Lysine methyltransferase G9a interacts with the transcription factor Sharp-1 and enhances transcriptional repression of muscle promoters. RNAi-mediated reduction of G9a expression or inhibition of its activity rescues myogenic differentiation in Sharp-1–overexpressing cells.
Abstract
Sharp-1, a basic helix-loop-helix transcription factor, is a potent repressor of skeletal muscle differentiation and is dysregulated in muscle pathologies. However, the mechanisms by which it inhibits myogenesis are not fully understood. Here we show that G9a, a lysine methyltransferase, is involved in Sharp-1–mediated inhibition of muscle differentiation. We demonstrate that G9a directly interacts with Sharp-1 and enhances its ability to transcriptionally repress the myogenin promoter. Concomitant with a differentiation block, G9a-dependent histone H3 lysine 9 dimethylation (H3K9me2) and MyoD methylation are apparent upon Sharp-1 overexpression in muscle cells. RNA interference–mediated reduction of G9a or pharmacological inhibition of its activity erases these repressive marks and rescues the differentiation defect imposed by Sharp-1. Our findings provide new insights into Sharp-1–dependent regulation of myogenesis and identify epigenetic mechanisms that could be targeted in myopathies characterized by elevated Sharp-1 levels.
INTRODUCTION
Myogenic regulatory factors (MRFs) MyoD, Myf5, myogenin, and MRF4 act together with epigenetic regulatory mechanisms to control skeletal muscle differentiation. All MRFs heterodimerize with ubiquitously expressed E-proteins (E12, E47) and bind to E-box sequences (CANNTG) in target gene promoters, thereby driving transcription of muscle-specific genes. MyoD and Myf5 are expressed in undifferentiated proliferating myoblasts. During differentiation, MyoD is activated and induces an irreversible cell cycle arrest by up-regulation of p21Cip/WAF1 expression, as well as activation of myogenin and MEF2, which are required for differentiation (Sabourin and Rudnicki, 2000; Sartorelli and Caretti, 2005; Tapscott, 2005). Because MyoD is expressed in myoblasts, its ability to induce cell cycle arrest and differentiation is tightly controlled by several mechanisms. For instance, high levels of Id in myoblasts sequester E-proteins, resulting in a block of MyoD transcriptional activity. Many DNA-binding transcription factors, including Hes1, Sharp-1, Mist, MyoR, and Twist, negatively affect MyoD function by competition for binding to E-box sites, formation of inactive heterodimers, and inhibition of its transcriptional activity. In addition, chromatin-modifying enzymes HDAC1, Ezh2, Suv39h1, and G9a, which are expressed in myoblasts, mediate repressive histone deacetylation and methylation marks on early and late muscle promoters, precluding MyoD transcriptional activity (Puri and Sartorelli, 2000; McKinsey et al., 2001; Perdiguero et al., 2009; Bharathy and Taneja, 2012). Among these, G9a mediates transcriptional repression by monomethylation and dimethylation of histone H3 lysine-9 (H3K9; Shinkai and Tachibana, 2011) and also methylates MyoD at Lys-104 (K104; Ling et al., 2012). Although increasing evidence demonstrates that repressive epigenetic modifications of histone and nonhistone substrates are important in the maintenance of an undifferentiated state of muscle cells, the mechanisms by which corepressors are recruited to muscle-specific promoters in myoblasts are largely unclear.
Sharp-1 is expressed widely in a number of cell types, including developing skeletal muscles (Fujimoto et al., 2001; Azmi and Taneja, 2002; Azmi et al., 2003), and has complex physiological functions in cellular differentiation programs, sleep length, circadian rhythms, tumor suppression, and Th2 lineage commitment (Honma et al., 2002; Azmi et al., 2004; Rossner et al., 2008; Gulbagci et al., 2009; Yang et al., 2009a; He et al., 2009). We previously demonstrated that Sharp-1 interacts with MyoD, resulting in the inhibition of its transcriptional activity and muscle differentiation (Azmi et al., 2004). Consistent with these findings, Sharp-1 is overexpressed in inclusion body myositis, which exhibits a differentiation defect, and is also associated with loss of skeletal muscle mass (Morosetti et al., 2006; Lecomte et al., 2010). Despite growing evidence of deregulated expression in pathologies, the mechanisms by which Sharp-1 functions as a repressor of differentiation have not been elucidated.
Here we identify G9a as a corepressor that mediates Sharp-1–dependent block of skeletal myogenesis. Sharp-1 and G9a physically associate, and inhibition of differentiation by Sharp-1 correlates with increased G9a-dependent H3K9me2 at the myogenin promoter, as well as with MyoD methylation. RNA interference–mediated reduction of G9a or inhibition of its activity in Sharp-1–overexpressing cells restores differentiation concomitant with removal of repressive methylation marks. Reexpression of wild-type MyoD and MyoD(K104R), where Lys-104 is mutated to arginine, underscores a role for G9a-dependent MyoD methylation in Sharp-1–dependent inhibition of myogenesis.
RESULTS
G9a associates with and enhances Sharp-1–mediated transcriptional repression
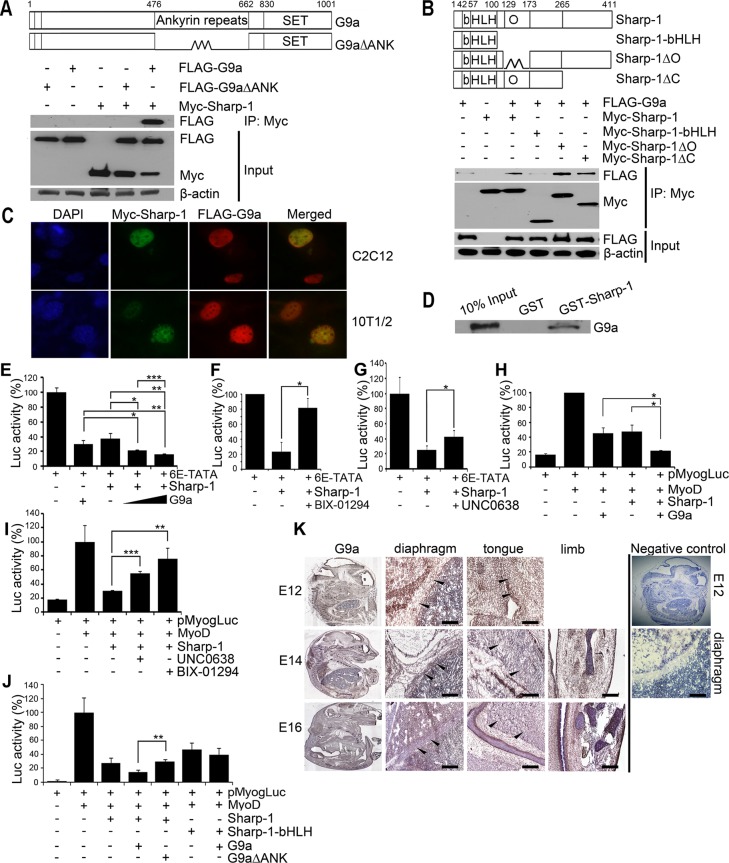
We previously demonstrated that Sharp-1 overexpression in preadipocytes inhibits their differentiation into adipocytes. Concomitantly, G9a and its hallmark repressive chromatin mark H3K9me2 were apparent on adipogenic promoters, suggesting that Sharp-1 may recruit G9a to inhibit cellular differentiation programs (Gulbagci et al., 2009). Indeed our recent studies showed that similar to Sharp-1, G9a overexpression in myoblasts impairs skeletal muscle differentiation (Azmi et al., 2004; Ling et al., 2012). To examine whether G9a is a corepressor involved in Sharp-1–dependent inhibition of skeletal myogenesis, we first investigated whether the two proteins interact by coimmunoprecipitation assays. Sharp-1 was expressed with full-length G9a or a deletion mutant lacking the ankyrin repeats (G9a∆ANK). Immunoprecipitation of Sharp-1 revealed that it interacts with G9a but not with G9a∆ANK, indicating that the ankyrin repeats are essential for association (Figure 1A). To identify the domain(s) in Sharp-1 needed for association, we transfected cells with Sharp-1 and its deletion mutants along with G9a (Figure 1B). Sharp-1 interacted with G9a through a region spanning amino acid residues 173–265 (Figure 1B), which was previously shown to be important for transcriptional repression (Garriga-Canut et al., 2001). Sharp-1 and G9a colocalization in the nucleus was apparent in C2C12 myoblasts, as well as in C3H10T1/2 (10T1/2) and NIH3T3 fibroblast cells (Figure 1C and unpublished data). Moreover, in glutathione S-transferase (GST) pull-down assays, G9a interacted with GST–Sharp-1 but not with GST, confirming a direct interaction between the two proteins (Figure 1D). We then tested whether G9a is involved in Sharp-1–mediated transcriptional repression. We transfected 10T1/2 fibroblast cells with 6E-TATA-Luc harboring Sharp-1–binding sites (Rossner et al., 2008). Coexpression of G9a enhanced transcriptional repression by Sharp-1 (Figure 1E), and, conversely, inhibition of G9a activity with BIX-01294 (Kubicek et al., 2007) or UNC0638 (Vedadi et al., 2011) reversed it (Figure 1, F and G). To investigate whether G9a contributes to Sharp-1–mediated repression of myogenesis, we analyzed its impact on the myogenin promoter reporter pMyog-Luc (Friday et al., 2000). Consistent with previous reports, MyoD-induced reporter activity was inhibited by Sharp-1 and G9a individually (Azmi et al., 2004; Ling et al., 2012). Coexpression of G9a enhanced Sharp-1–dependent repression of the myogenin promoter (Figure 1H), whereas treatment with BIX-01294 or UNC0638 abrogated it (Figure 1I). Moreover, G9a∆ANK did not enhance Sharp-1–dependent repression of the myogenin promoter, and Sharp-1 basic helix-loop-helix (bHLH)–dependent repression was not increased by G9a (Figure 1J). To further assess whether G9a is expressed in vivo in muscle cells, we analyzed its expression in mouse embryos at embryonic day 12 (E12) to E16 by immunohistochemistry. G9a was widely expressed at all stages, and its expression was apparent in developing skeletal muscles, including diaphragm, limb, and tongue (Figure 1K). We and others showed that Sharp-1 is also expressed in developing tongue and limb muscles in mouse and zebrafish embryos (Azmi and Taneja, 2002; Chen et al., 2010), further suggesting a potential regulatory connection between the two proteins.
FIGURE 1:
G9a interacts with and enhances Sharp-1–mediated transcriptional repression. (A) Schematic representation of full-length G9a and a deletion mutant lacking the ankyrin repeats (G9a∆ANK). Numbers indicate amino acid residues. Cells transfected with Myc–Sharp-1, FLAG-G9a, and FLAG-G9a∆ANK were immunoprecipitated with anti–c-Myc agarose beads and immunoblotted with anti-FLAG antibody. Lysates were analyzed for Sharp-1 and G9a expression by Western blot. β-Actin was used as a loading control. (B) Schematic representation of Sharp-1 with basic (b), helix-loop-helix (HLH), and Orange (O) domains. Deletion mutants of Sharp-1 are shown. Sharp-1 was immunoprecipitated and analyzed for interaction with G9a using anti-FLAG antibody. (C) Colocalization of Myc–Sharp-1(green) and FLAG-G9a (red) was visualized by immunofluorescence in C2C12 and 10T1/2 cells. Nuclei were stained with DAPI (blue). (D) In vitro–translated G9a was used for interaction with equivalent amounts of GST–Sharp-1 and GST proteins. A 10% input was run as a control. (E–G) 10T1/2 cells were transfected with 6E-TATA-Luc along with Sharp-1 and G9a (E), with Myc-Sharp-1 and 2.5 μM of BIX-01294 (F), or with 0.25 μM UNC0638 (G). (H, I) Cells were transfected with pMyog-Luc, MyoD, Sharp-1, and G9a (H) and, in the presence of MyoD, Sharp-1 with 2.5 μM BIX-01294 or 0.25 μM UNC0638 (I). Error bars indicate mean ± SD. (J) Cells were transfected with pMyogLuc along with Sharp-1 and deletion mutants together with G9a and G9a∆ANK. Error bars indicate mean ± SD. (K) Mouse embryo sections were immunostained with anti-G9a antibody at E12, E14, and E16. Arrowheads in magnified images indicate G9a expression (brown staining) in diaphragm, tongue, and limb muscles. Negative control shows E12 embryo section stained with secondary antibody only. A magnified image of the diaphragm is shown. Scale bar, 100 μm.
Sharp-1 overexpression correlates with increased H3K9me2 on the myogenin promoter
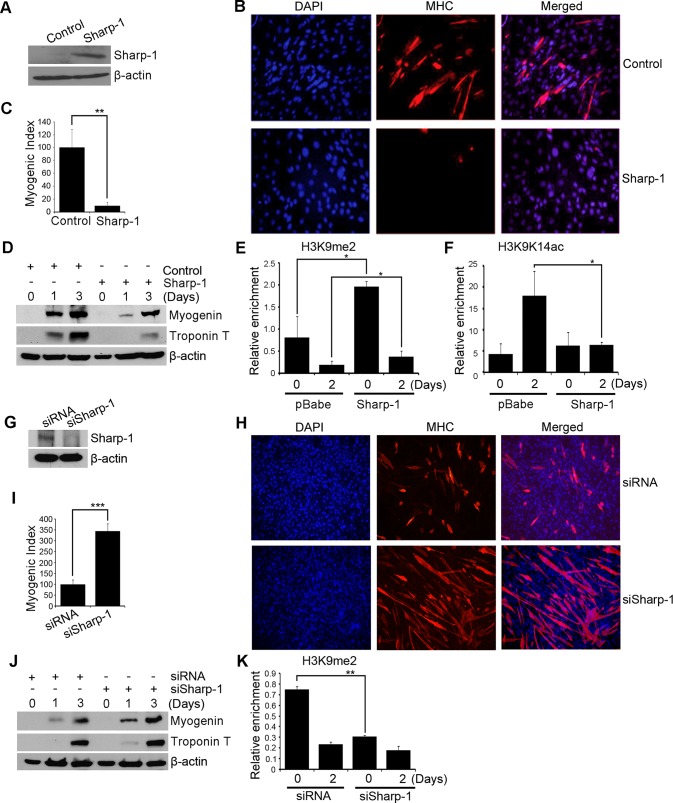
We reasoned that if G9a is central to repression of myogenin and muscle differentiation by Sharp-1, an increase in its activity should be apparent in Sharp-1–overexpressing cells. C2C12 cells were transduced with pBabe-Sharp-1 (Sharp-1) or with pBabe vector (control; Figure 2A). Consistent with previous reports (Azmi et al., 2004), Sharp-1 overexpression inhibited myogenic differentiation as assessed by reduced number of MHC+ myotubes and myogenic index (Figure 2, B and C). Moreover, the levels of myogenin and troponin T were strongly down-regulated in Sharp-1–overexpressing cells (Figure 2D). We then examined by chromatin immunoprecipitation (ChIP) assays G9a-mediated H3K9me2 on the myogenin promoter. Of interest, a significant increase in H3K9me2 mark was apparent in Sharp-1–overexpressing cells (Figure 2E) in a manner similar to G9a overexpression (Ling et al., 2012). Correspondingly, H3K9K14ac, a mark of transcriptional activation, was reduced upon Sharp-1 overexpression (Figure 2F). To further validate these findings, we inhibited endogenous Sharp-1 expression with small interfering RNA (siRNA; siSharp-1). Control cells (siRNA) were transfected with scrambled siRNA. Inhibition of endogenous Sharp-1 (Figure 2G) led to a marked increase in myogenic differentiation and expression of MyoD target genes (Figure 2, H–J). Moreover, a clear decline in H3K9me2 at the myogenin promoter was apparent in siSharp-1 cells relative to controls (Figure 2K), indicating that G9a is targeted to the myogenin promoter in a Sharp-1–dependent manner.
FIGURE 2:
Sharp-1 inhibits differentiation with increased H3K9me2. (A) Sharp-1 expression in pBabe (control) and pBabe-Sharp-1 C2C12 cells was analyzed by Western blot. (B–D) Differentiation of control and pBabe-Sharp-1 cells was analyzed with anti-MHC antibody. Nuclei were stained with DAPI (B). The myogenic index was determined and is represented as mean ± SD (C). Lysates from undifferentiated myoblasts (day 0) and after differentiation (days 1 and 3) were examined for myogenin and troponin T (D). (E, F) ChIP assays were performed at days 0 and 2 on the myogenin promoter, using H3K9me2 and H3K9K14ac antibodies. (G) C2C12 cells were transfected with scrambled siRNA or Sharp-1 siRNA (siSharp-1). The down-regulation of endogenous Sharp-1 was analyzed by Western blot. (H–J) Differentiation in siRNA and siSharp-1 cells was quantified by immunofluorescence analysis of MHC+ myotubes (H), myogenic index (I), and expression of myogenin and troponin T (J). (K) H3K9me2 enrichment was analyzed by ChIP in siRNA and siSharp-1 cells in undifferentiated (day 0) and differentiated cells (day 2).
G9a is critical for Sharp-1–mediated repression of myogenesis
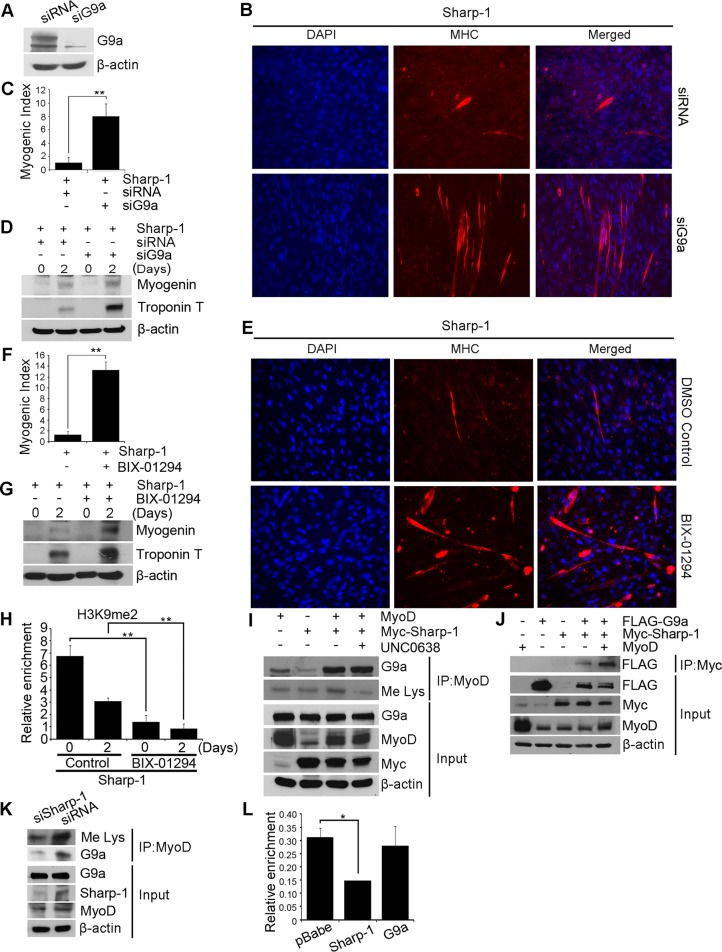
To test the role of G9a in Sharp-1–dependent inhibition of myogenesis, we transfected Sharp-1–overexpressing cells with G9a siRNA (siG9a) or as a control, with scrambled siRNA (siRNA). Down-regulation of G9a expression was apparent in siG9a cells relative to controls (Figure 3A). Of interest, siG9a cells exhibited enhanced myotube formation, myogenic index, and myogenic differentiation markers compared with siRNA cells (Figure 3, B–D). Moreover, treatment of pBabe-Sharp-1 cells with BIX-01294 rescued myogenic differentiation and expression of myogenin and troponin T similar to siG9a cells, indicating that G9a methyltransferase activity is essential for Sharp-1–mediated block of myogenic differentiation (Figure 3, E–G). Concomitant with a rescue of differentiation, H3K9me2 was reduced on the myogenin promoter in response to BIX-01294 treatment in Sharp-1–overexpressing cells (Figure 3H). Because Sharp-1 increased H3K9me2, we examined its impact on MyoD methylation, which is also mediated by G9a (Ling et al., 2012). C2C12 cells transfected with Sharp-1 and MyoD were left untreated or treated with UNC0638. Immunoprecipitation of MyoD revealed its association with endogenous G9a and was correspondingly methylated. Coexpression of Sharp-1 enhanced MyoD-G9a association and MyoD methylation. In the presence of UNC0638, MyoD methylation was reduced, without any impact on the MyoD–G9a complex (Figure 3I). Because both MyoD and Sharp-1 interact with the ankyrin-repeat domain in G9a, we tested whether MyoD had an impact on G9a–Sharp-1 association. In the presence of MyoD, the association of Sharp-1 and G9a was increased, indicating that both Sharp-1 and MyoD enhance association of all three proteins in a larger complex (Figure 3J). Consistent with this, MyoD–G9a interaction and MyoD methylation were reduced in siSharp-1 cells compared with control cells (Figure 3K). Taken together, these results suggest that Sharp-1 enhances both H3K9me2 and MyoD methylation likely via recruitment of G9a. We previously showed that Sharp-1 inhibits DNA-binding activity of MyoD (Azmi et al., 2004). To test whether G9a participates in inhibition of MyoD binding, we examined MyoD occupancy at the myogenin promoter in Sharp-1– and G9a–overexpressing cells. Consistent with our previous observations (Azmi et al., 2004), MyoD binding was reduced in Sharp-1–overexpressing cells but not in G9a-overexpressing cells (Figure 3L). These results suggest that Sharp-1–mediated displacement of MyoD occupancy occurs independent of G9a recruitment.
FIGURE 3:
Inhibition of G9a rescues Sharp-1–imposed differentiation block. (A) C2C12 cells overexpressing Sharp-1 were transfected with control siRNA or siG9a. G9a knockdown was determined by Western blot. (B) Cells were induced to differentiate and stained with anti-MHC antibody. Nuclei were stained with DAPI. (C) Differentiation was quantified by calculating myogenic index. (D) Myogenin and troponin T expression was determined by Western blot. (E–G) Sharp-1–overexpressing cells were incubated with DMSO (vehicle) or BIX-01294. Differentiation was assessed using anti-MHC antibody (E), myogenic index (F), and expression of myogenin and troponin T by Western blot (G). (H) Sharp-1–overexpressing cells were treated with DMSO or BIX-01294 for 0 and 2 d. ChIP assays were done using anti-H3K9me2 antibody on the myogenin promoter. (I) C2C12 cells were transfected with Myc–Sharp-1 and MyoD. MyoD was immunoprecipitated and analyzed for association with G9a and methylation in the absence and presence of UNC0638 treatment using anti-G9a and anti-Me Lys antibodies, respectively. Lysates were analyzed for expression of G9a, MyoD, and Sharp-1. (J) C2C12 cells were transfected with Myc–Sharp-1 and MyoD and G9a as indicated. The association of Sharp-1 with G9a was analyzed in the absence and presence of MyoD. (K) Endogenous MyoD methylation and association with G9a was examined by immunoprecipitation from siSharp-1 and siRNA cells using anti-Me Lys and anti-G9a antibodies. Input shows expression of G9a, Sharp-1, and MyoD in lysates. (L) MyoD occupancy on the myogenin promoter was analyzed by ChIP assays in myoblasts overexpressing pBabe, Sharp-1, or G9a. Error bars indicate mean ± SD.
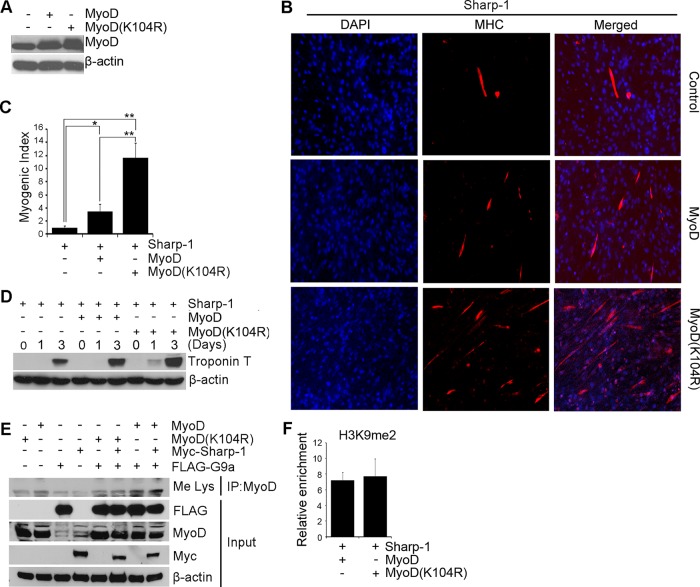
To investigate the significance of G9a-mediated H3K9me2 and MyoD methylation in Sharp-1–dependent repression of myogenesis, we reexpressed wild-type MyoD and MyoD(K104R) (Sartorelli et al., 1999) in Sharp-1–overexpressing C2C12 cells (Figure 4A). Consistent with our previous studies (Azmi et al., 2004), reexpression of MyoD partially rescued myogenic differentiation and troponin T expression (Figure 4, B–D). Of interest, at equivalent levels of expression, MyoD(K104R) was more effective in rescuing differentiation compared with wild-type MyoD. To examine the mechanisms underlying this effect, we examined the impact of Sharp-1 and G9a on MyoD and MyoD(K104R) (Figure 4E). In contrast to wild-type MyoD, the methylation of which was augmented in the presence of G9a alone and together with Sharp-1, MyoD(K104R) was insensitive to both proteins. The level of H3K9me2 was not altered by MyoD and MyoD(K104R) (Figure 4F), indicating that MyoD methylation plays a key role in the inhibition of myogenesis by Sharp-1.
FIGURE 4:
MyoD methylation is relevant in the inhibition of myogenesis by Sharp-1. (A) C2C12 cells overexpressing Sharp-1 were transfected with MyoD and MyoD(K104R). MyoD expression was analyzed by Western blot. (B-D) Control (vector), MyoD, and MyoD(K104R) cells were analyzed for differentiation with anti-MHC antibody (B), myogenic index (C), and troponin T expression by Western blot (D). (E) C3H10T1/2 cells were transfected with MyoD, MyoD(K104R), Myc-Sharp-1, and FLAG-G9a. MyoD was immunoprecipitated and probed with anti-Me Lys antibody. Expression of Sharp-1, MyoD, and G9a was analyzed in lysates. (F) ChIP assays were performed in Sharp-1–overexpressing C2C12 cells transfected with MyoD and MyoD(K104R). H3K9me2 was analyzed by ChIP assays on the myogenin promoter at D2 of differentiation. Error bars indicate mean ± SD.
DISCUSSION
In this study we provide novel insights that link Sharp-1 to epigenetic mechanisms for inhibition of skeletal muscle differentiation via recruitment of the corepressor G9a. We provide evidence that G9a is expressed in vivo in developing skeletal muscles. G9a interacts with and enhances Sharp-1–dependent repression of MyoD activity and target gene expression in a methyltransferase activity–dependent manner. In the absence of G9a function, both MyoD repression and the differentiation block imposed by Sharp-1 are rescued.
Sharp-1 is a member of the bHLH-Orange subfamily of transcription factors (Sun et al., 2007), which includes the Hes, Hey, Helt, and Stra13/Dec1 subfamilies. Sharp-1 binds with high affinity to E-box sites and also mediates repression by protein–protein interaction with various transcription factors, including MyoD and C/EBPβ (Garriga-Canut et al., 2001; Azmi et al., 2004; Fujimoto et al., 2007; Gulbagci et al., 2009). Moreover, Sharp-1 interacts with the corepressors HDAC1 and Sirt1 (Garriga-Canut et al., 2001; Fujimoto et al., 2007). However, the functional significance of association with these cofactors in Sharp-1–mediated biological functions in cellular differentiation, growth arrest, tumor cell quiescence, or circadian rhythms have not been documented.
We and others showed that Sharp-1 interacts with MyoD and inhibits its transcriptional activity and myogenic differentiation (Azmi et al., 2004; Fujimoto et al., 2007). The repression of MyoD and muscle differentiation by Sharp-1 likely involve multiple repression mechanisms. This includes formation of inactive heterodimers with MyoD and E-proteins that likely result in the inhibition of MyoD DNA binding. However, Sharp-1 inhibits tethered MyoD∼E47 heterodimers (Azmi et al., 2004), suggesting that additional mechanisms must be involved. Several lines of evidence in this study demonstrate that Sharp-1–dependent inhibition of MyoD and myogenesis is at least in part dependent on association with G9a and its recruitment at muscle promoters. 1) Sharp-1–overexpressing cells exhibit elevated G9a-mediated H3K9me2, which correlates with reduced myogenin expression and impaired muscle differentiation. Conversely, inhibition of endogenous Sharp-1 expression accelerates differentiation and is linked to reduced H3K9me2 at the myogenin promoter. 2) Reduction of G9a expression or pharmacological inhibition of its activity abrogates Sharp-1–dependent inhibition of differentiation concomitant with reduction of H3K9 and MyoD methylation. 3) G9a augments Sharp-1–dependent repression of MyoD activity at the myogenin promoter and, conversely, inhibition of its activity blocks it. Moreover, G9a∆ANK fails to associate with Sharp-1 and does not affect Sharp-1–dependent repression of myogenin. 4) The association of G9a with MyoD is enhanced in presence of Sharp-1 (Figure 3). This suggests that Sharp-1 could serve as an adaptor protein linking G9a to MyoD, thereby influencing MyoD transcriptional activity through epigenetic regulation of target genes. Consistently, Sharp-1 increases G9a-dependent MyoD methylation, which appears to be critical in the differentiation block imposed by Sharp-1. 5) Sharp-1–bHLH mutant, which fails to interact with G9a, represses MyoD to a lesser extent (∼50% inhibition) than full-length Sharp-1 (∼75% inhibition). The inhibition of MyoD by Sharp-1–bHLH is likely a reflection of heterodimerization with MyoD and E-proteins through the HLH domain, which may account for loss of MyoD DNA binding and can occur independent of G9a recruitment. On the other hand, recruitment of G9a results in H3K9me2 and MyoD methylation, resulting in loss of MyoD transcriptional activity independent of effects on MyoD DNA binding.
In addition to Sharp-1, G9a has been documented to interact with various transcription factors, including Snail, Gfi1, NF-kB, CDP, and REST (Nishio and Walsh, 2004; Roopra et al., 2004; Duan et al., 2005; Chen et al., 2009; Dong et al., 2012), which recruit it to distinct target promoters. Given its recruitment in muscle cells, targeting G9a may be therapeutically useful in myopathies with elevated Sharp-1 expression.
MATERIALS AND METHODS
Cell culture and differentiation assays
C2C12 cells were cultured in DMEM supplemented with 20% fetal bovine serum (FBS) and differentiated in DMEM supplemented with 2% horse serum. Phoenix, HEK293, and C3H10T1/2 cells were cultured in DMEM supplemented with 10% FBS. Differentiation in C2C12 cells was quantified by determining the ratio of nuclei in MHC+ myotubes over total nuclei (myogenic index). At least 600 nuclei were counted from three different fields.
Retroviral transduction and siRNA
C2C12 cells were transduced with pBabe (vector) or pBabe-Sharp-1 and selected with 2 μg/ml puromycin. pBabe-Sharp-1 cells were transfected with 100 nM scrambled siRNA or siG9a as described (Ling et al., 2012). For knockdown of Sharp-1, C2C12 cells were transfected with 100 nM Sharp-1 siRNA (Qiagen, Valencia, CA). G9a inhibitors BIX-01294 and UNC0638 (Sigma-Aldrich, St. Louis, MO) were used at 2.5 and 0.25 μM, respectively. Control cells were treated with dimethyl sulfoxide (vehicle).
Plasmids
FLAG-G9a, FLAG-G9a∆ANK, FLAG-MyoD, FLAG-MyoD(K104R), GST-Sharp-1, 6E-TATA, pMyogLuc, Myc-Sharp-1, and Myc-Sharp-1-bHLH have been described (Sartorelli et al., 1999; Friday et al., 2000; Azmi et al., 2004; Rossner et al., 2008; Gulbagci et al., 2009; Ling et al., 2012). Myc-Sharp-1∆O (∆ orange domain) and Myc-Sharp-1∆C (amino acids 1–265) were generated by PCR. Primer sequences are available upon request.
Immunoprecipitation and Western blot
A 500-μg amount of lysate was incubated with c-Myc agarose beads (Sigma-Aldrich) or 2 μg of anti-MyoD (Sigma-Aldrich) and analyzed by Western blot. The following antibodies were used: anti-MyoD and anti-myogenin (Santa Cruz Biotechnology, Santa Cruz, CA); anti-troponin T, anti-FLAG, anti-Myc, anti–Sharp-1, and anti–β-actin (Sigma-Aldrich); anti-G9a/EHMT2 (Cell Signaling, Beverly, MA) and anti-Me Lys (Abcam, Cambridge, MA).
GST pull-down assay
Glutathione–Sepharose beads were incubated with 10 μg of GST or GST-Sharp-1 and 20 μl of in vitro–translated FLAG-G9a prepared using TNT-coupled reticulocyte lysate system (Promega, Madison, WI). Eluted proteins were analyzed by Western blot using anti-Flag antibody.
Immunofluorescence analysis
Cells were incubated with anti-MHC (MY32), anti–Sharp-1, and anti–c-Myc (Sigma-Aldrich) antibodies, followed by Alexa Fluor 488 or with Alexa Fluor 568 antibodies (Molecular Probe, Eugene, OR). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA). Images were captured using a fluorescence microscope (Nikon Eclipse TE 2000-U; Nikon, Melville, NY) at 10× or 20× magnification using MetaMorph software, version 7.0r3 (Molecular Devices, Sunnyvale, CA).
Immunostaining of mouse embryo sections
Sagittal mouse embryo cryosections (Zyagen, San Diego, CA) were incubated with anti-G9a antibody, followed by detection with Vectastain ABC Kit (Vector Laboratories). Negative controls were performed by incubation with secondary antibody only. Slides were mounted using DePeX (Sigma-Aldrich) mounting media, and peroxidase staining was visualized under an Olympus slide scanner (Olympus, Tokyo, Japan) and photographed using an Aperio image scope viewer (Aperio, Vista, CA).
Luciferase assays
Cells were transfected with pMyog-Luc or 6E-TATA reporters with Myc–Sharp-1, FLAG-G9a, or FLAG-MyoD as indicated in the figure legends and 5 ng of Renilla luciferase. Luciferase activity was measured using the dual-luciferase reporter assay system (Promega). Experiments were performed at least twice, in triplicate.
Chromatin immunoprecipitation assay
ChIP assays were performed in C2C12 cells using 2 μg of H3K9me2 (Millipore, Billerica, MA), H3K9K14ac (Millipore), or MyoD (Santa Cruz Biotechnology) antibodies using the ChIP assay kit (Upstate, Millipore). DNA was amplified with primers specific to myogenin and β-actin promoters as described (Ling et al., 2012)
Statistical analysis
The p values were determined using Student's t test and presented as mean ± SD (*p < 0.05, **p < 0.01, ***p < 0.001).
Acknowledgments
We thank G. Pavlath, M. Rossner, V. Sartorelli, and M. Walsh for reagents. This research is supported by the Singapore Ministry of Health's National Medical Research Council under its Exploratory/Development Grant (R.T.).
Abbreviations used:
- bHLH
basic helix-loop-helix
- CDP
CCAAT displacement protein
- ChIP
chromatin immunoprecipitation
- DAPI
4’,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- Ezh2
enhancer of zeste homologue 2
- Gfi1
growth factor independent 1
- GST
glutathione S-transferase
- H3K9K14Ac
acetylated histone H3 at Lys-9 and Lys-14
- H3K9me2/3
di- or tri-methylated histone H3 at Lys-9
- HDAC1
histone deacetylase 1
- Hes1
hairy and enhancer of split 1
- Hesr
hairy and enhancer of split-related Hesr family
- Hey1
hairy/enhancer-of-split–related with YRPW motif 1
- Id
inhibitor of differentiation
- Luc activity
luciferase activity
- MEF2
myocyte enhancer factor 2
- Me Lys
methyl lysine
- MHC
myosin heavy chain
- MRF
myogenic regulatory factors
- MRF4
myogenic regulatory factor 4
- Myf5
myogenic factor 5
- MyoD(K104R)
MyoD with Lys-104 mutated to arginine
- MyoR
myogenic repressor
- REST
RE1-silencing transcription factor
- Sharp-1
enhancer-of-split and hairy-related protein 1
- siRNA
small interfering RNA
- SIRT1
silent mating type information regulation 2 homologue 1
- Su(var)3-9
suppressor of variegation 3-9
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-04-0311) on October 19, 2012.
REFERENCES
- Azmi S, Ozog A, Taneja R. Sharp-1/DEC2 inhibits skeletal muscle differentiation through repression of myogenic transcription factors. J Biol Chem. 2004;279:52643–52652. doi: 10.1074/jbc.M409188200. [DOI] [PubMed] [Google Scholar]
- Azmi S, Sun H, Ozog A, Taneja R. mSharp-1/DEC2, a basic helix-loop-helix protein functions as a transcriptional repressor of E box activity and Stra13 expression. J Biol Chem. 2003;278:20098–20109. doi: 10.1074/jbc.M210427200. [DOI] [PubMed] [Google Scholar]
- Azmi S, Taneja R. Embryonic expression of mSharp-1/mDEC2, which encodes a basic helix-loop-helix transcription factor. Mech Dev. 2002;114:181–185. doi: 10.1016/s0925-4773(02)00049-7. [DOI] [PubMed] [Google Scholar]
- Bharathy N, Taneja R. Methylation muscles into transcription factor silencing. Transcription. 2012;3:1–6. doi: 10.4161/trns.20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhou J, Xu H, Xu G, Xue J. Identification and developmental expression of Dec2 in zebrafish. Fish Physiol Biochem. 2010;36:667–675. doi: 10.1007/s10695-009-9341-7. [DOI] [PubMed] [Google Scholar]
- Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem. 2009;284:27857–27865. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, Evers BM, Zhou BP. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J Clin Invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Zarebski A, Montoya-Durango D, Grimes HL, Horwitz M. Gfi1 coordinates epigenetic repression of p21Cip/WAF1 by recruitment of histone lysine methyltransferase G9a and histone deacetylase 1. Mol Cell Biol. 2005;25:10338–10351. doi: 10.1128/MCB.25.23.10338-10351.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friday BB, Horsley V, Pavlath GK. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J Cell Biol. 2000;149:657–665. doi: 10.1083/jcb.149.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, Sato F, Noshiro M, Bhawal UK, Suardita K, Kato Y. Transcriptional repression by the basic helix-loop-helix protein Dec2: multiple mechanisms through E-box elements. Int J Mol Med. 2007;19:925–932. [PubMed] [Google Scholar]
- Fujimoto K, Shen M, Noshiro M, Matsubara K, Shingu S, Honda K, Yoshida E, Suardita K, Matsuda Y, Kato Y. Molecular cloning and characterization of DEC2, a new member of basic helix-loop-helix proteins. Biochem Biophys Res Commun. 2001;280:164–171. doi: 10.1006/bbrc.2000.4133. [DOI] [PubMed] [Google Scholar]
- Garriga-Canut M, Roopra A, Buckley NJ. The basic helix-loop-helix protein, sharp-1, represses transcription by a histone deacetylase-dependent and histone deacetylase-independent mechanism. J Biol Chem. 2001;276:14821–14828. doi: 10.1074/jbc.M011619200. [DOI] [PubMed] [Google Scholar]
- Gulbagci NT, Li L, Ling B, Gopinadhan S, Walsh M, Rossner M, Nave KA, Taneja R. SHARP1/DEC2 inhibits adipogenic differentiation by regulating the activity of C/EBP. EMBO Rep. 2009;10:79–86. doi: 10.1038/embor.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Jones CR, Fujiki N, Xu Y, Guo B, Holder JL Jr, Rossner MJ, Nishino S, Fu YH. The transcriptional repressor DEC2 regulates sleep length in mammals. Science. 2009;325:866–870. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- Kubicek S, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Lecomte V, Meugnier E, Euthine V, Durand C, Freyssenet D, Nemoz G, Rome S, Vidal H, Lefai E. A new role for sterol regulatory element binding protein 1 transcription factors in the regulation of muscle mass and muscle cell differentiation. Mol Cell Biol. 2010;30:1182–1198. doi: 10.1128/MCB.00690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling BM, et al. Lysine methyltransferase G9a methylates the transcription factor MyoD and regulates skeletal muscle differentiation. Proc Natl Acad Sci USA. 2012;109:841–846. doi: 10.1073/pnas.1111628109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- Morosetti R, et al. MyoD expression restores defective myogenic differentiation of human mesoangioblasts from inclusion-body myositis muscle. Proc Natl Acad Sci USA. 2006;103:16995–7000. doi: 10.1073/pnas.0603386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci USA. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E, Sousa-Victor P, Ballestar E, Muñoz-Cánoves P. Epigenetic regulation of myogenesis. Epigenetics. 2009;4:541–550.. doi: 10.4161/epi.4.8.10258. [DOI] [PubMed] [Google Scholar]
- Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J Cell Physiol. 2000;185:155–173. doi: 10.1002/1097-4652(200011)185:2<155::AID-JCP1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Roopra A, Qazi R, Schoenike B, Daley TJ, Morrison JF. Localized domains of G9a-mediated histone methylation are required for silencing of neuronal genes. Mol Cell. 2004;14:727–738. doi: 10.1016/j.molcel.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J, Eichele G, Taneja R, Nave KA. Disturbed clockwork resetting in Sharp-1 and Sharp-2 single and double mutant mice. PLoS One. 2008;3:e2762.. doi: 10.1371/journal.pone.0002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin LA, Rudnicki MA. The molecular regulation of myogenesis. Clin Genet. 2000;57:16–25. doi: 10.1034/j.1399-0004.2000.570103.x. [DOI] [PubMed] [Google Scholar]
- Sartorelli V, Caretti G. Mechanisms underlying the transcriptional regulation of skeletal myogenesis. Curr Opin Genet Dev. 2005;15:528–535. doi: 10.1016/j.gde.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Ghaffari S, Taneja R. bHLH-Orange transcription factors in development and cancer. Transl Oncogenomics. 2007;2:105–118. doi: 10.4137/tog.s436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Vedadi M, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, et al. Requirement for the basic helix-loop-helix transcription factor Dec2 in initial TH2 lineage commitment. Nat Immunol. 2009;10:1260–1266. doi: 10.1038/ni.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]