Abstract
Circadian rhythms show universally a 24-h oscillation pattern in metabolic, physiological and behavioral functions of almost all species. This pattern is due to a fundamental adaptation to the rotation of Earth around its own axis. Molecular mechanisms of generation of circadian rhythms organize a biochemical network in suprachiasmatic nucleus and peripheral tissues, building cell autonomous clock pacemakers. Rhythmicity is observed in transcriptional expression of a wide range of clock-controlled genes that regulate a variety of normal cell functions, such as cell division and proliferation. Desynchrony of this rhythmicity seems to be implicated in several pathologic conditions, including tumorigenesis and progression of cancer. In 2007, the International Agency for Research on Cancer (IARC) categorized “shiftwork that involves circadian disruption [as] probably carcinogenic to humans” (Group 2A in the IARC classification system of carcinogenic potency of an agentagent) (Painting, Firefighting, and Shiftwork; IARC; 2007). This review discusses the potential relation between disruptions of normal circadian rhythms with genetic driving machinery of cancer. Elucidation of the role of clockwork disruption, such as exposure to light at night and sleep disruption, in cancer biology could be important in developing new targeted anticancer therapies, optimizing individualized chronotherapy and modifying lighting environment in workplaces or homes.
INTRODUCTION
The term “circadian” is derived from the Latin phrase circa diem, which means “about a day.” The suprachiasmatic nucleus (SCN) in the anterior hypothalamus serves as a circadian master clock, or an endogenous biological oscillator, that controls biochemical, physiological and behavioral rhythms, entrained by light and other external signals (1). The first reference of periodicity in medicine was made by Hippocrates when describing the fever’s course: “The quotidian, tertian, and quartan fevers. . . .” (2). SCN also has the unique ability to provide critical stimulus for resetting its clock phase in direct response to a light signal, which is relayed from the retina via the retinohypothalamic tract (3).
Circadian rhythms are generated by a set of clock genes and proteins (4) regulating many functions, including the ability to fall asleep or to snap out of sleep into wakefulness (5), body temperature (6), blood pressure (7), hormone biosynthesis (8), digestive secretion (9) and immune responses (10). Further to the central nervous system, circadian rhythmicity is present in peripheral tissues, too. Individual normal cells and even cancer cells keep circadian time by expressing similar clock genes (11). Circadian clocks in peripheral tissues regulate the expression of specific genes and synthesis of products, such as thymidylate synthase, p21 and Wee-1, which control DNA synthesis, cell division cycle and cell proliferation, coordinating physiological processes in a circadian manner (12–14). It is noteworthy that, in the industrialized world, there is a change in the lighted environment from a sun-based system to an electricity-based system. Modern lifestyle forces more people to work late shifts, to change shifts regularly or to spend more time doing other activities, prolonging the exposure to light. Epidemiological studies correlate disruption of circadian rhythms with incidence of breast cancer and poorer prognosis of the disease (15,16). In mouse models, data show that disturbed circadian clock gene expression and disruption of circadian rhythms correlate with tumor development and tumor progression (17).
REGULATION OF CIRCADIAN CLOCK GENES
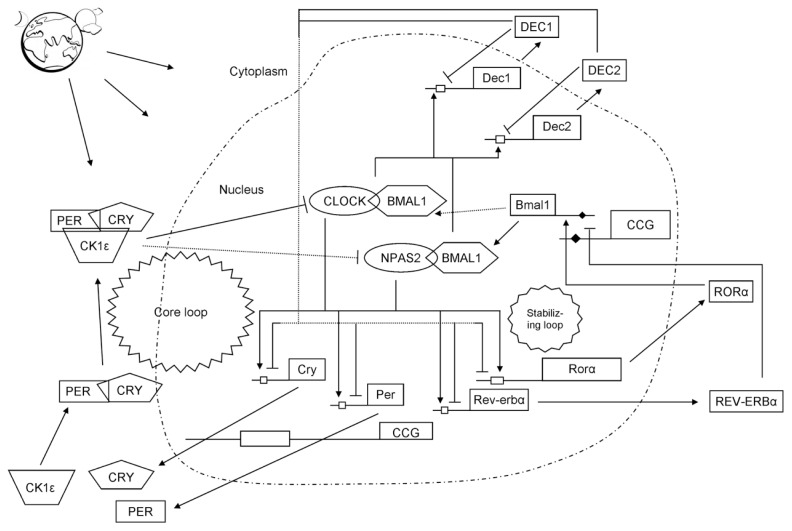
Timing of circadian clocks is established in a cell-autonomous manner by a self-sustaining molecular oscillator that consists of intertwined negative and positive transcription/translation-based feedback loops in SCN (18) and peripheral tissues (19,20). Human period (hPer1, -2 and -3) and cryptochrome (Cry1 and -2) clock genes are components of the SCN circadian clockwork (21–23). Protein products of these genes (PER and CRY) inhibit their own transcriptional activators: circadian locomotor output cycles kaput (CLOCK) and brain and muscle aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1) (24). For example, PER2 binds to both BMAL1 and CLOCK, whereas CRY1 and CRY2 are only able to interact with BMAL1 (25). CLOCK and BMAL1 form a heterodimer (CLOCK:BMAL1) that is involved in the rapid induction of mPer1 during phase resetting of the clock (26). CLOCK and BMAL1 are heterodimeric transcriptional activators consisting of two Per-Arnt/AhR-Sim basic helix-loop-helix (bHLH-PAS) domain protein subunits (27–29). A delay between production and action of inhibitory clock gene products is regulated by nuclear export of the PER protein, resulting in production of stable oscillations of gene expression with a period of 24 h (30). Dec1 and Dec2, basic helix-loop-helix transcription factors, are involved in the regulation of the mammalian circadian clock. CLOCK:BMAL2 heterodimer increases the expression of Dec1 and Dec2 genes. Deleted in esophageal cancer 1 (DEC1) and DEC2, products of Dec genes, suppress the expression of Per or Cry genes (31–34). As a result of these self-contained feedback loops, the circadian protein levels oscillate in a rhythmic manner (Figure 1). Light stimulus activates the expression of several genes in SCN, with different expression patterns. For example, the expression level of the circadian clock gene Per1 peaks 30–45 min after light pulse, with Per2 to show slower activation (35). Light also promotes binding of Cry1a to the transactivation domain of BMAL and blocks active dimerization of CLOCK and BMAL. Consequently, these actions inhibit CLOCK:BMAL function (36). Activity of the serotoninergic system possibly resets the circadian clock in SCN (37). The effect of light-at-night exposure to expression patterns of peripheral clock genes seems to be organ and time-of-day specific, in coordination with the autonomic nervous system that modifies this expression (38). However, a light pulse induces Dec1 expression in SCN and Per1 and Per2 in extra-SCN clocks (39,40). The exact role of the mammalian protein TIMELESS (TIM) in the circadian clock mechanism has not been fully elucidated. TIM forms a heterodimer with PER and translocates in the nucleus, where it inhibits the activity of CLOCK: BMAL1 on the mPer1 promoter (41). Another regulatory mechanism is controlled by microRNAs (miRNAs), small molecules that regulate gene expression, in the posttranscriptional level, via translational repression or direct destruction of their mRNA targets (42,43). miRNA- mediated translational control regulates the circadian clock, too. miR-132 is an miRNA that is induced in response to light stimulation in the murine SCN (44). The expression of miR-132 negatively regulates light-induced entrainment of the circadian clock through regulation of a number of target genes that are associated with chromatin remodeling (methyl-CpG-binding protein 2 [MeCP2], p300, JumonjiC (JmjC) and ARID domain-containing histone lysine demethylase 1a [JARID1A]) and protein translation (B-cell translocation gene 2 [BTG2], poly(A) binding protein interacting protein 2 [PAIP2A]) (45). However, there are other molecules that interact with clock genes and have important coeffects on circadian oscillation processes. For example, the receptor for activated protein kinase C-1 (RACK1) is a protein that mediates or regulates functions of PER1 (46). Activation of the MAPK cascade is able to trigger the induction and resetting of the circadian oscillation of gene expression (47). Fluctuating levels of circulating ovarian steroid hormones during the estrous cycle regulate the rhythm of clock gene expression in reproductive tissues (48).
Figure 1.
A simplified depiction of the mammalian molecular circadian clock machinery. Light perceived by the retina is the most potent synchronizer of the SCN clock. The circadian clock consists of positive and negative autoregulatory feedback loops. The oscillator is composed of interlocking transcription/translational feedback loops, controlling circadian timing. The CLOCK:BMAL1 or CLOCK:NPAS2 heterodimer (positive elements) is the “core loop” and induces E-box–mediated transcription of Per, Cry and Dec; their products are cyclically released in the cytoplasm. When PER and CRY proteins reach a critical concentration, they form heterodimers PER:CRY (negative elements), phosphorylate and translocate into the nucleus, where they inactivate the BMAL1:CLOCK or BMAL1:NPAS2 E-box–mediated transcription, including transcription of their own genes, which reduces their levels sufficiently to allow for the new transcription cycle. In addition, DECs bind to the E-box element of their promoter and inhibit their own transcription directly. CLOCK:BMAL1 also controls the levels of the nuclear receptors retinoid-related orphan receptor α (RORα) and Rev-erbα (known as nuclear receptor subfamily 1, group D, member 1 [NR1D1]), which constitute the “stabilizing/auxiliary loop” by repressing BMAL1 concentration via competitive actions on the retinoic acid–related orphan receptor response element (RORE) (black diamond shape) in the Bmal1 promoter. Cycling of clock components by the core and stabilizing/auxiliary loops also promotes cyclic accumulations of clock-controlled gene (CCG) mRNA species, thus achieving an oscillating pattern and generating rhythmic physiological outputs in a cell type–specific fashion (steroid biosynthesis, cell cycle progression/arrest, cell proliferation, apoptotic pathways, immune function, hormonal oscillations, body temperature, metabolism, DNA repair, response to anticancer drugs and so on). E-boxes (white rectangle shape): regulatory enhancer sequences present in the promoter regions of the genes to which CLOCK:BMAL1 heterodimer binds. Casein kinase (CK) isoforms phosphorylate PER and CRY proteins modulating the nucleocytoplasmic translocation of core clock elements and thereby their transcriptional activity.
ROLE OF CIRCADIAN CLOCK GENES IN CANCER
Core circadian genes seem to be important in tissue homeostasis and tumorigenesis. Disruption of circadian rhythms is associated with various forms of cancer in humans. There is increasing evidence that links dysfunction of the clockwork with the pathogenesis of cancer. The master circadian clock in SCN is an endogenous timekeeping system and controls multiple peripheral clocks in other peripheral tissues of the body. Many studies have shown that circadian clock gene deregulation is implicated in the development of cancer and other diseases. Disruption of circadian rhythms accelerates tumor progression, and potentially restoring circadian rhythms should improve prognosis. Decreased expression levels of Per1 and Per2 genes are observed in sporadic and familiar breast tumors when compared with normal breast tissues. The Per1 gene shows lower expression levels in familiar forms of breast cancer when compared with sporadic forms, suggesting that a potential deregulation of the circadian clock may contribute to the inherited form of the disease (49). Methylation of promoters of the Per1 and Cry1 genes may lead to survival of breast cancer cells through inactivation of expression of these genes and disruption of the circadian cell rhythm (50). Moreover, a significantly higher risk of breast cancer associated with clock genetic polymorphisms is observed in Chinese populations (51). Bmal1 epigenetic inactivation, via cytosine- guanine (CpG) island promoter hypermethylation, contributes to the development of hematologic malignancies, non–Hodgkin lymphoma and acute lymphocytic leukemia, by disrupting cellular circadian clock, leading to loss of circadian rhythmicity of target genes such as c-myc, catalase and p300 (52). Genetic variations in Cry2 and Ala394Thr functional polymorphism in the circadian gene neuronal PAS domain protein 2 (NPAS2) increase the inherited susceptibility to non–Hodgkin lymphoma (53,54). Methylation of CpG sites of the hPer3 gene is observed in patients with chronic myeloid leukemia (55). Prostate cancer is the most common cancer, excluding skin cancer, and the second leading cause of cancer- related death in men in the United States (56). The only well-established risk factors for prostate cancer are older age, family history of the disease and race. Circadian disruption may be a novel risk factor in prostate tumorigenesis. Results from a population-based case-control study provide evidence for an association of genetic variations in circadian genes with prostate tumorigenesis (53). Clock genes and the androgen receptor are expressed with circadian oscillations in the normal prostate. Per1 inhibits transcriptional activity of the androgen receptor, and downregulation of this clock gene seems to contribute to prostate tumorigenesis (57). Significant dysregulation of clock genes is one of the basic mechanisms driving the mesothelioma process (58). The expression of protooncogene c-myc is under circadian regulation, in the same phase with Per1, in the neuroblastoma cell line (59). Expression rates of Per1 and Per2 are lower in glioma cells when compared with non-malignant cells (60,61). Per1 and Per2 seem to be involved in suppressing the proliferation of pancreatic cancer cells (62,63).
Circadian disruption promotes liver carcinogenesis and possibly participates in its initiation, as observed in mice exposed to the hepatic carcinogen diethylnitrosamine (64). Hepatoma has the same circadian oscillation pattern with normal cells but is less sensitive to circadian timing signals, such as mealtimes, leading to dissociation of circadian rhythms in cancer and healthy cells (65).
The expression levels of Cry1 and Bmal1 core clock genes are correlated with clinicopathological parameters in epithelial ovarian cancer, and combination of low expression levels of both genes is an independent prognostic factor, as are stage and histological subtype (66). Disruption of the circadian clock, after methylation of the promoter CpG in Per1, Per2 or Cry1 circadian genes, is possibly involved in the development of endometrial cancers (67). The expression level of TIM was higher in the tumor tissue of colorectal cancer patients (68). The possible link of peripheral clock regulation in peripheral tissues with particular cancers is further supported by the following data. Several nuclear receptors are implicated in expression of peripheral clocks and constitute molecular links between clock genes and metabolic functions (reviewed in [69]). Expression levels of PER1–3, CRY1–2, CK1e and TIM are downregulated in patients with chronic myeloid leukemia (70). Tumor suppression through the ATM-p53 signaling is a clock-controlled physiological function, and disruption of this function leads to myc oncogenic activation in mice (71). β-Catenin increases β-TrCP (β-transducin repeat–containing protein) levels and shortens PER2 protein half-life, suggesting a possible mechanism for intestinal epithelial neoplastic transformation (72). Implication of circadian genes in various forms of cancer is supported by these data, and ongoing research will provide evidence to elucidate their biological role.
CIRCADIAN RHYTHMICITY IN CANCER GENETICS
DNA Repair and Circadian Rhythmicity
The circadian clock determines the strength of cellular responses to DNA damage, including DNA repair. DNA repair pathways maintain genetic stability, protecting DNA integrity from exogenous and/or endogenous stimuli (73,74). Several components of these pathways seem to be entrained by circadian oscillations. Nucleotide excision repair is a DNA repair mechanism that prevents genomes from damage caused by several sources, such as ultraviolet light irradiation and chemical mutagens (75). Nucleotide excision repair in the mouse brain seems to exhibit circadian periodicity, mainly mediated by xeroderma pigmentosum A, a DNA damage recognition protein (76,77). Tip60, a histone acetylase of chromatin, with DNA damage response and repair competency (78), is overexpressed in cisplatin-resistant cells and its silencing sensitizes cells to this cancer chemotherapeutic agent (79). In the same study, the expression of Tip60 is regulated by the circadian transcription factor Clock, providing evidence that DNA repair through histone acetylation is under circadian regulation. The high mobility group box 1 (Hmgb1) protein is involved in DNA mismatch repair (80) and shows a circadian rhythmic expression in rat retina (81). Apurinic/apyrimidinic endonuclease (APE) is an enzyme component of DNA base excision repair (82). The APE/Ref-1 gene is highly expressed in SCN, the main circadian pacemaker in mammals (83). However APE/Ref-1 mRNA levels do not show circadian patterns of expression (83).
Cell Proliferation and Cancer Cell Growth Are Under Clock Regulation
Proliferation rhythm of tumor cells follows a cyclical pattern different from that in normal tissues (84–87). Disruption of cellular circadian rhythm is associated with alterations in cancer cell proliferation. Downregulation of Per1 or Per2 increases cancer cell growth in vitro only at certain specific times of the day and enhances time-dependent tumor growth in vivo(88). Per1 has tumor suppressor function (89) and inhibits breast cancer cell proliferation and tumor growth in a circadian expression pattern (90). Down-regulation of Per2 accelerates breast cancer cell proliferation and tumor growth in a circadian time-dependent manner in vivo(91). Mammalian Per2 (mPer2)- deficient mice have tumor occurrences indicating tumor suppression function of mPer2(92). Clock mutation significantly inhibits cell growth and proliferation through upregulation of cell cycle inhibitory genes and reduced ability of mutant cells to respond to mitogenic signals (93). In contrast, disruption of the circadian clock, because of deficiency of the functional CLOCK protein, does not affect the rate of carcinogenesis in mice after exposure to ionizing radiation (94). These data suggest the existence of composite relations between genotoxic stress–induced carcinogenesis and the circadian clock. DNA synthesis in tumor cells seems to be modulated by several factors such as platelet-derived growth factor (PDGF) (95). PDGF signaling is activated during tumor development (96–98) and is related to tumor vascularization (99), adhesion, invasion (100) and aggressiveness (101) of cancer cells. Inhibition of this pathway synchronizes DNA synthesis in tumor cells with the rhythm of DNA synthesis in normal bone marrow cells (95). DNA synthesis and telomerase activity, which prevents cells from apoptosis (102), are expressed with a circadian pattern in hepatic cancer cells (103). Interferons (IFNs) are multi-functional cytokines that have antitumor activity, and their receptor shows a diurnal rhythm of expression in implanted-tumor cells (104), showing the importance of dosing time for IFN-β(105).
Mechanisms of Action of Circadian Genes in Cancer
Downregulation of Per2 increases β-catenin protein levels and its target cyclin D, leading to cell proliferation in colon cancer cell lines and intestinal and colonic polyp formation. This finding suggests that the Per2 gene product suppresses tumorigenesis in the small intestine and colon by downregulation of β-catenin and β-catenin target gene signaling pathways (106,107). Increased β-catenin affects the circadian clock and enhances PER2 clock protein degradation in colon cancer cells (107). Suppression of human β-catenin expression inhibits cellular proliferation in intestinal adenomas (108).
Disruption of the peripheral intestinal circadian clock may, in part, contribute to intestinal epithelial neoplastic transformation of human colorectal cancer. The circadian expression of dihydropyrimidine dehydrogenase (DPD), an enzyme that is implicated in the metabolism of the anticancer drug 5-fluorouracil, is possibly regulated by Per1 in high-grade colon tumors (109). Transferrin receptor 1 (TfR1) is a cell surface receptor required for iron delivery from transferrin to cells (110). Overexpression of TfR1 is associated with an increased rate of cell proliferation and malignant progression to colorectal cancer (111,112). TfR1 shows a 24-h rhythm of expression activated by the clock-controlled gene c-Myc in colon cancer–bearing mice (113). There is also a significant decrease of both estrogen receptor beta (ER-β) and Per1 in undifferentiated colorectal tumors (114). Casein kinase 1ɛ(CK1ɛ) phosphorylates PER2 protein, leading to degradation of PER2 by 26S proteasome (115,116). Thus, inhibition of CK1ɛby IC261, a kinase domain inhibitor, exerts a growth-suppressive effect of PER2 (117). Methylation of the Per gene promoters causes deregulation in the expression of PER proteins, resulting in proliferation of breast cancer cells (118). Per1 mediates inhibition of proliferation of a human pancreatic cancer cell line (MIA-PaCa2) by tumor necrosis factor (TNF)-α. The expression of Per1 is suppressed by TNF-α, and knockdown of Per1 decreases proliferation of MIA-PaCa2 cells (119). Chronic jetlag increases the risk of various cancers in mice, and circadian gene mutations make mutant mice more prone to cancer (71). Clock and Per2 protein levels are decreased, whereas Bmal1 protein levels are increased in prostate cancer (PCa) cells when compared with normal human prostate epithelial cells. Melatonin resynchronizes deregulated core clock genes in human PCa cells by upregulation of mRNA levels of Clock and Per2 and downregulation of Bmal1 mRNA (120), suggesting the preventive effects of melatonin against loss of rhythmicity, that is, observed during tumor progression (121).
Epigenetic Modifications of Circadian Clock Genes
Epigenetic modifications are heritable changes that take place independently of changes in the DNA sequence and are involved in regulation of gene transcription (122). DNA methylation and histone modifications are the main epigenetic mechanisms. In mammals, DNA methylation occurs primarily through addition of a methyl group to the 5′ carbon of cytosine located next to a guanine in CpG dinucleotides (123). Changes in DNA methylation accompany tumor initiation and progression (124,125). Promoter regions of tumor suppressor genes are methylated in cancer, resulting in gene silencing in contrast to normal cells, where most CpG islands are unmethylated (126). In addition, the consequence of hypomethylation leads to genomic instability through an opening of the chromatin and subsequent chromosomal breakage (127). Acetylation is the main, but not the only, posttranslational modification of nucleosomal histones that is involved in cancer initiation and progression (128,129). Histone acetylation is controlled by the opposing action of histone acetyltransferase (HAT) and histone deacetylases (HDAC) enzyme (130). Disruption of HAT or HDAC activity may play a key role in tumor invasion and metastasis (131).
INTERACTIONS BETWEEN ENVIRONMENT AND CLOCK GENES
Activation of aryl hydrocarbon receptor (AhR) by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibits Per1 gene expression by blocking CLOCK:BMAL1 heterodimer binding to enhancer boxes (E-boxes) in the Per1 promoter (132). This is a potential mechanism by which environmental pollutants may contribute to the development of carcinogenesis through disruption of circadian rhythm and repression of clock. Disruption of clock genes, Per1 and/or Per2, modifies mammary gland (133,134) and liver (134,135) responses to the environmental toxin TCDD by altering the inductive effects of TCDD on expression of cytochrome P450 genes (136). AhR activation by TCDD changes the circadian rhythms of murine hematopoietic stem progenitor cells (137) and mouse ovary (138). Per1 sensitizes cancer cells to activate their apoptotic machinery after DNA damage from double-strand break–causing agents, such as ionizing irradiation (89).
INTERACTIONS OF THE CLOCK WITH THE STEROID HORMONE RECEPTORS AND IMPACT ON CANCER
Glucocorticoid receptor (GR) is activated by glucocorticoids, a class of steroid hormones (139). Subsequent to, activation of GR involves (a) its nuclear translocation, (b) transactivation or binding to glucocorticoid-responsive element to regulate gene expression (140,141).
Stress has been associated with cancer progression through the glucocorticoid receptor system. GR expression is implicated in various forms of cancer such as prostate cancer and renal cell neoplasms (142,143). In human small cell lung cancer cells, GR expression is lost by DNA methylation, causing their increased survival (144). In contrast, high expression levels of GR are associated with shorter relapse-free survival in estrogen receptor–negative breast cancer (145). GR and core clock proteins (PER2 and CLOCK) are coexpressed in bronchiolar epithelial cells (146), and CLOCK-related genes regulate glucocorticoid action in all tissue through attenuation of the transcriptional activity of GR (147,148). A possible mechanism is the acetylation of several lysine residues of GR and concomitant attenuation of GR binding to glucocorticoid response elements (149). Desynchronization of circadian clock genes is possibly implicated in the role of the stress system in cancer progression through these mechanisms. As mentioned above, expression rhythms of circadian genes (Per1/2 and Bmal1) are modulated by the levels of ovarian steroid hormones in both reproductive and nonreproductive tissues (48). In addition, progesterone seems to cause acute increases of Per1, Per2 and Bmal1 expression in human breast cancer MCF-7 cells (48). Single nucleotide polymorphisms (SNPs) of the Clock gene are significantly associated with estrogen receptor/progesterone receptor (ER/PR)-negative cases of breast cancer (150). SNPs of NPAS2 have been linked to the risk of prostate cancer risk and hormone-related breast cancer (53,151). The possible mechanism implicates the aryl hydrocarbon receptor nuclear translocator-like (ARNTL)/NPAS2 heterodimer that suppresses transcription of the oncogene c-myc(152). Increased expression of Per2 in breast cancer cells leads to tumor apoptosis, possibly acting as an estrogen-inducible ER-α corepressor (153). CLOCK has histone acetyltransferase activity and acts as a coactivator of ERα, explaining the association between circadian rhythm disruption and breast cancer (154). Association of clock genes and proteins with various forms of cancers is summarized in Table 1.
Table 1.
Association of clock genes and proteins with various forms of cancers.
| Cancer type (and effect) | Trigger | Circadian genes/proteins | Reference |
|---|---|---|---|
| Sporadic and familiar breast tumors | Decreased expression levels | Per1, Per2 | 49 |
| Familiar breast tumors (than sporadic) | Lower expression levels | Per1 | 49 |
| Survival of breast cancer cells | Methylation of promoters | Per1, Cry1 | 50 |
| Proliferation of breast cancer cells | Methylation of promoters | Per | 118 |
| Higher risk of breast cancer | Polymorphisms | Clock | 51 |
| Inhibits breast cancer cell proliferation and tumor growth | Expression | Per1 | 89 |
| Breast cancer cell proliferation and tumor growth | Downregulation | Per2 | 91 |
| ER/PR-negative cases of breast cancer | SNPs | Clock | 150 |
| Breast cancer | Histone acetyltransferase activity | CLOCK (protein) | 154 |
| Tumor apoptosis in breast cancer | Increased expression | Per2 | 153 |
| Prostate cancer risk and hormone-related breast cancer | SNPs | NPAS2 | 53,151 |
| Non–Hodgkin lymphoma and acute lymphocytic leukemia | Epigenetic inactivation (via CpG hypermethylation) | Bmal1 | 52 |
| Non–Hodgkin lymphoma | Genetic variations, functional polymorphism | Cry2, NPAS2 | 53,54 |
| Chronic myeloid leukemia | Methylation | Per3 | 55 |
| Prostate cancer | Downregulation | Per1 | 57 |
| Prostate cancer | Increased expression level | Per2, Clock | 120 |
| Prostate cancer | Decreased expression levels | Bmal1 | 120 |
| Glioma | Lower expression rates | Per1, Per2 | 60,61 |
| Suppression of proliferation in pancreatic cancer | Expression | Per1, Per2 | 62,63 |
| Proliferation of human pancreatic cancer cell line | Knockdown | Per1 | 119 |
| Epithelial ovarian cancer | Low expression levels | Cry1, Bmal1 | 66 |
| Endometrial cancer | CpG methylation | Per1, Per2, Cry1 | 67 |
| Colorectal cancer | Increased expression level | Tim | 68 |
| Colon cancer | Downregulation | Per2 | 106,107 |
| Undifferentiated colorectal tumors | Decreased expression levels | Per1 | 114 |
| Chronic myeloid leukemia | Downregulation | Per1, Per2, Per3, CRY1–2, TIM | 70 |
| Intestinal epithelial neoplastic transformation | PER2 protein degradation | Per2 | 72 |
CLOCK GENES IN CANCER PROGRESSION, METASTASIS AND ANGIOGENESIS
Circadian disruption promotes tumor growth and angio-/stromagenesis, especially growth of both fibroblasts and vascular endothelial cells, induced by overexpression of wingless-type MMTV integration site family, member 10A (WNT10A) in tumor stroma cells as a result of increased levels of oxidative stress (155). Elevated expression of the mPer1 gene found in tumor stroma may affect interactions between cancer and stromal cells and is consequently involved in cancer progression and metastasis (156). The 24-h rhythm of methione aminopeptidases, which are involved in tumorigenesis (157) and tumor angiogenesis (157,158), is regulated by transcription of clock genes, enhanced by mCLOCK:mBMAL1 heterodimer and inhibited by mPER2 or mCRY1 (159). However, the effects of exogenous melatonin on tumor growth depend on the timing of administration (121). The laminin receptor 1 (Lamr1) is important in several physiological and pathological processes, including cell differentiation and viability, cancer development, invasion, migration and metastasis (160–163). Lamr1 interacts with human circadian clock protein hPer1 but does not have circadian pattern of expression (164). CLOCK:BMAL1 and sirtuin 1 (SIRT1) form a chromatin regulatory complex at promoters of clock-controlled genes (165). Sirtuins (SIRT proteins) are a unique class of type III (NAD)-dependent HDACs, which are important in the regulation of gene expression and especially in gene silencing (166). However, this activity shows oscillations in a circadian manner contributing to circadian control (165). SIRT1 is involved in the development of various cancers such as prostate (167), breast (168) and colorectal (169) and in chemotherapeutic drug resistance of cancer cells (reviewed in [170]).
MELATONIN AND CANCER
In 2007, the International Agency for Research on Cancer categorized “shift-work that involves circadian disruption [as] probably carcinogenic to humans” (Group 2A in the IARC classification system of carcinogenic potency of an agent) (171). Light during the night can suppress melatonin, disrupting circadian rhythms (172). Melatonin (5-methoxy-N-acetyltryptamine) is a hormone of the circadian system, synthesized in the pineal gland and retina (reviewed in [173,174]). In patients with untreated non–small-cell lung cancer (NSCLC) melatonin/cortisol mean nocturnal level ratio and melatonin nocturnal levels are decreased (175,176). These results may indicate a neuro-immune-endocrine system dysfunction. Melatonin concentrations progressively decrease after standard chemotherapy in NSCLC patients (176). Melatonin can resynchronize a rhythmic pattern of gene expression, correcting defects in expression patterns of various circadian rhythm genes responsible for cancer development (120). Melatonin inhibits myeloperoxidase catalytic activity (177), which is important in the pathogenesis of cancer (178,179). Melatonin has a protective effect against the DNA-damaging action of hydrogen peroxide, by chemical inactivation of this DNA-damaging agent and stimulation of DNA repair (180). Melatonin inhibits tumor signal transduction and metabolic activity of cancer cells, leading to suppression of growth of human breast cancer via activation of melatonin receptor MT1 (181). Disruption of nocturnal circadian melatonin signal by light at night upregulates tumor metabolism, stimulating its growth (182). Women with total visual blindness have a lower risk of breast cancer than blind women with light perception (183). The antiproliferative ability of melatonin is associated with its uptake into human androgen-dependent LNCaP and androgen-independent PC-3 prostate cancer cells, mainly mediated by an active transport (184).
Preventing low-wavelength light from reaching the retina, for example, by using optical filter goggles may protect shift workers from bright-light suppression of melatonin (185). If epidemiologic and basic science evidence leads to a “proof of causality” of adverse effects from light at night, then lighting standards and building designs should be developed with consideration of the circadian system both at night and during the day, to minimize or eliminate adverse consequences for human health (186–188).
CIRCADIAN RHYTHMS IN CANCER MANAGEMENT
The most important principle of chronomodulated therapeutics against various forms of cancer is to create a balance between effectiveness and adverse toxic effects of drugs. The circadian clock is responsible for rhythmicity of several physiological processes that in turn influence efficiency and tolerance of pharmacotherapy. Chronomodulated infusion of fluorouracil, leucovorin and oxaliplatin for 4 d achieves similar survival when compared with conventional 2-d delivery of the same drugs and acceptable tolerability, with more incidences of diarrhea with 4-d delivery and neutropenia with 2-d delivery. These results were observed in patients with metastatic colorectal cancer in a multi-center randomized phase III trial (189). However, chronomodulated hepatic arterial infusion multidrug chemotherapy shows antitumor activity and is well tolerated in patients with metastatic colorectal cancer after failure of several current standard therapeutic options (190). Combination of cetuximab, a chimeric monoclonal antibody directed against the extracellular domain of epidermal growth factor receptor, with circadian chronomodulated chemotherapy can be used effectively in initially unresectable residual metastatic colorectal cancer (191). In a phase II study, use of chronochemotherapy composed of 5-fluorouracil and leucovorin, and local hyperthermia combined with preoperative radiation therapy for locally advanced low-rectal adenocarcinoma had high antitumor activity rate and low incidence of adverse effects (192). Patients with ovarian cancer demonstrate altered diurnal cortisol rhythms, with significantly higher afternoon and nocturnal cortisol levels and lower cortisol variability when compared with patients with benign disease or healthy women (193). Dysregulation in rhythmic function of the hypothalamic-pituitary-adrenal axis is described in breast cancer survivors (194,195) or individuals with metastatic disease (196), in patients scheduled for lumbar disc surgery (197), in metastatic colorectal cancer patients (198) and in patients with cancer-related depression (193,199). Various anticancer agents are implicated in cancer therapeutics after circadian principles. Dietary methylselenocysteine (3 ppm selenium) given for 30 d significantly enhances circadian expression of circadian and growth- regulatory genes that are disrupted by nitrosomethylurea (200). Nitrosomethylurea-induced mammary carcinogenesis in rats is inhibited by methylselenocysteine, possibly through upregulation of circadian oscillations of Per2(201). As it mentioned above downregulation of Per2 accelerates breast cancer (91). The Per2 gene intratumoral delivery induces apoptosis and inhibits tumorigenesis in C57BL/6 mice transplanted with Lewis lung carcinoma (202). Antitumor effect and tolerability of Seliciclib, a cyclin-dependent kinase inhibitor in mice bearing Glasgow osteosarcoma, is found to depend on circadian rhythmicity (203). Biological parameters of a tumor, such as its growth kinetics, can affect the timing of optimal chronomodulated treatment, indicating the importance of tailoring these treatments for individual patients. The length of the cell cycle targeted by treatment and proliferation rate of cancer cells are important parameters to define the most effective time to administer cell cycle–specific drugs (204). The severity of acute gastrointestinal mucositis in patients undergoing radiotherapy is significantly increased when therapy is applied in the morning compared with the evening arm, implying that function of intestinal mucosa of the human intestine is possibly under circadian rhythmicity (205). In a retrospective review of local control, frequency of central nervous system–related cause of death and survival, in patients treated with gamma knife radiosurgery for metastatic NSCLC, had better outcomes in procedures earlier in the day versus later in the day (206). Various studies have indicated that toxicity and anticancer efficacy of anticancer drugs can be significantly modified by circadian stage of administration (reviewed in [203,207,208]). Drugs that target proliferation pathways and mimic or control rhythmicity increase susceptibility of cancer cells and thus improve their therapeutic index when given at specific times of day. Timing of any therapy targeting a cancer cell proliferation-related pathway will work substantially better if it is given at certain times within the day, when cancer cell proliferation is most active (reviewed in [209]). A way to optimize current therapies is the elucidation of links between clock genes and drug pharmacodynamic and pharmacokinetic parameters, resulting in development of new therapeutic strategies (210).
SUMMARY
Circadian genes have clock functions that regulate expression of other genes with circadian rhythmicity, resulting in daily oscillations of proteins. Therefore, disruption of clock damages organization of these gene and protein expressions, leading to deregulated cell proliferation and subsequent tumorigenesis. Circadian genes also have nonclock functions, which are important in regulation of cell cycle progression, DNA damage response and genomic stability. Clock and nonclock functions constitute the association between disruption of circadian rhythmicity and cancer.
A major consequence of modern lifestyle is disruption of circadian rhythms. Circadian disruptions induced by light at night, genetic or epigenetic variations in circadian genes and interactions between genes and environment form a set of data that propose that some cancer cases could be explained by these mechanisms. Elucidation of molecular mechanisms that form a link between disruption of circadian rhythm and cancer and determination of how a disrupted circadian peripheral clock contributes to neoplastic transformation is fundamental to provide essential leads developing future novel circadian clock–based strategies for cancer prevention, control and therapeutic intervention.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Albrecht U. Orchestration of gene expression and physiology by the circadian clock. J Physiol Paris. 2006;100:243–51. doi: 10.1016/j.jphysparis.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Adams CD, translator and editor. Hippocrates. On the Sacred Disease [Internet] Dover; New York: 1868. Available from: http://www.chlt.org/sandbox/dh/Adams/page.354.a.php. [Google Scholar]
- 3.Amir S, Stewart J. The effectiveness of light on the circadian clock is linked to its emotional value. Neuroscience. 1999;88:339–45. doi: 10.1016/s0306-4522(98)00477-1. [DOI] [PubMed] [Google Scholar]
- 4.Benca R, et al. Biological rhythms, higher brain function, and behavior: gaps, opportunities, and challenges. Brain Res Rev. 2009;62:57–70. doi: 10.1016/j.brainresrev.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wulff K, Porcheret K, Cussans E, Foster RG. Sleep and circadian rhythm disturbances: multiple genes and multiple phenotypes. Curr Opin Genet Dev. 2009;19:237–46. doi: 10.1016/j.gde.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Cajochen C, et al. High sensitivity of human melatonin, alertness, thermoregulation, and heart rate to short wavelength light. J Clin Endocrinol Metab. 2005;90:1311–6. doi: 10.1210/jc.2004-0957. [DOI] [PubMed] [Google Scholar]
- 7.Agarwal R. Regulation of circadian blood pressure: from mice to astronauts. Curr Opin Nephrol Hypertens. 2010;19:51–8. doi: 10.1097/MNH.0b013e3283336ddb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms. 2006;21:350–61. doi: 10.1177/0748730406293053. [DOI] [PubMed] [Google Scholar]
- 9.Froy O. Metabolism and circadian rhythms—implications for obesity. Endocr Rev. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 10.Keller M, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci U S A. 2009;106:21407–12. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood PA, Du-Quiton J, You S, Hrushesky WJM. Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther. 2006;5:2023–33. doi: 10.1158/1535-7163.MCT-06-0177. [DOI] [PubMed] [Google Scholar]
- 12.Lincoln DW, Hrushesky WJM, Wood PA. Circadian organization of thymidylate synthase activity in normal tissues: a possible basis for 5-fluorouracil chronotherapeutic advantage. Int J Cancer. 2000;88:479–85. [PubMed] [Google Scholar]
- 13.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–9. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 14.Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–42. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- 15.Stevens RG. Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology. 2005;16:254–8. doi: 10.1097/01.ede.0000152525.21924.54. [DOI] [PubMed] [Google Scholar]
- 16.Schernhammer ES, et al. Rotating night shifts and risk of breast cancer in women participating in the Nurses’ Health Study. J Natl Cancer Inst. 2001;93:1563–8. doi: 10.1093/jnci/93.20.1563. [DOI] [PubMed] [Google Scholar]
- 17.Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–31. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- 18.Kiyohara YB, et al. The BMAL1 C terminus regulates the circadian transcription feedback loop. Proc Natl Acad Sci U S A. 2006;103:10074–9. doi: 10.1073/pnas.0601416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 20.Oishi K, et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J Biol Chem. 2003;278:41519–27. doi: 10.1074/jbc.M304564200. [DOI] [PubMed] [Google Scholar]
- 21.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–36. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 22.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 23.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–9. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 24.Chen R, et al. Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell. 2009;36:417–30. doi: 10.1016/j.molcel.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmesser S, Tallone T, Bordon A, Rusconi S, Albrecht U. Interaction of circadian clock proteins PER2 and CRY with BMAL1 and CLOCK. BMC Mol Biol. 2008;9:41. doi: 10.1186/1471-2199-9-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung H, et al. Involvement of CLOCK: BMAL1 heterodimer in serum-responsive mPer1 induction. Neuroreport. 2003;14:15–9. doi: 10.1097/00001756-200301200-00003. [DOI] [PubMed] [Google Scholar]
- 27.Kamae Y, Tanaka F, Tomioka K. Molecular cloning and functional analysis of the clock genes, clock and cycle, in the firebrat Thermobia domestica. J Insect Physiol. 2010;56:1291–9. doi: 10.1016/j.jinsphys.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Kondratov RV, et al. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–32. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–53. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vielhaber EL, Duricka D, Ullman KS, Virshup DM. Nuclear export of mammalian PERIOD proteins. J Biol Chem. 2001;276:45921–7. doi: 10.1074/jbc.M107726200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawamoto T, et al. A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem Biophys Res Commun. 2004;313:117–24. doi: 10.1016/j.bbrc.2003.11.099. [DOI] [PubMed] [Google Scholar]
- 32.Butler MP, et al. Dec1 and Dec2 expression is disrupted in the suprachiasmatic nuclei of clock mutant mice. J Biol Rhythms. 2004;19:126–34. doi: 10.1177/0748730403262870. [DOI] [PubMed] [Google Scholar]
- 33.Hamaguchi H, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J. 2004;382:43–50. doi: 10.1042/BJ20031760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakashima A, et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol. 2008;28:4080–92. doi: 10.1128/MCB.02168-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porterfield VM, Mintz EM. Temporal patterns of light-induced immediate-early gene expression in the suprachiasmatic nucleus. Neurosci Lett. 2009;463:70–3. doi: 10.1016/j.neulet.2009.07.066. [DOI] [PubMed] [Google Scholar]
- 36.Tamai TK, Young LC, Whitmore D. Light signaling to the zebrafish circadian clock by Cryptochrome 1a. Proc Natl Acad Sci U S A. 2007;104:14712–7. doi: 10.1073/pnas.0704588104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cuesta M, Mendoza J, Clesse D, Pévet P, Challet E. Serotonergic activation potentiates light resetting of the main circadian clock and alters clock gene expression in a diurnal rodent. Exp Neurol. 2008;210:501–13. doi: 10.1016/j.expneurol.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 38.Cailotto C, et al. Effects of nocturnal light on (clock) gene expression in peripheral organs: a role for the autonomic innervation of the liver. PLoS One. 2009;4:e5650. doi: 10.1371/journal.pone.0005650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamada T, Honma S, Honma KI. Light responsiveness of clock genes, Per1 and Per2, in the olfactory bulb of mice. Biochem Biophys Res Commun. 2011;409:727–31. doi: 10.1016/j.bbrc.2011.05.076. [DOI] [PubMed] [Google Scholar]
- 40.Honma S, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–4. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 41.Sangoram AM, et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–13. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen AF, Gloggnitzer J, Martinez J. MicroRNAs cross the line: the battle for mRNA stability enters the coding sequence. Mol Cell. 2009;35:139–40. doi: 10.1016/j.molcel.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 43.Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–40. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Cheng H-YM, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–29. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez-Saavedra M, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Human Mol Genet. 2011;20:731–51. doi: 10.1093/hmg/ddq519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu L, et al. RACK1, a novel hPER1-interacting protein. J Mol Neurosci. 2006;29:55–63. doi: 10.1385/JMN:29:1:55. [DOI] [PubMed] [Google Scholar]
- 47.Akashi M, Nishida E. Involvement of the MAP kinase cascade in resetting of the mammalian circadian clock. Genes Dev. 2000;14:645–9. [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura TJ, et al. Influence of the estrous cycle on clock gene expression in reproductive tissues: effects of fluctuating ovarian steroid hormone levels. Steroids. 2010;75:203–12. doi: 10.1016/j.steroids.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter SL, Bosnoyan-Collins L, Pinnaduwage D, Andrulis IL. Expression of the circadian clock genes Per1 and Per2 in sporadic and familial breast tumors. Neoplasia. 2007;9:797–800. doi: 10.1593/neo.07595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuo S-J, et al. Disturbance of circadian gene expression in breast cancer. Virchows Arch. 2009;454:467–74. doi: 10.1007/s00428-009-0761-7. [DOI] [PubMed] [Google Scholar]
- 51.Dai H, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treatment. 2011;127:531–540. doi: 10.1007/s10549-010-1231-2. [DOI] [PubMed] [Google Scholar]
- 52.Taniguchi H, et al. Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res. 2009;69:8447–54. doi: 10.1158/0008-5472.CAN-09-0551. [DOI] [PubMed] [Google Scholar]
- 53.Zhu Y, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–22. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–5. doi: 10.1002/ijc.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang M-Y, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97:1298–307. doi: 10.1111/j.1349-7006.2006.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.A snapshot of prostate cancer. Bethesda (MD): National Cancer Institute; 2011. [updated 2011 Oct; cited 2012 Oct 26]. Available from: http://www.cancer.gov/cancertopics/types/prostate. [Google Scholar]
- 57.Cao Q, et al. A role for the clock gene Per1 in prostate cancer. Cancer Res. 2009;69:7619–25. doi: 10.1158/0008-5472.CAN-08-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Røe OD, et al. Genome-wide profile of pleural mesothelioma versus parietal and visceral pleura: the emerging gene portrait of the mesothelioma phenotype. PLoS One. 2009;4:e6554. doi: 10.1371/journal.pone.0006554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Repouskou A, Sourlingas TG, Sekeri-Pataryas KE, Prombona A. The circadian expression of c-MYC is modulated by the histone deacetylase inhibitor trichostatin A in synchronized murine neuroblastoma cells. Chronobiol Int. 2010;27:722–41. doi: 10.3109/07420521003786800. [DOI] [PubMed] [Google Scholar]
- 60.Fujioka A, Takashima N, Shigeyoshi Y. Circadian rhythm generation in a glioma cell line. Biochem Biophys Res Commun. 2006;346:169–74. doi: 10.1016/j.bbrc.2006.05.094. [DOI] [PubMed] [Google Scholar]
- 61.Xia HC, et al. Deregulated expression of the Per1 and Per2 in human gliomas. Can J Neurol Sci. 2010;37:365–70. doi: 10.1017/s031716710001026x. [DOI] [PubMed] [Google Scholar]
- 62.Pogue-Geile KL, Lyons-Weiler J, Whitcomb DC. Molecular overlap of fly circadian rhythms and human pancreatic cancer. Cancer Lett. 2006;243:55–7. doi: 10.1016/j.canlet.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 63.Oda A, et al. Clock gene mouse period2 overexpression inhibits growth of human pancreatic cancer cells and has synergistic effect with cisplatin. Anticancer Res. 2009;29:1201–9. [PubMed] [Google Scholar]
- 64.Filipski E, et al. Circadian disruption accelerates liver carcinogenesis in mice. Mutat Res. 2009;680:95–105. doi: 10.1016/j.mrgentox.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Davidson AJ, Straume M, Block GD, Menaker M. Daily timed meals dissociate circadian rhythms in hepatoma and healthy host liver. Int J Cancer. 2006;118:1623–7. doi: 10.1002/ijc.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tokunaga H, et al. Clinicopathological significance of circadian rhythm-related gene expression levels in patients with epithelial ovarian cancer. Acta Obstet Gynecol Scand. 2008;87:1060–70. doi: 10.1080/00016340802348286. [DOI] [PubMed] [Google Scholar]
- 67.Shih M-C, Yeh K-T, Tang K-P, Chen J-C, Chang JG. Promoter methylation in circadian genes of endometrial cancers detected by methylation-specific PCR. Mol Carcinog. 2006;45:732–40. doi: 10.1002/mc.20198. [DOI] [PubMed] [Google Scholar]
- 68.Mazzoccoli G, et al. Clock gene expression levels and relationship with clinical and pathological features in colorectal cancer patients. Chronobiol Int. 2011;28:841–51. doi: 10.3109/07420528.2011.615182. [DOI] [PubMed] [Google Scholar]
- 69.Teboul M, Guillaumond F, Gréchez-Cassiau A, Delaunay F. Minireview: the nuclear hormone receptor family round the clock. Mol Endocrinol. 2008;22:2573–82. doi: 10.1210/me.2007-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang M-Y, et al. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythms. 2011;26:136–48. doi: 10.1177/0748730410395527. [DOI] [PubMed] [Google Scholar]
- 71.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5:e10995. doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang X, et al. β-Catenin induces β-TrCP-mediated PER2 degradation altering circadian clock gene expression in intestinal mucosa of ApcMin/+ mice. J Biochem. 2009;145:289–97. doi: 10.1093/jb/mvn167. [DOI] [PubMed] [Google Scholar]
- 73.Pardo B, Gómez-González B, Aguilera A. DNA repair in mammalian cells. Cell Mol Life Sci. 2009;66:1039–56. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson A, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells. Cell. Mol. Life Sci. 2009;66:981–93. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu L, Lee J, Zhou P. Navigating the nucleotide excision repair threshold. J Cell Physiol. 2010;224:585–9. doi: 10.1002/jcp.22205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kang T-H, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci U S A. 2009;106:2864–7. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chronochemotherapy. Cell Cycle. 2009;8:1665–7. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 78.Ikura T, et al. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–73. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 79.Miyamoto N, et al. Tip60 is regulated by circadian transcription factor clock and is involved in cisplatin resistance. J Biol Chem. 2008;283:18218–26. doi: 10.1074/jbc.M802332200. [DOI] [PubMed] [Google Scholar]
- 80.Yuan F, Gu L, Guo S, Wang C, Li GM. Evidence for involvement of HMGB1 protein in human DNA mismatch repair. J Biol Chem. 2004;279:20935–40. doi: 10.1074/jbc.M401931200. [DOI] [PubMed] [Google Scholar]
- 81.Hoppe G, Rayborn ME, Sears JE. Diurnal rhythm of the chromatin protein Hmgb1 in rat photoreceptors is under circadian regulation. J Comp Neurol. 2007;501:219–30. doi: 10.1002/cne.21248. [DOI] [PubMed] [Google Scholar]
- 82.Warner HR, Demple BF, Deutsch WA, Kane CM, Linn S. Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc Natl Acad Sci U S A. 1980;77:4602–6. doi: 10.1073/pnas.77.8.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimura H, Dong X, Yagita K, Okamura H. Brain expression of apurinic/apyrimidinic endonuclease (APE/Ref-1) multifunctional DNA repair enzyme gene in the mouse with special reference to the suprachiasmatic nucleus. Neurosci Res. 2003;46:443–52. doi: 10.1016/s0168-0102(03)00124-x. [DOI] [PubMed] [Google Scholar]
- 84.Klevecz RR, Shymko RM, Blumenfeld D, Braly PS. Circadian gating of S phase in human ovarian cancer. Cancer Res. 1987;47:6267–71. [PubMed] [Google Scholar]
- 85.You S, et al. Daily coordination of cancer growth and circadian clock gene expression. Breast Cancer Res Treat. 2005;91:47–60. doi: 10.1007/s10549-004-6603-z. [DOI] [PubMed] [Google Scholar]
- 86.Brandi G, et al. Circadian variations of rectal cell proliferation in patients affected by advanced colorectal cancer. Cancer Lett. 2004;208:193–6. doi: 10.1016/j.canlet.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 87.Sedivy R, Thurner S, Budinsky AC, Köstler WJ, Zielinski CC. Short-term rhythmic proliferation of human breast cancer cell lines: surface effects and fractal growth patterns. J Pathol. 2002;197:163–9. doi: 10.1002/path.1118. [DOI] [PubMed] [Google Scholar]
- 88.Xiaoming Y, Wood PA, Ansell C, Hrushesky WJM. Circadian time-dependent tumor suppressor function of period genes. Integr Cancer Ther. 2009;8:309–16. doi: 10.1177/1534735409352083. [DOI] [PubMed] [Google Scholar]
- 89.Gery S, et al. The circadian gene Per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–82. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 90.Yang X, et al. The circadian clock gene Per1 suppresses cancer cell proliferation and tumor growth at specific times of day. Chronobiol Int. 2009;26:1323–39. doi: 10.3109/07420520903431301. [DOI] [PubMed] [Google Scholar]
- 91.Yang X, et al. Down regulation of circadian clock gene period 2 accelerates breast cancer growth by altering its daily growth rhythm. Breast Cancer Res Treat. 2009;117:423–31. doi: 10.1007/s10549-008-0133-z. [DOI] [PubMed] [Google Scholar]
- 92.Lee CC. The Circadian Clock and Tumor Suppression by Mammalian Period Genes. In: Michael WY, editor. Methods in Enzymology. Academic Press; 2005. pp. 852–861. [DOI] [PubMed] [Google Scholar]
- 93.Miller BH, et al. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–7. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Antoch MP, et al. Disruption of the circadian clock due to the clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakagawa H, et al. Modulation of circadian rhythm of DNA synthesis in tumor cells by inhibiting platelet-derived growth factor signaling. J Pharmacol Sci. 2008;107:401–7. doi: 10.1254/jphs.08080fp. [DOI] [PubMed] [Google Scholar]
- 96.van Zijl F, et al. Hepatic tumorstroma crosstalk guides epithelial to mesenchymal transition at the tumor edge. Oncogene. 2009;28:4022–33. doi: 10.1038/onc.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Soares R, Guerreiro S, Botelho M. Elucidating progesterone effects in breast cancer: cross talk with PDGF signaling pathway in smooth muscle cell. J Cell Biochem. 2007;100:174–83. doi: 10.1002/jcb.21045. [DOI] [PubMed] [Google Scholar]
- 98.Ustach CV, Kim H-RC. Platelet-derived growth factor D is activated by urokinase plasminogen activator in prostate carcinoma cells. Mol Cell Biol. 2005;25:6279–88. doi: 10.1128/MCB.25.14.6279-6288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki S, Heldin C-H, Heuchel R. Platelet-derived growth factor receptor-beta, carrying the activating mutation D849N, accelerates the establishment of B16 melanoma. BMC Cancer. 2007;7:224. doi: 10.1186/1471-2407-7-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kong D, et al. miR-200 regulates PDGF-D-mediated epithelial–mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–21. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ahmad A, et al. Platelet-derived growth factor-D contributes to aggressiveness of breast cancer cells by up-regulating Notch and NF-κB signaling pathways. Breast Cancer Res Treat. 2011;126:15–25. doi: 10.1007/s10549-010-0883-2. [DOI] [PubMed] [Google Scholar]
- 102.Fu W, Begley JG, Killen MW, Mattson MP. Anti-apoptotic role of telomerase in pheochromocytoma cells. J Biol Chem. 1999;274:7264–71. doi: 10.1074/jbc.274.11.7264. [DOI] [PubMed] [Google Scholar]
- 103.Qu Y, et al. Circadian telomerase activity and DNA synthesis for timing peptide administration. Peptides. 2003;24:363–9. doi: 10.1016/s0196-9781(03)00050-0. [DOI] [PubMed] [Google Scholar]
- 104.Takane H, et al. Relationship between diurnal rhythm of cell cycle and interferon receptor expression in implanted-tumor cells. Life Sci. 2001;68:1449–55. doi: 10.1016/s0024-3205(01)00943-2. [DOI] [PubMed] [Google Scholar]
- 105.Takane H, Ohdo S, Yamada T, Yukawa E, Higuchi S. Chronopharmacology of anti-tumor effect induced by interferon-β in tumor-bearing mice. J Pharmacol Exp Ther. 2000;294:746–52. [PubMed] [Google Scholar]
- 106.Faustino RS, et al. Ceramide regulation of nuclear protein import. J Lipid Res. 2008;49:654–62. doi: 10.1194/jlr.M700464-JLR200. [DOI] [PubMed] [Google Scholar]
- 107.Wood PA, Xiaoming Yang, Hrushesky WJM. Clock genes and cancer. Integr Cancer Ther. 2009;8:303–8. doi: 10.1177/1534735409355292. [DOI] [PubMed] [Google Scholar]
- 108.Foley P, et al. Targeted suppression of β-catenin blocks intestinal adenoma formation in APC Min mice. J Gastrointest Surg. 2008;12:1452–8. doi: 10.1007/s11605-008-0519-6. [DOI] [PubMed] [Google Scholar]
- 109.Krugluger W, et al. Regulation of genes of the circadian clock in human colon cancer: reduced period-1 and dihydropyrimidine dehydrogenase transcription correlates in high-grade tumors. Cancer Res. 2007;67:7917–22. doi: 10.1158/0008-5472.CAN-07-0133. [DOI] [PubMed] [Google Scholar]
- 110.Aisen P. Transferrin receptor 1. Int J Biochem Cell Biol. 2004;36:2137–43. doi: 10.1016/j.biocel.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 111.Boult J, et al. Overexpression of cellular iron import proteins is associated with malignant progression of esophageal adenocarcinoma. Clin Cancer Res. 2008;14:379–87. doi: 10.1158/1078-0432.CCR-07-1054. [DOI] [PubMed] [Google Scholar]
- 112.Brookes MJ, et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55:1449–60. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Okazaki F, et al. Circadian rhythm of transferrin receptor 1 gene expression controlled by c-Myc in colon cancer–bearing mice. Cancer Res. 2010;70:6238–46. doi: 10.1158/0008-5472.CAN-10-0184. [DOI] [PubMed] [Google Scholar]
- 114.Mostafaie N, et al. Correlated downregulation of estrogen receptor beta and the circadian clock gene Per1 in human colorectal cancer. Mol Carcinog. 2009;48:642–7. doi: 10.1002/mc.20510. [DOI] [PubMed] [Google Scholar]
- 115.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Meng Q-J, et al. Setting clock speed in mammals: the CK1tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang WS, Stockwell BR. Inhibition of casein kinase 1-epsilon induces cancer-cell-selective, PERIOD2-dependent growth arrest. Genome Biol. 2008;9:R92. doi: 10.1186/gb-2008-9-6-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen S-T, et al. Deregulated expression of the PER1, PER2 and PER3 genes in breast cancers. Carcinogenesis. 2005;26:1241–6. doi: 10.1093/carcin/bgi075. [DOI] [PubMed] [Google Scholar]
- 119.Suzuki T, et al. Period is involved in the proliferation of human pancreatic MIA-PaCa2 cancer cells by TNF-alpha. Biomed Res. 2008;29:99–103. doi: 10.2220/biomedres.29.99. [DOI] [PubMed] [Google Scholar]
- 120.Jung-Hynes B, Huang W, Reiter RJ, Ahmad N. Melatonin resynchronizes dysregulated circadian rhythm circuitry in human prostate cancer cells. J Pineal Res. 2010;49:60–8. doi: 10.1111/j.1600-079X.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Otálora BB, Madrid JA, Alvarez N, Vicente V, Rol MA. Effects of exogenous melatonin and circadian synchronization on tumor progression in melanoma-bearing C57BL6 mice. J Pineal Res. 2008;44:307–15. doi: 10.1111/j.1600-079X.2007.00531.x. [DOI] [PubMed] [Google Scholar]
- 122.Gasser SM, Paro R, Stewart F, Aasland R. The genetics of epigenetics. Cell Mol Life Sci. 1998;54:1–5. doi: 10.1007/s000180050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rottach A, Leonhardt H, Spada F. DNA methylation-mediated epigenetic control. J Cell Biochem. 2009;108:43–51. doi: 10.1002/jcb.22253. [DOI] [PubMed] [Google Scholar]
- 124.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:e67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kulis M, Esteller M. 2 - DNA Methylation and Cancer. In: Zdenko H, Toshikazu U, editors. Advances in Genetics. Academic Press; 2010. pp. 27–56. [DOI] [PubMed] [Google Scholar]
- 126.Sansam CG, Roberts CWM. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle. 2006;5:621–4. doi: 10.4161/cc.5.6.2579. [DOI] [PubMed] [Google Scholar]
- 127.Goodman JI, Counts JL. Hypomethylation of DNA: a possible nongenotoxic mechanism underlying the role of cell proliferation in carcinogenesis. Environ. Health Perspect. 1993;101(Suppl 5):169–72. doi: 10.1289/ehp.93101s5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Glaser KB, et al. Differential protein acetylation induced by novel histone deacetylase inhibitors. Biochem Biophys Res Commun. 2004;325:683–90. doi: 10.1016/j.bbrc.2004.10.082. [DOI] [PubMed] [Google Scholar]
- 129.Sawan C, Herceg Z. 3 - Histone Modifications and Cancer. In: Zdenko H, Toshikazu U, editors. Advances in Genetics. Academic Press; 2010. pp. 57–85. [DOI] [PubMed] [Google Scholar]
- 130.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 131.Mottet D, Castronovo V. Histone deacetylases: target enzymes for cancer therapy. Clin. Exp. Metastasis. 2008;25:183–9. doi: 10.1007/s10585-007-9131-5. [DOI] [PubMed] [Google Scholar]
- 132.Xu C-X, Krager SL, Liao D-F, Tischkau SA. Disruption of CLOCK-BMAL1 transcriptional activity is responsible for Aryl hydrocarbon receptor–mediated regulation of Period1 gene. Toxicol Sci. 2010;115:98–108. doi: 10.1093/toxsci/kfq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Qu X, Metz RP, Porter WW, Cassone VM, Earnest DJ. Disruption of clock gene expression alters responses of the Aryl hydrocarbon receptor signaling pathway in the mouse mammary gland. Mol Pharmacol. 2007;72:1349–58. doi: 10.1124/mol.107.039305. [DOI] [PubMed] [Google Scholar]
- 134.Qu X, et al. The clock genes period 1 and period 2 mediate diurnal rhythms in dioxin-induced Cyp1A1 expression in the mouse mammary gland and liver. Toxicol Lett. 2010;196:28–32. doi: 10.1016/j.toxlet.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qu X, Metz RP, Porter WW, Cassone VM, Earnest DJ. Disruption of period gene expression alters the inductive effects of dioxin on the AhR signaling pathway in the mouse liver. Toxicol Appl Pharmacol. 2009;234:370–7. doi: 10.1016/j.taap.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Le Vee M, Jouan E, Fardel O. Involvement of aryl hydrocarbon receptor in basal and 2,3,7,8-tetrachlorodibenzo-p-dioxin-induced expression of target genes in primary human hepatocytes. Toxicol In Vitro. 2010;24:1775–81. doi: 10.1016/j.tiv.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 137.Garrett RW, Gasiewicz TA. The Aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin alters the circadian rhythms, quiescence, and expression of clock genes in murine hematopoietic stem and progenitor cells. Mol Pharmacol. 2006;69:2076–83. doi: 10.1124/mol.105.021006. [DOI] [PubMed] [Google Scholar]
- 138.Tischkau SA, Jaeger CD, Krager SL. Circadian clock disruption in the mouse ovary in response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. Toxicol Lett. 2011;201:116–122. doi: 10.1016/j.toxlet.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wheeler RH, et al. Glucocorticoid receptor activation and inactivation in cultured human lymphocytes. J Biol Chem. 1981;256:434–41. [PubMed] [Google Scholar]
- 140.Robertson NM, Bodine PVN, Hsu T-C, Alnemri ES, Litwack G. Modulator inhibits nuclear translocation of the glucocorticoid receptor and inhibits glucocorticoid-induced apoptosis in the human leukemic cell line CEM C-7. Cancer Res. 1995;55:548–56. [PubMed] [Google Scholar]
- 141.Ronacher K, et al. Ligand-selective trans-activation and transrepression via the glucocorticoid receptor: role of cofactor interaction. Mol Cell Endocrinol. 2009;299:219–31. doi: 10.1016/j.mce.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 142.Dobos J, Kenessey I, Timar J, Ladanyi A. Glucocorticoid receptor expression and antiproliferative effect of dexamethasone on human melanoma cells. Pathol Oncol Res. 2011;17:729–34. doi: 10.1007/s12253-011-9377-8. [DOI] [PubMed] [Google Scholar]
- 143.Feng Y, Bai X, Yang Q, Wu H, Wang D. Downregulation of 15-lipoxygenase 2 by glucocorticoid receptor in prostate cancer cells. Int J Oncol. 2010;36:1541–9. doi: 10.3892/ijo_00000641. [DOI] [PubMed] [Google Scholar]
- 144.Kay P, et al. Loss of glucocorticoid receptor expression by DNA methylation prevents glucocorticoid induced apoptosis in human small cell lung cancer cells. PLoS One. 2011;6:e24839. doi: 10.1371/journal.pone.0024839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71:6360–70. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gibbs JE, et al. Circadian timing in the lung: a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150:268–76. doi: 10.1210/en.2008-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Charmandari E, et al. Peripheral CLOCK regulates target-tissue glucocorticoid receptor transcriptional activity in a circadian fashion in man. PLoS One. 2011;6:e25612. doi: 10.1371/journal.pone.0025612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kino T, Chrousos GP. Acetylation-mediated epigenetic regulation of glucocorticoid receptor activity: circadian rhythm-associated alterations of glucocorticoid actions in target tissues. Mol Cell Endocrinol. 2011;336:23–30. doi: 10.1016/j.mce.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009;23:1572–83. doi: 10.1096/fj.08-117697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hoffman AE, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70:1459–68. doi: 10.1158/0008-5472.CAN-09-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chu LW, et al. Correlation between circadian gene variants and serum levels of sex steroids and insulin-like growth factor-I. Cancer Epidemiol Biomarkers Prev. 2008;17:3268–73. doi: 10.1158/1055-9965.EPI-08-0073. [DOI] [PubMed] [Google Scholar]
- 152.Zhu Y, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5. doi: 10.1007/s10549-007-9565-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gery S, Virk RK, Chumakov K, Yu A, Koeffler HP. The clock gene Per2 links the circadian system to the estrogen receptor. Oncogene. 2007;26:7916–20. doi: 10.1038/sj.onc.1210585. [DOI] [PubMed] [Google Scholar]
- 154.Saha S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–31. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- 155.Yasuniwa Y, et al. Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS One. 2010;5:e15330. doi: 10.1371/journal.pone.0015330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Geusz ME, Blakely KT, Hiler DJ, Jamasbi RJ. Elevated mPer1 gene expression in tumor stroma imaged through bioluminescence. Int J Cancer. 2010;126:620–30. doi: 10.1002/ijc.24788. [DOI] [PubMed] [Google Scholar]
- 157.Wang J, et al. Correlation of tumor growth suppression and methionine aminopetidase-2 activity blockade using an orally active inhibitor. Proc Natl Acad Sci U S A. 2008;105:1838–43. doi: 10.1073/pnas.0708766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yeh J-RJ, et al. Targeted gene disruption of methionine aminopeptidase 2 results in an embryonic gastrulation defect and endothelial cell growth arrest. Proc Natl Acad Sci U S A. 2006;103:10379–84. doi: 10.1073/pnas.0511313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nakagawa H, et al. 24-Hour oscillation of mouse methionine aminopeptidase2, a regulator of tumor progression, is regulated by clock gene proteins. Cancer Res. 2004;64:8328–33. doi: 10.1158/0008-5472.CAN-04-2122. [DOI] [PubMed] [Google Scholar]
- 160.Maity G, Sen T, Chatterjee A. Laminin induces matrix metalloproteinase-9 expression and activation in human cervical cancer cell line (SiHa) J Cancer Res Clin Oncol. 2011;137:347–57. doi: 10.1007/s00432-010-0892-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Venticinque L, Jamieson KV, Meruelo D. Interactions between laminin receptor and the cytoskeleton during translation and cell motility. PLoS One. 2011;6:e15895. doi: 10.1371/journal.pone.0015895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Scheiman J, Jamieson KV, Ziello J, Tseng JC, Meruelo D. Extraribosomal functions associated with the C terminus of the 37/67 kDa laminin receptor are required for maintaining cell viability. Cell Death Dis. 2010;1:e42. doi: 10.1038/cddis.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Liu L, et al. Hypoxia promotes metastasis in human gastric cancer by up-regulating the 67-kDa laminin receptor. Cancer Sci. 2010;101:1653–60. doi: 10.1111/j.1349-7006.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ifuku M, et al. Bradykinin-induced microglial migration mediated by B1-bradykinin receptors depends on Ca2+ influx via reverse-mode activity of the Na+/Ca2+ exchanger. J Neurosci. 2007;27:13065–73. doi: 10.1523/JNEUROSCI.3467-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kyrylenko S, Kyrylenko O, Suuronen T, Salminen A. Differential regulation of the Sir2 histone deacetylase gene family by inhibitors of class I and II histone deacetylases. Cell Mol Life Sci. 2003;60:1990–7. doi: 10.1007/s00018-003-3090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. J Biol Chem. 2009;284:3823–32. doi: 10.1074/jbc.M807869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kim JE, Lou ZK, Chen JJ. Interactions between DBC1 and SIRT1 are deregulated in breast cancer cells. Cell Cycle. 2009;8:3784–5. doi: 10.4161/cc.8.22.10055. [DOI] [PubMed] [Google Scholar]
- 169.Nosho K, et al. SIRT1 histone deacetylase expression is associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol. 2009;22:922–32. doi: 10.1038/modpathol.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Olmos Y, Brosens JJ, Lam EWF. Interplay between SIRT proteins and tumour suppressor transcription factors in chemotherapeutic resistance of cancer. Drug Resistance Updates. 2011;14:35–44. doi: 10.1016/j.drup.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 171.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. IARC monographs on the evaluation of carcinogenic risks to humans. Vol. 98. Lyon (France): IARC; 2007. Painting, firefighting, and shiftwork. [PMC free article] [PubMed] [Google Scholar]
- 172.Davis S, Mirick D. Circadian disruption, shift work and the risk of cancer: a summary of the evidence and studies in Seattle. Cancer Causes Control. 2006;17:539–45. doi: 10.1007/s10552-005-9010-9. [DOI] [PubMed] [Google Scholar]
- 173.Dubocovich ML, et al. International Union of Basic and Clinical Pharmacology. LXXV. Nomenclature, classification, and pharmacology of G protein-coupled melatonin receptors. Pharmacol Rev. 2010;62:343–80. doi: 10.1124/pr.110.002832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reiter RJ, Tan D-X, Fuentes-Broto L. Chapter 8 - Melatonin: A Multitasking Molecule. In: Luciano M, editor. Progress in Brain Research. Elsevier; 2010. pp. 127–151. [DOI] [PubMed] [Google Scholar]
- 175.Mazzoccoli G, Vendemiale G, De Cata A, Carughi S, Tarquini R. Altered time structure of neuro-endocrine-immune system function in lung cancer patients. BMC Cancer. 2010;10:314. doi: 10.1186/1471-2407-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hu S, Shen G, Yin S, Xu W, Hu B. Melatonin and tryptophan circadian profiles in patients with advanced non-small cell lung cancer. Adv Ther. 2009;26:886–92. doi: 10.1007/s12325-009-0068-8. [DOI] [PubMed] [Google Scholar]
- 177.Galijasevic S, Abdulhamid I, Abu-Soud HM. Melatonin is a potent inhibitor for myeloperoxidase. Biochemistry. 2008;47:2668–77. doi: 10.1021/bi702016q. [DOI] [PubMed] [Google Scholar]
- 178.Roncucci L, et al. Myeloperoxidase- positive cell infiltration in colorectal carcinogenesis as indicator of colorectal cancer risk. Cancer Epidemiol Biomarkers Prev. 2008;17:2291–7. doi: 10.1158/1055-9965.EPI-08-0224. [DOI] [PubMed] [Google Scholar]
- 179.Wheatley-Price P, et al. Myeloperoxidase and superoxide dismutase polymorphisms are associated with an increased risk of developing pancreatic adenocarcinoma. Cancer. 2008;112:1037–42. doi: 10.1002/cncr.23267. [DOI] [PubMed] [Google Scholar]
- 180.Sliwinski T, et al. Protective action of melatonin against oxidative DNA damage: chemical inactivation versus base-excision repair. Mutat Res. 2007;634:220–7. doi: 10.1016/j.mrgentox.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 181.Hill SM, et al. Molecular mechanisms of melatonin anticancer effects. Integr Cancer Ther. 2009;8:337–46. doi: 10.1177/1534735409353332. [DOI] [PubMed] [Google Scholar]
- 182.Blask DE, Dauchy RT, Brainard GC, Hanifin JP. Circadian stage-dependent inhibition of human breast cancer metabolism and growth by the nocturnal melatonin signal: consequences of its disruption by light at night in rats and women. Integr Cancer Ther. 2009;8:347–53. doi: 10.1177/1534735409352320. [DOI] [PubMed] [Google Scholar]
- 183.Flynn-Evans E, Stevens R, Tabandeh H, Schernhammer E, Lockley S. Total visual blindness is protective against breast cancer. Cancer Causes Control. 2009;20:1753–6. doi: 10.1007/s10552-009-9405-0. [DOI] [PubMed] [Google Scholar]
- 184.Hevia D, et al. Melatonin uptake in prostate cancer cells: intracellular transport versus simple passive diffusion. J Pineal Res. 2008;45:247–57. doi: 10.1111/j.1600-079X.2008.00581.x. [DOI] [PubMed] [Google Scholar]
- 185.Kayumov L, Lowe A, Rahman SA, Casper RF, Shapiro CM. Prevention of melatonin suppression by nocturnal lighting: relevance to cancer. Eur J Cancer Prev. 2007;16:357–62. doi: 10.1097/01.cej.0000215622.59122.d4. [DOI] [PubMed] [Google Scholar]
- 186.Stevens RG. Light-at-night, circadian disruption and breast cancer: assessment of existing evidence. Int J Epidemiol. 2009;38:963–70. doi: 10.1093/ije/dyp178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Stevens R, Rea M. Light in the built environment: potential role of circadian disruption in endocrine disruption and breast cancer. Cancer Causes Control. 2001;12:279–87. doi: 10.1023/a:1011237000609. [DOI] [PubMed] [Google Scholar]
- 188.Stevens RG, et al. Meeting report: the role of environmental lighting and circadian disruption in cancer and other diseases. Environ Health Perspect. 2007;115:1357–62. doi: 10.1289/ehp.10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Giacchetti S, et al. Phase III trial comparing 4-day chronomodulated therapy versus 2-day conventional delivery of fluorouracil, leucovorin, and oxaliplatin as first-line chemotherapy of metastatic colorectal cancer: the European Organisation for Research and Treatment of Cancer Chronotherapy Group. J Clin Oncol. 2006;24:3562–9. doi: 10.1200/JCO.2006.06.1440. [DOI] [PubMed] [Google Scholar]
- 190.Bouchahda M, et al. Rescue chemotherapy using multidrug chronomodulated hepatic arterial infusion for patients with heavily pre-treated metastatic colorectal cancer. Cancer. 2009;115:4990–9. doi: 10.1002/cncr.24549. [DOI] [PubMed] [Google Scholar]
- 191.Lévi F, et al. Cetuximab and circadian chronomodulated chemotherapy as salvage treatment for metastatic colorectal cancer (mCRC): safety, efficacy and improved secondary surgical resectability. Cancer Chemother Pharmacol. 2011;67:339–48. doi: 10.1007/s00280-010-1327-8. [DOI] [PubMed] [Google Scholar]
- 192.Asao T, et al. The synchronization of chemotherapy to circadian rhythms and irradiation in pre-operative chemoradiation therapy with hyperthermia for local advanced rectal cancer. Int J Hyperthermia. 2006;22:399–406. doi: 10.1080/02656730600799873. [DOI] [PubMed] [Google Scholar]
- 193.Weinrib AZ, et al. Diurnal cortisol dysregulation, functional disability, and depression in women with ovarian cancer. Cancer. 2010;116:4410–9. doi: 10.1002/cncr.25299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Bower JE, et al. Diurnal cortisol rhythm and fatigue in breast cancer survivors. Psychoneuroendocrinology. 2005;30:92–100. doi: 10.1016/j.psyneuen.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 195.Bower JE, Ganz PA, Aziz N. Altered cortisol response to psychologic stress in breast cancer survivors with persistent Fatigue. Psychosom Med. 2005;67:277–80. doi: 10.1097/01.psy.0000155666.55034.c6. [DOI] [PubMed] [Google Scholar]
- 196.Abercrombie HC, et al. Flattened cortisol rhythms in metastatic breast cancer patients. Psychoneuroendocrinology. 2004;29:1082–92. doi: 10.1016/j.psyneuen.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 197.Johansson A-C, et al. Pain, disability and coping reflected in the diurnal cortisol variability in patients scheduled for lumbar disc surgery. Eur J Pain. 2008;12:633–40. doi: 10.1016/j.ejpain.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 198.Rich T, et al. Elevated serum cytokines correlated with altered behavior, serum cortisol rhythm, and dampened 24-hour rest-activity patterns in patients with metastatic colorectal cancer. Clin Cancer Res. 2005;11:1757–64. doi: 10.1158/1078-0432.CCR-04-2000. [DOI] [PubMed] [Google Scholar]
- 199.Sephton SE, et al. Depression, cortisol, and suppressed cell-mediated immunity in metastatic breast cancer. Brain Behav Immun. 2009;23:1148–55. doi: 10.1016/j.bbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 200.Fang MZ, Zhang X, Zarbl H. Methylselenocysteine resets the rhythmic expression of circadian and growth-regulatory genes disrupted by nitrosomethylurea in vivo. Cancer Prev Res. 2010;3:640–52. doi: 10.1158/1940-6207.CAPR-09-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhang X, Zarbl H. Chemopreventive doses of methylselenocysteine alter circadian rhythm in rat mammary tissue. Cancer Prev Res. 2008;1:119–27. doi: 10.1158/1940-6207.CAPR-08-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hua H, et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14:815–8. doi: 10.1038/sj.cgt.7701061. [DOI] [PubMed] [Google Scholar]
- 203.Iurisci I, et al. Improved tumor control through circadian clock induction by Seliciclib, a cyclin-dependent kinase inhibitor. Cancer Res. 2006;66:10720–8. doi: 10.1158/0008-5472.CAN-06-2086. [DOI] [PubMed] [Google Scholar]
- 204.Bernard S, Cajavec Bernard B, Lévi F, Herzel H. Tumor growth rate determines the timing of optimal chronomodulated treatment schedules. PLoS Comput. Biol. 2010;6:e1000712. doi: 10.1371/journal.pcbi.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shukla P, et al. Circadian variation in radiation-induced intestinal mucositis in patients with cervical carcinoma. Cancer. 2010;116:2031–5. doi: 10.1002/cncr.24867. [DOI] [PubMed] [Google Scholar]
- 206.Rahn DA, et al. Gamma knife radiosurgery for brain metastasis of nonsmall cell lung cancer: is there a difference in outcome between morning and afternoon treatment? Cancer. 2011;117:414–20. doi: 10.1002/cncr.25423. [DOI] [PubMed] [Google Scholar]
- 207.Innominato PF, Lévi FA, Bjarnason GA. Chronotherapy and the molecular clock: clinical implications in oncology. Adv Drug Delivery Rev. 2010;62:979–1001. doi: 10.1016/j.addr.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 208.Mormont M-C, Levi F. Cancer chronotherapy: principles, applications, and perspectives. Cancer. 2003;97:155–69. doi: 10.1002/cncr.11040. [DOI] [PubMed] [Google Scholar]
- 209.Hrushesky WJM, et al. Circadian clock manipulation for cancer prevention and control and the relief of cancer symptoms. Integr Cancer Ther. 2009;8:387–97. doi: 10.1177/1534735409352086. [DOI] [PubMed] [Google Scholar]
- 210.Ohdo S, Koyanagi S, Matsunaga N. Chronopharmacological strategies: intra- and inter-individual variability of molecular clock. Adv Drug Deliv Rev. 2010;62:885–97. doi: 10.1016/j.addr.2010.04.005. [DOI] [PubMed] [Google Scholar]