Abstract
Subgroups of patients with chronic lymphocytic leukemia (CLL) have distinct expression profiles of Toll-like receptor (TLR) pathway–associated genes. To test the hypothesis that signaling through innate immunity receptors may influence the behavior of the malignant clone, we investigated the functional response triggered by the stimulation of TLRs and NOD2 in 67 CLL cases assigned to different subgroups on the basis of immunoglobulin heavy variable (IGHV ) gene usage, IGHV gene mutational status or B-cell receptor (BcR) stereotypy. Differences in the induction of costimulatory molecules and/or apoptosis were observed in mutated versus unmutated CLL. Different responses were also identified in subsets with stereotyped BcRs, underscoring the idea that “subset-biased” innate immunity responses may occur independently of mutational status. Additionally, differential modulation of kinase activities was induced by TLR stimulation of different CLL subgroups, revealing a TLR7-tolerant state for cases belonging to stereotyped subset #4. The distinct patterns of TLR/NOD2 functional activity in cells from CLL subgroups defined by the molecular features of the clonotypic BcRs might prove relevant for elucidating the immune mechanisms underlying CLL natural history and for defining subgroups of patients who might benefit from treatment with specific TLR ligands.
INTRODUCTION
In recent years, chronic lymphocytic leukemia (CLL) has emerged as a prototype of cancers in which both genetic and microenvironmental factors concur in the onset, expansion and progression of the disease (1). Antigenic stimulation through the B-cell receptor (BcR) seems to be critically involved in CLL development and evolution as evidenced by: (i) restrictions in the immunoglobulin heavy variable (IGHV) gene repertoire (1–7); (ii) different prognosis of patients with different IGHV gene mutational status (8–10); and (iii) the existence of subsets of patients sharing BcRs with restricted, quasi-identical Ig sequences (stereotyped BcRs) (1,3–5,11) in roughly 30% of cases. Taken together, all these observations indicate that a correspondingly restricted set of antigens or structurally related epitopes might be implicated in the selection of clones carrying distinctive BcRs (1,12–16).
Emerging evidence suggests that CLL cells may also cross-talk with their (micro)environment via non–BcR-mediated modalities, including innate immunity receptors. Innate immune recognition relies on a restricted repertoire of receptors that recognize molecular structures (patterns) conserved in the microbial taxa but absent from evolutionarily advanced organisms (17,18). In mammals, the major class of pattern recognition receptors (PRRs) of the innate immune system is the Toll-like receptor (TLR) family (19). The NLR family (nucleotide-binding domain, leucine-rich repeat containing), including the NOD1 and NOD2 receptors, constitutes another important class of PRRs with various functions in the regulation of inflammatory and apoptotic responses (20).
In humans, naïve B cells express low to undetectable levels of all TLRs and rapidly upregulate TLR1, TLR6, TLR7, TLR9 and TLR10 upon crosslinking of the BcR, whereas memory B cells constitutively express these TLRs (21,22). It has been proposed that TLR stimulation acts as a direct third signal amplifying human B-cell responses to antigen (23,24), and a similar role may be invoked for NOD1 and NOD2 stimulation as well (25). Furthermore, recent evidence suggests that functional interactions between the BcR and TLR signaling pathways extend to the control of B-cell anergy (26,27) and/or TLR tolerance (28).
A number of studies have revealed that CLL cells exhibit a TLR expression profile similar to that of antigen-experienced B cells (29–32). Recently, we reported a comprehensive gene expression profiling of the TLR pathway in a large series of CLL patients, in whom we documented significant differences for selected genes in the TLR-signaling framework between cases carrying mutated versus unmutated IGHV genes or assigned to different subsets with stereotyped BcRs (32). On these grounds, we proposed that subgroups of CLL cases defined by BcR molecular features may exhibit distinctive activation patterns of the TLR signaling pathway.
Prompted by these findings, we conducted the present study with the aim of investigating the effects of TLR and NLR stimulation on the biological behavior of the malignant clones in a cohort of 67 CLL cases expressing BcRs with distinct molecular characteristics (that is, differential IGHV gene mutational status, different stereotyped antigen-binding sites). Our study significantly extends previously reported findings that the functional outcomes of TLR and NLR stimulation are quantitatively and qualitatively different between IGHV-unmutated or IGHV-mutated cases. Furthermore, it documents for the first time differential responses among subsets of CLL cases with different stereotyped BcRs, irrespective of the IGHV gene mutational status. These distinct patterns of TLR/NLR functionality might prove relevant for both elucidating the immune mechanisms underlying CLL natural history and defining subgroups of patients who might benefit from treatment with specific TLR ligands.
MATERIALS AND METHODS
Patients
Blood samples were collected from 67 patients with CLL diagnosed according to the revised guidelines of the International Workshop on Chronic Lymphocytic Leukemia/National Cancer Institute (33). All patients were either untreated or off therapy for at least 6 months before the study. Demographic, clinical and biological data for the patients included in the study, including detailed information on IGHV, IGHD and IGHJ gene repertoires and mutational status, are given in Table 1 and Supplementary Tables S1 and S2.
Table 1.
Clinical and biological data of the patient cohort.
| Parameter | Number |
|---|---|
| Sex | |
| Male | 39 |
| Female | 28 |
| Binet stage at diagnosis | |
| A | 54 |
| B | 7 |
| C | 2 |
| CD38 expression | |
| Positive | 23 |
| Negative | 4 |
| Surface Ig expression | |
| MD | 48 |
| G | 16 |
| IGHV gene mutational status | |
| Mutated | 44 |
| Unmutated | 23 |
| Disease progression | |
| Progressive | 25 |
| Stable | 3 |
The study was approved by the local ethics committee of each participating institution.
Polymerase Chain Reaction Amplification and Sequence Analysis of IGHV-IGHD-IGHJ Rearrangements
We performed reverse transcriptase–polymerase chain reaction of IGHV-IGHD-IGHJ rearrangements using IGHV leader primers along with appropriate IGHJ genes, as previously described (3). Purified polymerase chain reaction am-plicons were subjected to direct sequencing on both strands. Sequence data were analyzed with the IMGT® databases (34) and the IMGT/V-QUEST tool (http://www.imgt.org) (35).
Cell Enrichment
CD19+ B cells were negatively selected from peripheral blood samples by use of the RosetteSep kit (StemCell Technologies, Vancouver, BC, Canada) following the manufacturer’s instructions. The desired cells were collected as a highly enriched population by centrifugation on a gradient (Ficoll-Paque; GE Healthcare Bio-Sciences AB, Uppsala, Sweden). This procedure ensures that isolated B cells are not artificially stimulated through positive selection. The purity of all preparations was checked by flow cytometry and always exceeded 95% for CD19+ cells.
Stimulation of CLL Cells
Purified CD19+ B cells (3 × 106 cells/mL) were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mmol/L l-glutamine and 15 μg/mL gentamicin (Sigma-Aldrich, Taufkirchen, Germany) in 24-well plates in the presence or absence of specific TLR and NLR ligands including MALP-2 (mycoplasmal macrophage-activating lipopeptide-2 [Enzo Life Sciences AG, Lausen, Switzerland]) at 0.2 μg/mL, recognizing TLR2/6 heterodimer, Pam3CSK4 (1 μg/mL), a synthetic triacylated lipoprotein recognizing TLR1/2 heterodimer, lipopolysaccharide (LPS) (ultrapure Escherichia coli lipoprotein, 200 ng/mL); imiquimod and loxoribine, a synthetic and a natural ligand respectively, recognizing TLR7 (0.1 μg/mL and 1 mmol/L); ORN 06 (GU-rich oligonucleotide, 1 μg/mL) recognizing TLR8; CpG (ODN2006, stimulatory CpG-ODN type B, human specific) at 2.5 μg/mL, recognizing TLR9; and MDP (muramyl-dipeptide l isoform) at 10 μg/mL, recognizing NOD2, all from InvivoGen (San Diego, CA, USA). Cell cultures were maintained overnight at 37°C in a humidified atmosphere containing 5% CO2.
Analysis of CD25 and CD86 Expression by Flow Cytometry
After 12-h of culture, cells stimulated through the TLRs as described above as well as unstimulated control cells were collected, washed twice and stained for activation markers with anti-CD86-fluorescein isothiocyanate (FITC) (BD Bioscience, San Jose, CA, USA), anti-CD25-phycoerythrin (PE) (BD Bioscience) and 7-amino-actinomycin D (7-AAD) vital dye (Beckman Coulter, Marseille, France). Anti-mouse IgG1k-FITC and anti-mouse IgG1k-PE were also used as isotype controls. After staining, cells were washed and data acquisition followed on a BD FACS CANTO flow cytometer (BD Bioscience). The analysis was performed with BD FACS DIVA software.
Only 7-AAD–negative (viable) cells were analyzed for CD25 and CD86 expression. The percentage of live cells after stimulation differed less than 10% compared with untreated cells, indicating that none of the ligands was toxic to the cells after 12 h of culture at the concentrations used. The percentage of positive cells for each marker after stimulation was compared with the unstimulated control and the difference was estimated; differences >10% were considered significant. As a positive control, we used the TLR9 ligand CpG-ODN, which has been consistently shown to activate CLL cells in a similar setting (31,36).
Analysis of Apoptosis in Untreated and Stimulated CLL Cells by Flow Cytometry
After culture for 3, 6 and 9 d, cells stimulated through the TLRs, as described above, as well as unstimulated control cells were collected, washed twice and stained for annexin V and propidium iodide (PI) by use of the BD FITC Annexin V Apoptosis Detection Kit I (BD Bioscience) to measure cell viability. Data acquisition followed and was performed on a BD FACS CANTO flow cytometer. The analysis was performed with BD FACS DIVA software. The percentage of live cells after stimulation was compared with the unstimulated controls and the difference was estimated; differences >10% were considered significant.
Western Blot Analysis
Total cellular protein was isolated from purified B cells stimulated with Imiquimod or untreated for 1 h, run on a 10% NuPAGE Bis-Tris gel (Invitrogen, Paisley, UK) and transferred to poly-vinylidene difluoride membranes (Invitrogen) as detailed previously (32). Immunoblot analysis was performed with a phospho-JNK–specific mouse monoclonal antibody (Cell Signaling Technology Inc., Beverly, MA, USA). Membranes were stripped and reprobed with rabbit polyclonal antibodies specific for JNK1/3 or p38α (Santa Cruz Biotechnology Inc., Santa Cruz, CA), rabbit monoclonal anti-phospho-p38 antibody (Cell Signaling Technology) and β-actin (Sigma-Aldrich). Immunoreactivity was revealed by incubation with either goat anti-rabbit Ig or goat anti-mouse Ig (Upstate Biotechnology, Lake Placid, NY, USA) conjugated with horseradish peroxidase, and was followed by an enhanced chemiluminescence reaction (Pierce, Rockford, IL, USA) and film exposures.
Statistical Analysis
Descriptive statistics for categorical variables included frequency distributions, and those for quantitative variables included statistical measures like mean, median and standard deviation. Significance of bivariate relationships between factors and variables normally distributed was assessed with the use of a Student t test. In cases in which the underlying distribution was not normal, a Mann-Whitney test was performed. Correlation analysis was performed by use of the Spearman correlation coefficient. For all comparisons, a significance level of p < 0.05 was set. All statistical analyses were performed with the use of the Statistical Package SPSS Version 18.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA).
All supplementary materials are available online at www.molmed.org.
RESULTS
Heterogeneous Functional Responses after Stimulation of CLL cells via TLRs and/or NOD2
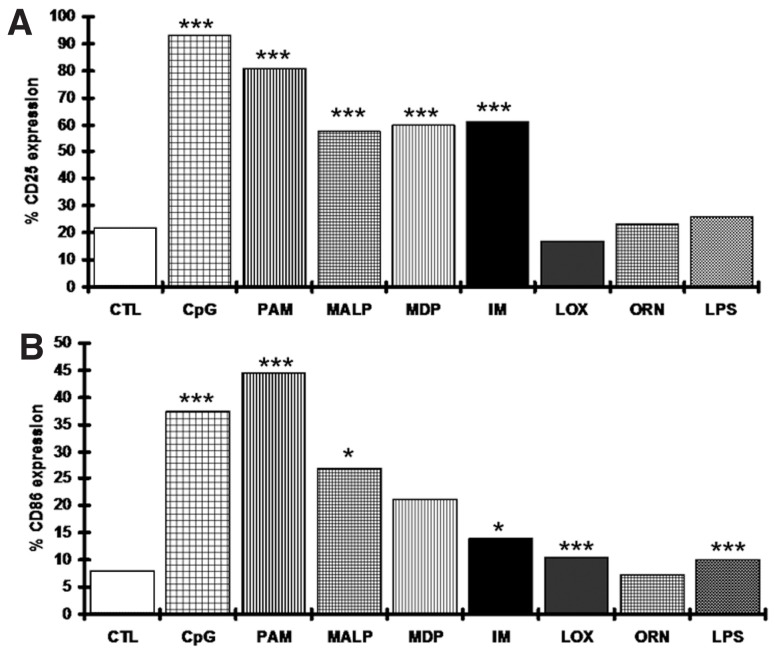
We analyzed the effects of TLR and NOD2 stimulation on both the induction of costimulatory molecules (CD25 and CD86) and the modulation of apoptosis in purified leukemic B cells from 67 and 30 CLL cases, respectively. Overall, we confirmed and significantly extended our previously reported findings (31), showing that: (i) TLR1/2, TLR2/6, TLR9 and NOD2 were functional in the great majority of cases, albeit in a heterogeneous fashion; (ii) TLR4, previously shown to be expressed at very low levels in a minority of CLL cases (32), was unresponsive to stimulation with LPS (in keeping with normal B cells); and (iii) TLR8, showing high protein expression in most CLL cases (31), was unresponsive to stimulation with ORN 08 (Figure 1).
Figure 1.
CLL cells respond differently to stimulation with ligands specific for different TLRs. Changes in CD25 (A) and CD86 (B) expression in all analyzed cases. The graphs represent median values from all cases unstimulated or stimulated with a certain ligand. Statistical analysis for the comparison to the untreated control was performed using a Student t test. Differences >10% from the untreated control with p < 0.05 were considered significant (*** and * indicate p values of <0.0005 and <0.05, respectively). CTL, unstimulated control; CpG, ODN 2006; PAM, Pam3CSK4; MALP, MALP-2; IM, imiquimod; LOX, loxoribine; ORN, ORN06.
Concerning the expression of CD25 and CD86 molecules after TLR stimulation, remarkable heterogeneity was observed, as illustrated for selected representative cases in Figure 2A and detailed in Supplementary Table S3. Furthermore, two novel general themes emerged. First, differential outcomes were obtained after TLR7 stimulation with loxoribine or imiquimod. In particular, whereas loxoribine had no significant effect, imiquimod induced the expression of CD25 and CD86 in a sizeable proportion of cases (39/67 and 21/67 cases, respectively), indicating that these TLR7 ligands induce intracellular signals through different downstream pathways (Supplementary Table S3). Second, concordant patterns of induction of CD25 and/or CD86 were observed after stimulation of TLR1/2, TLR2/6 and NOD2 with their respective ligands (Spearman correlation test, p < 0.05; Supplementary Figure S1).
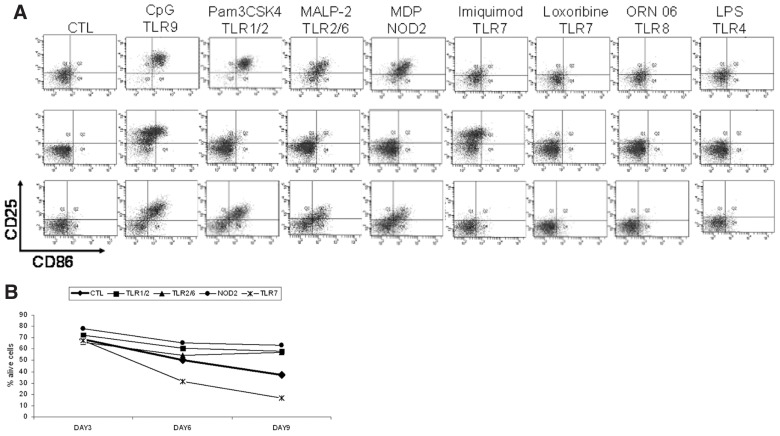
Figure 2.
Heterogeneous functional responses after stimulation of CLL cells via TLRs and/or NOD2. (A) TLR1/2, TLR2/6, TLR7, TLR9 and NOD2 stimulation variably induced the expression of CD25 and/or CD86. In contrast, CLL cells were unresponsive to TLR4 and TLR8 stimulation. Results from flow cytometry analysis for three representative cases. (B) Generally, TLR7 stimulation induced apoptosis on d 6 and 9, whereas TLR1/2, TLR2/6 and NOD2 stimulation had an antiapoptotic effect on d 9, albeit with interpatient variation. The graph shows median values of annexin V−/PI− cells. Before statistical analysis values were normalized to the unstimulated controls.
Concerning apoptosis, as expected, untreated leukemic cells underwent spontaneous apoptosis albeit with significant interpatient variability (Supplementary Table S4). Overall, compared with the unstimulated control, TLR7 stimulation induced significant apoptosis on d 6 and 9, whereas TLR1/2, TLR2/6 and NOD2 stimulation had an antiapoptotic effect only on d 9 (Figure 2B). Going along with the well-known variability in spontaneous apoptosis of CLL cells, large heterogeneity among different CLL cases was observed after TLR1/2, TLR2/6, TLR7, TLR9 and NOD2 stimulation (Supplementary Tables S4, S5).
CLL Clones with Distinct Clonotypic BcRs Respond Differently to TLR and NOD2 Stimulation
We recently reported that subgroups of CLL cases defined by the molecular features of the clonotypic BcRs have distinct expression profiles of the genes associated with the TLR signaling pathway (32). With this in mind, we next explored whether the pronounced heterogeneity of the functional responses after TLR and NOD2 stimulation reported above could be related to distinct BcR molecular features, in particular IGHV gene mutational status and BcR stereotypy.
To this end we normalized the percentages of CD25 and CD86 expression as well as the percentage of live cells at each time point to their respective unstimulated controls. We next calculated the median values for each group of cases and then evaluated the differences for each comparison below. Differences were considered significant at a level of 0.05. Detailed results from all comparisons are given in Supplementary Tables S3 and S5. Statistically significant differences are listed in Tables 2 (induction of costimulatory molecules) and 3 (apoptosis modulation).
Table 2.
Statistically significant differences in (A) CD25 and (B) CD86 expression between different CLL subgroups with distinct clonotypic BcRs after stimulation of TLRs and NOD2 with their respective ligands.a
| Evaluated TLR | Compared groups | Median values, % | p |
|---|---|---|---|
| A. CD25 | |||
| TLR1/2 | #1 versus #8 | 30.7 versus 82.8 | 0.050 |
| #4 versus non-#4/16 | 57.85 versus 7.9 | 0.007 | |
| UM versus #8 | 52.4 versus 82.8 | 0.050 | |
| TLR2/6 | #4 versus non-#4/16 | 35.05 versus 10.6 | 0.033 |
| #16 versus non-#4/16 | 47 versus 10.6 | 0.004 | |
| M versus #16 | 22.1 versus 47 | 0.021 | |
| TLR7 | M versus UM | 8.4 versus 50.1 | 0.001 |
| #4 versus #1 | 11 versus 46.8 | 0.026 | |
| UM versus #8 | 50.1 versus 75.6 | 0.028 | |
| TLR9 | #4 versus non-#4/16 | 61.9 versus 32.8 | 0.022 |
| NOD2 | #4 versus non-#4/16 | 27.8 versus 14.7 | 0.019 |
| B. CD86 | |||
| TLR1/2 | #4 versus #1 | 51 versus 11 | 0.034 |
| #4 versus non-#4/16 | 51 versus 9.8 | 0.003 | |
| #16 versus non-#4/16 | 55.5 versus 9.8 | 0.038 | |
| TLR2/6 | #4 versus #1 | 41.4 versus 6.3 | 0.049 |
| #4 versus non-#4/16 | 41.4 versus 15.3 | 0.022 | |
| #16 versus non-#4/16 | 41.1 versus 15.3 | 0.002 | |
| M versus #16 | 21.3 versus 41.1 | 0.004 | |
| TLR7 | UM versus #8 | 4.2 versus 18.3 | 0.034 |
| TLR9 | M versus UM | 40.4 versus 8.2 | 0.001 |
| #4 versus #1 | 56.8 versus 4.3 | 0.001 | |
| #4 versus #16 | 56.8 versus 24.9 | 0.031 | |
| #1 versus #8 | 4.3 versus 26.8 | 0.048 | |
| NOD2 | #4 versus #1 | 31.5 versus 2.8 | 0.001 |
| #4 versus non-#4/16 | 31.5 versus 16.2 | 0.019 | |
| M versus #4 | 15.2 versus 31.5 | 0.021 | |
Before statistical analysis values were normalized to the unstimulated controls.
M, mutated; UM, unmutated.
IGHV gene mutational status
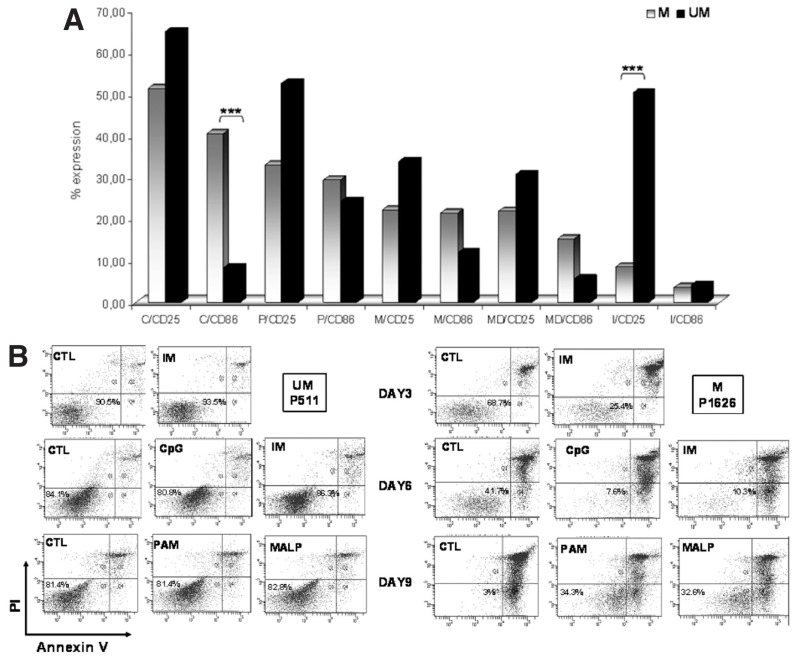
We first compared the functional responses obtained after TLR and NOD2 stimulation in M-CLL versus U-CLL (44 and 23 cases, respectively). TLR1/2, TLR2/6 and NOD2 stimulation induced CD25 and CD86 expression in both groups in a similar manner (Figure 3A). In contrast, differential responses were obtained after stimulation of TLR7 with Imiquimod and of TLR9 with CpG (Table 2, Figure 3A). In particular: (i) TLR7 stimulation induced CD25 expression preferentially in U-CLL versus M-CLL (p < 0.001) (CD86 was not affected by this treatment as shown above); and (ii) TLR9 stimulation induced CD86 expression preferentially in M-CLL versus U-CLL (p < 0.001) (CD25 induction was similar in both M-CLL and U-CLL).
Figure 3.
CLL clones exhibit differential responses to TLR and NOD2 stimulation depending on their IGHV gene mutational status. (A) Induction of CD25 and CD86 after stimulation of all functional receptors tested here (TLR1/2, TLR2/6, TLR7, TLR9 and NOD2). The graph shows median values (before statistical analysis values were normalized to the unstimulated controls). C, CpG ODN; P, Pam3CSK4; M (in x axis), MALP-2; MD, MDP; I, imiquimod; ***p < 0.001. Mutated cases (M [in upper corner]) are indicated in grey; unmutated cases (UM) are indicated in black. (B) TLR1/2, TLR2/6, TLR7 and TLR9 differentially regulate CLL cell apoptosis in mutated-CLL (M) versus unmutated-CLL (UM). Results from flow cytometry analysis for two representative cases are shown. CTL, control; IM, imiquimod; CpG, CpG ODN; PAM, Pam3CSK4; MALP, MALP-2.
Regarding cell viability, among unstimulated cells, similar rates were recorded for both M-CLL and UM-CLL (18 and 12 cases, respectively) on d 3 and 6; in contrast, significantly higher rates of viability were observed on d 9 in UM-CLL versus M-CLL (p < 0.05). Stimulation with various TLR ligands differentially modulated cell viability in M-CLL versus UM-CLL (Figure 3B). More specifically, TLR7 and TLR9 stimulation had a preferential proapoptotic effect on M-CLL versus U-CLL on d 3 and 6 (p < 0.05); an opposing effect was seen with TLR1/2 and TLR2/6 stimulation on d 9, both leading to protection from apoptosis selectively in M-CLL versus U-CLL (p < 0.05).
BcR stereotypy
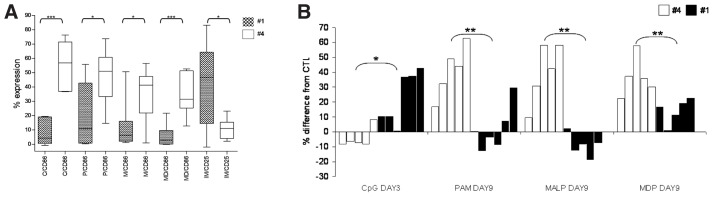
We next evaluated whether the clustering of CLL cases in subsets based on BcR stereotypy might be reflected in subset-biased responses to TLR and NOD2 stimulation. Special emphasis was given on subsets #1 (unmutated IGHV1/5/7-IGKV1(D)-39 BcRs) and #4 (mutated IGHV4-34/IGKV2-30 BcRs, IgG-switched) for the following reasons: (i) they are the largest subsets among UM-CLL and M-CLL, with reported frequencies by us and others in the order of 2.5%–3% and 1% (3,5,7), respectively; (ii) they differ significantly in terms of clinical presentation and outcome and, thus, can be considered as prototypes of “unmutated-bad prognosis” and “mutated-good prognosis” CLL subsets, respectively (3–5,11); (iii) they were the most populated subsets in our cohort (7 and 6 cases, respectively), enabling us to perform more meaningful comparisons; and (iv), perhaps most relevant for the purposes of the present analysis, as we recently documented, they exhibit “subset-biased” expression profiles of TLR pathway–associated genes (32).
Regarding the expression of the costimulatory molecules CD25 and CD86, TLR1/2, TLR2/6, TLR9 and NOD2 stimulation strongly upregulated CD86 expression almost exclusively in subset #4, with minimal effects on subset #1 (51% ± 19% versus 11% ± 22%, 41.4% ± 19% versus 6.3% ± 17%, 56.8% ± 16% versus 4.3% ± 8%, 31.5% ± 10% versus 2.8% ± 15%; p < 0.05, Figure 4A) (CD25 was similarly induced in both subsets, Supplementary Table S3). In contrast, TLR7 stimulation with Imiquimod induced CD25 upregulation selectively in subset #1 (11% ± 7% versus 46.8% ± 27%; p < 0.05, Figure 4A) (in both subsets, CD86 expression was unaffected by this treatment, Supplementary Table S3).
Figure 4.
Differential responses to TLR and NOD2 stimulation in stereotyped CLL subset #1 versus #4. (A) Stimulation of TLR9, TLR1/2, TLR2/6, and NOD2 with the respective ligands strongly upregulated CD86 expression in subset #4 cases, whereas the same treatment had minimal or no effects on subset #1 cases. In contrast, stimulation of the TLR7 with imiquimod led to a much stronger upregulation of CD25 expression in subset #1 versus subset #4 cases. Whisker boxes (min to max) for statistically significant differences. Before statistical analysis values were normalized to the unstimulated controls. C, CpG ODN 2006; P, Pam3CSK4; M, MALP-2; MD, MDP; IM, imiquimod. (B) TLR9, TLR1/2, TLR2/6 and NOD2 stimulation with the respective ligands differentially regulates CLL cell apoptosis in subset #1 versus subset #4 cases. Each column corresponds to a different case. CpG, ODN 2006; PAM, Pam3CSK4; MALP, MALP-2; *p < 0.05, **p < 0.005.
As for apoptosis, significantly different effects were identified for certain treatments at certain time points (Figure 4B, Supplementary Table S5). In detail, preferential antiapoptotic effects were seen: (i) on d 3 after TLR9 stimulation in subset #1 versus subset #4 (p < 0.05); and, (ii) on d 9 after TLR1/2, TLR2/6 and NOD2 stimulation in subset #4 versus subset #1 (p < 0.05).
Distinct Innate Immunity Responses in Stereotyped CLL Subsets Irrespectively of IGHV Gene Usage and/or Mutational Status
Finally, we sought to corroborate the concept of “subset-biased” innate immunity responses independently of IGHV gene usage and/or mutational status. To this end we compared the functional responses obtained after TLR and NOD2 stimulation in (i) mutated subset #4 versus M-CLL and (ii) subset #1 versus U-CLL, and we identified the following differences: (i) CD86 induction after NOD2 stimulation in subset #4 versus all other M-CLL (p < 0.05); (ii) protection from apoptosis on d 9 after TLR1/2 and TLR2/6 stimulation in subset #4 versus all other M-CLL (p < 0.05); and (iii) protection from apoptosis on d 3 after TLR9 and NOD2 stimulation in subset #1 versus all other U-CLL (p < 0.05) (Tables 2 and 3; Supplementary Figure S2).
Table 3.
Statistically significant differences in CLL cell viability between different CLL subgroups with distinct clonotypic BcRs after stimulation of TLRs and NOD2 with their respective ligands.a
| Evaluated TLR | Time point | Compared groups | Median values, % | p |
|---|---|---|---|---|
| TLR1/2 | d 9 | M versus UM | 16.9 versus −3.6 | 0.006 |
| #4 versus #1 | 44.3 versus −1.7 | 0.003 | ||
| M versus #4 | 16.9 versus 44.3 | 0.029 | ||
| TLR2/6 | d 9 | M versus UM | 17.1 versus −8.1 | 0.001 |
| #4 versus #1 | 42.2 versus −8.1 | 0.001 | ||
| #4 versus non-#4/16 | 42.2 versus 6.8 | 0.050 | ||
| M versus #4 | 17.1 versus 42.2 | 0.026 | ||
| TLR7 | d 3 | M versus UM | −21.5 versus 1.2 | 0.007 |
| d 6 | M versus UM | −27.3 versus −7.3 | 0.007 | |
| #4 versus non-#4/16 | −18 versus −43.2 | 0.050 | ||
| TLR9 | d 3 | #4 versus #1 | −7.5 versus 23.7 | 0.031 |
| UM versus #1 | 7.7 versus 23.7 | 0.050 | ||
| d 6 | M versus UM | −16.7 versus 1.7 | 0.006 | |
| NOD2 | d 3 | UM versus #1 | 5.5 versus 18.3 | 0.009 |
| d 9 | #4 versus #1 | 36.6 versus 16.6 | 0.016 |
Median values represent the percentage of live cells (annexin V−/PI−). Before statistical analysis values were normalized to the unstimulated controls.
M, mutated; UM, unmutated.
Our study also included three cases each belonging to stereotyped subsets #8 (IGHV4-39/IGKV1(D)-39, unmutated, IgG-switched [3,37]) and #16 (IGHV4-34/ IGKV3-20, mutated). Although caution is needed due to small numbers (reflecting their overall lower frequency in CLL—on the order of 0.4% and 0.2%, respectively [3,5,7]), it is relevant to mention that differential functional outcomes in terms of both costimulation and apoptosis regulation were noted when we compared un-mutated subset #8 versus other U-CLL and mutated subset #16 versus other M-CLL (Tables 2 and 3; Supplementary Figure S3). Distinct, subset-biased profiles were also identified when we limited the comparisons to subsets with similar mutational status (that is, subset #1 versus #8 and subset #4 versus #16), underscoring the idea of subset-biased innate immunity responses independently of IGHV gene usage and/or mutational status (Table 2; Supplementary Figure S3).
To further corroborate the concept that BcR stereotypy may underlie distinct profiles of functional responses to TLR ligands, we focused on cases using the IGHV4-34 gene, which were intentionally overrepresented in the cohort. Such cases, all expressing mutated BcRs, were classified into three groups: (i) subset #4 (n = 6); (ii) subset #16 (n = 3); and (iii) remaining cases (“non-#4/16,” n = 10). Interestingly, when comparing the two subsets to nonsubset cases, we still identified differential responses after certain treatments for both costimulation and apoptosis (Tables 2 and 3; Figure 5).
Figure 5.
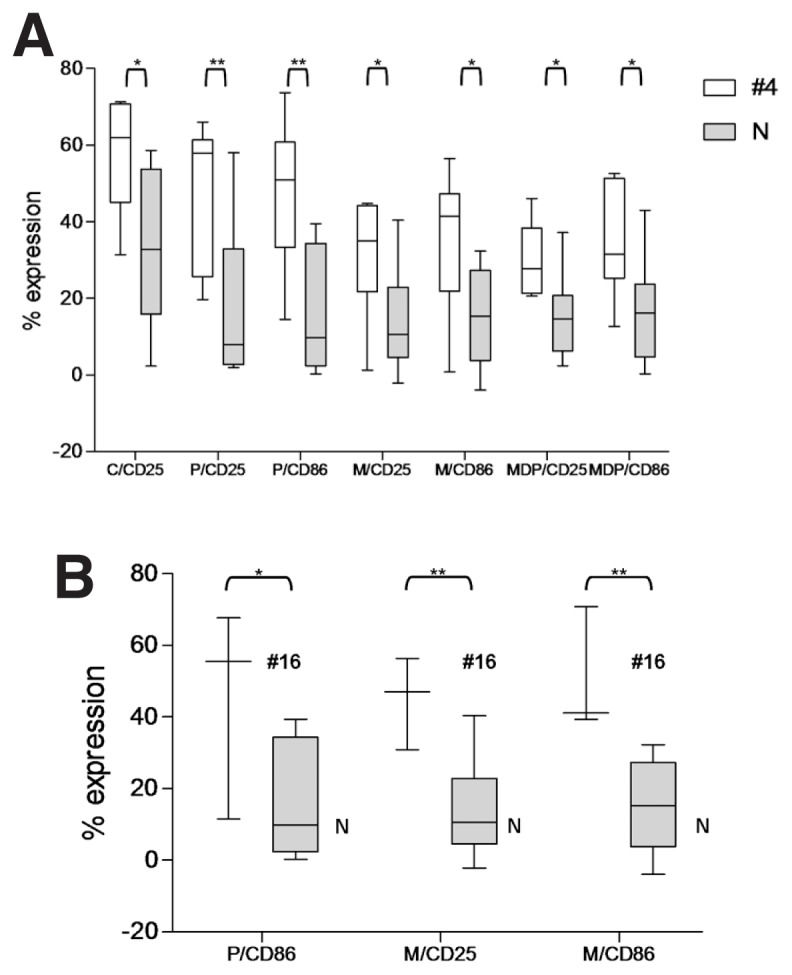
Differential responses to TLR and NOD2 stimulation in cases using the IGHV4-34 gene and assigned or not to certain stereotyped subsets. (A) Statistically significant differences were identified after stimulation of TLR1/2, TLR2/6, TLR9 and NOD2 with the respective ligands in different IGHV4-34–expressing CLL cases with stereotyped BCRs assigned to subsets #4 versus non-#4/16 BCRs (N). (B) Statistically significant differences were also identified after stimulation of TLR1/2 and TLR2/6 with the respective ligands in different IGHV4-34–expressing CLL cases with stereotyped BCRs assigned to subsets #16 versus non-#4/16 BCRs (N). Whisker boxes (minimum to maximum) represent only the statistically significant differences. Before statistical analysis values were normalized to the un-stimulated controls. C, CpG ODN 2006; P, Pam3CSK4; M, MALP-2; *p < 0.05, **p < 0.005.
Attenuated Kinase Activity in CLL Subset #4 after TLR7 Stimulation
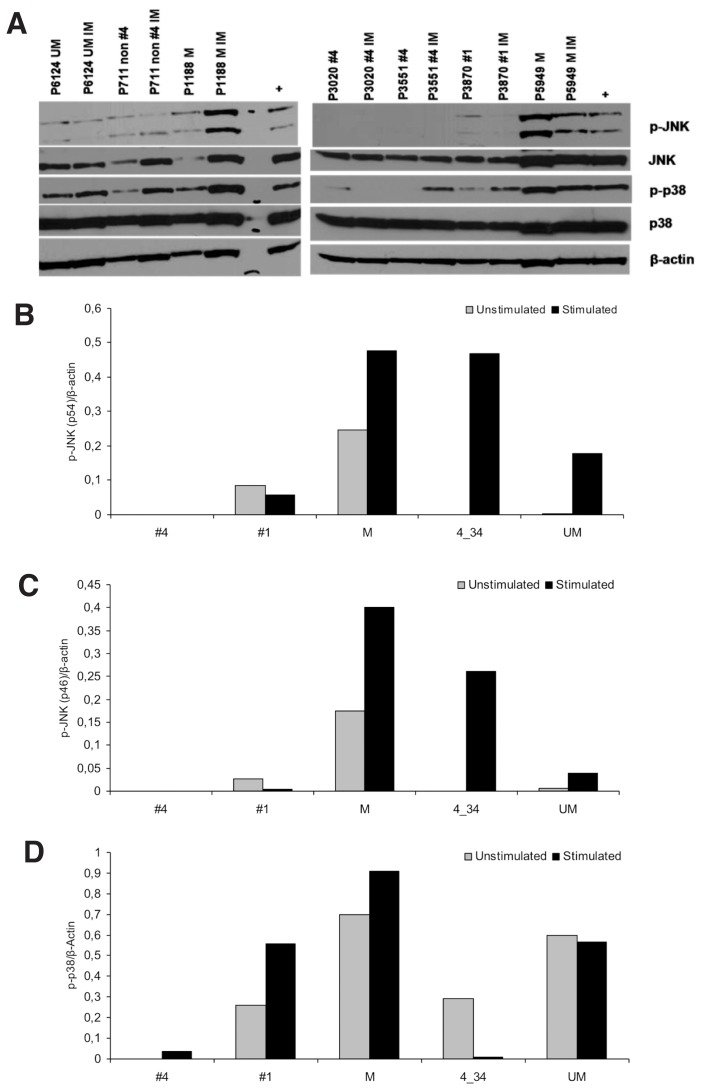
A recent study demonstrated that CLL B cells can become TLR7 tolerant after in vivo exposure to TLR7 ligands (38). Taking into account that tolerant cells are characterized by aberrant activation of JNK and p38 kinases, we investigated the phosphorylation status of JNK and p38 in the basal state and after stimulation of TLR7 with imiquimod in CLL cells from three cases each for subsets #1 and #4 and two cases each for M-CLL, U-CLL and non-#4/16 cases. In most cases, JNK and p38 phosphorylation was readily apparent under both unstimulated and stimulated conditions, although with heterogeneous patterns. The significant exception concerned subset #4 cases, in which patients showed no or low phosphorylation of either JNK or p38 (Figure 6), raising the possibility that subset #4 may have been TLR7 tolerant.
Figure 6.
TLR7 stimulation modulates p38 and JNK phosphorylation in CLL but has significantly less pronounced or no effect on stereotyped subset #4 cases. (A) Immunoblotting studies revealed that stimulation through TLR7 with imiquimod affects JNK and p38 phosphorylation in various CLL subgroups, both unmutated and mutated, with the significant exception of stereotyped subset #4 cases that are generally also nonresponsive to TLR7 stimulation in terms of costimulatory molecule or apoptosis induction. In this figure seven representative cases are represented. Densitometric analysis of (B) p-JNK(p54)/β-actin (C) p-JNK(p46)/β-actin and (C) p-p38/β-actin levels in the 12 CLL samples analyzed. The graphs represent the median values for each group which determined as the ratio of the optical density (OD) of p-protein and β-actin using the ImageJ software (National Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/ij/, 1997–2012). IIM, imiquimod; M: mutated; UM: unmutated; non #4 (4_34), cases expressing the IGHV4-34 gene in non-stereotyped rearrangements; #1, stereotyped subset #1; +, positive control (CLL cells treated for 1 h with CpG).
DISCUSSION
In the past decade, a wealth of new information has brought CLL to a crossroad between cancer and autoimmunity. Great emphasis has been ascribed to the BcRs expressed by malignant B cells, because molecular and functional evidence supports a critical role for BcR-mediated interactions with and selection by antigens in CLL immunopathogenesis (1,2,12–16).
The role of immune pathways operating independently of or superimposed to the BcR, including those activated by the stimulation of innate immunity receptors (for example, TLRs, NLRs), has been relatively overlooked, although several studies indicate their potential effects on B-cell functional status (27,28,32,39). Our recent finding that CLL subgroups defined by specific molecular characteristics of the clonotypic BcRs exhibit distinct expression profiles of TLR pathway–associated genes (32) led us to investigate whether these differences are also functionally relevant. We here show that TLRs and NLRs are functional in CLL cells in a heterogeneous fashion and that CLL subgroups expressing distinctive BcRs present distinct patterns of TLR/NLR functionality and/or TLR tolerance.
By analyzing the responses at the cohort level we found that TLR1/2, TLR2/6 and NOD2 stimulation showed similar patterns of functionality, evidenced by concordant responses to their respective ligands. Although not unexpected, given that ligands specific for different TLRs (for example, triacetylated or diacetylated bacterial lipoproteins for TLR1/2 or TLR2/6, peptidoglycan for NOD2) can coexist on the same type of pathogens, these findings reinforce the concept that a single pathogen may affect the behavior of CLL cells through the concerted stimulation of multiple immune receptors.
Significantly different responses were observed when CLL cells from cases with different IGHV gene mutational status were treated with specific TLR ligands. Imiquimod upregulated CD25 expression only in UM-CLL, whereas it promoted apoptosis only in M-CLL. CpG was more efficient in upregulating CD86 expression in M-CLL, in which it also induced apoptosis. These results are in keeping with those of a recent study showing that TLR9 stimulation selectively induces apoptosis in M-CLL (39). Therefore, M-CLL and U-CLL show different TLR functional responses, which, along with their well-established differential signaling through the BcR (40–42), likely indicate distinct modalities of BcR collaboration with specific TLRs. The net effect can be a significant impact on the behavior of the CLL clone.
Several lines of evidence indicate that the clustering of CLL cases into distinct subsets based on stereotyped primary Ig gene sequences is functionally and, likely, clinically relevant in addition to, but also independently of, IGHV gene mutational status (3,4,5,11,43). Indeed, CLL monoclonal antibodies from cases assigned to the same subset have been shown to exhibit similar antigen reactivity profiles, and preliminary evidence suggests that these profiles may underlie clinical behavior and outcome (15,16).
With this in mind, we investigated TLR functional profiles in relation to BcR stereotypy with a special focus on subsets #1 and #4, because they can be considered as prototypes of bad and good prognosis patients, respectively (3–5). Additionally, as we recently showed, these subsets exhibit distinct gene expression profiles of the TLR signaling pathway, suggesting subset-biased recognition of and selection by selective TLR ligands (32). Our present findings lend weight to this possibility. Indeed, we report subset-biased functional outcomes after specific TLR stimulation of subset #1 and subset #4, indicating that distinct immune pathways are operating in cases assigned to different stereotyped subsets.
In particular, markedly different effects on the expression of costimulatory molecules followed TLR1/2, TLR2/6, TLR9 and NOD2 stimulation, which strongly upregulated CD86 expression in subset #4, whereas the same treatment had minimal effects on subset #1. Opposite effects were seen after TLR7 stimulation with imiquimod, with significantly stronger upregulation of CD25 in subset #1 versus subset #4. Certain treatments also exerted a selective apoptosis-protective effect on either subset #1 or #4. Notably, we also identified different profiles when comparing subsets with similar mutational status (that is, subset #1 versus #8, subset #4 versus #16).
Furthermore, by focusing on cases utilizing the IGHV4-34 gene and assigned or not to different well-characterized stereotyped subsets, namely #4 and #16, we identified significantly different responses in subset #4 versus subset #16 versus non-subset#4/16 IGHV4-34 cases. Altogether, these findings underscore the idea of subset-biased innate immunity responses independently of IGHV gene usage or mutational status.
Stereotyped IGHV4-34 BcRs characteristic of subset #4 have distinctive molecular features, including: (i) long and positively charged VH CDR3s (3,4,44), reminiscent of pathogenic anti-DNA antibodies (45); (ii) precisely targeted somatic hypermutation, leading to shared (“stereotyped”) amino acid replacements, while at the same time sparing the VH FR1 motif responsible for super-antigenic-like interactions with self and microbial antigens that express the N-acetyllactosamine epitope (4); and (iii) pronounced intraclonal diversification likely within the context of ongoing interactions with antigen(s) even after transformation (46,47). Additionally, subset #4 CLL cases are distinctive for uniformly expressing IgG-switched BcRs (3,44,48), and they also exhibit subset-biased antigen reactivity (13,16), single-nucleotide polymorphism (49), gene expression (50), and TLR profiles (32) compared with either UM-CLL or M-CLL of both the “classic” IgM/IgD or the rare IgG variant. Altogether, these findings can be considered as evidence for discrete modalities of interaction with and selection by the cognate antigen(s).
The results of the present study lend support to the uniqueness of subset #4. We have already shown that these cases exhibit levels of TLR7 mRNA and protein expression that are high but are lower than those observed in other CLL subgroups, especially subset #1, and that they are also characterized by higher CD86 expression than both M-CLL and U-CLL, including subset #1 (32). We now show that subset #4 cases are unresposive to TLR7 stimulation, hence differing from both U-CLL, including subset #1, and M-CLL, including non-#4 IGHV4-34 CLL. Moreover, compared with all other analyzed cases, even of similar mutational status (that is, M-CLL), subset #4 cases have attenuated JNK and p38 phosphorylation. We propose that this constellation of features, taken together, indicates a TLR7-tolerant state for subset #4 possibly due to a prior in vivo exposure to TLR7 ligands, in keeping with what has been recently reported (38). Admittedly questions abound, especially concerning the precise nature and timing of the antigenic stimulation implicated in the development and evolution of subset #4 CLL clones. However, the fact that TLR7 recognizes single-stranded RNA and is therefore considered to be specialized in viral detection might eventually prove relevant.
CONCLUSION
In conclusion, we demonstrate that the biological behavior of CLL malignant B cells can be influenced by signals triggered by innate immunity receptors. It remains to be seen in larger studies if the distinct functional profiles obtained after TLR stimulation may be linked to clinical characteristics (for example, disease burden at diagnosis) or the genetic makeup of the CLL clones, including recurrent aberrations with prognostic significance (for example, disruption of the TP53 gene). That notwithstanding, on the basis of the present findings it is evident that CLL patient subgroups defined by specific molecular characteristics of their clonotypic BcRs have distinct patterns of TLR/NLR function and/or TLR tolerance. These differences support the hypothesis that specific modalities of BcR/TLR collaboration and/or regulation may eventually have an impact on the biological behavior of the malignant clones. Additionally, they might prove relevant both for elucidating the immune mechanisms that underlie CLL development and also for defining subgroups of patients who might benefit from treatment with specific TLR ligands, already used in the clinical setting (51).
Supplemental Data
ACKNOWLEDGMENTS
This project was supported by the ENosAI project (code 09SYN-13-880), cofunded by the EU and the Hellenic General Secretariat for Research and Technology; Cariplo Foundation (Milan, Italy); the Leukemia Research Foundation (Wilmette, IL, USA); Program Molecular Clinical Oncology-5 per mille no. 9965 and Investigator grants (to P Ghia and F Caligaris-Cappio), Associazione Italiana per la Ricerca sul Cancro (Italy); U.S./European Alliance for the Therapy of CLL, CLL Global Research Foundation (Houston, TX, USA); PRIN (Italy).
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2010;117:1781–91. doi: 10.1182/blood-2010-07-155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–94. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 3.Stamatopoulos K, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood. 2007;109:259–70. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 4.Murray F, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111:1524–33. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 5.Bomben R, et al. Molecular and clinical features of chronic lymphocytic leukaemia with stereotyped B cell receptors: results from an Italian multicentre study. Br J Haematol. 2009;144:492–506. doi: 10.1111/j.1365-2141.2008.07469.x. [DOI] [PubMed] [Google Scholar]
- 6.Fais F, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–25. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darzentas N, et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia. 2010;24:125–32. doi: 10.1038/leu.2009.186. [DOI] [PubMed] [Google Scholar]
- 8.Damle RN, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–47. [PubMed] [Google Scholar]
- 9.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 10.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 11.Belessi C, Stamatopoulos K. In: Stereotyped B-cell receptors in chronic lymphocytic leukemia. Stamatopoulos KGP, Rosenquist R, et al., editors. WK Health Books; Milan, Italy: 2010. pp. 119–34. [Google Scholar]
- 12.Lanemo Myhrinder A, et al. A new perspective: molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–48. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 13.Catera R, et al. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol Med. 2008;14:665–74. doi: 10.2119/2008-00102.Catera. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu CC, et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood. 2008;112:5122–9. doi: 10.1182/blood-2008-06-162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seiler T, et al. Characterization of structurally defined epitopes recognized by monoclonal antibodies produced by chronic lymphocytic leukemia B cells. Blood. 2009;114:3615–24. doi: 10.1182/blood-2009-01-197822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu CC, et al. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood. 2010;115:3907–15. doi: 10.1182/blood-2009-09-244251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 19.Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–44. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–7. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 21.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 22.Bourke E, Bosisio D, Golay J, Polentarutti N, Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–63. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- 23.Meyer-Bahlburg A, Khim S, Rawlings DJ. B cell intrinsic TLR signals amplify but are not required for humoral immunity. J Exp Med. 2007;204:3095–101. doi: 10.1084/jem.20071250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruprecht CR, Lanzavecchia A. Toll-like receptor stimulation as a third signal required for activation of human naive B cells. Eur J Immunol. 2006;36:810–6. doi: 10.1002/eji.200535744. [DOI] [PubMed] [Google Scholar]
- 25.Petterson T, et al. Effects of NOD-like receptors in human B lymphocytes and crosstalk between NOD1/NOD2 and Toll-like receptors. J Leukoc Biol. 2011;89:177–87. doi: 10.1189/jlb.0210061. [DOI] [PubMed] [Google Scholar]
- 26.Leadbetter EA, et al. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–7. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- 27.Lau CM, et al. RNA-associated autoantigens activate B cells by combined B cell antigen receptor/Toll-like receptor 7 engagement. J Exp Med. 2005;202:1171–7. doi: 10.1084/jem.20050630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poovassery JS, Vanden Bush TJ, Bishop GA. Antigen receptor signals rescue B cells from TLR tolerance. J Immunol. 2009;183:2974–83. doi: 10.4049/jimmunol.0900495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandjenette C, Kennel A, Faure GC, Bene MC, Feugier P. Expression of functional toll-like receptors by B-chronic lymphocytic leukemia cells. Haematologica. 2007;92:1279–81. doi: 10.3324/haematol.10975. [DOI] [PubMed] [Google Scholar]
- 30.Rozkova D, et al. Toll-like receptors on B-CLL cells: expression and functional consequences of their stimulation. Int J Cancer. 2010;126:1132–43. doi: 10.1002/ijc.24832. [DOI] [PubMed] [Google Scholar]
- 31.Muzio M, et al. Expression and function of toll like receptors in chronic lymphocytic leukaemia cells. Br J Haematol. 2009;144:507–16. doi: 10.1111/j.1365-2141.2008.07475.x. [DOI] [PubMed] [Google Scholar]
- 32.Arvaniti E, et al. Toll-like receptor signaling pathway in chronic lymphocytic leukemia: distinct gene expression profiles of potential pathogenetic significance in specific subsets of patients. Haematologica. 2011;96:1644–52. doi: 10.3324/haematol.2011.044792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallek M, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–56. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–12. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–8. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decker T, et al. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood. 2000;95:999–1006. [PubMed] [Google Scholar]
- 37.Ghiotto F, et al. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J Clin Invest. 2004;113:1008–16. doi: 10.1172/JCI19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y, White D, He L, Miller RL, Spaner DE. Toll-like receptor-7 tolerizes malignant B cells and enhances killing by cytotoxic agents. Cancer Res. 2007;67:1823–31. doi: 10.1158/0008-5472.CAN-06-2381. [DOI] [PubMed] [Google Scholar]
- 39.Longo PG, et al. The Akt signaling pathway determines the different proliferative capacity of chronic lymphocytic leukemia B-cells from patients with progressive and stable disease. Leukemia. 2007;21:110–20. doi: 10.1038/sj.leu.2404417. [DOI] [PubMed] [Google Scholar]
- 40.Lanham S, et al. Differential signaling via surface IgM is associated with VH gene mutational status and CD38 expression in chronic lymphocytic leukemia. Blood. 2003;101:1087–93. doi: 10.1182/blood-2002-06-1822. [DOI] [PubMed] [Google Scholar]
- 41.Mockridge CI, et al. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood. 2007;109:4424–31. doi: 10.1182/blood-2006-11-056648. [DOI] [PubMed] [Google Scholar]
- 42.Muzio M, et al. Constitutive activation of distinct BCR-signaling pathways in a subset of CLL patients: a molecular signature of anergy. Blood. 2008;112:188–95. doi: 10.1182/blood-2007-09-111344. [DOI] [PubMed] [Google Scholar]
- 43.Ghia P, et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3–21 gene. Blood. 2005;105:1678–85. doi: 10.1182/blood-2004-07-2606. [DOI] [PubMed] [Google Scholar]
- 44.Potter KN, et al. Structural and functional features of the B-cell receptor in IgG-positive chronic lymphocytic leukemia. Clin Cancer Res. 2006;12:1672–9. doi: 10.1158/1078-0432.CCR-05-2164. [DOI] [PubMed] [Google Scholar]
- 45.Jang YJ, Stollar BD. Anti-DNA antibodies: aspects of structure and pathogenicity. Cell Mol Life Sci. 2003;60:309–20. doi: 10.1007/s000180300026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kostareli E, et al. Molecular evidence for EBV and CMV persistence in a subset of patients with chronic lymphocytic leukemia expressing stereotyped IGHV4–34 B-cell receptors. Leukemia. 2009;23:919–24. doi: 10.1038/leu.2008.379. [DOI] [PubMed] [Google Scholar]
- 47.Sutton LA, et al. Extensive intraclonal diversification in a subgroup of chronic lymphocytic leukemia patients with stereotyped IGHV4–34 receptors: implications for ongoing interactions with antigen. Blood. 2009;114:4460–68. doi: 10.1182/blood-2009-05-221309. [DOI] [PubMed] [Google Scholar]
- 48.Messmer BT, et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200:519–25. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marincevic M, et al. High-density screening reveals a different spectrum of genomic aberrations in chronic lymphocytic leukemia patients with ‘stereotyped’ IGHV3–21 and IGHV4–34 B-cell receptors. Haematologica. 2010;95:1519–25. doi: 10.3324/haematol.2009.021014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marincevic M, et al. Distinct gene expression profiles in subsets of chronic lymphocytic leukemia expressing stereotyped IGHV4–34 B-cell receptors. Haematologica. 2010;95:2072–9. doi: 10.3324/haematol.2010.028639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spaner DE, Masellis A. Toll-like receptor agonists in the treatment of chronic lymphocytic leukemia. Leukemia. 2007;21:53–60. doi: 10.1038/sj.leu.2404456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.