Exploring the topics of bacterial antisense transcription and termination, Landick and colleagues find the Rho termination factor to have a major role in suppressing antisense transcription, in synergy with H-NS. Additionally, the Rho cofactor NusG collaborates with Rho and enhances termination at a subset of terminators with suboptimal sequences.
Keywords: RNA polymerase, Rho-dependent transcription termination, antisense transcription, H-NS, NusG, NusA
Abstract
Despite the prevalence of antisense transcripts in bacterial transcriptomes, little is known about how their synthesis is controlled. We report that a major function of the Escherichia coli termination factor Rho and its cofactor, NusG, is suppression of ubiquitous antisense transcription genome-wide. Rho binds C-rich unstructured nascent RNA (high C/G ratio) prior to its ATP-dependent dissociation of transcription complexes. NusG is required for efficient termination at minority subsets (∼20%) of both antisense and sense Rho-dependent terminators with lower C/G ratio sequences. In contrast, a widely studied nusA deletion proposed to compromise Rho-dependent termination had no effect on antisense or sense Rho-dependent terminators in vivo. Global colocalization of the histone-like nucleoid-structuring protein (H-NS) with Rho-dependent terminators and genetic interactions between hns and rho suggest that H-NS aids Rho in suppression of antisense transcription. The combined actions of Rho, NusG, and H-NS appear to be analogous to the Sen1–Nrd1–Nab3 and nucleosome systems that suppress antisense transcription in eukaryotes.
Antisense transcription is a common feature of both bacterial and eukaryotic transcriptomes. Antisense transcripts have been identified in diverse bacteria (Georg and Hess 2011), including Bacillus subtilis (Rasmussen et al. 2009; Irnov et al. 2010; Nicolas et al. 2012) and Escherichia coli (Peters et al. 2009; Dornenburg et al. 2010; Shinhara et al. 2011). With the exception of a few well-studied examples (e.g., λ OOP [Krinke and Wulff 1987] and E. coli GadY [Opdyke et al. 2004]), bacterial antisense transcripts remain largely uncharacterized. Although some antisense transcripts have specific regulatory functions, others may result from “transcriptional noise” generated by nonspecific transcription initiation or weak promoters that become fixed within genes by evolutionary constraints on the coding sequence (Struhl 2007). At high levels, however, even spurious antisense transcription could have deleterious effects by interfering with sense transcription, stimulating mRNA degradation, or diverting important cellular resources.
The bacterial Rho-dependent transcription termination pathway, which relies on the ATP-dependent translocase Rho (Roberts 1969; Banerjee et al. 2006; Boudvillain et al. 2010; Peters et al. 2011), targets untranslated RNAs including tRNAs and sRNAs and at least some antisense transcripts (Peters et al. 2009). Rho binds to ∼80 nucleotides (nt) of C-rich unstructured (G-depleted) nascent RNA segments known as Rho utilization (rut) sites and subsequently dissociates elongation complexes (ECs) via its RNA translocase activity (for review, see Peters et al. 2011). Rho targeting of untranslated RNA causes the polar effects of nonsense mutations on expression of downstream genes in bacterial operons (Adhya et al. 1974).
Bacteria contain two general elongation factors—NusA and NusG—that may modulate Rho-dependent termination (Fig. 1). In vitro, NusG enhances Rho termination through direct interactions with Rho and RNA polymerase (RNAP) (Li et al. 1993; Pasman and von Hippel 2000; Mooney et al. 2009b; Chalissery et al. 2011). In vivo, NusG is proposed to either aid all Rho-dependent termination (Cardinale et al. 2008) or assist just a subset of terminators (Sullivan and Gottesman 1992). In vitro, NusA exhibits variable effects on Rho-dependent termination (Ward and Gottesman 1981; Burns and Richardson 1995; Cardinale et al. 2008; Saxena and Gowrishankar 2011a) but is proposed to aid all Rho-dependent termination in vivo (Cardinale et al. 2008).
Figure 1.

Regulators of transcript elongation in bacteria. The EC is comprised of RNAP (β′βα2ω subunits), DNA template, and RNA transcript. The Rho hexamer binds nascent RNA in primary sites on each subunit and secondary site in the central pore. NusG contacts RNAP via its N-terminal domain (NTD) and Rho via a C-terminal domain (CTD) connected by a flexible linker. NusA binds RNAP via an NTD near the RNA exit channel and contains additional domains (KH1, KH2, S1, and a CTD present in some bacteria).
The bacterial histone-like nucleoid-structuring protein (H-NS) also can inhibit transcription. H-NS represses transcription initiation by binding directly or adjacent to promoter DNA, then oligomerizing into higher-order structures that occlude RNAP binding or trap RNAP at promoters (Fang and Rimsky 2008; Grainger and Busby 2008; Dorman 2009). H-NS may also affect transcription elongation by binding downstream from ECs and acting as a roadblock, although experimental evidence for such effects is minimal. Recent studies have identified a genetic link between Rho and H-NS-like proteins (Saxena and Gowrishankar 2011b; Tran et al. 2011). It is therefore possible that Rho and H-NS function together to silence transcription at the same loci. However, a comparison of the Rho termination sites to H-NS-binding locations has not been reported.
In a previous study, we identified 25 sites in the E. coli genome at which Rho terminated antisense transcription (Peters et al. 2009). These findings raised several important questions. Is suppression of antisense transcription a major, global function of Rho or is it particular to a relatively small set of transcripts? What are the contributions of the elongation factors NusG and NusA to suppression of antisense transcription and Rho termination in general? Finally, if bacterial Rho and eukaryotic Sen1 play analogous roles in terminating antisense transcripts (Arigo et al. 2006; Brow 2011), does H-NS also participate in silencing noncoding transcription in a manner similar to nucleosomes in eukaryotes?
In this study, we used two strand-specific, global RNA-profiling techniques—tiling microarray analysis (Perocchi et al. 2007) and RNA sequencing (RNA-seq) (Parkhomchuk et al. 2009)—to obtain high-resolution maps of Rho-dependent termination in E. coli and determine the functional relationships between Rho, H-NS, NusG, and NusA. Our results establish a major role for Rho in widespread suppression of antisense transcription; show that NusG, but not full-length NusA, plays a significant role in Rho-dependent termination; establish the sequence basis for NusG effects on Rho-dependent termination; and reveal synergy between Rho and H-NS in transcriptional silencing.
Results
A major function of Rho is suppression of antisense transcription
To investigate the effects of inhibiting Rho on the transcriptome of E. coli K-12, we first used tiling arrays to measure RNAs from wild-type E. coli grown with or without the specific Rho inhibitor bicyclomycin (BCM) at a concentration that reduces Rho function without affecting the rate of cell growth (Ederth et al. 2006). We defined sites of Rho termination as positions at which BCM treatment caused a statistically significant increase in downstream transcript levels (false discovery rate [FDR] ≤ 5%) (Supplemental Table S1). By these criteria, we identified a total of 1264 BCM significant transcripts (BSTs) (Fig. 2A) whose levels or lengths increased when Rho was inhibited. We next used RNA-seq to confirm the identification of Rho-dependent terminators. The majority (91%) of terminators detected using tiling arrays also exhibited a twofold or greater increase in RNA-seq-defined readthrough in BCM-treated cells (P < 10−4), which confirmed that these terminators were not the result of microarray artifacts.
Figure 2.
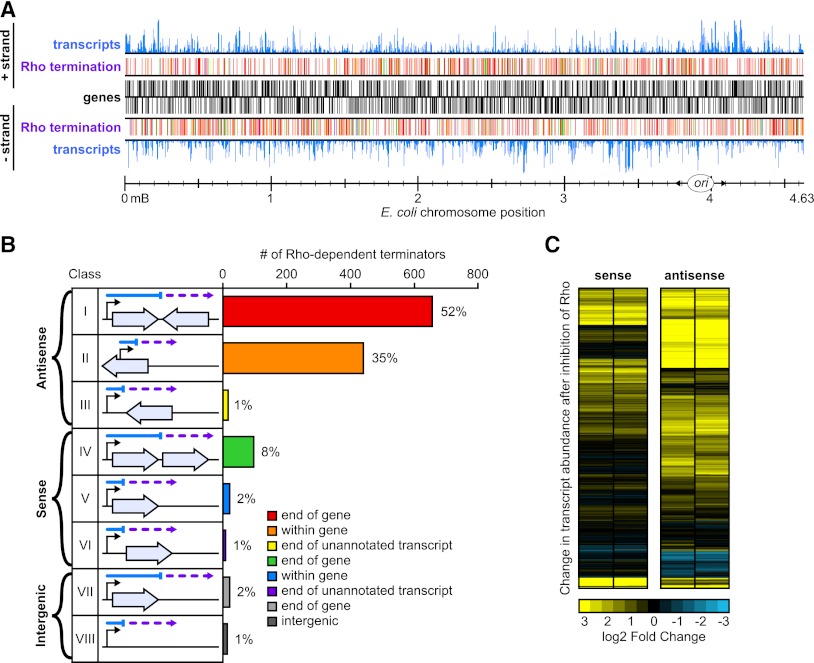
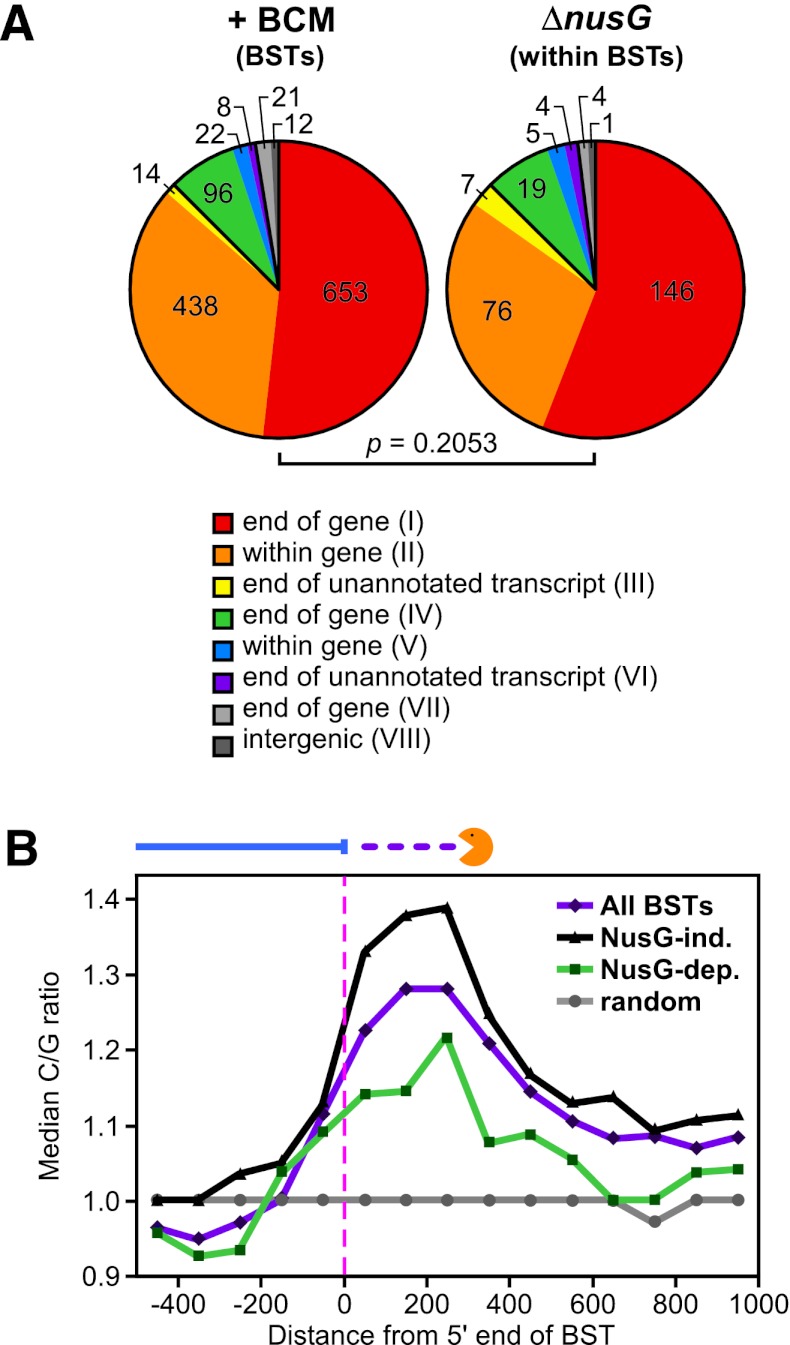
Genome-wide analysis of Rho-dependent transcription termination. (A) Distribution of Rho-dependent terminators in the E. coli genome as detected by BSTs. Features on the + strand are shown above the solid black lines, and features on the − strand are shown below the lines. Genes are depicted as black boxes, and transcript abundance detected by tiling arrays is shown as blue bar graphs. Rho-dependent termination sites are shown as colored boxes; the colors correspond to BST annotations from B. (B) Rho-dependent terminator annotations. Terminators are divided into eight classes based on their locations relative to annotated genes. The diagram immediately to the right of the class number illustrates the orientation of transcripts and termination sites relative to annotated genes. Light-blue arrows represent genes, and the arrows point in the direction of translation of the gene. The blue line indicates the length of the transcript in untreated cells, and the violet dashed line shows extension of the transcript in BCM-treated cells. The bar graph to the right of the gene diagram shows the number and percentage (rounded) of terminators in each class. (C) Effects of Rho inhibition on sense and antisense transcription. The log2 ratios of normalized read counts in BCM-treated versus untreated conditions are shown for the coding strand (sense) or the strand opposite the coding strand (antisense) for all E. coli K-12 genes in biological duplicate. Each row represents one gene. Fold increases in transcript abundance due to BCM treatment are shown in yellow, and decreases are shown in blue. The genes are clustered based on similar patterns of effects on antisense and sense transcription into arbitrarily ordered clusters by centroid linkage clustering using a Euclidean distance metric (see the Supplemental Material; Eisen et al. 1998).
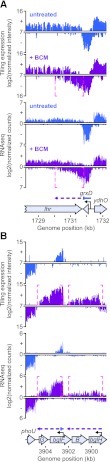
Analysis of the Rho-dependent terminator data set revealed a striking connection between Rho termination and antisense transcription. We divided terminators into three broad categories based on whether they affected antisense transcription, sense transcription, or intergenic transcription (Fig. 2B; Supplemental Material), using an updated E. coli K-12 genome annotation (http://www.ecocyc.org; Keseler et al. 2011). Antisense BSTs were caused by readthrough of Rho-dependent terminators either upstream of an oppositely oriented gene (classes I and III) or within a gene (class II). Sense BSTs resulted from readthrough of terminators at the ends of genes or unannotated transcripts (classes IV and VI) or within genes (class V). Intergenic BSTs resulted from readthrough of terminators at the ends of genes (class VII) or where no gene is known to exist (class VIII). The vast majority (88%) of Rho-dependent terminators controlled antisense transcription (Fig. 2B); 52% were class I (e.g., readthrough of grxD transcription into lhr) (Fig. 3A), 35% were class II (e.g., arising within bglF, or within bglH) (Fig. 3B), and a few (∼1%) were class III.
Figure 3.
Effects of Rho inhibition at class I and class II Rho-dependent terminators. (A) BCM effects on the class I Rho-dependent terminator at the grxD locus. A statistically significant increase in transcript levels (magenta dashed brackets) between untreated (blue bars) and BCM-treated (violet bars) cells occurs at the 3′ end of the grxD gene, indicating that the grxD transcript is terminated by Rho. (B) BCM effects on two class II Rho-dependent terminators at the bgl locus. Rho terminates antisense transcripts that arise from within the bglF and blgH genes. Note that the bglH antisense transcript is essentially undetectable in untreated conditions.
To quantify the occurrence of Rho-dependent termination in sense versus antisense transcription, we used RNA-seq to measure the levels of both sense and antisense transcripts for each gene in E. coli (Fig. 2C; Supplemental Fig. S1). Using a stringent cutoff (FDR ≤ 1%, and fivefold effect), we found that the antisense strands of 1555 genes (34% of all genes) were significantly up-regulated by BCM treatment, whereas only 416 genes were up-regulated to the same degree on the sense strand (Supplemental Table S2). Taken together, our results reveal a much greater increase in genome-wide antisense transcription after Rho inhibition than in sense transcription and thus establish that a major function of Rho is to suppress antisense transcription.
Rho-dependent termination of transcription units (TUs) with the potential to generate class I antisense transcripts relative to TUs lacking this potential was statistically significant. Of the 2054 genes at the end of TUs (Cho et al. 2009), 725 (35%) were terminated by Rho. Of the 1051 genes at the end of TUs at which the next gene is oriented in the opposite direction, 536 (51%) were terminated by Rho (P < 10−4), whereas only 189 of 1003 (19%) TUs for which the next gene is in the same orientation were terminated by Rho.
Sense transcription is not affected by moderate increases in antisense transcription
The functions of Rho-terminated antisense transcripts within genes are unknown. Proposed functions of the few antisense transcripts characterized to date include reduction of sense strand gene expression due to either pairing-mediated mRNA degradation or transcriptional interference (Georg and Hess 2011). In such cases, levels of sense and antisense transcripts should be anti-correlated; in other words, increases in antisense transcription would lead to decreases in sense expression. To determine whether increased antisense transcription in Rho-inhibited cells caused a decrease in sense strand transcription or an increase in RNA degradation, we calculated the correlation between BCM effects on sense and antisense transcription for annotated genes. We found no correlation between BCM-induced changes in sense and antisense transcript abundance at the gene level (r = −0.09) (Supplemental Fig. S2A). However, the effects of antisense transcription might be evident only at specific sites where antisense transcripts are up-regulated (i.e., BSTs), rather than at the gene level. To address this possibility, we conducted three additional correlation analyses by comparing the transcript levels from both strands of (1) all antisense BSTs, (2) antisense BSTs that occurred completely within genes (class II BSTs), and (3) BSTs at which the fold increase in antisense transcript levels following BCM treatment was greatest (the top 25% most up-regulated). Again, there was no significant correlation between sense and antisense transcript levels (r = −0.065, −0.014, and −0.079 for all antisense BSTs, class II BSTs, and most up-regulated antisense BSTs, respectively) (Supplemental Fig. S2B–D). Thus, significant increases in antisense transcription did not affect sense transcription at either the gene level or the subset of sites where antisense transcription increased most significantly. We conclude that an increase in antisense transcription caused by sublethal inhibition of Rho does not inhibit sense transcription, consistent with the idea that most antisense transcription is transcriptional noise. Greater increases in antisense transcription (caused by complete, and lethal, inhibition of Rho) could affect levels of sense RNAs but would be unlikely to reflect a physiologically relevant state of regulation in the cell.
Rho and H-NS silence transcription at the same genomic loci
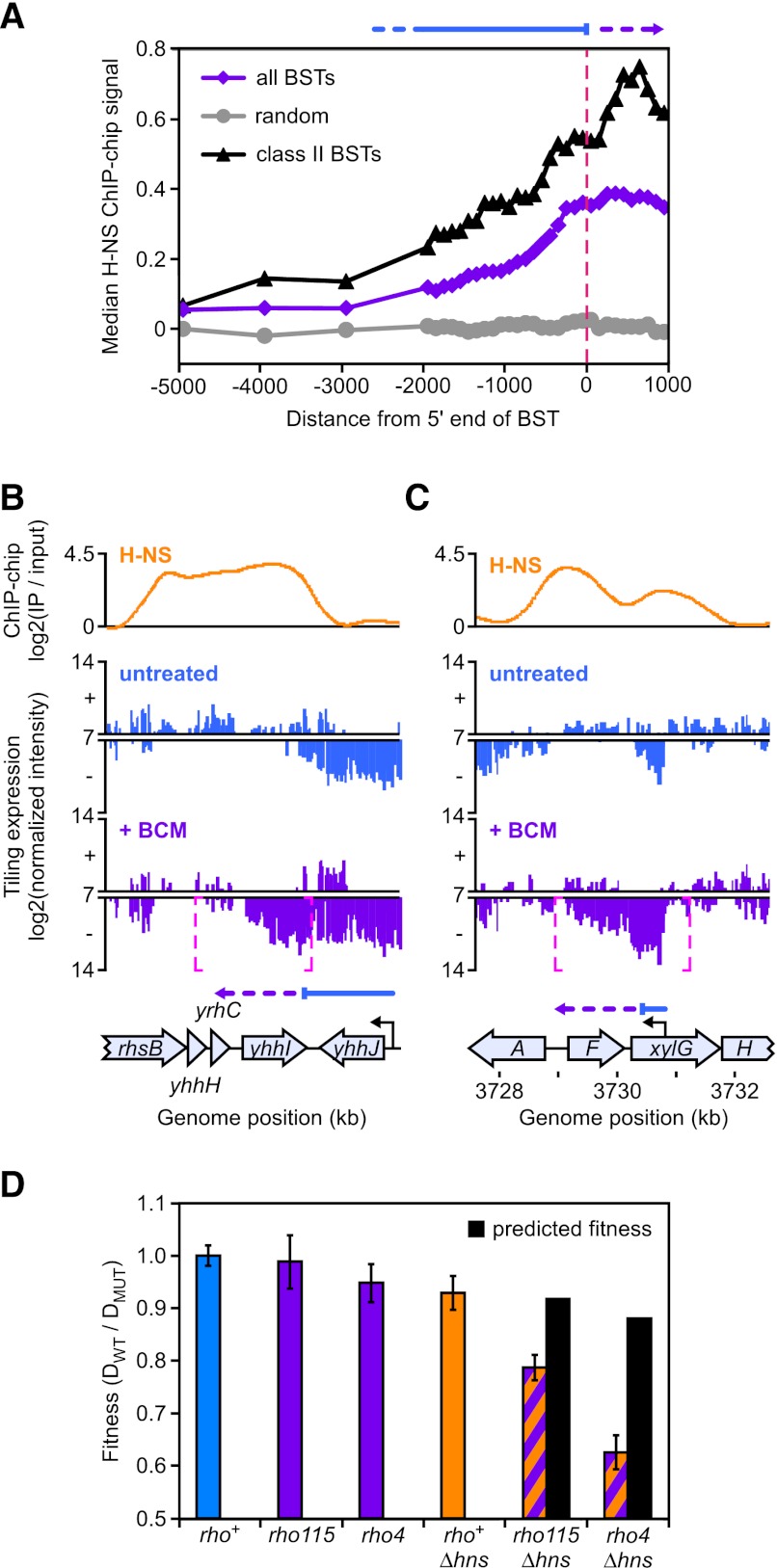
Recent studies have identified genetic interactions between Rho activity and genes encoding H-NS-like proteins (Saxena and Gowrishankar 2011b; Tran et al. 2011). These interactions may result from cooperative inhibition of deleterious transcription at certain loci by both Rho and H-NS. To test this hypothesis, we identified the genome-wide locations of H-NS binding using chromatin immunoprecipitation (ChIP) on high-density tiling microarrays (ChIP–chip) (Supplemental Table S3) under defined growth conditions; we then determined the overlap between these locations and the sites at which Rho terminated transcription. We found a strong association between H-NS binding sites and Rho-dependent terminators (Fig. 4A–C, cf. the sites of terminator readthrough in BCM-treated conditions and the occupancy of H-NS). Of the 1264 Rho-dependent terminators identified by tiling expression, 1066 (84%) were within 300 base pairs (bp) of sites with significant H-NS ChIP–chip signal. H-NS had a much higher probability of being bound near Rho-dependent terminators than at random locations in the genome (P < 10−4). Of the 1105 Rho-dependent terminators that suppress antisense transcription, 921 (83%) were associated with H-NS-binding sites. The occupancy of H-NS associated with Rho-dependent terminators was greatest near the sites of termination, indicating that Rho termination occurs within H-NS patches (Fig. 4A). Increased H-NS occupancy near Rho-dependent terminators was not simply due to H-NS binding in intergenic regions at the ends of genes; terminators that occurred within genes (class II BSTs) were bound to an even greater extent by H-NS (Fig. 4A). In addition, H-NS was specifically associated with Rho-dependent terminators rather than transcription terminations sites in general; H-NS was not statistically enriched at intrinsic (Rho-independent) terminators (P = 0.55) (intrinsic terminator coordinates from RegulonDB, http://regulondb.ccg.unam.mx). These results establish that H-NS generally occupies DNA at the sites of Rho termination and are consistent with a model in which Rho and H-NS act cooperatively to silence antisense transcription (see the Discussion).
Figure 4.
Spatial and functional associations between H-NS and Rho-dependent termination. (A) H-NS binding near Rho-dependent terminators. The median H-NS ChIP signal (see the Supplemental Material) is shown at specified distances from the 5′ end of all BSTs (violet), class II BSTs (black), or random chromosome positions (gray). (B) H-NS binding at the class I Rho-dependent terminator at the yhhJ locus. Colors are as in Figure 3, except that H-NS ChIP–chip data are shown in orange. Readthrough of the Rho-dependent terminator at the end of the yhhJ gene (indicated by the appearance of a BST shown as magenta dashed brackets) corresponds with a peak in H-NS ChIP–chip occupancy (orange). (C) H-NS binding at the class II Rho-dependent terminator within the xylG gene. Readthrough of the Rho-dependent terminator on the antisense strand of xylG corresponds with a peak in H-NS occupancy. (D) Genetic interactions between hns and rho. Fitness is expressed as the ratio of the doubling times (D) for wild-type (DWT = 24.75 ± 0.33 min) versus mutant cells in liquid LB medium. Wild-type fitness is set at one. Predicted fitness of double mutants is based on the multiplicative model (fitness of mutant #1 × fitness of mutant #2 = predicted fitness of double mutant) (St Onge et al. 2007).
The overlap between sites of Rho-dependent termination and H-NS binding suggests that both Rho and H-NS may suppress transcription at the same genomic loci. To address this possibility, we used ChIP–chip to identify loci at which RNAP occupancy significantly increased in cells deleted for the gene that encodes H-NS (Δhns) (Baba et al. 2006) and compared these loci with sites at which Rho terminated transcription. We found a significant overlap between loci at which transcription is repressed by H-NS and Rho-dependent termination. Of the 334 loci at which RNAP occupancy increased in Δhns cells (i.e., loci normally repressed by H-NS) (Supplemental Table S4), 222, or two-thirds, overlapped with Rho-dependent terminators (P < 10−4). The overlap between Rho- and H-NS-repressed loci further connected H-NS with suppression of antisense transcription. The majority of loci cosuppressed by Rho and H-NS were sites at which Rho suppressed antisense transcription (198 out of 222, or 89%), suggesting that H-NS may have a direct role in silencing antisense transcription at these loci. Increased RNAP occupancy in Δhns cells resulted directly from the loss of H-NS repression rather than from pleiotropic effects caused by Δhns, as the vast majority of loci at which RNAP occupancy increased overlapped with sites of H-NS binding (325 out of 334, or 97%; P < 10−4). We conclude that Rho and H-NS silence transcription at the same genomic loci.
Suppression of transcription at the same genomic loci by Rho-dependent termination and H-NS binding may reflect a functional relationship between Rho and H-NS. For instance, H-NS could slow elongating RNAP and allow time for Rho to terminate transcription of silenced genes. Functional relationships between genes can often be inferred from the growth phenotypes of double-mutant strains; a slower-than-expected growth rate for the double mutant indicates a genetic interaction that suggests shared function (St Onge et al. 2007). To test for a functional relationship between Rho and H-NS, we measured the growth rate of strains containing the defective rho alleles (rho4 [Morse and Guertin 1972] or rho115 [Guterman and Howitt 1979]) and a deletion of hns (Δhns) (Baba et al. 2006) in liquid LB medium (Fig. 4D). Double-mutant rho4 Δhns and rho115 Δhns strains grew more slowly than expected from a multiplicative model of fitness (Fig. 4D; St Onge et al. 2007), revealing genetic interactions between rho and hns. A third rho allele, rho15(Ts) (Das et al. 1976), was even more detrimental to cell viability when combined with Δhns. rho15(Ts) allows growth at 30°C but not 42°C. We could only construct a strain carrying both rho15(Ts) and Δhns in the presence of a rho+-complementing plasmid, regardless of the growth temperature, suggesting that the combination of rho15(Ts) and Δhns is lethal to cells. To test this possibility, we monitored retention of an unstable plasmid (Koop et al. 1987; Bernhardt and de Boer 2004) carrying a rho+ allele and a lacZ+ reporter gene by the rho15(Ts) Δhns double mutant on plates containing X-gal at 30°C (Supplemental Fig. S3). The rho15(Ts) Δhns strain formed only blue colonies, indicating that the rho+ plasmid was required for viability. These genetic interactions confirm cooperative action of Rho and H-NS in transcriptional silencing.
NusG principally assists termination at a minority subset of Rho-dependent terminators
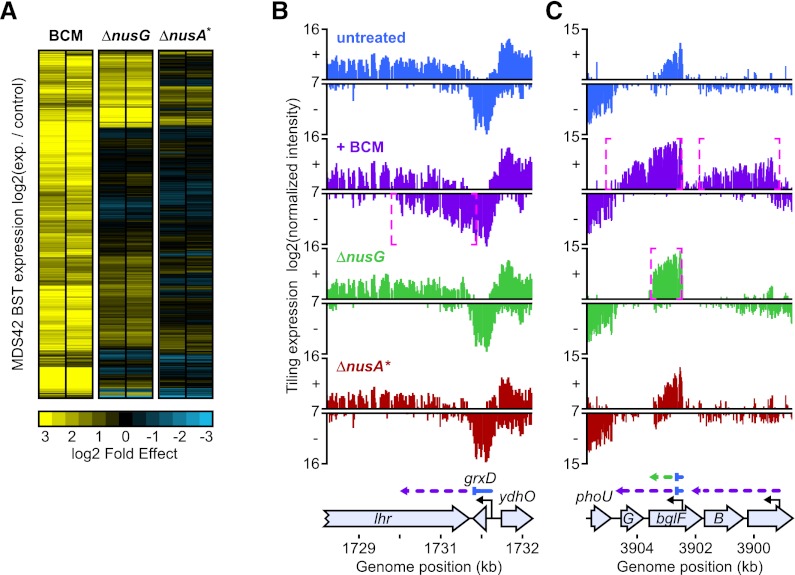
It is uncertain whether NusG enhancement is a significant requirement for termination in vivo at all Rho-dependent terminators (Cardinale et al. 2008; Burmann et al. 2010) or a subset of Rho-dependent terminators (Sullivan and Gottesman 1992). We used ChIP–chip to detect RNAP readthrough of Rho-dependent terminators in a multiple deletion strain lacking cryptic prophages (MDS42) (Posfai et al. 2006) and also lacking NusG (MDS42 ΔnusG) (Cardinale et al. 2008). We found a statistically significant overlap between Rho-dependent terminators and sites affected by deletion of nusG (P < 10−4). Of the 157 Rho-dependent terminators detectable by ChIP–chip (Peters et al. 2009) that are present in MDS42, 42 (27%) were within 300 bp of a statistically significant increase in RNAP occupancy in ΔnusG cells (Supplemental Table S5). These results indicate that NusG enhancement significantly affects termination efficiency at less than half of Rho-dependent terminators.
To investigate the contribution of NusG to suppression of antisense transcription, we performed tiling expression analysis on cells deleted for nusG and the rac prophage (Fig. 5A–C). Consistent with our ChIP–chip results, only a subset of the Rho-terminated transcripts was affected by deletion of nusG. Of 1264 Rho-dependent terminators, 247 (20%) exhibited greater readthrough in ΔnusG cells. Of the 1105 Rho-dependent terminators controlling antisense transcription, 229 (21%) exhibited greater readthrough when NusG was absent. These results demonstrate that NusG acts in concert with Rho to suppress antisense transcription at a minority subset of Rho-dependent terminators rather than at all terminators (Cardinale et al. 2008).
Figure 5.
Effects of ΔnusG and ΔnusA* on Rho-dependent termination. (A) Effects of ΔnusG and ΔnusA* on expression within MDS42 BSTs. The log2 ratio of median intensity within MDS42 BSTs in BCM-treated or mutant cells versus untreated cells is shown in biological duplicate. Each row represents one MDS42 BST. Fold increases in transcript abundance due to either BCM treatment—ΔnusG or ΔnusA*—are shown in yellow, and decreases are shown in blue. The genes are clustered based on similar patterns of effects on antisense and sense transcription into arbitrarily ordered clusters by centroid linkage clustering using a Euclidean distance metric (see the Supplemental Material; Eisen et al. 1998). (B) Effects of ΔnusG or ΔnusA* on the class I Rho-dependent terminator at the grxD locus. Neither ΔnusG nor ΔnusA* has a significant effect on transcript abundance at the 3′ end of the grxD gene. Colors are as in Figure 3, except that transcripts from ΔnusG cells are shown in green, and transcripts from ΔnusA* cells are shown in red. (C) Effects of ΔnusG or ΔnusA* on the class II Rho-dependent terminator within the bglF gene. ΔnusG had a significant effect on transcript abundance of the bglF antisense transcript, but ΔnusA* had no significant effect. Note that the ΔnusG effect on termination of the bglF antisense transcript was not as potent as direct inhibition of Rho with BCM.
The overlap between sites of Rho termination and either NusG enhancement or H-NS binding suggests that Rho, NusG, and H-NS may act synergistically at the same sites to suppress transcription. Alternatively, NusG may be required at terminators lacking H-NS. To distinguish between these possibilities, we compared Rho-dependent terminators that require NusG for efficient termination with those associated with H-NS. We found that the majority of NusG-dependent terminators were bound by H-NS (210 out of 247, or 85%; P < 10−4). Furthermore, H-NS association with NusG-affected terminators was statistically indistinguishable from H-NS association with terminators in general (P = 0.1). We conclude that NusG is not required at terminators lacking H-NS; instead, our data are consistent with the idea that Rho, NusG, and H-NS act coordinately at specific loci.
NusG is required at Rho-dependent terminators with suboptimal nucleic acid sequences
The features that distinguish Rho-dependent terminators that require NusG for efficient termination from those that do not are unknown. The NusG requirement at certain terminators is not explained by a lack of H-NS binding. In principle, NusG dependence could be associated with the type or function of the terminated gene (mRNA, tRNA, or sRNA), whether the gene downstream from the terminator is oriented in the same (sense) or opposite (antisense) direction as readthrough, or nucleic acid sequences present at the terminator. To determine whether NusG-dependent terminators are enriched at certain types of genes or classes of terminators, we compared gene associations and classes from the total set of BSTs to those BSTs with significant overlapping NusG effects. We found that NusG-dependent terminators were not significantly enriched at mRNAs, tRNAs, or sRNAs (P = 0.44). Furthermore, NusG enhancement was not differentially represented at any specific class (sense, antisense, within genes, at the end of genes, etc.) of Rho-dependent terminator (P = 0.21) (Fig. 6A). Finally, functional annotation analysis using the DAVID Bioinformatics Database (http://david.abcc.ncifcrf.gov) (Huang et al. 2009) failed to identify any particular set of genes that required NusG for efficient termination. We conclude that NusG stimulation of Rho termination is not associated with gene type or function or the location of the terminator relative to annotated genes.
Figure 6.
Basis for NusG effects on Rho-dependent termination. (A) NusG enhancement of termination is not associated with terminator class. All BSTs and BSTs with an overlapping significant ΔnusG effect were not statistically distinguishable by terminator class (Fisher's exact test; P = 0.21). Colors are as in Figure 2B. (B) NusG enhancement of termination is associated with sequences present at the termination site. The median C/G ratio (see the Supplemental Material) is shown at specified distances from the 5′ end of all BSTs (violet), NusG-independent BSTs (black), NusG-dependent BSTs (green), or random chromosome positions (gray). The orange “pacman” represents putative processing of transcripts by 3′ → 5′ exonucleases.
We next considered whether particular sequences distinguish NusG-dependent Rho terminators. Although upstream rut sites are present at some Rho-dependent terminators (e.g., λ tR1 [Chen and Richardson 1987] and E. coli trp t′ [Zalatan and Platt 1992]), sequences associated with Rho termination or NusG dependence have not been examined on a genome scale. To identify possible sequence patterns at Rho-dependent terminators, we calculated the C/G ratio in 100-bp windows from −500 bp upstream of to +1000 bp downstream from the first probe significantly affected by BCM treatment (high C/G ratio sequences will generate rut sites in RNA) (Fig. 6B). We found a peak in the C/G ratio centered at approximately +200 bp (P < 10−4) (Fig. 6B). This peak was due to both an increase in C (from 25%–27%) and a decrease in G (from 25%–21%) residues. Because we defined Rho-dependent terminators by the location of the transcript 3′ end in untreated cells and because Rho-terminated transcripts are typically processed by cellular 3′ → 5′ exonucleases (Mohanty and Kushner 2007), the elevated C/G ratio downstream from the mapped 3′ ends likely reflects Rho-dependent termination in or near the high C/G ratio sequences followed by processing of these transcripts to generate RNA 3′ ends that map upstream of these sequences (Fig. 6B). Interestingly, the median C/G ratio remained above the random baseline >1 kb downstream from the initial BCM effect (Supplemental Table S6), suggesting that sites capable of eliciting Rho termination are arranged consecutively in the genome to increase overall termination efficiency.
To investigate whether NusG effects on termination are related to nucleotide content at Rho-dependent terminators, we examined the C/G ratios of 300 terminators that were highly dependent on NusG for efficient termination and 300 that were NusG-independent (see the Supplemental Material). We found that terminators with strong NusG dependence exhibited a lower median C/G ratio than the total set of Rho-dependent terminators (Fig. 6B; Supplemental Table S6). In contrast, NusG-independent terminators displayed an even greater skew toward C and away from G than the majority of Rho-dependent terminators. Differences in the median C/G ratio for both NusG-dependent and NusG-independent terminators compared with all terminators were statistically significant within 300 bp of the 5′ end of the BST (Supplemental Table S6). We conclude that the NusG dependence of some Rho terminators results from the presence of sequences less likely to generate rut sites (i.e., lower C/G ratio sequences).
REP elements also are associated with Rho-dependent terminators
We used the MEME algorithm (Bailey and Elkan 1994) to identify potential motifs in the 300 bp upstream of Rho-dependent terminators (Supplemental Material). We found several long (20- to 29-nt) motifs with high information content (E-values from 1.7 × 10−36 to 3.7 × 10−108) at a subset of terminators (Supplemental Fig. S4A). The sequences and locations of the motifs matched those of REP elements, which are repetitive sequences found in intergenic regions near the 3′ ends of genes (Stern et al. 1984). Of the 695 REP elements present in the E. coli K-12 genome, 334 (48%) were within 300 bp of a Rho-dependent terminator (P < 10−4). REP elements contain palindromic units with dyad symmetry that form hairpin structures when transcribed into RNA (Supplemental Fig. S4B). REP elements near Rho-dependent terminators could potentially function as modulators of termination (Espeli et al. 2001) or as stabilizing hairpins that prevent RNA decay (Stern et al. 1984) in the absence of intrinsic terminator hairpins. To distinguish between these possibilities, we determined the distribution of REP elements from −500 bp upstream of to +1000 bp downstream from the first probe significantly affected by BCM treatment (Supplemental Fig. S4C). We found that the distribution of REP elements was centered at approximately −100, ∼300 bp upstream of high C/G ratios that represent Rho-binding sites. Thus, the majority of REP elements appear too far upstream to directly affect Rho binding and subsequent termination but optimally positioned to explain RNA 3′ ends present after exonuclease trimming. We suggest that the major function of REP elements found near Rho-dependent terminators is to inhibit RNA decay by impeding the processivity of 3′ → 5′ exonucleases (Stern et al. 1984) rather than to affect Rho termination efficiency.
A classic nusA deletion has no effect on Rho-dependent termination in vivo
The transcription elongation factor NusA has been proposed to enhance all Rho-dependent termination in vivo (Cardinale et al. 2008). We used RNAP ChIP–chip to identify sites of terminator readthrough in a strain that carried the same large nusA deletion studied previously in either a rho mutant strain containing unmapped mutations that allowed viability or MDS42 (Zheng and Friedman 1994; Cardinale et al. 2008). This deletion, which we term ΔnusA*, disrupts nusA at codon 128, leaving the NusA N-terminal domain (NTD) intact but removing the NusA S1, KH1, and KH2 domains. We found little or no overlap between Rho-dependent terminators and sites affected by ΔnusA*. Of the 157 MDS42 Rho-dependent terminators detectable by ChIP–chip (Peters et al. 2009), only nine (6%) were within 300 bp of a statistically significant increase in RNAP occupancy in ΔnusA* cells (P = 0.2756) (Supplemental Table S5). To rule out the possibility that our inability to detect a major effect of ΔnusA* on Rho termination was due to the low sensitivity of the ChIP–chip technique, we performed tiling expression analysis on ΔnusA* cells (Fig. 5A–C). Again, we found no significant overlap between BCM effects and ΔnusA* effects. Of the 1264 Rho-dependent terminators identified by tiling expression, only four (<1%) overlapped with a transcript significantly up-regulated in the ΔnusA* strain (P = 0.4139). Because our ΔnusA* results differed dramatically from those previously published using the same nusA allele (Cardinale et al. 2008), we confirmed that our strain contained the ΔnusA* allele by sequencing of the nusA locus (data not shown) and by visual inspection of nusA transcript data (Supplemental Fig. S5). We were unable to explain the discrepancy between our results and those reported by Cardinale et al. (2008). We conclude that the S1, KH1, and KH2 RNA-binding domains of NusA (the domains deleted in ΔnusA*) have little effect on Rho termination in vivo under standard E. coli growth conditions.
Discussion
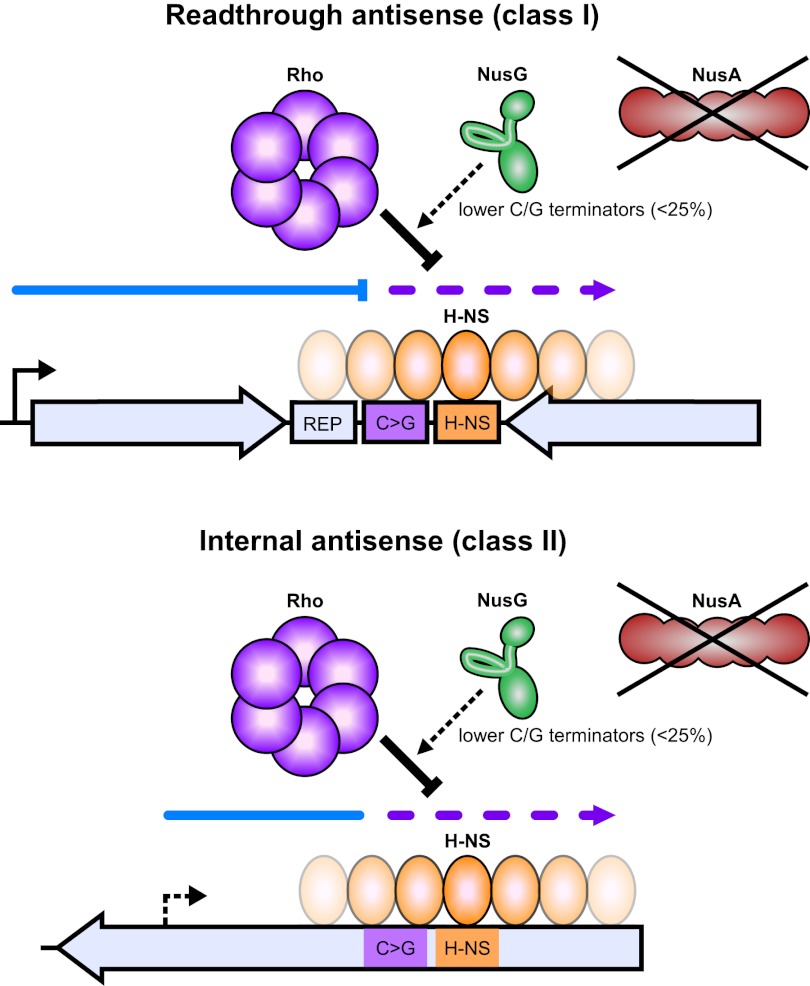
Our transcriptomic analysis of Rho termination establishes suppression of antisense transcription as a major role of Rho in bacteria. Most antisense transcription suppressed by Rho arises from a large and mostly uncharacterized set of antisense promoters within genes (internal antisense) (Fig. 7) and from continuation of sense transcription past the ends of genes into oppositely oriented downstream genes (readthrough antisense) (Fig. 7). Both H-NS and NusG contribute to the suppression of antisense transcription through apparently independent mechanisms. These findings raise several questions that merit discussion and further study. What impact does pervasive antisense transcription have on gene expression, and why has it evolved? Does H-NS affect Rho-dependent termination directly, possibly by slowing elongation by RNAP and allowing more time for Rho to terminate transcription? What is the mechanistic basis for NusG effects that depend on terminator sequence? Finally, how similar are the bacterial and eukaryotic systems that control antisense transcription?
Figure 7.
Models of antisense transcription termination by Rho. Rho terminates transcription at the end of genes, preventing antisense transcription into downstream genes (readthrough antisense, or class I terminators). Rho also terminates antisense transcription arising from within genes (internal antisense, or class II terminators). Termination sites are more C-rich and G-poor than random genomic DNA and thus facilitate Rho loading onto the nascent RNA (violet box labeled “C>G”). H-NS (orange ovals) is typically bound adjacent to sites of Rho termination (H-NS-binding sites are shown as orange boxes labeled “H-NS”) and is functionally synergistic with Rho. NusG enhances Rho termination at termination sites with reduced C and increased G content, which account for less than a quarter of all Rho-dependent terminators (dashed black arrow). The RNA-binding domains of NusA do not affect Rho termination of antisense transcription (black crossout). Class I terminators can also be associated with REP elements, which may stabilize the terminated mRNA (light-blue box labeled “REP”).
Impact of antisense transcription on sense transcription
We observed no apparent effect on sense transcription when Rho inhibition caused significantly elevated antisense transcription (Supplemental Fig. S2A–E,G). Although complete Rho inhibition, possibly causing higher levels of antisense transcription, could affect sense transcription, it is also lethal. This lethality could result in part from effects on sense transcription, RNA stability, or translation of even higher levels of antisense transcription than observed in our experiments, but such effects are unlikely to contribute to physiologically relevant regulation. If physiologically relevant effects on sense transcription occur by Rho modulation in wild-type cells, we should have detected them. This result suggests counterintuitively that most antisense transcription in wild-type cells has minimal effects on gene expression, even though specific effects of some antisense transcripts on gene expression are well known (Georg and Hess 2011).
Apparently, collisions between RNAPs as a result of antisense transcription are tolerated in normal cells as a form of transcriptional noise (Struhl 2007). The density of RNAP on most mRNA genes is low (less than one per cell) (Bon et al. 2006), so collisions between sense and antisense RNAP molecules are likely infrequent. Even when collisions do occur, transcription by sense but not antisense RNAP molecules will be aided by ribosomes translating nascent mRNA (Proshkin et al. 2010). Sense RNAPs may thus overpower antisense RNAPs, causing them to halt or backtrack and thereby facilitate Rho dissociation of antisense transcription complexes. From an evolutionary perspective, some level of antisense transcription may simply be tolerated as noise because eliminating it would adversely affect other essential processes (e.g., an antisense promoter may be the unavoidable consequence of encoding of an optimal protein sequence in the sense strand) or because the thermodynamic cost of completely eliminating it is too high (Lestas et al. 2010).
H-NS may assist Rho in silencing antisense transcription
Our findings that H-NS is generally associated with DNA at the sites of Rho termination and that hns and rho genetically interact suggest that Rho and H-NS act coordinately to silence transcription. One explanation for these synergistic effects is that H-NS increases the time window for effective Rho action by slowing transcript elongation or increasing pausing by RNAP. Several observations support this direct model of H-NS/Rho-synergy, although we cannot rule out indirect effects. First, H-NS occupancy of DNA increases as RNAP approaches the termination site, with maximal H-NS binding occurring at the position of termination (Fig. 4A). This is consistent with RNAP “running into” a downstream patch of oligomerized H-NS, which may serve as a block to elongation. Second, overexpression of H-NS lacking its C-terminal DNA-binding domain or of YdgT, which resembles the N-terminal H-NS oligomerization domain, can suppress defects in Rho-dependent termination caused by mutations in rho or nusG in hns+ cells (Williams et al. 1996; Saxena and Gowrishankar 2011b). These results suggest that changes to the H-NS nucleoprotein filament alter the efficiency of Rho termination. These effects are likely specific to sites of H-NS binding rather than reflecting general effects of YdgT or H-NS fragments on rates of transcript elongation because neither YdgT nor an H-NS fragment altered the rate of mRNA synthesis in a gene (lacZ) that does not bind H-NS (Grainger et al. 2006; Oshima et al. 2006; Kahramanoglou et al. 2011; Saxena and Gowrishankar 2011b). Finally, the direct model of H-NS and Rho synergy requires that sites of H-NS binding and Rho termination be coincident, but indirect models do not. Testing whether H-NS affects transcription elongation, pausing, and Rho termination directly will require carefully designed mechanistic and structure/function experiments.
Mechanism of NusG enhancement and relevance to polarity models
Our results establish that NusG affects a subset of Rho-dependent terminators and that these effects depend on sequences at the termination site. The simplest mechanistic explanation for these results is that NusG is required at terminators with suboptimal Rho-binding sites (rut sites). High C/G sequences that distinguish NusG-independent and NusG-dependent terminators should generate rut sites that bind Rho with high affinity. Furthermore, lower C/G sequences should generate RNA structures that further diminish unpaired C residues available for Rho binding. Logically, NusG may help Rho bind nascent RNA at terminators with lower C/G ratios by tethering Rho near the RNAP exit channel.
However, NusG stimulation of Rho–RNA binding challenges existing ideas about Rho termination. NusG does not affect the half-maximal concentration of Rho required for termination at a NusG-dependent terminator in lacZ (Burns and Richardson 1995). Furthermore, Sen and coworkers (Chalissery et al. 2011; Kalyani et al. 2011) argued that NusG stimulates the EC dissociation step rather than the RNA-binding step of Rho-dependent termination. Epshtein et al. (2010) argued that Rho is permanently associated with RNAP, obviating a role of NusG in tethering Rho to the EC. Conceivably, the sequence dependence of NusG effects on Rho-dependent terminators could reflect NusG modulation of a step other than RNA binding that also is favored by Rho–rut site interaction. Interestingly, Rho-dependent terminators that were more sensitive to BCM (i.e., had a higher percent readthrough in response to BCM) were modestly less dependent on NusG, suggesting that NusG activity may partially compensate for suboptimal termination conditions. Unambiguously determining the mechanistic basis of termination enhancement by NusG will require careful assays of the Rho concentration dependence of termination at a collection of Rho-dependent terminators.
Our finding that NusG affects a subset of Rho-dependent terminators also has implications for models of transcriptional polarity (Peters et al. 2011). In the RNA competition model, ribosomes block Rho binding to rut sites on the nascent RNA (Adhya et al. 1974). In the NusG competition model, ribosomes sequester the NusG C-terminal domain (CTD) in an interaction with ribosomal protein S10 (Burmann et al. 2010) and thereby prevent the NusG CTD from activating Rho-dependent termination. Since only a minority subset of Rho-dependent terminators requires NusG, however, most polarity suppression may not be explained by ribosomes sequestering the NusG CTD. Our results favor a mixed model in which both the RNA competition and NusG competition play roles. Mutations in the ribosome that disrupt the binding between S10 and NusG will be crucial to test the contribution of the S10–NusG interaction to polarity in vivo.
Analogous control of antisense transcription in bacteria and eukaryotes
Although the synergy among chromatin structure and elongation factors in terminating spurious transcription is more complex in eukaryotes, the helicase Sen1 plays an analogous role to bacterial Rho. Together with Nrd1 and Nab, which confer both RNA recognition (Carroll et al. 2007) and early elongation phase-specific RNAPII CTD recognition (Vasiljeva et al. 2008), Sen1, like Rho, terminates cryptic, untranslated transcripts, including antisense transcripts (Arigo et al. 2006; Brow 2011). Both histone modifications and nucleosome composition affect these processes in eukaryotes (Carrozza et al. 2005; Santisteban et al. 2011), and some histone modifications appear to aid Sen1 action (Terzi et al. 2011). Furthermore, the evolutionarily conserved Spt5 (NusG) elongation factor both aids transcript elongation (Hirtreiter et al. 2010) and, through its multiple C-terminal KOW domains, recruits elongation factors (Zhang et al. 2005), apparently including Nrd1 (Vasiljeva and Buratowski 2006; Lepore and Lafontaine 2011). Thus, in both bacteria and eukaryotes, nucleoprotein structure and a functionally analogous termination complex increase the overall fidelity of RNA synthesis by suppressing noncoding transcription.
Materials and methods
Strains, plasmids, and primers
The strains used are listed in Supplemental Table S7. The rho and hns strains were constructed by P1vir-mediated transduction (Thomason et al. 2007). The unstable plasmid pJP124 (pRC7-rho+) was generated by cloning a PCR product containing the rho promoter and rho gene between the ApaI and HindIII sites of pRC7 (Koop et al. 1987; Bernhardt and de Boer 2004) using the following primers: 5′-CTCTCTCTGGGCCCATAAGGGAATTTCATGTTCGG-3′ and 5′-CTCTCTAAGCTTATGAGCGTTTCATCATTT-3′. Transcription of rho+ and lacZ+ was driven by the rho promoter (Prho-rhoL+-rho+-lacZYA+).
RNA isolation
Cells were grown in MOPS minimal medium (Neidhardt et al. 1974) with 0.2% glucose at 37°C in gas-sparged Roux bottles to mid-log phase (OD600 ∼ 0.3–0.4). Culture samples were transferred directly into an ice-cold ethanol/phenol stop solution (Rhodius et al. 2006), which immediately inactivated cellular RNases. Cells were collected by centrifugation and stored at −80°C until RNA extraction. Total RNA was extracted from cell pellets by hot phenol extraction (Khodursky et al. 2003). The integrity of total RNA was determined from agarose gel or microchannel (Agilent Bioanalyzer) electrophoretograms. Ribosomal RNA (16S and 23S) was depleted prior to construction of RNA-seq libraries using MICROBExpress reagents (Ambion).
RNA-seq, data normalization, processing, and significant gene identification
RNA-seq was performed by the DOE Joint Genome Institute (Walnut Creek, CA) using the dUTP method (Parkhomchuk et al. 2009). Briefly, ribosome-depleted RNA was fragmented in a buffered zinc solution (Ambion), then purified using AMPure SPRI beads (Agencourt). First strand cDNAs were then synthesized from the fragmented RNA using SuperScript II reverse transcriptase (Invitrogen), followed by a second bead purification. dUTP was included in the second strand synthesis reaction in addition to dTTP to chemically mark the second strand. Two further bead purification steps using different ratios of beads to cDNA (85/100, then 140/100) selected cDNAs in a range between 150 and 350 bp. cDNAs were then A-tailed using Exo− Klenow, followed by ligation of sequencing adaptor oligos. Following bead purification, dUTP was cleaved from the second strand using AmpErase UNG (uracil N-glycosylase, Applied Biosystems), resulting in adaptor ligated single-stranded cDNAs. Deep sequencing of cDNAs was performed using the Illumina Tru-Seq sequencing platform. RNA-seq reads were mapped to the E. coli K-12 MG1655 genome (GenBank ID U00096.2) using short oligonucleotide alignment program (SOAP) (Supplemental Table S8; Li et al. 2008). The RNA-seq data depicted in Figures 2C and 3, A andB, were normalized by dividing either the total number of read counts per sense or antisense strand of genes (Fig. 2C) or the number of read counts at a given chromosome coordinate (Fig. 3A,B) by the total library size in millions (i.e., counts per million). Normalized biological replicates showed good agreement (r ≥ 0.99 at the count per gene level). The log2 ratios for BCM-treated versus untreated cells shown in Figure 2C were calculated after a constant of one read per million was first added to each gene to avoid divide by zero errors. These log2 ratios were quantile-normalized between biological replicates of the same strand (sense or antisense) using R (normalizequantiles) (Gautier et al. 2004). To identify genes with significant changes in expression in BCM-treated cells, raw (unnormalized) RNA-seq reads between the start and end coordinates of genes were summed for both sense and antisense strands. Library normalization and significant gene determination were carried out using an R implementation of the edgeR algorithm (Robinson et al. 2010). Fold effect and FDR values for each gene are listed in Supplemental Table S2.
Tiling expression, data normalization, processing, and significant transcript identification
Reverse transcription and cDNA labeling were performed as previously described (Cho et al. 2009), except that Cy3 was used instead of Cy5. Microarrays were designed using chipD (Dufour et al. 2010) and contained 378,408 probes that alternate strands with ≈12-bp spacing (Roche-Nimblegen). Hybridization and washing of microarrays were performed according to standard Nimblegen protocols (http://www.nimblegen.com). Microarrays were scanned at 532 nm using a GenePix 4000B scanner (Molecular Devices). Raw probe intensities were normalized across samples using robust multiarray (RMA) analysis implemented in the NimbleScan software package (Roche-Nimblegen). Data from Δrac and MDS42 strain backgrounds were normalized separately to avoid problems arising from the lack of probe intensity in MDS42-deleted regions. Normalized biological replicates showed good agreement (r ≥ 0.90 at the probe level). Normalized probe intensities were transformed to log2 and split into strands, and biological replicates were averaged. Transcripts that were significantly up-regulated in BCM-treated or mutant cells were identified using an R implementation of the CMARRT algorithm (Kuan et al. 2008). First, data from untreated cells were subtracted from data from BCM-treated or mutant cells. Second, the CMARRT was used to identify regions of at least three consecutive significantly up-regulated probes (FDR ≤ 5%) in the subtracted data. Probe data from MDS42 deletions were discarded prior to CMARRT analysis to avoid significant transcripts that spanned deletions being called as two distinct transcripts. The significance of overlap between two genome features (e.g., significantly up-regulated transcripts in BCM-treated cells and significantly up-regulated transcripts in ΔnusG cells) was determined by counting the number of overlapping features before and after rotation of the positions of one set of features by 1 Mb. Significance was then determined using the Mann-Whitney U-test.
ChIP–chip, data normalization, processing, and significant region identification
ChIP–chip was performed as previously described (Mooney et al. 2009a). The monoclonal antibody against RNAP (anti-β, NT63) was purchased from Neclone, and polyclonal antisera against H-NS (Harlan Laboratories) was purified by adsorbsion with cell power from an E. coli Δhns strain. ChIP–chip data were normalized and averaged as previously described (Mooney et al. 2009a). The H-NS ChIP–chip data shown in Figure 4 were smoothed using two rounds of sliding-window averaging over 300 bp. Significant increases in RNAP occupancy in ΔnusG and ΔnusA* cells were identified using CMARRT as previously described for BCM (Peters et al. 2009), except that probe data from MDS42 deletions were discarded prior to CMARRT analysis to avoid regions of increased RNAP occupancy that spanned deletions being called as two significant regions. Significant regions of H-NS occupancy were defined using CMARRT as regions of at least three consecutive probes that were significantly above background (FDR ≤ 5%).
Data sets
Tiling expression, RNA-seq, and ChIP–chip data sets were deposited at Gene Expression Omnibus (GEO) with the accession code GSE41940.
Acknowledgments
We thank members of the Landick laboratory, A.B. Banta (University of Wisconsin at Madison), and M.E. Gottesman (Columbia University) for critical reading of the manuscript; B.K. Cho and B.Ø. Palsson (University of California at San Diego) for sharing their tiling expression microarray protocol; D.H. Keating (Great Lakes Bioenergy Research Center), D.L. Court (National Cancer Institute at Frederick), and M E. Gottesman (Columbia University) for strains; the U.S. DOE Joint Genome Institute (Walnut Creek, CA) for RNA-seq library preparation and sequencing; and H. Yan (Great Lakes Bioenergy Research Center) for RNA-seq read mapping. This work was supported by the National Institutes of Health (GM38660). RNA-seq work was supported by the U.S. DOE Great Lakes Bioenergy Research Center (DOE Office of Science BER DE-FC02-07ER64494) and the U.S. DOE Joint Genome Institute (DOE Office of Science contract no. DE-AC02-05CH11231).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.196741.112.
References
- Adhya S, Gottesman M, De Crombrugghe B 1974. Termination and antitermination in transcription: Control of gene expression. Basic Life Sci 3: 213–221 [DOI] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2: 2006.0008 doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Banerjee S, Chalissery J, Bandey I, Sen R 2006. Rho-dependent transcription termination: More questions than answers. J Microbiol 44: 11–22 [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA 2004. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol Microbiol 52: 1255–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon M, McGowan SJ, Cook PR 2006. Many expressed genes in bacteria and yeast are transcribed only once per cell cycle. FASEB J 20: 1721–1723 [DOI] [PubMed] [Google Scholar]
- Boudvillain M, Nollmann M, Margeat E 2010. Keeping up to speed with the transcription termination factor Rho motor. Transcription 1: 70–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA 2011. Sen-sing RNA terminators. Mol Cell 42: 717–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, Rosch P 2010. A NusE:NusG complex links transcription and translation. Science 328: 501–504 [DOI] [PubMed] [Google Scholar]
- Burns CM, Richardson JP 1995. NusG is required to overcome a kinetic limitation to Rho function at an intragenic terminator. Proc Natl Acad Sci 92: 4738–4742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll KL, Ghirlando R, Ames JM, Corden JL 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Chalissery J, Muteeb G, Kalarickal NC, Mohan S, Jisha V, Sen R 2011. Interaction surface of the transcription terminator Rho required to form a complex with the C-terminal domain of the antiterminator NusG. J Mol Biol 405: 49–64 [DOI] [PubMed] [Google Scholar]
- Chen CY, Richardson JP 1987. Sequence elements essential for ρ-dependent transcription termination at λtR1. J Biol Chem 262: 11292–11299 [PubMed] [Google Scholar]
- Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO 2009. The transcription unit architecture of the Escherichia coli genome. Nat Biotechnol 27: 1043–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Court D, Adhya S 1976. Isolation and characterization of conditional lethal mutants of Escherichia coli defective in transcription termination factor rho. Proc Natl Acad Sci 73: 1959–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ 2009. Nucleoid-associated proteins and bacterial physiology. Adv Appl Microbiol 67: 47–64 [DOI] [PubMed] [Google Scholar]
- Dornenburg JE, Devita AM, Palumbo MJ, Wade JT 2010. Widespread antisense transcription in Escherichia coli. MBio 1: e00024-10 doi: 10.1128/mBio.00024-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour YS, Wesenberg GE, Tritt AJ, Glasner JD, Perna NT, Mitchell JC, Donohue TJ 2010. chipD: A Web tool to design oligonucleotide probes for high-density tiling arrays. Nucleic Acids Res 38: W321–W325 doi: 10.1093/nar/gkq517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ederth J, Mooney RA, Isaksson LA, Landick R 2006. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J Mol Biol 356: 1163–1179 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D 1998. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Dutta D, Wade J, Nudler E 2010. An allosteric mechanism of Rho-dependent transcription termination. Nature 463: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Moulin L, Boccard F 2001. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J Mol Biol 314: 375–386 [DOI] [PubMed] [Google Scholar]
- Fang FC, Rimsky S 2008. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol 11: 113–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA 2004. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Georg J, Hess WR 2011. Cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev 75: 286–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Busby SJ 2008. Global regulators of transcription in Escherichia coli: Mechanisms of action and methods for study. Adv Appl Microbiol 65: 93–113 [DOI] [PubMed] [Google Scholar]
- Grainger DC, Hurd D, Goldberg MD, Busby SJ 2006. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res 34: 4642–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guterman SK, Howitt CL 1979. Rifampicin supersensitivity of rho strains of E. coli, and suppression by sur mutation. Mol Gen Genet 169: 27–34 [DOI] [PubMed] [Google Scholar]
- Hirtreiter A, Damsma GE, Cheung AC, Klose D, Grohmann D, Vojnic E, Martin AC, Cramer P, Werner F 2010. Spt4/5 stimulates transcription elongation through the RNA polymerase clamp coiled-coil motif. Nucleic Acids Res 38: 4040–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irnov I, Sharma CM, Vogel J, Winkler WC 2010. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res 38: 6637–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahramanoglou C, Seshasayee AS, Prieto AI, Ibberson D, Schmidt S, Zimmermann J, Benes V, Fraser GM, Luscombe NM 2011. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res 39: 2073–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyani BS, Muteeb G, Qayyum MZ, Sen R 2011. Interaction with the nascent RNA is a prerequisite for the recruitment of Rho to the transcription elongation complex in vitro. J Mol Biol 413: 548–560 [DOI] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muniz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. 2011. EcoCyc: A comprehensive database of Escherichia coli biology. Nucleic Acids Res 39: D583–D590 doi: 10.1093/nar/gkq1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Bernstein JA, Peter BJ, Rhodius V, Wendisch VF, Zimmer DP 2003. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol Biol 224: 61–78 [DOI] [PubMed] [Google Scholar]
- Koop AH, Hartley ME, Bourgeois S 1987. A low-copy-number vector utilizing β-galactosidase for the analysis of gene control elements. Gene 52: 245–256 [DOI] [PubMed] [Google Scholar]
- Krinke L, Wulff DL 1987. OOP RNA, produced from multicopy plasmids, inhibits λ cII gene expression through an RNase III-dependent mechanism. Genes Dev 1: 1005–1013 [DOI] [PubMed] [Google Scholar]
- Kuan PF, Chun H, Keles S 2008. CMARRT: A tool for the analysis of ChIP–chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput 2008: 515–526 [PMC free article] [PubMed] [Google Scholar]
- Lepore N, Lafontaine DL 2011. A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast. PLoS ONE 6: e24962 doi: 10.1371/journal.pone.0024962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestas I, Vinnicombe G, Paulsson J 2010. Fundamental limits on the suppression of molecular fluctuations. Nature 467: 174–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mason SW, Greenblatt J 1993. Elongation factor NusG interacts with termination factor ρ to regulate termination and antitermination of transcription. Genes Dev 7: 161–172 [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J 2008. SOAP: Short oligonucleotide alignment program. Bioinformatics 24: 713–714 [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Kushner SR 2007. Ribonuclease P processes polycistronic tRNA transcripts in Escherichia coli independent of ribonuclease E. Nucleic Acids Res 35: 7614–7625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R 2009a. Regulator trafficking on bacterial transcription units in vivo. Mol Cell 33: 97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney RA, Schweimer K, Rosch P, Gottesman M, Landick R 2009b. Two structurally independent domains of E. coli NusG create regulatory plasticity via distinct interactions with RNA polymerase and regulators. J Mol Biol 391: 341–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse DE, Guertin M 1972. Amber suA mutations which relieve polarity. J Mol Biol 63: 605–608 [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF 1974. Culture medium for enterobacteria. J Bacteriol 119: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186: 6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N 2006. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res 13: 141–153 [DOI] [PubMed] [Google Scholar]
- Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H, Soldatov A 2009. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res 37: e123 doi: 10.1093/nar/gkp596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman Z, von Hippel PH 2000. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry 39: 5573–5585 [DOI] [PubMed] [Google Scholar]
- Perocchi F, Xu Z, Clauder-Munster S, Steinmetz LM 2007. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res 35: e128 doi: 10.1093/nar/gkm683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Mooney RA, Kuan PF, Rowland JL, Keles S, Landick R 2009. Rho directs widespread termination of intragenic and stable RNA transcription. Proc Natl Acad Sci 106: 15406–15411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM, Vangeloff AD, Landick R 2011. Bacterial transcription terminators: The RNA 3′-end chronicles. J Mol Biol 412: 793–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G, Plunkett G III, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312: 1044–1046 [DOI] [PubMed] [Google Scholar]
- Proshkin S, Rahmouni AR, Mironov A, Nudler E 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328: 504–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Nielsen HB, Jarmer H 2009. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol 73: 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius VA, Suh WC, Nonaka G, West J, Gross CA 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol 4: e2 doi: 10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JW 1969. Termination factor for RNA synthesis. Nature 224: 1168–1174 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Hang M, Smith MM 2011. Histone variant H2A.Z and RNA polymerase II transcription elongation. Mol Cell Biol 31: 1848–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Gowrishankar J 2011a. Compromised factor-dependent transcription termination in a nusA mutant of Escherichia coli: Spectrum of termination efficiencies generated by perturbations of Rho, NusG, NusA, and H-NS family proteins. J Bacteriol 193: 3842–3850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Gowrishankar J 2011b. Modulation of Rho-dependent transcription termination in Escherichia coli by the H-NS family of proteins. J Bacteriol 193: 3832–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinhara A, Matsui M, Hiraoka K, Nomura W, Hirano R, Nakahigashi K, Tomita M, Mori H, Kanai A 2011. Deep sequencing reveals as-yet-undiscovered small RNAs in Escherichia coli. BMC Genomics 12: 428 doi: 10.1186/1471-2164-12-428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MJ, Ames GF, Smith NH, Robinson EC, Higgins CF 1984. Repetitive extragenic palindromic sequences: A major component of the bacterial genome. Cell 37: 1015–1026 [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K 2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 14: 103–105 [DOI] [PubMed] [Google Scholar]
- Sullivan SL, Gottesman ME 1992. Requirement for E. coli NusG protein in factor-dependent transcription termination. Cell 68: 989–994 [DOI] [PubMed] [Google Scholar]
- Terzi N, Churchman LS, Vasiljeva L, Weissman J, Buratowski S 2011. H3K4 trimethylation by Set1 promotes efficient termination by the Nrd1–Nab3–Sen1 pathway. Mol Cell Biol 31: 3569–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomason LC, Costantino N, Court DL 2007. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol 79: 1.17.1–1.17.8 doi: 10.1002/0471142727.mb0117s79 [DOI] [PubMed] [Google Scholar]
- Tran L, van Baarsel JA, Washburn RS, Gottesman ME, Miller JH 2011. Single-gene deletion mutants of Escherichia coli with altered sensitivity to bicyclomycin, an inhibitor of transcription termination factor Rho. J Bacteriol 193: 2229–2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Buratowski S 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A 2008. The Nrd1–Nab3–Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat Struct Mol Biol 15: 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DF, Gottesman ME 1981. The nus mutations affect transcription termination in Escherichia coli. Nature 292: 212–215 [DOI] [PubMed] [Google Scholar]
- Williams RM, Rimsky S, Buc H 1996. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J Bacteriol 178: 4335–4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan F, Platt T 1992. Effects of decreased cytosine content on rho interaction with the rho-dependent terminator trp t′ in Escherichia coli. J Biol Chem 267: 19082–19088 [PubMed] [Google Scholar]
- Zhang Z, Fu J, Gilmour DS 2005. CTD-dependent dismantling of the RNA polymerase II elongation complex by the pre-mRNA 3′-end processing factor, Pcf11. Genes Dev 19: 1572–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Friedman DI 1994. Reduced Rho-dependent transcription termination permits NusA-independent growth of Escherichia coli. Proc Natl Acad Sci 91: 7543–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]