Abstract
Stress is a fundamental aspect of aging, as accumulated damage from a lifetime of stress can limit lifespan and protective responses to stress can extend lifespan. In this study, we identify a conserved Caenorhabditis elegans GATA transcription factor, egl-27, that is involved in several stress responses and aging. We found that overexpression of egl-27 extends the lifespan of wild-type animals. Furthermore, egl-27 is required for the pro-longevity effects from impaired insulin/IGF-1 like signaling (IIS), as reduced egl-27 activity fully suppresses the longevity of worms that are mutant for the IIS receptor, daf-2. egl-27 expression is inhibited by daf-2 and activated by pro-longevity factors daf-16/FOXO and elt-3/GATA, suggesting that egl-27 acts at the intersection of IIS and GATA pathways to extend lifespan. Consistent with its role in IIS signaling, we found that egl-27 is involved in stress response pathways. egl-27 expression is induced in the presence of multiple stresses, its targets are significantly enriched for many types of stress genes, and altering levels of egl-27 itself affects survival to heat and oxidative stress. Finally, we found that egl-27 expression increases between young and old animals, suggesting that increased levels of egl-27 in aged animals may act to promote stress resistance. These results identify egl-27 as a novel factor that links stress and aging pathways.
Author Summary
Stress is a fundamental aspect of aging, but it is unclear whether the molecular mechanisms underlying stress response become altered during normal aging and whether these alterations can affect the aging process. In this study, we found a GATA transcription factor called egl-27, whose targets are significantly enriched for age-dependent genes and stress response genes, and whose expression increases with age. In contrast to previous work describing factors that are causal for aging, we found that egl-27 activity is likely beneficial for survival since egl-27 overexpression extends lifespan. egl-27 promotes longevity by enhancing stress response; specifically, increased levels of egl-27 protect animals against heat stress, while reduced egl-27 activity impairs survival following heat and oxidative stress. These results suggest that aging is not simply a process of constant decline. Some factors, such as egl-27, are more active in old animals, working to restore organismal function and to improve survival.
Introduction
Responses to various forms of stress play an important role in aging and longevity. Several types of stress result in damage that can accumulate over time (e.g. oxidative stress results in damaged proteins that often accumulate with age) [1]–[3]. Responses to these stresses have protective effects that can alleviate the effects of damage accumulation. Consistent with this idea, previous studies have found that mutants with extended longevity often exhibit increased stress resistance [4]–[7]. For example, mutations that disrupt the insulin/IGF-1 like signaling (IIS) pathway not only extend longevity, but also increase resistance to many types of stress including heat, oxidative, and pathogenic stress [8]–[11].
As the first genetic pathway in Caenorhabditis elegans that was linked to longevity, the IIS pathway is a conserved endocrine component that controls important aspects of development, metabolism, and stress response [12]. Activation of the IIS receptor (DAF-2) causes phosphorylation of a phosphatidylinositol 3-kinase (AGE-1), which initiates a cascade of signals resulting in phosphorylation and inactivation of a FOXO transcription factor (DAF-16). Reduction of IIS through knockdown of daf-2 or through the presence of certain environmental stresses, results in activation of DAF-16/FOXO, which triggers a transcriptional program that promotes both stress resistance and longevity [10], [12]. Many genes in the DAF-16 transcriptional response are involved in various stress responses, and some of these also change in expression during aging. For example, heat shock proteins are induced by many types of stress including heat and pathogenic infection [13]–[17], and expression levels of certain heat shock genes are increased in C. elegans mutants with reduced IIS and extended longevity. Furthermore, heat shock proteins in C. elegans increase in expression between young and old animals, although expression is reduced in very old populations in which 90% of the population is dead [18], [19].
The increased expression of stress genes during aging is not confined to worms. Studies have shown that genes induced by oxidative stress increase with age in flies [20]; p53-related damage response genes increase with age in mice [21]; also, genes that are involved in immunological complement activation, which are generally induced in response to oxidative and pathogenic stress [22]–[26], increase expression in old age across four human tissues [27]. While these results suggest that stress response pathways become increasingly activated in old organisms, it is unclear whether this activation has a protective function and is beneficial for longevity or whether it represents a misregulation of stress pathways and is a contributor to organismal decline.
In C. elegans, the upstream regions of the genes that constitute the DAF-16 transcriptional program are enriched for both the DAF-16 binding site and a GATA-like transcription factor binding site [28]. One of the GATA factors that may be involved in the DAF-16 mediated IIS transcriptional program is ELT-3, as elt-3 expression is increased in age-1 mutants. Furthermore, elt-3 is required for the longevity phenotype of daf-2 mutants, suggesting that the elt-3/GATA transcription factor functions downstream of the IIS pathway [29].
The GATA family of transcription factors may also play important roles in regulating the molecular changes that accompany normal aging. Transcriptional profiling of young and old animals has revealed that the promoters of age-dependent genes are enriched for GATA motifs. The GATA transcription factor elt-3 is responsible for some of the age-dependent changes in gene expression. Expression of elt-3 declines as worms age, resulting in decreased expression of its downstream targets. Low levels of elt-3 have a deleterious effect on survival and stress response suggesting that this decline in elt-3 levels hastens the aging process [29].
In this work, we identify another GATA transcription factor, egl-27, that functions to promote stress survival and to delay aging. In addition to its homology to GATA factors, egl-27 is also homologous to the MTA1 component of NuRD chromatin remodeling complex [30]–[33]. Previous studies show that egl-27 is expressed in most somatic cells during development and in adult worms [30], [34]. We show that egl-27 expression increases with age and that increased levels of egl-27 through overexpression are sufficient to extend lifespan and to increase survival in response to heat stress. In contrast, reducing egl-27 activity suppresses the longevity and thermotolerance phenotypes of reduced insulin/IGF-1 like signaling. Moreover, egl-27 can respond to the presence of stress as its expression is induced by a variety of different stresses. EGL-27 binds upstream of genes involved in both stress and aging, but interestingly EGL-27 targets are enriched for genes whose expression decreases with age. Finally, egl-27 expression is regulated by the GATA transcription factor elt-3 and the IIS transducing gene daf-16. These results define egl-27 as a novel factor that promotes both longevity and stress response.
Results
egl-27 functions to promote longevity
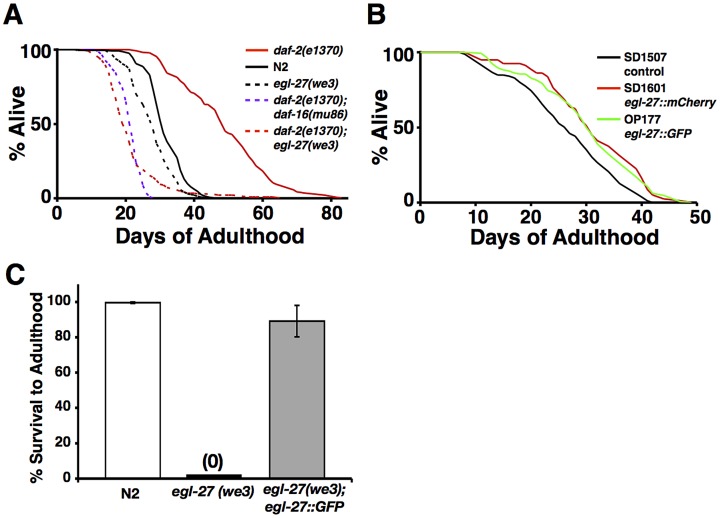
Reduction of egl-27 activity by RNAi knockdown was previously shown to partially suppress the longevity phenotype of the long-lived IIS receptor mutant daf-2(e1370) [29]. We extended this result by showing that the cold-sensitive egl-27(we3) allele could fully suppress the longevity phenotype of daf-2(e1370). Specifically, we found that daf-2(e1370); egl-27(we3) double mutants have a median lifespan that is 2.5 fold shorter than daf-2(e1370) single mutants and 35% shorter than wild-type worms (Figure 1A). egl-27(we3) mutants live only 10% shorter than wild-type worms, suggesting that the combination of the egl-27(we3) mutation and the daf-2(e1370) mutation results in a slight synthetic lethality (Figure 1A).
Figure 1. egl-27 functions to promote longevity.
(A) egl-27(we3) completely suppresses the longevity phenotype of daf-2(e1370) mutants. All worms were hatched at 20°C and shifted to 15°C at day 2 of adulthood. daf-2(e1370): n = 279, mean lifespan = 48.9 days, median lifespan = 49 days, 95% mortality = 70 days. daf-2(e1370); daf-16(mu86): n = 209, mean lifespan = 24.0 days, median lifespan = 25 days, 95% mortality = 30 days. daf-2(e1370); egl-27(we3): n = 234, mean lifespan = 21.5 days, median lifespan = 20 days, 95% mortality = 36 days. p = 0; log rank test comparing daf-2(e1370); egl-27(we3) vs. daf-2(e1370). N2: n = 246, mean lifespan = 31.6 days, median lifespan = 31 days, 95% mortality = 41 days. egl-27(we3): n = 232, mean lifespan = 27.8 days, median lifespan = 28 days, 95% mortality = 37 days. p = 7.8×10−10 vs. N2. Additional lifespan data can be found in Table S1. (B) OP177 and SD1601, two independent strains overexpressing egl-27 extend lifespan compared to SD1507, transgenic control worms. SD1507 (control): n = 165, mean lifespan = 26.3 days, median lifespan = 27 days, 95% mortality = 40 days. SD1601 (egl-27::mCherry): n = 80, mean lifespan = 31 days, median lifespan = 31 days, 95% mortality = 42 days. p-value = 4.0×10−5 vs. control. OP177 (egl-27::GFP): n = 146, mean lifespan = 30.1 days, median lifespan = 30.5 days, 95% mortality = 44 days. p-value = 4.9×10−5 vs. control. Additional lifespan data can be found in Table S1. (C) egl::GFP rescues the larval arrest and embryonic lethality phenotypes of egl-27(we3) mutants. Error bars indicate SEM across 10 independent experiments. Total number of eggs across all 10 experiments for N2 = 167, egl-27(we3) = 154, egl-27(we3); egl-27::GFP = 164.
As a control, we compared the extent of daf-2(e1370) suppression by egl-27(we3) to that of daf-16/FOXO, a well-characterized suppressor of daf-2 [35]. We found that daf-2(e1370); daf-16(mu86) double mutants have a median lifespan that is two-fold shorter than daf-2(e1370) single mutants and 19% shorter than wild-type worms (Figure 1A). These data show that egl-27(we3) suppresses daf-2(e1370) longevity to approximately the same extent as daf-16(mu86).
egl-27(we3) is cold-sensitive for lethality [30], and so we tested whether temperature affects the suppression of daf-2 longevity by egl-27(we3). We hatched worms at the developmentally permissive temperature (20°C) and then shifted them at day 2 of adulthood to 15°C, 20°C, or 25°C. We found that egl-27(we3) suppresses daf-2(e1370) longevity at all three temperatures; specifically, daf-2(e1370); egl-27(we3) worms have a median lifespan that is 2.9 fold shorter than daf-2(e1370) mutants at 15°C, 1.9 fold shorter at 20°C, and 1.7 fold shorter at 25°C (Figure S1A). These results show that egl-27(we3) is temperature-sensitive for developmental arrest but not for suppression of longevity by daf-2(e1370).
We next tested whether increased levels of egl-27 are sufficient to increase longevity. To do this, we engineered three different constructs containing egl-27 and generated strains overexpressing each construct. The first construct is from the modENCODE project and contains GFP-tagged egl-27 in a fosmid with 18 kb of sequence upstream and 8 kb of sequence downstream of egl-27. This fosmid also contains the full coding sequence for three other genes: F31E8.6, F31E8.1, and tbc-1. We found that worms expressing egl-27::GFP had a lifespan extension of 13% (Figure 1B). The second construct contains mCherry-tagged egl-27 with full intergenic regions covering 5 kb of sequence upstream and 152 bp of sequence downstream of egl-27. Worms overexpressing egl-27::mCherry had a lifespan extension of 15% (Figure 1B). Finally, we cloned the egl-27 genomic region containing the we3 temperature-sensitive mutation from egl-27(we3) worms. This construct also contains full intergenic regions. We generated three transgenic worm strains containing egl-27(we3) on an extrachromosomal array in order to create strains that conditionally overexpress egl-27 at the permissive temperature. We grew worms at either the non-permissive or permissive temperature starting at day two of adulthood, and then measured their lifespans. Interestingly, we found that overexpression of egl-27(we3) extended lifespan at both the permissive and non-permissive temperatures. Specifically, at 20°C, median lifespan was increased 23–31%(Figure S1B) and at 15°C, the median lifespan was increased 11–21% (Figure S1C). These results suggest that the addition of low levels of egl-27(we3) activity at 15°C is sufficient to extend lifespan or that egl-27(we3) is temperature sensitive for development but not for its life-extending functions.
To determine whether egl-27 is expressed at higher levels in these overexpression lines compared to control worms, we used qRT-PCR to measure levels of egl-27 mRNA expression in the different overexpression strains and in control worms. We found that egl-27 expression is increased 2.4 fold in the egl-27::GFP strain versus control worms (p = 0.0008, Figure S5E). egl-27 expression is increased 4.1 fold in the egl-27::mCherry strain (p = 0.02, Figure S1D). Finally, egl-27 expression is increased 23%, 11%, and 2.4 fold in the three egl-27(we3) overexpression strains although none of these increases are significant, possibly due to high expression variability caused by extrachromosomal array expression (Figure S1D). We did not observe any abnormal developmental phenotypes in any of the egl-27 overexpression lines, suggesting that these levels of increased egl-27 do not adversely affect development. Furthermore, we validated that transgenic egl-27::GFP can function like endogenous egl-27(+), as we showed that egl-27::GFP can rescue the egl-27(we3) lethal mutant phenotype (Figure 1C).
egl-27 functions to promote heat and oxidative stress survival
Increased longevity is strongly correlated with increased stress resistance [4], [36]. To determine whether egl-27 can promote stress survival, we assessed the relationship between egl-27 activity and survival to heat and oxidative stress. To assay the phenotype of an egl-27 reduction-of-function mutation, we used the hypomorphic mutation egl-27(we3). To assay the phenotype from overexpression of egl-27, we used the egl-27::GFP strain described above.
We assayed heat-stress survival by subjecting worms to 8 hours at 35°C and then measuring survival (Figure 2A). egl-27 reduction-of-function mutants die more quickly after heat stress than wild-type worms; egl-27(we3) has a median survival time following heat-stress that is 2.9 fold shorter than for wild-type worms (log rank p-value = 7.8×10−8). egl-27 gain-of-function worms survive longer after heat stress than control worms; they have a median survival time following heat stress that is 1.4 fold longer than for control worms (p = 5.8×10−6) and they also have a time to 95% mortality that is 2.6 fold longer than for control worms.
Figure 2. egl-27(+) promotes heat and oxidative stress survival.
For A and B, day 1 adult hermaphrodite worms worms were heat stressed at 35°C for 8 hours and survival was scored at 20°C as a function of time. Additional survival data can be found in Table S1. (A) egl-27(we3) worms have increased sensitivity to heat-stress while egl-27::GFP worms are more resistant to heat stress. N2: n = 129, mean survival = 19.6 hours, median survival = 23 hours, 95% mortality = 31 hours. egl-27(we3): n = 114, mean survival = 14.4 hours, median survival = 9 hours, 95% mortality = 31 hours. p = 7.8×10−8 vs. N2. Control: n = 108, mean survival = 24.6 hours, median survival = 23 hours, 95% mortality = 50 hours. egl-27::GFP: n = 78, mean survival = 52.4 hours, median survival = 31 hours, 95% mortality = 134 hours. p = 5.8×10−6 vs. control. (B) Loss of egl-27 activity partially suppresses the stress resistance of daf-2(e1370) mutants. daf-2(e1370) : n = 113, mean survival = 238 hours, median survival = 296 hours. daf-2(e1370); egl-27(we3): n = 118, mean survival = 183 hours, median survival = 172 hours. p = 7.8×10−10 vs. daf-2(e1370). daf-2(e1370); daf-16(mu86): n = 128, mean survival = 19.2 hours, median survival = 23 hours, 95% mortality = 31 hours. N2: n = 129, mean survival = 19.6 hours, median survival = 23 hours, 95% mortality = 31 hours. For C and D, day 1 adult hermaphrodite worms were grown on plates supplemented with 10 mM paraquat and 30 mM FuDR and survival was scored at 20°C as a function of time. Additional survival data can be found in Table S1. (C) egl-27 does not affect paraquat survival in wild-type and control worms. N2: n = 212, mean survival = 3.8 days, median survival = 4 days, 95% mortality = 5 days. egl-27(we3): n = 243, mean survival = 3.6 days, median survival = 4 days, 95% mortality = 5 days. Control: n = 133, mean survival = 5.4 days, median survival = 5 days, 95% mortality = 9 days. egl-27::GFP: n = 272, mean survival = 6.1 days, median survival = 5 days, 95% mortality = 14 days. (D) daf-2(e1370); egl-27(we3) double mutants are less resistant to paraquat-induced oxidative stress than daf-2(e1370) mutants. daf-2(e1370): n = 211, mean survival = 15.7 days, median survival = 14 days, 95% mortality = 29 days. daf-2(e1370); egl-27(we3): n = 284, mean survival = 8.2 days, median survival = 6 days, 95% mortality = 25 days (p-value vs. daf-2(e1370) = 0). daf-2(e1370); daf-16(mu86): n = 262, mean survival = 5.4 days, median = 6 days, 95% mortality = 7 days. N2: n = 212, mean survival = 3.8 days, median survival = 4 days, 95% mortality = 5 days.
daf-2 insulin-like receptor mutants are resistant to many types of stress [9], [11]. We showed that egl-27(we3) partially suppresses the heat resistance phenotype of daf-2. daf-2(e1370); egl-27(we3) double mutants have a median survival that is 1.7 fold shorter than daf-2(e1370) single mutants, but still 7.5 fold longer than wild-type worms. In contrast, a well-characterized suppressor of daf-2, the FOXO transcription factor daf-16(mu86) fully suppresses the heat stress resistance conferred by daf-2 mutants. daf-2(e1370); daf-16(mu86) double mutants have a median survival that is 12.9 fold shorter than daf-2(e1370) single mutants and that is the same as wild-type worms (Figure 2B). Additionally, egl-27(we3) has a less pronounced effect on the heat stress survival of daf-2(e1370) mutants than it does on wild-type worms (1.7 fold shorter vs. 2.9 fold shorter respectively) suggesting that egl-27 may be required for some but not all of the heat-stress resistance of daf-2(e1370) mutants.
We examined whether egl-27 has a functional role in mediating the response to oxidative stress. To do this, we grew worms on plates supplemented with 10 mM paraquat and then measured their survival time. We observed that altering levels of egl-27 did not have a large effect on oxidative stress survival as neither reduction nor gain of egl-27 activity affects median lifespan compared to control worms (Figure 2C). However, we found that egl-27(we3) partially suppresses the oxidative stress resistant phenotype of daf-2(e1370); specifically, daf-2(e1370); egl-27(we3) double mutants have a median lifespan that is 2.5-fold shorter than for daf-2(e1370) mutants and that is 1.4-fold longer than wild-type worms (Figure 2D). Although the median survival time for daf-2(e1370); egl-27(we3) double mutants is similar to that of daf-2(e1370); daf-16(mu86) double mutants (which have a median time of survival that is 2.3-fold shorter than for daf-2(e1370) mutants and that is 1.5-fold longer than for wild-type worms), the time to 95% mortality is different between the two lines. While daf-2(e1370); daf-16(mu86) double mutants have a time to 95% mortality that is 4.1 fold shorter than for daf-2(e1370) single mutants, daf-2(e1370); egl-27(we3) double mutants have a time to 95% mortality that is 16% shorter than for daf-2(e1370) single mutants (Figure 2D). These results suggest that egl-27 activity is important for some of the oxidative stress resistance conferred by reduced activity of the insulin-like receptor gene daf-2.
egl-27 acts downstream of daf-2/ILR and daf-16/FOXO in the IIS pathway, and downstream of the elt-3/GATA transcription factor
The results presented above suggest that egl-27 functions downstream of daf-2 in the IIS pathway. To test whether the IIS pathway can modulate egl-27 expression, we examined whether egl-27 expression is altered in daf-2 mutants. Using fluorescence microscopy, we compared the intestinal expression of an integrated egl-27::mCherry transcriptional reporter in a strain with reduced daf-2 activity to control worms in two day old adult, hermaphrodite worms (Figure 3A). We found that egl-27 expression increased 58% in daf-2(e1370) mutants compared to control worms, providing molecular evidence that egl-27 is regulated by daf-2 in the IIS pathway (Figure 3B).
Figure 3. egl-27 acts downstream of daf-2/ILR, daf-16/FOXO, and elt-3/GATA.
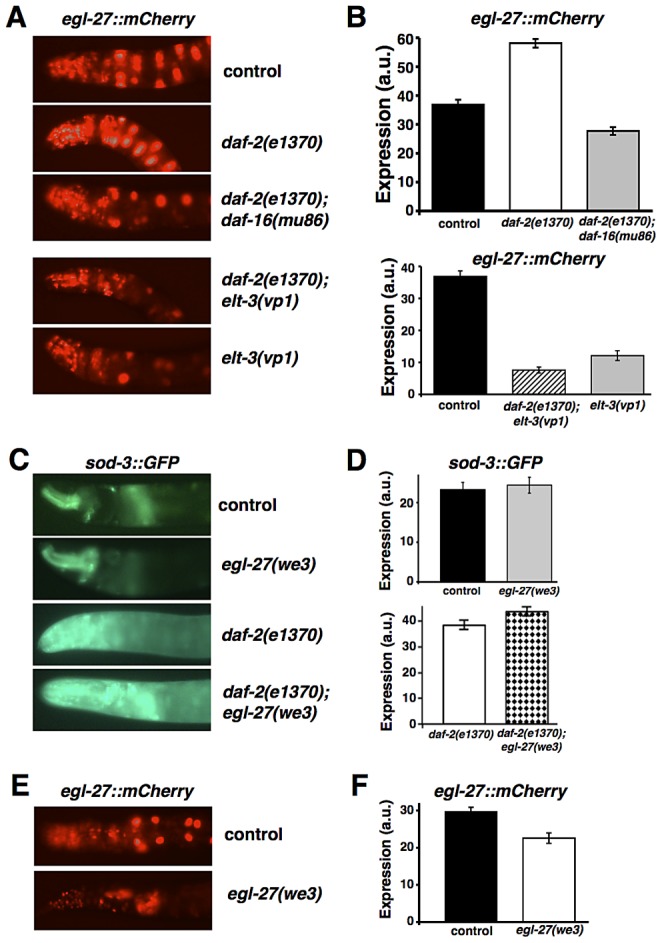
(A) representative images of egl-27::mCherry hermaphrodites at day 2 of adulthood showing median expression levels for each group. (B) egl-27 expression is increased in daf-2(e1370) mutants (p = 1.5×10−11 vs. control) and this increase is suppressed in daf-2(e1370); daf-16(mu86) double mutants (p = 2.9×10−17 vs. daf-2(e1370) worms). egl-27 expression is reduced in elt-3(vp1) mutants (p = 2.3×10−8 vs. control) and the addition of daf-2(e1370) in daf-2(e1370); elt-3(vp1) double mutants is not sufficient to revert this reduction (p = 1.5×10−18 vs. control, p = 9.6×10−29 vs. daf-2(1370) worms). Quantification of intestinal expression for each group in arbitrary units was achieved by measuring fluorescence levels in 20 images using ImageJ. All values indicate mean expression and error bars represent SEM. All p-values are t-test p-values. (C,D) egl-27 does not act upstream of daf-16 or elt-3, as indicated by expression of a sod-3::GFP transcriptional reporter. (C) Representative images showing day 2 adult hermaphrodites grown at 20°C expressing sod-3::GFP. (D) Quantification of intestinal expression for each group in arbitrary units using imageJ to measure fluorescence from 15 images. sod-3::GFP expression is increased in daf-2 mutants compared to wild-type. Levels of sod-3::GFP expression both in control and daf-2(e1370) worms are unaffected by egl-27(we3). (E,F) egl-27 regulates its own expression. (E) representative images showing egl-27::mCherry expression from day 2 adult hermaphrodites that were hatched at 20°C and grown at 15°C from day 1 of adulthood. (F) Quantification of intestinal expression for each group in arbitrary units using ImageJ to measure fluorescence intensity from 30 images. egl-27 expression is increased in egl-27(we3) mutants (p = 0.0015).
To further define where egl-27 acts in the IIS pathway, we examined whether egl-27 acts downstream or upstream of the IIS pathway modulator daf-16/FOXO. To do this, we tested whether a mutation in daf-16 suppresses the increased levels of egl-27 found in daf-2 mutants (Figure 3A). We found that levels of egl-27::mCherry in daf-2(e1370); daf-16(mu86) double mutants are 2.1 fold lower than in daf-2 mutants (Figure 3B). This indicates that daf-16 is required for increased egl-27 expression in daf-2 mutants, which suggests that egl-27/GATA is regulated by daf-16/FOXO in the insulin signaling pathway. Further supporting this idea, we found a canonical DAF-16/FOXO binding element (GTAAACA) [28], [37] 614 bps upstream of the egl-27 translational start site, suggesting that egl-27 may be a direct target of DAF-16.
In addition to daf-16/FOXO, the GATA transcription factor elt-3 modulates IIS, as elt-3 expression is increased in age-1 mutants with reduced IIS and elt-3 is partially required for the longevity phenotype of daf-2 mutants [29]. To determine whether egl-27 acts downstream of the GATA transcription factor elt-3, we tested whether egl-27 expression is affected by a null mutation in elt-3 (Figure 3A). We found that levels of egl-27::mCherry are 3.0 fold lower in elt-3(vp1) mutants compared to control worms, suggesting that egl-27 acts downstream of elt-3 (Figure 3B). We next tested whether elt-3 is required for increased levels of egl-27 expression found in daf-2 mutants. We found that levels of egl-27::mCherry in daf-2(e1370); elt-3(vp1) double mutants are 6.7 fold lower than in daf-2 mutants and 4.8 fold lower than control worms (Figure 3B). Furthermore, levels of egl-27::mCherry in daf-2(e1370); elt-3(vp1) double mutants are similar to levels in elt-3(vp1) mutants (Figure 3B), suggesting that elt-3 is necessary for heightened egl-27 expression in the context of reduced IIS.
In the expression experiments above, we measured egl-27::mCherry expression in the anterior intestine. To determine whether egl-27 is regulated in a tissue-specific manner, we also examined egl-27 expression in the head region, which is composed of several different cell types – hypodermal, neuronal, muscle, and pharyngeal cells. We found that egl-27::mCherry levels are 35% higher in daf-2(e1370) mutants and 34% lower in elt-3(vp1) mutants compared to control worms. egl-27::mCherry levels are 37% lower in daf-2(e1370); daf-16(mu86) double mutants and 68% lower in daf-2(e1370); elt-3(vp1) double mutants compared to daf-2(e1370) worms (Figure S2A). These results suggest that IIS and elt-3 regulate egl-27 expression across multiple tissues, although the magnitude of this regulation may vary slightly across tissues.
We also determined whether egl-27 regulation by IIS and elt-3 GATA transcription occurs only during adulthood, or whether the same regulation occurs during development. To do this, we examined whether egl-27 expression is affected by mutations in daf-2 and elt-3 at the L2 larval stage of development. We found that egl-27 levels are 80% higher in daf-2(e1370) mutants and 3.1 fold lower in elt-3(vp1) mutants compared to control worms (Figure S2B). Because egl-27 is regulated by daf-2 and elt-3 to approximately the same degree during development and adulthood, the genetic networks that regulate egl-27 expression are likely developmental programs that persist into adulthood. To confirm our fluorescent microscopy results, we also examined how endogenous egl-27 levels are affected in daf-2 and elt-3 mutants. Using qRT-PCR, we showed that egl-27 levels are 67% higher in daf-2(e1370) mutants (p = 0.009) and 2.0 fold lower in elt-3(vp1) mutants (p = 0.003) compared to control worms during the L2 larval stage of development (Figure S2C, S2D).
To determine whether egl-27 forms a feedback loop with daf-16 and elt-3, we examined whether egl-27 can act upstream of these regulators. To examine whether egl-27 can regulate either DAF-16 or ELT-3 activity, we examined how reduction of egl-27 affects levels of expression of sod-3::GFP, an established transcriptional reporter for DAF-16 [38], [39] and ELT-3 [29], [39] (Figure 3C). We found that levels of a sod-3::GFP transcriptional reporter are not significantly different between egl-27(we3) mutant worms and control worms (Figure 3D). However, DAF-16 activity is low in wild-type worms, so we examined whether reduction of egl-27 affects levels of sod-3::GFP expression in daf-2 mutants where DAF-16 is highly activated. Similar to previous reports [38], [39], we found using fluorescent microscopy that sod-3 expression is 4.3 fold higher in daf-2(e1370) worms compared to control worms, and that increased sod-3 expression is suppressed in daf-2(e1370); daf-16(mu86) double mutants (Figure S2E). We found that sod-3 levels are equally high in daf-2(e1370); egl-27(we3) double mutants (Figure 3D). These results indicate that reduction of egl-27 activity does not affect expression of sod-3::GFP, suggesting that neither DAF-16 nor ELT-3 activity are affected in egl-27(we3) mutants.
Finally, we examined whether egl-27 can regulate its own expression. To do this, we examined how reduction of egl-27 activity affects the expression of an egl-27::mCherry transcriptional reporter (Figure 3E). We found that egl-27::mCherry expression is reduced by 32% in egl-27(we3) mutants compared to control worms (Figure 3F). This suggests that egl-27 activates its own expression in a feed-forward loop. Supporting this, we found several GATA-like binding motifs in the promoter region of egl-27 (TTATC/GATAA 107 bps upstream, TATCA/TGATA 728 bps upstream, and CTTATCA/TGATAAG 800 bps upstream of the translational start site). These results suggest a role for GATA transcription factors such as ELT-3 or EGL-27 itself, in directly regulating egl-27 expression.
egl-27 expression is induced by multiple stresses
In response to many types of stress, DAF-16/FOXO becomes activated [40], [41]. This leads to increased protection from the stress itself, and may also lead to increased levels of egl-27 expression. According to this model, various types of stresses could also lead to changes in expression of egl-27, via activation of DAF-16/FOXO. To test this possibility, we exposed worms carrying an egl-27::mCherry transcriptional reporter to six different stresses (osmotic shock, gamma radiation, starvation, heat stress, oxidative damage, and UV damage). We then compared egl-27::mCherry expression under each stress condition to its expression in controls using fluorescent microscopy (Figure 4A). We found that egl-27::mCherry expression is induced after exposure to starvation, heat stress, oxidative damage, and UV stress (p value<0.001) (Figure 4B). Interestingly, none of the stresses increase egl-27 expression by more than two-fold. This suggests that egl-27 acts differently from canonical stress-induced genes, such as heat shock proteins, which are expressed at low levels under normal conditions but can be induced up to 100-fold following heat stress [42], [43].
Figure 4. egl-27 expression is induced in response to stress and aging.
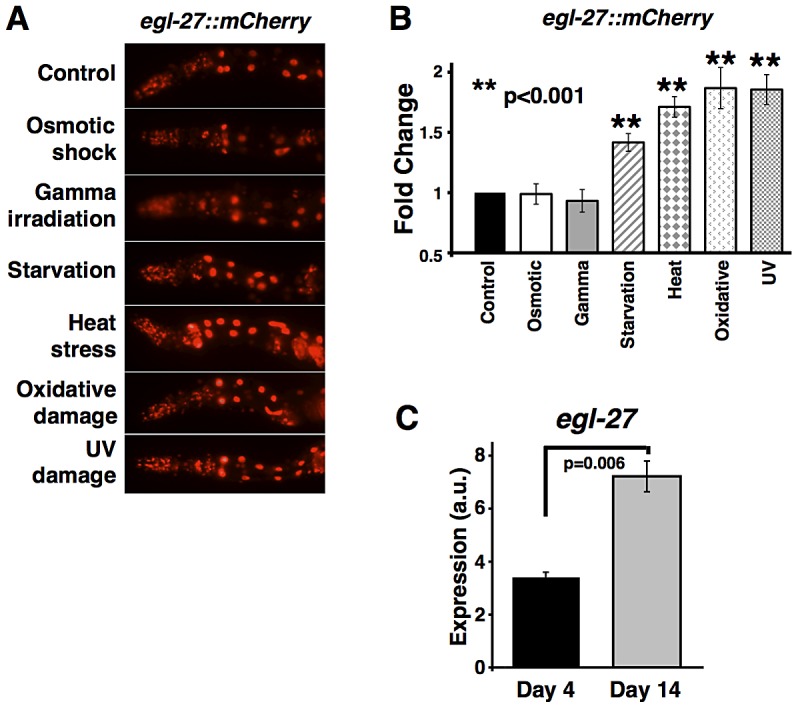
(A,B) egl-27::mCherry expression is induced in response to starvation, heat stress, oxidative stress, and UV stress. Day 1 adult hermaphrodites were exposed to indicated stresses and images of stressed and control worms were taken following stress exposure. (A) Representative images showing egl-27::mCherry expression in control and stressed worms. For all conditions, the worm with the median level of egl-27::mCherry expression is shown. (B) egl-27::mCherry expression was measured by quantification of fluorescence intensity of 15 images. Fold change in egl-27::mCherry expression for every condition was calculated in comparison to paired unstressed control. All values indicate mean expression and error bars represent SEM. (C) egl-27 expression increases with age by qRT-PCR. egl-27 expression levels were calculated using act-1 as a normalization control.
As a control, we showed that the increase in egl-27 expression is specific and not caused by a general increase in transcription in response to these stresses. We used fluorescence microscopy to measure expression levels of myo-3::GFP, a muscle-specific myosin gene (Figure S3A), and showed that its expression is not induced in response to starvation, heat stress, oxidative damage, or UV stress (Figure S3B).
egl-27 expression increases with age
To determine how egl-27 expression changes during aging, we used qRT-PCR to measure egl-27 expression in day 4 and day 14 adult worms. We found that egl-27 expression increases two-fold between young and old worms (Figure 4C). This is consistent with previous work showing that stress genes such as heat shock proteins increase in expression as worms age before declining in expression in extremely old worms [18], [44]. These data suggest that egl-27 expression, like the expression of other stress-related genes [18], [20], [21], [27], [45], increases in old worms.
ChIP–seq analysis identifies EGL-27 binding sites
To better understand the mechanism by which egl-27 promotes longevity and stress survival, we identified where EGL-27 binds in the genome. To do this, we prepared lysates of the egl-27::GFP worm strain described above, at the L2 larval stage of development. These lysates were used by the modENCODE consortium to generate binding site data for EGL-27, using a GFP antibody to immunoprecipate the GFP-tagged EGL-27. We chose to perform ChIP-seq using L2 larval stage worms rather than adult worms because the majority of datasets generated by modENCODE were for larval stages. Because we showed that egl-27::GFP can rescue the egl-27(we3) lethal mutant phenotype (Figure 1C), the sites that are bound by EGL-27::GFP are likely to be representative of the sites that are bound by endogenous EGL-27.
By examining ChIP-seq data from the modENCODE project, we identified 4113 DNA regions showing significant binding by EGL-27::GFP [46]. Previous work has shown that some DNA regions are bound by one or a few transcription factors (factor-specific) while other DNA sites are associated with a large number of transcription factors, and that the specific functions of each transcription factor are better defined by its factor-specific targets than by these redundantly bound targets [46]. Because we were interested in the factor-specific functions of EGL-27, we removed 2306 sites located within redundantly bound regions from further analysis. Of the factor-specific binding sites, 481 are located within gene promoters, defined as 1000 bps upstream to 500 bps downstream of a translational start site. 426 binding sites are located within exons and 466 binding sites are located within introns. 78 are located within 1000 bp downstream from the translational stop site. Finally, 516 are located in intergenic regions. Because we were interested in putative targets of EGL-27, we focused on the 481 peaks located within gene promoters.
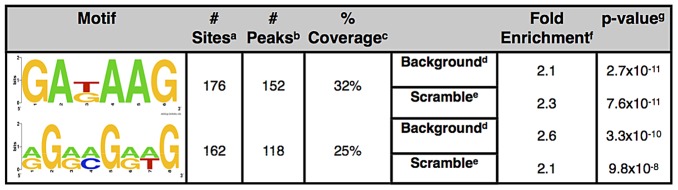
To identify the consensus sites that may be directly bound by EGL-27, we examined the 481 ChIP-seq peaks that fall within gene promoters for the presence of enriched DNA motifs. We used the Gibbs sampling program BioProspector [47] to perform a de novo motif search on the center 100 bp sequence of EGL-27 ChIP-seq identified binding sites. The top 10 motifs found by the program are variations of two motifs: the GATA motif and a novel RGRMGRWG motif (Table S2). The GATA motif (GAKAAG) is found in 32% of EGL-27 target peaks and the novel RGRMGRWG motif is found in 25% of target peaks. Both are significant when compared to background sets consisting of randomly generated 100 bp sequences centered from 1000 bp upstream to 500 bp downstream of translational start sites (Figure 5). Both motifs are also significant when compared to scrambled sequence derived from EGL-27 peaks that preserve nucleotide frequencies (Figure 5).
Figure 5. Motifs enriched in EGL-27 binding sites.
a Total number of times motif is found in EGL-27 binding site. b Number of peaks that contain at least one instance of the motif. c % of peaks that contain at least one instance of the motif. d Promoter sequences randomly sampled from genome. e Scrambling of original peak sequence. f Number of peaks that contain at least one instance of motif/average number of peaks containing at least one instance of motif across 1000 iterations of background or scrambled peaks. g chi-square p-value comparing observed to average number of peaks containing at least one instance of motif across 1000 iterations of background or scrambled peaks.
The novel RGRMGRWG motif is not a consensus-binding site for any known class of transcription factors. However, this motif was previously identified as enriched in the promoters of differentially-expressed genes in insulin signaling daf-2 mutants as well as sirtuin pathway mutants [48].
Since EGL-27 contains a GATA DNA-binding domain, the enrichment for GATA motifs in the EGL-27 binding sites supports its function as a GATA transcription factor. Previous studies have shown that GATA motifs are important for osmotic and pathogenic stress response [49]–[51] and aging [28], [29], which is consistent with our model that egl-27 regulates stress and aging genes by binding the GATA motif.
EGL-27 binds upstream of age- and stress-regulated genes
The 481 EGL-27 promoter peaks are located in the promoter regions of 501 unique genes (Table S3). We conducted gene ontology (GO) analysis [52], [53] on the set of 501 EGL-27 target genes in order to determine if the EGL-27 target genes are enriched for specific biological pathways. We found that EGL-27 targets are enriched for hypodermal genes that form the cuticle as well as intestinal genes involved in aromatic compound catabolic process (Table S4).
We examined whether target genes bound by EGL-27 significantly overlap genes that change expression with age. We obtained a list of 1132 age-regulated genes as previously defined by DNA microarrays [29]. Specifically, we computed the hypergeometric p-value for the overlap between the set of 501 EGL-27 target genes and the set of 1132 age-regulated genes. We found that 67 EGL-27 targets show age-dependent expression, which is a 2.8 fold enrichment over the number expected by chance (p = 9.5×10−15) (Table 1). Even though the ChIP-seq analysis was performed at the L2 larval stage of development, we were still able to find a strong enrichment for age-regulated genes. Interestingly, 61 of the 67 targets decline in expression with age (Figure S4) suggesting that increased levels of egl-27 during normal aging are insufficient to prevent the age-dependent decline of these genes. Using GO analysis, we found that these 67 age-dependent EGL-27 targets are even further enriched for hypodermal genes that form the cuticle as well as several metabolic categories including aromatic amino acid family metabolic process, oxoacid metabolic process, and organic acid metabolic process (Table S4).
Table 1. Hypergeometic p-value for EGL-27 targets and 16 stress conditions and aging.
| Condition | # EGL-27 targets | # Total genes | Fold Enrichment | p-value |
| E. carotovora | 29 | 959 | 1.31 | 0.0837 |
| E. faecalis | 45 | 1161 | 1.68 | 4.54×10−4 |
| P. aeruginosa | 46 | 634 | 4.36 | 6.98×10−17 |
| P. luminescens | 52 | 1146 | 1.97 | 2.32×10−6 |
| S. aureus | 25 | 386 | 3.89 | 9.80×10−9 |
| S. marcesens | 31 | 1120 | 1.2 | 0.169 |
| Cry5b | 53 | 1012 | 3.15 | 2.08×10−13 |
| Cadmium | 55 | 992 | 3.33 | 6.72×10−15 |
| Ethanol | 13 | 219 | 2.84 | 7.36×10−4 |
| Silver | 64 | 1519 | 2.53 | 9.29×10−12 |
| Heat | 37 | 614 | 3.62 | 2.11×10−11 |
| Osmotic | 25 | 313 | 4.8 | 1.31×10−10 |
| Oxidative | 73 | 1243 | 3.53 | 5.61×10−21 |
| Gamma | 48 | 1946 | 1.48 | 4.29×10−3 |
| X-ray | 28 | 607 | 2.77 | 1.50×10−6 |
| Starvation | 127 | 3479 | 2.19 | 4.25×10−18 |
| Aging | 67 | 1132 | 2.83 | 9.51×10−15 |
Because we previously showed that egl-27 expression is induced in response to several types of stress, we examined whether egl-27 targets are differentially-expressed in response to different types of stress. To do this, we acquired published transcriptional responses to 16 different stress conditions involving response to six pathogens, six environmental stresses, and four toxins (Table S5). We then compared each set of genes that are differentially-expressed in that particular stress condition to the set of EGL-27 targets to determine whether the two sets significantly overlap by a hypergeometric test. Even though the ChIP-seq experiment for EGL-27 was not performed under stress conditions, we found that EGL-27 target genes are significantly (p<10−5) enriched for differentially-expressed genes from 11 of the 16 different stress conditions (Table 1). This supports the idea that egl-27 is involved in the response to many types of stress.
egl-27 modulates heat stress gene expression in unstressed and heat-stressed worms
To better characterize how egl-27 mediates heat-stress survival, we assessed how egl-27 regulates heat-stress target genes. To do this, we examined the expression of four egl-27 target genes (grd-3, T14B1.1, Y37A1B.5, and lpr-3) that were previously shown by DNA microarray experiments to be differentially expressed following heat stress [54]. We first wanted to see that expression of these genes change as expected after heat stress in wild-type worms. To do this, we extracted RNA from wild-type second larval stage worms that were exposed to 90 minutes of heat at 35°C followed by a 2-hour recovery. We then used qRT-PCR to determine how the expression of each gene changes in heat stressed worms compared to unstressed worms. Similar to the DNA microarray data, we found that grd-3, T14B1.1, and Y37A1B.5 expression increases while lpr-3 expression decreases after heat stress (Figure 6A–6D).
Figure 6. Effects of egl-27(we3) on expression of heat-stress gene expression.
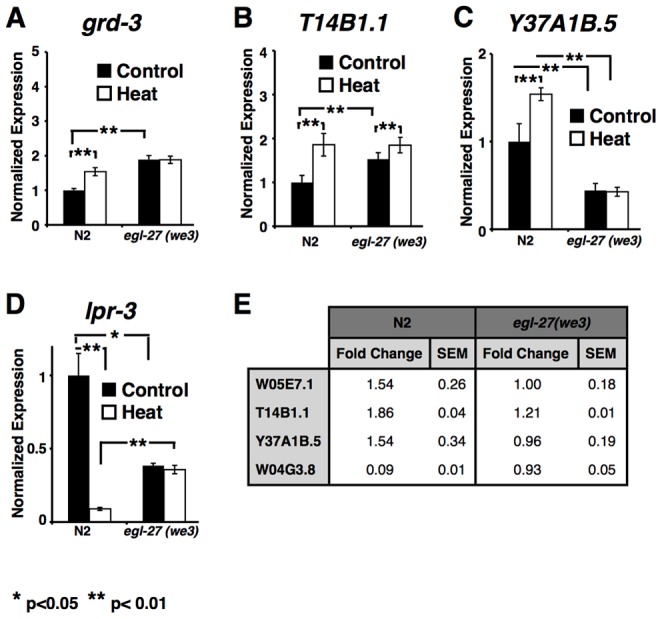
(A–D) qRT-PCR was used to measure levels of gene expression in wild-type (N2) and egl-27(we3) hermaphrodites at the L2 larval stage in unstressed (control) or heat-stressed (heat) conditions. act-1 was used as a normalization control. * p<0.05, ** p<0.01 (A) grd-3 (B) T14B1.1 (C) Y37A1B.5 (D) lpr-3 (E) Fold change of expression between heat-stressed and unstressed worms was calculated for each of the four genes in both N2 and egl-27(we3). SEM = standard error of the mean.
We examined how reduced egl-27 activity affects target gene expression before heat stress. To do this, we compared expression levels of EGL-27 target genes in egl-27(we3) mutants to their expression levels in wild-type worms. We found that two genes (grd-3 and T14B1.1) had higher expression while two genes (Y37A1B.5 and lpr-3) had lower expression in egl-27 loss-of-function worms than in wild-type worms (Figure 6A–6D). This shows that egl-27 plays a role in modulating target gene expression under normal conditions when worms are unstressed.
Next, we assessed how reduction of egl-27 activity affects the heat stress response of these four genes, where the heat stress response is defined as the ratio of expression after heat stress compared to before stress. We found that reduction of egl-27 activity resulted in attenuation of the heat-induced changes of all four genes (Figure 6E). Interestingly, the reduced heat stress response is not caused by lower levels of induced expression, but rather by higher levels of basal expression. Expression of all four target genes was significantly altered in egl-27 reduction-of-function mutants prior to heat stress (Figure 6A–6D). For example, in egl-27 reduction-of-function worms, the expression of two targets (grd-3 and T14B1.1) is induced even in pre-stressed animals. Although expression of these genes remains unchanged following stress, both pre-stress and post-stress expression levels in egl-27 reduction-of-function worms are similar to their levels in post-stress wild-type worms. In the case of these two genes, egl-27 may function to alter baseline expression levels in unstressed worms. However, expression levels of two other targets (Y37A1B.5 and lpr-3) are significantly different in egl-27 reduction-of-function worms compared to wild-type worms both pre- and post- heat stress. For these two genes, egl-27 may function to alter baseline expression levels in unstressed worms and is required for additional activation of expression following heat stress. These results suggest that endogenous egl-27 is required to reduce basal stress levels in wild-type worms.
To complement the reduction-of-function results, we examined how increased basal egl-27 activity in the egl-27::GFP overexpression strain affects differential expression of egl-27 in response to heat-stress (Figure S5). First, we used qRT-PCR to measure the combined RNA levels of egl-27::GFP and endogenous egl-27. As expected, the egl-27::GFP overexpression strain had both increased levels of combined egl-27 expression before and after stress when compared to control worms (Figure S5E). Additionally, we observed that the heat stress response of egl-27 is greater in egl-27::GFP worms compared to control worms (2.9 fold versus 2.2 fold respectively)(Figure S5E). This result suggests that worms with egl-27::GFP have increased levels of egl-27 activity, which may make the expression of egl-27 more sensitive to heat stress as part of a positive feedback loop.
However, the egl-27::GFP strain showed variable effects on the expression of EGL-27 target genes both before and after heat stress (Figure S5A–S5D). Expression of grd-3 was unaffected by increased levels of egl-27 before heat stress. Following heat stress, expression of grd-3 was higher in the egl-27::GFP strain than in the control strain, such that induction levels were 1.8 fold in the egl-27::GFP strain compared to 1.2 fold in the control strain. Expression of T14B1.1 was unaffected by increased levels of egl-27 both before and after heat stress. Expression of Y37A1B.5 was increased in egl-27 gain-of-function worms before heat stress but repressed in these worms following heat stress. Finally, expression of lpr-3 was 19-fold lower in egl-27 gain-of-function worms before heat-stress compared to control worms and 3.6-fold lower following heat stress. Whereas heat stress causes this gene to decrease expression slightly in control worms, heat stress causes its expression to increase in the egl-27::GFP strain. While expression of lpr-3 was significantly down-regulated following heat stress in wild-type worms, lpr-3 levels were not significantly changed in transgenic control worms, underscoring the background differences that can exist between wild-type and transgenic animals. These data suggest that increased levels of egl-27 affect the expression of some target genes both prior to and after heat stress.
Discussion
We have shown that a GATA/MTA1 transcription factor [29]–[33], [55], egl-27, is an important mediator of stress response and longevity in C. elegans. Using binding site data from chromatin immunoprecipitation followed by ultra high throughput sequencing (ChIP-seq), we have shown that EGL-27 binds upstream of genes that are enriched for those that increase with age and that change in response to diverse stresses.
We examined whether EGL-27 has a beneficial or detrimental effect on longevity and stress response. We found that overexpression of egl-27 increases both longevity as well as survival in response to heat stress. In contrast, egl-27 mutants have a shortened lifespan and reduced survival in response to heat stress. Furthermore, reduction of egl-27 activity suppresses the longevity phenotype as well as the heat and oxidative stress resistance phenotypes of daf-2 mutants. These results suggest that egl-27 may promote longevity through promoting stress resistance. This possibility is supported by other studies showing that increased expression of genes that confer resistance to specific stresses also extend lifespan. For example, increased levels of zebrafish lysosyme which confers antimicrobial defense [56], heat shock factor 1 which confers heat resistance [5], [56], and skn-1 which confers resistance to oxidative damage [6], [57], are all capable of extending lifespan when overexpressed in transgenic worms. Here, we identify a GATA transcription factor, egl-27, that plays a role in the response to multiple stresses and has a beneficial effect on longevity when overexpressed.
Altering levels of egl-27 activity affects the expression of heat stress genes both in unstressed as well as heat-stressed worms. From these experiments, we infer that egl-27 normally maintains a program to resolve cellular stress, and that altering levels of egl-27 alters baseline stress levels in worms. Similar to this, a previous study showed that certain stress response genes are expressed at lower levels in stress-resistant daf-2 mutants [58]. This suggests that that changes that alter baseline levels of stress can also alter baseline expression of stress response genes.
egl-27 expression is regulated by a variety of stress pathways. We found that egl-27 expression is induced by multiple stresses: heat stress, oxidative damage, UV irradiation, and starvation. Next, we showed that egl-27 acts downstream of the GATA transcription factor elt-3 and two IIS pathway components (IIS receptor daf-2 and the FOXO transcription factor daf-16). egl-27 expression is induced in long-lived daf-2(e1370) mutants and this induction is suppressed in daf-2(e1370); daf-16(mu86) and daf-2(e1370); elt-3(vp1) double mutants. These data support our finding that egl-27(we3) can fully suppress daf-2(e1370) longevity. Furthermore, previous work has shown that elt-3 is a pro-longevity factor whose expression is confined mainly to hypodermal cells [29], [59], [60]. Our finding that elt-3 can regulate egl-27 expression in several tissue types including the intestine suggests that elt-3 can affect gene expression in a cell non-autonomous manner.
Surprisingly, expression of sod-3, which acts as a readout for DAF-16 activity [38], [39], is unchanged in daf-2(e1370); egl-27(we3) double mutants compared to daf-2(e1370) single mutants, suggesting that activated DAF-16 is not sufficient for extended longevity in the absence of functional egl-27. Finally, we found that egl-27 can regulate its own expression in a feed-forward loop. This evidence for auto-regulation supports the idea that egl-27 may be involved in a complex circuit with feedback mechanism for regulating target gene expression.
Interestingly, egl-27 expression increases with age in wild-type worms. Our finding that increased egl-27 expression extends lifespan and improves stress resistance suggests that the way that egl-27 expression changes during aging is beneficial for the organism. In contrast, previous studies have focused on age-dependent changes in expression that are detrimental for the organism. For example, the GATA transcription factor elt-3 is an important regulator of the transcriptional changes that occur between young and old. elt-3 declines in expression with age and low levels of elt-3 have a deleterious effect on survival and stress response, suggesting that declining levels of elt-3 may act as a driver of aging [29]. Another study found that NF-κB acts as a key regulator of age-dependent gene expression differences in nine types of human and mouse tissues [61]. NF-κB expression increases in old animals, and this increase is detrimental as blocking NK-κB in old skin results in a partial reversal of the aging transcriptome and more youthful skin [61], [62].
In contrast to elt-3 in C. elegans and NF-κB in mice, our studies suggest that changes in egl-27 expression during aging may act to improve stress response and to promote longevity. However, most EGL-27 binding targets from ChIP-seq experiments decline in expression with age, suggesting that this increased expression is insufficient to prevent the age-dependent decline of these genes. Because increased levels of egl-27 extend lifespan, increased expression of egl-27 in old worms appears to delay the aging process instead of causing it. Our work offers novel insight into the interplay between stress and aging, and suggests that aging is not simply a process of moving from an ideal young transcriptome to an inadequate old transcriptome. Rather, age-dependent changes in gene expression are likely comprised of a mix of beneficial, detrimental, and neutral changes.
Materials and Methods
Strains
All C. elegans strains (Table S6) were handled and maintained as described previously [63]. Genotyping primers are described in Table S7.
Analysis of lifespan
Lifespan experiments were conducted as previously described [35], [64]. All experiments were done at 20°C unless otherwise noted. Age refers to days following adulthood and p-values were calculated using the log-rank (Mantel-Cox) method [65].
Genotyping
daf-2(e1370) and egl-27(we3) are single base pair change mutants, so we used the tetra-primer ARMS-PCR procedure [66] to design 4 primers for each SNP of interest. daf-16(mu86) and elt-3(vp1) are deletions so we used a combination of 3 primers (two flanking the deleted region and one inside of the deleted region) to probe for the deletion. To generate DNA for genotyping, 1–10 worms were lysed in 1× PCR buffer with 1.5 mM MgCl2 and Protease K. A single PCR reaction was setup using this DNA and all (three or four) primers and then visualized on a 2% agarose gel. For egl-27(we3), all reactions will produce a 396 bp product; mutant allele will produce a 213 bp product while wild-type allele will produce a 240 bp product. For daf-2(e1370), all reactions will produce a 300 bp product; mutant allele will produce 155 bp product while wild-type allele will produce a 203 bp product. For daf-16(mu86), mutant allele will produce a 400 bp product while wild-type allele will produce a 634 bp product. For elt-3(vp1), mutant allele will produce a 145 bp product while wild-type allele will produce a 401 bp product. Primers and temperatures are detailed in Table S7.
Rescue experiment
Worms carrying the integrated transgene egl-27::GFP (OP177) were crossed to egl-27(we3) (JA1194) to generate egl-27::GFP; egl-27(we3) (SD1751) in the F2 generation, which was identified by PCR genotyping (genotyping primers for egl-27(we3) detailed in Table S7; methods for single nucleotide genotyping above). N2, egl-27(we3), and egl-27::GFP; egl-27(we3) worms were synchronized using hypochlorite and grown to day 1 of adulthood. 10 hermaphrodites from each strain were individually placed onto NGM plates seeded with E. coli and allowed to lay eggs for 1 hour. The adult worms were removed and the number of eggs counted. The numbers of hatched worms were scored 1 day, 2 days (not shown), and 6 days (Figure 1C) after the beginning of egg laying. % survival was computed as the percentage of hatched worms divided by the number of total eggs. Error bars represent the standard error between the 10 replicates.
Imaging and quantification of Cherry and GFP expression
To quantify mCherry and GFP expression, we imaged at least 15 worms for each condition at 20× using a Zeiss Axioplan microscope. Images were analyzed using ImageJ [67].
Stress assays
All assays were performed using day 1 adult hermaphrodites. In all cases, E. coli refers to the OP50 strain. Control worms were always transferred the same number of times, and imaged at the same time as experimental worms. All strains used for imaging and survival assays are detailed in Table S6.
Osmotic shock
Worms were transferred to high salt NGM plates (200 mM NaCl) seeded with concentrated E. coli for 90 minutes and imaged immediately.
Gamma radiation
Worms were transferred to NGM plates without E. coli and irradiated with a 137Cs source at 40 Gy as described previously [29]. Worms were then transferred to NGM plates seeded with E. coli and allowed to recover for 24 hours before imaging. Control worms were transferred to unseeded NGM plates, allowed to recover on seeded NGM plates, and imaged at the same time as experimental worms.
Starvation
Worms were transferred to unseeded NGM plates for 2 hours and imaged immediately.
UV radiation
Worms were transferred to NGM plates without E. coli and irradiated with 30 J/m2 UV using a UV Stratalinker (Stratagene) as described previously [68]. Worms were then transferred to NGM plates seeded with E. coli and allowed to recover for 5 hours before imaging. Control worms were transferred to unseeded NGM plates, allowed to recover on seeded NGM plates, and imaged at the same time as experimental worms.
Heat stress
Worms were incubated at 35°C for 90 minutes, and allowed to recover at 20°C for 1 hour before imaging. For the survival assay, worms were incubated at 35°C for 8 hours before scoring.
Oxidative stress
Worms were transferred to NGM plates containing 10 mM Paraquat, and imaged 24 hours later. For survival assay, worms were transferred onto NGM plates containing 10 mM Paraquat supplemented with 30 mM FuDR to prevent growth of progeny. Plates were seeded with concentrated E. coli. Death was scored as described above.
Identification of ChIP–seq targets
Two independent cultures of worms expressing egl-27::GFP (OP177) were synchronized using sodium hypochlorite to isolate eggs followed by hatching in S basal overnight. Arrested L1 stage larva were collected and grown on NGM plates seeded with E. coli until mid L2 stage. ChIP-seq was performed by the modENCODE consortium [46], [69], [70]. The program PeakSeq [71] was used to identify EGL-27::GFP binding sites (q value<0.00001). Peaks bound by five or more out of the original 23 transcription factors were removed from further analysis. Significant, factor-specific peaks were then compared to the C. elegans genome to identify putative EGL-27 target genes. A gene is identified as a target if the center of a peak occurs 1.5 kb upstream or 500 bp downstream of its translational start site.
Motif analysis
The center 100 bp sequences from the 481 promoter peaks were filtered for low complexity regions using RepeatMasker [72] and then submitted to BioProspector to identify overrepresented cis regulatory sequences [47]. To eliminate motifs or amino acid distributions that are generally enriched in all promoters, a random set containing 481 masked 100 bp promoter sequences was submitted to BioProspector as background input. We report the highest 10 motifs from BioProspector (Table S2), but since only 2 are unique, we ran further analysis on these two.
To determine the fold enrichment and probability of these two motifs occurring in our dataset compared to the probability expected by chance, 1000 datasets containing 481 randomly-generated 100 bp promoter sequences (to match the number of promoter peaks) were created. We scored the number of times these two motifs were found in our promoter sequences and the average number of times they were found in each of the 1000 background datasets. An enrichment factor for each motif was calculated using (# of ChIP-seq peaks containing at least one instance of the motif)/(average # of background peaks containing at least one instance of the motif). A chi-squared p-value for each motif was calculated using the # of ChIP-seq peaks containing at least one instance of the motif as observed and # of background peaks containing at least one instance of the motif as expected (Figure 5).
As a second control, we generated a set containing the 481 original sequences but scrambled the order of the nucleotides in order to create a random sequence while preserving the nucleotide frequency. We generated 1000 iterations of this set of 481 scrambled sequences and for each motif, scored the average number of times they were found in this scrambled sequences. Again, we calculated an enrichment factor and a p-value for each motif in comparison to the scrambled sequences (Figure 5).
Gene Ontology analysis
EGL-27 target genes and age-regulated EGL-27 target genes were submitted to GOrilla [52], [53] for Gene Ontology (GO) analysis. A background gene list was also submitted. For EGL-27 target genes, the background list consisted of all annotated C. elegans genes. For age-regulated EGL-27 target genes, the background list consisted of all genes represented on the Stanford C. elegans microarray platform. GOrilla outputs a FDR q-value and an enrichment score for each GO category. The FDR q-value is the hypergeometric p-value corrected for multiple hypotheses testing using the Benjamini and Hochberg method [73]. Enrichment score is computed as (# of inputted genes in GO category/# of total inputted genes)/(# of genes in each GO category/# of background genes) [52], [53].
Comparison with aging and stress microarrays
Age-regulated genes were obtained from [29] and probe IDs were remapped to WormBase gene IDs (WS228) using annotations from Stanford Microarray Database. Probes that could not be mapped to gene IDs were removed from further analysis. Differentially-expressed stress response genes for each of 16 stress conditions were obtained from publications detailed in Table S5. When probe names were supplied, probe IDs were remapped to WormBase gene IDs (WS228) using annotations from Affymetrix, Stanford Microarray Database, and Washington University Genome Sequencing Center. Probes that match multiple genes were assigned all associated gene IDs. Probes that could not be mapped to gene IDs were removed from further analysis. If only gene names were supplied, gene IDs were filtered using WormBase gene annotations (WS228). Unmatched probe IDs or gene IDs were removed from further analysis.
To determine whether EGL-27 target genes were significantly enriched for age-regulated or stress-response gene sets, we first determined the number of EGL-27 targets that are significantly differentially-expressed in each age or stress response set. For each comparison, the background gene count is the number of genes included in the platform for that microarray. Using this number, we computed, both a hypergeometric p-value and an enrichment score for each comparison, where enrichment score is defined as (# of EGL-27 targets/# of differentially expressed genes)/(# EGL-27 targets/# of background genes).
Quantitative RT–PCR
N2, egl-27(we3), control, and egl-27::GFP (OP177) worms (full genotypes and strain information in Table S6) were synchronized using hypochlorite to isolate eggs followed by hatching in S basal overnight. Arrested L1 stage larva were collected and grown on NGM plates seeded with E. coli until mid-L2 stage before splitting into an experimental and control set. Experimental worms were then incubated at 35°C for 90 minutes and allowed to recover at 20°C for 2 hours before collecting in Trizol (Invitrogen). Control worms were kept at 20°C and collected in Trizol at the same time. Total RNA was isolated using Trizol reagent, treated with DNaseI to degrade genomic DNA, and purified using RNeasy kit (Qiagen). cDNA was synthesized using oligo dT primers and SuperScript II First Strand Kit (Invitrogen). qPCR reactions were performed using RT2SYBR Green qPCR Mastermix (Qiagen) and the 7900HT Fast Real-Time PCR System (ABI). Melting curve analysis was performed with each primer pair to ensure that quantification is the result of only one product. A serial dilution was performed for each primer pair to generate a standard curve. act-1 was used as an internal control to normalize expression levels as previously described [74], [75]. All primers are detailed in Table S7.
Supporting Information
The cold-sensitive effect of egl-27(we3) allele has only a mild effect on longevity. (A) egl-27(we3) suppresses daf-2(e1370) longevity at all temperatures. All worms were hatched at 20°C and shifted to the indicated temperatures at day 2 of adulthood. daf-2(e1370) (15°C): n = 153, mean lifespan = 34.2, median lifespan = 35, 95% mortality lifespan = 44.4. daf-2(e1370); egl-27(we3) (15°C): n = 168, mean lifespan = 17.6, median lifespan = 12, 95% mortality = 43. daf-2(e1370) (20°C): n = 162, mean lifespan = 31.3, median lifespan = 31, 95% mortality = 47. daf-2(e1370); egl-27(we3) (20°C): n = 134, mean lifespan = 19.8, median lifespan = 16, 95% mortality = 43.4. daf-2(e1370) (25°C): n = 216, mean lifespan = 21.8, median lifespan = 22, 95% mortality = 34. daf-2(e1370); egl-27(we3) (25°C): n = 113, mean lifespan = 14.0, median lifespan = 13, 95% mortality = 23.2. (B,C) Three strains overexpressing the we3 allele of egl-27 extend lifespan compared to control. (B) Worms were hatched at 15°C and shifted to 20°C at day 2 of adulthood. Control: n = 61, mean lifespan = 14.3, median lifespan = 13, 95% mortality = 23. egl-27(we3) OE #1: n = 128, mean lifespan = 16.9, median lifespan = 16, 95% mortality = 30. egl-27(we3) OE #2: n = 106, mean lifespan = 17.5, median lifespan = 16.5, 95% mortality = 30.8. egl-27(we3) OE #3: n = 137, mean lifespan = 18.3, median lifespan = 17, 95% mortality = 30. (C) Worms were hatched at 15°C and maintained at 15°C throughout adulthood. Control: n = 54, mean lifespan = 21.4, median lifespan = 19, 95% mortality = 38. egl-27(we3) OE #1: n = 131, mean lifespan = 23.1, median lifespan = 21, 95% mortality = 38. egl-27(we3) OE #2: n = 92, mean lifespan = 23.9, median lifespan = 23, 95% mortality = 38. egl-27(we3) OE #3: n = 99, mean lifespan = 23.4, median lifespan = 21, 95% mortality = 37.1. (D) qRT-PCR analysis of egl-27 levels in egl-27 overexpression lines. act-1 was used as a normalization control.
(TIF)
egl-27 acts downstream of IIS and elt-3 GATA transcription in multiple tissues and stages. (A) ImageJ quantification of egl-27::mCherry expression in the head region of 20 day 2 hermaphrodites shows that egl-27 expression is increased in daf-2(e1370) mutants (p = 6.6×10−6 compared to control worms) and that this increase is suppressed in daf-2(e1370); daf-16(mu86) double mutants (p = 5.5×10−7 compared to daf-2(e1370) mutants). egl-27 expression is reduced in both elt-3(vp1) mutants (p = 4.9×10−6 compared to control) and daf-2(e1370); elt-3(vp1) double mutants (p = 0.0016 compared to control, p = 8.6×10−12 compared to daf-2(e1370) mutants). (B) ImageJ quantification of egl-27::mCherry expression in the anterior intestinal region of 20 L2 larval stage hermaphrodites shows that egl-27 expression is increased in daf-2(e1370) mutants (p = 1.6×10−5 compared to control) and reduced in elt-3(vp1) mutants (p = 1.8×10−5 compared to control). (C, D) qRT-PCR shows that endogenous egl-27 levels are altered in daf-2 and elt-3 mutants in hermaphrodites at the L2 stage of development. act-1 was used as a normalization control. (C) egl-27 expression is increased in daf-2(e1370) mutants compared to wild-type worms (p = 0.0088) (D) egl-27 expression is reduced in elt-3(vp1) mutants compared to wild-type worms(p = 0.0029) (E) sod-3::GFP is highly activated in daf-2 mutants. Quantification is of intestinal expression for each group in arbitrary units using ImageJ to measure fluorescence from 15 images. Levels of sod-3::GFP transcriptional reporter are 4.3 fold higher in daf-2(e1370) worms compared to control worms, and this increase is abolished in daf-2(e1370); daf-16(mu86) double mutants.
(TIF)
myo-3 expression levels are not affected by most stresses. (A,B) myo-3::GFP serves as a negative control to show that gene expression is not generally induced under conditions of starvation, heat stress, oxidative stress, and UV damage. myo-3::GFP expression is slightly induced after osmotic stress. (A) representative images showing myo-3::GFP expression in control and stressed worms. For all conditions, the worm with median levels of myo-3 expression is shown. (B) myo-3::GFP expression was measured by quantification of fluorescence intensity of 15 images. Fold change in myo-3::GFP expression for every condition was calculated in comparison to paired unstressed control.
(TIF)
Age-dependent EGL-27 targets primarily decline in expression with age. Heatmap showing expression changes of 67 age-dependent EGL-27 target genes during aging. Expression change for a given day X is represented as −log2(Expression on day X/Expression on day 4). Expression data from [29].
(TIF)
egl-27 regulation of heat stress gene expression. (A–E) qRT-PCR measured gene expression in control and egl-27::GFP (OP177) overexpression worms in unstressed (control) or heat stressed (heat) conditions. act-1 was used as a normalization control. (A) grd-3 (B) T14B1.1 (C) Y37A1B.5 (D) lpr-3 (E) egl-27.
(TIF)
Additional lifespan data.
(DOCX)
Top ten motifs in EGL-27 binding sites found by BioProspector.
(DOCX)
List of EGL-27 targets.
(XLSX)
Top GO categories enriched in EGL-27 targets.
(DOCX)
Description of all stress microarray datasets used.
(DOCX)
Strains.
(DOCX)
Primers.
(DOCX)
Acknowledgments
We thank Cindie Slightam and Beijing Wu for help generating the egl-27::GFP strain and for help in preparing extracts for ChIP–seq. We thank members of the modENCODE consortium for providing ChIP–seq data for EGL-27. We thank Dror Sagi for help generating the egl-27(we3) overexpression strains, Adolfo Sánchez-Blanco for technical advice on the sod-3::GFP expression experiments, and Eric Van Nostrand for help on the computational analysis. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center. We thank members of the Kim lab for helpful discussion, for suggestions and improvements, and for critical reading of the manuscript.
Funding Statement
This work was supported by grants from the National Institute on Aging to SKK (R01AG025941) and by the Stanford Cancer Biology Training Grant to XX. Research in the laboratory of SKK is also supported by the NHGRI, NIGMS, and the Glenn Foundation. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Finkel T, Holbrook NJ (2000) Oxidants, oxidative stress and the biology of ageing. Nature 408: 239–247 doi:10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 2.Kregel KC (2002) Invited Review: Heat shock proteins: modifying factors in physiological stress responses and acquired thermotolerance. [DOI] [PubMed]
- 3. Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, et al. (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature 447: 725–729 doi:10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 4. Johnson TE, de Castro E, Hegi de Castro S, Cypser J, Henderson S, et al. (2001) Relationship between increased longevity and stress resistance as assessed through gerontogene mutations in Caenorhabditis elegans. Exp Gerontol 36: 1609–1617. [DOI] [PubMed] [Google Scholar]
- 5. Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15: 657–664 doi:10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, et al. (2008) Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell 132: 1025–1038 doi:10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hansen M, Taubert S, Crawford D, Libina N, Lee S-J, et al. (2007) Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6: 95–110 doi:10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 8. Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, et al. (2003) Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300: 1921 doi:10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 9. Honda Y, Honda S (1999) The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. Faseb J 13: 1385–1393. [PubMed] [Google Scholar]
- 10. Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 doi:10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 11. Lithgow GJ, White TM, Hinerfeld DA, Johnson TE (1994) Thermotolerance of a long-lived mutant of Caenorhabditis elegans. J Gerontol 49: B270–B276. [DOI] [PubMed] [Google Scholar]
- 12. Kenyon C (2005) The plasticity of aging: insights from long-lived mutants. Cell 120: 449–460 doi:10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 13. Burg MB (1995) Molecular basis of osmotic regulation. Am J Physiol 268: F983–F996. [DOI] [PubMed] [Google Scholar]
- 14. Kalmar B, Greensmith L (2009) Induction of heat shock proteins for protection against oxidative stress. Adv Drug Deliv Rev 61: 310–318 doi:10.1016/j.addr.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 15. Lindquist S, Craig EA (1988) The heat-shock proteins. Annu Rev Genet 22: 631–677 doi:10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 16. Petersen R, Lindquist S (1988) The Drosophila hsp70 message is rapidly degraded at normal temperatures and stabilized by heat shock. Gene 72: 161–168. [DOI] [PubMed] [Google Scholar]
- 17. Zugel U, Kaufmann SH (1999) Role of heat shock proteins in protection from and pathogenesis of infectious diseases. Clin Microbiol Rev 12: 19–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lund J, Tedesco P, Duke K, Wang J, Kim SK, et al. (2002) Transcriptional profile of aging in C. elegans. Curr Biol 12: 1566–1573. [DOI] [PubMed] [Google Scholar]
- 19. McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin C-S, et al. (2004) Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat Genet 36: 197–204 doi:10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- 20. Landis GN, Abdueva D, Skvortsov D, Yang J, Rabin BE, et al. (2004) Similar gene expression patterns characterize aging and oxidative stress in Drosophila melanogaster. Proc Natl Acad Sci USA 101: 7663–7668 doi:10.1073/pnas.0307605101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Edwards MG, Anderson RM, Yuan M, Kendziorski CM, Weindruch R, et al. (2007) Gene expression profiling of aging reveals activation of a p53-mediated transcriptional program. BMC Genomics 8: 80 doi:10.1186/1471-2164-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HPN, et al. (2011) Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature 478: 76–81 doi:10.1038/nature10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Collard CD, Lekowski R, Jordan JE, Agah A, Stahl GL (1999) Complement activation following oxidative stress. Mol Immunol 36: 941–948. [DOI] [PubMed] [Google Scholar]
- 24. Walport MJ (2001) Complement. First of two parts. N Engl J Med 344: 1058–1066 doi:10.1056/NEJM200104053441406. [DOI] [PubMed] [Google Scholar]
- 25. Walport MJ (2001) Complement. Second of two parts. N Engl J Med 344: 1140–1144 doi:10.1056/NEJM200104123441506. [DOI] [PubMed] [Google Scholar]
- 26. Speth C, Rambach G, Lass-Flörl C, Dierich MP, Würzner R (2004) The role of complement in invasive fungal infections. Mycoses 47: 93–103 doi:10.1111/j.1439-0507.2004.00979.x. [DOI] [PubMed] [Google Scholar]
- 27. Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, et al. (2006) Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet 2: e115 doi:10.1371/journal.pgen.0020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, et al. (2003) Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature 424: 277–283 doi:10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 29. Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, et al. (2008) An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134: 291–303 doi:10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Solari F, Bateman A, Ahringer J (1999) The Caenorhabditis elegans genes egl-27 and egr-1 are similar to MTA1, a member of a chromatin regulatory complex, and are redundantly required for embryonic patterning. Development 126: 2483–2494. [DOI] [PubMed] [Google Scholar]
- 31. Solari F, Ahringer J (2000) NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol 10: 223–226. [DOI] [PubMed] [Google Scholar]
- 32. Herman MA, Ch'ng Q, Hettenbach SM, Ratliff TM, Kenyon C, et al. (1999) EGL-27 is similar to a metastasis-associated factor and controls cell polarity and cell migration in C. elegans. Development 126: 1055–1064. [DOI] [PubMed] [Google Scholar]
- 33. Kagias K, Ahier A, Fischer N, Jarriault S (2012) Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc Natl Acad Sci USA 109: 6596–6601 doi:10.1073/pnas.1117031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu X, Long F, Peng H, Aerni SJ, Jiang M, et al. (2009) Analysis of cell fate from single-cell gene expression profiles in C. elegans. Cell 139: 623–633 doi:10.1016/j.cell.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 doi:10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 36. Johnson TE, Cypser J, de Castro E, de Castro S, Henderson S, et al. (2000) Gerontogenes mediate health and longevity in nematodes through increasing resistance to environmental toxins and stressors. Exp Gerontol 35: 687–694. [DOI] [PubMed] [Google Scholar]
- 37. Furuyama T, Nakazawa T, Nakano I, Mori N (2000) Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J 349: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Libina N, Berman JR, Kenyon C (2003) Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell 115: 489–502. [DOI] [PubMed] [Google Scholar]
- 39. Sánchez-Blanco A, Kim SK (2011) Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS Genet 7: e1002047 doi:10.1371/journal.pgen.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lin K, Hsin H, Libina N, Kenyon C (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145 doi:10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 41. Henderson ST, Johnson TE (2001) daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol 11: 1975–1980. [DOI] [PubMed] [Google Scholar]
- 42. Snutch TP, Heschl MF, Baillie DL (1988) The Caenorhabditis elegans hsp70 gene family: a molecular genetic characterization. Gene 64: 241–255. [DOI] [PubMed] [Google Scholar]
- 43. Prahlad V, Cornelius T, Morimoto RI (2008) Regulation of the cellular heat shock response in Caenorhabditis elegans by thermosensory neurons. Science 320: 811–814 doi:10.1126/science.1156093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Golden TR, Melov S (2004) Microarray analysis of gene expression with age in individual nematodes. Aging Cell 3: 111–124 doi:10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- 45. McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D (2004) Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem 279: 44533–44543 doi:10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- 46. Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 doi:10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu X, Brutlag DL, Liu JS (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac Symp Biocomput 127–138. [PubMed] [Google Scholar]
- 48. Rizki G, Iwata TN, Li J, Riedel CG, Picard CL, et al. (2011) The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet 7: e1002235 doi:10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, et al. (2006) A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci USA 103: 14086–14091 doi:10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kerry S, TeKippe M, Gaddis NC, Aballay A (2006) GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS ONE 1: e77 doi:10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rohlfing A-K, Miteva Y, Hannenhalli S, Lamitina T (2010) Genetic and physiological activation of osmosensitive gene expression mimics transcriptional signatures of pathogen infection in C. elegans. PLoS ONE 5: e9010 doi:10.1371/journal.pone.0009010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eden E, Lipson D, Yogev S, Yakhini Z (2007) Discovering motifs in ranked lists of DNA sequences. PLoS Comput Biol 3: e39 doi:10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009) GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 doi:10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mongkoldhumrongkul N, Swain SC, Jayasinghe SN, Stürzenbaum S (2010) Bio-electrospraying the nematode Caenorhabditis elegans: studying whole-genome transcriptional responses and key life cycle parameters. J R Soc Interface 7: 595–601 doi:10.1098/rsif.2009.0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ch'ng Q, Kenyon C (1999) egl-27 generates anteroposterior patterns of cell fusion in C. elegans by regulating Hox gene expression and Hox protein function. Development 126: 3303–3312. [DOI] [PubMed] [Google Scholar]
- 56. Sagi D, Kim SK (2012) An Engineering Approach to Extending Lifespan in C. elegans. PLoS Genet 8: e1002780 doi:doi:10.1371/journal.pgen.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Park S-K, Tedesco PM, Johnson TE (2009) Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell 8: 258–269 doi:10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henis-Korenblit S, Zhang P, Hansen M, McCormick M, Lee S-J, et al. (2010) Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci USA 107: 9730–9735 doi:10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tonsaker T, Pratt RM, McGhee JD (2012) Re-evaluating the role of ELT-3 in a GATA transcription factor circuit proposed to guide aging in C. elegans. Mech Ageing Dev 133: 50–53 doi:10.1016/j.mad.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 60. Kim SK, Budovskaya YV, Johnson TE (2012) Response to Tonsaker et al. Mech Ageing Dev 133: 54–6–discussion57–8 doi:10.1016/j.mad.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Adler AS, Sinha S, Kawahara TLA, Zhang JY, Segal E, et al. (2007) Motif module map reveals enforcement of aging by continual NF-kappaB activity. Genes Dev 21: 3244–3257 doi:10.1101/gad.1588507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Adler AS, Kawahara TLA, Segal E, Chang HY (2008) Reversal of aging by NFkappaB blockade. Cell Cycle 7: 556–559. [DOI] [PubMed] [Google Scholar]
- 63. Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Apfeld J, Kenyon C (1999) Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809 doi:10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 65.Lawless JF, Lawless JF (1982) Statistical models and methods for lifetime data.
- 66. Ye S, Dhillon S, Ke X, Collins AR, Day IN (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29: E88–E88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abramoff MD, Magalhaes PJ, Ram SJ (2004) Image processing with ImageJ. Biophotonics international 11: 36–42. [Google Scholar]
- 68. Murakami S, Johnson TE (1996) A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics 143: 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, et al. (2011) Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21: 245–254 doi:10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, et al. (2010) Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6: e1000848 doi:10.1371/journal.pgen.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, et al. (2009) PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 27: 66–75 doi:10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smit A, Hubley R, Green P, editors (1996) RepeatMasker Open-3.0 Available:http://www.repeatmasker.org. Accessed 26 June 2012.
- 73. Hochberg Y, Benjamini Y (1990) More powerful procedures for multiple significance testing. Stat Med 9: 811–818. [DOI] [PubMed] [Google Scholar]
- 74. Li J, Ebata A, Dong Y, Rizki G, Iwata T, et al. (2008) Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol 6: e233 doi:10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, et al. (2008) Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog 4: e1000105 doi:10.1371/journal.ppat.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The cold-sensitive effect of egl-27(we3) allele has only a mild effect on longevity. (A) egl-27(we3) suppresses daf-2(e1370) longevity at all temperatures. All worms were hatched at 20°C and shifted to the indicated temperatures at day 2 of adulthood. daf-2(e1370) (15°C): n = 153, mean lifespan = 34.2, median lifespan = 35, 95% mortality lifespan = 44.4. daf-2(e1370); egl-27(we3) (15°C): n = 168, mean lifespan = 17.6, median lifespan = 12, 95% mortality = 43. daf-2(e1370) (20°C): n = 162, mean lifespan = 31.3, median lifespan = 31, 95% mortality = 47. daf-2(e1370); egl-27(we3) (20°C): n = 134, mean lifespan = 19.8, median lifespan = 16, 95% mortality = 43.4. daf-2(e1370) (25°C): n = 216, mean lifespan = 21.8, median lifespan = 22, 95% mortality = 34. daf-2(e1370); egl-27(we3) (25°C): n = 113, mean lifespan = 14.0, median lifespan = 13, 95% mortality = 23.2. (B,C) Three strains overexpressing the we3 allele of egl-27 extend lifespan compared to control. (B) Worms were hatched at 15°C and shifted to 20°C at day 2 of adulthood. Control: n = 61, mean lifespan = 14.3, median lifespan = 13, 95% mortality = 23. egl-27(we3) OE #1: n = 128, mean lifespan = 16.9, median lifespan = 16, 95% mortality = 30. egl-27(we3) OE #2: n = 106, mean lifespan = 17.5, median lifespan = 16.5, 95% mortality = 30.8. egl-27(we3) OE #3: n = 137, mean lifespan = 18.3, median lifespan = 17, 95% mortality = 30. (C) Worms were hatched at 15°C and maintained at 15°C throughout adulthood. Control: n = 54, mean lifespan = 21.4, median lifespan = 19, 95% mortality = 38. egl-27(we3) OE #1: n = 131, mean lifespan = 23.1, median lifespan = 21, 95% mortality = 38. egl-27(we3) OE #2: n = 92, mean lifespan = 23.9, median lifespan = 23, 95% mortality = 38. egl-27(we3) OE #3: n = 99, mean lifespan = 23.4, median lifespan = 21, 95% mortality = 37.1. (D) qRT-PCR analysis of egl-27 levels in egl-27 overexpression lines. act-1 was used as a normalization control.
(TIF)
egl-27 acts downstream of IIS and elt-3 GATA transcription in multiple tissues and stages. (A) ImageJ quantification of egl-27::mCherry expression in the head region of 20 day 2 hermaphrodites shows that egl-27 expression is increased in daf-2(e1370) mutants (p = 6.6×10−6 compared to control worms) and that this increase is suppressed in daf-2(e1370); daf-16(mu86) double mutants (p = 5.5×10−7 compared to daf-2(e1370) mutants). egl-27 expression is reduced in both elt-3(vp1) mutants (p = 4.9×10−6 compared to control) and daf-2(e1370); elt-3(vp1) double mutants (p = 0.0016 compared to control, p = 8.6×10−12 compared to daf-2(e1370) mutants). (B) ImageJ quantification of egl-27::mCherry expression in the anterior intestinal region of 20 L2 larval stage hermaphrodites shows that egl-27 expression is increased in daf-2(e1370) mutants (p = 1.6×10−5 compared to control) and reduced in elt-3(vp1) mutants (p = 1.8×10−5 compared to control). (C, D) qRT-PCR shows that endogenous egl-27 levels are altered in daf-2 and elt-3 mutants in hermaphrodites at the L2 stage of development. act-1 was used as a normalization control. (C) egl-27 expression is increased in daf-2(e1370) mutants compared to wild-type worms (p = 0.0088) (D) egl-27 expression is reduced in elt-3(vp1) mutants compared to wild-type worms(p = 0.0029) (E) sod-3::GFP is highly activated in daf-2 mutants. Quantification is of intestinal expression for each group in arbitrary units using ImageJ to measure fluorescence from 15 images. Levels of sod-3::GFP transcriptional reporter are 4.3 fold higher in daf-2(e1370) worms compared to control worms, and this increase is abolished in daf-2(e1370); daf-16(mu86) double mutants.
(TIF)
myo-3 expression levels are not affected by most stresses. (A,B) myo-3::GFP serves as a negative control to show that gene expression is not generally induced under conditions of starvation, heat stress, oxidative stress, and UV damage. myo-3::GFP expression is slightly induced after osmotic stress. (A) representative images showing myo-3::GFP expression in control and stressed worms. For all conditions, the worm with median levels of myo-3 expression is shown. (B) myo-3::GFP expression was measured by quantification of fluorescence intensity of 15 images. Fold change in myo-3::GFP expression for every condition was calculated in comparison to paired unstressed control.
(TIF)
Age-dependent EGL-27 targets primarily decline in expression with age. Heatmap showing expression changes of 67 age-dependent EGL-27 target genes during aging. Expression change for a given day X is represented as −log2(Expression on day X/Expression on day 4). Expression data from [29].
(TIF)
egl-27 regulation of heat stress gene expression. (A–E) qRT-PCR measured gene expression in control and egl-27::GFP (OP177) overexpression worms in unstressed (control) or heat stressed (heat) conditions. act-1 was used as a normalization control. (A) grd-3 (B) T14B1.1 (C) Y37A1B.5 (D) lpr-3 (E) egl-27.
(TIF)
Additional lifespan data.
(DOCX)
Top ten motifs in EGL-27 binding sites found by BioProspector.
(DOCX)
List of EGL-27 targets.
(XLSX)
Top GO categories enriched in EGL-27 targets.
(DOCX)
Description of all stress microarray datasets used.
(DOCX)
Strains.
(DOCX)
Primers.
(DOCX)