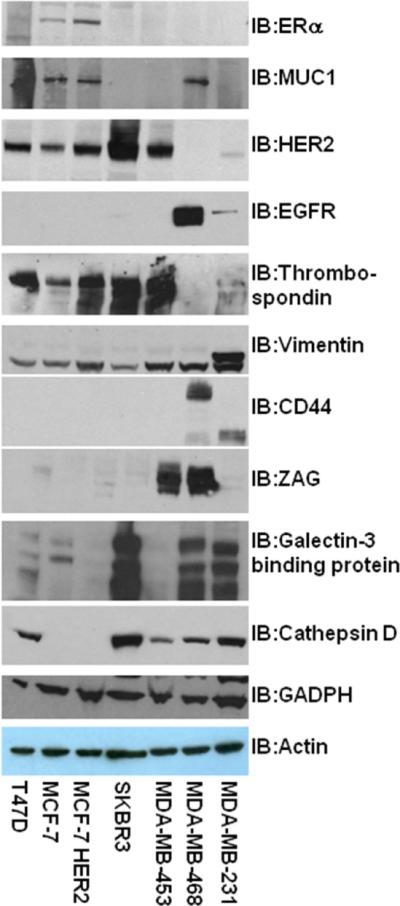
Western blot comparison of 7 potential biomarkers identified the LC-MS/MS study from whole cell lysates of 7 different breast cancer cell lines. Three known biomarkers, ERα, MUC1, and HER2 were also included in the Western blot study as controls. Minimally three experiments were performed.
Proteins identified by mass spectrometry in serum free media were then confirmed by dot blot analysis (Figure 3). The dot blots analysis allowed us to analyze multiple samples in triplicate and compare relevant levels of protein in the serum free media using sensitive and specific antibodies. Consistent in almost every case, levels of each proteins found by mass spectrometry coincided with the results of dot blot analysis of the serum free media, except from MDAMB-231 for the expression of thrombospondin 1 and the absence of cathepsin D from MCF-7 and MCF-7HER2 cells. Though there was some non-specific background observed in a few dot blots, development of IgY antibodies in chickens may give cleaner results. We were able to detect these proteins at 1 to 500 dilution of the sample (data not shown).
Figure 3.
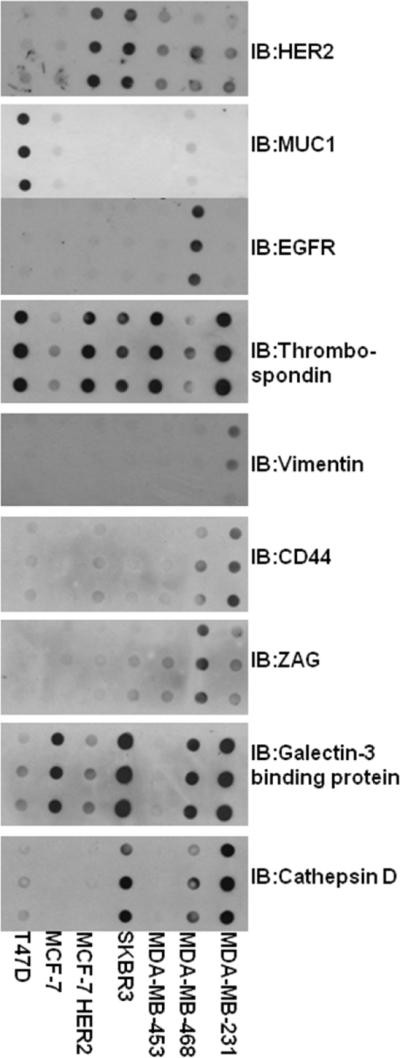
Dot blot comparison of 7 LC-MS/MS identified potential biomarkers present in the proximal fluid of 7 different breast cancer cell lines. Serum free media was collected from 7 different breast cancer cell lines as described. Cells were counted and equivalent ratios of media was loaded and analyzed for each breast cancer cell line. Each experiment consisted of three repeats.
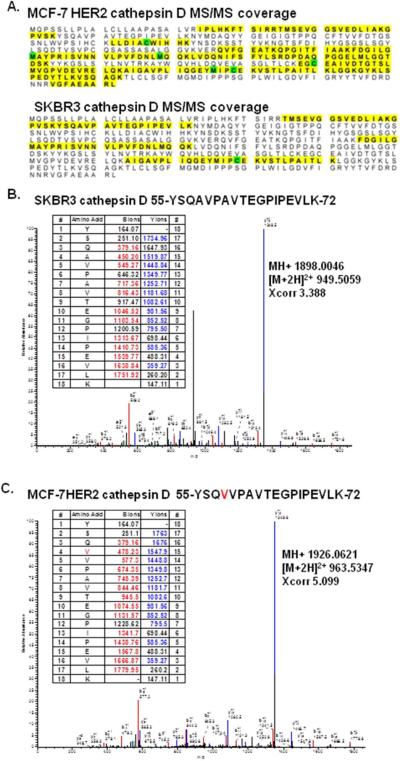
The lack of cathepsin D detection in MCF-7 and MCF-7HER2 proximal fluid was consistent with the Western blot data of their whole cell lysates. Interestingly, the mass spectrometry coverage of MCF-7HER2 cathepsin D (MS/MS coverage of 24–54 amino acids) did not completely span the antigenic region (first 75 amino acids) recognized by the monoclonal antibody (Figure 4A). While in SKBR-3 cells the mass spectrometry coverage only included amino acids 45–82 of the cathepsin D, strong immunoreactivity was seen in both Western blot analysis of the whole cell lysate and proximal fluid (Figure 4A). Therefore, the Universal Protein knowledgebase (UniProtKB) was used to search for possible posttranslational modifications and/or amino acid mutations on cathepsin D that would block the antibody interaction. Since there are 4 potential single-nucleotide polymorphisms (SNPs) that could occur in cathepsin D (58A →V; 229F→I; 282G→R; 383W→C)36, 37, a database was manually constructed containing all potential cathepsin D SNPs and mass spectrometry data of all seven cell lines was searched. Consistent with the Western blot findings of whole cell lysate and the proximal fluid analysis we detected the wild type amino acid sequence 55-YSQAVPAVTEGPIPEVLK-72 in cell lines MDA-MB-231 and SKBR-3 (Figure 4B). However, only the MCF-7 and MCF-7HER2 proximal fluid cathepsin D was found to have a SNP at amino acid 58A→V (Figure 4C). The identification of a SNP in cathepsin D may allow for the development of a new breast cancer cell line specific biomarker antibody.
Figure 4.
Single-nucleotide polymorphism (SNP) identified in cathepsin D by LC-MS/MS. A. MS/MS coverage of cathepsin D in the breast cancer cell lines of SKBR-3 and MCF-7HER2. B. MS/MS spectra of the wild type cathepsin D peptide 55-YSQAVPAVTEGPIPEVLK-72 in SKBR3 (Calc. MH+1898.0270; Exp MH+1898.0046). C. MS/MS spectra of the SNP in cathepsin D peptide 55-YSQVVPAVTEGPIPEVLK-72 in MCF-7HER2 (Calc MH+1926.0582; Exp MH+1926.0621).
Clinical grouping of breast cancer currently is based on three major protein biomarkers, HER2, estrogen receptor, and progesterone receptor. However, many other proteins defining unique features of a cancer cell may also prove important in breast cancer stratification and targeted treatment. Mass spectrometry identified 249 proteins from the seven breast cancer cell lines in triplicate quantitative spectral Scaffold analysis, thus resulting in 21 observations for each protein. These seven cell lines were classified into four groups HER2 positive, hormone receptor positive and HER2 negative, triple hormone receptor negative, and MCF-7 transfected with HER2. A blind statistical analysis (without protein names) was conducted by the UCLA statistical biomathematical consulting clinic (SBCC). Univariate comparison of mean values was used for each protein across groups via one-way analysis of variance methods (Supplemental Table 2). The mean comparisons of all 229 proteins were ranked from most statistically significant to least statistically significant. Using all four breast cancer groups including MCF7 HER2, 145 of the 229 proteins have a p value less than 0.05 in at least one of the 6 comparisons (Supplemental Table 2). Multivariate analysis of all seven cell lines was computed for all 229 proteins. A plot of first versus second principle component is the projection (shadow) of the 229 dimensional data shown in a two dimensional plot (Figure 5). Reproducibility of the principle components can be seen in the mass spectrometry data for each sample. In addition, there are differences between each individual cell line regardless of breast cancer group demonstrating the potential for breast cancer stratification.
Figure 5. Two Principle Component Dimension Analysis.

A plot of the first versus second principle component is the projection of the 229 dimensional data in a two dimensional plot. Triplicate replicates of all seven cell lines cluster appropriately, however clusters from each of the two different cell lines in the same group do not cluster together. Each breast cancer cell line is represented by a different symbol in triplicate (see legend).
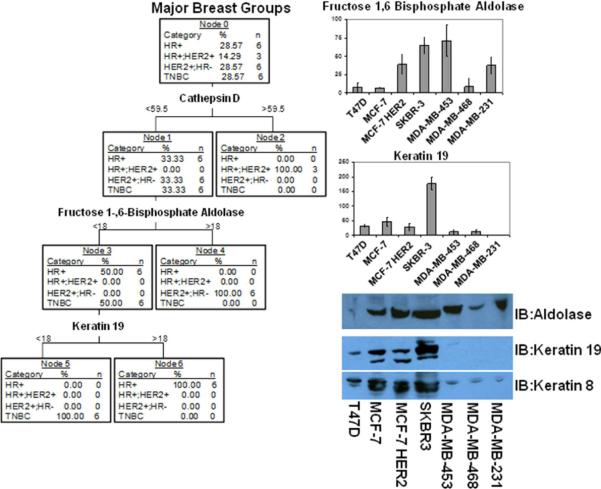
Predictive software was used to determine if a cell line may be grouped into HER2 positive, hormone receptor positive and HER2 negative, hormone receptor positive and HER2 over-expressed or the triple hormone receptor negative cohorts by analyzing a set of unique biosignature proteins. The Tree test required only a maximum of 3 proteins, cathepsin D, fructose 1,6-bisphosphate aldolase and keratin 19, to differentiate between each cell group (Figure 6). Tree results clearly identified cathepsin D as an indicator of MCF-7 cells over-expressing HER2 with a spectral count of greater than 59.5. Secondly fructose 1,6-bisphosphate aldolase at greater than 18 spectral counts classified the cell line as HER2 positive while less than 18 spectral counts required a second protein, keratin 19, to further distinguish the remaining 2 groups. Spectral counts less than 18 for keratin 19 indicated triple negative cell lines. Spectral counts greater than 18 indicated hormone positive and HER2 negative cell lines. Interestingly, Keratin 8 is also found in higher quantities of hormone positive and HER2 negative cell lines and may be another useful biomarker in conjunction with keratin 19. The keratin 19 data correlated with our other studies in primary breast cancer tissue by both mass spectrometry analysis and Western blot validation38, 39. Western blot analysis of fructose 1,6-bisphosphate aldolase demonstrated it was present in 4 of the 5 HER2+, 2 out of 5 in the ER+PR+ while found in low levels of TNBC tissue (Figure 7). While Western blot analysis of keratin 8 in five patient samples per hormone receptor positive and HER2 negative (3 of 5 positive), HER2 positive and hormone receptor negative (1 of 5 positive) and triple negative (0 of 5 positive) breast cancer tissue was conducted (Figure 7). We further tested the potential usefulness of keratin 19 as a biomarker in the serum of 10 hormone positive, 10 HER2 positive and triple negative breast cancer patients by ELISA (Supplemental Figure 5). Although the data was not considered experimentally significant the trend of increased levels of keratin 19 in circulation was consistent with the data reflected in mass spectrometry analysis and affinity blots of the breast cancer media and whole cell extracts. Interestingly, in our recent published work (Jianbo et al.39) galectin-3 binding protein was found in higher concentrations of triple negative breast cancer tissue than HER2+ tissue. A number of other proteins may also be used in combination for identification of TNBC such as annexin A1, annexin A5, CD44, EGFR, and vimentin. These observations have not been validated extensively and is provisional, but they are encouraging for the application of mass spectrometry data to differentiating breast cancer groups. However, we plan to test a panel of biomarkers selected from this manuscript and our other breast cancer studies32, 33, 38, 39 in 250 cases by tissue microarray. In addition, a list of the top 100 biomarker candidates will also be screened in the serum of 20 hormone receptor positive, 20 HER2 positive, 20 TNBC and 20 control patients.
Figure 6.
Tree test predicts that only cathepsin D, muscle fructose 1,6-bisphosphate aldolase and keratin 19 are necessary to predict breast cancer cell line group as hormone receptor positive and HER2 positive (HR+ and HER2+), hormone receptor positive and HER2 negative (HR+ and HER2−), HER2 positive and hormone receptor negative (HER2+ and HR−), and triple negative breast cancer (TNBC). In the right column is a comparison of MS/MS spectral counts across all seven breast cancer cell lines for muscle fructose 1,6-bisphosphate aldolase and keratin 19. In lower right column is a Western blot analysis of fructose 1,6-bisphosphate aldolase, keratin 19 and keratin 8 of breast cancer cell line total extracts. Experiments were performed in triplicate.
Figure 7.

Tree Test validation of fructose 1,6 bisphosphate aldolase A and keratin 8 presence in hormone HER2 positive and hormone receptor negative, hormone receptor positive and HER2 negative, triple negative breast cancer tissue. Western blot analysis using fructose 1,6 bisphosphate aldolase A and keratin 8 antibody was conducted on a total of 15 breast cancer tissue samples, five of each type of breast cancer.
Discussion
Early detection remains a key to improved breast cancer survival rates. Despite decades of research, little progress has been made in the development of an effective new blood assay for the early detection of any cancer. The study of blood serum for biomarker discovery has been hindered by the enormous number of serum proteins and large volume of circulating blood in cancer patients making direct identification of new breast cancer related proteins in blood to be an impossible task. In addition, 99% of serum proteins consists of 20 highly abundant serum proteins that overwhelm most methods of fractionation before detecting low abundance biomarkers of disease including human breast cancer.
Therefore, to improve the chance of developing a novel blood assay for breast cancer screening, biomarker discovery must be conducted on the source of breast cancer and nearby interstitial fluid. To circumvent the variability caused by heterogeneous breast cancer tissue we focused our study on breast cancer cell lines and their proximal fluids. Using an LTQ Orbitrap mass spectrometer in conjunction with Scaffold we were able to identify proteins with a dynamic range of 3–4 orders of magnitude and quantitatively compare the protein signatures of each of the seven breast cancer cell lines. Although identifying low abundance proteins are needed to further characterize breast cancer, it is unlikely they would be at high enough concentrations to be first detected by mass spectrometry in serum of a patient. Therefore, in this study we focused on the secreted, shed or leaked protein biosignatures of breast cancer cells as potential biomarkers for the future development of a blood assay.
Exactly 249 proteins were identified with 99% confidence from a panel of seven breast cancer cell lines representing four clinically different types of breast cancer. Selective validation of biomarkers differentially expressed in these breast cancer cells and their proximal fluid samples were performed. Seven highly expressed candidates, EGFR, vimentin, thrombospondin 1, CD44, ZAG, galectin-3 binding protein and cathepsin D, were selected for their clearly distinct biosignatures in each cancer cell line. Western blot validation of these proteins from whole cell lysates suggested their biosignatures of the cell lines were closely matched to the mass spectrometer expression levels. Furthermore, we were able to validate most of these proteins in the proximal fluid of each cell line by dot blot analysis confirming the quantitative spectral count results found by LC-MS/MS analysis. However, a few exceptions including the level of ZAG protein in the proximal fluid of MDA-MB-453 was lower than that found by mass spectrometry possibly due to digestion by proteases in the proximal fluid or differential posttranslational modifications interfering with the antibody-antigen binding.
In addition, cathepsin D was not detected by Western blots in the whole cell lysates or proximal fluid of either MCF-7 or MCF-7HER2, but found by mass spectrometry. By creating a database of all potential cathepsin D SNPs, the mutation 58A→V was identified from the mass spectrometry data. This unique mutation found only in MCF-7 cells may explain why the cathepsin D was not detected by the monoclonal antibody in the Western blot or dot blot analysis while it was clearly identified by mass spectrometry. SNPs naturally occur by a single nucleotide mutation in DNA resulting in the translation of an amino acid that is different from the wild type.
Whether the cathepsin D SNP found in MCF-7 has any biological significance is unknown, but SNPs play a pathogenic role in a number of diseases including Alzheimer's disease, Crohn's disease, autism, psoriasis, Parkinson's disease, schizophrenia and cancer40. Intriguingly, the 58Ala to 58Val polymorphism may affect the intracellular trafficking and maturation of this pro-enzyme in cancer41 and the level of beta-amyloid and tau increasing the risk of Alzhemer's disease36. The alteration in proenzyme routing in several breast cancer cell lines leads to its hypersecretion and also makes cathepsin D an excellent candidate for a blood assay. Moreover, procathepsin D (pCD), is secreted from cancer cells, acts as a mitogen on the cancer cell, stromal cells, and endothelial cells by stimulating their pro-invasive and pro-metastatic properties42. Others have also shown that over-expression of cathepsin D in human breast cancers is associated with a higher risk of relapse and metastasis41, 42. Interestingly, the Tree test designated cathepsin D as the number one biomarker in MCF-7 HER2 cell lines due to its high expression.
Another discordant finding was that the proximal fluid of MDA-MB-231 had significantly higher quantities of thrombospondin than seen by mass spectrometry analysis of whole cell lysates. This may be due to a combination of factors including posttranslational modifications, SNPs, and/or incomplete reduction and alkylation of the many disulfide bonds. Thrombospondin is also heavily glycosylated and this could also interfere with mass spectrometry identification. Since high quantities of thrombospondin are also found in blood, the identification of mutations such as the SNP found in cathepsin D or posttranslational modifications unique to breast cancer cells would need further investigation before becoming a candidate biomarkers. Monoclonal antibodies can be raised against SNPs or posttranslational modifications such as phosphorylation or glycosylation sites on normal versus disease specific biomarkers43, 44, giving rise to a highly selective tool in the detection of disease modified proteins.
All the proteins validated in this study except vimentin, may be enriched by their N-linked glycosylation sites using the hydrazide method as we described in our previous study32. The hydrazide method or a lectin column may be used to enrich for disease specific glycosylated protein biomarkers. Knowing the glycosylation patterns of these candidate biomarker will also allow for the development of antibodies to recognize the absence or presence of disease specific glycosylation. In addition, the recent discovery of O-GlcNAc modified vimentin by Slawson et al. allows for the development of site specific antibody45. Though the mass spectrometer reproducibly and consistently detected the expression levels of most protein biosignatures validated by antibody-based platforms in whole cell lysate and the proximal fluid, the value of this instrument is mainly in discovery while affinity assays are more important for later implementation of sensitive diagnostic tests.
As a positive control we also validated the presence of HER2 in the breast cancer whole cell lysates and proximal fluid despite not being detected by the LTQ mass spectrometer. The HER2 protein was found to be expressed at high levels in MCF-7HER2, SKBR3, and MDA-MB-453, but at a significantly lower level in MCF-7 and T47D breast cancer cell lines by Western blots. HER2 could also be clearly identified by an antibody-based assay in the proximal fluids of MCF-7HER2 and SKBR3 cell lines. In addition, to the specificity and sensitivity of antibody based detection systems, these antibodies may also be attached covalently to resin to enrich low abundant biomarkers from larger volumes of blood by immunoprecipitation. Theoretically, an immunoprecipitation of 10 mL of serum from a breast cancer patient with an average circulating volume of 3500–4500 mL would allow the detection of biomarkers with a dilution of 1 to 350–450 of the analyte, well within working range of dilution 1 to 500 that we observed in the Western dot blot analysis of proximal fluids. Still the key to the success of a viable blood assay is the identification of the true biosignatures of cancer from the discovery phase using mass spectrometry followed by the validation of biomarkers by affinity assays.
Although 249 proteins were detected by mass spectrometry analysis from the proximal fluids of seven cell lines we have selectively validated and analyzed several candidate biomarkers. It is necessary to look at a wider scope of individual cancer's biosignatures to properly identify every candidate. Although a larger cohort needs to be analyzed, the principle component plot of each breast cancer cell line demonstrates the reproducibility of the mass spectrometry analysis for the proximal fluid proteome and the unique biosignatures of each cell line. Each potential biomarker proteins selected for Western blot validation was found to be statistically significant in the univariate analysis across all breast cancer groups except zinc-α2-glycoprotein. Therefore, any number of these proteins could lead to the development of a multi-biomarker affinity assays for grouping breast cancer cells. Interestingly, the Tree test only required 3 biosignature proteins to successfully group HER2 positive, hormone receptor positive and HER2 negative, hormone receptor positive over-expressing HER2 and triple negative breast cancer cell lines.
The first qualifying protein used in the Tree test was the SNP containing cathepsin D which is an excellent candidate for creating and antibody that specifically recognizes a specific amino acid sequence containing the SNP mutation. The second qualifiying protein is muscle fructose 1,6-bisphosphate aldolase, a key protein in glycolysis. Fructose 1,6-bisphosphate aldolase as well as other metabolic proteins have been implicated as potential biomarker's in a number of diseases such as pancreatic ductal adenocarcinoma46, melanoma47 and Schizophrenia48. Metabolic upregulation and high glucose consumption is common in cancer cells, known as the Warburg effect, allowing their aggressive growth. A number of other key glycolysis enzymes including glucose-6-phosphate isomerase, phosphoglycerate kinase, enolase 1, and pyruvate kinase 3 were differentially secreted among the seven breast cancer cell lines (Supplemental Table 1 and 3). When the proximal fluid protein profiles were compared to the expression levels in the nucleocytoplasmic fractions of the same cell lines there was a significant degree of variability between each cell line. In addition, the number 1 protein 60kDa heat shock protein (mitochondrial) found in the nucleocytoplasmic fractions was found in significantly less quantities in the proximal fluid fractions suggestion the proximal fluid profiles were unique to each breast cancer cell line and minimal cell death occurred (Supplemental Table 3). Any combination of these enzymes may help in the stratification of human breast tumors and may be key anti-cancer drug target points. Pyruvate kinase 3 (also known as pyruvate kinase 2) is abnormally expressed in breast cancer tissue while in normal breast cells pyruvate kinase 1 is expressed49, making it a potential drug target. Any one of these metabolic enzymes may contain single-nucleotide polymorphisms that affect its function. The third qualifying protein from the Tree test was keratin 19, a well studied biomarker for breast cancer50–52. Interestingly, keratin 8 was also found to be in high quantities in similar cell lines as keratin 19, which may also lead to it being a candidate biomarker. Analysis of 10 hormone positive, 10 HER2 positive and 10 triple negative serum samples for the presence of keratin 19 by ELISA was consistent with mass spectrometry and affinity blot data of the breast cancer media and cell lines. Further analysis and validation of these biosignatures will be important in stratification, diagnosis, systemic treatment, response, and/or metastasis of human cancer.
Since breast cancer is highly heterogeneous, it is necessary to look at a larger pool of candidate biomarkers beyond HER2, PR, ER proteins in defining breast cancer in order to successfully deliver personalized cancer treatment. The combination of mass spectrometry in the discovery phase to study primary cancer cells and proximal fluid followed by use of sensitive affinity assays in validation will allow us a better chance to develop simple and non-invasive multi-biomarker blood assays for characterizing breast cancer. In addition, our study introduces a new opportunity to develop antibodies recognizing cell specific SNPs or posttranslational protein modifications which may also become a tool in biomarker validation.
Supplementary Material
Acknowledgments
We would like to thank and acknowledge Jefferey Gornbein of the Statistical Biomathematical Consulting Clinic (SBCC) from the UCLA Biomathematics Department for his assistance in the statistical analysis of our data. This work was supported in part by the California Breast Cancer Research Program (6JB-0013), the Department of Defense (DAMD17-01-1-0179), the National Institute of Health (1RO1CA93736), the Gonda Foundation, the EIF-Women Cancer Research Fund and Friends of the Breast Program at UCLA.
Abbreviations
- SNP
single nucleotide polymorphism
- ER
estrogen receptor
- PR
progesterone receptor
- EGFR
epidermal growth factor receptor
- ZAG
zinc-α2-glycoprotein
- MUC1
Mucin 1
- CEA
carcinoembryonic antigen
- CA27.29
Carcinoma antigen 27.29
- PSA
prostrate specific antigen
- CA125
cancer antigen 125
- CEA
carcinoembryonic antigen
- AFP
alpha-fetoprotein
References
- 1.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nature reviews. 2003;3(4):243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 2.Pan S, Aebersold R, Chen R, Rush J, Goodlett DR, McIntosh MW, Zhang J, Brentnall TA. Mass spectrometry based targeted protein quantification: methods and applications. Journal of proteome research. 2009;8(2):787–97. doi: 10.1021/pr800538n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong K, Moye E, Williams S, Berlin JA, Reynolds EE. Screening mammography in women 40 to 49 years of age: a systematic review for the American College of Physicians. Annals of internal medicine. 2007;146(7):516–26. doi: 10.7326/0003-4819-146-7-200704030-00008. [DOI] [PubMed] [Google Scholar]
- 4.Glick SJ, Breast CT. Annu Rev Biomed Eng. 2006 doi: 10.1146/annurev.bioeng.9.060906.151924. [DOI] [PubMed] [Google Scholar]
- 5.Karellas A, Vedantham S. Breast cancer imaging: a perspective for the next decade. Medical physics. 2008;35(11):4878–97. doi: 10.1118/1.2986144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peppercorn J. Breast cancer in women under 40. Oncology (Williston Park, N.Y. 2009;23(6):465–74. [PubMed] [Google Scholar]
- 7.Houssami N, Irwig L, Simpson JM, McKessar M, Blome S, Noakes J. The influence of knowledge of mammography findings on the accuracy of breast ultrasound in symptomatic women. The breast journal. 2005;11(3):167–72. doi: 10.1111/j.1075-122X.2005.21643.x. [DOI] [PubMed] [Google Scholar]
- 8.Arslan AA, Formenti SC. Mammography in developing countries: the risks associated with globalizing the experiences of the Western world. Nature clinical practice. 2009;6(3):136–7. doi: 10.1038/ncponc1282. [DOI] [PubMed] [Google Scholar]
- 9.Jorgensen KJ, Gotzsche PC. Who evaluates public health programmes? A review of the NHS Breast Screening Programme. Journal of the Royal Society of Medicine. 103(1):14–20. doi: 10.1258/jrsm.2009.090342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gotzsche PC, Nielsen M. Screening for breast cancer with mammography. Cochrane database of systematic reviews (Online) 2009;(4):CD001877. doi: 10.1002/14651858.CD001877.pub3. [DOI] [PubMed] [Google Scholar]
- 11.Kohn EC, Azad N, Annunziata C, Dhamoon AS, Whiteley G. Proteomics as a tool for biomarker discovery. Disease markers. 2007;23(5–6):411–7. doi: 10.1155/2007/967023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamandis EP. Point: Proteomic patterns in biological fluids: do they represent the future of cancer diagnostics? Clinical chemistry. 2003;49(8):1272–5. doi: 10.1373/49.8.1272. [DOI] [PubMed] [Google Scholar]
- 13.Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nature biotechnology. 2006;24(8):971–83. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 14.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics. 2004;3(4):367–78. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kulasingam V, Diamandis EP. Proteomic analysis of conditioned media from three breast cancer cell lines: A mine for biomarkers and therapeutic targets. Mol Cell Proteomics. 2007 doi: 10.1074/mcp.M600465-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Bast RC, Jr., Ravdin P, Hayes DF, Bates S, Fritsche H, Jr., Jessup JM, Kemeny N, Locker GY, Mennel RG, Somerfield MR. 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol. 2001;19(6):1865–78. doi: 10.1200/JCO.2001.19.6.1865. [DOI] [PubMed] [Google Scholar]
- 17.Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412(6849):822–6. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- 18.Mathelin C, Koehl C, Rio MC. Circulating proteinic biomarkers and breast cancer. Gynecologie, obstetrique & fertilite. 2006;34(7–8):638–46. doi: 10.1016/j.gyobfe.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Lauro S, Trasatti L, Bordin F, Lanzetta G, Bria E, Gelibter A, Reale MG, Vecchione A. Comparison of CEA, MCA, CA 15–3 and CA 27–29 in follow-up and monitoring therapeutic response in breast cancer patients. Anticancer research. 1999;19(4C):3511–5. [PubMed] [Google Scholar]
- 20.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nature reviews. 2005;5(11):845–56. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 21.Yates JR, Ruse CI, Nakorchevsky A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu Rev Biomed Eng. 2009;11:49–79. doi: 10.1146/annurev-bioeng-061008-124934. [DOI] [PubMed] [Google Scholar]
- 22.Perry RH, Cooks RG, Noll RJ. Orbitrap mass spectrometry: instrumentation, ion motion and applications. Mass spectrometry reviews. 2008;27(6):661–99. doi: 10.1002/mas.20186. [DOI] [PubMed] [Google Scholar]
- 23.Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol Cell Proteomics. 2011;10(9):M111 011015. doi: 10.1074/mcp.M111.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Irvin WJ, Jr., Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44(18):2799–805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 25.Bedard PL, de Azambuja E, Cardoso F. Beyond trastuzumab: overcoming resistance to targeted HER-2 therapy in breast cancer. Current cancer drug targets. 2009;9(2):148–62. doi: 10.2174/156800909787581024. [DOI] [PubMed] [Google Scholar]
- 26.Colleoni M, Zahrieh D, Gelber RD, Viale G, Luini A, Veronesi P, Intra M, Galimberti V, Renne G, Goldhirsch A. Preoperative systemic treatment: prediction of responsiveness. Breast (Edinburgh, Scotland) 2003;12(6):538–42. doi: 10.1016/s0960-9776(03)00163-2. [DOI] [PubMed] [Google Scholar]
- 27.Cleator S, Heller W, Coombes RC. Triple-negative breast cancer: therapeutic options. The lancet oncology. 2007;8(3):235–44. doi: 10.1016/S1470-2045(07)70074-8. [DOI] [PubMed] [Google Scholar]
- 28.Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clinical chemistry. 2006;52(3):345–51. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 29.Al-azawi D, Kelly G, Myers E, McDermott EW, Hill AD, Duffy MJ, Higgins NO. CA 15–3 is predictive of response and disease recurrence following treatment in locally advanced breast cancer. BMC cancer. 2006;6:220. doi: 10.1186/1471-2407-6-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harris L, Fritsche H, Mennel R, Norton L, Ravdin P, Taube S, Somerfield MR, Hayes DF, Bast RC., Jr. American Society of Clinical Oncology 2007 update of recommendations for the use of tumor markers in breast cancer. J Clin Oncol. 2007;25(33):5287–312. doi: 10.1200/JCO.2007.14.2364. [DOI] [PubMed] [Google Scholar]
- 31.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, Bertucci F. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–84. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 32.Whelan SA, Lu M, He J, Yan W, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Mass spectrometry (LC-MS/MS) site-mapping of N-glycosylated membrane proteins for breast cancer biomarkers. Journal of proteome research. 2009;8(8):4151–60. doi: 10.1021/pr900322g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, Whitelegge JP, Whelan SA, He J, Saxton RE, Faull KF, Chang HR. Hydrophobic Fractionation Enhances Novel Protein Detection by Mass Spectrometry in Triple Negative Breast Cancer. J Proteomics Bioinformatics. 2010;3:029–038. doi: 10.4172/jpb.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17(20):2310–6. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 35.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 36.Papassotiropoulos A, Bagli M, Kurz A, Kornhuber J, Forstl H, Maier W, Pauls J, Lautenschlager N, Heun R. A genetic variation of cathepsin D is a major risk factor for Alzheimer's disease. Annals of neurology. 2000;47(3):399–403. [PubMed] [Google Scholar]
- 37.Steinfeld R, Reinhardt K, Schreiber K, Hillebrand M, Kraetzner R, Bruck W, Saftig P, Gartner J. Cathepsin D deficiency is associated with a human neurodegenerative disorder. American journal of human genetics. 2006;78(6):988–98. doi: 10.1086/504159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu M, Whelan SA, He J, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Hydrophobic Proteome Analysis of Triple Negative and Hormone-Receptor-Positive-Her2-Negative Breast Cancer by Mass Spectrometer. Clinical proteomics. 2010;6(3):93–103. doi: 10.1007/s12014-010-9052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He J, Whelan SA, Lu M, Shen D, Chung DU, Saxton RE, Faull KF, Whitelegge JP, Chang HR. Mass spectrometry identified protein biosignatures for classifying breast cancer and predicting responses to chemotherapy. submitted to Breast Cancer Research. 2010 [Google Scholar]
- 40.LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic acids research. 2009;37(13):4181–93. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Touitou I, Capony F, Brouillet JP, Rochefort H. Missense polymorphism (C/T224) in the human cathepsin D pro-fragment determined by polymerase chain reaction--single strand conformational polymorphism analysis and possible consequences in cancer cells. Eur J Cancer. 1994;30A(3):390–4. doi: 10.1016/0959-8049(94)90261-5. [DOI] [PubMed] [Google Scholar]
- 42.Benes P, Vetvicka V, Fusek M. Cathepsin D--many functions of one aspartic protease. Critical reviews in oncology/hematology. 2008;68(1):12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baeckstrom D, Hansson GC, Nilsson O, Johansson C, Gendler SJ, Lindholm L. Purification and characterization of a membrane-bound and a secreted mucin-type glycoprotein carrying the carcinoma-associated sialyl-Lea epitope on distinct core proteins. The Journal of biological chemistry. 1991;266(32):21537–47. [PubMed] [Google Scholar]
- 44.Schmitz B, Peter-Katalinic J, Egge H, Schachner M. Monoclonal antibodies raised against membrane glycoproteins from mouse brain recognize N-linked oligomannosidic glycans. Glycobiology. 1993;3(6):609–17. doi: 10.1093/glycob/3.6.609. [DOI] [PubMed] [Google Scholar]
- 45.Slawson C, Lakshmanan T, Knapp S, Hart GW. A mitotic GlcNAcylation/phosphorylation signaling complex alters the posttranslational state of the cytoskeletal protein vimentin. Molecular biology of the cell. 2008;19(10):4130–40. doi: 10.1091/mbc.E07-11-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cui Y, Tian M, Zong M, Teng M, Chen Y, Lu J, Jiang J, Liu X, Han J. Proteomic analysis of pancreatic ductal adenocarcinoma compared with normal adjacent pancreatic tissue and pancreatic benign cystadenoma. Pancreatology. 2009;9(1–2):89–98. doi: 10.1159/000178879. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki A, Iizuka A, Komiyama M, Takikawa M, Kume A, Tai S, Ohshita C, Kurusu A, Nakamura Y, Yamamoto A, Yamazaki N, Yoshikawa S, Kiyohara Y, Akiyama Y. Identification of melanoma antigens using a Serological Proteome Approach (SERPA) Cancer genomics & proteomics. 7(1):17–23. [PubMed] [Google Scholar]
- 48.Gomez A, Ferrer I. Increased oxidation of certain glycolysis and energy metabolism enzymes in the frontal cortex in Lewy body diseases. Journal of neuroscience research. 2009;87(4):1002–13. doi: 10.1002/jnr.21904. [DOI] [PubMed] [Google Scholar]
- 49.Ibsen KH, Orlando RA, Garratt KN, Hernandez AM, Giorlando S, Nungaray G. Expression of multimolecular forms of pyruvate kinase in normal, benign, and malignant human breast tissue. Cancer research. 1982;42(3):888–92. [PubMed] [Google Scholar]
- 50.Chen Y, Zou TN, Wu ZP, Zhou YC, Gu YL, Liu X, Jin CG, Wang XC. Detection of cytokeratin 19, human mammaglobin, and carcinoembryonic antigen-positive circulating tumor cells by three-marker reverse transcription-PCR assay and its relation to clinical outcome in early breast cancer. The International journal of biological markers. 25(2):59–68. doi: 10.1177/172460081002500201. [DOI] [PubMed] [Google Scholar]
- 51.Ignatiadis M, Georgoulias V, Mavroudis D. Circulating tumor cells in breast cancer. Current opinion in obstetrics & gynecology. 2008;20(1):55–60. doi: 10.1097/GCO.0b013e3282f22b2e. [DOI] [PubMed] [Google Scholar]
- 52.Bonin S, Brunetti D, Benedetti E, Dotti I, Gorji N, Stanta G. Molecular characterisation of breast cancer patients at high and low recurrence risk. Virchows Arch. 2008;452(3):241–50. doi: 10.1007/s00428-007-0570-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.