Basal bodies act as anchorage sites for cilia and resist forces generated by ciliary beating. Bld10/Cep135 is a basal body assembly factor. A novel role is found for Bld10 in stabilizing basal bodies to resist cilia-generated forces. Bld10 achieves this by stabilizing the A- and C-tubules of the basal body triplet microtubule blades.
Abstract
Basal bodies nucleate, anchor, and organize cilia. As the anchor for motile cilia, basal bodies must be resistant to the forces directed toward the cell as a consequence of ciliary beating. The molecules and generalized mechanisms that contribute to the maintenance of basal bodies remain to be discovered. Bld10/Cep135 is a basal body outer cartwheel domain protein that has established roles in the assembly of nascent basal bodies. We find that Bld10 protein first incorporates stably at basal bodies early during new assembly. Bld10 protein continues to accumulate at basal bodies after assembly, and we hypothesize that the full complement of Bld10 is required to stabilize basal bodies. We identify a novel mechanism for Bld10/Cep135 in basal body maintenance so that basal bodies can withstand the forces produced by motile cilia. Bld10 stabilizes basal bodies by promoting the stability of the A- and C-tubules of the basal body triplet microtubules and by properly positioning the triplet microtubule blades. The forces generated by ciliary beating promote basal body disassembly in bld10Δ cells. Thus Bld10/Cep135 acts to maintain the structural integrity of basal bodies against the forces of ciliary beating in addition to its separable role in basal body assembly.
INTRODUCTION
Centrioles and basal bodies (CBBs) function as microtubule-organizing centers in eukaryotic cells. Centrioles are part of the centrosome that organizes the interphase microtubule aster and the poles of spindles for chromosome segregation. During the G0/G1 phase of the cell cycle, the centriole converts its function from a centriole to a basal body, which organizes the microtubules of the ciliary axoneme. Cilia are cellular extensions that perform diverse roles in signaling and motility. Ciliary dysfunction causes human disorders, including syndromes known as ciliopathies, which exhibit a wide range of symptoms, including cystic kidneys, mental retardation, microcephaly, polydactyly, respiratory illness, and retinal degeneration (Saeki et al., 1984; Marszalek et al., 2000; Nachury et al., 2007; Kuijpers and Hoogenraad, 2011; Hussain et al., 2012). Many of the gene mutations that cause ciliopathies encode proteins that localize to and function at the basal body (Nigg and Raff, 2009; Bettencourt-Dias et al., 2011; Garcia-Gonzalo and Reiter, 2012). Moreover, it remains to be determined whether these mutations affect ciliary function or whether they are also important in centriolar functions (Delaval et al., 2011). Thus understanding the mechanics for how CBBs assemble and nucleate centrosomes and cilia, respectively, is important to understanding this class of human maladies.
CBBs are anchorage sites for both centrosomes and cilia. Centrioles withstand mechanical forces to facilitate spindle positioning in cells and to segregate the duplicated genome during anaphase. This is evident when cells are injected with function-blocking antibodies that abrogate centriole stability, causing centrosome disruption (Bobinnec et al., 1998). Moreover, the imbalance of microtubule-generated forces causes the fragmentation of centrosomes (Abal et al., 2005). Consistent with a role for centrioles in withstanding mechanical forces, basal bodies resist mechanical forces from ciliary beating (Kunimoto et al., 2012). This is facilitated by the anchorage of basal bodies within the plasma membrane and the cortical architecture. Despite the important role for CBBs in withstanding mechanical forces, the molecular and structural mechanisms by which this stabilization occurs are poorly defined.
The major structural component of CBBs is microtubules. These dynamic polymers form into nine modified triplet microtubule blades that are organized into a stable cylindrical structure. The stabilization of CBBs begins during new CBB formation. CBB assembly is initiated by the formation of the cartwheel. The cartwheel forms nine symmetrically spaced spokes that radiate outward from a central hub and attach to the triplet microtubules (Dippell, 1968; Sorokin, 1968; Allen, 1969; Dirksen, 1971; Cavalier-Smith, 1974). The inner domain containing the central hub and spokes is hypothesized to establish the ninefold symmetry (Kitagawa et al., 2011b; van Breugel et al., 2011). The outer domain, which links to the inner domain, is responsible for the nucleation of the triplet microtubules (Inclan and Nogales, 2001; Raynaud-Messina et al., 2004; Guichard et al., 2010). A limited number of protein components are known to associate with these two domains. Sas6 and Sas5/Ana2/STIL are required to assemble the inner cartwheel domain (Habedanck et al., 2005; Stevens et al., 2010; Kitagawa et al., 2011a; Arquint et al., 2012; Vulprecht et al., 2012). Bld10/Cep135 and Poc1 are the only known outer cartwheel proteins (Matsuura et al., 2004; Hiraki et al., 2007; Pearson et al., 2009b). Both domains are absolutely essential for the assembly of the CBB. Triplet microtubules are organized so that the microtubule minus ends orient toward the proximal cartwheel structure and the plus ends are positioned at a distal cap called the terminal plate or transition zone (Dippell, 1968; Sorokin, 1968; Allen, 1969; Dirksen, 1971). This cap, containing a protein complex of CP110 and interacting proteins, ensures proper centriolar length and stabilization (Chen et al., 2002; Pearson et al., 2007; Spektor et al., 2007; Tsang et al., 2008; Schmidt et al., 2009).
A limited number of protein components are known to stabilize these structures. Centrobin is essential for new assembly and the stabilization of existing centrioles by promoting centriole microtubule stability (Zou et al., 2005; Jeong et al., 2007; Gudi et al., 2011). Tetrahymena γ-tubulin loss causes the instability of basal bodies (Shang et al., 2002). In addition, mouse spermatocyte centriole disintegration correlates with centrin loss from the CBB, and mutations in the C-terminal domain of TtCen1 cause basal body instability (Manandhar et al., 1999; Stemm-Wolf et al., 2005; Vonderfecht et al., 2011). Function-blocking antibodies that target glutamylation modifications on centriolar tubulin disrupt centriole and centrosome stability (Abal et al., 2005). In Tetrahymena, glutamylation of microtubules is required for efficient basal body assembly (Wloga et al., 2008). These studies suggest that the stability of the CBB microtubules is important for the maintenance of these structures. The conserved CBB microtubule cylinder wall and outer cartwheel domain protein, Poc1, is also required to maintain centrioles (Pearson et al., 2009b). However, centriole assembly occurs without Poc1, suggesting that Poc1 and its localization domains on CBBs have important functions in CBB maintenance.
We searched for additional basal body stability components. In particular, we explored the possibility that outer cartwheel domain proteins stabilize CBBs. Bld10/Cep135 is a conserved outer cartwheel domain protein that is required for basal body assembly and for stable integration of Sas6 protein at basal bodies (Hiraki et al., 2007; Kleylein-Sohn et al., 2007; Nakazawa et al., 2007; Carvalho-Santos et al., 2010; Jerka-Dziadosz et al., 2010; Roque et al., 2012). Chlamydomonas and Paramecium Bld10 makes up the tips of the cartwheel spokes (Hiraki et al., 2007; Jerka-Dziadosz et al., 2010). In addition, Drosophila Bld10 is a microtubule-associated protein that stabilizes microtubules and is required for assembly of the axoneme central doublet microtubules, suggesting that this component has multiple roles during the basal body life cycle (Blachon et al., 2009; Mottier-Pavie and Megraw, 2009; Carvalho-Santos et al., 2010; Carvalho-Santos et al., 2012). Because Bld10/Cep135 is hypothesized to connect the inner cartwheel to the outer cartwheel and binds and stabilizes microtubules, we hypothesized that, like Poc1, it also functions to stabilize the entire CBB. Tetrahymena Bld10/Cep135 (TtBld10) is required to stabilize and maintain existing basal bodies in addition to its established role in assembly of basal bodies. We identify a novel role for Bld10/Cep135 in stabilizing existing basal bodies to resist the forces produced by ciliary beating. In summary, TtBld10 has two separable and important functions in CBB assembly and maintenance.
RESULTS
TtBld10 is a conserved basal body cartwheel outer domain protein
The outer cartwheel domain protein Poc1 stabilizes basal bodies (Pearson et al., 2009b). Because CBB stability is essential for its function, we searched for other outer cartwheel proteins that act as basal body stability factors. Bld10/Cep135 was a good candidate because it also localizes to the outer cartwheel domain of Chlamydomonas and Paramecium basal bodies (Hiraki et al., 2007; Jerka-Dziadosz et al., 2010). A single BLD10/CEP135 orthologue exists in the Tetrahymena thermophila genome and will be referred to as TtBLD10. TtBLD10 encodes a 171-kDa protein, TtBld10. As with other Bld10 family members, the protein contains extensive coiled-coil domains with two conserved regions called conserved region 1 (CR1) and conserved region 2 (CR2; Carvalho-Santos et al., 2010; Hodges et al., 2010; Supplemental Figure S1A). TtBld10 shares 42% protein sequence similarity with the human Bld10 homologue, Cep135 (Supplemental Figure S1, B and C). Consistent with a role in basal body assembly, TtBLD10 is expressed similarly to other core Tetrahymena basal body components (Miao et al., 2009).
We localized TtBld10 to determine whether TtBld10 shares a similar localization profile to that observed in other organisms. TtBld10-mCherry was expressed under the control of its native TtBLD10 promoter in Tetrahymena cells. We found that TtBld10-mCherry localizes with TtCen1 (Stemm-Wolf et al., 2005) at all basal bodies and remains localized to basal bodies at all stages of the cell cycle (Figure 1A). Moreover, TtBld10-mCherry did not localize in cilia (Figure 1B). Similar to other organisms tested, TtBld10 is a CBB protein.
FIGURE 1:
TtBld10 localizes to the basal body outer cartwheel domain. (A) Both TtBld10 and TtCen1 colocalize at basal bodies. T. thermophila cells expressing TtBld10-mCherry (red) were stained for the basal body marker TtCen1 (green; Stemm-Wolf et al., 2005). (B) TtBld10-mCherry (red) localizes to the base of cilia labeled using α-tubulin antibodies (green). (C) Colocalization of TtBld10 and TtCen1 within a single basal body. Longitudinal view of TtBld10-mCherry (red) stained with basal body marker TtCen1 (green). Schematic drawing represents the known TtCen1 ultrastructural localization based on immuno–electron microscopy (Stemm-Wolf et al., 2005; green) and the predicted TtBld10 (red) localization based on its localization relative to TtCen1 (green). TtCen1 at the basal body proximal end is asymmetrically localized relative to the longitudinal axis of the basal body, and this generates an offset between TtCen1 and TtBld10 at the cartwheel. Scale bar, 10 μm (A, B), 1 μm (C). (D) IEM localization of TtBld10 at the basal body outer cartwheel domain. Micrographs of TtBld10-GFP localization in longitudinal sections through a basal body (left) at 15,500×. Right, TtBld10 localization within the cartwheel domain of cross-sectional views from the proximal end of the basal body. Arrows denote sites of TtBld10 localization. Red dots represent localization based on the relative distribution of 25 gold particles to represent the total quantified gold label. Longitudinal section, n = 254 gold particles in 90 basal bodies; cross section, n = 64 gold particles in 14 basal bodies. Numbers on schematics denote the domains used to quantify TtBld10 localization.
To determine where TtBld10 localizes within the basal body architecture, we colocalized TtBld10-mCherry relative to TtCen1, which localizes asymmetrically to the proximal end and to the site of kinetodesmal fiber attachment of basal bodies (Stemm-Wolf et al., 2005). TtBld10 localized to the proximal end of the basal body, coincident with the site of the cartwheel (Figure 1C), and was not found along the length of the basal body. We then localized TtBld10-mCherry relative to green fluorescent protein (GFP)–TtSas6a, which localizes to the central hub of the cartwheel (Kilburn et al., 2007; Supplemental Figure S1D). TtBld10 localizes peripherally to TtSas6a, consistent with its localization to the outer cartwheel domain. Next, we used immuno–electron microscopy (IEM) to determine the ultrastructural localization of TtBld10-GFP. Consistent with our fluorescence data, the majority (73%) of TtBld10 immuno-gold label localized to the basal body cartwheel (Figure 1D). We found that a small fraction (14%) of TtBld10 localizes to the terminal plate (Figure 1D). Drosophila Bld10 localizes to the distal end of basal bodies and is required to form the central doublet microtubules of motile cilia (Blachon et al., 2009; Mottier-Pavie and Megraw, 2009; Carvalho-Santos et al., 2010, 2012). This raises the possibility that TtBld10, like DmBld10, may be required for central doublet formation. However, axoneme central doublet microtubules were normal in Ttbld10Δ cells (unpublished data), suggesting that TtBld10 does not regulate the axoneme central pair microtubules in Tetrahymena as it does in Drosophila. To determine where TtBld10-GFP localizes within the cartwheel, we quantified the relative immuno-gold distribution in cross-sectional views of the cartwheel. TtBld10 associates with the ends of the cartwheel spokes (39%) and triplet microtubules (44%; Figure 1E). This is consistent with Chlamydomonas and Paramecium Bld10, which also localizes to the outer cartwheel and spoke tips (Hiraki et al., 2007; Jerka-Dziadosz et al., 2010). Taken together, the results indicate that TtBld10 localization is predominantly restricted to the basal body outer cartwheel.
Ttbld10Δ causes the loss of basal bodies
Prior studies addressing the function of Bld10/Cep135 were limited to using hypomorphic alleles and knockdowns because a complete BLD10 genomic knockout was not accessible. Here we created, for the first time, a complete genomic knockout of BLD10 (Ttbld10Δ). Ttbld10Δ was induced by mating two Ttbld10Δ heterokaryon knockout strains to produce progeny with complete macronuclear Ttbld10Δ (Hai et al., 2000). Control cells were generated by mating wild-type cells with either heterokaryon knockout strain, which results in phenotypically normal cells. These control cells are referred to as TtBLD10. Ttbld10Δ cells exhibit deleterious phenotypes that are common among basal body and ciliary mutants (Brown et al., 1999; Pearson and Winey, 2009). Ttbld10Δ causes cellular lethality. To determine the number of cellular divisions that Ttbld10Δ cells underwent before death, we quantified growth rates of Ttbld10Δ cell populations. Ttbld10Δ cells averaged 3.1 ± 0.7 divisions before division ceased (n = 3; Supplemental Figure S2). In addition to a reduced rate of cellular growth, the qualitative rate of cellular swimming was reduced in Ttbld10Δ cells. Moreover, Ttbld10Δ cells exhibited a decrease in directed forward motility, as seen by an increase in lateral cellular movement relative to forward movement. In summary, TtBld10 is required for cell viability, motility, and normal cell cycle progression.
Because Ttbld10Δ cells exhibited similar, albeit stronger, mutant phenotypes compared with Ttpoc1Δ cells, we next asked whether TtBld10 loss, like TtPoc1 loss, affects the total number of basal bodies per cell (Pearson et al., 2009b). We quantified the frequency of α-TtCen1–stained basal bodies at 0, 12, 24, and 48 h after TtBLD10 knockout. Basal body number per cell declined and organization was increasingly disrupted with time after TtBLD10 knockout (Figure 2, A and C). Moreover, the basal bodies of the oral apparatus disassembled in Ttbld10Δ cells (Figure 2A). To confirm that the loss of TtBLD10 was responsible for the observed phenotypes, we rescued the Ttbld10Δ cells by reintroducing the wild-type TtBLD10 gene after knockout. Basal body number and organization were both restored by the reintroduction of TtBLD10 (Figure 2B). Thus TtBld10, like other basal body components, is required for normal basal body frequency and organization (Stemm-Wolf et al., 2005; Culver et al., 2009; Pearson et al., 2009b; Pearson and Winey, 2009).
FIGURE 2:
TtBLD10 knockout causes a loss of basal bodies. (A) TtBld10 is required to maintain the normal number and organization of basal bodies. Images describe a time course of basal bodies after Ttbld10Δ. Basal bodies are visualized by anti-TtCen1 staining. By 12 h after knockout, a significant number of basal bodies are diminished. The remaining basal bodies are less organized. This phenotype is exacerbated with time. Scale bar, 10 μm. (B) Ttbld10Δ phenotypes are rescued by reintroducing the TtBLD10 gene into cells. (C) The frequency of basal bodies per unit length significantly dropped after Ttbld10Δ knockout. The number of basal bodies was quantified per 10 μm in TtBLD10 control and Ttbld10Δ cells. *p < 0.0001, n = 3 separate experiments of 100 basal body rows (20 cells) for each condition.
Bld10 is required for new basal body assembly
The inhibition of new basal body assembly causes a progressive reduction of basal bodies at each cell division. This is because basal bodies are segregated to the future cells without producing new ones to maintain the normal complement of basal bodies. In Tetrahymena, new basal bodies form anteriorly to existing basal bodies. These basal bodies form as doublets after assembly (one new and one old). The newly assembled basal body then moves anteriorly away from the old basal body while maturing into a basal body that nucleates a cilium (Allen, 1969; Ng and Frankel, 1977). To determine whether new basal bodies are formed in Ttbld10Δ cells, we visualized both old and new basal bodies. Old basal bodies were labeled with a marker that surrounds mature basal bodies that resembles the K-antigen (Williams et al., 1990), here called K-like antigen (Kl-Ag). This is colocalized with the panspecific basal body marker centrin (TtCen1; Stemm-Wolf et al., 2005). Kl-Ag levels increase with basal body maturity (Supplemental Figure S3A). New basal body assembly is evident as basal body doublets with TtCen1 staining but no Kl-Ag staining at the anteriorly positioned basal body (Figure 3, A and B). Approximately 18% of the total basal bodies were newly duplicated at both 0 and 24 h for control cells (Figure 3; green arrow). The proportion of newly assembled basal bodies dramatically decreased from 18% (0 h) to 3% (24 h) in Ttbld10Δ cells (Figure 3C). Moreover, we were unable to identify new basal body assembly in Ttbld10Δ cells at later time points (36 and 48 h). We predict that the basal body assembly observed in Ttbld10Δ cells (0, 12, and 24 h) is the result of residual, yet reduced, TtBld10 protein after knockout. The amount of new assembly decreases and is not detectable by 36 h. Thus TtBld10 is required for new basal body assembly.
FIGURE 3:

TtBld10 is required for basal body assembly. (A) Fluorescence images of all basal bodies (anti-TtCen1; green) and mature basal bodies (Kl-Ag; red) in control and Ttbld10Δ cells 12 h after knockout. Newly assembled basal bodies are evident as the anteriorly positioned basal body of two closely positioned basal bodies (doublet) that is only labeled by TtCen1 (green arrows; no Kl-Ag). Mature basal bodies are posteriorly positioned and labeled with both TtCen1 and Kl-Ag. Basal body disassembly is evident by Kl-Ag–stained foci without associated TtCen1 (red arrowhead). Scale bar, 10 μm. (B) Insets are from images in A at 0 and 12 h after TtBLD10 knockout. New basal bodies are denoted with green arrows. Panel width, 2 μm. (C) The frequency of new basal body assembly decreases with time after TtBLD10 knockout. Quantification of the relative frequency of new basal body assembly after TtBld10 loss. *p < 0.0001, n = 3 separate experiments of 20 cells each, counting 50 basal bodies per cell for each condition.
Of interest, we observed Kl-Ag–stained foci without TtCen1 staining in Ttbld10Δ cells (Figure 3A, red arrowhead). The original K-Ag antibody recognizes domains within the membrane skeleton surrounding basal bodies but does not directly stain basal bodies (Williams et al., 1990). Kl-Ag accumulates at these sites with time after basal body assembly and remains at these sites even in the absence of basal bodies in cycling cells (Figure 3A, red arrowhead, and Supplemental Figure S3A). Thus loss of basal bodies does not result in the loss of Kl-Ag staining in cycling cells. We find Kl-Ag staining in the absence of TtCen1, and these foci mark locations where basal bodies once existed and are now sites of basal body disassembly. Furthermore, basal body disassembly occurred at immature basal bodies, as judged by the reduced level of Kl-Ag staining relative to Kl-Ag levels in mature basal bodies. This suggests that the basal bodies disassembled before their complete maturation. TtBld10 is, therefore, required not only for new basal body assembly, but also to stabilize developing basal bodies.
Bld10 is required to stabilize and maintain basal bodies
Basal bodies in G1-arrested cells have full levels of Kl-Ag, which indicates that they are mature. We tested whether mature basal bodies (as judged by Kl-Ag) disassemble in the absence of TtBld10. Ttbld10Δ cells were arrested in G1, so that cell division and new basal body assembly was repressed. A reduced number of basal bodies was observed in G1-arrested Ttbld10Δ cells compared with control cells (Figure 4, A and B). Moreover, the decrease in basal body number was time dependent, suggesting that basal bodies did not immediately disassemble, but instead there was a temporal loss in basal bodies. These results further indicate that TtBld10 has an important role in maintaining and stabilizing existing basal bodies.
FIGURE 4:

Bld10 is necessary for the maintenance of basal bodies. (A) Basal bodies disassemble in G1 cell cycle–arrested Ttbld10Δ cells. Immunofluorescence images of cell cycle–arrested Ttbld10Δ cells stained for the basal body marker anti-TtCen1 at 0 and 12 h after TtBld10 loss. (B) The number of basal bodies temporally decreases in Ttbld10Δ but not control cells. The number of basal bodies was quantified per 10 μm in TtBLD10 control and Ttbld10Δ cells. *p < 0.0001, n = 3 separate experiments of 100 basal body rows (20 cells) for each condition. (C) Kl-Ag remains at basal bodies after basal body disassembly and marks basal body disassembly sites. Insets, G1-arrested TtBLD10 control and Ttbld10Δ cells at 0 and 12 h post–TtBLD10 knockout. Cells are stained with the basal body marker TtCen1 (green) and the mature basal body marker Kl-Ag (red). Red arrow denotes site of basal body disassembly. Inset length, 10 μm. (D) Frequency of basal body disassembly quantified at 0, 12, and 24 h. Disassembly events are identified as Kl-Ag foci without TtCen1 staining. No disassembly was observed in TtBLD10 control cells *p < 0.0001, n = 3 separate experiments of 20 cells each counting 50 basal bodies per cell for each condition.
To directly visualize basal body disassembly, we used Kl-Ag to mark the site of basal bodies that existed before TtBld10 knockout. We colocalized Kl-Ag with TtCen1 in Ttbld10Δ cells that were arrested in G1. Disassembly events (Kl-Ag staining without TtCen1 staining) in Ttbld10Δ cells were observed in a low but significant (p < 0.001) fraction of the basal body pool relative to TtBLD10 cells (Figure 4, C and D). We hypothesize that this low fraction is due to a transient Kl-Ag signal after basal body disassembly in G1-arrested cells, and this makes disassembly events difficult to capture. Moreover, the progression of Kl-Ag disassembly was visualized in G1-arrested Ttbld10Δ cells (Supplemental Figure S3B). Basal body and Kl-Ag disassembly was not observed in control cells. Thus TtBld10 is required to maintain both immature and mature basal bodies.
TtBld10 promotes triplet microtubule stability
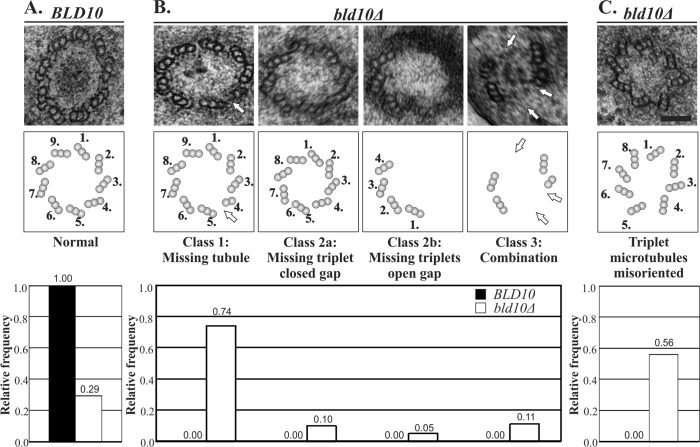
Because TtBld10 is required for the assembly of new basal bodies and the stability of existing basal bodies, we hypothesized that TtBld10 regulates the core CBB structure. In particular, we postulated that TtBld10 regulates the triplet microtubules that comprise CBBs. The A-tubules of triplet microtubule blades are attached to the central hub of the cartwheel via a spoke linkage. After assembly and attachment of the A-tubule to the cartwheels, the B- and C-tubules are then sequentially added (Dippell, 1968; Guichard et al., 2010). To determine whether this organization is affected by TtBld10 loss, we visualized the basal body ultrastructure in Ttbld10Δ cells. Ttbld10Δ cells at 12 h postknockout were prepared for transmission electron microscopy (TEM; Dahl and Staehelin, 1989; Meehl et al., 2009; Winey et al., 2012). Seventy-one percent of the Ttbld10Δ basal bodies exhibited defects that were not found in control basal bodies (Figure 5; n = 100 basal bodies). The Ttbld10Δ-associated defects in triplet microtubules were categorized into three classes. Seventy-four percent of the microtubule-defective Ttbld10Δ basal bodies were missing a single or multiple tubules of the microtubule triplet blade, causing basal bodies to have only doublet or singlet microtubules in at least one of the basal body triplet microtubule positions (Figure 5B, class 1). In cases in which a doublet was present instead of a triplet the C-tubule was missing most commonly (65% of class 1 mutants); however, a significant fraction of A-tubules were also missing (22% of class 1 mutants). In cases in which a singlet was present instead of a triplet the B-and C-tubules were always missing. In 56% of class 1 basal bodies, the missing tubule was lost from the entire basal body length based on serial sections. In the remaining class 1 samples, the basal body proximal end contained all three tubules of the microtubule triplet, and the segment distal to the cartwheel exhibited a decreased number of tubules, generating doublet or singlet morphology. The instability of tubules of the basal body triplet (class 1) was the major defect found in Ttbld10Δ basal bodies.
FIGURE 5:
Triplet microtubule assembly and stability defects in Ttbld10Δ cells. (A) Electron micrograph image of a cross-sectional view of a TtBLD10 control cell (top). Middle, representative schematic of the basal body structure. Bottom, relative frequency observed for the normal morphology class for TtBLD10 (black bars) and Ttbld10Δ (white bars) basal bodies. The relative frequency is indicated above each bar. (B) Ttbld10Δ mutant phenotypes are distributed among three classes that were observed 12 h after Ttbld10Δ knockout. Top, representative electron micrographs of basal body cross-sectional views in bld10Δ cells. Class 1 consists of basal bodies with missing tubules of the triplet microtubules where an A-, C-, A-B-, or B-C-tubule is missing. Class 2 is characterized by missing microtubule triplets through the entire length of the basal body. Class 2a contains basal bodies with missing triplet microtubule blades that do not leave a gap but reduce the basal body diameter. Class 2b contains basal bodies that have a gap at the site of the missing triplet microtubule blade. Class 3 is a combination of class 1 and class 2 phenotypes. None of the mutant phenotypes were observed in control TtBLD10 cells. Middle, schematics of the mutant phenotypes. Bottom, relative frequency observed for each mutant class. (C) The triplet microtubule blades are commonly misoriented in Ttbld10Δ mutant basal bodies at 12 h post–TtBLD10 knockout. Magnification, 30,000×. Scale bar, 0.1 μm. n = 100 basal bodies.
Fifteen percent of defective Ttbld10Δ basal bodies were missing at least one triplet microtubule through the entire length of the basal body (Figure 5B, class 2). A similar basal body phenotype was found in Paramecium cells where PtBld10a was depleted by RNA interference (Jerka-Dziadosz et al., 2010). The Tetrahymena phenotype was separated into two subcategories. Class 2a consists of basal bodies with triplet microtubules that conform to the missing gap, thereby decreasing the basal body diameter (Figure 5B, class 2a; 10% of defective basal bodies). Class 2b consists of a missing triplet microtubule that produces a gap in the ninefold symmetry (Figure 5B, class 2b; 5% of defective basal bodies). In class 2a, the ninefold symmetry was likely never established, and this represents a basal body assembly defect. In class 2b, the missing triplet microtubule likely established correct ninefold symmetry; however, microtubule attachment and stability was disrupted.
The third class of Ttbld10Δ basal body defects (class 3; 11% of defective Ttbld10Δ basal bodies) consisted of a combination of the first two classes, in which tubules of microtubule triplets and complete triplets are missing (Figure 5B). Thus triplet microtubule stabilization and organization are lost in Ttbld10Δ cells.
In addition to disruption of the individual triplet microtubule structure, we find that the majority (56%) of basal bodies in Ttbld10Δ cells display triplet microtubule orientation defects. These defects are characterized by off-axis positioning of entire triplet microtubule blades (Figure 5C). Moreover, 77% of basal bodies that possess class 1–3 phenotypes also exhibit triplet microtubule orientation defects. These defects, in conjunction with the triplet microtubule structural defects (classes 1–3), suggest that TtBld10 stabilizes the structure and orientation of the basal body triplet microtubules.
TtBld10 protein stably incorporates during basal body assembly and maturation
The disassembly of both immature (new, daughter) basal bodies in Ttbld10Δ cycling cells and mature (old, mother) basal bodies in Ttbld10Δ G1-arrested cells led us to ask when TtBld10 is incorporated at basal bodies to perform its functions. We quantified the relative amounts and the timing of when TtBld10 protein incorporates during the assembly of new basal bodies and the maintenance of existing basal bodies. TtBld10-mCherry levels were variable, depending on the age of the basal body. The basal body age was estimated based on distance between the daughter and mother basal bodies. Newly assembled, daughter basal bodies were closely positioned near the mother basal body, whereas older daughter basal bodies were more physically separated from their mother basal bodies. Mature basal bodies had increased levels of TtBld10 protein compared with immature basal bodies (Figure 6, A and B). At the time when the separation of mother and daughter basal bodies can be resolved, newly assembled basal bodies have a mean Bld10-mCherry fluorescence intensity of ∼40% of the mother TtBld10-mCherry fluorescence intensity. As the basal body increases in separation from its mother, an increased level of TtBld10-mCherry is observed until a maximum protein level is reached that is equal to that of the mother basal body. Mature basal bodies did not increase or decrease in fluorescence with time, suggesting that once the basal body reaches the maximum level of TtBld10-mCherry, this level remains constant (Figure 6B). A similar incorporation behavior was observed using GFP-TtBld10 (Supplemental Figure S4, A and B). Thus TtBld10 protein levels accumulate as basal bodies temporally mature.
FIGURE 6:

TtBld10 protein stably accumulates at basal bodies. (A) TtBld10-mCherry fluorescence intensities are low at newly formed basal bodies and increase as the daughter basal bodies separate from their mother basal bodies. The separation distance corresponds to basal body age, so that as basal bodies mature, the level of TtBld10 increases. Insets show mature TtBld10-mCherry–labeled basal bodies (top) and a newly assembled basal body (middle basal body; bottom). Arrows and arrowheads denote mature and newly formed basal bodies, respectively. Scale bar, 5 μm. (B) Quantification of the basal body–localized TtBld10-mCherry fluorescence intensity relative to the separation of newly assembled daughter basal bodies from their mature mother basal body. Fluorescence intensity is defined as the ratio between the daughter and the mother basal body fluorescence. (C) TtBld10 is a stable basal body component. FRAP of TtBld10-mCherry. A mature basal body (arrow) was photobleached, and fluorescence recovery was visualized over time. Low fluorescence recovery (<0.05) was observed, indicating that TtBld10-mCherry stably assembles at basal bodies. The relative fluorescence intensity was quantified over time. (D) TtBld10-mCherry matures to its maximum level before the basal body becomes ciliated. Arrows and arrowheads denote ciliated and unciliated basal bodies, respectively. Schematic shows that only basal bodies with full levels of TtBld10 nucleate a cilium. Scale bar, 1 μm. n = 50 basal bodies.
Fluorescence recovery after photobleaching (FRAP) was used to determine whether the incorporated TtBld10 protein is stably associated with basal bodies. Single, TtBld10-mCherry–labeled mature basal bodies were photobleached, and live-cell imaging was used to visualize the kinetics of protein redistribution and fluorescence recovery. A low level of TtBld10-mCherry fluorescence recovery was observed, indicating that TtBld10 is stably bound to the basal body (percentage recovery, <5%; Figure 6C). When we performed longer recovery experiments of up to 10 min, we also did not observe a significant fluorescence recovery (unpublished data). This supports the model that once TtBld10 protein incorporates, it remains at basal bodies.
To determine whether TtBld10 levels were recruited to maximum levels before ciliogenesis, we quantified the levels of TtBld10-mCherry at basal bodies possessing a cilium. TtBld10 protein is at its maximum level in ciliated basal bodies (Figure 6D, arrows). This suggests that only mature basal bodies, as judged by TtBld10 levels, produce a cilium. Because mature basal bodies disassemble in arrested Ttbld10Δ cells, we hypothesized that disassembly is due to ciliary beating. The instability of basal bodies without TtBld10 led us to two simplified models for TtBld10 function. First, TtBld10 is required early during new basal body assembly for assembly and stabilization. Second, TtBld10 stabilizes mature basal bodies to resist forces generated by beating cilia.
Decreased ciliary beating rescues basal body instability in Ttbld10Δ cells
TtBld10 loss causes mature basal bodies to disassemble, and ciliated basal bodies have a maximum level of TtBld10 protein (Figure 4, A and B, and Figure 6). This suggests that TtBld10 is required to resist the forces created by ciliogenesis or ciliary beating. We assessed whether ciliary beating promotes the disassembly of basal bodies in Ttbld10Δ cells. Tetrahymena cells use cilia-dependent forces to move. To test whether Ttbld10Δ basal bodies disassemble as a result of the forces produced by ciliary beating, we inhibited ciliary beating in G1 cell cycle–arrested Ttbld10Δ cells at 12 and 24 h post–TtBLD10 knockout by treatment with NiCl2. We found that inhibition of ciliary beating significantly rescued basal body frequency and organization in Ttbld10Δ cells compared with control cells (Figure 7A). These data suggest that TtBld10 stabilizes basal bodies to resist cilia-dependent forces.
FIGURE 7:
Cilia-generated forces destabilize basal bodies in Ttbld10Δ cells. (A) Ciliary inhibition rescues basal body disassembly in Ttbld10Δ. G1-arrested TtBLD10 and Ttbld10Δ cells at 12 and 24 h postknockout and treated with NiCl2 to inhibit ciliary beating. Quantification of the number of basal bodies per 10 μm is shown below for control, 12 and 24 h post–TtBLD10 knockout. Scale bar, 5 μm. n = 3 separate experiments of 100 basal body rows (20 cells) for each condition. *p < 0.001. (B) Increased viscosity of media causes an increased basal body disassembly in Ttbld10Δ. G1-arrested TtBLD10 and Ttbld10Δ cells at 24 h postknockout and treated with 5% PEO to increase the viscosity of the media. Quantification of the number of basal bodies per 10 μm is shown below. Scale bar, 5 μm. n = 3 separate experiments of 100 basal body rows (20 cells) for each condition. *p < 0.01.
In addition to inhibiting ciliary beating, we increased the physical resistance or drag force of the media in which cells swim by increasing the media viscosity with 5% polyethylene oxide (PEO). G1 cell cycle–arrested Ttbld10Δ cells in 5% PEO disassemble basal bodies at a significantly greater level compared with Ttbld10Δ cells swimming in normal media (Figure 7B). The frequency of basal bodies was not affected in control cells treated with 5% PEO, suggesting that TtBld10 is necessary to stabilize basal bodies from the increased drag force.
DISCUSSION
Basal bodies are organizing centers for the axoneme microtubules of cilia. To resist the extreme mechanical forces on them, basal bodies must stabilize the microtubules that comprise these structures. The triplet microtubule blades are important targets for basal body maintenance. We demonstrate that TtBld10 localizes to the base of the triplet microtubules at the outer cartwheel domain and that TtBld10 acts as a stability factor for basal bodies. TtBld10 is required to stabilize both immature and mature basal bodies. Our data indicate that an early-incorporating population of TtBld10 protein is necessary to assemble and stabilize immature basal bodies. Subsequently a later-incorporating population of TtBld10 protects mature basal bodies against the forces of ciliary beating.
Bld10 stabilizes basal bodies
Both immature and mature basal bodies disassemble in the absence of TtBld10. We hypothesize that immature basal bodies disassemble in cells progressing through the cell cycle because they do not have a full complement of TtBld10 protein at the time of TtBLD10 knockout. In contrast, G1-arrested cells have a full complement of TtBld10 and Kl-Ag and are ciliated at the time of TtBLD10 knockout, suggesting that mature basal bodies also disassemble in Ttbld10Δ cells (Figures 4 and 6D). However, in order to completely mature, CBBs progress through more than a single cell cycle (Nigg, 2007). At the time of starvation there is a heterogeneous population of basal bodies in each cell. Some basal bodies have progressed through one or more cell cycles, whereas a second population of basal bodies has not progressed through a single cell cycle. Thus the number of cell cycles through which the basal bodies have progressed may affect TtBld10-dependent basal body maintenance. This suggests that TtBld10 requires additional cell cycle progression to become competent to fulfill its stabilization duties.
Alternatively, TtBld10 may exhibit protein turnover that was not detected by our FRAP experiments. Loss of TtBld10 protein at mature basal bodies by turnover may render Ttbld10Δ basal bodies unstable. The latter possibility is unlikely because we do not observe disassembly of all basal bodies over time. Instead, the level of basal body disassembly plateaus after ∼36 h (Figure 4D and unpublished data). Another alternative is that basal bodies disassemble by an age-dependent turnover mechanism that is accelerated in Ttbld10Δ cells. Our data indicate that the disassembly of basal bodies is not caused by increased basal body turnover. If basal bodies were to disassemble as a result of normal basal body turnover, we would expect continued reduction of basal bodies to zero after Ttbld10Δ knockout. Moreover, basal bodies do not disassemble in control cells that are arrested in G1, as would be expected if there was age-dependent basal body turnover (Figure 4). These studies show that basal bodies are stable structures that rarely turn over or disassemble in normal Tetrahymena cells. Once assembled, basal bodies survive for many cell generations.
Bld10 is required for the triplet microtubule structure
Our electron microscopy (EM) analyses of Tetrahymena cells at 12 h after TtBLD10 knockout captured several classes of basal body abnormalities. These defects range from loss of tubules of the triplet microtubule blades (class 1), to loss of complete triplet microtubule blades (class 2), to loss of triplet microtubule blade orientation (Figure 5). All of these phenotypes suggest that TtBld10 is important to establish and stabilize the basal body triplet microtubules. Seventy-four percent of the defective Ttbld10Δ basal bodies were missing either the A- or C-tubules of the triplet microtubules. It is interesting to note that C. elegans centrioles lack triplet microtubules and possess a ninefold array of singlet microtubules (Wolf et al., 1978). A BLD10/CEP135 orthologue is not found in the C. elegans genome (Carvalho-Santos et al., 2010; Hodges et al., 2010), suggesting that triplet microtubules, and in particular C-tubules, require Bld10/Cep135 for their formation and/or maintenance. Chlamydomonas δ- and ε-tubulin mutants disrupt the tubules of triplet microtubule blades (Dutcher et al., 2002; Fromherz et al., 2004). These phenotypes were specific to the B- and C-tubules, and the majority (65%) of the tubules lost in Ttbld10Δ cells were C-tubules. Of interest, the A-tubule was missing in 22% of the tubule-missing basal bodies (class 1). The A-tubule loss is surprising because this tubule forms first during CBB assembly (Dippell, 1968; Guichard et al., 2010). Subsequently the B- and C-tubules form from the A-tubule, and the B-tubule shares protofilaments with the A-tubule (Tilney et al., 1973; Li et al., 2012). This argues that the basal body triplet microtubules formed first but that our images capture a state of A- and C-tubule disassembly. This may be an intermediate step toward basal body disassembly in the absence of TtBld10. Carvalho-Santos et al. (2012) showed that Drosophila Bld10 binds to and stabilizes the central pair microtubules of the ciliary axoneme. We hypothesize that cartwheel-localized Tetrahymena Bld10 binds to and stabilizes the tubules of the triplet microtubule blades that make up the basal body structure.
Paramecium PtBld10a knockdown by RNA interference causes the loss of entire triplet microtubule blades, similar to the Ttbld10Δ class 2 phenotype (Figure 5; Jerka-Dziadosz et al., 2010). However, the loss of individual tubules within triplet microtubule blades of Paramecium basal bodies was not observed. This is the major phenotype observed in Tetrahymena basal bodies 12 h after TtBLD10 knockout. This distinction between phenotypes in these two ciliate organisms might be explained by incomplete knockdown of PtBld10 or unique functions for the second Paramecium Bld10 (PtBld10b) paralogue in regulating individual microtubule stability. Like Paramecium, Chlamydomonas Bld10 stabilizes entire triplet microtubule blades, and individual tubule loss was not observed in Chlamydomonas Bld10 truncation mutants (Hiraki et al., 2007). An alternative explanation is that our ultrastructural studies captured an earlier phenotype (loss of individual tubules) after TtBld10 loss that was not observed in the prior studies. Ultimately, all of these studies indicate that Bld10/Cep135 is necessary to stabilize the triplet microtubule blades.
The orientation of the remaining intact microtubule blades is abrogated in the majority of Ttbld10Δ basal bodies (Figure 5C). Perturbation of blade orientation may be caused by disruption of the linkage between triplet microtubule blades and the inner cartwheel and/or by disruption of the linkage between microtubule blades themselves. Bld10 comprises the spoke tip, which connects the inner cartwheel domain to the outer cartwheel domain (Figure 1; Hiraki et al., 2007; Jerka-Dziadosz et al., 2010). Although we do not see shorter cartwheel spokes in Ttbld10Δ basal bodies, many Ttbld10Δ basal bodies are oval shaped instead of circular when viewed as a cross section through the cartwheel (unpublished data). The oval shape of Ttbld10Δ basal bodies suggests that there is a loss of the support between the inner and the outer cartwheel domains. In addition, TtBld10 may function as a linker between the A- and C-tubules of adjacent triplet microtubule blades (A–C linker) that are required to connect one triplet microtubule blade to its nearest neighbor. The coiled-coil domain of TtBld10, when fully elongated, can stretch from the spoke tips to well beyond the C-tubule of an adjacent microtubule blade, and it is possible that TtBld10 acts at both the A–C microtubule linkage and the cartwheel spoke tip as a scaffold to organize the outer cartwheel domain and to link with the inner cartwheel.
In summary, our ultrastructural analyses suggest that Bld10/Cep135 has a unique function in regulating microtubule stabilization and orientation at the basal body cartwheel. This provides the foundation for basal body stability, and when defective the tubules disassemble, leading to a collapse of the system.
Bld10 stabilizes basal bodies to resist cilia-generated forces
Basal bodies require TtBld10 to resist forces produced by ciliary beating (Figure 7A). This is evident by the rescue of G1-arrested Ttbld10Δ cells to normal basal body numbers by reducing ciliary beating with NiCl2 (Larsen and Satir, 1991). Thus TtBld10, in addition to promoting new basal body assembly, is necessary to stabilize basal bodies as they anchor and counteract mechanical forces generated from beating cilia. On the basis of studies showing that Drosophila Bld10 binds to and stabilizes the axoneme central pair microtubules (Carvalho-Santos et al., 2012), we hypothesize that TtBld10 functions to stabilize the basal body triplet microtubules. Moreover, it remains to be determined whether Bld10/Cep135 stabilizes centrioles in mitotic cells to render them resistant to the mechanical forces produced by the mitotic spindle.
We next tested whether increasing the drag force on cilia and, therefore, basal bodies increased the level of basal body disassembly in Ttbld10Δ cells. By increasing the viscosity of the media in which the cells swim, we increased basal body disassembly in Ttbld10Δ cells but not in control cells (Figure 7B). The increased viscosity elevates the total drag force and load on cilia, causing increased basal body disassembly. The amplified force load from increased viscosity is complicated by a decrease in ciliary beat frequency that decreases the viscous load on cilia and basal bodies. However, we postulate that increased load is still experienced by these basal bodies. Moreover, metachronal beating is disrupted by increasing the viscosity of the medium, and this might somehow contribute to basal body disassembly (Machemer, 1972). We predict that a significant force increase disrupts TtBld10-dependent basal body stabilization, which is important for efficient ciliary beating, which requires basal body anchorage and mechanical force resistance. In metazoans, this is important for mucociliary clearance, in which cilia of multiciliated epithelial cells beat in a metachronal manner to move a highly viscous mucus layer (Hill et al., 2010). We predict that the loss of basal body resistance to such forces may contribute to respiratory illness.
In summary, Bld10/Cep135 performs two distinct roles at basal bodies. Bld10 is essential for the assembly and stabilization of basal bodies. We show here that Bld10 stabilizes basal bodies so that they can withstand the mechanical forces exerted by undulating cilia.
MATERIALS AND METHODS
T. thermophila cell culture
All strains used were grown in 2% SPP media (2% protease peptone, 0.2% glucose, 0.1% yeast extract, and 0.003% Fe-EDTA) to mid–log phase at 30°C, unless otherwise indicated. Cells were considered mid–log phase at a density of ∼3 × 105 cells/ml as determined using a Coulter Counter Z1 (Beckman Coulter, Brea, CA) or a hemocytometer. To arrest cells in G1 of the cell cycle, cells were washed and resuspended in starvation media (10 mM Tris-HCl, pH 7.4). A small fraction of cells become arrested in G2; however, the majority of cells that are the focus of this study arrest in G1.
Perturbations that affect the rate of ciliary-based swimming (0.1 and 0.5 mM NiCl2, dependent on the experiment; Larsen and Satir, 1991; or 5% PEO) were introduced at the time of Ttbld10Δ with drug selection (0 h). Ciliary inhibition by treatment of cells with NiCl2 was visually confirmed by decreased cellular swimming rate.
TtBLD10 identification and conservation
The T. thermophila BLD10 gene was identified by searching the genome for the reciprocal best BLAST hit to human CEP135 and Chlamydomonas BLD10. We identified TTHERM_01164140 to fit this criterion. A Phylogenic tree was generated using Web-based phylogeny tree generation (www.phylogeny.fr; Dereeper et al., 2008). Sequences were aligned using MUSCLE (Edgar, 2004). After alignment, gaps were removed, and the phylogenetic tree was generated using the maximum-likelihood method in the PhyML program (version 3.0 aLRT; Castresana, 2000; Guindon and Gascuel, 2003; Anisimova and Gascuel, 2006). One hundred bootstrap replicates were performed. Graphical representation was generated using TreeDyn (Chevenet et al., 2006; Supplemental Figure S1).
Plasmids
A TtBld10-mCherry strain was constructed by transforming cells with p4T2-1:BLD10:mCherry. This cassette integrates at the endogenous TtBLD10 locus and remains under the control of the endogenous promoter. p4T2-1:BLD10:mCherry was generated by PCR amplifying (5′-CGggtaccGAAGTTGATAACTGTAAGTATAC and 5′-CGgaattcATTATTATTTTTAGATTTAGTAGAGCTTGGAGG) and cloning the final 1.5 kb of TtBLD10 without the TGA stop codon into p4T2-1-mCherryLAP (Winey et al., 2012). A 0.9-kb fragment downstream of the TGA stop codon (5′-CGggatccTGCTCCATTCATATTTCTAT and 5′-CGgagctcAATATATCTACTCTAGCTTC) was then cloned into the plasmid to create p4T2-1:BLD10:mCherry. This plasmid contains NEO2 drug selection. A similar strain was also created using the same strategy to produce a C-terminal GFP fusion called p4T2-1:BLD10:GFP.
The bld10Δ strain was generated using a BLD10-knockout cassette (pbld10::NEO2). This construct was created by inserting 0.9 kb downstream of the TGA stop codon (5′-CGggatccTGCTCCATTCATATTTCTAT and 5′-CGgagctcAATATATCTACTCTAGCTTC) as described into p4T2-1-mCherryLAP to make p4T2-1-mCherryLAP:BLD10DS. A 1.3-kb fragment upstream of the ATG start codon was then PCR amplified (5′-CGggtaccGACTTTGGACAATTTTGCTG and 5′-CGctcgagGTGATGCTAATTTGCTTCG) and cloned into p4T2-1-mCherryLAP that contains a NEO2-knockout cassette. This was cloned into a site that removes the mCherryLAP sequence but maintains the NEO2 cassette for drug selection.
To rescue the Ttbld10Δ strain, we generated a genomic clone of TtBLD10 with DNA flanking the BLD10 open reading frame (pBS:BLD10). A genomic TtBLD10 fragment with flanking sequence of 7.7 kb was PCR amplified (5′-CGggatccGACTTTGGACAATTTTGCTG and 5′-CGgagctcAATATATCTACTCTAGCTTC) and cloned into pBluescript (Studier and Moffatt, 1986). The rescue cassette was then released from the vector backbone using a SacI and XhoI double digest before transformation into Tetrahymena cells.
Macronuclear transformation
GFP and mCherry fusion proteins were inserted into the macronucleus by biolistic transformation (Bruns and Cassidy-Hanley, 2000). Transformed cells were selected by using paromomycin (200 μg/ml) to select for the NEO2 gene (Gaertig et al., 1994; Hai et al., 2000). To increase the copy number of TtBld10-mCherry, the cells were selectively assorted by incrementally increasing the dose of paromomycin.
Generation of the Ttbld10Δ strain
Complete genomic knockout of TtBLD10 (Ttbld10Δ) was achieved by using targeted homologous recombination to delete the germline micronuclear TtBLD10 gene using biolistic bombardment (Bruns and Cassidy-Hanley, 2000). The TtBLD10 locus was targeted using a cassette in which the entire open reading frame was replaced by the NEO2 (Hai et al., 2000). After confirming the generation of a heterozygous micronuclear Ttbld10Δ by genetic and PCR-based strategies, we performed star crosses to generate two homozygous micronuclear knockout strains of different mating types (MATII and MATVI). Two confirmed micronuclear knockout heterokaryon strains, Bld10KO1A.4 and Bld10KO 1A.4.1A, were created. Mating of Bld10KO1A.4 and Bld10KO1A.4.1A results in resistance of the progeny to paromomycin, and this is coincident with the removal of TtBLD10 from the expressed macronucleus. Moreover, drug selection with paromomycin ensures that cells that did not pass through mating to knockout TtBLD10 were eliminated. For these studies, there is a small fraction of contaminating wild-type cells (∼5%) that do not mate and die from paromomycin treatment after ∼24 h post–drug treatment. These cells were distinguishable from the bld10Δ cells and were excluded from the experiments in both fluorescence and immuno-EM studies.
BLD10 knockout
The described homozygous heterokaryon strains Bld10KO1A.4 (MATVI) and Bld10KO 1A.4.1A (MATII) were grown to mid–log phase (∼3 × 105 cells/ml) and then washed and maintained in starvation medium (10 mM Tris-HCl, pH.7.4) for 14 h. Equal numbers of cells were mixed in large flasks to induce mating. At 10 h after mating initiation an equal volume of 2× SPP medium was added to the mating cells (>90% mating efficiency). Paromomycin (200 μg/ml) was then added 7 h after media addition, and this time point was the starting or 0-h time point of TtBLD10 knockout.
Light microscopy
Fluorescence imaging and FRAP were performed as previously described (Pearson et al., 2009b). In this study, a Nikon Ti Eclipse inverted microscope (Nikon, Melville, NY) with a Nikon 100× PlanApo numerical aperture 1.4 objective was used. Images were captured with an Andor iXon electron-multiplying charge-coupled device (CCD) 888E camera (Andor Technologies, Belfast, United Kingdom). Image analysis and quantification was performed using NIS Elements imaging software (Nikon). All images taken were acquired at room temperature. Acquisition times for images ranged between 50 and 500 ms, depending on the experiment.
The number of basal bodies per unit length or basal body frequency was quantified by counting the number of basal bodies (marked by anti–TtCen1 staining) along a 10-μm segment of a ciliary row or kinety within the medial half of the Tetrahymena cell (Pearson et al., 2009a). Basal body frequency was quantified in interphase cells only to ensure that the number of basal bodies per 10 μm was constant. A minimum of 100 data points were quantified for each condition by measuring at least five ciliary rows and a minimum of 20 cells. All experiments were performed in triplicate.
Fluorescence intensities of TtBld10-mCherry at basal bodies for the maturation of TtBld10 levels and FRAP were quantified as previously described (Pearson et al., 2009b). Briefly, a 5 × 5 pixel region was placed over the basal body of interest, and then four surrounding 5 × 5 pixel regions were used to quantify the background fluorescence levels. The mean of the four background regions was measured, and this was subtracted from the basal body fluorescence value. This corrected value was determined for each data point.
We used FRAP to quantify the dynamics of TtBld10 protein association with Tetrahymena basal bodies using methods similar to those described in Pearson et al. (2009a). Briefly, TtBld10-mCherry–labeled basal bodies were photobleached by administering a focused 561-nm laser light pulsed on for a 45-ms exposure to bleach ∼75% of the total basal body–localized TtBld10-mCherry signal. The recovery of fluorescence was then followed in a subsequent time course using fluorescence imaging. The limited recovery that was observed with TtBld10-mCherry precluded us from measuring a final rate of recovery or an accurate assessment of the recovery amount (∼5% recovery). The photobleaching caused by image acquisition was corrected for by quantifying the loss in fluorescence intensity of a neighboring, unbleached basal body and correcting the bleached levels. To further account for photobleaching, we also performed longer-interval recovery studies by taking the time course out to 5–10 min with limited image acquisition until the end of the time course. Under these conditions, we still did not observe additional recovery of TtBld10-mCherry. These results suggest that TtBld10 stably incorporates and remains at the basal body.
Immunofluorescence
Immunofluorescence was performed as previously described (Cole et al., 2002). Cells were washed once in PHEM buffer (60 mM 1,4-piperazinediethanesulfonic acid, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM EGTA, 2 mM MgCl2, pH 6.9) and then fixed in formaldehyde fixative (1% paraformaldehyde, 0.2% Triton X-100, PHEM buffer) for 1 min. Cells then were washed three times in either 0.5% boiled goat serum (BDS) or 0.1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS; BDS-PBS or BSA-PBS, respectively) and incubated in primary antibody in 5% BDS-PBS or 1% BSA-PBS for 24 h at 4°C. The primary antibodies that we used were anti-TtCen1 (Stemm-Wolf et al., 2005), anti-centrin clone (20H5; Millipore; Uzawa et al., 1995), anti–Kl antigen (10D12; Shang et al., 2002; Pearson et al., 2009b), and anti–α-tubulin (DM1A; Sigma-Aldrich, St. Louis, MO; Blose et al., 1984). After primary antibody incubation, cells were washed three times and then incubated in secondary antibody (Alexa Fluor 594 or 488 goat anti–rabbit immunoglobulin G [IgG], Alexa Fluor 594 or 488 goat anti–mouse IgG; Invitrogen, Carlsbad, CA) diluted in 5% BDS-PBS or 1% BSA-PBS. After secondary incubation, cells were washed three times and adhered to poly-l-lysine–coated coverslips. Coverslips were mounted using Citifluor mounting media (Ted Pella, Redding, CA) and sealed using clear nail polish.
Transmission electron microscopy
A Tetrahymena strain expressing an endogenous C-terminal TtBld10-GFP fusion was grown to mid–log phase and then prepared for IEM using high-pressure freezing and freeze substitution (HPF-FS; Dahl and Staehelin, 1989; Meehl et al., 2009). Rabbit-generated anti-GFP antibodies were used to localize TtBld10-GFP, followed by incubation with anti-rabbit secondary antibodies conjugated to 15-nm gold particles. TtBld10 was then localized in 60-nm sections using TEM. Images were collected using a Philips CM10 electron microscope (Philips, Eindhoven, Netherlands) equipped with a Gatan BioScan2 CCD camera (Gatan, Pleasanton, CA).
For structural analyses of Ttbld10Δ basal body defects, Ttbld10Δ and control cells were subjected to HPF-FS after 12 h of TtBLD10 knockout. Samples were prepared as previously described (Pearson et al., 2009b). Images were acquired using an FEI Tecnai G2 (FEI, Hillsboro, OR) equipped with a Gatan Ultrascan digital camera. All images were processed for figures using Corel Draw (Corel, Mountain View, CA).
Supplementary Material
Acknowledgments
We thank Joe Frankel for his insightful discussions and important contributions to the manuscript, Jennifer Deluca and Brian Mitchell for critical comments on the manuscript, Kristin Dahl for comments and discussion, and Dot Dill for EM work. We thank Alex Stemm-Wolf for the anti-TtCen1 antibody and discussions and Joe Frankel for the Kl-antigen antibody (10D12). M.W. is funded by National Institute of General Medical Sciences Grant GM074746 and C.G.P. is funded by National Institute of General Medical Sciences Grant GM099820, the Pew Biomedical Scholars Program, the Boettcher Webb-Waring Foundation, and the American Cancer Society.
Abbreviations used:
- CBB
centriole and basal body
- FRAP
fluorescence recovery after photobleaching
- IEM
immuno–electron microscopy
- Kl-Ag
K-like antigen
- TEM
transmission electron microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-08-0577) on October 31, 2012.
REFERENCES
- Abal M, Keryer G, Bornens M. Centrioles resist forces applied on centrosomes during G2/M transition. Biol Cell. 2005;97:425–434. doi: 10.1042/BC20040112. [DOI] [PubMed] [Google Scholar]
- Allen RD. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol. 1969;40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Arquint C, Sonnen KF, Stierhof YD, Nigg EA. Cell-cycle-regulated expression of STIL controls centriole number in human cells. J Cell Sci. 2012;125:1342–1352. doi: 10.1242/jcs.099887. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, Avidor-Reiss T. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182:133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blose SH, Meltzer DI, Feramisco JR. 10-nm filaments are induced to collapse in living cells microinjected with monoclonal and polyclonal antibodies against tubulin. J Cell Biol. 1984;98:847–858. doi: 10.1083/jcb.98.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Marsala C, Kosoy R, Gaertig J. Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol Biol Cell. 1999;10:3081–3096. doi: 10.1091/mbc.10.10.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns PJ, Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000;62:501–512. doi: 10.1016/s0091-679x(08)61553-8. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, et al. BLD10/CEP135 is a microtubule-associated protein that controls the formation of the flagellum central microtubule pair. Dev Cell. 2012;23:412–424. doi: 10.1016/j.devcel.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M. Stepwise evolution of the centriole-assembly pathway. J Cell Sci. 2010;123:1414–1426. doi: 10.1242/jcs.064931. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evolution. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ES, Stuart KR, Marsh TC, Aufderheide K, Ringlien W. Confocal fluorescence microscopy for Tetrahymena thermophila. Methods Cell Biol. 2002;70:337–359. doi: 10.1016/s0091-679x(02)70009-5. [DOI] [PubMed] [Google Scholar]
- Culver BP, Meehl JB, Giddings TH, Jr, Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol Biol Cell. 2009;20:1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl R, Staehelin LA. High-pressure freezing for the preservation of biological structure: theory and practice. J Electron Microsc Technique. 1989;13:165–174. doi: 10.1002/jemt.1060130305. [DOI] [PubMed] [Google Scholar]
- Delaval B, Bright A, Lawson ND, Doxsey S. The cilia protein IFT88 is required for spindle orientation in mitosis. Nat Cell Biol. 2011;13:461–468. doi: 10.1038/ncb2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen ER. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J Cell Biol. 1971;51:286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE, multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromherz S, Giddings TH, Jr, Gomez-Ospina N, Dutcher SK. Mutations in alpha-tubulin promote basal body maturation and flagellar assembly in the absence of delta-tubulin. J Cell Sci. 2004;117:303–314. doi: 10.1242/jcs.00859. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Reiter JF. Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol. 2012;197:697–709. doi: 10.1083/jcb.201111146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudi R, Zou C, Li J, Gao Q. Centrobin-tubulin interaction is required for centriole elongation and stability. J Cell Biol. 2011;193:711–725. doi: 10.1083/jcb.201006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard P, Chretien D, Marco S, Tassin AM. Procentriole assembly revealed by cryo-electron tomography. EMBO J. 2010;29:1565–1572. doi: 10.1038/emboj.2010.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hai B, Gaertig J, Gorovsky MA. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 2000;62:513–531. doi: 10.1016/s0091-679x(08)61554-x. [DOI] [PubMed] [Google Scholar]
- Hill DB, Swaminathan V, Estes A, Cribb J, O'Brien ET, Davis CW, Superfine R. Force generation and dynamics of individual cilia under external loading. Biophys J. 2010;98:57–66. doi: 10.1016/j.bpj.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain MS, et al. A truncating mutation of CEP135 causes primary microcephaly and disturbed centrosomal function. Am J Hum Genet. 2012;90:871–878. doi: 10.1016/j.ajhg.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inclan YF, Nogales E. Structural models for the self-assembly and microtubule interactions of gamma-, delta- and epsilon-tubulin. J Cell Sci. 2001;114:413–422. doi: 10.1242/jcs.114.2.413. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Lee J, Kim K, Yoo JC, Rhee K. Characterization of NIP2/centrobin, a novel substrate of Nek2, and its potential role in microtubule stabilization. J Cell Sci. 2007;120:2106–2116. doi: 10.1242/jcs.03458. [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M, Gogendeau D, Klotz C, Cohen J, Beisson J, Koll F. Basal body duplication in Paramecium, the key role of Bld10 in assembly and stability of the cartwheel. Cytoskeleton (Hoboken) 2010;67:161–171. doi: 10.1002/cm.20433. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Fluckiger I, Gonczy P. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci. 2011a;124:3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011b;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kuijpers M, Hoogenraad CC. Centrosomes, microtubules and neuronal development. Mol Cell Neurosci. 2011;48:349–358. doi: 10.1016/j.mcn.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Kunimoto K, et al. Coordinated ciliary beating requires Odf2-mediated polarization of basal bodies via basal feet. Cell. 2012;148:189–200. doi: 10.1016/j.cell.2011.10.052. [DOI] [PubMed] [Google Scholar]
- Larsen J, Satir P. Analysis of Ni(2+)-induced arrest of Paramecium axonemes. J Cell Sci. 1991;99:33–40. doi: 10.1242/jcs.99.1.33. [DOI] [PubMed] [Google Scholar]
- Li S, Fernandez JJ, Marshall WF, Agard DA. Three-dimensional structure of basal body triplet revealed by electron cryo-tomography. EMBO J. 2012;31:552–562. doi: 10.1038/emboj.2011.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machemer H. Ciliary activity and the origin of metachrony in Paramecium, effects of increased viscosity. J Exp Biol. 1972;57:239–259. doi: 10.1242/jeb.57.1.239. [DOI] [PubMed] [Google Scholar]
- Manandhar G, Simerly C, Salisbury JL, Schatten G. Centriole and centrin degeneration during mouse spermiogenesis. Cell Motil Cytoskeleton. 1999;43:137–144. doi: 10.1002/(SICI)1097-0169(1999)43:2<137::AID-CM5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Liu X, Roberts EA, Chui D, Marth JD, Williams DS, Goldstein LS. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl JB, Giddings TH, Jr, Winey M. High pressure freezing, electron microscopy, and immuno-electron microscopy of Tetrahymena thermophila basal bodies. Methods Mol Biol. 2009;586:227–241. doi: 10.1007/978-1-60761-376-3_12. [DOI] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PloS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20:2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Ng SF, Frankel J. 180 degrees rotation of ciliary rows and its morphogenetic implications in Tetrahymena pyriformis. Proc Natl Acad Sci USA. 1977;74:1115–1119. doi: 10.1073/pnas.74.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication, of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Culver BP, Winey M. Centrioles want to move out and make cilia. Dev Cell. 2007;13:319–321. doi: 10.1016/j.devcel.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Giddings TH, Jr, Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol Biol Cell. 2009a;20:904–914. doi: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Osborn DP, Giddings TH, Jr, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J Cell Biol. 2009b;187:905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Winey M. Basal body assembly in ciliates, the power of numbers. Traffic. 2009;10:461–471. doi: 10.1111/j.1600-0854.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud-Messina B, Mazzolini L, Moisand A, Cirinesi AM, Wright M. Elongation of centriolar microtubule triplets contributes to the formation of the mitotic spindle in gamma-tubulin-depleted cells. J Cell Sci. 2004;117:5497–5507. doi: 10.1242/jcs.01401. [DOI] [PubMed] [Google Scholar]
- Roque H, Wainman A, Richens J, Kozyrska K, Franz A, Raff JW. Drosophila Cep135/Bld10 maintains proper centriole structure but is dispensable for cartwheel formation. J Cell Sci. 2012 doi: 10.1242/jcs.113506. DOI 10.1242/jcs.113506. [DOI] [PubMed] [Google Scholar]
- Saeki H, Kondo S, Morita T, Sasagawa I, Ishizuka G, Koizumi Y. Immotile cilia syndrome associated with polycystic kidney. J Urol. 1984;132:1165–1166. doi: 10.1016/s0022-5347(17)50080-4. [DOI] [PubMed] [Google Scholar]
- Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin SP. Centriole formation and ciliogenesis. Aspen Emphysema Conf. 1968;11:213–216. [PubMed] [Google Scholar]
- Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Bryan J, Bush DJ, Fujiwara K, Mooseker MS, Murphy DB, Snyder DH. Microtubules: evidence for 13 protofilaments. J Cell Biol. 1973;59:267–275. doi: 10.1083/jcb.59.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. CP110 suppresses primary cilia formation through its interaction with CEP290: a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa M, Grams J, Madden B, Toft D, Salisbury JL. Identification of a complex between centrin and heat shock proteins in CSF-arrested Xenopus oocytes and dissociation of the complex following oocyte activation. Dev Biol. 1995;171:51–59. doi: 10.1006/dbio.1995.1259. [DOI] [PubMed] [Google Scholar]
- van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- Vonderfecht T, Stemm-Wolf AJ, Hendershott M, Giddings TH, Jr, Meehl JB, Winey M. The two domains of centrin have distinct basal body functions in Tetrahymena. Mol Biol Cell. 2011;22:2221–2234. doi: 10.1091/mbc.E11-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulprecht J, et al. STIL is required for centriole duplication in human cells. J Cell Sci. 2012;125:1353–1362. doi: 10.1242/jcs.104109. [DOI] [PubMed] [Google Scholar]
- Williams NE, Honts JE, Kaczanowska J. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 1990;109:935–942. doi: 10.1242/dev.109.4.935. [DOI] [PubMed] [Google Scholar]
- Winey M, Stemm-Wolf AJ, Giddings TH, Jr, Pearson CG. Cytological analysis of Tetrahymena thermophila. Methods Cell Biol. 2012;109:357–378. doi: 10.1016/B978-0-12-385967-9.00013-X. [DOI] [PubMed] [Google Scholar]
- Wloga D, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryotic Cell. 2008;7:1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf N, Hirsh D, McIntosh JR. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans. J Ultrastructure Res. 1978;63:155–169. doi: 10.1016/s0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- Zou C, Li J, Bai Y, Gunning WT, Wazer DE, Band V, Gao Q. Centrobin, a novel daughter centriole-associated protein that is required for centriole duplication. J Cell Biol. 2005;171:437–445. doi: 10.1083/jcb.200506185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.