Germ layer segregation is the first differentiation event in the embryo. Colas et al. now report that two conserved microRNA families, let-7 and miR-18, bias the decision to become endoderm versus mesoderm. let-7 knockdown converts presumptive ectoderm and mesoderm to develop as gut endoderm. let-7 and miR-18 down-regulate components of the Nodal signaling pathway. Thus, let-7 and miR-18 translate a gradient of secreted Nodal protein into a sharp border between germ layers.
Keywords: microRNA, let-7, mir-18, endoderm, germ layer, high-throughput screening, embryonic stem cells
Abstract
Tight control over the segregation of endoderm, mesoderm, and ectoderm is essential for normal embryonic development of all species, yet how neighboring embryonic blastomeres can contribute to different germ layers has never been fully explained. We postulated that microRNAs, which fine-tune many biological processes, might modulate the response of embryonic blastomeres to growth factors and other signals that govern germ layer fate. A systematic screen of a whole-genome microRNA library revealed that the let-7 and miR-18 families increase mesoderm at the expense of endoderm in mouse embryonic stem cells. Both families are expressed in ectoderm and mesoderm, but not endoderm, as these tissues become distinct during mouse and frog embryogenesis. Blocking let-7 function in vivo dramatically affected cell fate, diverting presumptive mesoderm and ectoderm into endoderm. siRNA knockdown of computationally predicted targets followed by mutational analyses revealed that let-7 and miR-18 down-regulate Acvr1b and Smad2, respectively, to attenuate Nodal responsiveness and bias blastomeres to ectoderm and mesoderm fates. These findings suggest a crucial role for the let-7 and miR-18 families in germ layer specification and reveal a remarkable conservation of function from amphibians to mammals.
The allocation and segregation of pluripotent cells into ectoderm, mesoderm, and endoderm is the first step in forming the body plan during embryogenesis and is critical for all subsequent development. In nonchordates, this process depends heavily on the segregation of maternally encoded factors that specify germ layer fates, which become manifest as the embryo undergoes the morphological process of gastrulation when the ectoderm, mesoderm, and endoderm become distinct. Germ layer allocation in vertebrates depends on diffusible signals that induce mesoderm and endoderm from pluripotent epiblast cells preceding or coincident with early gastrulation movements. Research over the past 30 years has established that the nature, intensity, and duration of exposure to different signaling molecules, such as Nodal, Wnts, and bone morphogenetic proterins (BMPs), orchestrate the commitment to individual germ layers (Green et al. 1992; Dosch et al. 1997; Schier et al. 1997; Stathopoulos and Levine 2002; Dougan et al. 2003; Dunn et al. 2004; Tabata and Takei 2004). Positive and negative feedback circuits as well as secreted antagonists of these factors also play essential roles to provide tight spatiotemporal control over signaling (Re'em-Kalma et al. 1995; Piccolo et al. 1999; Perea-Gomez et al. 2002). Despite this knowledge, many key phenomena are incompletely understood; in particular, how signaling is modulated so that two adjacent, even daughter, cells can adopt distinct germ layer fates.
MicroRNAs (miRs) are endogenous, ∼22-nucleotide (nt) ssRNAs that directly bind and suppress multiple mRNA targets within the context of the RNA-induced silencing complex (RISC). miRs bind mRNAs through base-pairing of their “seed” sequences to the 3′ untranslated region (UTR) or, less commonly, the coding regions, thus targeting mRNAs for translational inhibition and degradation within the RISC. By governing translation, they have been found to influence nearly every normal and pathological process examined (Filipowicz et al. 2008; Bartel 2009). We postulated that miRs might act at the earliest stages of embryonic development to control the specification of mesoderm and endoderm from common embryonic blastomeres.
We systematically screened most human miRs for the ability to bias the segregation of mesoderm and endoderm derivatives of mouse embryonic stem cells (mESCs), which have been used successfully for detailed molecular analyses of signaling processes that guide cell fate allocation (Tada et al. 2005; Murry and Keller 2008). High-throughput functional screening revealed two families, let-7 and miR-18, that promote mesoderm differentiation at the expense of endoderm. let-7 and miR-18 control germ layer fate by negatively regulating TGFβ/Nodal signaling by directly targeting Acvr1b and Smad2, respectively. Interestingly, the function of let-7 is not restricted to mesoderm; it is also expressed in the emerging ectoderm and mesoderm of mouse and Xenopus embryos, where it prevents these tissues from becoming endoderm. We conclude that let-7 family members play an evolutionarily conserved role as repressors of endoderm formation and, moreover, act nonredundantly with secreted antagonists of Nodal such as Cerberus and Lefty proteins. By acting cell-autonomously, they provide a means to translate a gradient of secreted Nodal signaling into a sharp border between germ layers.
Results
Screen for miRs that control endoderm and mesoderm fate
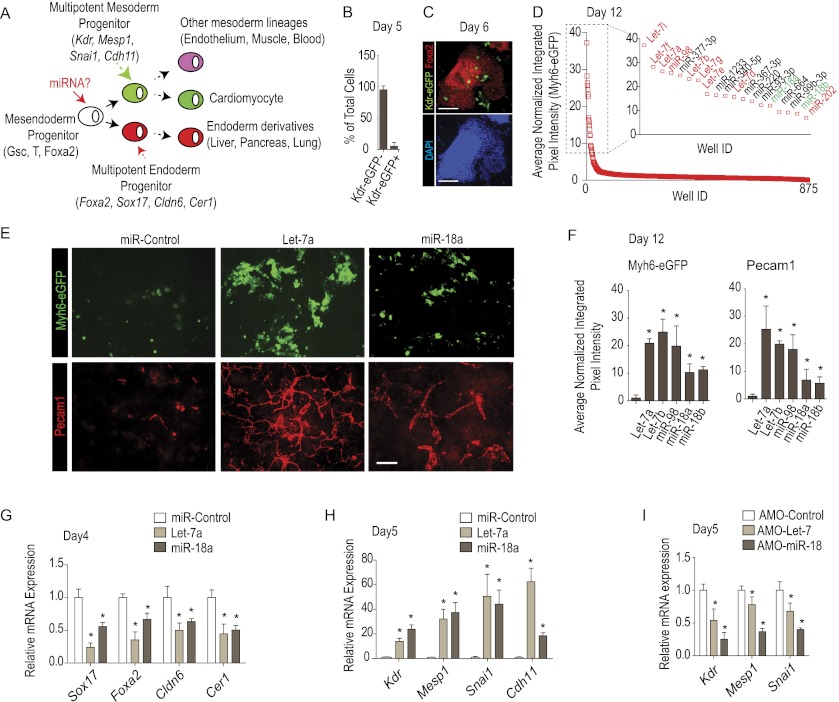
In order to identify miRs involved in germ layer diversification, we first designed a mESC-based differentiation protocol that is sensitive to the differentiation of mesoderm and endoderm (Fig. 1A). Molecular characterization of differentiating mESCs under basal conditions showed that mesendoderm genes (Tada et al. 2005) such as Gsc, Foxa2, and T (Bra) peak at day 4 of differentiation, while the expression of genetic markers of differentiating mesoderm, including Kdr and Pdgfra, and endoderm, including Sox17 and Cer1, peak at day 5 of differentiation (Supplemental Fig. 1A), consistent with day 4 mesendoderm progenitors segregating into mesoderm and endoderm lineages by day 5. To ascertain when cardiogenic progenitors arise, we took advantage of a transgenic mESC line with eGFP under the control of the endogenous Kdr locus (Ema et al. 2006). Fluorescence-activated cell sorting (FACS) analysis revealed that ∼5.0% of cells are Kdr-eGFP+ mesoderm at day 5 without addition of exogenous miRs (Fig. 1B). These Kdr-eGFP+ cells express Mesp1, Cdh11, Snai1, and Pdgfra, validating their mesodermal nature (Supplemental Fig. 1B). Conversely, the Kdr-eGFP− cells are enriched for endodermal marker genes, including Sox17, Foxa2, Cldn6, and Cer1 (Supplemental Fig. 1C). Furthermore, specific immunostaining confirms that the vast majority of Kdr-eGFP− cells express Foxa2, consistent with an early endoderm identity (Fig. 1C). Together, these results show that days 4–5 span the developmental window when mesendoderm progenitors give rise to mesoderm and endoderm and that most cells under basal conditions form endoderm.
Figure 1.
Screen for miRs that control endoderm and mesoderm fate. (A) Schematic representation of the screening strategy. (B,C) FACS analysis of differentiating mESCs with eGFP under the control of Kdr promoter (n = 3) revealed that most cells are Kdr-eGFP− (shown in B). (C) Immunostaining confirmed that most cells express the endoderm marker Foxa2 at day 6. See Supplemental Figure 1 for additional marker profiling showing endodermal and mesodermal marker expression in the differentiating cells. (D) Ranking of 875 miRs screened for induction of the Myh6-eGFP reporter indicative of cardiomyogenic cells. The inset shows the top 19 hits. (E,F) Confirmation that let-7 and miR-18 bias Myh6-eGFP and Alexa Fluor568-Pecam1 fluorescence levels (integrated pixel intensity) (see the Materials and Methods) in mESCs transfected at day 3 and assayed at day 12. Fluorescence values of responses to let-7 (let-7a, let-7b, and miR-98) and miR-18 (miR-18a and miR-18b) plotted relative to that for the control (scrambled sequence) miR. (F) Representative images of Myh6-eGFP and Alexa Fluor568-Pecam1. (G–I) qRT–PCR of early endoderm (G,I) and mesoderm (H) marker genes in response to transfection with let-7, miR-18 (G,H), or specific anti-miRs (I). Note that miRs repress endoderm and stimulate mesoderm marker gene expression. The anti-miRs repress mesoderm markers; endodermal markers were not examined, since the assay baseline has maximal endodermal marker expression and is insensitive to any increase. All qRT–PCR data were normalized to β-actin mRNA levels. All data are presented as mean ± SD. (*) P < 0.05.
In order to identify miRs that bias cultures toward mesoderm, we performed the screen using a transgenic mESC line expressing eGFP under the control of the cardiac-specific Myh6 promoter (Takahashi et al. 2003). Reading out cardiomyocyte differentiation ensured that mesoderm cells affected by the miRs could form a differentiated cell type. We reverse-transfected wells (384-well plate format) with individual synthetic oligonucleotides from a human miR library (875 pre-miRNAs; Ambion) at day 3, 1 d prior to the mesendoderm lineage diversification window defined above. Cells were then cultured for an additional 9 d followed by fixation, automatic imaging, and quantification of Myh6-eGFP fluorescence using a custom algorithm in CyteSeer that measures the integrated fluorescence intensity (average pixel intensity × area) of the eGFP-positive area, which provides an indication of the number of differentiating cells (see the Materials and Methods). Primary hits were ranked for Myh6-eGFP induction relative to a control (inert sequence) miR (Fig. 1D). Hits were selected based on representation within the top fifth percentile (Fig. 1D, inset) and the additional criteria that all family members should exhibit positive activity and that the miR seed sequence be conserved among vertebrate (human, mouse, and Xenopus) species. let-7 and miR-18 families (Fig. 1D, inset, red and green, respectively) increased Myh6-eGFP activity 10-fold to 35-fold over control miRs. Each member of the let-7 family, (comprising 10 members—let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let-7i, miR-98, and miR-202—in mice) and both members of the miR-18 family (miR-18a and miR-18b) were within the top fifth percentile, with the exception of let-7c. let-7a, let-7b, miR-98, miR-18a, and miR-18b were reordered and individually tested to confirm the screen results (Fig. 1E,F). Furthermore, staining cultures with anti-Pecam1 revealed that both let-7 and miR-18 markedly enhanced endothelial differentiation in addition to elevating Myh6-eGFP. Thus, we identified two families of evolutionarily conserved miRs that promote cardiomyocyte and endothelial cell differentiation.
let-7 and miR-18 promote mesoderm at the expense of endoderm
To gain insight into the biological activity of let-7 and miR-18, we assessed the expression of early mesoderm and endoderm markers by quantitative RT–PCR (qRT–PCR). The first phenotypic effect of the miRs is to markedly down-regulate endoderm-specific genes (Sox17, Cer1, Foxa2, and Cldn6) at day 4 of differentiation (Fig. 1G). Subsequently, at day 5, the miRs substantially up-regulated the mesoderm markers Kdr, Mesp1, Snai1, and Cdh11 (Fig. 1H) and, more modestly, increased Pdgfra and Pdgfrb expression (data not shown). Conversely, specific anti-miRs (AMO) to let-7 and miR-18 decreased mesoderm marker expression (Fig. 1I), suggesting the involvement of endogenous miR-18 and let-7 family members, a possibility that is examined in more detail below. Collectively, these data show that the miRs bias differentiation toward mesoderm at the expense of endoderm in the mESC assay.
let-7 and miR-18 modulate cell fate through inhibition of Nodal signaling
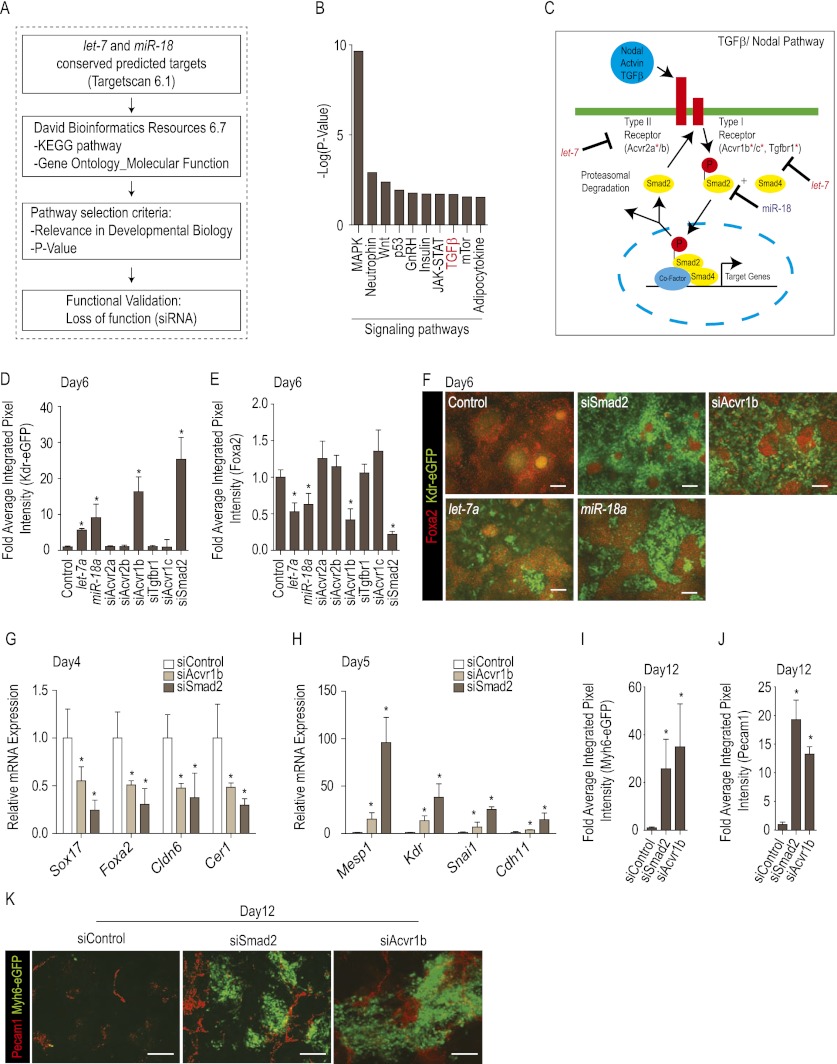
Next, we sought to identify the targets through which let-7 and miR-18 promote mesoderm differentiation. Our first approach was to mimic let-7 and miR-18's effect using siRNAs directed against previously validated targets such as lin28a/b (Rybak et al. 2008) and Hmga2 (high-mobility group AT-hook-2) (Mayr et al. 2007) for let-7 and Esr1 (estrogen receptor-α) (Castellano et al. 2009) and Runx1 (Ben-Ami et al. 2009) for miR-18. None of the specific siRNAs, either alone or in combination, significantly enhanced mesoderm differentiation (data not shown). We then took a systems approach to identify candidate signaling pathways, hypothesizing that let-7 and miR-18 might exert their action by targeting multiple components of the same signaling pathway. We performed Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway analysis on the computationally predicted targets of let-7 and miR-18, representing 1200 genes (Fig. 2A). Visualization of the 10 most prominently affected signaling pathways (Fig. 2B) and molecular functions led us to investigate the TGFβ/Nodal signal transduction pathway, which is capable of inducing mesoderm and endoderm in Xenopus and mouse embryos (Armes and Smith 1997; Piccolo et al. 1999; Iratni et al. 2002). let-7 was predicted to target most receptors of the Nodal pathway (Acvr1b, Acvr1c, Acvr2a, and Tgfbr1), and miR-18 was predicted to target the intracellular mediator Smad2 (Fig. 2C). However, neither miR was predicted to target components of the closely related BMP pathway.
Figure 2.
let-7 and miR-18 modulate cell fate through inhibition of Nodal signaling. (A) miR target identification strategy (see the Materials and Methods). (B) Top 10 computationally predicted signaling pathways targeted by let-7 and miR-18 ranked by −log(P-value). (C) Predicted let-7 and miR-18 targets in the Nodal/TGFβ signaling pathway (red asterisk). (D–F) Kdr-eGFP (D) and Alexa Fluor568-Foxa2 (E) fluorescence measurements at day 6 in mESCs transfected with siRNAs against Nodal/TGFβ signaling pathway components, quantified by automated microscopy, and plotted relative to values obtained with control (scrambled sequence) siRNA. (F) Representative images of Kdr-eGFP and Alexa Fluor568-Foxa2 fluorescence as in D and E. Note that siRNAs to Acvr1b and Smad2 specifically enhanced Kdr-eGFP while reducing Foxa2 levels. (G,H) Effect of Smad2 and Acvr1b knockdown on early endoderm (G) and mesoderm (H) marker genes by qRT–PCR. Note repression of endoderm and stimulation of mesoderm marker genes. (I–K) Myh6-eGFP (I) and Alexa Fluor568-Pecam1 (J) at day 12 of differentiation after transfection of siRNAs against Smad2 or Acvr1b at day 3, relative to control (scrambled) siRNA. Representative image of Myh6-eGFP and Alexa Fluor568-Pecam1, as in I and J. Quantitative data are presented as means ± SD. (*) P < 0.05.
In order to validate the bioinformatics-derived hypothesis, siRNAs against the predicted targets were evaluated individually for their ability to bias Kdr-eGFP and Foxa2 expression at day 6 of differentiation. siRNAs against Acvr1b and Smad2, but none of the other predicted targets, enhanced Kdr-eGFP and repressed Foxa2, as quantified cytometrically by automated microscopy (Fig. 2D–F). Additionally, qRT–PCR analysis confirmed the activity of these siRNAs, revealing a down-regulation of endodermal genetic markers at day 4 and a dramatic up-regulation of mesodermal markers at day 5 (Fig. 2G,H) as was seen with let-7 and miR-18 (Fig. 1G,H). We examined subsequent differentiation and found that Acvr1b and Smad2 siRNAs transfected at day 3 strongly enhanced cardiomyocyte and endothelial differentiation by day 12 (Fig. 2I–K), indicating that modulating this pathway leads to the formation of differentiated derivatives. These data implicate Acvr1b and Smad2 as the biologically relevant molecular targets.
let-7 and miR-18 directly target Acvr1b and Smad2 3′ UTRs
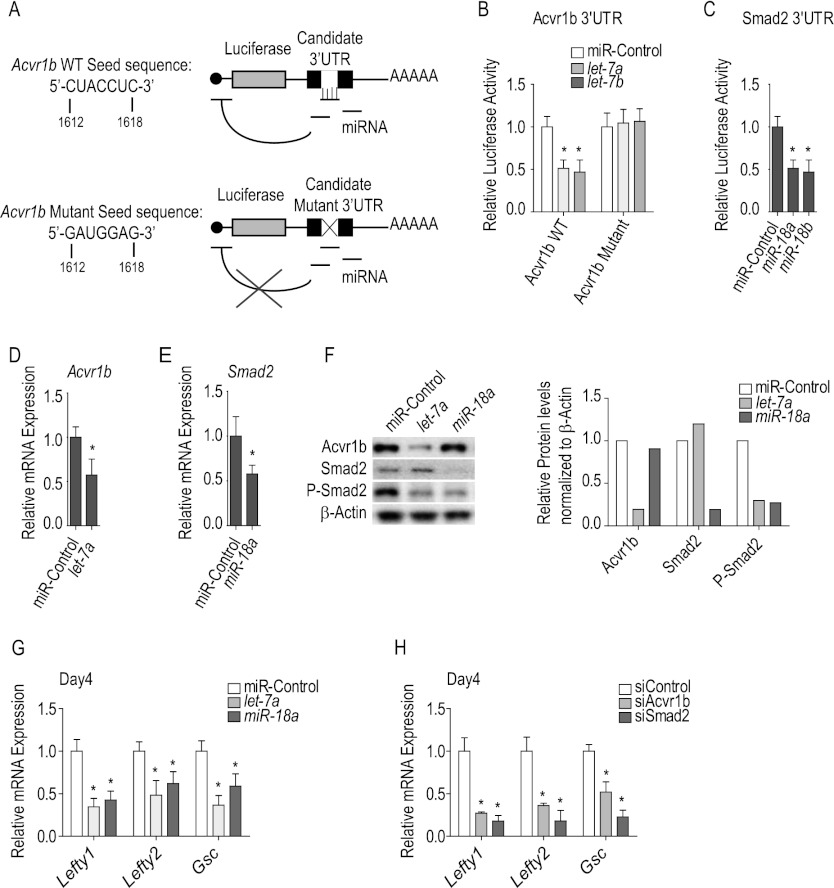
Next, we tested whether let-7 and miR-18 can directly target the computationally predicted recognition sites in the 3′ UTRs of Acvr1b and Smad2 mRNAs. The entire 3′ UTRs were placed downstream from a luciferase ORF (luc) under the control of the cytomegalovirus (CMV) promoter (Fig. 3A). In transfected HEK 293T cells, let-7a and let-7b repressed luciferase activity from the CMV-luc-Acvr1b 3′ UTR construct (Fig. 3B). Repression was abolished if the putative recognition site was mutated (5′-1612CUACCUC1618-3′ mutated to 5′-1612GAUGGAG1618-3′) (Fig. 3B). Similarly, miR-18 repressed the luciferase activity from the CMV-luc-Smad2 3′ UTR construct (Fig. 3C), as previously reported (Li et al. 2011).
Figure 3.
let-7 and miR-18 directly target Acvr1b and Smad2. (A) Schematic showing that mutation of the mRE is predicted to abolish the effect of the miR against the luc reporter-3′ UTR fusion construct, validating dependence on a specific miR:mRE interaction. (B,C) Mutation of the putative recognition elements in Acvr1b (B) and Smad2 (C) 3′ UTRs. Note the reduction of luciferase activity in HEK293T cells 24 h after transfection with specific miR relative to control miR. (D,E) Decrease in endogenous Acvr1b (D) or Smad2 (E) transcript levels after transfection of mESCs at day 3 with let-7a or miR-18a, respectively, relative to control miR. (F) Western blot analysis of Acvr1b, Smad2, P-Smad2, and β-Actin protein levels in mESCs transfected at day 3 with let-7a, miR-18a, or control miRs. (G,H) qRT–PCR of transcriptional targets (Lefty1, Lefty2, and Gsc) in mESCs transfected at day 3 with either with let-7a or miR-18a relative to control miR (G) and siRNAs against Acvr1b or Smad2 relative to control siRNA (H).
We subsequently verified that both miRs affect the endogenous versions of their candidate targets. Transfection of mESCs with let-7 and miR-18 at day 3 of differentiation (Fig. 3D–H) revealed miR-specific repression of endogenous Acvr1b and Smad2 transcripts (Fig. 3D,E) and proteins (Fig. 3F). Consequently, both miRs down-regulated the level of phosphorylated Smad2 in mESCs, indicating that endogenous Nodal signaling activity had been attenuated. This hypothesis was confirmed by a dramatic decrease in Lefty1, Lefty2, and Goosecoid (Gsc) mRNAs (Fig. 3G), which are direct transcriptional targets of Nodal/Acvr1b/Smad2 signaling (Fig. 3H). Together, these results establish that let-7 and miR-18 are capable of repressing Nodal/Acvr1b/Smad2 signaling.
miR control of germ layer segregation in embryos
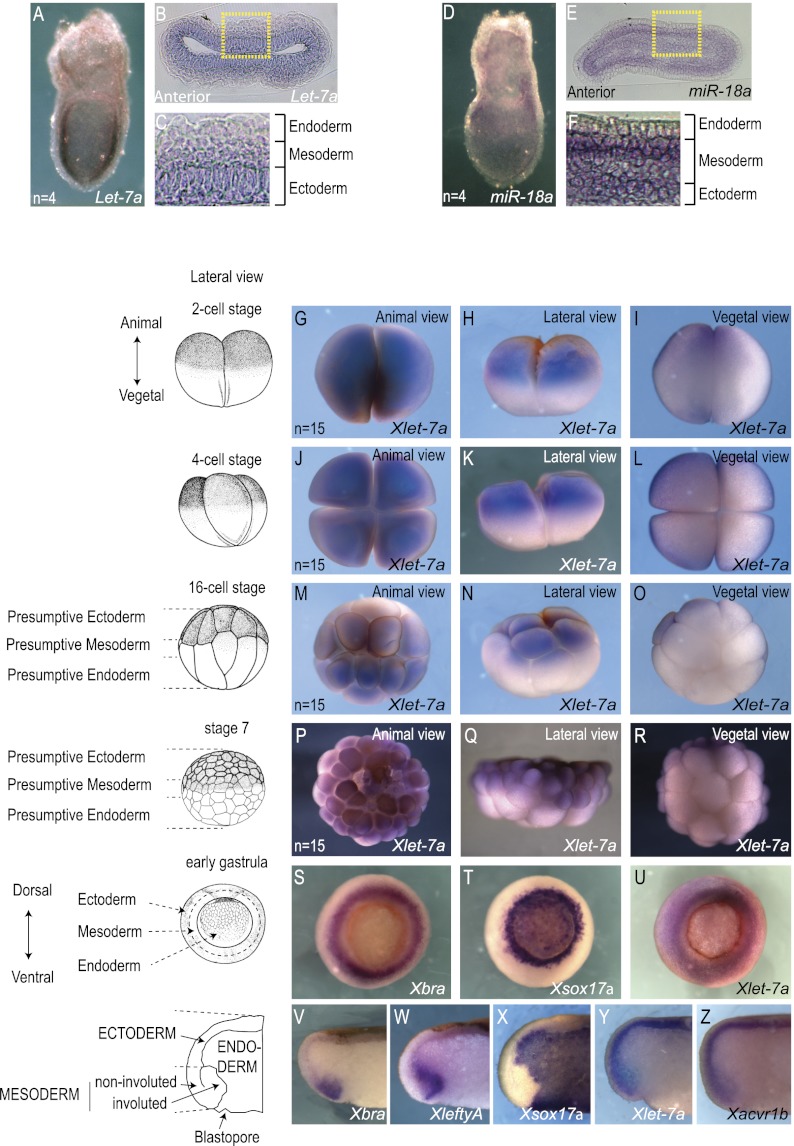
The preceding experiments established miR-18 and let-7 as capable of modulating germ layer segregation in mESC culture. Profiling let-7 and miR-18 family members during the course of cardiogenic mESC differentiation, we found that let-7a, let-7b, let-7c, let-7e, let-7f, let-7g, let-7i, miR-18a, and miR-18b show peak expression at day 5 of differentiation (let-7d and miR-98 were undetected) (Supplemental Fig. 2A), when mesoderm and endoderm segregation starts to occur. Moreover, let-7a, let-7b, let-7c, miR-18a, and miR-18b were enriched in Kdr-eGFP+ cells versus Kdr-eGFP− cells. Few Kdr-eGFP+ cells are produced at the baseline in this assay, so the other let-7 family members were beneath the detection threshold (Supplemental Fig. 2B). The expression profiling is consistent with a role in the segregation of mesoderm and endoderm in vivo. Of the expressed family members, let-7a and miR-18a are abundantly expressed in embryonic day 7 (E7) mouse embryos and so were selected for further analysis. Both miRs are preferentially expressed in ectoderm and mesoderm of E7 embryos, contrasting with their low or undetectable expression in endoderm (Fig. 4A–F), consistent with the hypothesis that they suppress endoderm formation.
Figure 4.
Endogenous localization of let-7a and miR-18a in early vertebrate embryos. (A–F) In situ hybridization showing endogenous let-7a and miR-18a in E7 mouse embryos viewed in whole-mount (A,D) or transverse histological section (B,E). (C,F) High-magnification view distinguishing epiblast mesoderm and endoderm layers. Note the abundant expression of let-7a (A–C) and miR-18a (D–F) in ectoderm and mesoderm but not in endoderm. (G–R) Endogenous Xlet-7a in cleavage stage Xenopus embryos showing expression in the animal hemisphere (presumptive ectoderm and mesoderm). (S–Z) Endogenous Xlet-7a expression in gastrula stage (stage 10.5) Xenopus embryos. View of the mesoderm in the equatorial region marked by Xbra (S) and involuting endoderm marked by Xsox17α (T) reveals overlap with Xlet-7a prominently in mesoderm (U). (V–Z) Transverse sections show expression domains of Xbra (V), XleftyA (W), and Xsox17α (X) relative to Xlet-7a (Y), revealing that the miR is present in both ectoderm and mesoderm but not in endoderm and is nonoverlapping with XleftyA but coincident with Xacvr1b (Z) mRNAs.
Xenopus laevis is an excellent model system to study inductive signals that establish the vertebrate body plan. As in mammalian embryos, Nodal signaling induces mesoderm and endoderm (Smith et al. 1990; Armes and Smith 1997; Agius et al. 2000). The let-7 recognition site (Fig. 3) is conserved in the 3′ UTR of Acvr1b in X. laevis as well as in zebrafish (Danio rerio). Although the miR-18 recognition site is conserved in Xenopus tropicalis, it is not present in X. laevis Smad2 sequences in the NCBI nucleotide database, so we focused on examining Xlet-7 function during germ layer segregation in Xenopus. Whole-mount in situ hybridization localized Xlet-7a during cleavage stages (two-cell, four-cell, 16-cell, and 64-cell) to animal hemisphere cells (Fig. 4G–R) that give rise to ectoderm and mesoderm but not endoderm (Dale and Slack 1987). Expression persisted as development progressed, such that Xlet-7a was observed within the ectoderm and mesoderm domains but was completely absent in the endoderm of gastrula stage (stage 10.5) embryos (Fig. 4S–Z). Other let-7 family members (let-7b, let-7c, let-7e, let-7g, and let-7i) are also enriched in mesoderm as compared with endoderm (Supplemental Fig. 3A,B). Close examination of transversely bisected embryos showed that Xlet-7a is strongly expressed in the noninvoluted mesoderm (Fig. 4Y) overlapping Xbra expression (mesoderm) but is excluded from the involuted mesoderm marked by XleftyA (also expressed by adjacent endoderm) (Fig. 4V–Y). The pattern of Xlet-7a expression is coincident with that of Xacvr1b (Figure 4Y,Z; Chen et al. 2005), consistent with the preceding biochemical evidence of direct interaction.
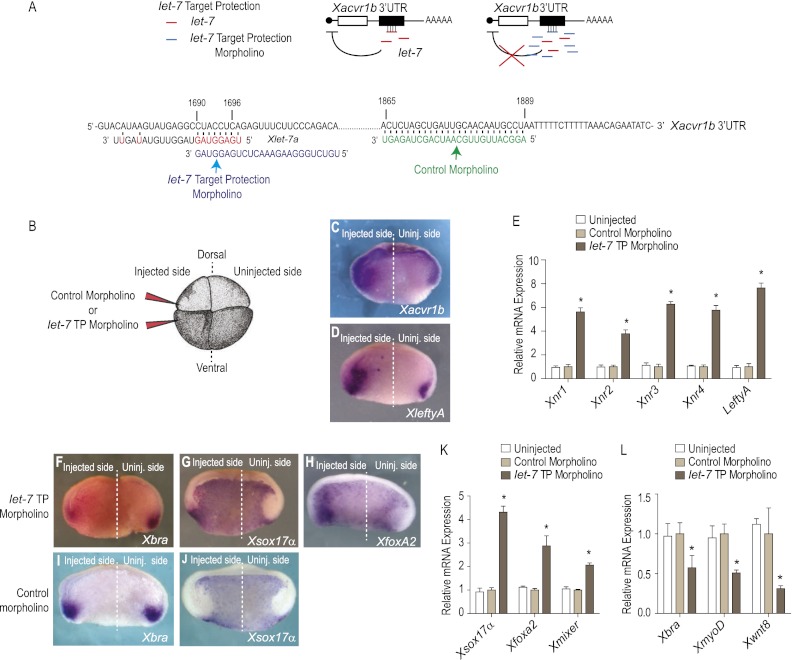
The analogous expression pattern in mesoderm and ectoderm of mouse and Xenopus embryos suggests that the let-7 family plays an evolutionarily conserved role in distinguishing these tissues from endoderm. However, the function of let-7 family members has not been tested previously because the 10 different members distributed across 14 genomic locations in the mouse genome and, similarly, in other vertebrates (Roush and Slack 2008) complicate genetic knockdown or deletion studies. Since we identified a single potential target, Xacvr1b (Fig. 2), it is possible to probe function by microinjecting a morpholino to mask the miR recognition element (mRE) and prevent base-pairing with Xlet-7a (Fig. 5A). Injection of a target protection (TP) morpholino oligoribonucleotide complementary to the Xacvr1b mRE (plus 17 base pairs [bp] 5′) (diagrammed in Fig. 5A) into the equatorial region of four-cell embryos (Fig. 5B) dramatically increased the abundance of Xacvr1b transcripts detectable by in situ hybridization (cf. the TP morpholino-injected and uninjected sides of the embryo in Fig. 5C; Table 1). In addition to increased transcript levels, the domains of detectable expression also appeared to extend to the involuted mesoderm and adjacent endoderm. The TP morpholino also up-regulated expression of XleftyA (Fig. 5D; Table 1). as well as other Nodal/TGFβ transcriptional targets (Xnr1, Xnr2, Xnr3, and Xnr4) (Fig. 5E; Luxardi et al. 2010). As a control, a morpholino of similar base-pair composition but complementary to another region of the Acvr1b 3′ UTR (control morpholino diagrammed in Fig. 5A) had no effect on Xnr genes (Fig. 5E). Collectively, these data indicate that disrupting the interaction between Xlet-7 and its mRE in Xacvr1b 3′ UTR leads to overactive Nodal signaling.
Figure 5.
Endogenous Xlet-7 discriminates mesoderm and endoderm by repressing Xacvr1b. (A) Diagram of the TP loss-of-function strategy to block interaction of the miR with the mRE in vivo. (B) Either Xlet-7 TP or control morpholinos were injected equatorially into two blastomeres on one side of four-cell stage embryos. (C,D) Unilaterally injected embryos (as in B) cultured to gastrula stage (stage 10.5), bisected transversely, and probed for Xacrv1b (C) and XleftyA (D) expression. Injection of Xlet-7 TP morpholino up-regulated and expanded the domains of Xacrv1b and XleftyA expression in the marginal zone mesoderm and underlying deep endoderm. (E) Xlet-7 TP injection up-regulated Xnr-1, Xnr-2, Xnr-3, and Xnr-4 as well as XleftyA by qRT–PCR on dissected lateral marginal zone explants relative to the control morpholino, which has no effect on development or gene expression. (F–H) Unilateral Xlet-7 TP morpholino injection repressed a marker of mesoderm, Xbra (F), while concomitantly expanding the domain of endodermal marker genes Xsox17α (G) and Xfoxa2 (H) into the mesoderm domain. (I,J) Unilateral control morpholino injection had no effect on both mesodermal (I) and endodermal (J) markers. (K,L) qRT–PCR analyses on dissected lateral marginal zone explants show that the Xlet-7 TP morpholino up-regulated expression of endodermal genes (XSox17α, Xfoxa2, and Xmixer) (K) and repressed mesodermal genes (Xbra, XmyoD, and Xwnt8) (L) as compared with the control morpholino.
Table 1.
Effect of TP morpholino on marker expression
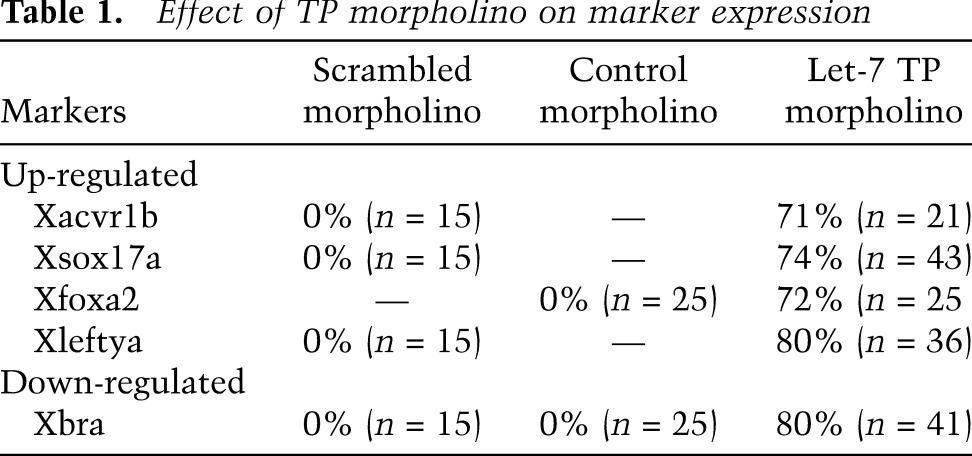
To define the embryological role of let-7, we examined whether the TP morpholino altered mesoderm and endoderm fate as visualized by genetic markers. The TP morpholino, injected into the equatorial region of the embryo at the four-cell stage as above, expanded the domains of Xsox17α and Xfoxa2 expression into the equatorial region of the gastrula stage (stage 10.5) embryo that is normally specified as mesoderm (Fig. 5G,H). Conversely, the Xbra domain was markedly reduced (Fig. 5F). Similarly, endodermal genes (Xsox17α, Xfoxa2, and Xmixer) were up-regulated, while mesodermal genes (Xbra, XmyoD, and Xwnt8) were down-regulated (Fig. 5K,L). As above, the control morpholino had no effect on gene expression (Fig. 5I,J). Thus, preventing Xlet-7 from binding to the Xavr1b 3′ UTR was sufficient to cause presumptive mesodermal cells to differentiate as endoderm. See Table 1 for quantificaton.
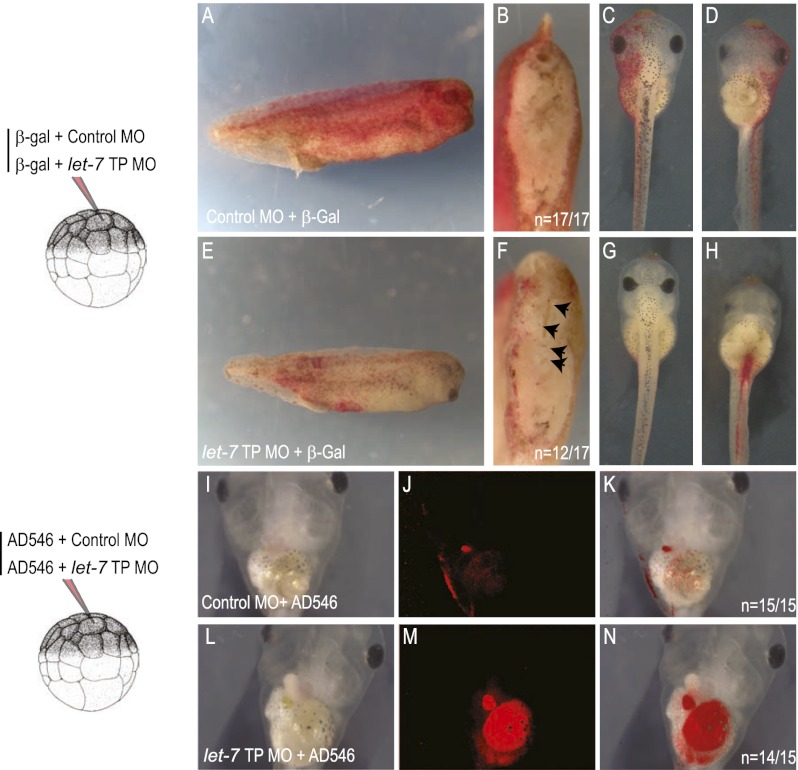
In addition to controlling mesodermal fate, the coincident expression of Xlet-7 with Xacvr1b throughout the presumptive ectoderm suggests that it might control ectoderm fate as well. Despite receptor expression, ectoderm is thought to lack endogenous Nodal signaling, although overactivation of activin type 1 receptors Taram-A in zebrafish (Peyrieras et al. 1998) and Xacvr1c in Xenopus (Reissmann et al. 2001) can divert presumptive ectoderm cells to adopt an endoderm fate. Postulating that Xlet-7 might suppress ectodermal Xacvr1b, we asked whether the TP morpholino would affect cell fate. Single presumptive ectodermal blastomeres at the 32-cell stage (stage 5) were injected with LacZ mRNA as a lineage tracer together with either let-7 TP or control morpholino, and the progeny of injected cells were examined in late tailbud embryos (stage 35). While control (scrambled) morpholino-injected cells contributed to patches of epidermis and never to the inner endoderm cells (Fig. 6A–D), specific TP morpholino-injected cells gave only a few ectodermal cells but contributed instead to gut endoderm (Fig. 6E–H). We repeated the experiment (this time coinjecting an Alexa546-conjugated fluorescent dextran as a lineage label) and examined the progeny of the injected blastomeres in early tadpoles (stages 40–45). The diversion of presumptive ectoderm to endoderm was even more striking at the later stages: The ectodermal cells were brightly visible as patches in the epidermis, while the progeny of TP morpholino-injected cells were equally visible in the coiled gut (Fig. 6I–N). Thus, as in mesoderm, endogenous Xlet-7 maintains Xacvr1b levels sufficiently low in ectoderm to prevent endogenous Nodal from directing the cells to become endoderm.
Figure 6.
Blocking endogenous let-7 converts ectoderm to endoderm in embryos. (A–H) Embryos at the 16-cell stage were injected in one animal blastomere (presumptive ectoderm) together with Xlet-7 TP or control morpholino and LacZ mRNA dextran as a lineage label. Whereas the progeny of control morpholino-injected blastomeres contributed exclusively to epidermis (A–D), Xlet-7 TP morpholino-injected blastomeres contributed to endoderm, examined at the tailbud stage (stage 35) in lateral view (A,D), transverse bisection (B,F), dorsal view (C,G), and ventral view (D,H). (I–N) The same as in A–H, but examined at the tadpole stage (stage 45) and injected with Alexa Fluor546 dextran as a lineage label. Note the pronounced diversion of ectoderm to gut cell fate conversion, viewed ventrally.
Discussion
A large number of embryological studies have concluded that the intensity and duration of exposure to inductive signals specifies germ layers from pluripotent cells of the early embryo. Nodal plays a central role in this process, as modeled by the demonstration that increasing concentrations of Activin progressively induce ectoderm, mesoderm, and endoderm in Xenopus and amniote embryos as well as in mouse and human ESCs (Green and Smith 1990; Gurdon et al. 1994, 1999; Latinkic et al. 1997; Chang and Harland 2007; Willems and Leyns 2008; Lee et al. 2009). Although small concentration differences can induce different cell types when manipulated experimentally, it has never been entirely clear how neighboring cells in the embryo can adopt two different fates. Contributing mechanisms include extracellular molecules that limit diffusion and antagonize function in order to sharpen an initially shallow gradient of Nodal activity, fulfilling a prediction of the Turing/Meinhardt reaction diffusion model for biological pattern generation (Marjoram and Wright 2011; Muller et al. 2012). In addition, intercellular communication by Notch can refine mesoderm and endoderm specification, at least in zebrafish (Kikuchi et al. 2004). Whether these are sufficient to appropriately specify germ layers in vivo has not been resolved. Our findings—initially from functionally screening a whole-genome miR library—that let-7 and miR-18 target Acvr1b and Smad2 to control mesoderm at the expense of endoderm reveal an unanticipated and evolutionarily conserved role for miRs that is nonredundant with known regulators of germ layer specification.
The transcriptional response to Nodal signaling is proportionate to the number of occupied receptors on the cell surface (Dyson and Gurdon 1998). For example, activation of 100 receptors in Xenopus ectoderm induces mesoderm marked by Xbra, whereas activation of 300 or more induces the mesendoderm marked by Xgsc. Occupancy is similarly important, and the high affinity and avidity of ligands such as Nodal or TGFβ for their cognate receptors means that a cell cannot rapidly shift its response to declining concentrations of ligand, such as when titrated by antagonists or when cells migrate away from the ligand source (Dyson and Gurdon 1998; Green 2002). Therefore, mechanisms that regulate the absolute number of receptors on the cell surface and/or intracellular factors that control responsiveness are of particular importance to specifying cell fate. However, crucial components of Nodal responsiveness, such as the Nodal receptor Acvr1b or the intracellular effector Smad2, do not exhibit graded or regionally localized patterns of mRNA expression as germ layers become distinct in mouse gastrula stage embryos, arguing against transcriptional regulation. In contrast, we found that let-7a and miR-18a are present in the epiblast and nascent mesoderm but absent in endoderm (Figs. 2, 4). Since these miRs were found to down-regulate Acvr1b and Smad2, respectively (Fig. 3), we postulated that the miRs would dampen Nodal responsiveness in the epiblast and nascent mesoderm while permitting signaling in endoderm, thereby patterning the border separating endoderm from ectoderm and mesoderm (as diagrammed in Fig. 7). To test this model, we mapped the recognition element of let-7 to the 3′ UTR of mouse and Xenopus Acvr1b/Xacvr1b transcripts (Fig. 3) and then injected morpholino ribonucleotides complementary to this sequence into the presumptive mesoderm and ectoderm of Xenopus embryos to block the interaction of endogenous let-7, effectively diverting the fate of both tissues to endoderm (Figs. 5, 6). An advantage of TP over antisense or sponge approaches for knocking down let-7 function is that it tests the relevance of Xacvr1b as the direct miR target and also disrupts binding of all let-7 family members. Since disrupting the interaction between let-7 and Acvr1b dramatically diverted the cell fate of both presumptive mesoderm and ectoderm, the interaction must be essential to maintain the cell fate of both tissues against the challenge of endogenous Nodal. Our findings support the model that let-7 and miR-18 down-regulate the responsiveness of cells to a shallow gradient of endogenous Nodal (Jones et al. 1995; Varlet et al. 1997) such that high-level signaling occurs only in the endoderm, while intermediate- and low-level signaling occurs in mesoderm and ectoderm, respectively. Although we documented that disrupting the interaction between Xlet-7 and Xacvr1b mRNA was sufficient to direct presumptive ectoderm cells to become gut endoderm (Fig. 6), additional proteins, such as ectodermin (Dupont et al. 2005), and potentially other miRs influence the establishment of the ectoderm–mesoderm border.
Figure 7.
miR control of germ layer segregation in vertebrate embryos. let-7 and miR-18 family members repress Acvr1b and Smad2 protein production, rendering cells less sensitive to Nodal signaling and sharpening a border between endoderm versus ectoderm and mesoderm. By acting cell-autonomously to titrate Nodal responsiveness, the miRs function nonredundantly with protein inhibitors (e.g., Lefty and Cerberus proteins) and the miR-302/427/430 family, previously shown to influence mesendoderm specification by repressing secreted modulators of Nodal signaling (see the text).
What dictates the spatial pattern of let-7 that presages the mesoderm–endoderm border? In mice, 14 different genomic loci encode for different let-7 family members (Roush and Slack 2008), which allows for a very diverse repertoire of transcriptional regulation. Surprisingly, only a few reports have identified transcriptional regulators of let-7 family members' primary transcripts, including Myc, which negatively regulates let-7a, let-7d, and let-7g in human P493-6 cells (Chang et al. 2008), and NF-κB, which activates let-7a-3 and let-7b (Wang et al. 2012). However, the presence of primary transcripts is not always predictive of let-7 activity, since potent negative post-transcriptional regulators, such as lin-28, can prevent maturation of primary transcripts (Newman et al. 2008; Viswanathan et al. 2008). Interestingly, the similar spatial localization of let-7a with respect to the germ layers of mouse and Xenopus embryos is likely to arise from very different mechanisms. In Xenopus, zygotic transcription begins at the mid-blastula stage when the embryo has ∼1000 cells, whereas the mouse zygotic genome becomes active at the two-cell stage (for review, see Tadros and Lipshitz 2009). Transcripts in cleavage and early blastula stage embryos are maternally encoded and regionally deposited in the egg and can be redistributed as oogenesis proceeds (King et al. 2005). Therefore, the germ layer-specific patterns of let-7a and miR-18a expression are likely to involve zygotic transcription in mice but localized deposition and distribution of maternally encoded miRs in Xenopus. Additional studies elucidating how let-7 transcription and function are regulated during the early steps of embryogenesis will be critical to fully understand germ layer specification.
The observation that knocking down the interaction of Xlet-7 with Xacvr1b had a dramatic effect on diverting ectoderm and mesoderm to endoderm was unexpected given the presence of endogenous secreted Nodal antagonists LeftyA, LeftyB, and Cerberus. However, in early gastrula stage embryos, Lefty proteins are expressed in the involuted mesoderm and adjacent deep endoderm (Fig. 4; Branford and Yost 2002), consistent with induction by the highest levels of Nodal signaling. Cerberus is expressed in the deep endoderm on the dorsoanterior side of the embryo (Schneider and Mercola 1999). Thus, let-7 is spatially and functionally nonredundant with the secreted antagonists by cell-autonomously titering Nodal responsiveness. Moreover, let-7 also functions nonredundantly with miR-427, which is an ortholog of miR-302 in humans and miR-430 in zebrafish, and down-regulates Lefty and Nodal proteins (Choi et al. 2007; Rosa et al. 2009). Loss of miR-427 function caused a generalized reduction of anterior structures in Xenopus, consistent with the prominent function of miR-427 to repress Lefty proteins, thereby increasing peak levels of Nodal signaling. In contrast, let-7, given its distinct target and localization, cell-autonomously inhibits Nodal responsiveness at sites distal to Lefty expression to impart a prepattern that biases cells to mesodermal and ectodermal lineages. Although let-7 functions earlier, it is mechanistically similar to miR-15 and miR-16, which down-regulate Acvr2a in Xenopus to restrict the size of the organizer within the dorsal mesoderm (Choi et al. 2007).
In summary, Nodal plays an evolutionarily conserved role in germ layer specification such that increasing levels progressively induce mesoderm then endoderm (for review, see Shen 2007). Our findings suggest that the let-7 and miR-18 families provide a means to transduce the levels of endogenous Nodal protein (depicted in Fig. 7 as a shallow gradient) into a sharply delineated pattern of signaling that dictates whether cells become ectoderm and mesoderm versus endoderm (Fig. 7). Despite major differences in the architecture and developmental strategies that distinguish amphibians and mammals, the interaction between let-7 and Nodal responsiveness define a miR/antagonist–Nodal/agonist module that was selected and evolutionarily conserved in order to shape distinct domains of Nodal activity at this highly dynamic and critical stage of embryogenesis.
Materials and methods
mESC culture
mESCs with Myh6-eGFP (Takahashi et al. 2003) and Kdr-eGFP (Ema et al. 2006) were maintained in DMEM high-glucose (HyClone) medium supplemented with 10% fetal bovine serum (FBS), 1 mM sodium pyruvate (Sigma), 1× MEM NEAA (Gibco), 2 mM L-glutamine (Gibco), 100 U/mL and 100 μg/mL penicillin-streptomycin (HyClone), 50 μM β-mercaptoethanol (Sigma), and U/mL LIF 1000 (Millipore). For differentiation, mESCs were seeded in 10-cm low-attachment tissue culture dishes at a density of 500,000 cells per dish in a chemically defined medium (CDM) (Gadue et al. 2006) where they formed embryoid bodies (EBs) over a period of 2 d. EBs were then dissociated using 0.25% Trypsin EDTA (Gibco) and replated in CDM supplemented with 30 ng/mL ActivinA (R&D Systems) in 10-cm low-attachment tissue culture dishes.
High-content screening assay
On day 3 of differentiation, EBs were collected and dissociated using 0.25% Trypsin EDTA, and 5000 cells per well in 100 μL of CDM containing ActivinA (30 ng/mL) were plated into gelatin-coated 384-well optical tissue culture plates (Greiner Bio-One) prespotted with miRNAs at a concentration of 10 nM in 0.2 μL of Lipofectamine RNAiMax in 14.8 μL of OPTI-MEM I (Gibco). On day 5 of differentiation, the medium was replaced with CDM without ActivinA. Subsequently, CDM was replaced every other day until fixation at day 12. Fixation was performed using 4% paraformaldehyde. Imaging was performed using an automated microscope (InCell Analyzer 1000, GE Healthcare), and image analysis was done using Cyteseer image analysis software (Vala Sciences) by measuring the integrated pixel intensity (pixel area × average pixel intensity) of eGFP or Alexa Fluor568 signal within an eGFP-positive area (Bushway and Mercola 2006). The top 5% of miRs was considered hits and confirmed by reordering (Supplemental Table 2) and retesting at 10 nM in the primary assay.
For specific anti-miR treatment of mESCs, control and specific anti-miRs for let-7 and miR-18 were provided by Regulus, Inc. and transfected as above. Regulus anti-miRs are a high-affinity oligonucleotide complementary to the active site of miR-18 or let-7 with a phosphorothioate backbone containing modifications (fluoro, methoxyethyl, and bicyclic sugar) at the sugar 2′ position.
Immunostaining
Cells grown on gelatin-coated 384-well optical tissue culture plates (Greiner Bio-One) were fixed using 4% paraformaldehyde and immunostained by incubating in block solution (10% horse serum, 0.5% Triton X-100, 0.01% gelatin in phosphate-buffered saline [PBS] [Cellgro]) for 30 min at room temperature followed by incubation with either goat anti-HNF3β (Santa Cruz Biotechnology, sc-6554) or goat anti-Pecam1 (Santa Cruz Biotechnology, sc-1506) for 1 h at room temperature in the block solution. The cells were then washed three times with PBS and incubated with Alexa568 anti-goat IgG (Life Technologies, A-11057) in block solution for 1 h at room temperature. The cells were then washed three times with PBS and stored in 50% glycerol (v/v) in PBS for high-throughput image analysis.
Western blotting
mESCs were lysed using 0.5% Triton X-100 (Sigma) diluted in PBS (Cellgro), vortexed vigorously, incubated for 10 min on ice, and centrifuged at 18,000 relative centrifugal force (RCF) for 1.5 min to remove the membranous material. Clarified lysates were then run on a 4%–12% Tris-glycine NuPAGE gel (Life Technologies) and transferred onto polyvinylidene difluoride membranes (Millipore). The membranes were blocked in 5% skim milk diluted in 1× Tris-buffered saline (pH 7.4) and 0.1% Tween-20 (TBST) for 30 min at room temperature. After blocking, membranes were incubated with primary antibodies against Acvr1b (AbCam, ab77049), Smad2 (Cell Signaling, 3103), phospho-Smad2 (Cell Signaling, 3101S), or β-actin (Santa Cruz Biotechnology, sc-53142) overnight at 4°C in 5% skim milk. After incubation with primary antibodies, the membranes were washed three times with 1× TBST and incubated with HRP-conjugated anti-rabbit IgG (Jackson Immunoresearch, 711-036-152) or HRP-conjugated anti-mouse IgG (Jackson Immunoresearch, 715-035-151) in 5% skim milk for 1 h at room temperature followed by autoradiography using the ECL Plus Western blotting detection system (GE Healthcare).
qRT–PCR
Total RNA was extracted with TRIzol (Invitrogen) and reverse-transcribed to cDNA with QuantiTect reverse transcription kit (Qiagen) according to the manufacturer's instructions. cDNA samples synthesized from 1 μg of total RNA were subjected to qRT–PCR with 7900HT Fast Real-Time PCR system (Applied Biosystems) using the iTaq SYBR Green Supermix with ROX (Bio-Rad). The data were analyzed with the ΔΔCt method, applying β-Actin as a normalization control.
Target sensor luciferase assay
Plasmids containing 3′ UTRs of Acvr1b and Smad2 sequences encompassing the predicted binding sites of both let-7 and miR-18, respectively, were purchased from GeneCopoeia (MmiT029638 and MmiT026495). The luciferase assay was performed by transfecting HEK293T cells with 100 ng of plasmid and 10 nM relevant miRNA using Lipofectamine 2000 (Life Technologies). Luciferase activity was determined the following day using Dual-Glo (Promega) and EnVision plate reader (PerkinElmer). Renilla luciferase activity was used to normalize transfection efficiency.
Flow cytometry
Cells were dissociated using 0.25% Trypsin EDTA for flow cytometry in PBS containing 0.1% FBS using either LSRFortessa or FACSAria flow cytometers (BD Biosciences).
Bioinformatics analysis
The conserved predicted miR targets were obtained from TargetScan Mouse (version 6.1; http://www.targetscan.org/mmu_61/mmu_61_data_download/Predicted_Targets_Info.txt.zip). A functional annotation analysis was performed using DAVID Bioinformatics Resource (version 6.7) for KEGG pathway and gene ontology (GO) molecular function analysis.
X. laevis embryo culture
Embryos were fertilized in vitro, dejellied in 2% cysteine–HCl (pH7.8), and maintained in 0.1× MMR (Peng 1991). Embryos were staged according to Nieuwkoop and Faber (Nieuwkoop 1967). For gene expression analysis, whole embryos were frozen at −20°C in buffer RLT (Qiagen RNeasy lipid tissue minikit) for RNA isolation or fixed in MEMFA for in situ hybridization as below. For lineage tracing, one dorsoanterior blastomere of a 16-cell stage embryo was injected with either Xlet-7 TP or control morpholinos + β-gal mRNA or Alexa Fluor546 dextran (10,000 molecular weight). The analysis of the progeny distribution was performed at stages 35 and 40–45. β-Gal staining was performed as previously described (Vize et al. 1991) using Red Gal (Research Organics) as the substrate.
Morpholino and mRNA injection in X. laevis
Synthetic capped mRNA for injection was transcribed from pSP6-nls-gal plasmid using SP6 mMessage kit (Ambion). Xlet-7 TP (UGUCUGGGAAGAAACUCUGAGGUAG), scrambled (CCUCCUACCUCAGUUACAAUUUAUA), and control (AGGCAUUGUUGCAAUCAGCUAGAGU) morpholinos were ordered from GeneTools and injected at 40 ng per blastomere into four-cell stage or 16-cell stage embryos. No miRs were predicted to bind the sequence blocked by let-7 TP morpholino other than let-7 (targetscanFish, targetscanMouse release 6.2).
In situ hybridization in X. laevis embryos
In situ hybridization for Xbra (Colas et al. 2008), Xsox17α (Hudson et al. 1997), XleftyA (Branford et al. 2000), Xfoxa2 (Ruiz i Altaba et al. 1993), and Xacvr1b (Open Biosystems, EXL1051-7575674) was carried out as previously described (Djiane et al. 2000). In situ hybridization with Xlet-7a probe (Exiqon, catalog no. 300500-15, hsa-let-7a) was performed following the vendor's recommendations (Sweetman et al. 2008). Hybridization temperature for the Xlet-7a probe was 60°C.
Mouse embryos
Mouse embryos were dissected into DEPC-treated PBS, fixed overnight in 4% PFA, and dehydrated into MeOH. In situ hybridization used let-7a (Exiqon, catalog no. 300500-15) and miR-18a (Exiqon, catalog no. 300500-15) probes (60°C hybridization) as described (Wilkinson and Nieto 1993; Sweetman et al. 2008). Stained embryos were equilibrated in a gelatin–BSA solution (0.5% gelatin, 30% BSA, 20% sucrose in 1× PBS; Sigma) for 1 h and embedded in fresh gelatin–BSA solution, polymerized with 1.75% glutaraldehyde (Sigma), and vibratome-sectioned at 35 μm.
Statistics
Each experiment represents at least three biological replicates per condition. Statistical analysis was performed with an unpaired Student's t-test. An asterisk in figures represents a significant difference, P < 0.05. Error bars indicate the standard deviation.
Acknowledgments
We gratefully acknowledge Pedro Aza-Blanc and the SBMRI Functional Genomics Core (supported by P30 CA030199-30) for screening support, and Dimitrios G. Zisoulis (Regulus Therapeutics, San Diego, CA) for providing anti-miRs. This research was funded by the NIH (R33 HL088266 and R01 HL113601 to M.M., and R33 HL087375 to S.S.), the California Institute for Regenerative Medicine (RC1-000132 to M.M.), and the American Heart Association for post-doctoral fellowship to A.C. (11POST7610096)
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.200758.112.
References
- Agius E, Oelgeschlager M, Wessely O, Kemp C, De Robertis EM 2000. Endodermal Nodal-related signals and mesoderm induction in Xenopus. Development 127: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armes NA, Smith JC 1997. The ALK-2 and ALK-4 activin receptors transduce distinct mesoderm-inducing signals during early Xenopus development but do not co-operate to establish thresholds. Development 124: 3797–3804 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2009. MicroRNAs: Target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y 2009. A regulatory interplay between miR-27a and Runx1 during megakaryopoiesis. Proc Natl Acad Sci 106: 238–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branford WW, Yost HJ 2002. Lefty-dependent inhibition of Nodal- and Wnt-responsive organizer gene expression is essential for normal gastrulation. Curr Biol 12: 2136–2141 [DOI] [PubMed] [Google Scholar]
- Branford WW, Essner JJ, Yost HJ 2000. Regulation of gut and heart left-right asymmetry by context-dependent interactions between Xenopus lefty and BMP4 signaling. Dev Biol 223: 291–306 [DOI] [PubMed] [Google Scholar]
- Bushway PJ, Mercola M 2006. High-throughput screening for modulators of stem cell differentiation. Methods Enzymol 414: 300–316 [DOI] [PubMed] [Google Scholar]
- Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J, et al. 2009. The estrogen receptor-α-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci 106: 15732–15737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Harland RM 2007. Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development 134: 3861–3872 [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Whitaker LL, Ramsdell AF 2005. Developmental analysis of activin-like kinase receptor-4 (ALK4) expression in Xenopus laevis. Dev Dyn 232: 393–398 [DOI] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318: 271–274 [DOI] [PubMed] [Google Scholar]
- Colas A, Cartry J, Buisson I, Umbhauer M, Smith JC, Riou JF 2008. Mix.1/2-dependent control of FGF availability during gastrulation is essential for pronephros development in Xenopus. Dev Biol 320: 351–365 [DOI] [PubMed] [Google Scholar]
- Dale L, Slack JM 1987. Fate map for the 32-cell stage of Xenopus laevis. Development 99: 527–551 [DOI] [PubMed] [Google Scholar]
- Djiane A, Riou J, Umbhauer M, Boucaut J, Shi D 2000. Role of frizzled 7 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development 127: 3091–3100 [DOI] [PubMed] [Google Scholar]
- Dosch R, Gawantka V, Delius H, Blumenstock C, Niehrs C 1997. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development 124: 2325–2334 [DOI] [PubMed] [Google Scholar]
- Dougan ST, Warga RM, Kane DA, Schier AF, Talbot WS 2003. The role of the zebrafish nodal-related genes squint and cyclops in patterning of mesendoderm. Development 130: 1837–1851 [DOI] [PubMed] [Google Scholar]
- Dunn NR, Vincent SD, Oxburgh L, Robertson EJ, Bikoff EK 2004. Combinatorial activities of Smad2 and Smad3 regulate mesoderm formation and patterning in the mouse embryo. Development 131: 1717–1728 [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S 2005. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121: 87–99 [DOI] [PubMed] [Google Scholar]
- Dyson S, Gurdon JB 1998. The interpretation of position in a morphogen gradient as revealed by occupancy of activin receptors. Cell 93: 557–568 [DOI] [PubMed] [Google Scholar]
- Ema M, Takahashi S, Rossant J 2006. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 107: 111–117 [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N 2008. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet 9: 102–114 [DOI] [PubMed] [Google Scholar]
- Gadue P, Huber TL, Paddison PJ, Keller GM 2006. Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci 103: 16806–16811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J 2002. Morphogen gradients, positional information, and Xenopus: Interplay of theory and experiment. Dev Dyn 225: 392–408 [DOI] [PubMed] [Google Scholar]
- Green JB, Smith JC 1990. Graded changes in dose of a Xenopus activin A homologue elicit stepwise transitions in embryonic cell fate. Nature 347: 391–394 [DOI] [PubMed] [Google Scholar]
- Green JB, New HV, Smith JC 1992. Responses of embryonic Xenopus cells to activin and FGF are separated by multiple dose thresholds and correspond to distinct axes of the mesoderm. Cell 71: 731–739 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Harger P, Mitchell A, Lemaire P 1994. Activin signalling and response to a morphogen gradient. Nature 371: 487–492 [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Standley H, Dyson S, Butler K, Langon T, Ryan K, Stennard F, Shimizu K, Zorn A 1999. Single cells can sense their position in a morphogen gradient. Development 126: 5309–5317 [DOI] [PubMed] [Google Scholar]
- Hudson C, Clements D, Friday RV, Stott D, Woodland HR 1997. Xsox17α and -β mediate endoderm formation in Xenopus. Cell 91: 397–405 [DOI] [PubMed] [Google Scholar]
- Iratni R, Yan YT, Chen C, Ding J, Zhang Y, Price SM, Reinberg D, Shen MM 2002. Inhibition of excess nodal signaling during mouse gastrulation by the transcriptional corepressor DRAP1. Science 298: 1996–1999 [DOI] [PubMed] [Google Scholar]
- Jones CM, Kuehn MR, Hogan BL, Smith JC, Wright CV 1995. Nodal-related signals induce axial mesoderm and dorsalize mesoderm during gastrulation. Development 121: 3651–3662 [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Verkade H, Reiter JF, Kim CH, Chitnis AB, Kuroiwa A, Stainier DY 2004. Notch signaling can regulate endoderm formation in zebrafish. Dev Dyn 229: 756–762 [DOI] [PubMed] [Google Scholar]
- King ML, Messitt TJ, Mowry KL 2005. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell 97: 19–33 [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Umbhauer M, Neal KA, Lerchner W, Smith JC, Cunliffe V 1997. The Xenopus Brachyury promoter is activated by FGF and low concentrations of activin and suppressed by high concentrations of activin and by paired-type homeodomain proteins. Genes Dev 11: 3265–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LH, Peerani R, Ungrin M, Joshi C, Kumacheva E, Zandstra P 2009. Micropatterning of human embryonic stem cells dissects the mesoderm and endoderm lineages. Stem Cell Res (Amst) 2: 155–162 [DOI] [PubMed] [Google Scholar]
- Li L, Shi JY, Zhu GQ, Shi B 2011. miR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. J Cell Biochem 113: 1235–1244 [DOI] [PubMed] [Google Scholar]
- Luxardi G, Marchal L, Thome V, Kodjabachian L 2010. Distinct Xenopus Nodal ligands sequentially induce mesendoderm and control gastrulation movements in parallel to the Wnt/PCP pathway. Development 137: 417–426 [DOI] [PubMed] [Google Scholar]
- Marjoram L, Wright C 2011. Rapid differential transport of Nodal and Lefty on sulfated proteoglycan-rich extracellular matrix regulates left-right asymmetry in Xenopus. Development 138: 475–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Hemann MT, Bartel DP 2007. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 315: 1576–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Rogers KW, Jordan BM, Lee JS, Robson D, Ramanathan S, Schier AF 2012. Differential diffusivity of Nodal and Lefty underlies a reaction-diffusion patterning system. Science 336: 721–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G 2008. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell 132: 661–680 [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD 1967. The ‘organization centre.’ 3. Segregation and pattern formation in morphogenetic fields. Acta Biotheor 17: 178–194 [DOI] [PubMed] [Google Scholar]
- Peng HB 1991. Xenopus laevis: Practical uses in cell and molecular biology. Solutions and protocols. Methods Cell Biol 36: 657–662 [PubMed] [Google Scholar]
- Perea-Gomez A, Vella FD, Shawlot W, Oulad-Abdelghani M, Chazaud C, Meno C, Pfister V, Chen L, Robertson E, Hamada H, et al. 2002. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev Cell 3: 745–756 [DOI] [PubMed] [Google Scholar]
- Peyrieras N, Strahle U, Rosa F 1998. Conversion of zebrafish blastomeres to an endodermal fate by TGF-β-related signaling. Curr Biol 8: 783–786 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM 1999. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature 397: 707–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Re'em-Kalma Y, Lamb T, Frank D 1995. Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during Xenopus development. Proc Natl Acad Sci 92: 12141–12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH 2001. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes Dev 15: 2010–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH 2009. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527 [DOI] [PubMed] [Google Scholar]
- Roush S, Slack FJ 2008. The let-7 family of microRNAs. Trends Cell Biol 18: 505–516 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Prezioso VR, Darnell JE, Jessell TM 1993. Sequential expression of HNF-3β and HNF-3α by embryonic organizing centers: The dorsal lip/node, notochord and floor plate. Mech Dev 44: 91–108 [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG 2008. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10: 987–993 [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W 1997. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development 124: 327–342 [DOI] [PubMed] [Google Scholar]
- Schneider VA, Mercola M 1999. Spatially distinct head and heart inducers within the Xenopus organizer region. Curr Biol 9: 800–809 [DOI] [PubMed] [Google Scholar]
- Shen MM 2007. Nodal signaling: Developmental roles and regulation. Development 134: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Smith JC, Price BM, Van Nimmen K, Huylebroeck D 1990. Identification of a potent Xenopus mesoderm-inducing factor as a homologue of activin A. Nature 345: 729–731 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2002. Dorsal gradient networks in the Drosophila embryo. Dev Biol 246: 57–67 [DOI] [PubMed] [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Munsterberg A 2008. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol 321: 491–499 [DOI] [PubMed] [Google Scholar]
- Tabata T, Takei Y 2004. Morphogens, their identification and regulation. Development 131: 703–712 [DOI] [PubMed] [Google Scholar]
- Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T 2005. Characterization of mesendoderm: A diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development 132: 4363–4374 [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD 2009. The maternal-to-zygotic transition: A play in two acts. Development 136: 3033–3042 [DOI] [PubMed] [Google Scholar]
- Takahashi T, Lord B, Schulze PC, Fryer RM, Sarang SS, Gullans SR, Lee RT 2003. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation 107: 1912–1916 [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ 1997. nodal expression in the primitive endoderm is required for specification of the anterior axis during mouse gastrulation. Development 124: 1033–1044 [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI 2008. Selective blockade of microRNA processing by Lin28. Science 320: 97–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vize PD, Melton DA, Hemmati-Brivanlou A, Harland RM 1991. Assays for gene function in developing Xenopus embryos. Methods Cell Biol 36: 367–387 [DOI] [PubMed] [Google Scholar]
- Wang DJ, Legesse-Miller A, Johnson EL, Coller HA 2012. Regulation of the let-7a-3 promoter by NF-κB. PLoS ONE 7: e31240 doi: 10.1371/journal.pone.0031240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson DG, Nieto MA 1993. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 225: 361–373 [DOI] [PubMed] [Google Scholar]
- Willems E, Leyns L 2008. Patterning of mouse embryonic stem cell-derived pan-mesoderm by Activin A/Nodal and Bmp4 signaling requires fibroblast growth factor activity. Differentiation 76: 745–759 [DOI] [PubMed] [Google Scholar]