The N-terminus of the kinesin-13 family (Kif2a, Kif2b, Kif2c) is the primary localization determinant. However, the C-terminus of Kif2b associates with Cep170 and Cep170R to create targeting specificity. Cep170 has microtubule-binding properties in vitro and provides a second microtubule-binding site to Kif2b to target it to the spindle.
Abstract
Microtubule dynamics are essential throughout mitosis to ensure correct chromosome segregation. Microtubule depolymerization is controlled in part by microtubule depolymerases, including the kinesin-13 family of proteins. In humans, there are three closely related kinesin-13 isoforms (Kif2a, Kif2b, and Kif2c/MCAK), which are highly conserved in their primary sequences but display distinct localization and nonoverlapping functions. Here we demonstrate that the N-terminus is a primary determinant of kinesin-13 localization. However, we also find that differences in the C-terminus alter the properties of kinesin-13, in part by facilitating unique protein–protein interactions. We identify the spindle-localized proteins Cep170 and Cep170R (KIAA0284) as specifically associating with Kif2b. Cep170 binds to microtubules in vitro and provides Kif2b with a second microtubule-binding site to target it to the spindle. Thus the intrinsic properties of kinesin-13s and extrinsic factors such as their associated proteins result in the diversity and specificity within the kinesin-13 depolymerase family.
INTRODUCTION
The mitotic spindle is a complex structure composed of dynamic microtubule polymers that is essential to ensure correct chromosome segregation. The dynamic instability of microtubules is modulated by microtubule-associated proteins that impart stabilizing or destabilizing properties. The control of these microtubule-associated proteins is critical for the formation and maintenance of a bipolar spindle, the alignment and oscillation of the sister chromatids, and the movement of chromosomes to daughter cells in anaphase (Goshima and Scholey, 2010). The kinesin-13 family of microtubule depolymerases, composed of Kif2a, Kif2b, and Kif2c/MCAK in humans, plays an important role in each of these microtubule-based mitotic processes to spatially and temporally control microtubule dynamics. Kinesin-13 depletion results in defects in spindle bipolarity and length, chromosome segregation, and microtubule dynamics (Maney et al., 1998; Goshima and Vale, 2003; Goshima et al., 2005, 2007; Ganem and Compton, 2004; Kline-Smith et al., 2004; Rogers et al., 2004; Manning et al., 2007; Bakhoum et al., 2009).
Despite roughly similar sequences, Kif2a, Kif2b, and Kif2c display distinct localization and nonoverlapping functions during mitosis. Kif2a localizes to centrosomes (Ganem and Compton, 2004) and has been observed weakly at kinetochores (Cameron et al., 2006). Kif2b localizes to spindles, centrosomes, and unaligned kinetochores (Manning et al., 2007). MCAK/Kif2c localizes primarily to kinetochores and weakly to centrosomes during mitosis (Walczak et al., 1996, 2002; Maney et al., 1998). In addition, MCAK/Kif2c associates with EB1 and Kif18b to target to microtubule plus ends (Honnappa et al., 2009; Tanenbaum et al., 2011). In contrast, the mechanisms controlling Kif2a and Kif2b localization remain less clear. Although the general roles and functions of kinesin motor domains and their interactions with microtubules are well established (reviewed in Hirokawa and Takemura, 2004; Miki et al., 2005; Wordeman, 2010), the molecular basis for their differences in targeting to the correct subcellular locations remains ill defined. In addition, it is not known whether these three closely related proteins display functional specificity beyond their differential localization.
Here we conduct a comparative analysis of the kinesin-13 proteins in human cells. We define the divergent regions flanking the motor domains of Kif2a, Kif2b, and Kif2c as the regions responsible for specifying kinesin-13 targeting to distinct microtubule-based structures. In addition, we find that Kif2b, but not Kif2a or Kif2c, stably associates with the microtubule-binding protein Cep170. The Cep170–Kif2b interaction provides a second extrinsic microtubule-binding activity to Kif2b to target it to the mitotic spindle. Thus the intrinsic features of a kinesin, as well as specific extrinsic associations, define its localization and activities.
RESULTS
The N-terminal region of kinesin-13 proteins specifies their localization
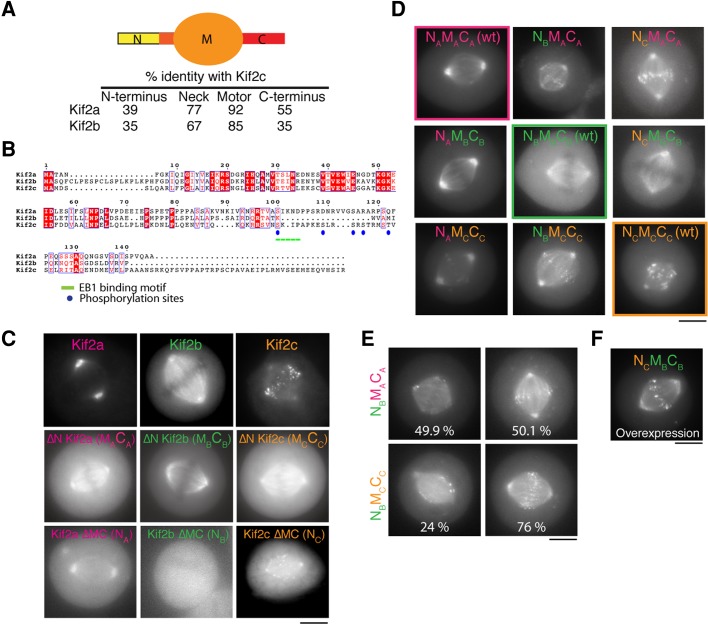
Each of the three closely related kinesin-13 proteins plays a distinct role in chromosome segregation. The enzymatic depolymerase kinesin “motor” domain and the neck region (together denoted as M in this article) are well conserved among the kinesin-13 isoforms (Figure 1A), whereas the N- and C-terminal regions (denoted N and C, respectively) show greater diversity (Figure 1, A and B). To assess the relative contributions of these domains, we first generated clonal cell lines stably expressing low to moderate levels of Kif2a, Kif2b, and Kif2c/MCAK as green fluorescent protein (GFP) fusions (Figure 1C). Each kinesin targeted to distinct microtubule-based structures throughout mitosis (Supplemental Figure S1A), similar to previous reports (Wordeman and Mitchison, 1995; Ganem and Compton, 2004; Manning et al., 2007). To dissect the contributions of the N, M, and C regions to the localization of each kinesin-13 during mitosis, we first expressed GFP fusions lacking the N-terminus. The three kinesin-13 MC constructs (also denoted as ∆NT) each displayed identical localization to microtubules during interphase (unpublished data) and to spindle microtubules during mitosis with an accumulation at centrosomes (Figure 1C). Thus the localization of kinesin-13 proteins to specific cellular sites and microtubule subpopulations requires their N-terminal regions.
FIGURE 1:
The N-terminus of kinesin-13 is a primary determinant of kinesin localization. (A) Schematic diagram showing the kinesin-13 domains and their percentage similarity with respect to Kif2c/MCAK. (B) Sequence alignment of the N-terminus of human Kif2a, Kif2b, and Kif2c. The sequences were aligned using the program T-coffee (EBI) and formatted with ESPRIPT (Gouet et al., 1999). Known phosphorylation sites for Kif2c/MCAK and the EB1 binding motif are highlighted. (C) Images of HeLa cells transiently expressing GFP fusions to full-length and domains of Kif2a, Kif2b, and Kif2c. The ΔN and ΔMC domains represent GFP fusions lacking the N-terminus or the motor and C-terminal domains of the kinesins, respectively. (D) Images of mitotic HeLa cells transiently expressing the indicated GFP-tagged kinesin-13 chimeras. (E) Images of chimeric kinesin-13 GFP fusion constructs that display heterogeneous localization in the observed cells. Percentages represent the frequency with which the chimera shows localization to only spindle microtubules (left) or spindle microtubules and kinetochores (right). n = 100 cells. (F) Image of a cell transiently overexpressing GFP-NCMBCB. Scale bars, 10 μm.
Although the kinesin-13 catalytic domains are highly conserved among the isoforms, the N- and C-terminal regions show greater divergence (Figure 1A) and are likely to be involved in creating specificity and diversity among the kinesin-13 family. The essential contribution of the N-terminus of MCAK/Kif2c to its localization to kinetochores is well established (Maney et al., 1998; Walczak et al., 2002). The N-terminal region of MCAK also contains the majority of the established regulatory sites in MCAK/Kif2c (Andrews et al., 2004; Lan et al., 2004) and the EB1 binding motif that targets MCAK to microtubule plus ends (Honnappa et al., 2009; Figure 1B). To test the contribution of the N-terminal region of the kinesin-13 proteins to their distinct localization, we expressed their N-terminal regions alone (N, also labeled ∆MC). In contrast to previous studies (Maney et al., 1998), we found that the N-terminal region of Kif2c was sufficient to target to kinetochores in human cells (Figure 1C). Like full-length Kif2a, the N-terminal region of Kif2a localized to centrosomes (Figure 1C). In contrast, the N-terminal region of Kif2b did not localize to specific structures (Figure 1C). These results indicate that the divergent N-terminal regions define the localization of kinesin-13 proteins but that Kif2b localization requires additional contributions from its motor and C-terminal domains.
Sequences in the enzymatic and C-terminal regions alter targeting of the kinesin-13 N-terminus
Although the N-terminal region is an important determinant for kinesin-13 localization, we next sought to test whether there is additional functional specificity among the three kinesin-13 proteins arising from the catalytic or C-terminal domains. To test this, we generated chimeric constructs in which the N-terminal domain of each kinesin was fused to the enzymatic and C-terminal domains of the three isoforms (Figure 1D). To designate these nine fusion proteins, we refer to these as NxMyCz, with x, y, and z indicating the corresponding kinesin isoform. Of importance, after transient transfection in HeLa cells, each control chimera constructed using this cloning strategy (NAMACA, NCMCCC, and NBMBCB) displayed localization identical to that for the corresponding wild-type protein (Figure 1D). The subcellular localization of each chimera and fusion protein is summarized in Supplemental Figure S1B. In the majority of cases, we found that the localization of the chimera was determined primarily by the identity of the N-terminal region and displayed similar localization to the control kinesins. For example, NAMBCB and NAMCCC both displayed strong localization to the spindle poles and very weak localization to the kinetochores, similar to Kif2a (Figure 1D). In addition to its recruitment to kinetochores in mitosis, NCMACA could track the plus end of microtubules in a similar manner to Kif2c/MCAK in interphase (Supplemental Movie 1). In addition, NCMACA was enriched at microtubule plus ends, similar to EB1 (Supplemental Figure S1C).
However, we also observed some notable differences with respect to the localization of other chimeras. Unlike Kif2b (NBMBCB), which localized only to the spindle during metaphase (Figure 1D), NBMACA and NBMCCC also localized to metaphase kinetochores and centrosomes in the majority of cells (Figure 1, D and E, and Supplemental Figure S1D). Despite the EB1 binding motif present in the NC domain, NCMBCB did not track microtubule plus ends during interphase but instead bound to microtubules along their length (Supplemental Figure S1E). In addition, when moderately expressed, NCMBCB localized primarily to the spindle in metaphase (Figure 1D) and targeted to astral microtubules in anaphase and telophase (Supplemental Movie 2), whereas NCMCCC/Kif2c did not, relocating from kinetochores to the midzone (Figure 1D and Supplemental Figure S1A). When overexpressed, NCMBCB localized to kinetochores, centrosomes, and spindles (Figure 1F), similar to NCMCCC/Kif2c (Figure 1D).
Taken together, these results indicate that chimeras containing the enzymatic domain and C-terminal region of Kif2b localize more robustly to microtubules regardless of the presence of a domain that would normally target the protein to kinetochores or microtubule plus ends. This suggests that the N-terminal region of kinesin-13s is a major determinant for targeting the catalytic domain but that the C-terminal and enzymatic domains also contribute to its localization.
Kif2b stably associates with unique interacting partners
As described, intrinsic factors related to specific kinesin sequences contribute to the specificity of each kinesin's localization. However, extrinsic factors such as association with distinct partners could also result in differences in kinesin-13 regulation or activity. To define the unique associations of kinesin-13s, we isolated Kif2a, Kif2b, and Kif2c in separate one-step affinity purifications from human cells using our established procedures (Cheeseman and Desai, 2005). We did not identify any stable interacting partners for Kif2c/MCAK (Supplemental Figure S2A), suggesting that the previously identified interactions with EB1 and Sgo2 (Honnappa et al., 2009; Tanno et al., 2010) do not persist under our purification conditions. In reciprocal purifications, we found that GFP-EB3 interacted with EB1 and EB2 as established by pervious work (De Groot et al., 2010), but we did not isolate Kif2c (Supplemental Figure S2B). Similarly, we did not identify interacting partners for Kif2a (Supplemental Figure S2A). Kif2a has been reported to bind to DDa3 (Jang et al., 2008). However, we also did not identify Kif2a in reciprocal affinity purifications using GFP-DDa3 (Supplemental Figure S2A). We note that DDa3 and Kif2a do not display identical subcellular localization (Supplemental Figure S2C), suggesting that these proteins may also function independently of each other.
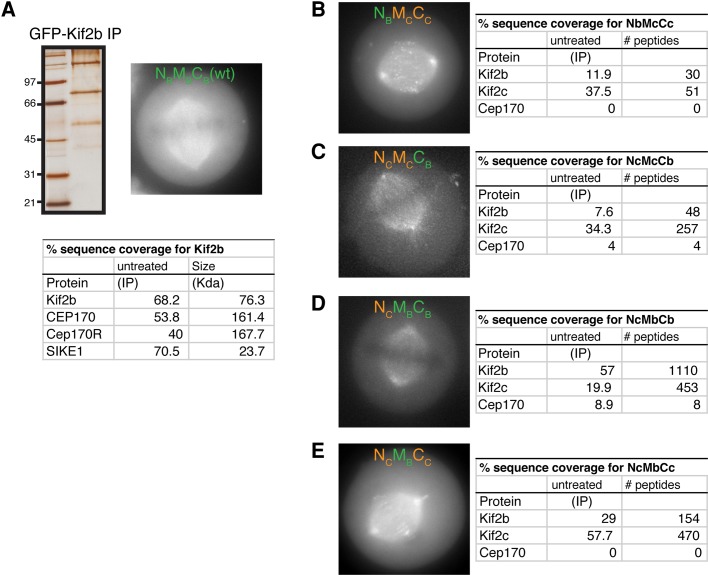
In contrast, GFP-Kif2b purifications isolated three interacting proteins (Figure 2A): Cep170, an uncharacterized protein KIAA0284/Fam68C that shows strong sequence similarity to Cep170, and suppressor of IKK epsilon (SIKE1; Huang et al., 2005). In contrast to recent reports (Manning et al., 2010), we did not detect CLASP1 in Kif2b purifications. Although we consistently isolated SIKE1 in Kif2b purifications, we were unable to validate the significance of this interaction in downstream analyses (unpublished data). Thus we chose to focus on the interaction of Kif2b with Cep170 and KIAA0284.
FIGURE 2:
Kif2b associates with Cep170 and Cep170R. GFPLAP-tagged fusions were used to isolate Kif2b and kinesin-13 chimeras from stable cell lines using one-step immunoprecipitations as described in Cheeseman and Desai (2005). (A) Top left, silver-stained gel of the GFP-Kif2b immunoprecipitation. Top right, image of cell expressing GFP-Kif2b. Bottom, percentage sequence coverage from the mass spectrometric analysis of the indicated samples showing the proteins identified in the Kif2b complex purifications but not in unrelated control samples (including diverse samples that our lab has tested under identical similar conditions; for example, see Schmidt et al., 2010; Welburn et al., 2010). (C–F) Left, images of the indicated kinesin-13 chimeras showing their localization during metaphase. Right, percentage sequence coverage from the mass spectrometric analysis of the indicated samples showing the proteins identified in each complex and the number of peptides recovered. Note that both Kif2b and Kif2c were identified in these samples due to the peptides present in the chimeric proteins. The association of Cep170 with Kif2b requires the C-terminus. Scale bars, 10 μm.
Cep170 and KIAA0284 possess the same domain structure with an FHA phospho-binding domain at the N-terminus and uncharacterized but similar C-termini (Supplemental Figure S3). Because of their sequence similarity, we will refer to KIAA0284/Fam68C as Cep170-related (Cep170R). Cep170 was shown previously to localize to centrosomes and microtubules (Guarguaglini et al., 2005). Recent large-scale analyses also identified Cep170 as interacting with the kinesin-14 Kifc3 (Hutchins et al., 2010), an interaction that we confirmed using affinity purifications of GFP-Kifc3 from HeLa cells (Supplemental Figure S2A).
To determine which domains of Kif2b support the interaction with Cep170 and Cep170R, we purified the Kif2b/Kif2c chimeras from human cells. We did not identify any associated proteins from NBMCCC purifications (Figure 2B), similar to Kif2c purifications. However, Cep170 copurified with NCMCCB and NCMBCB (Figure 2, D and E) but not NCMBCC (Figure 2F), suggesting that Cep170 interacts with Kif2b via the C-terminal domain. We note that we did not identify Cep170R in any of the purifications with the chimeric Kif2b proteins, suggesting either that multiple regions are required for this interaction or that additional regulation of this interaction is occurring. Thus Kif2b displays specific interactions with Cep170.
The Kif2b-interacting proteins Cep170 and Cep170R localize to microtubules
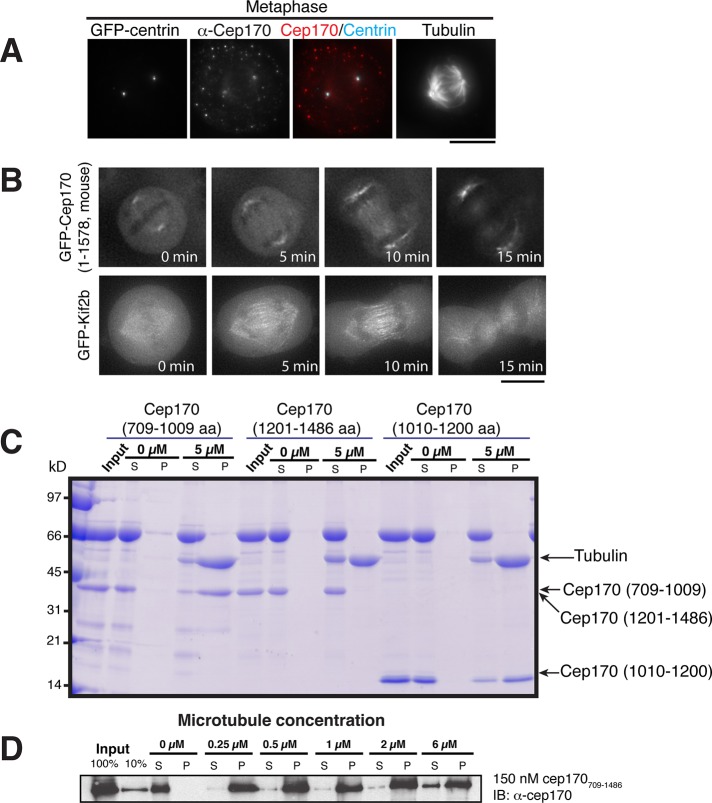
We next analyzed the localization of Cep170 and Cep170R. Cep170 has been reported to localize to centrosomes and the mitotic spindle (Guarguaglini et al., 2005; Hutchins et al., 2010), similar to Kif2b (Figure 1C). Consistent with this, on the basis of antibodies against endogenous Cep170 and a Cep170-GFP fusion, we found that Cep170 localized to microtubules and centriole structures during interphase (Supplemental Figure S4A and unpublished data) and to spindle microtubules during metaphase (Figure 3, A and B). Based on antibodies against the endogenous protein and GFP fusions to both the full-length protein and C-terminus (CT; residues 921–1553), Cep170R also localized to microtubule-based structures, including microtubules and centrioles during interphase (Supplemental Figure S4A), centrosomes during metaphase (Supplemental Figure S4B), and the midzone during telophase (Supplemental Figure S4C). However, in contrast to Cep170, Cep170R localized only weakly to the spindle in metaphase (Supplemental Figure S4, B and D) but became strongly enriched on microtubules in the late anaphase (Supplemental Figure S4D).
FIGURE 3:
Cep170 targets to the spindle and has microtubule-binding activity. (A) Immunofluorescence images of metaphase cells expressing GFP-centrin stained for Cep170 and microtubules. (B) Images of live cells expressing GFP-Kif2b and GFP-Cep170 at the indicated time points throughout mitosis. Cep170 and Kif2b localize to microtubules throughout mitosis. (C) Cep170 has two microtubule-binding domains. Coomassie gel showing the cosedimentation of the indicated Cep170 domains in the absence or presence of 5 μM microtubules. Cep170 fragments containing amino acids 709–1009 or 1010–1200 bound to microtubules. (D) Western blot showing the cosedimentation of 250 nM histidine-Cep170-CT (amino acids 709–1486) at the indicated microtubule concentrations. The Western blot was probed with anti-Cep170 antibodies.
The change in Cep170R localization at anaphase onset suggests that it maybe controlled by CDK activity. To test this, we added the CDK inhibitor flavopiridol to cells that had been arrested in mitosis using the MG132 protease inhibitor. Cep170R rapidly localized to spindle and astral microtubules upon the addition of flavopiridol (Supplemental Figure S4E), suggesting that Cep170R localization to microtubules is negatively regulated by CDK1. Both Cep170 and Cep170R have multiple potential S/T-P CDK consensus sites in their C-terminus, which are known to be phosphorylated in vivo based on large-scale analyses (Nousiainen et al., 2006; Dephoure et al., 2008; Malik et al., 2009) and the identification of these phosphorylation sites in our affinity purifications (unpublished data). Consistent with this, we found that purified Cep170 (709–1486) is directly phosphorylated by CDK2/cyclinA in vitro (unpublished data). However, although both proteins are likely to be phosphorylated by CDK, only the localization of Cep170R is altered by this phosphorylation. Thus, despite their primary sequence similarity, the association of Cep170 and Cep170R with microtubules is differentially regulated during mitosis.
Cep170 binds to microtubules with high affinity
To determine the functional contribution of the Kif2b-interacting proteins, we next tested their biochemical properties. Overexpression of Cep170 leads to microtubule bundling in interphase cells (Guarguaglini et al., 2005). Based on the localization of Cep170 to microtubules and this bundling activity, we tested the microtubule-binding properties of Cep170. The C-terminus of Cep170 comprises three predicted globular domains. Two of the three domains—Cep170709-1009 and Cep1701010-1209—displayed independent binding to microtubules in microtubule cosedimentation assays (Figure 3C). To determine the affinity of Cep170 for microtubules, we expressed the entire C-terminus of Cep170709-1486 containing both microtubule-binding domains. Under the tested conditions, Cep170709-1486 fully bound to 0.25 μM microtubules, indicating that the apparent affinity was significantly stronger than 0.25 μM (Figure 3D). This high affinity is likely due to the cooperativity between the two microtubule-binding domains. We were unable to purify a similar fragment of Cep170R, preventing us from directly testing its microtubule-binding activity. However, based on the sequence similarity between Cep170 and Cep170R and the localization of both proteins to microtubules, we suspect that both proteins bind microtubules. Thus Cep170 possesses an intrinsic microtubule-binding activity, thereby providing a second, nonmotor microtubule-binding site to the Kif2b complex.
Cep170 association with the C-terminus of Kif2b enhances localization of Kif2b to the spindle
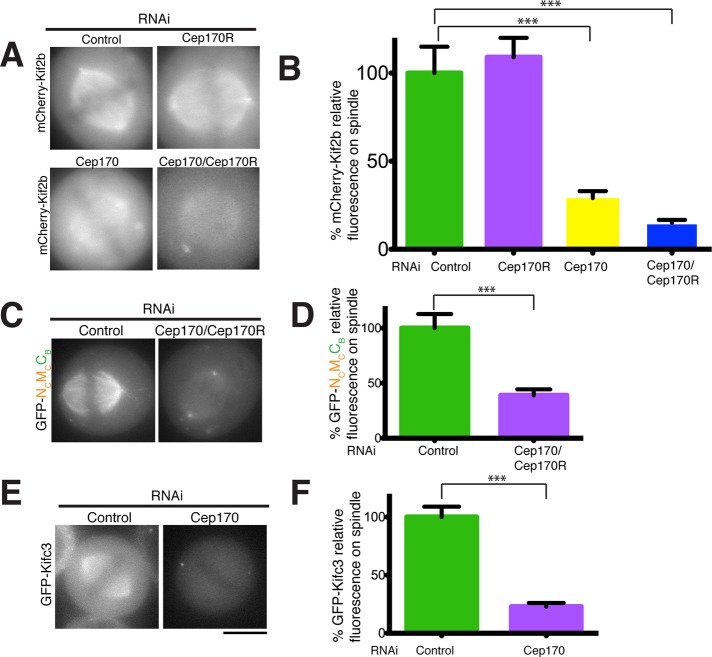
Based on the localization of the kinesin-13 chimeras described earlier, the C-terminal region of Kif2b is important for its localization to the spindle (Figure 1). The Kif2b C-terminus is also the region that interacts with Cep170 (Figure 2), which possesses its own microtubule-binding activity (Figure 4, C and D). We hypothesized that Cep170 may provide a second extrinsic microtubule-binding activity to target Kif2b to spindle microtubules. To test this, we codepleted Cep170 and Cep170R from cell lines expressing low to moderate amounts of mCherry-Kif2b. Indeed, we observed that the levels of Kif2b on the spindle were decreased by 80% upon individual depletion of Cep170 (p = 1.1 × 10−5; Figure 4, A and B) or codepletion of Cep170/Cep170R (p = 4.3 × 10−17; Figure 4, A and B). Similarly, upon Cep170/Cep170R codepletion, the targeting of the NCMCCB chimera, which associates with Cep170 (Figure 2C), to spindles was also severely reduced (p = 3 × 10−4; Figure 4, C and D). In contrast, the protein levels of mCherry-Kif2b as assessed by Western blotting were unaffected after depletion of Cep170 or Cep170R (Supplemental Figure S5A). These data suggest that Cep170 is required to target Kif2b to the mitotic spindle.
FIGURE 4:
Kif2b and Kifc3 spindle localization requires Cep170. (A) Images of live mitotic cells expressing mCherry-Kif2b under control conditions or after Cep170 or Cep170R depletion or Cep170/Cep170R codepletion by RNAi. (B) Quantification of mCherry-Kif2b fluorescence on the mitotic spindle from the cells in A. (C) Images of live mitotic cells expressing GFP-NCMCCB under control conditions or after Cep170/Cep170R codepletion by RNAi. (D) Quantification of GFP-NCMCCB fluorescence on the mitotic spindle from the cells in C. (E) Images of live mitotic cells expressing GFP-Kifc3 under control conditions or after Cep170 depletion by RNAi. (F) Quantification of GFP-Kifc3 spindle-localized fluorescence in controls and Cep170-depleted cells. For all fluorescence localization experiments in this figure, each experiment was repeated three times, with 50 cells analyzed for each experiment. The fluorescence intensity values indicate the average percentage quantified fluorescence intensity (±SEM) relative to controls. Statistical i tests were performed for each experiment, and the confidence interval is indicated by and asterisk. ***p < 0.001. Scale bars, 10 μm.
In reciprocal experiments, Cep170 localization to the spindle was not affected by the depletion of Kif2b or the other Kif2b-interacting proteins (Supplemental Figure S5B), suggesting that these proteins function upstream of Kif2b for spindle localization. Of importance, the localization of Kifc3, which also interacts with Cep170 (Supplemental Figure S2A; Hutchins et al., 2010), to spindles was also strongly reduced upon depletion of Cep170 (Figure 4, E and F; p = 5.4 × 10−10). However, Kifc3 depletion did not affect Cep170 localization (Supplemental Figure S5C). Taken together, these results suggest that Cep170 targets at least the two kinesins Kif2b and Kifc3 to the spindle by providing a second microtubule-binding activity to these complexes.
Cep170 is required for Kif2b-induced microtubule depolymerization
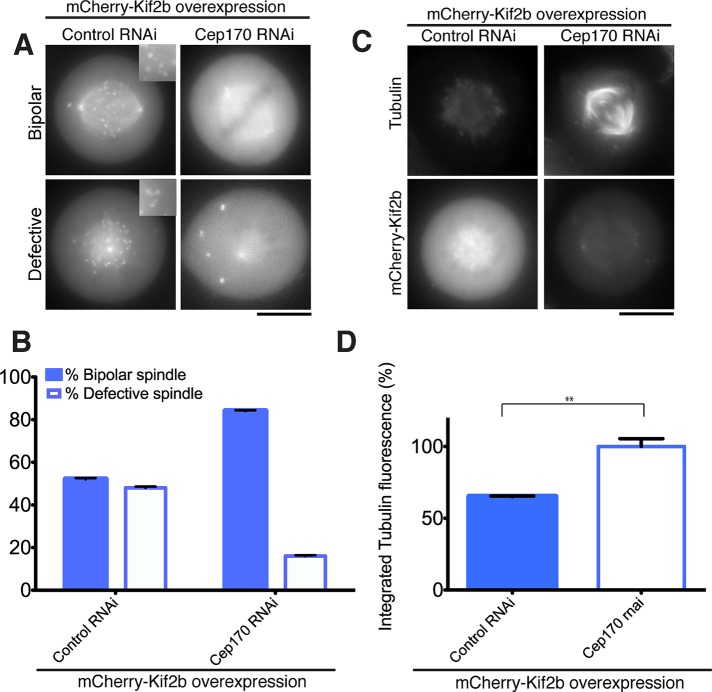
The microtubule-associated protein Cep170 is necessary to target Kif2b to the mitotic spindle (Figure 4). Next we sought to test whether Cep170 contributes to Kif2b-induced microtubule depolymerization. Endogenous Kif2b is present at as a low-abundance protein in HeLa cells and other tissue culture systems but shows similar localization to that of GFP-Kif2b (Manning et al., 2007, 2010). Its low abundance and potential redundancy with other pathways prevented us from assessing the activity of endogenous Kif2b. Thus we examined the depolymerase activity of Kif2b in a Kif2b-inducible cell line. On overexpression, Kif2b accumulated at kinetochores of bioriented chromosomes (Supplemental Figure S6A) and resulted in spindle defects such as monopolar spindles and short spindles (Figure 5, A and B, and unpublished data). These observations are a hallmark of kinesin-13 hyperactivity and are similar to the spindle defects caused by MCAK overexpression (Supplemental Figure S6, B and C; Maney et al., 1998). When Cep170 was depleted from cells overexpressing Kif2b, we observed a significant decrease in spindle defects relative to control cells overexpressing Kif2b (Figure 5, A and B). To test whether the spindle defects were due to excessive Kif2b depolymerase activity, we quantified the levels of polymerized tubulin in cells overexpressing Kif2b in the presence or absence of Cep170. We found that there were increased levels of tubulin polymer when Cep170 was depleted (Figure 5, C and D), consistent with reduced levels of depolymerase activity in the absence of Cep170. Taken together, these data suggest that Cep170 targets Kif2b to spindles. Future biochemical analyses will be required to determine whether Cep170 modulates Kif2b activity in addition to promoting its spindle localization.
FIGURE 5:
Cep170 promotes Kif2b activity in vivo. (A) Images of HeLa cells transiently overexpressing mCherry-Kif2b in the presence or absence of Cep170. Representative examples of normal bipolar and defective spindle structures present in these cells are shown. (B) Graph showing the frequency of cells displaying a bipolar or defective spindle morphology when Kif2b is overexpressed in control cells or after Cep170 depletion as in A. Error bars represent the SEM. (C) Immunofluorescence images showing the localization of tubulin/microtubules (using DM1α antibodies) in a cell line overexpressing mCherry-Kif2b in controls or cells depleted for Cep170 by RNAi. (D) Quantification of microtubule intensity for the cells in C. Numbers indicate the average percentage quantified spindle fluorescence intensity in cells overexpressing mCherry-Kif2b (±SEM) relative to cells overexpressing mCherry-Kif2b in the absence of Cep170. Statistical t tests were performed for each experiment, and the confidence interval is indicated by and asterisk. **p < 0.01. Each experiment in this figure was repeated three times, with 30 cells counted each time. Scale bars, 10 μm.
DISCUSSION
Extrinsic factors control the targeting and depolymerase activity of Kif2b
Here we demonstrated that Kif2b, but not other kinesin-13 proteins, interacts with Cep170 to target to the mitotic spindle. The identification of Kif2b-associated proteins that display their own microtubule-binding activity presents a paradigm for the control of kinesin microtubule depolymerases. The EB1 family of proteins has been shown to target Kif2c/MCAK to microtubule plus ends by interacting with the Kif2c N-terminal region. Our work demonstrates that Cep170 uses its microtubule-binding properties to increase the association of Kif2b with microtubules via the C-terminal region of Kif2b. On association with Cep170, the enhanced association of Kif2b with the microtubule lattice could facilitate its depolymerase activity by increasing its concentration on microtubules.
The function of Kif2b and the context for its microtubule depolymerase activity have been investigated primarily in tissue culture systems and remain under debate, due in part to its low level of expression and the redundancy with other pathways controlling microtubule dynamics during mitosis (Tanenbaum et al., 2009; Manning et al., 2010). In addition to controlling spindle assembly and chromosome movement, kinesin-13 family proteins have also been reported to contribute to regulating centriole length (Kobayashi et al., 2011; Delgehyr et al., 2012), and it remains possible that Kif2b could contribute to diverse processes in humans. Our work suggests that Kif2b does possess a microtubule depolymerase activity based on the decreased levels of microtubules observed when Kif2b is overexpressed and demonstrates that extrinsic interacting partners contribute to the proper localization of Kif2b to the spindle. Cep170 is widely expressed in diverse cell types (Novartis Gene Atlas, http://biogps.org) and can act as a potent activator of Kif2b depolymerase based on the work presented here. Low levels of Kif2b are found in most cell types, with higher levels in testes (Su et al., 2004; Wu et al., 2009). To prevent excessive microtubule depolymerase activity, controlling Kif2b expression levels may be critical. It remains to be determined how the Kif2b–Cep170 interaction is regulated in cells. For example, MCAK levels are higher than Kif2b in cells (Wu et al., 2009), but MCAK activity and localization are tightly regulated by protein interactions and phosphorylation to prevent spindle abnormalities. Elevated levels of MCAK in cancer cells are associated with resistance to Taxol (Ganguly et al., 2011). For Kif2b, the expression or protein levels of Kif2b appear to be a rate-limiting factor. Therefore ensuring low levels of Kif2b may prevent microtubule defects that would be associated with the activity of the Kif2b depolymerase that is promoted by targeting to the spindle by Cep170. It has also been suggested that Plk1 phosphorylation of Kif2b regulates its activity in vivo (Hood et al., 2012). Future work is needed to determine under which conditions Kif2b acts as a microtubule depolymerase and whether Cep170 interactions or phosphorylation contributes to Kif2b activation using biochemical approaches.
Addition of a second microtubule-binding site alters the targeting and functional properties of kinesins
The kinesin-13 depolymerases play important and nonredundant roles in regulating microtubule dynamics (Rogers et al., 2004; Mennella et al., 2005). Here we dissected the properties that make each member of the kinesin-13 family unique. Kinesins have an intrinsic microtubule-binding site in the catalytic domain. Most kinesin studies have focused exclusively on the motor domain and its interactions with microtubules. However, recent work has identified cases in which a second, nonmotor microtubule-binding site is necessary for correct kinesin function. For example, the kinesin-5 Eg5 has been reported to have a second microtubule-binding site in addition to the motor domain (Weinger et al., 2011). This second site in the C-terminal tail increases the association of kinesin-5 with microtubules to ultimately increase the processivity of the motor. Similarly, kinesin-8 uses a second microtubule-binding site to target to the correct subcellular structure (Stumpff et al., 2011; Su et al., 2011; Weaver et al., 2011). This nonmotor microtubule-binding domain enhances the processivity to allow targeting to microtubule plus ends. Kinesins also associate with other microtubule-binding proteins, such as EB1, to promote recruitment of the kinesin to a particular cellular localization (Honnappa et al., 2009; Stout et al., 2011; Tanenbaum et al., 2011). The microtubule-binding protein CHICA has been shown to associate with Kid/Kif22 to target it to the spindle (Santamaria et al., 2008). Here we demonstrated that Cep170 can impart its microtubule-binding activity to Kif2b and Kifc3 to target these kinesins to the mitotic spindle. Future work will be required to test the role of Cep170 as a general microtubule-targeting factor for kinesins. The Cep170 interaction partner Kifc3 is overexpressed in cancer cell lines that are resistant to Taxol (De et al., 2009). Therefore it will be interesting to test whether the stability of Kifc3 and the resistance to Taxol in these cancer types can be modulated by Cep170 function. This work points to a generalized mechanism for how kinesins use a second intrinsic or extrinsic microtubule-binding site to function appropriately.
MATERIALS AND METHODS
Molecular biology and cell culture
cDNAs were obtained as IMAGE clones. Stable clonal cells lines expressing GFPLAP fusions were generated in HeLa cells as described previously (Cheeseman et al., 2004). To generate the mCherry-Kif2b HeLa cell line, we used the inducible Flp-In system (Invitrogen, Carlsbad, CA). The Kif2b cDNA was inserted into a pcDNA5/FRT/TO-based mCherry-tagged vector and cotransfected into the HeLa FRT line along with a plasmid expressing the FLP recombinase (pOG44, Invitrogen) using Lipofectamine Plus (Invitrogen). Cells were selected in 400 μg/ml hygromycin B (Roche, Indianapolis, IN), and colonies were pooled and expanded. Protein expression was induced with 0.5–1 μg/ml tetracycline overnight. To achieve Kif2b overexpression, we transiently transfected mCherry-Kif2b into an mCherry-Kif2b–expressing HeLa cell line.
A HeLa Kyoto cell line expressing Cep170-GFP (mouse) was obtained from MitoCheck (Hutchins et al., 2010). RNA interference (RNAi) experiments were conducted using RNAi MAX transfection reagent (Invitrogen) according to the manufacturer's guidelines. For experiments with small-molecule inhibitors, the following final concentrations were used: MG132 (10 μM), flavopiridol (5 μM), STLC (10 μM), and ZM447439 (2 μM).
To evaluate the depletion efficiency of Kif2b siRNA, we tested two sets of small interfering RNAs (siRNAs). With previously published Kif2b siRNAs (Manning et al., 2007; Bakhoum et al., 2009) we could not rescue the phenotype using a GFP-Kif2b resistant to the siRNAs (Supplemental Figure S5D). We therefore used a different set of Kif2b siRNA oligos (Dharmacon SMART-pool siRNA, CGAAAUGGGUUGCGAUGAU, GCUCAGAAACUCCACAUAU, GCACAUGAUCGAAGAGUAU, and CAAGGUGUAUGAUUUGUUG) and obtained full depletion of GFP-Kif2b. Pools of siRNAs for Cep170 (GAAGUAAAGUAACGAAAUC, CGUAACAUCUCUCGGAUUUG, GAUUAUAAUAGGCCUGUUA, and CGAUGUAGCAGGAGAGAUA) and Cep170R (GUACGGCGCUCAGCCAUAA, GGCAAGAGAGCUUCACUAA, GAAUGGGGACGCUGUGUUA, and GCUAGGUUCUCGCCGGAA) were obtained from Dharmacon (Lafayette, CO). Successful depletion of Cep170 and Cep170R (Dharmacon SMART-pool siRNA) was confirmed by immunofluorescence cells using anti-Cep170 and Cep170R antibodies, respectively (Supplemental Figure S5B). Cep170 depletion (Dharmacon SMART-pool siRNA) was also confirmed by Western blotting using anti-Cep170 antibodies (Supplemental Figure S5E). Although the phenotypes observed after depletion of Cep170 appear specific and are consistent with previously published work (Guarguaglini et al., 2005), we note that we were unable to test the ability of an RNAi-resistant version of Cep170 to complement these defects due to the large size of this cDNA, which prevented the generation of a full-length “hardened” version of this cDNA and corresponding stable cell line.
Immunofluorescence and microscopy
Immunofluorescence was conducted as described previously (Kline et al., 2006). For immunofluorescence against microtubules, cells were fixed in methanol for 5 min, and DM1α (Sigma-Aldrich, St. Louis, MO) was used at 1:1000. For visualization of kinetochore proteins, we used mouse EB1 (BD Biosciences, San Diego, CA), mouse anti-HEC1 (9G3; Abcam, Cambridge, MA), and human anti-centromere antibodies (ACA; Antibodies Incorporated, Davis, CA). Affinity-purified rabbit polyclonal antibodies were generated against Cep1701010-1200 and Cep170R1341-1553 as described previously (Desai et al., 2003). For time-lapse imaging, cells were maintained in CO2-independent media (Invitrogen) at 37ºC and imaged every 4–5 min. To visualize DNA in live cells, Hoechst stain was used at 100 ng/ml. Images were acquired on a DeltaVision Core microscope (Applied Precision, Issaquah, WA) equipped with a CoolSnap HQ2 camera (Photometrics, Tucson, AZ). Thirty z-sections were acquired at 0.2-μm steps using a 100×/1.3 numerical aperture Olympus U-Plan Apochromat objective lens with 1 × 1 binning (Olympus, Tokyo, Japan). For live-cell imaging, 6–12 z-sections were acquired at 0.5- to 1-μm steps. Images were deconvolved using the DeltaVision software. Images shown represent maximal-intensity projections. Equivalent exposure conditions and scaling were used between controls and RNAi-depleted cells. To quantitate the fluorescence intensity of Kif2b on spindles, we measured the integrated intensity of Kif2b over a 21 × 21 pixel box on the spindle and in the background, using a mCherry-Kif2b cell line or transiently transfected GFP-NCMCCB. Rare cells for which the mean spindle intensity was three times greater than the average spindle average intensity in either controls or test conditions were removed from the analysis. Each experiment was repeated three times. To quantitate the fluorescence intensity, we measured the integrated fluorescence intensity of tubulin over the entire cell, as described previously (Stumpff et al., 2007). From 40 to 50 cells were examined for each experiment. Each experiment was repeated three times.
Protein purification and biochemical assays
GFPLAP-tagged Kif2a, DDa3, Kif2b, Kif2c, Cep170FL, Cep170755-1486, Cep170R259-1553, Cep170R982-1553, and NCMBCC, NCMBCB, and NCMCCB chimeras were isolated from HeLa cells as described previously (Cheeseman and Desai, 2005). Purified proteins were identified by mass spectrometry using an LTQ XL ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA) using SEQUEST software as described previously (Washburn et al., 2001).
For the expression and purification of the recombinant Cep170 fragments (709–1009, 1010–1200, 1200–1486, 709–1486), hexahistidine-Cep170 fusions were generated in pET28a. Histidine-Cep170R1341-1553 was purified and used for antibody production. Proteins were purified using nickel-nitriloacetic acid agarose (Qiagen, Valencia, CA) according to the manufacturer's instructions, further purified by gel filtration and then desalted into S buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.0, 50 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol). Microtubule-binding assays using the purified proteins were conducted as described previously (Cheeseman et al., 2006) using equal volumes of microtubules in BRB80 and test protein in S buffer.
Supplementary Material
Acknowledgments
We thank members of the Cheeseman laboratory and Sébastien Besson for discussions and critical reading of the manuscript. We also thank Sébastien Besson for help with MATLAB data analysis. This work was supported by awards to I.M.C. from the Searle Scholars Program and the Human Frontiers Science Foundation and a grant from the National Institutes of Health/National Institute of General Medical Sciences (GM088313). I.M.C. is a Thomas D. and Virginia W. Cabot Career Development Professor of Biology. J.W. is currently supported by a Career Development Award from Cancer Research UK.
Abbreviations used:
- CDK
cyclin-dependent kinase
- CT
C terminus
- GFP
green fluorescent protein
- MCAK
mitotic centromere-associated kinesin
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-03-0214) on October 19, 2012.
*Present address: Wellcome Trust Center for Cell Biology, School of Biological Sciences, University of Edinburgh, Edinburgh EH9 3JR, Scotland, United Kingdom.
REFERENCES
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/s1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron LA, Yang G, Cimini D, Canman JC, Kisurina-Evgenieva O, Khodjakov A, Danuser G, Salmon ED. Kinesin 5-independent poleward flux of kinetochore microtubules in PtK1 cells. J Cell Biol. 2006;173:173–179. doi: 10.1083/jcb.200601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Cipriano R, Jackson MW, Stark GR. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009;69:8035–8042. doi: 10.1158/0008-5472.CAN-09-1224. [DOI] [PubMed] [Google Scholar]
- De Groot CO, et al. Molecular insights into mammalian end-binding protein heterodimerization. J Biol Chem. 2010;285:5802–5814. doi: 10.1074/jbc.M109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, Glover DM. Klp10A, a microtubule-depolymerizing kinesin-13, cooperates with CP110 to control Drosophila centriole length. Curr Biol. 2012;22:502–509. doi: 10.1016/j.cub.2012.01.046. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Compton DA. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J Cell Biol. 2004;166:473–478. doi: 10.1083/jcb.200404012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly A, Yang H, Pedroza M, Bhattacharya R, Cabral F. Mitotic centromere-associated kinesin (MCAK) mediates paclitaxel resistance. J Biol Chem. 2011;286:36378–36384. doi: 10.1074/jbc.M111.296483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. doi: 10.1016/j.cub.2005.09.054. [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16:1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Honnappa S, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Hood EA, Kettenbach AN, Gerber SA, Compton DA. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol Biol Cell. 2012;23:2264–2274. doi: 10.1091/mbc.E11-12-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Liu T, Xu LG, Chen D, Zhai Z, Shu HB. SIKE is an IKK epsilon/TBK1-associated suppressor of TLR3- and virus-triggered IRF-3 activation pathways. EMBO J. 2005;24:4018–4028. doi: 10.1038/sj.emboj.7600863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins JR, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CY, Wong J, Coppinger JA, Seki A, Yates JR 3rd, Fang G. DDA3 recruits microtubule depolymerase Kif2a to spindle poles and controls spindle dynamics and mitotic chromosome movement. J Cell Biol. 2008;181:255–267. doi: 10.1083/jcb.200711032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline SL, Cheeseman IM, Hori T, Fukagawa T, Desai A. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin Kif24 interacts with CP110 to remodel microtubules and regulate ciliogenesis. Cell. 2011;145:914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]
- Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- Malik R, Lenobel R, Santamaria A, Ries A, Nigg EA, Korner R. Quantitative analysis of the human spindle phosphoproteome at distinct mitotic stages. J Proteome Res. 2009;8:4553–4563. doi: 10.1021/pr9003773. [DOI] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Sillje HH, Sauer G, Nigg EA, Korner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Santamaria A, Nagel S, Sillje HH, Nigg EA. The spindle protein CHICA mediates localization of the chromokinesin Kid to the mitotic spindle. Curr Biol. 2008;18:723–729. doi: 10.1016/j.cub.2008.04.041. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Cooper J, Domnitz S, Moore AT, Rankin KE, Wagenbach M, Wordeman L. In vitro and in vivo analysis of microtubule-destabilizing kinesins. Methods Mol Biol. 2007;392:37–49. doi: 10.1007/978-1-59745-490-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24:2169–2179. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Gan EC, Desai A, Mitchison TJ, Kline-Smith SL. The microtubule-destabilizing kinesin XKCM1 is required for chromosome positioning during spindle assembly. Curr Biol. 2002;12:1885–1889. doi: 10.1016/s0960-9822(02)01227-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopus kinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR 3rd. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, Leblanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinger JS, Qiu M, Yang G, Kapoor TM. A nonmotor microtubule binding site in kinesin-5 is required for filament crosslinking and sliding. Curr Biol. 2011;21:154–160. doi: 10.1016/j.cub.2010.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JP, Vleugel M, Liu D, Yates JR 3rd, Lampson MA, Fukagawa T, Cheeseman IM. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L. How kinesin motor proteins drive mitotic spindle function: lessons from molecular assays. Semin Cell Dev Biol. 2010;21:260–268. doi: 10.1016/j.semcdb.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.