Reinberg and colleagues investigate the link between histone H4K20 methylation status and DNA replication licensing. H4K20 monomethylated by PR-Set7 is sequentially converted to H4K20me2/3 by Suv-20h. These marks are found to be important for ORC recognition and recruitment. Turnover of PR-Set7 by CRL4-Cdt2 is thus required to prevent rereplication. The work highlights the importance of coordinating H4K20 methylation status in replication origin firing.
Keywords: H4K20, ORC, PR-Set7, Suv4-20, cell cycle, chromatin
Abstract
PR-Set7 is the sole monomethyltransferase responsible for H4K20 monomethylation (H4K20me1) that is the substrate for further methylation by Suv4-20h1/h2. PR-Set7 is required for proper cell cycle progression and is subject to degradation by the CRL4Cdt2 ubiquitin ligase complex as a function of the cell cycle and DNA damage. This report demonstrates that PR-Set7 is an important downstream effector of CRL4Cdt2 function during origin of DNA replication licensing, dependent on Suv4-20h1/2 activity. Aberrant rereplication correlates with decreased levels of H4K20me1 and increased levels of H4K20 trimethylation (H4K20me3). Expression of a degradation-resistant PR-Set7 mutant in the mouse embryo that is normally devoid of Suv4-20 does not compromise development or cell cycle progression unless Suv4-20h is coexpressed. PR-Set7 targeting to an artificial locus results in recruitment of the origin recognition complex (ORC) in a manner dependent on Suv4-20h and H4K20me3. Consistent with this, H4K20 methylation status plays a direct role in recruiting ORC through the binding properties of ORC1 and ORCA/LRWD1. Thus, coordinating the status of H4K20 methylation is pivotal for the proper selection of DNA replication origins in higher eukaryotes.
The proper maintenance and control of genome integrity is an important and well-coordinated process. Many factors function to coordinate the proper progression of the cell cycle, including those that directly regulate the chromatin states critical for replication, mitotic condensation, and cell division. Central among these chromatin-associated cell cycle regulators is PR-Set7/Set8/Setd8, the sole mammalian histone H4K20 monomethyltransferase. In embryonic stem (ES) and differentiated cells, PR-Set7 deletion results in massive DNA damage and defects in cell cycle progression (Beck et al. 2012), leading to an accumulation of cells in G2/M phase (Jorgensen et al. 2007; Tardat et al. 2007; Houston et al. 2008; Oda et al. 2009). Moreover, PR-Set7 is degraded prior to S phase and after UV damage by a PCNA/CRL4Cdt2-dependent mechanism, a conserved means to regulate origins of DNA replication licensing (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010; Tardat et al. 2010). Thus, the oscillation of PR-Set7 levels during the cell cycle suggests a means by which H4K20 methylation (H4K20me) regulates cell cycle progression. To date, there are only two known substrates for PR-Set7—H4K20 and p53K382—with most functions attributed to PR-Set7 resting on its catalysis of H4K20 monomethylation (H4K20me1) (Beck et al. 2012).
H4K20me is unique among histone modifications in that it oscillates during the cell cycle. The levels of H4K20me are directly controlled by three sets of enzymes. PR-Set7 catalyzes H4K20me1, which is required for subsequent catalysis of H4K20 di/trimethylation (H4K20me2/3) (Nishioka et al. 2002; Rice et al. 2002) via the activity of Suv4-20h1/h2 (Schotta et al. 2008), or is subject to demethylation, along with H3K9me1, by PHF8 activity (Liu et al. 2010; Qi et al. 2010 ). Misregulation of PR-Set7 or Suv4-20h1/h2 (Suv4-20h) causes increased sensitivity to DNA damage and defects in the cell cycle (Schotta et al. 2008; Oda et al. 2009).
How PR-Set7 and H4K20me1 function to regulate the cell cycle remains an open question. PR-Set7 is normally absent during DNA replication and peaks during mitosis. In addition, the deletion or knockdown of PR-Set7 gives rise to marked defects in both mitosis and S phase. More recently, gain-of-function experiments have suggested a role for PR-Set7 in proper DNA replication (Abbas et al. 2010; Tardat et al. 2010). Expression of a nondegradable PR-Set7 mutant, PR-Set7PIPM2 (PCNA-interacting peptide [PIP] degron mutant), is highly toxic, and this toxicity is dependent on PR-Set7 catalytic activity (Abbas et al. 2010; Centore et al. 2010). While this phenomenon may be due to a multitude of reported defects, a leading culprit is the uncontrolled rereplication of the genome upon expression of PR-Set7PIPM2. Consistent with this, loss of PR-Set7 enzymatic activity causes defects in origin of replication firing (Jorgensen et al. 2007; Tardat et al. 2007).
The mechanisms behind origin of replication firing in higher eukaryotes are poorly understood. In contrast to the case of lower eukaryotes, there is no evidence of a sequence-specific directed mechanism for origin recognition and firing (Gilbert 2010). For this reason, other features of chromatin are candidates as the targets for recruitment of the DNA replication machinery, although, as yet, no specific histone post-translational modification has correlated with origins of replication (Karnani et al. 2010; Eaton et al. 2011; Mesner et al. 2011). Because origin of replication firing does correlate with PR-Set7 activity, PR-Set7 and/or H4K20me1 may potentially provide a chromatin-dependent mechanism for recruitment of the origin recognition complex (ORC).
In this study, we sought to determine how PR-Set7 functions in the origin of replication licensing pathway. We found that the rereplication phenotype caused by loss of the CRL4Cdt2 ubiquitin ligase complex is fully rescued upon codepletion of PR-Set7. Either Cdt2 knockdown or PR-Set7 stabilization results in increased global levels of H4K20me3, suggesting that H4K20me3 is a key effector in the rereplication pathway. In line with this, we show that the defects in cell cycle progression and massive DNA damage that result when cells are exposed to a degradation-resistant version of PR-Set7 are a consequence of Suv4-20h activity. Forced targeting of PR-Set7 using a GAL4-UAS system results in elevated H4K20me3 at this locus and increased ORC recruitment, while depletion of Suv4-20h1 led to a drastic reduction in ORC recruitment. This recruitment involves two different components of ORC—ORC1 and ORCA (LRWD1)—that bind directly to H4K20me2/3 in vitro. Our results suggest that PR-Set7 modulates the chromatin environment such that the chromatin is permissive for replication firing through the downstream action of Suv4-20.
Results
PR-Set7 is a major downstream effector of Cdt2 function
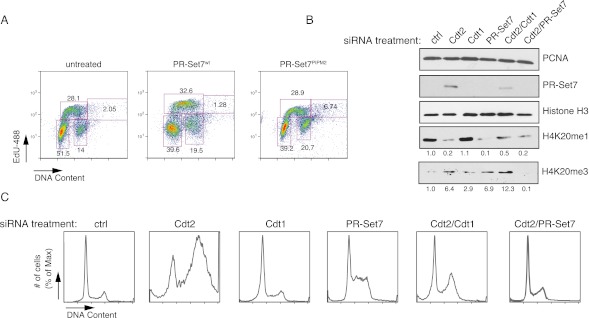
During our initial characterization of PR-Set7PIPM2, we noticed that expression of this mutant elicited toxic effects in a variety of cell lines (Supplemental Fig. 1A). PR-Set7PIPM2 contains mutations in the second PIP domain, PIP degron, that inhibit PR-Set7 binding to PCNA and thus blocks the normal process of PCNA-dependent CRL4Cdt2-mediated degradation of PR-Set7 after UV damage and during S phase (Abbas et al. 2010; Centore et al. 2010; Oda et al. 2010; Tardat et al. 2010). PR-Set7 also contains another PIP domain (PIP1) that is not required for PCNA binding or CRL4Cdt2-mediated degradation (Supplemental Fig. 1A). We first readdressed whether expression of PR-Set7PIPM2 has an effect on cell cycle progression. We expressed a retrovirus encoding PR-Set7PIPM2 in HeLa and U2OS cells that led to sustained PR-Set7 levels, as expected (Supplemental Fig. 1B; data not shown). HeLa cells expressing PR-Set7PIPM2 displayed an altered cell cycle profile with increased population of cells within G2/M phase along with an increased number of cells containing +4n DNA content and increased nuclear volume that are replicating DNA (EdU+) (Fig. 1A; Supplemental Fig. 1C), suggesting rereplication or aberrant origin of replication firing in agreement with previous observations (Abbas et al. 2010; Tardat et al. 2010). Thus, stabilization of PR-Set7 during S phase leads to increased rereplication and G2/M arrest, a unique characteristic of proteins that regulate origin of replication licensing (Arias and Walter 2007; Abbas and Dutta 2011; Havens and Walter 2011), and suggests that the abnormal persistence of PR-Set7 throughout the cell cycle leads to cell cycle alterations.
Figure 1.
PR-Set7 is a critical substrate of CRL4Cdt2 that regulates replication licensing. (A) Cell cycle progression of HeLa cells expressing PR-Set7wt or PR-Set7PIPM2 after FACS analysis measuring DNA and EdU incorporation. (B) Western blot of protein expression after various siRNA treatments in HeLa cells; H3 and PCNA served as loading controls. (C) FACS analysis showing the cell cycle profile of HeLa cells after treatment with various siRNAs, including control, Cdt2, Cdt1, and PR-Set7.
To address this hypothesis directly, we next sought to determine whether the cell cycle defects seen in cells lacking Cdt2 were due to excess PR-Set7 using RNAi-mediated knockdown of Cdt2, PR-Set7 itself, and another Cdt2 degradation target, Cdt1. Cdt2 has been previously shown to participate in cell cycle-regulated degradation of PR-Set7, and we found that upon Cdt2 knockdown, PR-Set7 was stabilized (Fig. 1B; Oda et al. 2010 ). Importantly, loss of Cdt2 also gave rise to robust defects in cell cycle progression, including G2/M arrest and rereplication (Fig. 1C; Supplemental Figs. 2, 3A). Because this phenotype might potentially be due to other Cdt2 targets, we tested whether the stabilization of Cdt1 might contribute to this effect by performing double-knockdown experiments. Depletion of PR-Set7 or Cdt1 rescued the cell cycle arrest observed in cells lacking Cdt2, suggesting that Cdt2 regulates cell cycle progression through control of both PR-Set7 and Cdt1 levels (Fig. 1C; Supplemental Figs. 2, 3A). On the other hand, PR-Set7 siRNA treatment was unable to rescue the cell cycle arrest observed upon knockdown of another, more promiscuous member of the CRL4Cdt2 complex, DDB1 (Supplemental Fig. 4A,B). These findings indicate that PR-Set7 is a specific, critical downstream effector of CRL4Cdt2-regulated origin of replication licensing.
CRL4 Cdt2 has a well-characterized role in regulating the dynamics of origins of DNA replication. To determine the mechanism by which PR-Set7 and Cdt2 may regulate origin of replication licensing, we examined the effects of loss of Cdt2 and PR-Set7 on H4K20 methylation. Loss of Cdt2 alone resulted in PR-Set7 stabilization along with a decrease in H4K20me1 levels and a concomitant increase in H4K20me3 (Fig. 1B). The elevated levels of PR-Set7 likely led to increased levels of H4K20me1 that was efficiently converted to H4K20me3 (Abbas et al. 2010). In keeping with this, the increase in H4K20me3 levels was reversed upon codepletion of PR-Set7, suggesting that a potential cause of rereplication is the overabundance of H4K20me3. In contrast, codepletion of Cdt1 did not restore H4K20me3 levels (Fig. 1B). Although the observed alterations in H4K20me3 levels likely reflect changes in PR-Set7 levels, we cannot exclude the possibility that changes in the cell cycle may contribute. Our findings are partially consistent with a recent report indicating that H4K20me3 levels are elevated during G1/S phase (Zee et al. 2012).
Suv4-20h1/h2 are required for PR-Set7PIPM2-mediated cell cycle defects in differentiated cells
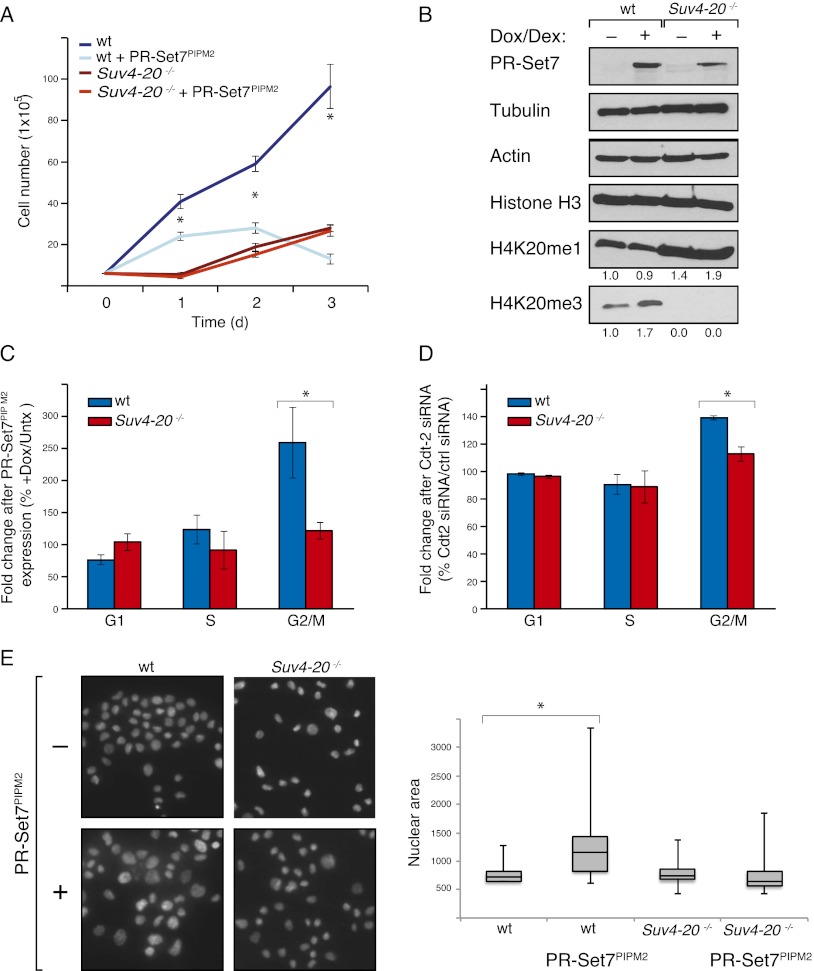
Despite the defined role of H4K20me1 in mitotic condensation, it is not clear how this modification is biologically relevant during DNA replication, since both PR-Set7 and H4K20me1 are mostly absent during S phase. As a first step toward understanding this enigma, we sought to determine whether the function of PR-Set7 during DNA replication was dependent on the downstream di- and trimethylation states of H4K20me catalyzed by Suv4-20h1/h2 (Schotta et al. 2008), given that H4K20me2/3 are at their highest levels during G1 and S phases (Abbas et al. 2010). To test this possibility, we generated wild-type and Suv4-20h1/h2−/− mouse embryonic fibroblasts (MEFs) (Suv4-20 dn) in which the PR-Set7PIPM2 is expressed under the control of an inducible doxycycline/dexamethasone-responsive promoter (Fig. 2B). Expression of PR-Set7PIPM2 in wild-type MEFs caused a striking reduction in growth rate (Fig. 2A) but was ineffectual in the case of Suv4-20 dn MEFs (Fig. 2A). Expression of the PR-Set7PIPM2 led to both a slight decrease in H4K20me1 levels and an increase in H4K20me3 levels in wild-type MEFs, neither of which was observed in the case of Suv4-20 dn MEFs (Fig. 2B). These results implicate a globally increased transition from the mono- to the trimethylated state of H4K20 in the pathway leading to cell cycle defects mediated by PR-Set7PIPM2.
Figure 2.
PR-Set7PIPM2 toxicity is dependent on Suv4-20 and H4K20me2/3 in MEFs. (A) Proliferation of wild-type (wt) and Suv4-20−/− MEFs after inducible expression of Flag-HA PR-Set7PIPM2. Induction of PR-Set7PIPM2 after addition of doxycycline and dexamethasone at day 0. (B) Western blot of wild-type and Suv4-20−/− MEF lysates after inducible expression of PR-Set7PIPM2. (C) FACS analysis of cell cycle distribution of wild-type and Suv4-20−/− MEFs before and after induction of PR-Set7PIPM2. (D) FACS analysis of cell cycle distribution of wild-type and Suv4-20−/− MEFs before and after treatment with siRNA against Cdt2. (E) DAPI staining and quantification of the nuclear size of wild-type and Suv4-20−/− MEFs after induction of PR-Set7PIPM2. n = 3 for proliferation and cell cycle experiments. (*) Statistical significance with P < .05.
We next compared the cell cycle profile of wild-type and Suv4-20 dn MEFs as a function of PR-Set7PIPM2 expression. In contrast to wild-type MEFs in which PR-Set7PIPM2 expression led to a 2.5-fold increase in G2/M-phase cells, similar to our observation in HeLa cells and earlier work by other groups (Centore et al. 2010; Tardat et al. 2010), Suv4-20 dn MEFs did not display changes in the cell cycle profile upon expression of PR-Set7PIPM2 (Fig. 2C). Similarly, Cdt2 siRNA treatment of wild-type but not Suv4-20 dn MEFs caused an accumulation of cells in G2/M phase (Fig. 2D; Supplemental Fig. 3B). Importantly, untreated wild-type and Suv4-20 dn MEFs have similar amounts of cells in G2/M phase (Supplemental Fig. 5). Finally, while wild-type MEFs showed increased nuclear volume upon induction of PR-Set7PIPM2 expression, this was not the case for Suv4-20 dn MEFs (Fig. 2E). These combined results demonstrate that the cell cycle progression defects caused by PR-Set7 stabilization during S phase are dependent on Suv4-20h and its products, H4K20me2/3.
Absence of H4K20me3 prevents toxicity of PR-Set7PIPmt and DNA damage in PR-Set7−/− early embryos
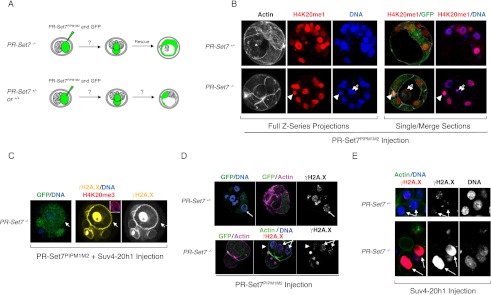
We previously showed that deletion of PR-Set7 in mice results in embryonic death at the transition between the four- and eight-cell stage (Oda et al. 2009). This lethality can be rescued upon reintroduction of a catalytically active PR-Set7 into PR-Set7−/− embryos. In contrast to other cell types in which PR-Set7 is depleted, there is no accumulation of DNA damage in the PR-Set7−/− embryo (Oda et al. 2009). To better understand the aberrant functioning of PR-Set7PIP mutants in vivo, we first investigated whether PR-Set7PIPM1M2 expression could rescue the PR-Set7−/− embryos. We collected two-cell stage embryos from PR-Set7−/+ crosses and injected one of the two cells with mRNA encoding PR-Set7PIPM1M2 along with mRNA encoding a fluorescent marker as a lineage tracer (Fig. 3A). In this way, if PR-Set7PIPM1M2 is able to rescue the lethality caused upon loss of PR-Set7, the injected cell of the two-cell stage embryo of PR-Set7−/− embryos is expected to develop as normal and generate a blastocyst, while the other cell would serve as an internal control (Fig. 3A; Oda et al. 2009). Also, these experiments should reveal whether wild-type and heterozygous embryos would tolerate PR-Set7PIPM1M2 expression (Fig. 3A). Surprisingly, expression of PR-Set7PIPM1M2 was able to completely rescue the PR-Set7−/− embryos (Fig. 3B). Moreover, PR-Set7PIPM1M2 was tolerated in wild-type and heterozygous embryos (Fig. 3B). Importantly, expression of PR-Set7PIPM1M2 in embryonic nuclei did not lead to increased accumulation of γH2A.X (Fig. 3D). These results are in stark contrast to the phenotype observed upon expression of PR-Set7PIPM1M2 in differentiated cells, which led to the accumulation of DNA damage as well as multiple cell cycle defects, including G2/M-phase arrest and rereplication (Abbas et al. 2010; Tardat et al. 2010). Thus, the early embryo appears to contain control mechanisms for cell cycle progression and checkpoint control that are distinct from those within differentiated cell lines.
Figure 3.
Both PR-Set7PIPM2 and PR-Set7−/− phenotypes are dependent on Suv4-20 in the early embryo. (A) Two-cell stage embryo injection experiment schematic depicting the possible outcomes for wild-type and heterozygous PR-Set7 embryos versus the null embryos. (B) PR-Set7+/− and PR-Set7−/− two-cell stage embryos were injected with PR-Set7PIPM1M2 mRNA. Injected cells are marked with GFP; the polar body is labeled by arrowheads, and GFP+ cells are indicated by arrows. Embryos were monitored for growth and stained with H4K20me1 (red). Full Z-series projections and single sections are depicted. (C) PR-Set7−/+ and PR-Set7−/− were injected with mRNA for PR-Set7PIPM1M2 in combination with Suv4-20h1 mRNA and GFP as a tracer. The arrows point to one of the injected cells. Embryos were stained with H4K20me3 (red) and γH2A.X (yellow). Insets show an injected cell stained for H4K20me3 (red) and DAPI (blue). (D) PR-Set7−/+ and PR-Set7−/− were injected with PR-Set7PIPM1M2. Arrows point to injected cells, which were identified by the presence of GFP; the polar body is marked by arrowheads. Embryos were stained with γH2A.X, cortical actin, and DAPI (blue). (E) PR-Set7−/− and PR-Set7−/+ embryos were injected with Suv4-20h1 mRNA alone and stained for cortical actin (green) and γH2A.X (red) accumulation. Injected cells (marked by arrows) were identified by the presence of GFP (not shown). The genotypes of individual embryos were determined after confocal acquisition as described previously.
A particular feature of early embryonic chromatin is the absence of most typical heterochromatic marks, including H4K20me3 (Kourmouli et al. 2004; Burton and Torres-Padilla 2010). Given our results above, the lack of Suv4-20h activity in the early embryo may explain why PR-Set7PIPM1M2 does not result in growth arrest and elevated DNA damage. Thus, we next investigated whether coexpression of Suv4-20h1 in the embryo would lead to the toxicity associated with PR-Set7PIPM1M2 in differentiated cells. Indeed, in contrast to injection of the PR-Set7PIPM1M2 transcript alone, coinjection with the Suv4-20h1 transcript failed to rescue the PR-Set7-null embryos. Instead, expression of PR-Set7PIPM1M2 and Suv4-20h1 caused growth arrest and high levels of γH2A.X (Fig. 3C). This toxicity occurred independently of the PR-Set7 embryo genotype. These findings show that the toxic effects caused by stabilization of PR-Set7 during S phase are mediated through Suv4-20 and, likely, H4K20me2/3.
Contrary to ES cells and transformed cells, loss of PR-Set7 in the early embryo does not trigger DNA damage (Jorgensen et al. 2007; Oda et al. 2009). The results above suggest that the difference between PR-Set7 phenotypes in the early embryo, compared with ES cells and transformed cell lines, is dependent on Suv4-20. In support of this hypothesis, PR-Set7−/− embryos show massive DNA damage only when injected with Suv4-20h1 mRNA, as demonstrated by γH2A.X accumulation (Fig. 3E). Importantly, expression of Suv4-20h1 in wild-type or heterozygous embryos did not elicit DNA damage (Fig. 3E). Altogether, the lack of Suv4-20h activity in the normal embryo revealed two insights into PR-Set7 biology during S phase: The PR-Set7PIPM2-mediated toxicity and the DNA damage incurred upon loss of PR-Set7 are both dependent on Suv4-20h and H4K20me2/3. These results confirm that PR-Set7 has essential functions in the embryo unrelated to Suv4-20h, but that those related to S phase in differentiated cells can be recapitulated in embryos forced to express Suv4-20h.
PR-Set7 recruits the ORC through direct binding of ORC1 and ORCA to H4K20me2/3
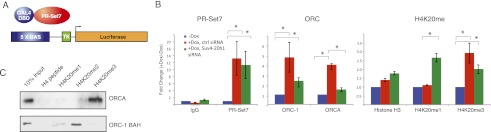
Given our findings that PR-Set7 misregulation leads to disruption in the cell cycle and abnormal DNA replication through Suv4-20h and H4K20me2/3, we next explored the mechanistic basis for the link between H4K20me2/3 and appropriate DNA replication. Our previous work, along with that of others, has shown that the ORC binds to H4K20me2/3 in vitro (Oda et al. 2010; Vermeulen et al. 2010). We hypothesized that H4K20me2/3 may serve as a recruitment mechanism for specific components of the ORC. To test this hypothesis, we first examined whether artificial recruitment of GAL4-PR-Set7 to a UAS site would lead to ORC recruitment. Using a 293T-REx cell line containing a stably integrated 5xUAS site directing luciferase expression, we expressed GAL-4 PR-Set7 under an inducible doxycycline-responsive promoter (Fig. 4A; Li et al. 2010). Chromatin immunoprecipitation (ChIP) analysis was performed at the UAS site as a function of GAL4-PR-Set7 expression. PR-Set7 recruitment to the artificial UAS locus led to an approximately fourfold increase in ORCA and an approximately fivefold increase in ORC1 recruitment relative to untreated cells (Fig. 4B). Coincident with this recruitment was a statistically significant accumulation of H4K20me3 at the UAS site, which was generated from an earlier accumulation of H4K20me1 (Fig. 4B). Upon siRNA treatment for Suv4-20h1, H4K20me3 decreased at the UAS site with a concomitant reduction in ORC-1 and ORCA recruitment (Fig. 4B; Supplemental Fig. 3). These data confirm that PR-Set7 participates in ORC recruitment at specific loci, without the requirement for any upstream factors, through Suv4-20 and H4K20me3 (Tardat et al. 2010).
Figure 4.
PR-Set7 recruits the ORC to chromatin through H4K20me. (A) Diagram depicting the GAL4-UAS system in 293T-REx cells. (B) ChIP analysis at an integrated UAS-binding site in 293T-REx cells before and after inducible expression of GAL4-PR-Set7 using GAL4, PR-Set7, H3, H4K20me1, H4K20me3, ORC1, and ORCA antibodies. (C) Peptide pull-down for the GST-ORC1 BAH domain and His-ORCA binding to unmethylated, H4K20me1, H4K20me2, and H4K20me3 peptides. (*) Statistical significance with P < 0.05.
After determining that ORC can be directly recruited to chromatin by PR-Set7, we next tested whether any ORC components can directly bind H4K20me2/3. After analyzing predicted chromatin-binding domains (http://www.uniprot.org), we found that only two members of ORC could potentially accomplish this function: ORC1 with its BAH domain and ORCA with its WD40 domain. Recently, the ORC1 BAH domain was shown to specifically bind H4K20me2, further supporting this connection (Kuo et al. 2012). We used the BAH domain of ORC1 (amino acids 1–185) and full length ORCA in peptide-binding studies. The ORC-1 BAH domain bound primarily H4K20me2 in vitro (Fig. 4C). In contrast, ORCA showed a preference for H4K20me3. Thus, in combination with our previous unbiased peptide-binding experiments (Oda et al. 2010), these findings demonstrate that both ORC1 and ORCA are involved in recruitment of the ORC to H4K20me2/3 sites. Together, our results show a direct mechanistic connection between PR-Set7 and the proper recruitment of the ORC through the products of Suv4-20 activity to effectively promote DNA replication.
Discussion
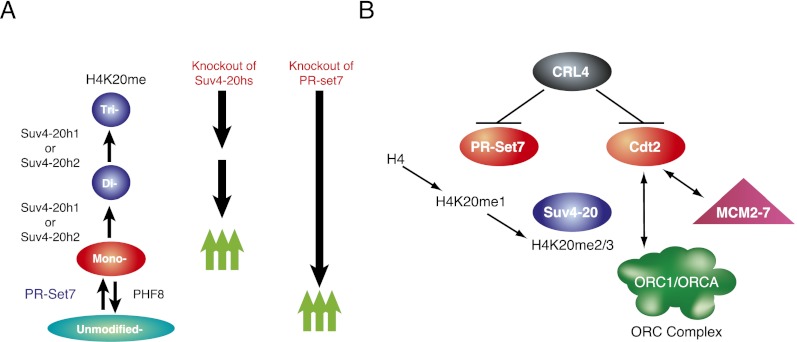
In this study, we showed that the oscillation in PR-Set7 protein levels during the cell cycle has direct repercussions on the proper recruitment of the ORC through H4K20me2/3 and Suv4-20h (Fig. 5). The disruption in PR-Set7 oscillation and the associated catalysis of H4K20me1 lead to faulty DNA replication. Since the initial identification of PR-Set7 and Suv4-20, H4K20me has been functionally linked to proper cell cycle progression, specifically during mitosis and DNA replication (Fig. 5). This study places PR-Set7 and Suv4-20h as key chromatin mediators of DNA replication and specifies how their activities function in conjunction to ensure bona fide ORC recruitment to chromatin in higher eukaryotic cells.
Figure 5.
Summary of function of PR-Set7 and Suv4-20 (A) along with their role in regulation of origin of replication licensing through CRL4Cdt2 (B).
We found that aberrant PR-Set7 stabilization during S phase causes robust rereplication. Rereplication was previously shown to be caused by the loss of the ubiquitin ligase Cdt2 (Arias and Walter 2007), and this abnormality can be completely suppressed by codepletion of PR-Set7. These results place PR-Set7 as a major effector of Cdt2 function. We also found that cell cycle defects in MEFs that express PR-Set7PIPM2 are dependent on Suv4-20h and H4K20me2/3. Surprisingly, expression of PR-Set7PIPM1M2 is not toxic during early embryogenesis and can even rescue PR-Set7−/− embryos to an extent similar to wild-type PR-Set7. This is due to the minimal amount of H4K20me2/3 inherent to early embryogenesis. Consistent with this interpretation, coexpression of PR-Set7PIPM1M2 along with Suv4-20h1, as opposed to expression of either gene alone, caused growth arrest in the embryo. Together, these results connect PR-Set7 and Suv4-20h with regulation of replication during S phase and DNA damage and accentuate that different regulatory processes guide the cell cycle between the early embryo and differentiated cells. Finally, we determined how H4K20me affects origin licensing by demonstrating that PR-Set7 can trigger recruitment of the ORC to chromatin and that ORC1 and ORCA bind H4K20me directly.
The sequence and determinants of origins of DNA replication in higher eukaryotes have remained elusive despite extensive investigation (Gilbert 2010). The metazoan ORC does not contain any intrinsic DNA-binding specificity and thus must require additional factors to be properly recruited. A leading hypothesis in the field has been that chromatin may be involved in this process. Our study provides mechanistic insights into how H4K20me may function during this critical process. Taken together, the regulation and timing of H4K20me1/2/3 is critical for proper origin of replication firing.
In vivo, H4K20 is primarily found in both the mono- and dimethylation state, raising the question of how these highly ubiquitous modifications affect the selection of DNA replication origins. Our findings, along with the previously demonstrated importance of regulation and timing of H4K20 methylation, suggest that H4K20me3, rather than H4K20me2, acts as a more specific guide for origin of replication selection, although both marks may contribute to this process. This situates H4K20me and the degree of such methylation as key components in an oscillating control mechanism for proper cell cycle progression, controlled in part by the degradation of PR-Set7. In addition, differential regulation of these modifications between the early embryo and later lineage-committed cells demonstrates the contrasting requirements for cell cycle progression during development.
Materials and methods
Microinjection and embryo analysis
Two-cell stage mouse embryos were isolated from PR-Set7−/+ crosses at 46 h post-hCG hormonal stimulation. Embryos were microinjected with 1–2 pL of mRNA transcribed in vitro from the pRN3P expression vector that contains 5′ and 3′ untranslated regions (UTRs) of the β-globin as described previously (Oda et al. 2009). Embryos were cultured in KSOM droplets under mineral oil under a 5% CO2 atmosphere at 37°C and monitored for developmental progression daily. For immunostaining, embryos were fixed as described earlier (permeabilization with 0.5% Triton and washed three times in PBS-T [0.1% Tween in PBS]), blocked, and incubated with the primary antibodies in 3% BSA and 0.1% Tween in PBS for ∼12 h at 4°C. Embryos were then washed twice in PBS-T, blocked for 30 min, and incubated with the corresponding secondary antibodies and 4′-6-diamidino-2-phenylindole. Embryos were then analyzed in individual drops using a TCS SP5 confocal microscope (Leica) and genotyped after acquisition.
Cell culture and flow cytometry
Suv4-20−/− and wild-type MEFs were kindly provided by Drs. Gunnar Schotta and Thomas Jenuwein. 293T-REx cells were purchased from Invitrogen, and HeLa and U2OS were acquired from the American Type Culture Collection (ATCC). Cells were grown using DMEM, 10% FBS, 1% penicillin, 1% streptomycin, and 1% nonessential amino acids. For proliferation assays, cells were counted using a hemacytometer or cyto60 dye (Invitrogen). For flow cytometry, cells were trypsinized, washed once in cold PBS, and fixed in 70% ethanol for 20 min on ice. For cell cycle analyses, cells were washed and resuspended in PBS containing RNase A (100 μg/mL) and propidium iodide (40 μg/mL). EdU labeling was performed using the Click-iT EDU Alexa Fluor 488 flow cytometry assay kit (Invitrogen). Samples were run on a FACScalibur (Becton Dickinson), and data analysis was performed using FloJo (Dean-Jett-Fox cell cycle analysis platform) and ModFit LT 3.2.1.
Protein extracts and microscopy
Cells were lysed with buffer A (10 mM HEPES at pH 7.9, 0.1% NP-40, 10 mM MgCl2, 250 mM sucrose) containing protease inhibitors (Roche) and centrifuged. Nuclear pellets were lysed with buffer B (25 mM HEPES at pH 7.9, 20% glycerol, 0.7 M NaCl, 1.5 mM MgCl2, 0.1 mM EDTA), sonicated, and centrifuged. Proteins were separated on sodium dodecyl sulfate–polyacrylamide gels. For microscopy, cells were grown on coverslips, fixed with 4% paraformaldehyde, and stained using DAPI dye.
Gene silencing by siRNA and quantitative PCR (qPCR)-based detection
siRNA transfections were performed using RNAiMAX (Invitrogen) according to the manufacturer's specifications. The nucleotide sequences of siRNA and the primers used for detection are listed below. siRNAs for Hs PR-Set7 and si-AllStars as a negative control were purchased from Qiagen. siRNAs for Hs Cdt2 and Hs DDB1 were obtained from Invitrogen. siRNAs for Hs Cdt1, Hs Suv4-20h1, and Mm Cdt2 were ordered from Dharmacon.
siRNA oligos used were as follows: Hs PR-Set7-7 (target sequence: CTGCAGTCTGAAgAAAGGAAA), Hs PR-Set7-8 (sense, CCUGUUGAUUGCCAAAAACTTdTdT; antisense, GUUUUUGGCAAUCAACAGGdTdT), Hs Cdt2-1 (sense, GAAUUAUACUGCUUAUCGAdTdT; antisense, UCGAUAAGCAGUAUAAUUCdTdT), Hs DDB1-1 (sense, ACAGAGUGGCGAGAGCAUUdTdT; antisense, AAUGCUCUCGCCACUCUGUdTdT), Hs DDB1-2 (sense, CCUGUUGAUUGCCAAAAACTTdTdT; antisense, GUUUUUGGCAAUCAACAGGdTdT), Hs Cdt-1 (Dharmacon si genome pool), and Mm Cdt-2 (Dharmacon si genome pool).
Detection primers used were as follows: Hs Cdt1 1508 S (GCTGTTGTACTATCATGAGCCC), Hs Cdt1 1626 AS (GTCCAGCTTGACGTAGGTGTC), Hs Cdt2 308 S (GACCCTGATGCTCTTCAATTC), Hs Cdt2 402 AS (ACCACTGCACTGATAACCAGTC), Mm Cdt2 24 S (ACGATGAACACACGTCTTATGG), Mm Cdt2 357 AS (AGCCTTCTTCATTAGCAACTGC), Mm mGapdh S (CAAGCTCATTTCCTGGTATGAC), Mm Gapdh AS (CTCCTGTTATTATGGGGGTCTG), Hs Gapdh S (AACCATGAGAAGTATGACAACAGC), and Hs Gapdh AS (CAGTCTTCTGGGTGGCAGTG).
Plasmids
Human PR-Set7 wild-type and mutants were subcloned into the pMSCV Puro retroviral vector (Invitrogen). pINTO GAL4 PR-Set7 and 3xFlag-tagged PR-Set7 were cloned as an XhoI/BamHI fragment into the pMSCV TtIGPP vector (S. Lowe Laboratory, Cold Spring Harbor Laboratory [CSHL]). iTetONGR* (Anastassiadis et al. 2002) was cloned as an EcoRI/BamHI fragment into pMSCV MB (M. Huebner, CSHL). Additional Suv4-20h1 and PR-Set7PIPM1M2 were cloned into the pRNP3 vector. ORC-1 and ORCA domains were cloned from HeLa cDNA into pGEX6P1 (GE Life Sciences) and pET30a (Novagen), respectively, using BamHI and NotI restriction enzymes. Retrovirus was produced in 293T cells according to standard methods and then used to infect MEFs.
Antibodies and reagents
The sources for the following reagents were as follows: PR-Set7 (custom), H3 (Abcam), H4K20me1 (Abcam), H4K20me3 (Abcam), γH2A.X (Millipore), DDB1 antibody (Invitrogen), β-tubulin (Abcam), PCNA (Santa Cruz Biotechnology), actin (Abcam), ORC1 (Abcam), ORCA (Bethyl), and GAL4 (Millipore).
ChIP
ChIP was performed using 293T-REx inducible cell lines with a stably inserted 6xUAS-TK-LUC. ChIP was performed 48 h after induction of GAL4-PR-Set7 expression using 1 μg/mL doxycycline. Detection was performed using primers against the TK Luc promoter (5′-CACCGAGCGACCCTGCATAAG-3′ and 5′-GCTTCTGCCAACCGAACGGAC-3′). ChIP protocol was performed as described previously (Gao et al. 2012).
Peptides and recombinant pull-downs
GST-ORC1 BAH domain (amino acids 1–185) and His-ORCA (full length) were incubated with biotinylated H4 peptides (amino acids 9–28) with specified post-translational modifications. One microgram of the above recombinant protein was incubated with 1 μg of peptide (preincubated with Streptactin macroprep beads [IBA]) for 4 h to overnight and then washed with 10mM Tris (pH 7.9), 500 mM KCl, 0.2mM EDTA, and 5% Sarkosyl detergent prior to binding analysis by Western blotting.
Acknowledgments
We are grateful to Stephanie Kim, Deborah Hernandez, and Brent Kopenhaver for technical assistance, and members of the Reinberg laboratory for helpful discussions. We also thank Drs. Gunnar Schotta and Thomas Jenuwein for Suv4-20 dn MEFs, Gary Felsenfeld for insulator DNA sequences, Pascal Eberling for preparing H4K20me peptides, Michael Huebner and David Spector for retroviral inducible system, Zhiguo Zhang for the ORCA baculovirus, and Lynne Vales for comments on the manuscript. This work was supported by grants from the NIH (GM64844 to D.R.) and HHMI (to D.R.). Research in the Torres-Padilla laboratory was supported by ANR-09-Blanc-0114, FP7 EpiGeneSys NoE, and an ERC-Stg Grant (“NuclearPotency”). A.B. is supported by a post-doctoral fellowship from the Fondation pour la Recherche Medicale.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.195636.112
References
- Abbas T, Dutta A 2011. CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle 10: 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas T, Shibata E, Park J, Jha S, Karnani N, Dutta A 2010. CRL4(Cdt2) regulates cell proliferation and histone gene expression by targeting PR-Set7/Set8 for degradation. Mol Cell 40: 9–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Kim J, Daigle N, Sprengel R, Scholer HR, Stewart AF 2002. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene 298: 159–172 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Beck DB, Oda H, Shen SS, Reinberg D 2012. PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev 26: 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton A, Torres-Padilla ME 2010. Epigenetic reprogramming and development: A unique heterochromatin organization in the preimplantation mouse embryo. Brief Funct Genomics 9: 444–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centore RC, Havens CG, Manning AL, Li JM, Flynn RL, Tse A, Jin J, Dyson NJ, Walter JC, Zou L 2010. CRL4(Cdt2)-mediated destruction of the histone methyltransferase Set8 prevents premature chromatin compaction in S phase. Mol Cell 40: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45: 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM 2010. Evaluating genome-scale approaches to eukaryotic DNA replication. Nat Rev Genet 11: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havens CG, Walter JC 2011. Mechanism of CRL4(Cdt2), a PCNA-dependent E3 ubiquitin ligase. Genes Dev 25: 1568–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC 2008. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem 283: 19478–19488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS 2007. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol 179: 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnani N, Taylor CM, Malhotra A, Dutta A 2010. Genomic study of replication initiation in human chromosomes reveals the influence of transcription regulation and chromatin structure on origin selection. Mol Biol Cell 21: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Jeppesen P, Mahadevhaiah S, Burgoyne P, Wu R, Gilbert DM, Bongiorni S, Prantera G, Fanti L, Pimpinelli S, et al. 2004. Heterochromatin and tri-methylated lysine 20 of histone H4 in animals. J Cell Sci 117: 2491–2501 [DOI] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Margueron R, Ku M, Chambon P, Bernstein BE, Reinberg D 2010. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev 24: 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. 2010. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature 466: 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, Bekiranov S 2011. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res 21: 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka K, Rice JC, Sarma K, Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P, Tempst P, Steward R, et al. 2002. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell 9: 1201–1213 [DOI] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D 2009. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol 29: 2278–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D 2010. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell 40: 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Sarkissian M, Hu GQ, Wang Z, Bhattacharjee A, Gordon DB, Gonzales M, Lan F, Ongusaha PP, Huarte M, et al. 2010. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature 466: 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. 2008. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev 22: 2048–2061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E 2007. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol 179: 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Zee BM, Britton LM, Wolle D, Haberman DM, Garcia BA 2012. Origins and formation of histone methylation across the human cell cycle. Mol Cell Biol 32: 2503–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]