Abstract
All cellular proteins are subject to quality control “decisions”, which helps prevent or delay a myriad of diseases. Quality control within the secretory pathway creates a special challenge, as aberrant polypeptides are recognized and returned to the cytoplasm for proteasomal degradation. This process is termed ER associated degradation (ERAD).
The textbook view of the endoplasmic reticulum (ER) consists of an undulating form, situated between the nucleus and Golgi stacks and freckled with dark circles that depict bound ribosomes. In the early years, those of us who obsessed over the inner workings of the ER emphasized that roughly one-third of newly synthesized proteins enter this organelle, an event that serves as the first stop in the journey taken by secreted and membrane proteins. Nevertheless, the ER was frequently considered as a simple way-station in an efficient assembly line. Indeed, under most experimental conditions the ER was thought to dutifully fold and process nascent proteins before they are encapsulated into Golgi-targeted vesicles. The realization only came later that we had been fooled: perturbations in protein folding and ER homeostasis trigger a signaling cascade—the unfolded protein response (UPR)—that profoundly impacts cellular and organismal health. Another surprise was the sizeable fraction of proteins that fail to pass through the ER folding combine. These polypeptides, which may be misfolded or incompletely processed, are ejected from the ER and returned to the cytoplasm. During or soon after entering the cytoplasm the polypeptides are ubiquitinated and degraded by the 26S proteasome. These events are collectively referred to as ER associated degradation, or ERAD, and to date >60 human diseases have been linked to the ERAD pathway (Guerriero and Brodsky, 2012). Studies on ERAD have benefited from a range of experimental strategies in yeast and mammalian cells, and recent reports have shed light on the mechanisms underlying the ERAD pathway. In this Minireview, the focus is on some of these discoveries and the open questions that remain in this field.
Are you a good protein or a bad protein?
Polypeptides entering the secretory pathway are received at the ER membrane by the Sec61 protein translocation channel in a non-native state (Braakman and Bulleid, 2011). Most of these polypeptides are posttranslationally modified by the signal peptidase, by the glycosylation machinery, and/or by lipid conjugation. The efficiencies of these events can be altered by the UPR. In addition, each nascent protein transits between a significant and variable number of intermediate folding states. Protein folding, which requires a cadre of ER associated molecular chaperones, is also compromised by stress as well as by perturbations in the ER’s specialized environment, which is more oxidizing than the cytoplasm and is calcium-rich. Furthermore, metabolic signal transduction pathways and developmental processes can induce the UPR. Combined with the fact that genetic mutations are not that uncommon—and that errors in transcription and translation arise—a significant number of newly synthesized proteins in the ER never attain their native states, or do so quite slowly.
Can the cell risk the threat that these aberrant species pose? Absolutely not, as unfolded proteins can aggregate or illegitimately bind other proteins and exert dominant negative effects. Misfolded proteins exhibiting subtly altered conformations can be secreted and form extracellular amyloids, as evidenced by some cases of transthyretin amyloidosis (Sekijima et al., 2005). However, ER chaperones and chaperone-like proteins more commonly survey the conformations of nascent polypeptides in the ER with high fidelity. Key members of these chaperone classes are Hsp70s (the Hsp70 in the ER lumen is BiP) and lectins that bind to and in some cases modify the appended N-glycan while facilitating polypeptide chain folding (Aebi et al., 2010).
The branched N-glycan moiety that is added onto most secreted and membrane proteins is built from two N-acetylglucosamines, nine mannoses, and three terminal glucose residues at the end of one branch. Glucose trimming favors client protein interactions with calnexin and calreticulin, which in turn facilitate protein folding. If folding is attenuated, sequential rounds of re-glucosylation and calnexin/calreticulin re-association occur, which sometimes leads to successful folding. The trimming of mannose residues from one of the three branches competes with folding and triggers ERAD, which prevents a futile and unproductive folding cycle. Critical mediators of mannose trimming and ERAD substrate selection are an ER mannosidase and the ER degradation enhancing α-mannosidase-like protein-1, or EDEM1. Although this sequence of events is well-supported, recent reports have offered new views of how some of these components function. For example, EDEM1 is not only a lectin but it can directly recognize non-native proteins, thereby exhibiting chaperone activity; in fact, most ER lectins exhibit peptide-binding activity. Instead of trimming mannose residues on ERAD substrates, EDEM1’s mannosidase domain may instead be utilized to interact with SEL1, an adapter that links substrates to other components of the ERAD machinery (Cormier et al., 2009). Moreover, EDEM1 overexpression—as occurs during UPR induction—accelerates the degradation of both glycosylated and unglycosylated proteins, overriding the need for mannose trimming (Ron et al., 2011). Thus, the UPR short-circuits a critical event that is otherwise imperative during glycoprotein quality control. Further, based on the examination of a relatively small number of substrates, it remains unknown whether all glycosylated proteins are subjected to the calnexin cycle. Finally, unglycosylated proteins that bind to BiP and function outside of the lectin folding cycle employ another factor, Herp, to aid in targeting ERAD substrates to the proteasome (Okuda-Shimizu and Hendershot, 2007). Our understanding of the mechanisms underlying the selection of glycosylated and unglycosylated ERAD substrates will certainly continue to evolve.
In contrast to the selection of soluble proteins within the ER, the recognition of misfolded cytoplasmic domains in membrane proteins appears at first glance to be simpler. By definition, these proteins can access cytoplasmic chaperones that play well-defined roles in protein folding and quality control. Hsp70 chaperones may bridge or help recruit distinct E3 ubiquitin ligases to membrane proteins that fail to fold, and some ligases can operate sequentially (Nakatsukasa et al., 2008; Younger et al., 2006). Why there are so many cytoplasmic chaperones is less clear (the yeast and human cytoplasm play host to 7 and 8 Hsp70s, respectively, plus an enlarged number of Hsp70 co-chaperones), and the rules governing substrate specificity between chaperone classes and amongst members of even the same class are undefined.
Shoot first, ask questions later
In some instances proteins are destroyed that might—under the right circumstances—fold into their native structures. A prominent example is the cystic fibrosis transmembrane conductance regulator (CFTR), the protein that is linked to the most common, lethal inherited disease in Caucasians (Lukacs and Verkman, 2012). A significant fraction of the wild type protein is targeted for ERAD as a consequence of CFTR’s complex and inefficient folding pathway and because it may take 30 minutes for the protein to be translated and attain its native conformation. It is not surprising, then, that the deletion of a single amino acid, F508, pushes CFTR over the edge so that nearly the entire protein pool is destroyed, which results in cystic fibrosis.
The CFTR folding pathway has been the focus of intense research efforts. Recent data indicate the existence of two thermodynamic peaks in the CFTR folding pathway that must be surmounted: the first is the folding of a nucleotide binding domain, and the second is the association between this domain and the fourth intracellular loop (Mendoza et al., 2012; Rabeh et al., 2012). Consequently, the cure for cystic fibrosis might require two drugs that each target one step. Based on this knowledge and the development of technologies that monitor CFTR folding and function, screens for ΔF508-CFTR correctors have been performed. One effort led to the isolation of a potent and efficacious compound that restored the function of the most frequent disease-causing mutant in cultured cells (Van Goor et al., 2011). The compound is now in advanced clinical trials (www.clinicaltrials.gov). Other compounds have shown efficacy in cellular models of different diseases associated with the ERAD pathway (Guerriero and Brodsky, 2012), but in most cases it is unknown how they function and clinical trials are few and far between.
Because ΔF508-CFTR can fold and then function at the plasma membrane, cystic fibrosis may represent an example where ERAD is overzealous. In contrast, liver disease associated with antitrypsin deficiency appears to arise from aggregation or polymerization of the antitrypsin Z allele (ATZ), which in its soluble form is an ERAD substrate. Here, an increase in ERAD would lead to disease amelioration. Unfortunately, specific small molecule activators of ERAD are not yet available. However, the administration of an autophagy activator lessened the pathological consequences of ATZ in a murine liver disease model (Hidvegi et al., 2010). These data are in-line with several studies indicating that the ERAD and autophagy pathways cooperate, and opens up the possibility of using autophagy modulators to treat select diseases associated with ER dysfunction.
Is the retrotranslocation channel a Jack-of-all-trades?
Once selected for degradation, soluble ERAD substrates in the lumen must somehow gain access to the cytoplasmic proteasome. Integral membrane proteins that are ERAD substrates present a unique challenge: how are embedded membrane-spanning domains discharged from the lipid bilayer? One scenario is that Sec61 functions bidirectionally, facilitating both nascent protein translocation and “retrotranslocation”. Evidence in support of this model continues to emerge from genetic tools and through the use of model substrates in yeast (see for example (Schafer and Wolf, 2009)). Alternatively, members of a family of membrane proteins (Der1 in yeast and Derlin-1, -2, or -3 in mammals) that organize several ERAD-requiring components were proposed to function as retrotranslocation channels. Intriguingly, members of the Derlin family are similar to rhomboid proteases, but a catalytic dyad required for activity is absent (Greenblatt et al., 2011). These and other data (Horn et al., 2009) suggest that the Derlins instead bind unfolded ERAD substrates in the membrane as they pass into the cytoplasm and/or regulate the activities of other integral membrane components of the ERAD machinery.
Although evidence indicates that some ERAD substrates with folded domains efficiently transit into the cytoplasm (Tirosh et al., 2003), other ERAD substrates containing residual structure may have to unfold to fit through the confines of the retrotranslocation channel. This event would require the breaking of disulfide bonds. A candidate for the necessary disulfide reductase in mammals is ERdj5. ERdj5 possesses four thioredoxin motifs and also binds EDEM1, as well as BiP. Based on ERdj5’s structure and a series of biochemical studies, it was proposed that a substrate passes sequentially from calnexin to the EDEM1-ERdj5 complex and then on to the retrotranslocation channel, an event that may be chaperoned by BiP (Hagiwara et al., 2011). Over time, it will be exciting to discover whether ERdj5 acts on a diverse ensemble of ERAD substrates. It is also curious that yeast lack an ERdj5 homolog. How disulfide bonds in ERAD substrates are broken in the yeast ER—or whether this is needed—is an open question.
Another black box in the field is the mechanism by which soluble ERAD substrates initially enter the cytoplasm. As there is no obvious pushing force generated from the ER, a series of handoffs between ERAD mediators and a substrate must ultimately favor substrate transit into and then through a retrotranslocation channel. Once a polypeptide enters the cytoplasm, and once misfolded membrane proteins are selected for ERAD, they are polyubiquitinated; therefore, the acquisition of a polyubiquitin chain may provide the necessary Brownian ratchet to facilitate retrotranslocation. Based on these considerations and other data, the field has increasingly focused on the relationship between ubiquitin ligases and unique steps in the ERAD pathway.
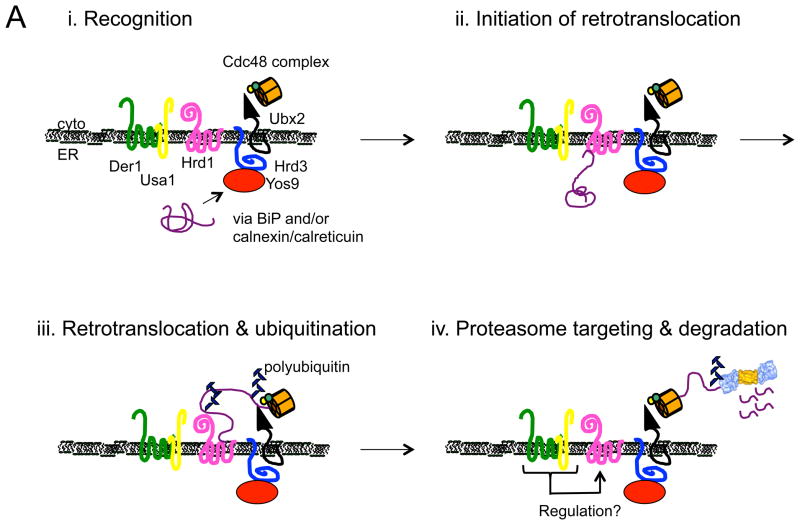
In contrast with the large number of ubiquitin ligases that exist in eukaryotic cells, relatively few of these enzymes are associated with the ER (Claessen et al., 2012). In yeast, the two ER ligases are Hrd1 and Doa10. For many years, it was curious why Hrd1 and Doa10, whose catalytic domains reside in the cytoplasm, also possess multiple membrane spanning domains. An exciting discovery was that Hrd1’s membrane spanning segments, along with soluble domains, appear to recognize misfolded integral membrane regions in ERAD substrates (Sato et al., 2009). Moreover, this ER resident E3 might even function as the long sought retrotranslocation channel for soluble substrates, thus coupling protein export with the acquisition of a polyubiquitin tag. Crosslinking experiments examining the early stages of substrate retrotranslocation are consistent with the view that Hrd1 ushers soluble lumenal substrates to the cytoplasm; moreover, cycles of Hrd1 oligomerization and monomerization are coupled to substrate binding, ubiquitination, and degradation, and Hrd1 associates with factors that play critical roles during each of these events (Carvalho et al., 2010; Horn et al., 2009) (Fig. 1A). If protein translocation into the ER is slowed, Hrd1 can even steal substrates that otherwise use Doa10 (Rubenstein et al., 2012), further implicating Hrd1 as a central player during ERAD. Still, the inability to identify a universal channel for soluble substrates is baffling: Might there be multiple retrotranslocation channels, such that different substrates utilize different channels (e.g., Sec61 versus the Hrd1 complex)? In turn, do integral membrane proteins need a channel, or might they access the proteasome directly or via a lipid body intermediate (Hartman et al., 2010)? Stay tuned.
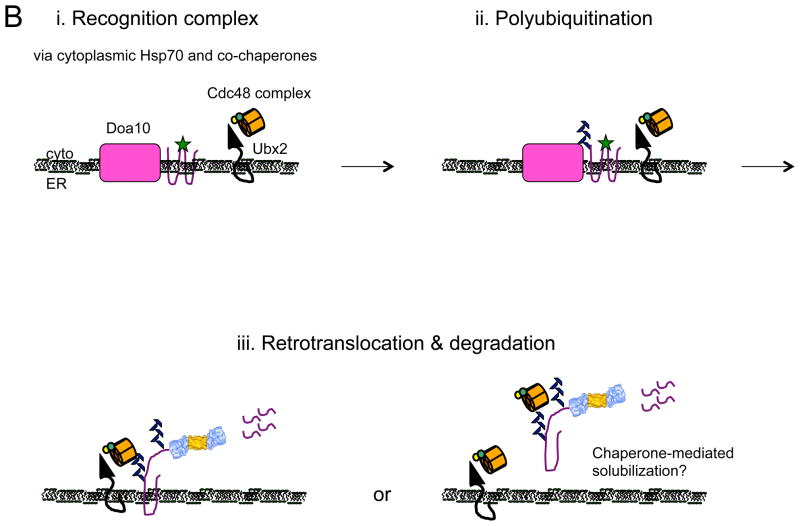
Figure 1.
Models for the recognition, targeting, and degradation of (A) soluble and (B) integral membrane ERAD substrates. (A) The ERAD of soluble, lumenal substrates in yeast requires the action of the Hrd1 complex, whose members are depicted in this figure. Recognition (i) of a substrate by this complex follows selection by BiP and/or by lectins with chaperone-like properties (e.g., calnexin/calreticulin). Yos9 is also an ER lectin that binds to Hrd3 (SEL1 in higher eukaryotes), and lectins that act similarly in mammals include Os9 and XTP-3B. Hrd1 is most intimately linked to substrate retrotranslocation (ii) and ubiquitinates and delivers substrates to the Cdc48 complex, which harbors hexameric Cdc48 (p97 in mammals) and single copies of Ufd1 and Npl4 (iii). Cdc48 couples ATP hydrolysis with subsequent delivery to the proteasome (iv). Cdc48 may also help unfold and disaggregate substrates prior to degradation, and appears to be tethered to the ER via Ubx2. Der1 (Derlin-1, 2, and 3 in mammals) and Usa1 (the mammalian homolog is Herp) like function as Hrd1 regulatory factors. (B) During the ERAD of an integral membrane protein that contains a misfolded cytoplasmic domain (depicted with a green star), a substrate is recognized by cytoplasmic chaperones (i) and is then ubiquitinated by Doa10 (ii) in yeast. Doa10 contains 14 transmembrane segments and like Hrd1 has also been proposed to act as a retrotranslocation channel. The Cdc48 complex extracts and maintains the solubility of the ERAD substrate (iii), before or concomitant with proteasome-mediated degradation. Not shown are E2 ubiquitin conjugating enzymes, which are integral membrane proteins or are tethered to the membrane, as well as proteasome adaptors that aid in the final targeting of substrates to the proteasome. Also not shown is the pathway that leads to the degradation of a protein with a misfolded lesion residing in a membrane-spanning segment, a process that also requires Hrd1. The ubiquitin ligase activities of Hrd1 and Doa10 are mediated by the RING domain that resides in the cytoplasm. In both panels, the ERAD substrate is depicted in purple. See text for additional details and (Xie and Ng, 2010).
The ERAD engine: Extraction and delivery to the proteasome
Most ERAD substrates require a AAA protein, Cdc48 (in yeast) or p97 (in mammals) to be extracted from the ER. Divergent models depict substrates threading through the aperture of the AAA hexamer, concomitant with ATP hydrolysis, or view the protein acting as a segregase that dissolves stable membrane-associated entities. Cdc48/p97 also serves as a platform on which a variety of ERAD facilitators sit, including ubiquitin binding proteins, factors that link Cdc48/p97 to the proteasome, and enzymes that can extend or reduce the length of the polyubiquitin chain (Stolz et al., 2011).
Surprisingly, integral membrane proteins can be fully solubilized by Cdc48 and reside in the cytoplasm prior to degradation (Fig. 1B); in yeast, maximal solubilization requires polyubiquitin chain extension, which may reflect increased avidity between Cdc48 and the substrate (Garza et al., 2009; Nakatsukasa et al., 2008). In mammalian cells, membrane protein solubilization is aided by components of a complex that also target tail anchor protein insertion into the ER (Wang et al., 2011), an event that similarly requires the transport of hydrophobic species in the cytoplasm. It is likely that other components help solubilize integral membrane ERAD substrates, and efficient solubilization is critical as cytoplasmic aggregates are toxic and prominent in several diseases.
Open questions and future research directions
As uniformly evident in other research fields, studies on the ERAD pathway have yielded more questions than answers. Several of these questions fall into the following categories:
First, proteins that facilitate ERAD have been isolated through genetic and biochemical attacks. The continued employment of this approach—in different cell types, under conditions in which the delivery of a substrate is blocked so that intermediates accumulate, and in animals—is vital. Undiscovered contributing factors and substrate-specific ERAD modifiers most certainly exist, and to date next to nothing is known about the function and regulation of the ERAD machinery in animals.
Second, ERAD is often viewed as a constitutive process, or at least one whose efficiency is modulated by the UPR (Jonikas et al., 2009). Intriguingly, ERAD efficiency may be “tuned” via the packaging of EDEM1 and Os-9 (another lectin that contributes to substrate selection) into vesicles targeted for lysosomal degradation (Bernasconi et al., 2012). Surprisingly, this regulatory circuit occurs independent of UPR activation. These data suggest that there may be other novel ways to modulate ERAD.
Third, a growing number of diseases are associated with the ERAD pathway because a mutated protein is destroyed, because a component of the ERAD machinery is defective, or because the pathway is coopted by pathogens. The ERAD pathway is also used as a metabolic regulator, especially with regard to events underlying lipid metabolism. In fact, the ERAD of misfolded proteins might have evolved as a byproduct of a more primal need to regulate the degradation of enzymes and lipid carriers that reside in or pass through the ER. The penetrance and expressivity of ERAD-related diseases, especially those involved in lipid metabolism, may be linked to genetic polymorphisms. To date, few studies have correlated polymorphisms in ERAD substrates with disease presentation. Undoubtedly, this pursuit will accelerate as genome sequencing and personalized medicine become commonplace.
Acknowledgments
I apologize to and thank the many colleagues and authors who helped shape this field, but whose contributions could not be cited due to space limitations. Work in the Brodsky laboratory on ERAD is supported by grants from the National Institutes of Health and Cystic Fibrosis Foundation Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aebi M, Bernasconi R, Clerc S, Molinari M. N-glycan structures: recognition and processing in the ER. Trends in biochemical sciences. 2010;35:74–82. doi: 10.1016/j.tibs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M. Role of the SEL1L:LC3-I Complex as an ERAD Tuning Receptor in the Mammalian ER. Molecular cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annual review of biochemistry. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen JH, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends in cell biology. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier JH, Tamura T, Sunryd JC, Hebert DN. EDEM1 recognition and delivery of misfolded proteins to the SEL1L-containing ERAD complex. Mol Cell. 2009;34:627–633. doi: 10.1016/j.molcel.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J Biol Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nature structural & molecular biology. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiological reviews. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Molecular cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Hartman IZ, Liu P, Zehmer JK, Luby-Phelps K, Jo Y, Anderson RG, DeBose-Boyd RA. Sterol-induced dislocation of 3-hydroxy-3-methylglutaryl coenzyme A reductase from endoplasmic reticulum membranes into the cytosol through a subcellular compartment resembling lipid droplets. The Journal of biological chemistry. 2010;285:19288–19298. doi: 10.1074/jbc.M110.134213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Horn SC, Hanna J, Hirsch C, Volkwein C, Schutz A, Heinemann U, Sommer T, Jarosch E. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs GL, Verkman AS. CFTR: folding, misfolding and correcting the DeltaF508 conformational defect. Trends Mol Med. 2012;18:81–91. doi: 10.1016/j.molmed.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, Feranchak AP, Brautigam CA, Thomas PJ. Requirements for efficient correction of DeltaF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabeh WM, Bossard F, Xu H, Okiyoneda T, Bagdany M, Mulvihill CM, Du K, di Bernardo S, Liu Y, Konermann L, et al. Correction of both NBD1 energetics and domain interface is required to restore DeltaF508 CFTR folding and function. Cell. 2012;148:150–163. doi: 10.1016/j.cell.2011.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron E, Shenkman M, Groisman B, Izenshtein Y, Leitman J, Lederkremer GZ. Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Molecular biology of the cell. 2011;22:3945–3954. doi: 10.1091/mbc.E10-12-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein EM, Kreft SG, Greenblatt W, Swanson R, Hochstrasser M. Aberrant substrate engagement of the ER translocon triggers degradation by the Hrd1 ubiquitin ligase. The Journal of cell biology. 2012;197:761–773. doi: 10.1083/jcb.201203061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Wolf DH. Sec61p is part of the endoplasmic reticulum-associated degradation machinery. EMBO J. 2009;28:2874–2884. doi: 10.1038/emboj.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekijima Y, Wiseman RL, Matteson J, Hammarstrom P, Miller SR, Sawkar AR, Balch WE, Kelly JW. The biological and chemical basis for tissue-selective amyloid disease. Cell. 2005;121:73–85. doi: 10.1016/j.cell.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Stolz A, Hilt W, Buchberger A, Wolf DH. Cdc48: a power machine in protein degradation. Trends in biochemical sciences. 2011;36:515–523. doi: 10.1016/j.tibs.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Tirosh B, Furman MH, Tortorella D, Ploegh HL. Protein unfolding is not a prerequisite for endoplasmic reticulum-to-cytosol dislocation. The Journal of biological chemistry. 2003;278:6664–6672. doi: 10.1074/jbc.M210158200. [DOI] [PubMed] [Google Scholar]
- Van Goor F, Hadida S, Grootenhuis PD, Burton B, Stack JH, Straley KS, Decker CJ, Miller M, McCartney J, Olson ER, et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18843–18848. doi: 10.1073/pnas.1105787108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Molecular cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Ng DT. ERAD substrate recognition in budding yeast. Seminars in cell & developmental biology. 2010;21:533–539. doi: 10.1016/j.semcdb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]