The E. coli transcription factor DksA mediates the effects of nutritional stress response via directly binding RNA polymerase (RNAP) and regulating the transcription of hundreds of target genes. Using cutting-edge site-specific photo-cross-linking and distance-restrained docking approaches, Gourse and colleagues generate and then functionally validate a structural model of DksA's interaction with RNAP, elucidating how DksA exerts its effects on the polymerase.
Keywords: RNA polymerase, promoter, DksA, ppGpp, transcription initiation, ribosome synthesis
Abstract
Escherichia coli DksA is a transcription factor that binds to RNA polymerase (RNAP) without binding to DNA, destabilizing RNAP–promoter interactions, sensitizing RNAP to the global regulator ppGpp, and regulating transcription of several hundred target genes, including those encoding rRNA. Previously, we described promoter sequences and kinetic properties that account for DksA's promoter specificity, but how DksA exerts its effects on RNAP has remained unclear. To better understand DksA's mechanism of action, we incorporated benzoyl-phenylalanine at specific positions in DksA and mapped its cross-links to RNAP, constraining computational docking of the two proteins. The resulting evidence-based model of the DksA–RNAP complex as well as additional genetic and biochemical approaches confirmed that DksA binds to the RNAP secondary channel, defined the orientation of DksA in the channel, and predicted a network of DksA interactions with RNAP that includes the rim helices and the mobile trigger loop (TL) domain. Engineered cysteine substitutions in the TL and DksA coiled-coil tip generated a disulfide bond between them, and the interacting residues were absolutely required for DksA function. We suggest that DksA traps the TL in a conformation that destabilizes promoter complexes, an interaction explaining the requirement for the DksA tip and its effects on transcription.
Most bacterial transcription factors are DNA-binding proteins that recognize a particular DNA sequence, thereby regulating transcription of nearby promoters. However, the 151-amino-acid (17.5-kDa) DksA protein does not bind to DNA, but rather has been proposed to bind to the RNA polymerase (RNAP) secondary channel (Perederina et al. 2004; Lennon et al. 2009; Rutherford et al. 2009). DksA regulates transcription in conjunction with the small molecules guanosine tetraphosphate and pentaphosphate (referred to here as ppGpp) and the transcript's initiating nucleotide (iNTP) by modifying the kinetic properties of the promoter–RNAP complex (Barker et al. 2001; Paul et al. 2004). Although the concentration of DksA remains relatively constant during growth of Escherichia coli (Paul et al. 2004; Rutherford et al. 2007; Chandrangsu et al. 2011), the concentrations of ppGpp and NTPs fluctuate, accounting for the dynamic effects of DksA on transcriptional responses to changing nutritional conditions (Murray et al. 2003; Paul et al. 2004).
DksA and ppGpp together inhibit transcription from rRNA promoters, many ribosomal protein and tRNA promoters, the promoter for FlhDC (the master regulator of flagella synthesis), the Fis promoter, DksA's own promoter, and as many as 300 others and directly activate promoters for amino acid biosynthesis and/or transport, virulence, the sRNA-binding protein Hfq, σE-dependent transcription, and as many as 400 others (for review,see Haugen et al. 2008; see also Potrykus et al. 2006; Magnusson et al. 2007; Durfee et al. 2008; Traxler et al. 2008; Lemke et al. 2009, 2011; Chandrangsu et al. 2011; J Lemke and RL Gourse, unpubl.). However, the mechanisms responsible for inhibition and activation remain ill-defined.
Transcription initiation begins with the binding of RNAP holoenzyme (subunit composition α2ββ′ωσ) to promoter DNA to form an initial closed complex (RPC) (Haugen et al. 2008; Saecker et al. 2011). This complex isomerizes to additional intermediates, abbreviated here as RPI, before formation of a transcriptionally competent open complex (RPO) and RNA synthesis (Haugen et al. 2008; Saecker et al. 2011). DksA inhibits the transition from RPC to RPI (Rutherford et al. 2009). We proposed that by binding in the RNAP secondary channel, DksA causes an allosteric change in the bridge helix (BH) and/or the trigger loop (TL) that ultimately affects the switch regions of RNAP. The switches interact with the promoter near the transcription start site and control RNAP clamp opening, thereby affecting DNA contacts further downstream in RPO (Rutherford et al. 2009).
Rather than achieving promoter specificity by binding to specific DNA sites, DksA exploits promoter-specific variation in the kinetics of transcription initiation (Paul et al. 2004, 2005; Rutherford et al. 2009). At most promoters, the RNAP–promoter complex is long-lived. However, at rRNA promoters, this complex is intrinsically short-lived, such that dissociation of RNAP from rRNA promoters is in competition with NTP addition (Barker et al. 2001). DksA/ppGpp decrease the lifetime of the RNAP–promoter complex, inhibiting rRNA transcription (Barker et al. 2001; Paul et al. 2004; Haugen et al. 2008), whereas long-lived promoters are not inhibited by DksA/ppGpp because RNAP escapes from the complex before dissociation significantly affects transcriptional output (Barker et al. 2001; Paul et al. 2004, 2005).
Although we have a kinetic framework for understanding DksA's effects on transcription initiation (Rutherford et al. 2009) and there is a high-resolution structure of DksA (Perederina et al. 2004), a molecular understanding of its mechanism of action has been hampered by the absence of a structure of a DksA–RNAP complex. Structural information is available for the secondary channel-binding factors GreB, TFIIS, and Gfh1 in complex with RNAP (Kettenberger et al. 2003; Opalka et al. 2003; Tagami et al. 2010). However, these factors primarily affect transcription elongation rather than initiation, and their amino acid sequences bear little resemblance to DksA. Furthermore, a DksA homolog has not been identified in the thermophilic organisms from which all reported bacterial RNAP structures to date have been obtained. An RNAP–DksA complex structure has not been reported, and E. coli DksA does not appear to bind to, or function on, Thermus RNAP (T Gaal, CE Vrentas, and RL Gourse, unpubl.).
Here we present a model for the interaction of DksA with E. coli RNAP developed by site-specific incorporation of the cross-linkable amino acid benzoyl-phenylalanine (Bpa) (Chin et al. 2002) into DksA, mapping of cross-linked sites in RNAP, and then computational docking of DksA on RNAP. The model, in conjunction with the results of hydroxyl radical protein–protein footprinting and characterization of variants of RNAP that affect DksA binding and/or function, unambiguously positions DksA in the secondary channel and identifies interactions with both the rim helices (RHs), located at the entrance to the secondary channel, and the TL of the β′ subunit. Engineered cysteine residues in the DksA coiled-coil tip (cc-tip) and the TL resulted in formation of a disulfide bond between them. We propose that a TL–DksA interaction provides an explanation for the sequence requirement of the cc-tip for regulating transcriptional output and plays a central role in the mechanism of DksA action.
Results
Bpa incorporates site-specifically into DksA and cross-links to the β′ subunit of RNAP
To build an evidence-based model of DksA bound to RNAP, we used site-specific incorporation of the nonnatural, UV-cross-linkable amino acid Bpa into DksA, a technique that relies on in vivo expression of an evolved tRNA and tRNA synthetase pair and incorporation of Bpa at an amber stop codon (Chin et al. 2002; Ryu and Schultz 2006). After excitation with UV light, Bpa side chains form covalent cross-links in vitro with carbons within ∼3 Å (Kauer et al. 1986).
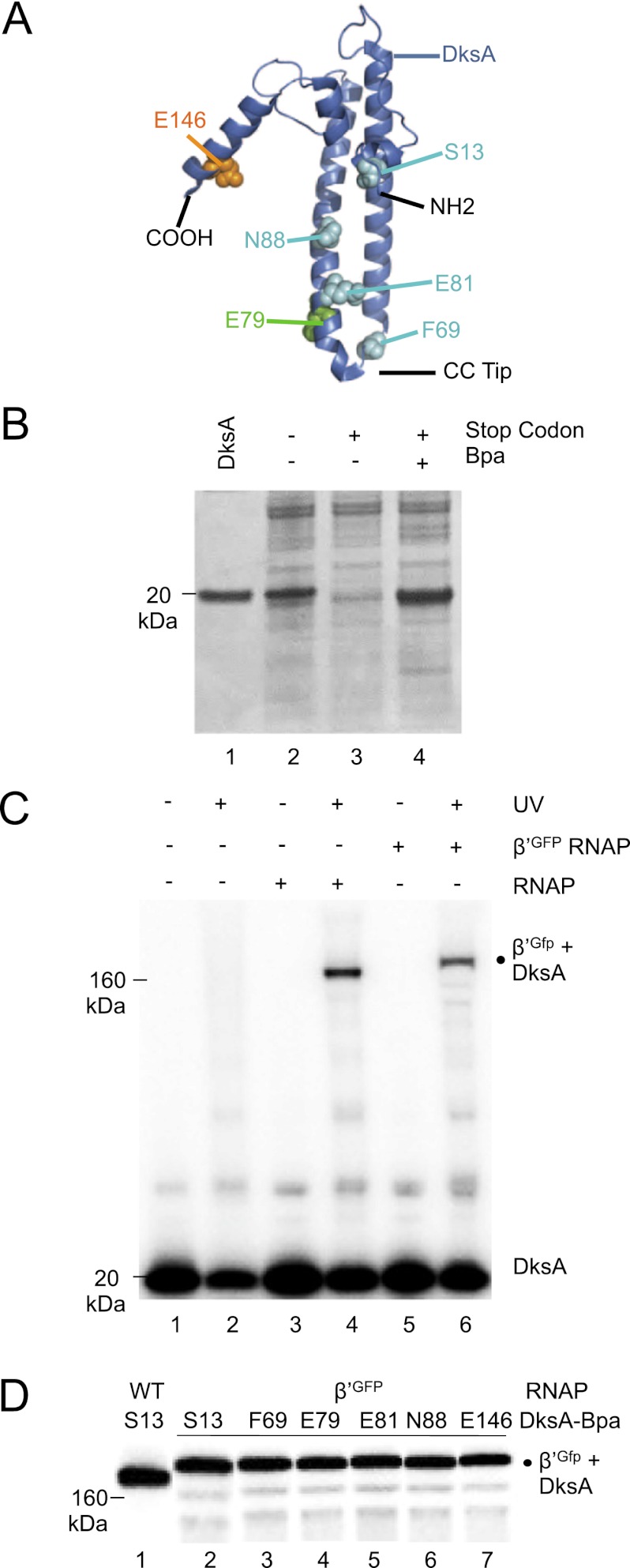
DksA has two domains: a long coiled-coil consisting of residues from the central portion of the protein, with two acidic residues at the tip, and a globular domain consisting of both N-terminal and C-terminal segments of the protein, including four cysteines that coordinate a zinc atom (Fig. 1A; Perederina et al. 2004). DksA variants containing single Bpa substitutions were created at 12 surface-accessible positions (codons S13, A18, R52, D64, F69, E79, E81, R87, N88, K94, E146, and Q148) chosen because they were unlikely to participate in protein folding. Overexpressed DksA-Bpa proteins comigrated with purified wild-type DksA (Fig. 1B, cf. lanes 1 and 4, Bpa is at residue 69 in the example shown) and with overexpressed wild-type DksA (Fig. 1B; lane 2). Little or no full-length DksA was obtained when the growth medium lacked Bpa (Fig. 1B; lane 3). The yield of DksA proteins containing Bpa was similar to that from expression of wild-type DksA (Fig. 1B, cf. lanes 2 and 4; data not shown). When Bpa was not added to the growth medium, truncated DksA products were observed only from the constructs with stop codons at E146 or Q148 (data not shown), suggesting that the other truncated DksA products were unstable.
Figure 1.
Bpa incorporates site-specifically into DksA and cross-links to the β′ subunit of RNAP. (A) Structure of DksA (PDB: 1TJL) (Perederina et al. 2004). N and C termini and cc-tip region are indicated. Positions where Bpa substitutions were created are shown in spacefill. (B) Expression of DksA F69-Bpa. Protein lysates were examined on a 4%–12% SDS gel stained with Coomassie blue. (Lane 1) Purified H6-HMK DksA (5 μg). (Lane 2) DH10B containing pSUPT/BpF and pRLG9502 (DksA wild type [WT]), no Bpa. (Lane 3) DH10B containing pSUPT/BpF and pRLG9505 (DksA with UAG at codon 69), no Bpa. (Lane 4) DH10B containing pSUPT/BpF and pRLG9505, 1 mM Bpa. Bpa was incorporated at UAG codons at S13, F69, E79, E81, N88, and E146 with similar efficiency. (C) 32P-DksA E79-Bpa was incubated in the absence of RNAP (lanes 1,2), with wild-type RNAP (lanes 3,4), or with RNAP containing a β′-GFP fusion (lanes 5,6) and either exposed (lanes 2,4,6) or not exposed (lanes 1,3,5) to UV light. Samples were electrophoresed on a 4%–12% SDS gel. Cross-linked β′-DksA (lane 4) and β′-GFP-DksA (lane 6) are indicated. (D) 32P-DksA-Bpa proteins were cross-linked to β′-GFP RNAP (lanes 2–7), or 32P DksA S13-Bpa was cross-linked to wild-type RNAP (lane 1), and samples were electrophoresed as in C.
Specific cross-links to RNAP were observed with 32P-labeled HMK-tagged DksA proteins with a Bpa at six positions: S13, F69, E79, E81, N88, and E146 (Fig. 1C, E79-Bpa is shown as an example) and were used for modeling (below). (The other six Bpa substitutions did not form specific cross-links [Supplemental Material] and were not used further.) The cross-links were UV-dependent (Fig. 1C, lanes 3,4), and the electrophoretic mobilities of the cross-linked complexes on SDS-PAGE corresponded to those expected for HMK-DksA (18 kDa) and one of the large RNAP subunits, β (151 kDa) or β′ (155 kDa) (Fig. 1C). The DksA proteins that cross-linked specifically to RNAP retained function: Each reduced transcription from rrnB P1 but not from the plasmid-encoded promoter RNA-1 (Supplemental Fig. S1).
To determine whether the DksA cross-link was to β or β′, we repeated the reaction with each of the six DksA-Bpa proteins and RNAP containing a β′ subunit with a C-terminal GFP fusion adding ∼27 kDa to β′ (Bratton et al. 2011). In all six cases, ∼90%–95% of the radiolabeled cross-linked species migrated slightly slower than with wild-type RNAP to a position consistent with the molecular weight (MW) of a complex containing β′-GFP (155 kDa plus 27 kDa) cross-linked to DksA (18 kDa) (Fig. 1C, [lane 6], D [lanes 2–7]). This is also consistent with the fact that the secondary channel of RNAP is composed almost entirely of β′ (Vassylyev et al. 2002).
We demonstrated previously that core RNAP and holoenzyme bind DksA with the same affinity (Lennon et al. 2009), consistent with a location for DksA far from where σ binds to RNAP. Additionally, we did not observe any differences in DksA cross-linking in pilot experiments in the presence or absence of σ (data not shown). Therefore, we used core RNAP to conserve material in the studies described below.
Mapping the DksA cross-links within β′
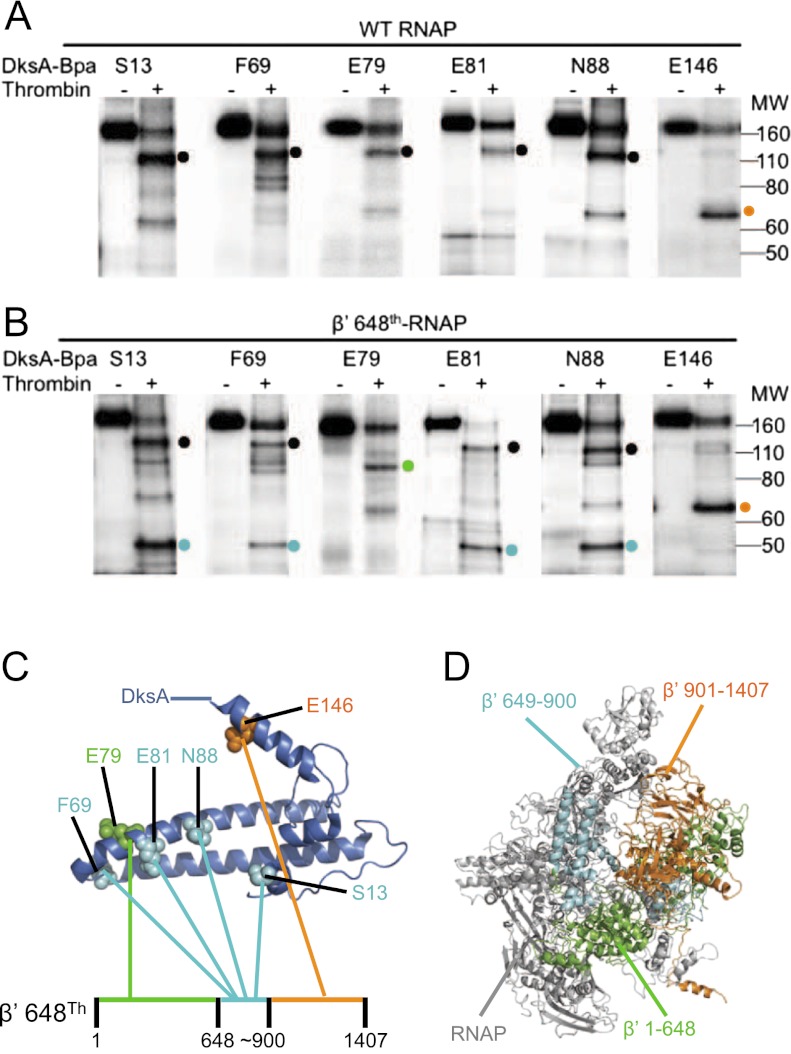
DksA cross-links were mapped within β′ using thrombin, which does not cleave DksA under these conditions. Thrombin cut β′ primarily at one site, corresponding to position ∼900 (Fig. 2A, right lane for each DksA variant ; see also the Supplemental Material). The major radioactive digestion product of the cross-linked RNAP–DksA complex containing the S13, F69, E79, E81, or N88 DksA-Bpa variants migrated slightly above a 110-kDa size marker, consistent with the predicted size of a complex containing β′ 1–900 (101 kDa) and HMK-DksA (18 kDa) (Fig. 2A, black dot). The cross-linked complex containing E146 DksA-Bpa migrated between the 60-kDa and 80-kDa size markers, consistent with the expected size of β′ 901–1407 (54 kDa) cross-linked to HMK-DksA (18 kDa) (Fig. 2A, orange dot).
Figure 2.
Mapping DksA cross-links within β′. 32P HMK DksA proteins with S13-Bpa, F69-Bpa, E79-Bpa, E81-Bpa, N88-Bpa, or E146-Bpa were cross-linked to wild-type (WT) RNAP (native thrombin site at residue ∼900 of β′) (A) or RNAP with an additional thrombin site engineered at β′ residue 648 (648Th-RNAP) (B), digested with thrombin, and electrophoresed on 4%–12% SDS gels. The major thrombin cleavage product containing the cross-linked DksA in each reaction is indicated by a dot. (Black dots) β′ Fragment 1–900; (orange dots) β′ 901–1407; (green dot) β′ 1–648; (cyan dots) β′ 649–900. (C) Summary of DksA cross-links with β′ 648Th thrombin fragments. DksA-Bpa residues are color-coded to match the β′ fragments to which they cross-linked. (D) Model of E. coli core RNAP (PDB: 3LU0) (Opalka et al. 2010) with β′ thrombin-fragment intervals containing cross-links colored as in Figures 1A and 2C.
To confirm and further map the positions of the cross-links, we used an RNAP containing β′ with a thrombin site engineered at position 648 (β′ 648Th) in addition to the one at ∼900 (Supplemental Material), resulting in cleavage of β′ 1–900 into β′ 1–648 and β′ 649–900 fragments (Fig. 2B,C). When the cross-linked complexes formed by β′ 648Th RNAP and the S13, F69, E79, E81, or N88 DksA-Bpa variants were treated with thrombin, the predominant labeled digestion product in each case migrated as a smaller complex than that formed with wild-type RNAP, confirming that the cross-link occurred within the β′ 1–900 fragment (Fig. 2B). In four cases (S13, F69, E81, and N88 DksA-Bpa), a product (Fig. 2B, cyan dots) migrated at approximately the position of the ∼50-kDa size marker, consistent with a complex consisting of HMK-DksA (18 kDa) and β′ 649–900 (28 kDa). An E79 DksA-Bpa cleavage product migrated between the 80-kDa and 110-kDa size markers, consistent with a cross-link of HMK-DksA (18 kDa) to β′ 1–648 (73 kDa) (Fig. 2B, green dot). Treatment of the E146 DksA-Bpa-β′ 648Th RNAP complex with thrombin did not change the mobility of the cross-linked fragment (Fig. 2B, orange dot) compared with that observed with wild-type RNAP, consistent with the conclusion that E146 DksA-Bpa cross-linked to the C-terminal thrombin fragment of β′ (901–1407; 54 kDa). This was confirmed by cross-linking β′-GFP RNAP with E146 DksA-Bpa followed by digestion with thrombin, resulting in a complex that migrated slower than the same complex derived from wild-type β′ (data not shown).
In summary, DksA residues S13, F69, E81, and N88 cross-linked to β′ 649–900; E79 cross-linked to β′ 1–648; and E146 cross-linked to β′ 901–1407 (Fig. 2C). Figure 2D illustrates the locations of these fragments on E. coli core RNAP (Opalka et al. 2010), color-coded as in Figure 2C.
Construction of our model of the DksA–RNAP complex depended on one further constraint—namely that Fe2+ substitution for the active site (AS) Mg2+ in RNAP results in cleavage of DksA upon binding to RNAP (Perederina et al. 2004; Lennon et al. 2009). Creation of genetically truncated DksAs as size markers for comparison with the DksA cleavage product allowed us to refine the DksA–RNAP interaction. The 32P-HMK DksA N-terminal Fe2+ cleavage product and a 32P-HMK DksA genetically truncated at residue 73 migrated similarly on gels (Supplemental Fig. S2, cf. lanes 2 and 3), whereas full-length DksA (Supplemental Fig. S2, lane 1) or a DksA genetically truncated at residue 85 (Supplemental Fig. S2, lane 4) migrated more slowly. We conclude that the cc-tip of DksA (residues 66–76) is within 10 Ǻ of the catalytic Mg2+ in RNAP.
Model of DksA bound to RNAP
Modeling of the DksA–RNAP complex was performed using Haddock version 2.0 (Dominguez et al. 2003; de Vries et al. 2007), a molecular docking program that can incorporate biochemical and genetic information in addition to energetics and shape complementarity. We started with the structure of DksA (Protein Data Bank [PDB]: 1TJL) (Perederina et al. 2004) and the model of E. coli core RNAP (PDB: 3LU0, which in turn is based on the high-resolution Thermus thermophilus RNAP structure) (Opalka et al. 2010; Vassylyev et al. 2002). The model was forced to satisfy the constraints imposed by the six cross-links to β′ and the proximity of the cc-tip to the AS Mg2+ (see the Supplemental Material), but DksA was purposely not constrained to the secondary channel in order to build the model. The biochemical data thus limited the potential ways that DksA could dock on the surface of RNAP. The 24 best models were refined (see Supplemental Fig. S3 for an overlay), and the lowest-energy model is shown in Figure 3 (see the Supplemental Material for details).
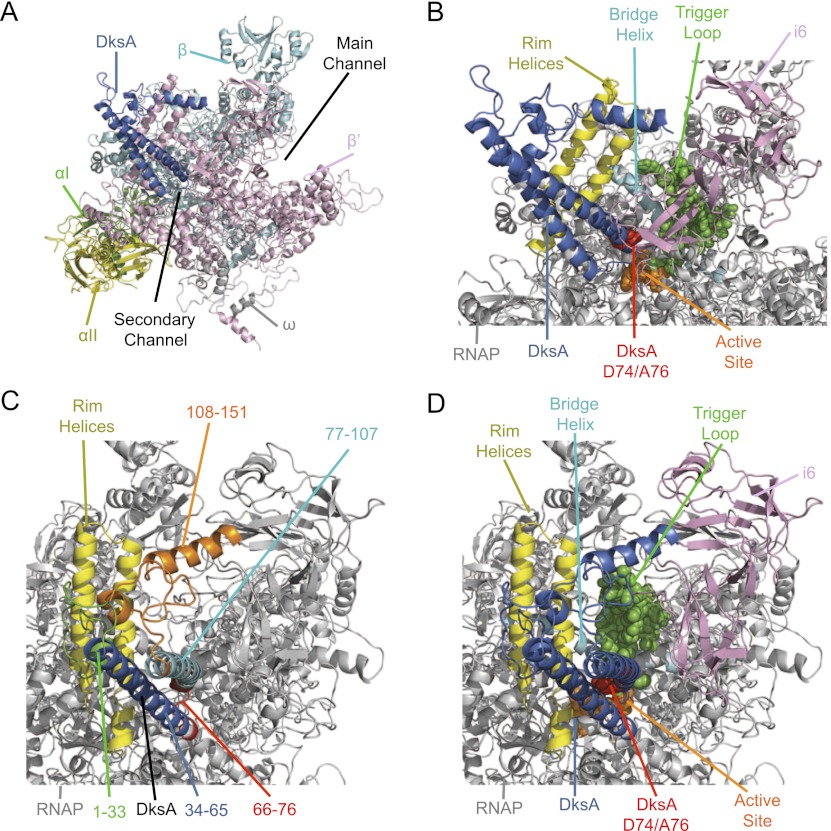
Figure 3.
Haddock model of E. coli DksA bound to E. coli core RNAP, with different segments of the complex colored for emphasis. A–D are different views of the RNAP–DksA complex. (A) Position of DksA relative to the RNAP subunits and the main and secondary channels. (Blue) DksA; (green) αI subunit; (yellow) αII; (cyan) β; (pink) β′; (gray) ω. (B) Position of DksA relative to specific features in RNAP. (Blue) DksA; (red spacefill) DksA tip residues D74 and A76; (yellow) RHs; (cyan) BH; (green spacefill) TL; (orange spacefill) AS; (pink) β′i6. (C) Positions of segments of DksA relative to RNAP. The orientation has been rotated to look down the secondary channel. (Green) DksA residues 1–33 (N-terminal globular domain); (blue) DksA 34–65 (coiled-coil helix 1); (red) DksA 66–76 (tip region); (cyan) DksA 77–107 (coiled-coil helix 2); (orange) DksA 108–151 (C-terminal globular domain); (yellow) RH. (D) Position of DksA relative to specific features of RNAP, colored as in B but from the angle shown in C.
As predicted from previous studies of DksA (Perederina et al. 2004; Rutherford et al. 2009) and from the location of the structurally similar Gre factors (Opalka et al. 2003), DksA binds in the secondary channel of RNAP in our model, with helix 2 of DksA (residues 77–107) rather than helix 1 (residues 34–65) closer to β′i6 (an E. coli lineage-specific sequence insertion previously called sequence insertion 3 [SI3]) (Fig. 3B, pink), the TL (Fig. 3B, green), and the BH (Fig. 3B, cyan). Conversely, helix 1 is closer to the RH and to the back of the channel than helix 2. The N-terminal part of the globular domain of DksA (residues 1–33) appears to interact or even potentially clash with the RH, and the C-terminal section in the globular domain is between the RH and β′i6 (Fig. 3C,D). Segments in both helices of the coiled-coil domain of DksA interact with the RH (Fig. 3B,D, yellow).
The functionally critical residues at the tip of the DksA coiled-coil (D74 and A76) (Fig. 3B, red; Lee et al. 2012) are oriented toward the TL. We showed previously that deletion of the TL eliminates DksA function (Rutherford et al. 2009). In the structure of RNAP on which our Haddock model is based, the TL is in the unfolded conformation (Fig. 3B,D, green spacefill; Opalka et al. 2010), but as discussed below, the TL is mobile and oscillates between an unstructured loop conformation and folded or partially folded α-helical hairpin conformations (trigger helices [THs]; TL/THs) during the catalytic cycle (Vassylyev et al. 2007; Zhang et al. 2010). There is little or no information about the state of the TL during transcription initiation.
Our Haddock model is physically plausible and evidence-based. The cross-links allowed us to fix the relative orientation of different regions of DksA relative to those of RNAP but is not a high-resolution structure, and interactions can only be inferred. Therefore, additional approaches were taken to confirm and extend the predictions of the model.
Biochemical and genetic support for the Haddock model
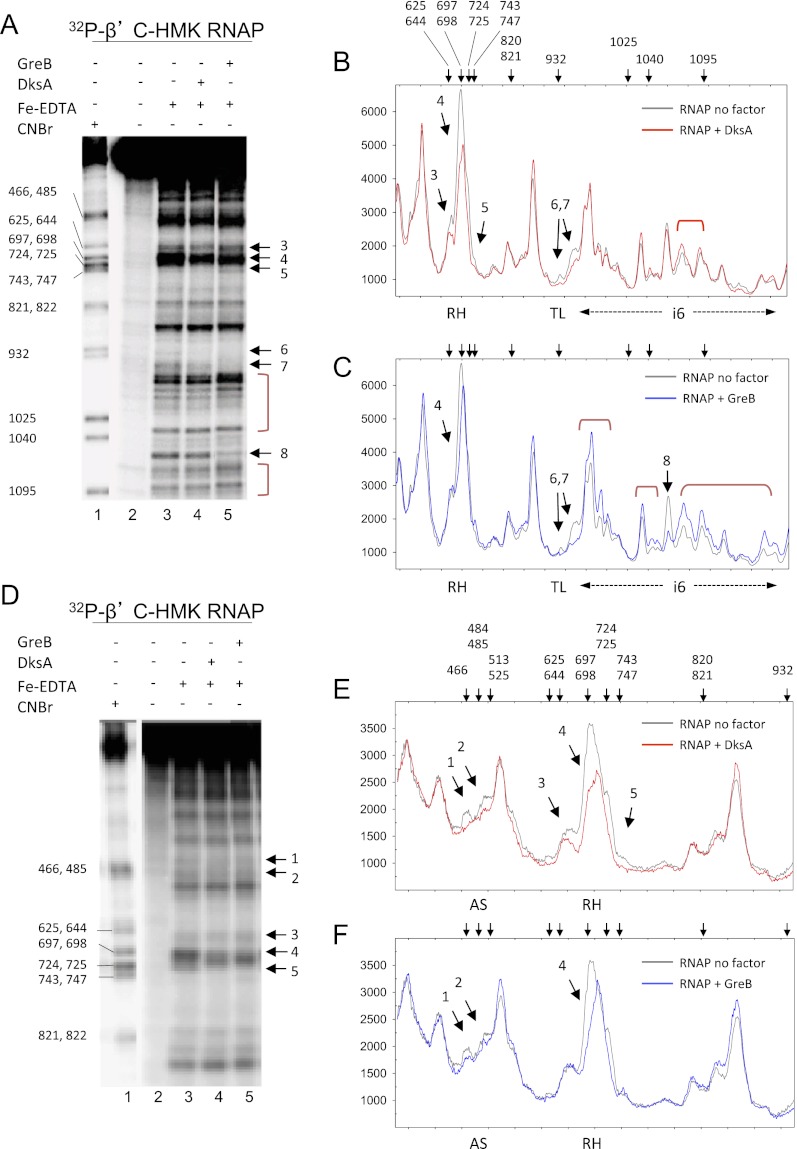
Protein–protein footprinting provided an independent means to assess interactions between RNAP and DksA. Complexes formed with DksA (or GreB for comparison) and RNAP (32P-labeled at an HMK site on the C terminus of β′) were cleaved by hydroxyl radicals generated in solution by Fe2+-EDTA, and the cleaved fragments were separated by electrophoresis. The reactions shown in Figure 4A were electrophoresed on a lower-percentage gel to resolve additional regions in β′ (Fig. 4D).
Figure 4.
Fe2+-EDTA protein–protein footprints of RNAP with DksA and GreB. (A) RNAP was 32P-labeled at a C-terminal HMK site on β′. (Lane 1) CNBr-digested RNAP size markers. (Lane 2) RNAP (140 nM), no Fe-EDTA. (Lane 3) RNAP only (140 nM). (Lane 4) RNAP (140 nM) plus DksA (3.5 μM). (Lane 5) RNAP (140 nM) plus GreB (3.5 μM). Samples were electrophoresed on an 8%–16% acrylamide Tris-glycine SDS gel. The numbered arrows to the right of gel images in A and D indicate positions in/near the RH (arrows 3–5), TL (arrows 6,7) and β′i6 (arrow 8) protected by DksA or GreB and correspond to the numbered peaks in B, C, E, and F. Brackets represent regions with enhanced cleavages. (B) Graph of lane 3 (RNAP, gray line) versus lane 4 (RNAP plus DksA, red line) from gel shown in A. Arrows above the panels in B, C, E, and F indicate the positions of markers generated by CNBr cleavage at the indicated positions in lane 1. (C) Graph of lane 3 (RNAP, gray line) versus lane 5 (RNAP plus GreB, blue line). (D) Same samples as in A were electrophoresed on an 8% gel to better resolve the AS region (arrows 1,2) and the RH region (arrows 3–5). (E) Graph of lane 3 (RNAP, gray) versus lane 4 (RNAP plus DksA, red). (F) Graph of lane 3 (RNAP, gray) versus lane 4 (RNAP plus GreB, blue).
Sites protected by DksA and/or GreB are indicated by arrows. Graphs of individual lanes in the gel images from Figure 4, A and D, are shown in Figure 4, B and E, for DksA and for GreB in Figure 4, C and F. DksA reduced cleavage at several positions in and near the secondary channel, including the region flanking the cyanogen bromide-generated marker band at position 466, corresponding to the AS region (Fig. 4E, peak 1), and nearby residues in the secondary channel (Fig. 4E, peak 2, β′ ∼485–510). DksA also protected an extensive region corresponding to the RH and part of the segment of β′ connecting the RH to the BH (Fig. 4E, peaks 3–5, β′ ∼648–699 and ∼700–750). The reduced cleavage could result from direct protection by DksA or from conformational changes in RNAP induced by DksA binding.
The cleavage pattern in the presence of GreB (Fig. 4C,F) was distinctly different from that observed in the presence of DksA. Like DksA, GreB protected the AS region (peaks 1 and 2), but GreB did not protect peak 3 in the RH (β′ 648–660) and protected peaks 4 and 5 in the RH less well than DksA. Conversely, GreB affected multiple sites in β′i6 much more than DksA, protecting some residues (peak 8) and enhancing others (peaks 6 and 7). Both DksA and GreB protected the TL region near residue 932, although inefficient cleavage made the protected positions difficult to visualize.
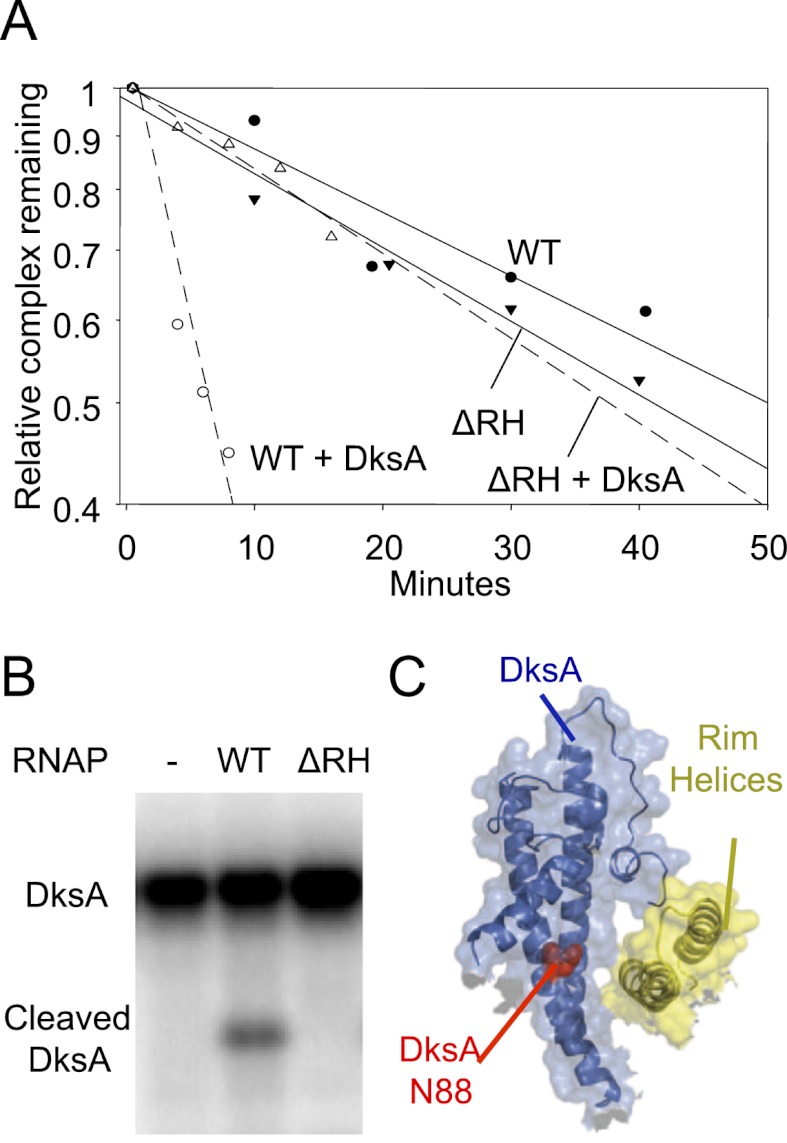
We tested the importance of the proposed interaction of DksA with the RH by monitoring its effects on promoter complex lifetime. The half-life of a promoter with RNAP lacking the RH (β′Δ645–718, β′ΔRH) was similar to that containing wild-type RNAP (Fig. 5A). However, DksA decreased the lifetime of promoter complexes containing wild-type RNAP about sevenfold under the conditions tested, whereas it had little or no effect on complexes containing β′ΔRH RNAP (Fig. 5A). We conclude that DksA does not function on RNAP lacking the β′ RH.
Figure 5.
The β′ RHs are required for DksA function and binding. (A) Lifetime of RNAP–lacUV5 promoter complex, measured by filter retention in the presence or absence of 1.6 μM DksA. Plots show the fraction of promoter–RNAP complex remaining at times following heparin addition. (WT) Wild-type RNAP; (ΔRH) RNAP lacking residues β′ 645–718. (B) ΔRH RNAP does not bind DksA. Binding was measured by RNAP-localized Fe2+-mediated cleavage of DksA (Lennon et al. 2009). (C) Haddock model illustrating the DksA interaction with RH. (Blue) DksA; (yellow) RH; (red spacefill) N88. The image was generated using PyMol with a surface transparency setting of 60%.
Using the Fe2+-mediated cleavage assay (Lennon et al. 2009), we measured binding of DksA to β′ΔRH RNAP. No DksA cleavage was detected, even when the RNAP concentration was 1 μM (Fig. 5B), ∼10-fold higher than the apparent binding constant for wild-type RNAP (100 nM) (Lennon et al. 2009). These results support the prediction of the Haddock model that DksA docks on the RH. The model also predicted that DksA residue N88 is in close proximity to, and points toward, the RH surface (Fig. 5C), consistent with the effect of an N88I substitution on DksA binding and function (Blankschien et al. 2009).
The Haddock model also suggested some residues in the secondary channel that might interact with DksA, including β′ K598/K599 and β′ R731 (Supplemental Fig. S4A), positions at which we previously constructed substitutions (Vrentas et al. 2008). β′ K598A/K599A and β′ R731A RNAPs exhibited twofold to threefold reduced affinity for DksA relative to wild-type RNAP in the Fe2+-mediated cleavage assay (Supplemental Fig. S4B). DksA inhibited transcription from rrnB P1 by the mutant RNAPs twofold or less in vitro, whereas it reduced transcription by wild-type RNAP >30-fold (Supplemental Fig. S4C). Therefore, the substitutions may affect steps in the DksA mechanism in addition to binding to RNAP. Although the responses of wild-type and β′ R731A RNAP to GreB were similar, GreB affected β′ K598A/K599A RNAP more strongly than wild-type RNAP (Supplemental Fig. S4D), consistent with the overlapping but distinct modes of interaction of DksA and GreB with RNAP and with their overlapping but distinct functions (Fig. 4; Rutherford et al. 2007; Lee et al. 2012).
The cc-tip of DksA interacts directly with the TL domain of RNAP
Deletion of the TL domain in β′ or substitutions at two positions in the DksA cc-tip, D74 or A76, eliminated DksA function (Rutherford et al. 2009; Lee et al. 2012). Even the conservative change of an aspartate to a glutamate at DksA residue 74 eliminated the effects of DksA on transcription (Lee et al. 2012). Furthermore, the DksA-dependent protection of the TL observed in our protein–protein footprints (Fig. 4) suggested that DksA might interact with the TL. However, the TL domain did not appear to be in direct contact with the cc-tip of DksA in our Haddock model (Fig. 3).
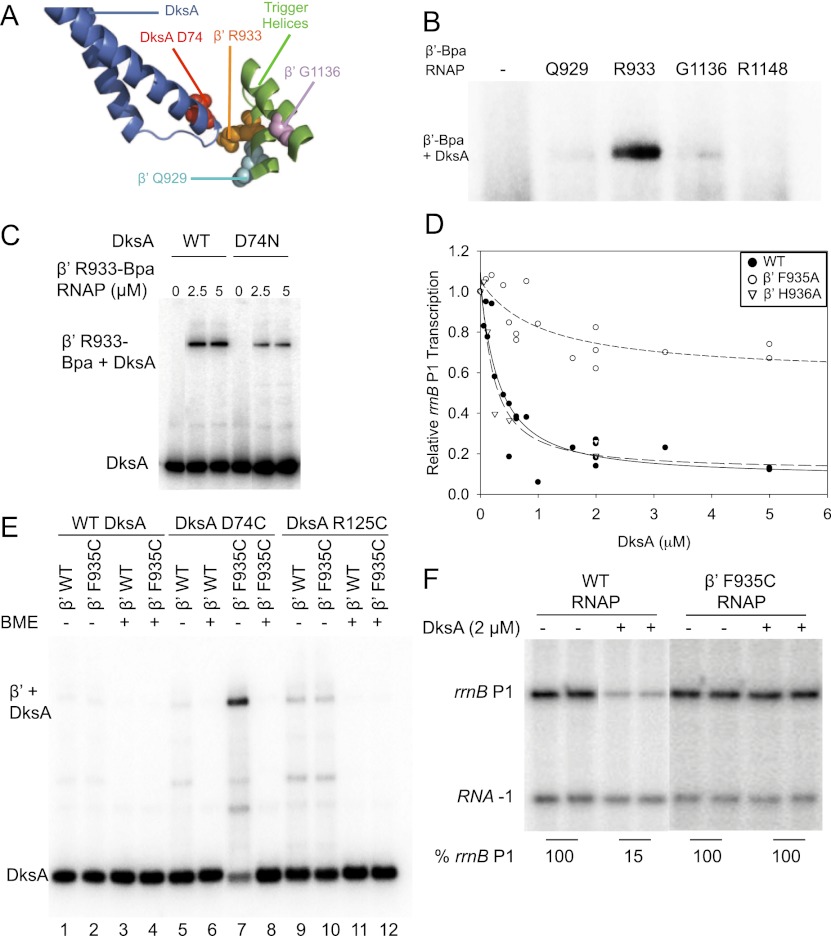
Because the TL in this model is in the unfolded conformation, we speculated that perhaps a different conformation of the TL might interact directly with DksA; i.e., that a fully folded (TH) or a partially folded (TL/TH) conformation of the TL domain might happen during transcription initiation as in transcription elongation (Vassylyev et al. 2007; Zhang et al. 2010) and might interact with DksA. When the entire β′ subunit with the fully folded (TH) structure of the TL domain was used to replace β′ in the Haddock model using the Align function of PyMol, residues in the TL approached DksA (Fig. 6A).
Figure 6.
Interaction of the RNAP TL with the DksA cc-tip is required for DksA function. (A) Residues where Bpa was inserted in the β′ TL are shown in spacefill in the Haddock model. The fully folded TH conformation of the TL was substituted for the unfolded TL domain (see Fig. 7). (Cyan) β′ Q929; (orange) β′ R933; (pink) β′G1136; (blue) DksA; (red) DksA cc-tip residue D74. (B) β′ R933-Bpa cross-links to 32P-DksA. Cross-linked complexes were analyzed by SDS-PAGE as in Figure 1. (C) D74N reduces the amount of DksA cross-linked to β′ R933-Bpa. Cross-linking was measured at different RNAP concentrations; 2.5 and 5 μM were saturating. (D) β′TL substitution F935A RNAP reduces DksA function as measured by inhibition of rrnB P1 promoter activity. A plasmid containing rrnB P1 was transcribed with wild-type (WT) RNAP, β′ F935A RNAP, or β′ H936A RNAP at a range of DksA concentrations. Products were analyzed by gel electrophoresis and PhosphorImaging, and the fraction of transcription relative to that without DksA is plotted for each enzyme. (E) DksA tip residue D74C forms a disulfide bond with TL β′ F935C. 32P-DksA (wild-type, D74C, or R125C) was incubated with RNAP (wild-type or β′ F935C) under oxidizing (without BME) or reducing (+BME) conditions, and the products were examined by SDS-PAGE (see the Supplemental Material). (F) β′ F935C RNAP does not respond to DksA. Transcription was performed as in C. Average transcription from rrnB P1 relative to that without DksA is indicated below the duplicate gel lanes.
To address experimentally whether the TL is in close proximity to DksA, we engineered Bpa substitutions at three different positions in the TL (β′ Q929, R933, and G1136) as well as at a position in β′ outside of the TL as a control (R1148). RNAP with Bpa at β′ R933 cross-linked most efficiently to DksA (Fig. 6B), and β′ R933-Bpa cross-linking was greatly reduced with D74N-DksA compared with wild-type DksA, even at high RNAP concentrations (Fig. 6C). These results implicate the cc-tip as the potential target of the cross-link.
To assess the importance of residues in this section of the TL for DksA function, we used RNAP variants containing β′ M932A, F935A, or H936A (β′ R933A RNAP was not recovered for unknown reasons). β′ M932A RNAP was transcriptionally inactive, as expected from previous studies (Zhang et al. 2010). However, RNAPs containing β′ F935A or H936A purified efficiently and retained transcription initiation activity. β′ H936A RNAP responded to DksA similarly to wild-type RNAP, but β′ F935A RNAP was severely defective in responding to DksA (Fig. 6D). We conclude that β′ F935, an amino acid near the position of the R933-Bpa cross-link to DksA, plays an important role in DksA function.
To further determine whether the TL domain is in close proximity to the DksA cc-tip, cysteines were introduced into β′ at F935 and at the residues in the DksA tip most critical for function, D74 or A76. Under oxidizing conditions, D74C-DksA and β′ F935C formed a complex that migrated to the position of β′-DksA, consistent with formation of a disulfide bond, indicating the two cysteines were within ∼2 Å of each other (Fig. 6E, lane 7). When β-mercaptoethanol (BME) was added to the SDS loading solution, the complex was no longer observed (Fig. 6E, lane 8). D74C-DksA did not form a disulfide with wild-type RNAP (Fig. 6E, lane 5), and β′ F935C did not form a disulfide with wild-type DksA (Fig. 6E, lane 2). As expected, R125C (a cysteine substitution in the globular domain of DksA) did not form a disulfide with β′ F935C RNAP (Fig. 6E, lane 10) or with wild-type RNAP (Fig. 6E, lane 9). A disulfide bond was also obtained with β′ F935C and A76C-DksA (Supplemental Fig. S5). These data suggest that the cc-tip of DksA at or very near residues D74 and A76 interacts directly with the β′ TL at or near residue F935. β′ F935C RNAP transcribed efficiently but, consistent with the importance of the DksA tip–TL interaction for DksA function and with the effect of the β′ F935A substitution (Fig. 6D), did not respond to DksA (Fig. 6F).
Role of the TL–DksA tip interaction in the mechanism of DksA action
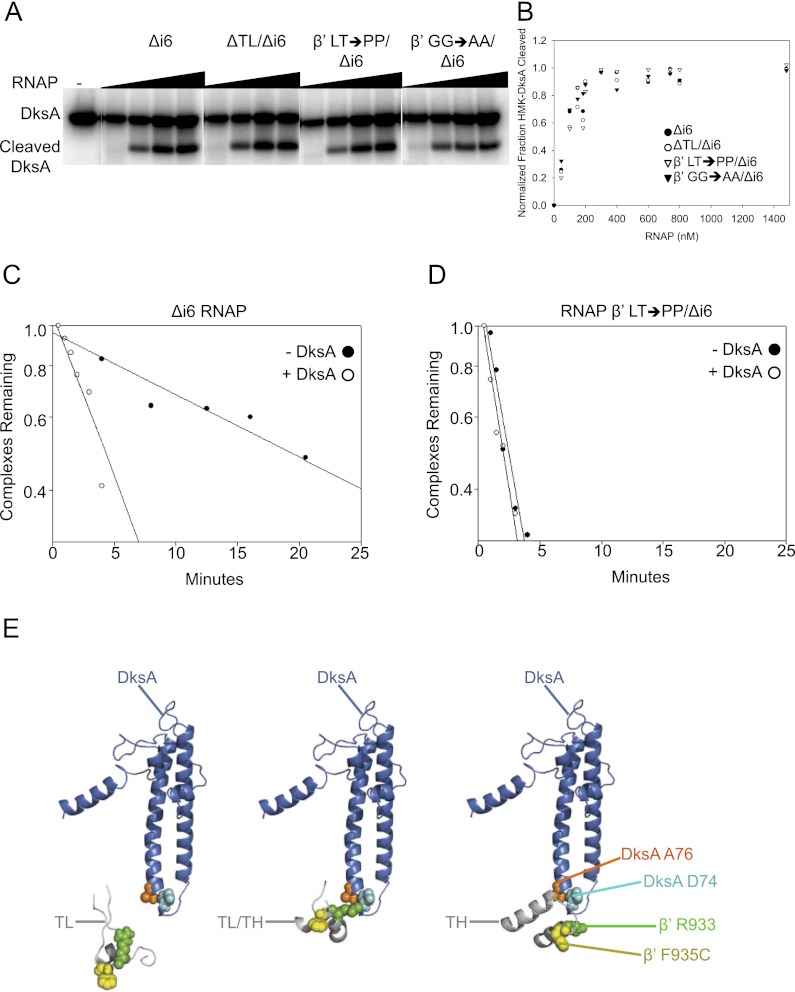
RNAP containing a deletion of the β′ TL does not respond to DksA (Rutherford et al. 2009). To address whether the TL–DksA tip interaction is important for DksA function because it contributes to the overall affinity of DksA for RNAP or rather because it affects a later step in the mechanism, we measured the effects of the TL deletion on DksA affinity using the Fe2+-mediated cleavage assay (Lennon et al. 2009). E. coli RNAP contains the lineage-specific insertion β′i6 (residues 943–1130) in the TL (residues 926–942 and 1131–1143). Therefore, the TL deletion construct lacked β′i6 as well (ΔTL/Δi6). We compared the effects of a β′i6 deletion alone (Δi6) with the effects of the ΔTL/Δi6 deletion. The apparent binding constants for DksA were about the same for the Δi6 and ΔTL/Δi6 RNAPs (∼100 nM) (Fig. 7A,B) and for wild-type RNAP (Lennon et al. 2009; Lee et al. 2012), suggesting that the interaction of the TL domain with the DksA cc-tip is not a major determinant of the overall affinity of DksA for RNAP. Likewise, tip mutants D74N-DksA and A76T-DksA eliminate DksA function without affecting the overall affinity of DksA for RNAP (Lee et al. 2012).
Figure 7.
The β′ TL is required for DksA function but not for DksA binding to RNAP. (A) Binding of DksA to Δi6 RNAP (deletion of β′ residues 943–1130), ΔTL/Δi6 RNAP (deletion of β′ residues 931–1137), β′ 930,931 LT → PP/Δi6 RNAP (i.e., β′TL LT → PP/Δi6 RNAP), and β′ 938,939 GG → AA/Δi6 RNAP (i.e., β′TL GG → AA/Δi6) was measured by localized Fe2+-mediated cleavage of DksA at different RNAP concentrations (Lennon et al. 2009). (B) Binding curves for TL variants shown in A. Overall apparent binding affinities of mutant RNAPs for DksA were the same within error (Kd ∼100 ± 15 nM for each RNAP). (C) Effect of DksA on Δi6 RNAP. Lifetimes of lacUV5 promoter–RNAP complexes were determined in the presence/absence of 1.6 μM DksA. Each data point represents the fraction of 32P-promoter DNA retained on nitrocellulose filters at times following competitor addition (Rutherford et al. 2009). (D) β′TL LT → PP RNAP is resistant to the effects of DksA. Half-life measurements were performed as in C. (E) Models of DksA interactions with the TL domain in three different conformations: unfolded TL (see Fig. 3), partially folded TL (TL/TH) (PDB: 2PPB) (Vassylyev et al. 2007), and folded THs (PDB: 2O5J) (Vassylyev et al. 2007). The entire β′ subunit with the partially folded (TL/TH) or fully folded (TH) structures was used to replace β′ from the Haddock model using the Align function of PyMol. Only the TL domain and DksA are shown. (Blue) DksA; (cyan spacefill) DksA residue D74; (orange spacefill) DksA A76; (gray) TL domain; (green spacefill) β′ R933; (yellow spacefill) β′ F935.
It has been predicted that a double substitution in the TL, β′TL L930P/T931P/Δi6 (referred to below as β′TL LT → PP) blocks folding of the THs (Vassylyev et al. 2007; Zhang et al. 2010). Because the TL is needed for nucleotide addition, we used a filter-binding assay to assess DksA function with this RNAP because this assay measures open complex formation or decay without requiring RNA formation. Consistent with our previous results (Rutherford et al. 2009), DksA reduced the half-life of the control Δi6 promoter complexes ∼4.5-fold (from 18 min to 4 min) (Fig. 7C). In contrast, β′TL LT → PP RNAP formed a ninefold less stable promoter complex than the control, Δi6 RNAP (∼2-min vs. 18-min half-life), and the mutant complex was completely resistant to the effects of DksA (Fig. 7D).
RNAP with a different two-residue substitution in the TL, β′G938A/G939A/Δi6 (referred to below as β′TL GG → AA) is hypersensitive to DksA (Rutherford et al. 2009). The β′TL GG → AA substitution is predicted to restrict the turn between the two THs, locking the enzyme in a partially folded state (Vassylyev et al. 2007; Zhang et al. 2010). Thus, both the β′TL GG → AA and the β′TL LT → PP substitutions block full folding of the THs but are proposed to favor different TL conformations (Toulokhonov et al. 2007; Zhang et al. 2010). Although neither the β′TL GG → AA nor the β′TL LT → PP substitutions affected the apparent affinity of DksA for RNAP, consistent with the lack of an effect of the β′ΔTL on overall DksA binding (Fig. 7A,B), our results suggest that the conformation of the TL has a major impact on DksA function. In our Haddock model, β′TL in the fully folded (TH) conformation (from Vassylyev et al. 2007) appeared closer to the DksA tip than β′ with an unfolded TL (models created using the Align function of PyMol; see above), and the model with a partially folded (TL/TH) conformation was even closer to the DksA tip (Fig. 7E; see the Discussion).
Discussion
Conformational state of the TL and DksA function
To understand how DksA functions, we took biochemical and genetic approaches to probe its interactions with RNAP. Our results demonstrate that DksA binds in the RNAP secondary channel (as predicted previously), that residues in the N-terminal (and possibly C-terminal) segment of the globular domain of DksA and both helices of the DksA coiled-coil bind to the RHs, and that the cc-tip of DksA interacts directly with the TL domain of β′. We propose that this latter interaction explains the precise sequence requirement of the DksA tip residues for DksA function.
Our evidence-based model of the DksA–RNAP complex was made possible by introduction of the nonnatural amino acid Bpa at precise positions in DksA. The model placed the DksA tip near the TL domain but not close enough to explain the disulfide bond formed between D74C or A76C in the DksA tip and β′ F935C in the TL. However, in the models containing the fully folded conformation of the TL (THs) or the partially folded conformation of the TL trapped by the antibiotic streptolydigin (TL/TH) (see Fig. 7E; Vassylyev et al. 2007), it appears that β′TL residue F935 is considerably closer to the DksA cc-tip (within a few angstroms of D74 and A76) than when the TL is in the unfolded conformation.
Although there is strong evidence that TL–TH transitions occur during transcription elongation (Vassylyev et al. 2007; Miropolskaya et al. 2010), there is little or no information about whether such transitions occur during transcription initiation. DksA's effect on transcription initiation was eliminated by either deletion of the TL or substitutions in the TL predicted to favor its unfolded form (β′TL LT → PP RNAP). In contrast, DksA function was enhanced by substitutions in the TL predicted to favor a partially folded state (β′TL GG → AA RNAP) (Rutherford et al. 2009). Taken together, our data suggest that the favored conformation for DksA activity is a partially folded state of the TL. One model for DksA function is that it traps the TL in this partially folded state, that the β′TL GG → AA substitutions facilitate this TL conformation, and that the β′TL LT → PP substitutions preclude the interaction of the DksA cc-tip with the TL. Because the β′TL LT → PP RNAP forms a short-lived promoter complex in the absence of DksA (Fig. 7D), an alternative explanation for the observed resistance of the β′TL LT → PP complex to DksA is that the substitutions result in the same change in RNAP conformation that results from DksA interaction, precluding a further effect of DksA.
DksA disfavors contacts between RNAP and promoter DNA downstream from the transcription start site, inhibiting progression of initiation past the closed complex stage in negatively regulated promoters (Rutherford et al. 2009). If DksA primarily occupies RPC and functions by capturing a TL/TH form of the TL domain, then it seems reasonable that the TL/TH interaction with the tip of DksA takes place in RPC. Substitutions in RNAP that bypass the DksA requirement were found predominantly in the TL and BH and in four of the five switch regions of RNAP (Rutherford et al. 2009). These genetic results suggested a model in which the 60-Ǻ-long BH (β′ 769–804), which forms a three-helix bundle with the TL, might link DksA to promoter DNA and the RNAP switch regions (Rutherford et al. 2009). By affecting the switch regions allosterically, DksA could affect DNA loading, DNA unwinding, and clamp closure following RPC formation (see Chakraborty et al. 2012). Thus, we propose that the DksA cc-tip interaction with the TL is a critical step in DksA function.
Comparisons of DksA with other secondary channel-binding factors
A number of small proteins, including DksA, GreA, GreB, Rnk, Gfh1, TraR, and TFIIS, bind in the RNAP secondary channel and affect transcription in different ways. Although there are high-resolution structures of TFIIS with RNAP (Kettenberger et al. 2003) and of Gfh1 with RNAP (Tagami et al. 2010) and a lower-resolution structure of GreB with RNAP (Opalka et al. 2003), details about interactions between RNAP and the other factors can only be inferred. Most of the different factors bear little sequence similarity to each other, and our data suggest that even very subtle changes in amino acid sequence can have dramatic effects on the outcomes of the interactions of the factors with RNAP (Lee et al. 2012). Thus, conclusions based only on structural analogy are of limited utility.
DksA has specific functions that are not shared by the other secondary channel-binding proteins. Its most explored effects are on the control of transcription initiation, although it clearly also affects elongation, DNA repair, and RNAP–DNA polymerase collisions (Trautinger et al. 2005; Tehranchi et al. 2010). No DksA homologs are present in the thermophilic organisms from which we have high-resolution structural information of RNAP, and E. coli DksA does not affect Thermus RNAP like it does E. coli RNAP (T Gaal, CE Vrentas, and RL Gourse, unpubl.).
Like DksA, GreB binds in the secondary channel of RNAP (Opalka et al. 2003), can inhibit rrnB P1 promoter activity (when overproduced to DksA-like levels in vivo or provided at the same concentration as DksA in vitro) (Rutherford et al. 2007), and requires a conserved acidic tip residue for function (D74 for DksA, and E44 for GreB). However, DksA and GreB do not share all functions. Gre factors can cleave RNA in backtracked transcription complexes (Laptenko et al. 2003) but DksA cannot (Furman et al. 2012), and DksA can positively regulate transcription from a number of promoters directly but Gre factors cannot (Rutherford et al. 2007). Recently, we reported that DksA residue D74 is required for both positive and negative transcription regulation and that changing the corresponding residue in either GreA or GreB (E44) to an aspartate resulted in acquisition of partial DksA function in the positive control (Lee et al. 2012). Taken as a whole, it is likely that DksA and the Gre factors interact with RNAP in a similar manner overall but that the network of interactions that DksA makes with RNAP, including its interaction with the TL, is finely tuned for its specific role in transcription initiation.
An X-ray structure of the T. thermophilus secondary channel-binding protein Gfh1 was reported in complex with T. thermophilus RNAP (Tagami et al. 2010). In the cocrystal, RNAP is in a so-called “ratcheted state,” the TL is unstructured, and it appears that Gfh1 would clash with the TL if the TL were in its fully folded conformation. Tagami et al. (2010) proposed that Gfh1 alters the equilibrium between different conformations of RNAP (the ratcheted vs. nonratcheted states) in which the orientation between two rigid body modules (the core and shelf) is altered. The ratcheted conformation is proposed to affect RNAP function by widening the main and secondary channels, thereby weakening interactions with DNA. We recently found that ppGpp binds at the junction of the core and shelf modules (W Ross and RL Gourse, in prep.), potentially explaining how ppGpp and DksA might work together. Thus, identification of the interactions between DksA and RNAP has implications not only for understanding the mechanism of DksA function, but also for how ppGpp and DksA might work together to control transcription.
In the ratcheted T. thermophilus RNAP–Gfh1 complex, Gfh1 appears to extend further into the secondary channel than DksA in our nonratcheted E. coli RNAP model. To examine whether DksA might extend further into the secondary channel in the ratcheted state of RNAP, we created a Haddock model using E. coli DksA and the ratcheted form of T. thermophilus RNAP from the Gfh1 cocrystal (Tagami et al. 2010). The model of the ratcheted T. thermophilus RNAP–E. coli DksA complex is similar to the RNAP–DksA complex shown in Figure 3. However, the tip region of DksA in this model does extend further into the channel than with the nonratcheted form of the enzyme, albeit not as far as the tip of Gfh1 in the ratcheted RNAP complex (Supplemental Fig. S6). In the ratcheted RNAP–DksA complex, the cc-tip of DksA would be predicted to interact with a partially folded TL, but there would be a greater potential for a clash with the fully folded form of the TL. DksA is slightly rotated and further from the RH compared with what was observed in our nonratcheted RNAP–DksA model (Supplemental Fig. S6). Whether the E. coli enzyme has a ratcheted state and whether DksA binds to it remain to be determined.
Prospect
In summary, our work provides the first evidence-based model for the DksA–RNAP complex. We suggest that a DksA cc-tip interaction with a partially folded state of the TL occurs in RPC and is a critical component of the network of interactions that is required for the specific effects of DksA on transcription initiation. Determining how the DksA tip–TL interaction contributes to inhibiting the progression from the closed to the open complex and whether this interaction is limited to specific steps in the transcription mechanism remains a challenge for the future.
Materials and methods
Bacterial strains and plasmids
E. coli strains and plasmids are listed in the Supplemental Material (Supplemental Table S1). Plasmids were constructed using standard procedures for cloning and mutagenesis and were verified by DNA sequencing.
Expression and purification of Bpa-substituted proteins
Plasmids containing genes encoding RNAP subunits or DksA were engineered with a stop codon (UAG) at the position chosen for Bpa incorporation (see Supplemental Table S1 for plasmids and strains). Single UAG substitutions were constructed at multiple positions within each of the five sections of DksA (N-terminal helix, helix 1, cc-tip, helix 2, and C-terminal helix). We chose positions, based on the structure of DksA (Perederina et al. 2004), that were likely to be surface-exposed, unlikely to participate in protein folding, and unlikely to be required for DksA function based on previous information (Lee et al. 2012). DksA proteins containing Bpa at specific positions were overexpressed from the araBAD promoter on pRLG9502 and its derivatives in DH10B. pRLG9502 was constructed by inserting an NcoI–HindIII fragment coding for His6-HMK-DksA from pRLG8150 into pRLG6640 (pBAD24/His6-DksA) (Webb et al. 1999) digested with NcoI and HindIII. We note that Bpa incorporation into DksA was unsuccessful when we used the T7 promoter in vector pET33 in BL21(DE3) cells.
Bpa-substituted RNAPs were overexpressed from the T7 promoter on a multisubunit vector in BL21(DE3) cells induced with 1 mM IPTG. The multisubunit RNAP vectors pIA299 and pIA423 do not contain the gene coding for ω, so strains used for overexpression of RNAP also carried pCDFω (Vrentas et al. 2005).
Strains for overexpression of Bpa-containing proteins also contained pSUPT/BpF which encodes a tRNA and tRNA synthetase pair evolved for Bpa incorporation at UAG codons (Ryu and Schultz 2006). Positive controls were always performed when establishing conditions for incorporation of Bpa into previously untested positions or proteins.
For protein purification, cotransformation of the overexpression plasmid and the tRNA/tRNA synthetase plasmid was performed fresh for each experiment by electroporation, selecting for both antibiotic resistances. Fresh transformants were scraped from plates for use as an inoculum, generating a relatively high starting culture density (OD600 ∼0.3), and grown at 37°C in LB with Bpa (1 mM), ampicillin (100 μg/mL), and chloramphenicol (25 μg/mL). Using a high inoculum from plates avoided suppressor accumulation from extended growth in liquid culture. The culture medium was prepared by addition of Bpa to LB medium dropwise from a freshly made 100 mM Bpa stock in 1 M NaOH, and an equal volume of 1 M HCl was added to produce a final pH of ∼7.3. After 1 h of growth with Bpa, expression was induced with 1 mM IPTG or 0.2% L-arabinose, depending on the expression system, and growth was continued for ∼20 h. Bpa incorporation was idiosyncratic: Optimal temperature, inducer concentration, and time of expression varied with the protein of interest. Furthermore, UAG codons were not equally efficient at all positions, and it was critical to ensure that full-length protein was made by stop codon readthrough and only when Bpa was present.
RNAPs with a His6 or His10 tag at the C terminus of the β′ subunit were purified using Ni-agarose and heparin affinity chromatography sequentially. For cell resuspension and Ni-agarose chromatography, buffer A contained 40 mM Tris-Cl (pH 7.9), 5 mM imidazole, and either 300 mM NaCl (for His6 β′ RNAPs) or 1M NaCl and 10% ethanol (for His10 β′ RNAPs). Cell lysis buffer also included 10 μg/mL lysozyme and 23 μg/mL phenylmethylsullfonyl fluoride (PMSF). For culture volumes of 100 mL to 1 L, cells were collected by centrifugation, resuspended in lysis buffer, and lysed by sonication. After centrifugation, the cleared lysate was added to pre-equilibrated Ni resin (∼0.001 times the volume of the original culture), the column was washed with buffer A, and the protein was eluted with buffer A containing 150 mM imidazole. The eluate was diluted to 200 mM NaCl with TGED (Burgess and Jendrisak 1975), bound to heparin resin (∼0.001 times the volume of the original culture pre-equilibrated in TGED plus 200 mM NaCl), and washed with the equilibration buffer, and RNAP was eluted with TGED plus 600 mM NaCl. RNAPs were concentrated using 0.5-mL Microcon centrifugal filtration units, diluted into storage buffer (50% glycerol, 10 mM Tris-Cl at pH 7.9, 100 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA), and stored at −20°C. Protein concentrations were measured using the Bradford assay reagent (Bio-Rad) using bovine serum albumen (BSA) as a standard.
Following purification of His-HMK-DksA from Ni-agarose as described above for RNAP, the His6 tag was cleaved off using biotinylated thrombin (EMD Biosciences), leaving eight residues (including the five-residue HMK site) on the DksA N terminus for labeling with 32P. This N-terminal extension did not affect the activity of DksA (Lennon et al. 2009). Biotinylated thrombin was removed using streptavidin agarose (EMD Biosciences), and DksA was passed over Ni-NTA agarose again, collecting the flow-through to remove any impurities that coeluted with DksA during the first purification step.
Expression and purification of mutant proteins not containing Bpa
Non-Bpa-containing DksAs and RNAPs were produced using overexpression vectors, as described previously (Paul et al. 2004; Rutherford et al. 2009; Lee et al. 2012) and in the Supplemental Material.
Bpa cross-link mapping
32P-labeled HMK-DksA (0.01–1.0 μM) was incubated for 5–15 min at room temperature with RNAP (0.01–0.05 μM) in 20 mM HEPES (pH 8), 20 mM NaCl, and 10 mM MgCl2 in a microfuge tube. The tube was placed directly on the filter of a 365-nm UV source for 3 min. The cross-linked region in β′ was mapped by digestion with thrombin and SDS-PAGE. Where indicated, the β′ subunit contained an engineered thrombin site at position 648. Procedures are described further in the Results and in the Supplemental Material.
Modeling the DksA–RNAP complex
The structure of the complex between DksA and RNAP was modeled using Haddock version 2.0 (Dominguez et al. 2003; de Vries et al. 2007) starting from PDB structures of DksA (1TJL) (Perederina et al. 2004) and the model of E. coli core RNAP (3LU0) (Opalka et al. 2010), which in turn is based on the high-resolution T. thermophilus RNAP structure (Vassylyev et al. 2002). See the Results the and Supplemental Material for further details.
Localized RNAP-mediated Fe2+ cleavage of DksA
We employed a method in which hydroxyl radicals generated by replacement of Mg2+ with Fe2+ in the RNAP active center cleaved the cc-tip of DksA (Supplemental Material; Lennon et al. 2009).
Fe-EDTA protein–protein footprinting
Protein–protein footprinting was performed with hydroxyl radicals generated in solution by Fe-EDTA as described (see the Supplemental Material; Laptenko et al. 2003).
In vitro transcription
Single-round and multiple-round in vitro transcription reactions were performed essentially as described previously (Lee et al. 2012) with additional details in the Supplemental Material.
Disulfide formation
Cysteine residues were introduced into DksA and RNAP at positions predicted to be in close proximity in the complex. 32P-labeled wild-type, D74C, or R125C DksA (5 nM) was incubated for 10 min at room temperature with wild-type or β′ F935C RNAP (2.5 μM) in 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, 10 mM MgCl2, and 10 mM BME. Reactions were then exchanged into 20 mM Tris-HCl (pH 7.9), 100 mM NaCl, and 10 mM MgCl2 with no BME using G-50 QuickSpin columns (GE Healthcare) and allowed to incubate for 5 min at room temperature. Reactions were stopped using twofold-concentrated LDS loading buffer with or without BME, as indicated in Figure 6. 32P-labeled products were separated using 4%–12% Bis-Tris NuPAGE gels and MOPS gel running buffer (Invitrogen).
RNAP–promoter complex lifetime assay
Competitor-resistant promoter complex lifetimes were measured by nitrocellulose filter retention in the presence of 10 μg/mL heparin with a DNA fragment containing the lacUV5 promoter (endpoints −60 to +38 from pRLG4264) as described previously (Barker et al. 2001).
Acknowledgments
We thank L. Li and D. Ma for mass spectrometry of thrombin fragments, and J. Winkelman and other members of the Gourse laboratory, R. Mooney, J. Zhang, R. Landick, I. Artsimovitch, and P. Schultz for kindly providing strains, plasmids, and helpful comments. This work was supported by grants from the National Institutes of Health (R37 GM37048 to R.L.G., and a predoctoral fellowship to C.W.L.). Computational modeling resources were provided by the National Magnetic Resonance Facility at Madison, which is supported by NIH grants P41RR02301 (BRTP/NCRR) and P41GM66326 (NIGMS), and a PyMol script for analysis and display of models was generously provided by L. Clos II.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.204693.112.
References
- Barker M, Gaal T, Josaitis C, Gourse R 2001. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol 305: 673–688 [DOI] [PubMed] [Google Scholar]
- Blankschien M, Lee J, Grace E, Lennon C, Halliday J, Ross W, Gourse R, Herman C 2009. Super DksAs: Substitutions in DksA enhancing its effects on transcription initiation. EMBO J 28: 1720–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton B, Mooney R, Weisshaar J 2011. Spatial distribution and diffusive motion of RNA polymerase in live Escherichia coli. J Bacteriol 193: 5138–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess R, Jendrisak J 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatrography. Biochemistry 14: 4634–4638 [DOI] [PubMed] [Google Scholar]
- Chakraborty A, Wang D, Ebright Y, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, et al. 2012. Opening and closing of the bacterial RNA polymerase clamp. Science 337: 591–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrangsu P, Lemke J, Gourse R 2011. The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol Microbiol 80: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J, Martin A, King D, Wang P, Schultz P 2002. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc Natl Acad Sci 99: 11020–11024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries S, van Dijk A, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin A 2007. Haddock versus Haddock: New features and performance of Haddock 2.0 on the CAPRI targets. Proteins 69: 726–733 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin A 2003. Haddock: A protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Durfee T, Hansen A, Zhi H, Blattner F, Jin D 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190: 1084–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman R, Sevostyanova A, Artsimovitch I 2012. Transcription initiation factor DksA has diverse effects on RNA chain elongation. Nucleic Acids Res 40: 3392–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugen S, Ross W, Gourse R 2008. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol 6: 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer J, Erickson-Viitanen S, Wolfe H, DeGrado W 1986. p-Benzoyl-L-phenylalanine, a new photoreactive amino acid. Photolabeling of calmodulin with a synthetic calmodulin-binding peptide. J Biol Chem 261: 10695–10700 [PubMed] [Google Scholar]
- Kettenberger H, Armache K, Cramer P 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114: 347–357 [DOI] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S 2003. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J 22: 6322–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Lennon C, Ross W, Gourse R 2012. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J Mol Biol 416: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke J, Durfee T, Gourse R 2009. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol 74: 1368–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke J, Sanchez-Vazquez P, Burgos H, Hedberg G, Ross W, Gourse R 2011. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci 108: 5712–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon C, Gaal T, Ross W, Gourse R 2009. Escherichia coli DksA binds to free RNA Polymerase with higher affinity than to RNA Polymerase in an open complex. J Bacteriol 191: 5854–5858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson L, Gummesson B, Jaksimović P, Farewell A, Nyström T 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol 189: 5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miropolskaya N, Nikiforov V, Klimasauskas S, Artsimovitch I, Kulbachinskiy A 2010. Modulation of RNA polymerase activity through the trigger loop folding. Transcription 1: 89–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray H, Schneider D, Gourse R 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell 12: 125–134 [DOI] [PubMed] [Google Scholar]
- Opalka N, Chlenov M, Chacon P, Rice W, Wriggers W, Darst S 2003. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell 114: 335–345 [DOI] [PubMed] [Google Scholar]
- Opalka N, Brown J, Lane W, Twist K, Landick R, Asturias F, Darst S 2010. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol 8: e1000483 doi: 10.1371/journal.pbio.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B, Barker M, Ross W, Schneider D, Webb C, Foster J, Gourse R 2004. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118: 311–322 [DOI] [PubMed] [Google Scholar]
- Paul B, Barker M, Gourse R 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci 102: 7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perederina A, Svetlov A, Vassylyeva M, Tahirov T, Yokoyama S, Artsimovitch I, Vassylyev D 2004. Regulation through the secondary channel-structural framework for ppGpp-DksA synergism during transcription. Cell 118: 297–309 [DOI] [PubMed] [Google Scholar]
- Potrykus D, Vinella D, Murphy H, Szalewska-Palasz A, D'Ari R, Cashel M 2006. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J Biol Chem 281: 15238–15248 [DOI] [PubMed] [Google Scholar]
- Rutherford S, Lemke J, Vrentas C, Gaal T, Ross W, Gourse R 2007. Effects of DksA, GreA, and GreB on transcription initiation: Insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol 366: 1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Villers C, Lee J, Ross W, Gourse R 2009. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev 23: 236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu Y, Schultz P 2006. Efficient incorporation of unnatural amino acids into proteins of Escherichia coli. Nat Methods 3: 263–265 [DOI] [PubMed] [Google Scholar]
- Saecker R, Record M, deHaseth P 2011. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J Mol Biol 412: 754–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S 2010. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature 468: 978–982 [DOI] [PubMed] [Google Scholar]
- Tehranchi A, Blankschien M, Zhang Y, Halliday J, Srivatsan A, Peng J, Herman C, Wang J 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141: 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov I, Zhang J, Palangat M, Landick R 2007. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell 27: 406–419 [DOI] [PubMed] [Google Scholar]
- Trautinger B, Jaktaji R, Ruskova E, Lloyd R 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19: 247–258 [DOI] [PubMed] [Google Scholar]
- Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol 68: 1128–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D, Sekine S, Laptenko O, Lee J, Vassylyeva M, Borukhov S, Yokoyama S 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature 417: 712–719 [DOI] [PubMed] [Google Scholar]
- Vassylyev D, Vassylyeva M, Zhang J, Palangat M, Artsimovitch I, Landick R 2007. Structural basis for substrate loading in bacterial RNA polymerase. Nature 448: 163–168 [DOI] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL 2005. Response of RNA polymerase to ppGpp: Requirement for the ω subunit and relief of this requirement by DksA. Genes Dev 19: 2378–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas C, Gaal T, Barker M, Rutherford S, Haugen S, Vassylyev D, Ross W, Gourse R 2008. Still looking for the magic spot: The crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol 377: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C, Moreno M, Wilmes-Riesenberg M, Curtiss R, Foster J 1999. Effects of DksA and ClpP protease on σS production and virulence in Salmonella typhimurium. Mol Microbiol 34: 112–123 [DOI] [PubMed] [Google Scholar]
- Zhang J, Palangat M, Landick R 2010. Role of the RNA polymerase trigger loop in catalysis and pausing. Nat Struct Mol Biol 17: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]