Using macrophages overexpressing or reducing SNAP-23, this study shows that SNAP-23 is implicated in phagosome formation and maturation, presumably by mediating SNARE-based membrane traffic. Indeed, a conformational change in SNAP-23 structure based on FRET signal is observed on the phagosome membrane of cells overexpressing the lysosomal SNARE VAMP7.
Abstract
Synaptosomal associated protein of 23 kDa (SNAP-23), a plasma membrane–localized soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE), has been implicated in phagocytosis by macrophages. For elucidation of its precise role in this process, a macrophage line overexpressing monomeric Venus–tagged SNAP-23 was established. These cells showed enhanced Fc receptor–mediated phagocytosis. Detailed analyses of each process of phagocytosis revealed a marked increase in the production of reactive oxygen species within phagosomes. Also, enhanced accumulation of a lysotropic dye, as well as augmented quenching of a pH-sensitive fluorophore were observed. Analyses of isolated phagosomes indicated the critical role of SNAP-23 in the functional recruitment of the NADPH oxidase complex and vacuolar-type H+-ATPase to phagosomes. The data from the overexpression experiments were confirmed by SNAP-23 knockdown, which demonstrated a significant delay in phagosome maturation and a reduction in uptake activity. Finally, for analyzing whether phagosomal SNAP-23 entails a structural change in the protein, an intramolecular Förster resonance energy transfer (FRET) probe was constructed, in which the distance within a TagGFP2-TagRFP was altered upon close approximation of the N-termini of its two SNARE motifs. FRET efficiency on phagosomes was markedly enhanced only when VAMP7, a lysosomal SNARE, was coexpressed. Taken together, our results strongly suggest the involvement of SNAP-23 in both phagosome formation and maturation in macrophages, presumably by mediating SNARE-based membrane traffic.
INTRODUCTION
Professional phagocytes, particularly macrophages, neutrophils, and dendritic cells, play a significant role in host defense to internalize, kill, and digest many species of bacteria and other immunoglobulin (Ig)-opsonized targets. Phagocytic processing by these cells is composed of phagosome formation and maturation. In addition, with the exception of neutrophils, phagocytes exhibit antigen-presenting properties by major histocompatibility complex (MHC) class I and II molecules (Jutras and Desjardins, 2005; Haas, 2007). Phagocytosis is triggered by the activation of various types of cell-surface receptors. For example, in Ig-mediated phagocytosis, Fc receptors (FcRs) cluster at sites at which they contact the Ig-opsonized surface of large foreign particles; this encounter induces cytoskeletal reorganization, resulting in the formation of pseudopod extensions and phagocytic cups that engulf the particle (Swanson and Hoppe, 2004). The newly formed phagosomes progressively undergo maturation to generate phagolysosomes by acquiring reactive oxygen species (ROS) that kill pathogens and an acidic environment to digest them by several lysosomal hydrolases (Haas, 2007). These are complicated processes involving the fusion and fission of various types of membranes (Vieira et al., 2002; Desjardins, 2003) and are mediated by as yet poorly understood molecular mechanisms.
In eukaryotic cells, biological membrane fusion is generally executed by fusogenic soluble N-ethylmaleimide–sensitive factor (NSF) attachment protein (SNAP) receptors (SNAREs). These membrane-anchoring proteins localize to and function in diverse endomembrane systems in which docking and fusion between membranes take place (Chen and Scheller, 2001). SNARE proteins are characterized by a common stretch of 60–70 amino acids known as the SNARE motif, located adjacent to the membrane anchor domain. Two SNARE subgroups are distinguished. The syntaxin and SNAP-25 (which contains two SNARE motifs) families contain a conserved glutamine (Q) residue at a central position of the SNARE motif and are therefore called Qa- and Qbc-SNAREs, respectively. Members of the vesicle-associated membrane protein (VAMP, also called synaptobrevin) family contain a conserved arginine (R) residue at the same position and are therefore called R-SNAREs (Fasshauer et al., 1998). The proper assembly of SNARE motifs (Qa, Qbc, and R), which are extended from the SNARE proteins in both the vesicle and the target membranes, leads to the formation of the SNARE complex (a parallel four-helix bundle) and finally to membrane fusion and cargo delivery (Fasshauer et al., 1998; Jahn and Scheller, 2006).
During phagocytosis in macrophages, membrane fusions between the plasma membrane and endocytic organelles or the endoplasmic reticulum (ER) are mediated by VAMP3 (recycling endosomes; Bajno et al., 2000), VAMP7 (late endosomes and lysosomes; Braun et al., 2004), and syntaxin 18, D12, and Sec22b (ER; Hatsuzawa et al., 2006, 2009). VAMP8 (late endosomes) plays an inhibitory role in phagocytic function of dendritic cells (Ho et al., 2008). Syntaxin 4 is a cognate SNARE partner (a functional pairing partner for the formation of a SNARE complex) of VAMP3 during phagocytosis and in association with tumor necrosis factor (TNF)-α secretion in activated macrophages (Murray et al., 2005). After internalizing the foreign particle, the phagosome continuously fuses with vesicles and/or those compartments related to endocytic organelles. Syntaxin 13 (recycling endosomes) and syntaxin 7 (late endosomes and lysosomes) are required for the early and late stage of phagosome maturation, respectively (Collins et al., 2002). However, a specific cognate SNARE partner on the plasma membrane and phagosome has yet to be determined, and the functional involvement of SNARE proteins in phagosome formation and maturation in macrophages remains to be elucidated (Stow et al., 2006).
SNAP-23 is a ubiquitously expressed SNARE protein that belongs to the SNAP-25 family (Jahn and Scheller, 2006). Like SNAP-25, SNAP-23 is present in many types of cells at the plasma membrane domain, with a little intracellular localization, and mediates exocytosis of secretory vesicles. However, in neutrophils, SNAP-23 is mainly distributed on cytoplasmic granules (specific and gelatinase-rich tertiary granules) and mediates the secretion of these granules (Martin-Martin et al., 2000; Mollinedo et al., 2006). More recently, it was reported that membrane fusion between specific granules and phagosomes is involved in a SNAP-23–mediated trafficking pathway in human neutrophils (Uriarte et al., 2011).
In macrophages, SNAP-23 is a major candidate Qbc-SNARE protein located on the plasma membrane, but little is known about its involvement in membrane fusion with intracellular organelles during phagocytosis (Stow et al., 2006). In this paper, we report that the overexpression of SNAP-23 critically regulates the entire process of phagocytosis through enhanced ROS production and acidification within phagosome in J774 macrophages. These observations were supported by the results of SNAP-23 knockdown experiments, which showed a delay in phagosome maturation and a reduction in uptake activity. Interestingly, we found that SNAP-23 assumes distinct conformations on phagosomal membranes, as determined by intramolecular Förster resonance energy transfer (FRET) analyses in cells overexpressing VAMP7. Thus we conclude that SNAP-23 regulates phagosome formation and maturation by mediating membrane fusion in macrophages.
RESULTS
Stable overexpression of mVenus-SNAP-23 in J774 macrophages
In macrophages, SNAP-23, a plasma membrane-localized SNARE protein (Supplemental Figure S1A), together with syntaxin 4 was shown to be involved in phagocytosis associated with TNF-α secretion (Murray et al., 2005). However, as there is no direct evidence that SNAP-23 participates in phagocytosis, we established a line of J774 macrophages stably expressing SNAP-23 N-terminally fused to the fluorescent protein mVenus (J774/mV-S23 cells). It is well known that SNAP-25 is a target of the neurotoxin botulinum type A (BoNT/A), which cleaves a nine–amino acid peptide off the protein's C-terminus, resulting in the inhibition of neurotransmitter release (Schiavo et al., 1993). Although BoNT/A does not cleave SNAP-23 at the corresponding site, SNAP-23ΔC8, in which eight C-terminal amino acids are cleaved from SNAP-23, thus mimicking the action of BoNT/A on SNAP-25, has been analyzed as a dominant-negative mutant in exocytosis (Kawanishi et al., 2000; Huang et al., 2001). Western blot analysis revealed that the expression of mV-S23 and mVenus-SNAP-23ΔC8 (mV-S23Δ8) was nearly twice as high as that of endogenous SNAP-23 without alterations in the expression levels of CD64 (FcR Ia) and syntaxin 4 (Figure 1A). Immunofluorescence analysis showed that both of the tagged proteins were predominantly located at the plasma membrane (Figure S1B) quite similarly to endogenous SNAP-23 (Figure S1A) and around phagosomes containing Texas Red–conjugated zymosan particles opsonized with anti-zymosan rabbit IgGs (Figure 1B). The presence of those molecules was confirmed at a higher resolution, using structured illumination microscopy (SIM). In that experiment, the phagocytized particles were nonfluorescent beads previously opsonized with IgGs. Incubation of the beads with the fluorescently labeled secondary antibody allowed only beads outside the cells to be visualized with Alexa Fluor 594 dye (Figure 1C).
FIGURE 1:
Stable overexpression of mVenus-SNAP-23 in J774 macrophages. (A) Total lysates from J774 macrophages stably expressing mVenus (mV), mVenus-SNAP-23 (mV-S23), and mVenus-SNAP-23ΔC8 (mV-S23ΔC8) were analyzed by Western blotting using the indicated antibodies. (B) Confocal live-cell imaging of J774 cells expressing mV-S23 or mV-S23ΔC8 during the ingestion of Texas Red–conjugated zymosan A (TR-zymosan) particles previously opsonized with rabbit anti-zymosan IgGs. The asterisks denote the phagosomes containing Texas Red–conjugated zymosan A particles. Scale bar: 10 μm. (C) Three-dimensional SIM image of J774/mV-S23 cells during the ingestion of IgG-opsonized latex beads (3.0 μm in diameter). Beads outside the cells were stained with an anti-IgG Alexa Fluor 594 antibody. The asterisks denote the bead-containing phagosomes. (D) J774/mV, J774/mV-S23, and J774/mV-S23ΔC8 cells were incubated with IgG-opsonized FITC-zymosan particles, and the efficiency of association and FcR-mediated phagocytosis were then measured as described in Materials and Methods. Arbitrary fluorescence units were normalized to the maximum value obtained for mV cells within the same experiment, defined as 100%. Data presented are the mean ± SE of three independent experiments.
We then examined the effects of mV-S23 overexpression on FcR-mediated phagocytosis by measuring the uptake of fluorescein isothiocyanate (FITC)-labeled zymosan particles opsonized with IgG (see Materials and Methods). Overexpression of either mV-S23 or mV-S23ΔC8 had no effect on the efficiency of phagocytosis (Figure 1D, bottom), nor was there a change in the association of IgG-opsonized FITC-zymosan particles in J774 cells (Figure 1D, top) compared with that of mVenus (mV). Because mV-S23 and mV-S23ΔC8 are suspected to not be functional in J774 cells, we investigated whether these cells are stimulated in ROS release (caused by assembly and activation of NADPH oxidase 2 [NOX2] components on the plasma membrane) from the cells by phorbol 12-myristate 13-acetate (PMA). As shown in Figure S2A, mV-S23 strongly enhanced ROS release compared with control mV, while mV-S23ΔC8 inhibited ROS release in a dominant-negative manner, indicating that these mVenus-tagged proteins are functional in J774 cells and that the PMA-stimulated ROS release results from a type of exocytosis mediated by SNAP-23 (Figure S3). These results suggested that either overexpressing SNAP-23 plays no role in phagocytosis or the amount of endogenous SNAP-23 already expressed in J774 cells is sufficient for phagocytosis.
Overexpression of mVenus-SNAP-23 enhances phagosome maturation processes and phagocytic activity
Because the overexpression of mV-S23 did not invoke changes in uptake activity of IgG-opsonized FITC-zymosan particles (Figure 1D), we next measured the kinetics of ROS production within the phagosome, using nonopsonized luminol-bound microbeads. Prior to that, we checked whether these cells affected uptake activity of nonopsonized synthetic beads. Consistent with the case of opsonized FITC-zymosan particles, we found that those cells expressing tagged proteins did not show a statistically significant difference in uptake activity of FITC-conjugated microbeads (Figure 2A). Exposure of the luminol beads to ROS produced within the phagosome results in their chemiluminescence. Surprisingly, as shown in Figure 2B, the rate of ROS production in J774/mV-S23 cells was fourfold higher than in control J774/mV cells, in contrast to a slight decrease in ROS production in J774/mV-S23ΔC8 cells (Figure 2B). It is known that within phagosomes, ROS are produced by the NOX2 complex. This enzyme, referred to as the NOX2 system, is a complex of two membrane-associated components (gp91phox and p22phox) and four cytoplasmic components (p67phox, p47phox, p40phox, and Rac) that is associated with phagosomes in macrophages (Sumimoto, 2008). As shown in Figure S2B, overexpression of mVenus-tagged proteins in J774 cells caused little change in the expression of NOX2 components or of SNARE proteins either localized on the plasma membrane (syntaxin 2 and syntaxin 3) or involved in phagocytosis (syntaxin 18, D12, Sec22b, VAMP3, VAMP7, and VAMP8; Bajno et al., 2000; Braun et al., 2004; Hatsuzawa et al., 2006; Ho et al., 2008). Together with the finding that mV-S23 overexpression did not alter the uptake of FITC particles by J774 cells (Figures 1D and 2A), these results suggest that mV-S23 enhances ROS production within phagosomes.
FIGURE 2:
mVenus-SNAP-23 enhances not only phagosome maturation processes but also uptake activity. (A) J774/mV, J774/mV-S23, and J774/mV-S23ΔC8 cells were incubated with nonopsonized FITC-conjugated microbeads to quantify a phagocytosis efficiency for synthetic particles, as described in Materials and Methods. Arbitrary fluorescence units were normalized to the maximum value obtained for mV cells within the same experiment, defined as 100%. Data presented are the mean ± SE of six independent experiments. (B) The J774 cells expressing mVenus-tagged proteins were incubated with luminol-bound microbeads. The efficiency of ROS production within the phagosomes of the cells was determined by measuring chemiluminescence from cells that had ingested the beads. Chemiluminescence was measured on a GloMax 20/20n luminometer every 1 min for up to 15 min. Relative light units were normalized to the maximum value obtained for mV cells within the same experiment, defined as 100%. The amount of signal from mV and mV-S23 cells incubated with beads in the presence of cytochalasin B (cyto B; final: 10 μM) was not significant. Data presented are the mean ± SE of three independent experiments. (C) Left, the cells were stained with LysoTracker Red DND-99 (final: 50 nM) for 20 min. After being washed in HBSS, each cell line's fluorescence intensity was measured, as described in Materials and Methods. Arbitrary fluorescence units were normalized to the value obtained for mV cells within the same experiment, defined as 100%. Data presented are the mean ± SE of five independent experiments. Right, these LysoTracker-treated cells were incubated first with IgG-opsonized latex beads (3.0 μm in diameter) for 5 min at 30°C to allow phagosome formation, and the cells were incubated at 30°C in the presence of cytochalasin B (final: 20 μM) to halt the initiation of phagocytosis. At the indicated time points, the cells were cooled on ice, and then LysoTracker-positive phagosome and nonlabeled phagosome were analyzed under a microscope, as described in Materials and Methods. The results are expressed as the percentage of LysoTracker-positive phagosome (more than 30 individual phagosomes from at least 30 different cells for each experiment) at the indicated time points. Data presented are the mean ± SE of five independent experiments. *, p < 0.02, compared with control mV cells using Student's paired t test, one-tailed. (D) The cells were incubated with IgG-opsonized Texas Red–zymosan particles, and the efficiency of FcR-mediated phagocytosis was then measured, as described in Materials and Methods. Arbitrary fluorescence units were normalized to the maximum value obtained for mV cells within the same experiment, defined as 100%. Data presented are the mean ± SE of three to six independent experiments. *, p < 0.005, compared with control mV cells using Student's paired t test, one-tailed.
On the basis of the results described above, we hypothesized that SNAP-23 is involved in phagosome maturation, such as ROS production within the phagosome, rather than in phagosome formation. To address this possibility, we analyzed the phagosomal environment in J774 cells expressing mVenus-tagged proteins using LysoTracker Red DND-99 dye, which is a weak base conjugated to a red fluorophore, as a marker that accumulates in acidic organelles, as described in Materials and Methods. Although almost no difference was observed in the total staining intensity with LysoTracker (Figure 2C, left), the number of LysoTracker-positive phagosomes in J774/mV-S23 cells was clearly increased compared with the control mV cells (Figure 2C, right), indicating that phagosomal acidification was enhanced, presumably due to the accelerated recruitment of factors such as H+-ATPase during phagosome maturation. In contrast, almost no inhibitory effect on this phagosomal acidification was observed in J774/mV-S23ΔC8 cells. These results suggest that the domain of eight C-terminal amino acids is critical for binding to a SNARE partner(s) in phagosome maturation processes (Figure S3).
The results raised the possibility that the FITC-conjugated particles used in the uptake analysis (Figures 1D, bottom, and 2A) could be partially quenched within the phagosome of the J774/mV-S23 cells by enhanced acidification. Therefore we reassessed the uptake activity of the cells expressing mVenus-tagged proteins using Texas Red (a pH-insensitive dye)-conjugated zymosan. Indeed, we found that the uptake activity in J774/mV-S23 cells was enhanced nearly twofold compared with that in J774/mV cells (Figure 2D). The apparent discrepancy of uptake activity (Figures 1D and 2A vs. Figure 2D), which should have been caused by the quenching of FITC fluorescence, strongly supports that overexpression of mV-S23 up-regulates phagosomal acidification.
Together these findings provide evidence of enhanced phagosome formation and maturation in macrophages expressing mV-S23, suggesting that SNAP-23 is involved in phagosomal acquisition of some endosomal and/or lysosomal proteins.
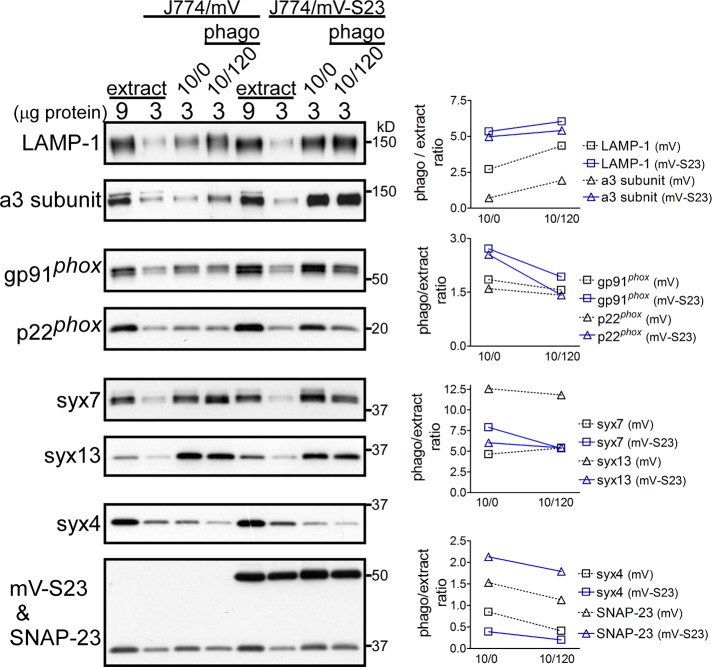
NOX2 components and H+-ATPase are highly enriched in phagosomes from J774 cells expressing mVenus-SNAP-23
During phagosome maturation, the NOX2 components and the vacuolar-type proton-transporting ATPase (V-ATPase) are assembled and/or recruited on phagosomes, resulting in phagosomal ROS production and acidification, respectively (Savina and Amigorena, 2007; Sun-Wada et al., 2009). We therefore asked whether the above-described enhancements of these activities in J774/mV-S23 cells were due to the increased recruitment of related molecules on phagosomes. Phagosomes were isolated from postnuclear supernatants (extracts) prepared from J774/mV and J774/mV-S23 cells that had ingested IgG-opsonized latex beads (0.8 μm in diameter). For obtaining phagosomes of different maturation levels, the cells were incubated with the beads for 10 min (pulse incubation) and, after the excess beads had been washed away, either harvested immediately (phago 10/0) or chased for 120 min (phago 10/120). As shown in Figure 3, the a3 subunit of V-ATPase, one of the a-subunit isoforms localized on lysosome-related organelles (Sun-Wada et al., 2009), and lysosome-associated membrane glycoprotein 1 (LAMP-1), a lysosomal membrane protein, were more rapidly enriched on the phago10/0 and 10/120 of J774/mV-S23 cells than on the corresponding phagosomes of J774/mV cells. In addition, NOX2 components, including gp91phox and p22phox, were already recruited on phago10/0 of J774/mV-S23 (Figure 3), although the amounts of these proteins were similar to those of J774/mV cells after a 2-h chase (Figure 3, right). These results are consistent with enhanced ROS production and acidification within phagosomes of J774/mV-S23 cells, as shown on the right in Figure 2, B and C. Among the SNARE proteins, the distribution of syntaxin 7 (late endosomes/lysosomes) and syntaxin 13 (recycling endosomes), both of which are reported to be involved in phagosome maturation, on phagosomes did not markedly differ between J774/mV and J774/mV-S23 cells (Figure 3). A significant amount of mV-S23 was recruited to the phagosomes of J774/mV-S23 cells, with little or no effect on the phagosomal distribution of endogenous SNAP-23 and syntaxin 4. Our results therefore suggest the involvement of mV-S23 in enhanced membrane fusion with organelles (or vesicles) during phagosome maturation.
FIGURE 3:
mVenus-SNAP-23 recruits the NOX2 complex and H+-ATPase to phagosomes. J774/mV and J774/mV-S23 cells were incubated with IgG-opsonized latex beads for a 10-min pulse at 37°C (10/0). After a 120-min chase incubation (10/120), the phagosome fraction was isolated from the cells by sucrose density-gradient centrifugation. The total cell extract (9 and 3 μg) and the isolated phagosome fraction (3 μg) were analyzed by SDS–PAGE and subsequently by Western blotting with the indicated antibodies (left). LAMP-1 is a lysosomal marker protein. The intensity of the signals on the Western blot (left) was quantified by densitometry using ImageJ Version 1.44 (National Institutes of Health, Bethesda, MD), with the values for each protein expressed as a ratio of phagosome (10/0 or 10/120) to extract containing 3 μg of protein (right).
SNAP-23 interacts with plasmalemmal and endocytic SNARE proteins involved in phagosome formation and maturation
The results thus far suggested that the difference in the extent of phagosome maturation between cells expressing mV-S23 and those expressing mV-S23ΔC8 reflected differences in the interactions of these two proteins with other SNARE proteins. Accordingly, we asked whether SNAP-23 (Qbc-SNARE protein) interacts with plasmalemmal SNARE proteins, such as syntaxins 2, 3, 4, and 11 (Qa-SNARE proteins) and VAMP5 (R-SNARE protein; Figure S8A; Hackam et al., 1996; Zeng et al., 1998; Zhang et al., 2008), and with endocytic SNARE proteins, such as VAMP3, VAMP7, and VAMP8 (R-SNARE proteins) and syntaxins 7 and 13 (Qa-SNARE proteins), to form a SNARE complex. In macrophages, these SNARE proteins are localized in the phagosome membrane. Among them, syntaxins 7 and 13 have been specifically implicated in phagosome maturation (Collins et al., 2002), while other SNARE proteins, except syntaxin 2, syntaxin 3, and VAMP5, are involved in phagosome formation (Bajno et al., 2000; Braun et al., 2004; Ho et al., 2008). The functional differences suggested that these SNARE proteins could cooperate with SNAP-23 in phagosome formation and maturation as cognate SNARE partners. For addressing this possibility, lysates from J774 cells stably expressing mV, mV-S23, or mV-S23ΔC8 were incubated with anti–enhanced green fluorescent protein (anti-EGFP) antibodies, and the immunoprecipitates were subjected to SDS–PAGE followed by Western blot analysis with antibodies against the SNARE proteins described above and anti-EGFP antibodies (Figure 4). With the exception of syntaxin 2, all of the examined SNARE proteins coprecipitated with mV-S23. In particular, syntaxin 11 and the endosomal SNARE proteins interacted much more strongly with mV-S23 than with mV-S23ΔC8 (Figure 4, right), possibly mirroring the differences in the phagosome maturation profiles of J774/mV-S23 and J774/mV-S23ΔC8 cells (Figure 2, B and C, right). These results indicated that SNAP-23 could mediate phagosome formation and maturation by forming several distinct SNARE complexes on the plasma membrane and/or the phagosome membrane.
FIGURE 4:

SNAP-23 interacts with syntaxin 11 and/or endocytic SNARE proteins more effectively than its C-terminal truncated form. J774 cells stably expressing mV, mV-S23, or mV-S23ΔC8 were lysed, and the lysates were immunoprecipitated (IP) with anti-EGFP antibodies. The immunocomplexes were subjected to SDS–PAGE, which was followed by Western blot analysis using the indicated antibodies.
The knockdown of SNAP-23 expression reduces phagocytosis
Next, to determine whether the enhanced phagosome maturation observed in J774/mV-S23 cells was the result of enhanced SNAP-23 function, we made use of small interfering RNAs (siRNAs) directed against SNAP-23, thereby suppressing its endogenous expression in J774 cells. As shown in Figure 5A, transfection of the cells with SNAP-23 siRNA#1, which is targeted to the open reading frame of SNAP-23 mRNA (Figure S5A), decreased SNAP-23 expression to ∼40% of the control value without any change in the expression of endogenous proteins such as gp91phox, CD64, a3 subunit, and syntaxin 4 (Figure S4). Immunofluorescence analysis showed a reduction of SNAP-23 in most cells compared with the control siRNA cells (Figure 5B). A luminol bead assay performed in SNAP-23 siRNA#1 cells showed a decrease in ROS production within the phagosome to ∼50% of the level in control siRNA cells (Figure 5C), implying that the transfection of SNAP-23 siRNA#1 impairs phagosome formation, maturation, or both in J774 cells. These results were explored further by analyzing siRNA-transfected cells using the opsonized Texas Red-zymosan assay. This experiment showed a decrease in phagocytosis in the SNAP-23 siRNA#1 cells to ∼50% of the control activity (Figure 5D, right), although siRNA treatment had no effect on the association of zymosan particles with the cell surface (Figure 5D, left). As shown in Figure S5, SNAP-23 siRNA#2, which is targeted to the 5′-untranslated region (UTR) of SNAP-23 mRNA, had similar effects to siRNA#1 on immunofluorescence and phagocytic activity. Thus, consistent with Figure 2D, SNAP-23 appears to be one of the essential SNARE proteins for membrane fusion events during phagosome formation.
FIGURE 5:
Knockdown of SNAP-23 expression inhibits FcR-mediated phagocytosis. (A) J774 cells were transfected with SNAP-23 siRNAs (#1 and #2) or a control nonspecific siRNA. Total cell lysates from siRNA-transfected cells were analyzed by Western blotting using the indicated antibodies (see also Figure S4). The bands of the Western blotting experiment were quantified using ImageJ. The value for each protein was expressed as a ratio of SNAP-23 siRNA#1 or siRNA#2 cells to control siRNA cells and then normalized to the GAPDH internal control, defined as 100%. Data presented are the mean ± SE of three independent experiments. *, p < 0.01, **, p < 0.02, compared with GAPDH in each cells using Student's paired t test, one-tailed. (B) J774 cells transfected with siRNAs were fixed and then double-stained with antibodies against SNAP-23 (green) and GM130 (red), a Golgi marker protein. SNAP-23 expression was efficiently reduced in almost all cells. Scale bar: 10 μm. (C) J774 cells transfected with siRNAs were analyzed by the luminol bead assay described in Materials and Methods. Relative light units were normalized to the maximal value obtained for control siRNA cells within the same experiment, defined as 100%. Transfection with the SNAP-23 siRNA reduced the efficiency of phagosomal ROS production. Data presented are the mean ± SE of three independent experiments. (D) siRNA-transfected cells were incubated with IgG-opsonized Texas Red–zymosan particles and the efficiencies of association (left) and phagocytosis (right) were measured, as described in Figure 1D. Arbitrary fluorescence units of each cell line were normalized to the maximal value obtained for control siRNA cells within the same experiment, defined as 100%. Data presented are the mean ± SE of three independent experiments. *, p < 0.005, compared with control siRNA cells using Student's paired t test, one-tailed.
SNAP-23 depletion causes a delay in phagosome maturation
The zymosan and luminol bead assays are conventional phagocytosis analyses that reliably estimate uptake efficiency and provide information on phagosome maturation, respectively. However, to directly monitor the progress of phagosome maturation, we developed a new assay system that allowed the tracking of a single phagosome. First, to identify and label a formed phagosome, we established a J774 cell line stably expressing FcγRIIA C-terminally tagged TagRFP (RIIa-TagRFP). FcγRIIA is an FcR that is predominately localized at the plasma membrane (Figure 6B, left, bottom; Hatsuzawa et al., 2006). We then prepared IgG-opsonized EGFP-bound beads, in which the fluorescence signal is quenched by protonation (Sawano and Miyawaki, 2000) and/or ROS (Schwartz et al., 2009), and incubated them with the cells. Fusion of the phagosome with lysosomes or lysosome-related vesicles was expected to quench the fluorescence due to the resulting acidification and/or ROS production within the phagosome (see Materials and Methods). The siRNA-transfected J774/RIIa-TagRFP cells were loaded with IgG-opsonized EGFP-bound beads and then incubated for 5 min to allow phagosome formation; this was followed by a chase for the indicated times in the presence of cytochalasin B to halt the initiation of phagocytosis (Figure 6B, right). After the chase, the incubation buffer was replaced with ice-cold citrate buffer (pH 4.0) to quench the fluorescence signal of beads outside the cells. The RIIa-TagRFP-labeled phagosomes (Figure 6B, left, bottom) were then classified accordingly as type A, type B, type C, and type D to denote whether phagosomes containing EGFP beads were fully fluorescent, slightly fluorescent, faintly fluorescent, or almost completely quenched, respectively (Figure 6B, left, middle). Based on the measurements of more than 30 individual phagosomes from at least 30 different cells for each experiment, increases in the type D phagosome population by as much as 30% were detected in cells transfected with control siRNA and chased for 30 min, whereas in SNAP-23 siRNA#1-transfected cells, the increase was only 15% (Figures 6B, right, and S6), indicating that phagosome maturation is significantly delayed by the knockdown of SNAP-23.
FIGURE 6:
Knockdown of SNAP-23 expression delays phagosome maturation. (A) J774 cells stably expressing FcγRIIA fused to TagRFP at its C-terminus (J774/RIIa-TagRFP) were transfected with control siRNA or SNAP-23 siRNA#1. These cells were incubated first with IgG-opsonized EGFP-bound beads for 5 min to allow phagosome formation (phagocytosis) and then in the presence of cytochalasin B at a final concentration of 20 μM (chase incubation). At the indicated time points, the cells were cooled on ice and subsequently treated with sodium citrate buffer (pH 4.0) to quench the EGFP signal of beads outside the cell, as described in Materials and Methods. Microscopic analysis of the phagosome enclosed with RIIa-TagRFP is presented as follows. (B) Left, arrows denote phagosomes enclosed with RIIa-TagRFP (bottom, red fluorescence) and containing an EGFP-bound bead. These phagosomes were classified into four types depending on the EGFP signal intensity (middle). Scale bar: 10 μm. Right, each type of phagosome was quantified over time. The results are expressed as a percent of the total number of the four phagosome types at the indicated time points. Data presented are the mean ± SE of seven independent experiments. The fraction of type D phagosomes in SNAP-23 siRNA cells was significantly reduced at 30 min (Figure S6) compared with control siRNA cells. (C) At 2 d after transfection with siRNAs, J774 cells were incubated with RB-dextran (final: 50 μg/ml) for 12 h. The medium was replaced with fresh dextran-free growth medium, and the cells were chased for 5 h. These cells were incubated first with IgG-opsonized beads for 5 min at 30°C to allow phagosome formation. After the beads outside the cells were stained with Alexa Fluor 488–conjugated secondary antibodies on ice, the cells were incubated at 30°C in the presence of cytochalasin B (final: 20 μM). At the indicated time points, the cells were cooled on ice and then RB-dextran–positive phagosome and nonlabeled phagosome were analyzed under a microscope, as described in Materials and Methods. The results are expressed as the percentage of RB-dextran–positive phagosomes (more than 30 individual phagosomes from at least 30 different cells for each experiment) at the indicated time points. Data presented are the mean ± SE of four independent experiments. *, p < 0.005, **, p < 0.001, compared with control siRNA cells using Student's paired t test, one-tailed. (D) Rescue effects of SNAP-23 expression. The cells transfected with control siRNA or SNAP-23 siRNA#2, which targets to 5′ UTR of SNAP-23 mRNA, were incubated with RB-dextran for 8 h, which was followed by replacing and chasing in dextran-free growth medium for an additional 5 h prior to overnight transfection with plasmids of mVenus-tagged proteins. The cells were analyzed as described above and in Materials and Methods. The results are expressed as the percentage of RB-dextran–positive phagosomes at 15-min chase time. Data presented are the mean ± SE of four independent experiments. Student's paired t test, one-tailed.
To further confirm these results, we examined the effect of SNAP-23 knockdown on phagosome–lysosome fusion by transfection with siRNAs in cells whose late endosomes and lysosomes were preloaded with a fluid-phase marker, rhodamine B–conjugated dextran (RB-dextran). Almost no difference was observed in labeling efficiency with RB-dextran between the cells transfected siRNAs (Figure S7A, right panel). After being washed and incubated in dextran-free growth medium for 5 h, the cells were loaded with IgG-opsonized beads and then incubated for 5 min to allow phagosome formation; this was followed by a chase for the periods indicated in Figure 6C. After the chase, the beads containing phagosomes (RB-dextran–positive phagosomes and unlabeled phagosomes) were counted under a microscope (see Materials and Methods). As shown in Figure 6C, the number of RB-dextran–positive phagosomes, even at 15 min of chase incubation, in SNAP-23 siRNA-transfected cells was significantly decreased compared with control siRNA cells (Figure 6C), indicating that the fusion efficiency of the phagosome with lysosomes and/or lysosome-related organelles is reduced by depletion of SNAP-23. To ascertain the specificity of the effects observed upon depletion of SNAP-23, we next performed rescue experiments in which the siRNA-treated cells were transiently overexpressed with mVenus-tagged proteins. The cells transfected with control siRNA and SNAP-23 siRNA#2 (Figure S5A) were labeled by preloading with RB-dextran for 8 h. After being washed, the cells were transfected with plasmids of mVenus-tagged proteins overnight in the dextran-free growth medium. The phagosomes at 15 min of chase incubation in the cells expressing mVenus-tagged proteins were counted under a microscope. As shown in Figure 6D, mV-S23 remarkably restored the number of RB-dextran–positive phagosomes, although phagosome–lysosome fusion efficiency was not recovered by mV in SNAP-23 siRNA#2 cells. Interestingly, mV-S23ΔC8 also reversed the reduction in the fusion efficiency to some extent, while mV-S23 significantly (p = 0.0056) recovered more than mV-S23ΔC8 did (Figure 6D), indicating that SNAP-23ΔC8 is weakly functional, but apparently less competent, compared with mV-S23. Similar results were obtained from the phagosomal acidification assay using LysoTracker (Figure S7, B and C).
Overexpression of VAMP7 is associated with a conformational change in the structure of SNAP-23 on the phagosome membrane
If, as a component of the SNARE machinery, SNAP-23 is involved in membrane reorganization during phagosome formation and maturation, it should undergo a structural change to form a SNARE complex. To determine whether this is indeed the case, we designed a set of intramolecular FRET probes of SNAP-23. Structural analyses of SNARE proteins (Sutton et al., 1998) identified two SNARE motifs in SNAP-23, SN1(1-75) and SN2(148-211). During membrane fusion, the N-terminal regions of these motifs move closer together subsequent to the formation of a SNARE complex with other SNARE proteins (Figure 7A, inset). In previous work, the use of an intramolecular FRET probe of the SNAP-23 isoform SNAP-25 showed an enhanced FRET signal on the plasma membrane during exocytosis (An and Almers, 2004; Wang et al., 2008; Takahashi et al., 2010). On the basis of the design of SNAP-25 FRET, we constructed a SNAP-23 FRET probe (tG-S1-tR-S2) modified such that TagGFP2 (tG) and TagRFP (tR) were adopted as donor and acceptor fluorescence probes at the N-termini of SN1 and SN2, respectively (Figure 7A; Shcherbo et al., 2009). As a negative control, tG-S1-S2-tR was constructed, introducing TagRFP at the C-terminus of SN2. If SNAP-23 functions as part of the membrane fusion machinery during phagosome maturation, as described above, a structural alteration of SNAP-23 should result, namely, the close approximation of the N-terminal domains of SN1 and SN2 accompanied by the detection of the FRET signal from tG-S1-tR-S2 folded within a SNARE complex on the phagosome membrane (Figure 7A, inset).
FIGURE 7:
The FRET signal of SNAP-23 probe (tG-S1-tR-S2) on the phagosome membrane is increased by the overexpression of VAMP7. (A) Schematic representation of various SNAP-23 constructs (see Materials and Methods). Approximation of the N-termini of the two SNARE motifs (SN1(1-75) and SN2(148-211)) is expected when the complete SNARE complex is formed with other SNARE proteins for membrane fusion. For restriction of the flexibility of the fluorescent proteins at the connection, the C-terminal 11 residues were truncated from TagGFP2, a truncated form of TagGFP2 (Shimozono et al., 2006), and fused to SNAP-23. The inset shows the predicted conformation of the SNAP-23–based FRET construct upon forming fusogenic SNARE complexes. The asterisk indicates a potential lipid-modified cysteine residue. (B) Transiently expressed tG-S1-tR-S2 and tG-S1-S2-tR were predominately localized at the plasma membrane of J774 cells. Spectroscopic data were obtained using the N-SIM system. The asterisks denote the phagosomes containing IgG-opsonized latex beads (3.0 μm in diameter). Scale bar: 10 μm. (C–E) J774 cells were transiently cotransfected with SNAP-23 FRET probes and the Myc-tagged SNARE proteins as indicated. The spectroscopic function of a laser-scanning microscope was used to excite the plasma membrane of living cells at 458 nm, and the resulting emission spectra were analyzed (see Figure S9). Emission at 580 nm was divided by that at 505 nm (TagRFP/TagGFP) and normalized to the value obtained for cells cotransfected with the Myc-vector in the same experiment. This was set to 1.00. Data presented are the mean ± SE of three to six independent experiments. Student's paired t test, two-tailed. (F) J774 cells cotransfected with SNAP-23 FRET probes and the indicated Myc-tagged constructs were incubated with IgG-opsonized zymosan particles at 37°C for 20 min. Extra particles were removed in a washing step, and the FRET efficiency on the phagosome membrane of living cells was then analyzed as described above. Student's paired t test, two-tailed.
Expression of the SNAP-23 FRET probes in J774 cells resulted in their predominant localization at the plasma membrane and on the membranes of formed phagosomes (Figure 7B). However, co-overexpression of additional SNARE partners might be required for detection of FRET signal from the tG-S1-tR-S2 probe (Wang et al., 2008), because of the limited number of endogenous partner molecules. Thus we first examined whether the probe increased the FRET signal on the plasma membrane when cotransfected with plasmalemmal SNARE proteins.
To quantify the amount of energy transferred to TagRFP from TagGFP2, we measured the emission spectrum of the probe (Figure S9, A and B). FRET efficiency was represented by an emission ratio of 580/505 nm (Figure 7C). A 458-nm laser line was used for excitation to minimize the contribution of TagRFP fluorescence in directly exciting illumination. As shown in Figure 7C, an enhanced FRET signal from tG-S1-tR-S2 was fortunately observed on the plasma membrane only when it was coexpressed with Myc-VAMP5, a plasma membrane–localized SNARE protein (Figure S8A) that interacts with SNAP-23 (Figure 4, left). This result confirmed that tG-S1-tR-S2 is the first useful intramolecular FRET probe to detect functional and structural changes in SNAP-23 on the phagosomal membrane. Moreover, this signal was synergistically enhanced on the plasma membrane when the probe was coexpressed with Myc-VAMP5 and Myc-syntaxin 3 (Figure 7D), implying that SNAP-23 forms a novel SNARE complex with these two SNARE proteins in macrophages. In contrast, the VAMP5-induced FRET signal was repressed in cells coexpressing syntaxin 4 or syntaxin 11 (Figure 7D), consistent with an association of these syntaxins with tG-S1-tR-S2 through a mutually exclusive interaction with respect to VAMP5. The enhanced FRET signal by VAMP5 and syntaxin 3 was not detected in cells coexpressing the negative control tG-S1-S2-tR or a combination of tG-S1-S2 and Myc-S1-S2-tR (Figure 7D). The coexpression of endosomal VAMP proteins, such as VAMP3, VAMP7, and VAMP8 (Figure S8B), also had little effect on the FRET efficiency of tG-S1-tR-S2 at the plasma membrane (Figure 7E).
Finally, on the basis of the results obtained above, we analyzed the FRET efficiency of tG-S1-tR-S2 selectively on the phagosomes in living cells. In this experiment, phagosome formation was induced using IgG-opsonized zymosan particles, because the reflection of latex beads interferes with spectrum measurements, whereas zymosan is nonreflecting. The tG-S1-tR-S2 probe was coexpressed with Myc-VAMP3 or Myc-VAMP7 to examine whether an enhanced FRET signal on the phagosome membrane was obtained, since VAMP3 and VAMP7 interact more efficiently with mV-S23 than with mV-S23ΔC8 (Figure 4, right). In J774 cells incubated for 20 min with the opsonized zymosan particles, a significantly enhanced FRET signal was observed on the phagosome membrane of cells coexpressing Myc-VAMP7 (Figures 7F and S9B), in contrast to the weak FRET signal on the phagosome membrane in cells coexpressing either the negative control tG-S1-S2-tR probe and Myc-VAMP7 (Figure 7F) or tG-S1-tR-S2 and other SNARE proteins (Figure S9C). These results strongly suggested that SNAP-23 on the phagosome pairs with endosomal SNARE proteins, such as VAMP7, to play a direct role in membrane fusion during phagosome maturation.
Taken together, these findings provide strong evidence that SNAP-23 mediates phagosome maturation in macrophages by playing a role in SNARE-based membrane trafficking.
DISCUSSION
As part of the biophylactic strategy of mammals, macrophages ingest foreign particles by dynamically arranging endomembranes. Integral to phagosome formation (phagocytosis) is membrane fusion between cytoplasmic organelles (or vesicles derived from them) and the plasma membrane. This step is followed by phagosome maturation, during which endocytic organelles fuse with the phagosome. Little is known about the specific role of plasma membrane–localized SNARE proteins in phagocytosis, and only a limited number of studies have been published addressing how these proteins contribute to phagosome maturation (Collins et al., 2002; Becken et al., 2010). SNAP-23 is a ubiquitously expressed SNARE protein responsible for exocytosis and is mostly present on the plasma membrane with some endomembrane localization (Sadoul et al., 1997; Jahn and Scheller, 2006). In J774 macrophages, SNAP-23 is mainly localized at the plasma membrane, judging from immunofluorescence experiments (Figures 5B and S1A). In the present work, we demonstrated that uptake activity (Figure 2D) and maturation processes, such as phagosomal ROS production and acidification (Figure 2, B and C), are enhanced in J774 cells overexpressing SNAP-23 (J774/mV-S23 cells). In J774 cells, knockdown of SNAP-23 by specific siRNAs caused a reduction in uptake activity (Figure 5D, right) and a delay in phagosome maturation (Figure 6). Using intramolecular FRET probes of SNAP-23, we showed that, in J774 cells coexpressing with VAMP7, SNAP-23 underwent a structural change on the phagosome membrane (Figure 7F), suggesting that it forms one or more SNARE complexes to mediate membrane fusion with endocytic organelles, such as late endosomes and lysosomes. Our results strongly implicate SNAP-23 in mediating phagosome formation and maturation in macrophages. This is the first report of a functional role for a SNARE protein in membrane fusion on the phagosome.
Based on the analysis of isolated phagosomes (Figure 3), the excess mVenus-SNAP-23 distributed in phagosomes appears to be responsible for the subsequent fusion events required for phagosome maturation under the condition of the experiment. During phagocytosis, overexpression of mVenus-SNAP-23 enhanced uptake activity of the IgG-opsonized particles, indicating that SNAP-23 is apparently rate-limiting for its related pathway. In previous work, we showed that the overexpression of syntaxin 18, an ER-localized SNARE protein, enhances phagocytosis (Hatsuzawa et al., 2006), suggesting that a sufficient amount of its cognate SNARE protein(s) must be present on the plasma membrane. Thus SNAP-23 is not likely to be a candidate for SNARE partner in the ER-mediated phagocytosis.
After engulfing foreign particles, phagosomes sequestered from the plasma membrane gradually mature to become phagolysosomes, acquiring, for example, ROS production activity, an acidic milieu, and several lysosomal hydrolases. ROS within phagosomes are produced by the NOX2 complex, which is composed of membrane (gp91phox and p22phox) and cytosolic (p67phox, p47phox, p40phox, and Rac) proteins (Sumimoto, 2008). In the assembly of these components on the phagosome, p40phox is recruited to early endosome-like vesicular structures through its phox domain and then localizes to the phagosomes in macrophages (Ellson et al., 2001; Ueyama et al., 2007). In dendritic cells, gp91phox localized at lysosome-related organelles is recruited to the phagosome by the action of the monomeric small GTPase Rab27a (Jancic et al., 2007). The acidic milieu of the phagosome is ensured by the recruitment of V-ATPase, which is localized to late endosomes and lysosomes. In macrophages, lysosomal V-ATPase is recruited to the phagosome by membrane fusion directly with the tubulated lysosomes (the “kiss and run” mechanism; Sun-Wada et al., 2009). The results of our study suggest the possibility that SNAP-23 is implicated in these multiple steps of membrane fusion during phagosome maturation.
In this study, we found that SNAP-23 in macrophages function in various host-defense processes, such as phagosome formation and maturation, as well as in PMA-stimulated ROS release. It is likely that SNAP-23 forms several distinct SNARE complexes with a specific SNARE partner(s) in each process. Interestingly, overexpression of mVenus-tagged SNAP-23ΔC8 (mV-S23ΔC8), which is reported to be a dominant-negative mutant in exocytosis of some secretion pathways (Kawanishi et al., 2000; Huang et al., 2001), indeed caused inhibition of PMA-stimulated ROS release in J774 macrophages (Figure S2A). In this process, mV-S23ΔC8 would bind to the same set of SNARE partners as endogenous SNAP-23 but could not form a functional fusion complex, thereby competing with the SNAP-23 function (Figure S3). However, mV-S23ΔC8 overexpression had no effect on phagosome formation (Figure 2D, left), phagosomal ROS production (Figure 2B), and phagosomal acidification (Figure 2C, right). These data clearly indicate that interacting domains of SNAP-23 with SNARE partners for phagosome formation and maturation are distinct from those for PMA-stimulated ROS release. Probably the truncated C-terminal domain would be required for forming the stable complex with its SNARE partners in phagocytosis processes. The weak restoration activities of mV-S23ΔC8 in the knockdown cells (Figures 6D and S7C) are consistent with these observations. The different immunoprecipitation recoveries of various SNARE proteins with either mV-S23 or mV-S23ΔC8 (Figure 4) also support a notion that SNAP-23 could use at least two types of configuration to form a SNARE complex (Figure S3).
Syntaxins 13 and 7 reportedly function at distinct fusion steps in phagosome maturation (Collins et al., 2002). According to that study, syntaxin 13, an early endosome– and/or recycling endosome–localized SNARE protein, is transiently recruited to the phagosome in one of the early steps of phagosome maturation, while syntaxin 7, a late endosome– and lysosome-localized SNARE protein, is required during later steps of maturation (Collins et al., 2002). However, the only early phagosomal localization of syntaxin 13 is not consistent with our phagosome fractionation results (Figure 3), perhaps because the fluorescence of syntaxin 13 fused to N-terminal EGFP (Collins et al., 2002) is immediately quenched upon exposure to the increased ROS and acidic pH within phagosomes, as EGFP fluorescence is known to be damaged by ROS (Schwartz et al., 2009) and be highly acid sensitive (Sawano and Miyawaki, 2000). The involvement of syntaxin 7, as well as syntaxin 13, in phagosome maturation is also supported by the results of the in vitro cell-free phagosome–lysosome fusion assay (Becken et al., 2010). In any case, these SNARE proteins are likely to be Qa-SNARE candidates for SNAP-23. In addition, our FRET analyses (Figure 7F) identified VAMP7 as a candidate for R-SNARE during phagosome maturation. Why then was a conformational change of SNAP-23 (enhanced FRET signal) observed on the phagosomes by VAMP7 overexpression? The answer may be that the overexpression of VAMP7 caused a delayed disassembly or accelerated assembly of the SNARE complex containing SNAP-23 on the phagosome. Thus, while our FRET analyses strongly support the occurrence of a structural change in SNAP-23 on both the cell surface and phagosomes, further rigorous studies are required to determine the functional SNARE partner(s) for SNAP-23 during phagosome formation and maturation.
The phagosome is critically important in bacterial killing, the degradation of foreign particles, and the generation of peptides for antigen presentation during maturation of the phagosome to form the phagolysosome (Jutras and Desjardins, 2005; Haas, 2007). These activities of resting-state macrophages are physiologically distinct from those of dendritic cells (Savina and Amigorena, 2007) and interferon-γ– and lipopolysaccharide-stimulated macrophages (Yates et al., 2007), resulting in differences in antigen presentation to T-cells. Thus resting-state macrophages do not have enough capability to recruit NOX2 components and/or lysosomal proteins to the phagosome (Savina et al., 2006; Yates et al., 2007) and to modify the luminal redox potential of phagosomes (Rybicka et al., 2010) compared with the stimulated macrophages. These events are among those regulated by membrane trafficking between endocytic organelles and phagosome, but their molecular basis is poorly understood. Our results suggest that phagosomal SNAP-23 is one of the key players regulating the phagosomal environment in macrophages, although further studies are needed to understand its precise function in membrane trafficking leading to phagosome maturation.
MATERIALS AND METHODS
Antibodies
Polyclonal antibodies to syntaxin 18, D12, Sec22b, and EGFP and a monoclonal antibody to V-ATPase a3 subunit were prepared as described previously (Hatsuzawa et al., 2009; Sun-Wada et al., 2011). The polyclonal antibodies to syntaxin 7, syntaxin 11, VAMP3, VAMP5, and VAMP7 were raised against the respective bacterially expressed glutathione S-transferase–tagged proteins. The remaining antibodies were obtained from commercial sources: syntaxin 2 and syntaxin 13 from Stressgen (Victoria, Canada); syntaxin 3, syntaxin 4, and SNAP-23 from Sigma-Aldrich (St. Louis, MO); VAMP8 from Synaptic Systems (Göttingen, Germany); GM130 and gp91phox from BD Transduction Laboratories (San Jose, CA); p22phox, p67phox, p47phox, p40phox, CD64, and LAMP-1 from Santa Cruz Biotechnology (Santa Cruz, CA); Rac from Cell Signaling Technology (Beverly, MA); glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from Ambion (Austin, TX); and fluorescently labeled secondary antibodies from Invitrogen (Tokyo, Japan).
Cell culture
J774 cells were obtained from the Riken Cell Bank (Tsukuba, Japan) and grown in RPMI 1640 medium (Wako Pure Chemical Industries, Osaka, Japan) supplemented with 10% fetal calf serum (FCS). J774 cells stably expressing mVenus-tagged or TagRFP-tagged proteins were maintained in RPMI with 10% FCS supplemented with puromycin (2 μM).
Expression vectors and establishment of stable transformants
SNAP-23 cDNA was obtained by PCR from a MATCHMAKER human leukocyte cDNA library (Clontech, Mountain View, CA). SNAP-23 and FcγRIIA expression plasmids were constructed by subcloning the PCR-generated cDNA fragments into the pmVenus-C1 vector (Hatsuzawa et al., 2006) and pTagRFP-N (Evrogen, Moscow, Russia), respectively. J774 cell lines expressing mVenus-tagged proteins (J774/mVenus, J774/mVenus-SNAP-23 (1-211), and J774/mVenus-SNAP-23ΔC8 (1-203)) and the TagRFP-tagged protein (J774/FcγRIIA-TagRFP) were established by infection with recombinant retrovirus generated using cDNAs of fluorescence probe–tagged proteins cloned into the pCX4puro vector, as previously described (Akagi et al., 2000; Hatsuzawa et al., 2006, 2009).
Constructions of FRET Probes of SNAP-23 and other SNARE proteins
To construct the pTagGFPΔC11 (1-227) vector, which lacks 11 C-terminal residues from TagGFP2 (Shimozono et al., 2006), we deleted the sequence between the codon for C228 and the unique PstI site in the pTagGFP2-C vector (Evrogen, Moscow, Russia) by PCR. The probe's photostability was increased by modifying the codon for S162 in the pTagRFP-N vector to T162 (TagRFP-t) using the QuickChange protocol (Stratagene, La|Jolla, CA). The tG-S1-tR-S2 probe (TagGFP2ΔC11 (1-227)-SNAP-23 (1-147)-TagRFP-t (1-237)-SNAP-23 (148-211)) was prepared by subcloning the PCR-amplified inserts of SNAP-23 (1-147) and TagRFP-t (1-237)-SNAP-23 (148-211) into the pTagGFPΔC11 vector. The tG-S1-S2 probe (TagGFP2ΔC11 (1-227)-SNAP-23 (1-211)) was prepared by subcloning the full-length cDNA of SNAP-23 into the pTagGFPΔC11 vector. The tG-S1-S2-tR probe (TagGFP2ΔC11 (1-227)-SNAP-23 (1-211)-TagRFP-t (1-237)) was prepared by subcloning the PCR-amplified inserts of SNAP-23 (1-211) and TagRFP-t (1-237) into the pTagGFPΔC11 vector. The Myc-S1-S2-tR probe (Myc-SNAP-23 (1-211)-TagRFP-t (1-237)) was prepared by subcloning the PCR-amplified inserts of SNAP-23 (1-211) and TagRFP-t (1-237) into the pcDNA-Myc-C1 vector.
Syntaxin 11, VAMP5, and VAMP8 cDNAs were obtained by PCR using total RNA extracted from J774 cells. The cDNAs were cloned into the pcDNA-Myc-C1 vector, and their sequences were confirmed with a DNA sequencer. The cDNA inserts from pFLAG-syntaxin 3, pFLAG-syntaxin 4, pFLAG-VAMP3, and pFLAG-VAMP7 (Hatsuzawa et al., 2006) were subcloned into the pcDNA-Myc-C1 vector.
Opsonized fluorescent dye–conjugated zymosan assay
The opsonized fluorescent dye–conjugated zymosan assay was performed as described previously (Hatsuzawa et al., 2009). Briefly, J774 cells stably expressing mVenus-tagged proteins or transfected with siRNAs were incubated for 1 h in the presence or absence of an ∼30-fold excess of FITC- or Texas Red–conjugated zymosan A particles (cat. no. Z2841 or 2843; Invitrogen) treated with opsonizing reagent (rabbit anti-zymosan IgG, cat. no. Z2850; Invitrogen). The cells were washed with phosphate-buffered saline (PBS) to remove the free particles and then incubated with trypan blue to quench the fluorescence of noninternalized particles. Cellular fluorescence was quantified in a Plate Chameleon V microplate reader (Hidex, Turku, Finland) at excitation 485 nm/emission 535 nm, or in a Varioskan Flash microplate reader (Thermo Fisher Scientific, Milford, MA) at excitation 565 nm/emission 615 nm. Arbitrary fluorescence units were obtained by subtracting the fluorescence intensity observed in the absence of IgG-opsonized fluorescent zymosan particles from that observed in the presence of the particles. Alternatively, the cells were incubated for 30 min on ice with IgG-opsonized fluorescent zymosan particles and then washed thoroughly with PBS to remove the free particles. The fluorescence of the particles associated with the cells was then measured as described above.
Luminol bead assay
The luminol bead assay was conducted essentially as described previously (Hatsuzawa et al., 2006, 2009). Briefly, J774 cells stably expressing mVenus-tagged proteins or transfected with siRNAs were plated at a density of (3–4) × 106 cells per 35-mm–diameter culture dish (cat. no. 3294; Corning, Corning, NY) and cultured overnight. The medium was replaced with Hank's balanced salt solutions (HBSS) containing 1.08 × 108 luminol-bound microbeads (1.5–2.0 μm in diameter; Kamakura Techno-Science, Kamakura, Japan; Uchida et al., 1985; Savina et al., 2006). Chemiluminescence was generated from the microbeads by the reactive oxygen within phagosomes incubated at 30°C, and the signal intensity of each dish was quantified in a GloMax 20/20n luminometer (Promega, Madison, WI) every 1 min.
Because it is difficult to determine the exact number of phagocytized luminol beads, FITC-conjugated microbeads (1.87 μm in diameter [mean size], cat. no. F1CP-20-2; Spherotech, Lake Forest, IL) were used for quantitating phagocytosis of synthetic particles. J774 cells stably expressing mVenus-tagged proteins were incubated for 1 h in the presence of an ∼10-fold excess of nonopsonized FITC beads. The cells were washed with PBS to remove the free beads and then incubated with trypan blue solution. Cellular fluorescence was quantified in a Plate Chameleon V microplate reader at excitation and emission wavelengths of 485 and 535 nm, respectively. Arbitrary fluorescence units were obtained by subtracting the fluorescence intensity observed in the absence of FITC beads from that observed in the presence of the beads.
Phagosomal acidification assay
The J774 cells (at 0.75 × 106 cells on 35-mm–diameter glass-bottom dishes) stably expressing mVenus-tagged proteins and/or J774 cells transfected with siRNAs were incubated for 20 min in the presence of LysoTracker Red DND-99 (final concentration 50 nM; Invitrogen) to stain acidic compartments. For rescue experiments (Figure S7C), the cells treated with SNAP-23 siRNA#2 were transfected with plasmids of mVenus-tagged proteins on the day before analysis and then stained with LysoTracker. The cells were washed in ice-cold HBSS, incubated for 30 min on ice in the presence of an ∼20-fold excess of IgG-opsonized latex beads (3.0 μm in diameter, Sigma-Aldrich), and incubated at 30°C for 5 min to initiate phagocytosis. The cells were washed three times in ice-cold HBSS and incubated for 20 min on ice in the presence of Alexa Fluor 488 (or Alexa Fluor 633)–conjugated goat anti-rabbit secondary antibodies (Invitrogen) to stain the beads outside the cells. The cells were then incubated in HBSS containing cytochalasin B (final concentration: 20 μM) for the indicated times at 30°C. Images were captured on an LSM510meta laser-scanning microscope using a Plan-Apochromat 63×/1.4 numerical aperture (NA) oil-immersion objective (Carl-Zeiss, Oberkochen, Germany) under low-temperature conditions (∼6°C). More than 30 phagosomes were then counted for each experiment and categorized into two types (LysoTracker-positive phagosome or unlabeled phagosome) based on the presence or absence of detectable LysoTracker Red fluorescence signal. For estimation of the amount of acidic organelles such as lysosomes, the cells were stained with LysoTracker Red DND-99 for 20 min. After washing in HBSS, the fluorescence intensity from the cells was quantified in a Varioskan Flash microplate reader (Thermo Fisher Scientific) at excitation and emission wavelengths of 564 and 590 nm, respectively.
Isolation of phagosomes
A 10% suspension of latex beads (0.8 μm in diameter, dyed deep blue; Sigma-Aldrich) opsonized with IgGs was added to J774 cells at a dilution of 1:200 in RPMI medium; this was followed by incubation for 10 min at 37°C to allow bead ingestion. The cells were then washed with ice-cold PBS to remove free beads and homogenized either directly or after further incubation for 120 min at 37°C to prepare a postnuclear supernatant (PNS). Intact mitochondria and small ER debris from the phagosome fraction were removed by treating the PNS with 10 mM ATP/4 mM MgCl2 for 15 min on ice prior to sucrose density-gradient centrifugation. Phagosomes were then isolated from the PNS by flotation in a discontinuous sucrose gradient, as described previously (Desjardins et al., 1994; Hatsuzawa et al., 2006).
Small interfering RNA experiment
An siRNA duplex with a 52% GC content (5′-GUACCGCACGUCAUUCGUAUC-3′; RNAi, Co., Tokyo, Japan) was used as the control. The RNA duplexes used for targeting were SNAP-23 siRNA#1 (5′-CAUUAAACGUAUAACUAAUGA-3′) and siRNA#2 (5′-CGGGACAGAGUAUCCGUAUUU-3′) corresponding to the open reading frame and the 5′-untranslated region (UTR) of mouse SNAP-23 mRNA, respectively (Figure S5A). J774 cells or J774/FcγRIIA-TagRFP cells were transfected with either the control or the SNAP-23 siRNAs using HiPerFect transfection reagent (Qiagen, Valencia, CA). Forty-eight hours after transfection, the cells were split into two or three dishes for Western blot analysis, immunofluorescence observation, or other assays.
Opsonized EGFP-bound bead assay
Latex beads (3.0 μm in diameter) were washed in PBS and incubated in 1.5 mg purified EGFP solution/ml for 12 h at room temperature with gentle rotation under light-shielding conditions. The beads were washed in PBS and incubated with anti-EGFP antibodies at 37°C for 2 h. Opsonized EGFP-bound beads were washed three times and resuspended in HBSS. Forty-eight hours after the above-described siRNA transfection, J774 cells stably expressing FcγRIIA-TagRFP were plated onto the center of 35-mm–diameter glass-bottom dishes at a density of 0.75 × 106 cells and cultured overnight. The cells were washed in ice-cold HBSS, incubated for 30 min on ice in the presence of 1.5 × 107 opsonized EGFP-bound beads, and incubated again at 32°C for 5 min to initiate phagocytosis. The cells were then washed three times in warm (32°C) HBSS and immediately incubated in HBSS containing cytochalasin B (final concentration 20 μM) for the indicated times at 32°C. At the end of the incubation period, the cells were washed with HBSS, and ice-cold 150 mM NaCl/50 mM citrate buffer (pH 4.0) was added to quench the EGFP signal from beads outside the cells. Images were captured on an LSM510meta laser-scanning microscope under low-temperature conditions (∼6°C). Phagosomes surrounded by FcγRIIA-TagRFP were categorized into four types (A–D) based on the fluorescence intensity.
Phagosome–lysosome fusion assay
Forty-eight hours after the above-described siRNA transfection, J774 cells were plated onto the center of 35-mm–diameter glass-bottom dishes at a density of 0.75 × 106 cells and labeled overnight at 37°C with 50 μg/ml RB-dextran (mol. wt.: 10,000; Invitrogen). The labeling medium was then removed, and cells were chased for 5 h before analysis. On the other hand, for rescue effects of SNAP-23 expression on J774 cells transfected with siRNAs of control and SNAP-23#2, cells were labeled with RB-dextran for 8 h and plated onto the glass-bottom dishes after washing. Next cells were transfected with plasmids of mVenus-tagged proteins overnight in the dextran-free growth medium. The cells were washed in ice-cold HBSS, incubated for 30 min on ice in the presence of an ∼20-fold excess of IgG-opsonized latex beads, and incubated at 30°C for 5 min to initiate phagocytosis. The cells were washed three times in ice-cold HBSS and incubated for 20 min on ice in the presence of Alexa Fluor 488 (or Alexa Fluor 633)–conjugated goat anti-rabbit secondary antibodies (Invitrogen) to stain the beads outside the cells. The cells were then incubated in HBSS containing cytochalasin B (final 20 μM) for the indicated times at 30°C. Images were captured on an LSM510meta laser-scanning microscope under low-temperature conditions (∼6°C). More than 30 phagosomes were then counted for each experiment and categorized into two types (RB-dextran–positive phagosome or unlabeled phagosome) based on the presence or absence of detectable rhodamine B fluorescence signal. For estimating the amount of RB-dextran–positive organelles, the cells stained with RB-dextran overnight were washed in HBSS, and then the fluorescence intensity from the cells was quantified in a Varioskan Flash microplate reader at excitation and emission wavelengths of 543 and 590 nm, respectively.
Immunoprecipitation
Lysates from J774 cells stably expressing mVenus-tagged proteins were incubated with an anti-EGFP antibody for 30 min at 4°C. Protein A-Sepharose (GE Healthcare Bio-Sciences, Tokyo, Japan) was then added, and the mixture was allowed to incubate for 16 h at 4°C with gentle rotation. The beads were washed four times with extraction buffer (20 mM HEPES-KOH, pH 7.2, 100 mM KCl, 2 mM EDTA, 1% Triton X-100, 1 mM phenylmethysulfonyl fluoride, 1 mM dithiothreitol, and a protease inhibitor cocktail; Nakarai Chemicals, Kyoto, Japan), and the immune complexes were subsequently eluted from the Sepharose beads with SDS–PAGE sample buffer. In addition, 10% of the total lysate volume was mixed with 5× SDS–PAGE sample buffer and heated at 95°C for 5 min. After SDS–PAGE, the samples were analyzed by Western blotting using Clean-Blot IP Detection Reagent (Thermo Fisher Scientific), according to the manufacturer's instructions. Immunoreactive proteins were visualized using ECL Western Blotting Detection Reagents (GE Healthcare Bio-Sciences).
FRET analysis
FRET probes of SNAP-23 were expressed in J774 cells together with Myc-tagged members of the syntaxin family (syntaxin3, syntaxin4, or syntaxin11) and/or Myc-tagged members of the VAMP family (VAMP3, VAMP5, VAMP7, or VAMP8) using X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, Indianapolis, IN). Fluorescence spectra of the probes on the plasma and phagosome membranes of living cells were obtained 20 h after transfection using an LSM510meta laser scanning microscope with a C-Apochromat 40×/1.2 W objective (Carl-Zeiss) at an excitation wavelength of 458 nm. Prior to the measurements, the dynamics range at each wavelength was calibrated using the standard solution according to the manufacturer's protocol. The spectrum with a fluorescence intensity of around 3000 arbitrary units (a.u.) at 505 nm was used for the analysis. FRET efficiency was represented as the 580/505-nm emission ratio.
Imaging analyses
J774 cells expressing mVenus-SNAP-23 or mVenus-SNAP-23ΔC8 were incubated at 37°C for 10 min in the presence of Texas Red–conjugated zymosan A (Invitrogen) previously treated with opsonizing reagent. The cells were then washed in HBSS, and the images were captured on an LSM510meta laser-scanning microscope, using a Plan-Apochromat 63×/ 1.4 NA oil-immersion objective (Carl-Zeiss).
J774 cells expressing mVenus-SNAP-23 were incubated at 37°C for 10 min in the presence of IgG-opsonized latex beads (3.0 μm in diameter) and then washed in ice-cold PBS. Beads outside the cells were visualized with an anti–rabbit-IgG-conjugated Alexa Fluor 594 dye (Invitrogen) and then fixed with 4% paraformaldehyde/PBS. Images were taken with a Nikon structured illumination microscopy (N-SIM) system using a CFI Plan-Apo total internal reflection fluorescence 100×/1.49 NA oil-immersion objective (Nikon). Z-stacks obtained from scanning whole cells were used to construct three-dimensional images, based on a 200-nm step size.
Statistics
Data are presented as the mean ± SE for the number of experiments indicated in the figure legends. Statistical significance was determined with Student's paired t test (one-tailed or two-tailed) using GraphPad Prism (GraphPad Software, San Diego, CA). Differences between the analyzed samples were considered significant at p < 0.05.
Supplementary Material
Acknowledgments
We thank Ka-ai Hirata, Hiromi Hashimoto, and Mayumi Takeuchi for excellent technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research (C) (#22570189) from the Japan Society for the Promotion of Science to K.H.
Abbreviations used:
- BoNT/A
botulinum type A toxin
- ECL
enhanced chemiluminescence
- EGFP
enhanced green fluorescent protein
- ER
endoplasmic reticulum
- FcR
Fc receptor
- FCS
fetal calf serum
- FITC
fluorescein isothiocyanate
- FRET
Förster resonance energy transfer
- HBSS
Hanks’ balanced salt solution
- Ig
immunoglobulin
- LAMP-1
lysosome-associated membrane glycoprotein 1
- MHC
major histocompatibility complex
- mVenus
monomeric Venus
- NA
numerical aperture
- NOX
NADPH oxidase
- NSF
N-ethylmaleimide–sensitive factor
- PBS
phosphate-buffered saline
- PMA
phorbol 12-myristate 13-acetate
- PNS
postnuclear supernatant
- RB-dextran
rhodamine B–conjugated dextran
- ROS
reactive oxygen species
- SIM
structured illumination microscopy
- siRNA
small interfering RNA
- SNAP-23
synaptosomal associated protein of 23 kDa
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
- TNF
tumor necrosis factor
- UTR
untranslated region
- VAMP
vesicle-associated membrane protein
- V-ATPase
vacuolar-type proton-transporting ATPase
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-01-0069) on October 19, 2012.
*Present address: School of Biotechnology, Jiangnan University, Wuxi, Jiangsu 214122, China.
REFERENCES
- Akagi T, Shishido T, Murata K, Hanafusa H. v-Crk activates the phosphoinositide 3-kinase/AKT pathway in transformation. Proc Natl Acad Sci USA. 2000;97:7290–7295. doi: 10.1073/pnas.140210297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An SJ, Almers W. Tracking SNARE complex formation in live endocrine cells. Science. 2004;306:1042–1046. doi: 10.1126/science.1102559. [DOI] [PubMed] [Google Scholar]
- Bajno L, Peng XR, Schreiber AD, Moore HP, Trimble WS, Grinstein S. Focal exocytosis of VAMP3-containing vesicles at sites of phagosome formation. J Cell Biol. 2000;149:697–706. doi: 10.1083/jcb.149.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becken U, Jeschke A, Veltman K, Haas A. Cell-free fusion of bacteria-containing phagosomes with endocytic compartments. Proc Natl Acad Sci USA. 2010;107:20726–20731. doi: 10.1073/pnas.1007295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Fraisier V, Raposo G, Hurbain I, Sibarita JB, Chavrier P, Galli T, Niedergang F. TI-VAMP/VAMP7 is required for optimal phagocytosis of opsonised particles in macrophages. EMBO J. 2004;23:4166–4176. doi: 10.1038/sj.emboj.7600427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Collins RF, Schreiber AD, Grinstein S, Trimble WS. Syntaxins 13 and 7 function at distinct steps during phagocytosis. J Immunol. 2002;169:3250–3256. doi: 10.4049/jimmunol.169.6.3250. [DOI] [PubMed] [Google Scholar]
- Desjardins M. ER-mediated phagocytosis: a new membrane for new functions. Nat Rev Immunol. 2003;3:280–291. doi: 10.1038/nri1053. [DOI] [PubMed] [Google Scholar]
- Desjardins M, Celis JE, van Meer G, Dieplinger H, Jahraus A, Griffiths G, Huber LA. Molecular characterization of phagosomes. J Biol Chem. 1994;269:32194–32200. [PubMed] [Google Scholar]
- Ellson CD, Anderson KE, Morgan G, Chilvers ER, Lipp P, Stephens LR, Hawkins PT. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol. 2001;11:1631–1635. doi: 10.1016/s0960-9822(01)00447-x. [DOI] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. The phagosome: compartment with a license to kill. Traffic. 2007;8:311–330. doi: 10.1111/j.1600-0854.2006.00531.x. [DOI] [PubMed] [Google Scholar]
- Hackam DJ, Rotstein OD, Bennett MK, Klip A, Grinstein S, Manolson MF. Characterization and subcellular localization of target membrane soluble NSF attachment protein receptors (t-SNAREs) in macrophages. Syntaxins 2, 3, and 4 are present on phagosomal membranes. J Immunol. 1996;156:4377–4383. [PubMed] [Google Scholar]
- Hatsuzawa K, Hashimoto H, Arai S, Tamura T, Higa-Nishiyama A, Wada I. Sec22b is a negative regulator of phagocytosis in macrophages. Mol Biol Cell. 2009;20:4435–4443. doi: 10.1091/mbc.E09-03-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatsuzawa K, Tamura T, Hashimoto H, Yokoya S, Miura M, Nagaya H, Wada I. Involvement of syntaxin 18, an endoplasmic reticulum (ER)-localized SNARE protein, in ER-mediated phagocytosis. Mol Biol Cell. 2006;17:3964–3977. doi: 10.1091/mbc.E05-12-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YH, Cai DT, Wang CC, Huang D, Wong SH. Vesicle-associated membrane protein-8/endobrevin negatively regulates phagocytosis of bacteria in dendritic cells. J Immunol. 2008;180:3148–3157. doi: 10.4049/jimmunol.180.5.3148. [DOI] [PubMed] [Google Scholar]
- Huang X, Sheu L, Tamori Y, Trimble WS, Gaisano HY. Cholecystokinin-regulated exocytosis in rat pancreatic acinar cells is inhibited by a C-terminus truncated mutant of SNAP-23. Pancreas. 2001;23:125–133. doi: 10.1097/00006676-200108000-00002. [DOI] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Jancic C, et al. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat Cell Biol. 2007;9:367–378. doi: 10.1038/ncb1552. [DOI] [PubMed] [Google Scholar]
- Jutras I, Desjardins M. Phagocytosis: at the crossroads of innate and adaptive immunity. Annu Rev Cell Dev Biol. 2005;21:511–527. doi: 10.1146/annurev.cellbio.20.010403.102755. [DOI] [PubMed] [Google Scholar]
- Kawanishi M, Tamori Y, Okazawa H, Araki S, Shinoda H, Kasuga M. Role of SNAP23 in insulin-induced translocation of GLUT4 in 3T3-L1 adipocytes. Mediation of complex formation between syntaxin4 and VAMP2. J Biol Chem. 2000;275:8240–8247. doi: 10.1074/jbc.275.11.8240. [DOI] [PubMed] [Google Scholar]
- Martin-Martin B, Nabokina SM, Blasi J, Lazo PA, Mollinedo F. Involvement of SNAP-23 and syntaxin 6 in human neutrophil exocytosis. Blood. 2000;96:2574–2583. [PubMed] [Google Scholar]
- Mollinedo F, Calafat J, Janssen H, Martin-Martin B, Canchado J, Nabokina SM, Gajate C. Combinatorial SNARE complexes modulate the secretion of cytoplasmic granules in human neutrophils. J Immunol. 2006;177:2831–2841. doi: 10.4049/jimmunol.177.5.2831. [DOI] [PubMed] [Google Scholar]
- Murray RZ, Kay JG, Sangermani DG, Stow JL. A role for the phagosome in cytokine secretion. Science. 2005;310:1492–1495. doi: 10.1126/science.1120225. [DOI] [PubMed] [Google Scholar]
- Rybicka JM, Balce DR, Khan MF, Krohn RM, Yates RM. NADPH oxidase activity controls phagosomal proteolysis in macrophages through modulation of the lumenal redox environment of phagosomes. Proc Natl Acad Sci USA. 2010;107:10496–10501. doi: 10.1073/pnas.0914867107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoul K, Berger A, Niemann H, Weller U, Roche PA, Klip A, Trimble WS, Regazzi R, Catsicas S, Halban PA. SNAP-23 is not cleaved by botulinum neurotoxin E and can replace SNAP-25 in the process of insulin secretion. J Biol Chem. 1997;272:33023–33027. doi: 10.1074/jbc.272.52.33023. [DOI] [PubMed] [Google Scholar]
- Savina A, Amigorena S. Phagocytosis and antigen presentation in dendritic cells. Immunol Rev. 2007;219:143–156. doi: 10.1111/j.1600-065X.2007.00552.x. [DOI] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavo G, Santucci A, Dasgupta BR, Mehta PP, Jontes J, Benfenati F, Wilson MC, Montecucco C. Botulinum neurotoxins serotypes A and E cleave SNAP-25 at distinct COOH-terminal peptide bonds. FEBS Lett. 1993;335:99–103. doi: 10.1016/0014-5793(93)80448-4. [DOI] [PubMed] [Google Scholar]
- Schwartz J, Leidal KG, Femling JK, Weiss JP, Nauseef WM. Neutrophil bleaching of GFP-expressing Staphylococci: probing the intraphagosomal fate of individual bacteria. J Immunol. 2009;183:2632–2641. doi: 10.4049/jimmunol.0804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Souslova EA, Goedhart J, Chepurnykh TV, Gaintzeva A, Shemiakina II, Gadella TW, Lukyanov S, Chudakov DM. Practical and reliable FRET/FLIM pair of fluorescent proteins. BMC Biotechnol. 2009;9:24. doi: 10.1186/1472-6750-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimozono S, Hosoi H, Mizuno H, Fukano T, Tahara T, Miyawaki A. Concatenation of cyan and yellow fluorescent proteins for efficient resonance energy transfer. Biochemistry. 2006;45:6267–6271. doi: 10.1021/bi060093i. [DOI] [PubMed] [Google Scholar]
- Stow JL, Manderson AP, Murray RZ. SNAREing immunity: the role of SNAREs in the immune system. Nat Rev Immunol. 2006;6:919–929. doi: 10.1038/nri1980. [DOI] [PubMed] [Google Scholar]
- Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- Sun-Wada GH, Tabata H, Kawamura N, Aoyama M, Wada Y. Direct recruitment of H+-ATPase from lysosomes for phagosomal acidification. J Cell Sci. 2009;122:2504–2513. doi: 10.1242/jcs.050443. [DOI] [PubMed] [Google Scholar]
- Sun-Wada GH, Tabata H, Kuhara M, Kitahara I, Takashima Y, Wada Y. Generation of chicken monoclonal antibodies against the a1, a2, and a3 subunit isoforms of vacuolar-type proton ATPase. Hybridoma (Larchmt) 2011;30:199–203. doi: 10.1089/hyb.2010.0087. [DOI] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Swanson JA, Hoppe AD. The coordination of signaling during Fc receptor-mediated phagocytosis. J Leukoc Biol. 2004;76:1093–1103. doi: 10.1189/jlb.0804439. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Hatakeyama H, Okado H, Noguchi J, Ohno M, Kasai H. SNARE conformational changes that prepare vesicles for exocytosis. Cell Metab. 2010;12:19–29. doi: 10.1016/j.cmet.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Uchida T, Kanno T, Hosaka S. Direct measurement of phagosomal reactive oxygen by luminol-binding microspheres. J Immunol Methods. 1985;77:55–61. doi: 10.1016/0022-1759(85)90183-8. [DOI] [PubMed] [Google Scholar]
- Ueyama T, Tatsuno T, Kawasaki T, Tsujibe S, Shirai Y, Sumimoto H, Leto TL, Saito N. A regulated adaptor function of p40phox: distinct p67phox membrane targeting by p40phox and by p47phox. Mol Biol Cell. 2007;18:441–454. doi: 10.1091/mbc.E06-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uriarte SM, Rane MJ, Luerman GC, Barati MT, Ward RA, Nauseef WM, McLeish KR. Granule exocytosis contributes to priming and activation of the human neutrophil respiratory burst. J Immunol. 2011;187:391–400. doi: 10.4049/jimmunol.1003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J. 2002;366:689–704. doi: 10.1042/BJ20020691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Bittner MA, Axelrod D, Holz RW. The structural and functional implications of linked SNARE motifs in SNAP25. Mol Biol Cell. 2008;19:3944–3955. doi: 10.1091/mbc.E08-04-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates RM, Hermetter A, Taylor GA, Russell DG. Macrophage activation downregulates the degradative capacity of the phagosome. Traffic. 2007;8:241–250. doi: 10.1111/j.1600-0854.2006.00528.x. [DOI] [PubMed] [Google Scholar]
- Zeng Q, Subramaniam VN, Wong SH, Tang BL, Parton RG, Rea S, James DE, Hong W. A novel synaptobrevin/VAMP homologous protein (VAMP5) is increased during in vitro myogenesis and present in the plasma membrane. Mol Biol Cell. 1998;9:2423–2437. doi: 10.1091/mbc.9.9.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Ma D, Wang X, Celkan T, Nordenskjold M, Henter JI, Fadeel B, Zheng C. Syntaxin-11 is expressed in primary human monocytes/macrophages and acts as a negative regulator of macrophage engulfment of apoptotic cells and IgG-opsonized target cells. Br J Haematol. 2008;142:469–479. doi: 10.1111/j.1365-2141.2008.07191.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.