Abstract
OBJECTIVE:
Enterobacteriaceae bacteria harboring Klebsiella pneumoniae carbapenemase are a serious worldwide threat. The molecular identification of these pathogens is not routine in Brazilian hospitals, and a rapid phenotypic screening test is desirable. This study aims to evaluate the modified Hodge test as a phenotypic screening test for Klebsiella pneumoniae carbapenemase.
METHOD:
From April 2009 to July 2011, all Enterobacteriaceae bacteria that were not susceptible to ertapenem according to Vitek2 analysis were analyzed with the modified Hodge test. All positive isolates and a random subset of negative isolates were also assayed for the presence of blaKPC. Isolates that were positive in modified Hodge tests were sub-classified as true-positives (E. coli touched the ertapenem disk) or inconclusive (distortion of the inhibition zone of E. coli, but growth did not reach the ertapenem disk). Negative results were defined as samples with no distortion of the inhibition zone around the ertapenem disk.
RESULTS:
Among the 1521 isolates of Enterobacteriaceae bacteria that were not susceptible to ertapenem, 30% were positive for blaKPC, and 35% were positive according to the modified Hodge test (81% specificity). Under the proposed sub-classification, true positives showed a 98% agreement with the blaKPC results. The negative predictive value of the modified Hodge test for detection was 100%. KPC producers showed high antimicrobial resistance rates, but 90% and 77% of these isolates were susceptible to aminoglycoside and tigecycline, respectively.
CONCLUSION:
Standardizing the modified Hodge test interpretation may improve the specificity of KPC detection. In this study, negative test results ruled out 100% of the isolates harboring Klebsiella pneumoniae carbapenemase-2. The test may therefore be regarded as a good epidemiological tool.
Keywords: Modified Hodge Test, KPC, Carbapenemase, Ertapenem
INTRODUCTION
Carbapenems are one of the last remaining antimicrobial treatments for multidrug-resistant Enterobacteriaceae, and resistance to this class of antibiotics is becoming a threat worldwide (1). Impermeability due to porin loss concomitant with the hyperproduction of cephalosporinases is often the cause of carbapenem resistance, but serine beta-lactamases, such as Klebsiella pneumoniae carbapenemase (KPC), are spreading fast and becoming prevalent worldwide (1-3). The genes encoding the KPC enzyme are usually flanked by transposon-related sequences that have been identified on transferable plasmids, giving these genes the potential to disseminate to other genera and species (4). Currently, KPC producers are found all over the world, with reports from Greece (5), Israel (6), France (7), China (8), and South America (4).
In Brazil, KPC was first described in the northwestern region in 2006 and has become an important mechanism of resistance to carbapenems since then (9-11). In our institution, carbapenem resistance among Enterobacteriaceae was only sporadically noted prior to 2005. However, starting in 2008, KPC producers became an epidemiological challenge.
The early recognition of KPC-producing isolates has become necessary due to the clinical and epidemiological implications of the presence of KPC. The routine laboratory detection of KPC is a challenge, especially for laboratories without molecular diagnostic resources, as is common in Brazilian hospitals.
The modified Hodge test (MHT) is a phenotypic screening test for carbapenemases that is used for epidemiological purposes, and its use is currently proposed by the Clinical and Laboratory Standards Institute (CLSI) (12). The MHT is easy to perform, but divergent specificity values have been reported, and false-positive results are a concern (13,14).
The aim of this study was to evaluate the performance of a new proposal for interpreting the modified Hodge test and detect the blaKPC gene among Enterobacteriaceae isolates, which were collected in our 2500–bed hospital, that exhibited ertapenem MICs ≥1 µg/ml.
MATERIALS AND METHODS
From April 2009 to July 2011, a total of 1521 Enterobacteriaceae isolates that were not susceptible to ertapenem (ENSEs), with MICs ≥1 µg/ml from any clinical specimen, were selected from among the patient cultures of the Hospital das Clínicas da Faculdade de Medicina da Universidade de São Paulo. When more than one isolate was recovered from a patient, the second isolate was only included if it was another species or the same species with a different susceptibility pattern.
Identification and antimicrobial susceptibility testing were performed using a Vitek2 system (bioMerieux Marcy-L'ile, France) with the AST-N105 and AST-N104 cards. Categorical interpretations were performed according to CLSI M100–S21 (12). For colistin, the CLSI breakpoints for P. aeruginosa were the reference for categorical interpretation, and the U.S. Food and Drug Administration (FDA) breakpoints were used for tigecycline. All isolates were tested by the MHT according to the CLSI recommendations and quality control.
The results of the modified Hodge test were categorized according to the CLSI recommendations as follows: negative when there was no distortion of the inhibition zone around the ertapenem disk, positive when any distortion of the E. coli ATCC 25922 (strain indicator) inhibition zone was noted around the ertapenem disk, and indeterminate when the inhibition of E. coli ATCC 25922 growth around the streak (tested strain) was evidenced by a clear area.
All isolates with positive modified Hodge test results were classified using a new sub-classification proposed by our group: A) a result was considered a true-positive when E. coli ATCC 25922 touched the ertapenem disk; and B) a result was considered inconclusive when there was distortion of the E. coli ATCC 25922 inhibition zone around the ertapenem disk, but the bacteria did not touch the edge of the disk. See Figure 1 for the modified Hodge test sub-classification definition in our study.
Figure 1.
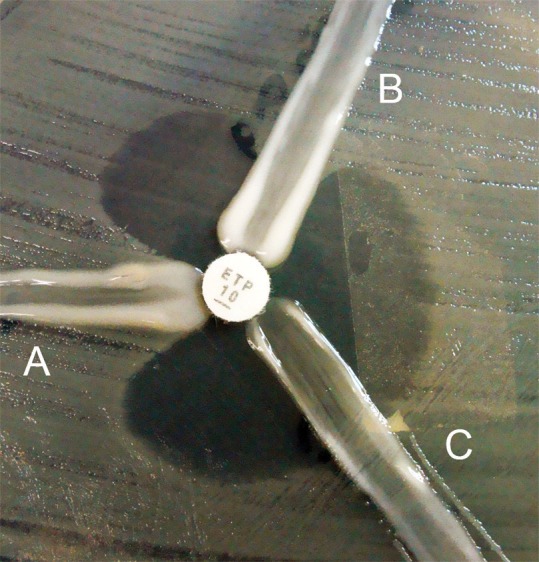
Modified Hodge test (MHT) with ertapenem disks. Interpretation of the MHT results proposed in this study: A) true-positive MHT, B) inconclusive MHT and C) negative MHT.- Modified Hodge test (MHT) with ertapenem disks. Interpretation of the MHT results proposed in this study: A) true-positive MHT, B) inconclusive MHT and C) negative MHT.
All isolates with positive and indeterminate modified Hodge test results were submitted to molecular detection of the blaKPC gene by PCR. Isolates with negative test results were also randomly chosen for molecular KPC testing. The primers used for PCR amplification and the reaction conditions were described previously (15). The PCR products were visualized by electrophoresis on 2% agarose gels stained with ethidium bromide and visualized with UV light.
Two samples of blaKPC amplicons (859 bp) produced by PCR were selected and submitted to DNA sequencing with a BigDye Terminator cycle sequencing kit v3.1 (Applied Biosystems Inc., Foster City, CA) in an MJ Research PTC-200 DNA Engine thermal cycler (Bio-Rad Laboratories, Waltham, USA). The sequencing reaction products were purified by ethanol precipitation, separated, and analyzed using an ABI Prism 3100 genetic analyzer (ABI, Foster City, USA) following the manufacturer's protocols. The sequenced products were compared using software available at the National Center for Biotechnology Information website (http://blast.ncbi.nlm.nih.gov).
The antibiograms for KPC producers and non-producers were analyzed. The MIC50 and MIC90 values were calculated using Whonet version 5.4.
The sensitivity, specificity, positive predictive values (PPVs), and negative predictive values (NPVs) were calculated according to the method of Ilstrup (16).
RESULTS
We analyzed 1521 Enterobacteriaceae isolates that were not susceptible to ertapenem, of which 30% (458) were positive for blaKPC. Among the blaKPC-positive isolates, 93% were Klebsiella pneumoniae. The modified Hodge test results are summarized according to the species distribution and molecular detection of blaKPC in Table 1. The modified Hodge test was positive in 35.5% of Enterobacteriaceae isolates that were not susceptible to ertapenem (540), and according to our proposed sub-classification, 71% (386) of those isolates showed true-positive results, and 29% (154) had inconclusive results. The true-positive results showed 98% agreement with the molecular KPC results, whereas the level of agreement for the inconclusive results was only 51%.
Table 1.
Distribution of 1521 Enterobacteriaceae isolates that were not susceptible to ertapenem (ENSE) according to the modified Hodge test (MHT) results and the presence of blaKPC.
| MHT resultsa)N° ENSE | Organism | N° ENSE(blaKPC tested) | blaKPCNegative | blaKPCPositive | |
| K. pneumoniae | 481 (191) | 191 | 0 | ||
| E. cloacae | 293 (111) | 111 | 0 | ||
| E. aerogenes | 85 (16) | 16 | 0 | ||
| E. coli | 62 (22) | 22 | 0 | ||
| S. marcescens | 15 (8) | 8 | 0 | ||
| Negative | P. mirabilis | 12 (1) | 1 | 0 | |
| 975 (64.1%) | M. morgannii | 8 (0) | 0 | 0 | |
| C. freundii | 7 (4) | 4 | 0 | ||
| E. gergoviae | 7 (2) | 2 | 0 | ||
| K. oxytoca | 2 (0) | 0 | 0 | ||
| P. stuartii | 2 (0) | 0 | 0 | ||
| P. vulgaris | 1 (0) | 0 | 0 | ||
| Total | 975 (355) | 355 (100%) | 0 (0) | ||
| True-positiveb) | K. pneumoniae | (354) | 1 | 353 | |
| 386 (71%) | E. cloacae | (10) | 3 | 7 | |
| E. aerogenes | (3) | 0 | 3 | ||
| E. coli | (6) | 0 | 6 | ||
| S. marcescens | (6) | 3 | 3 | ||
| C. koseri | (1) | 0 | 1 | ||
| Positive | C. freundii | (1) | 0 | 1 | |
| 540 (35.5%) | E. asburiae | (2) | 0 | 2 | |
| K. oxytoca | (2) | 0 | 2 | ||
| Total | (386) | 7 (2%) | 379 (98%) | ||
| Inconclusiveb) | K. pneumoniae | (111) | 40 | 71 | |
| 154 (29%) | E. cloacae | (26) | 24 | 2 | |
| E. aerogenes | (10) | 10 | 0 | ||
| E. coli | (2) | 0 | 2 | ||
| E. gergoviae | (2) | 2 | 0 | ||
| E. asburiae | (1) | 0 | 1 | ||
| S. marcescens | (2) | 0 | 2 | ||
| Total | (154) | 76 (49%) | 78 (51%) | ||
| Indeterminate | E. cloacae | (4) | 3 | 1 | |
| 6 (0.4%) | E. coli | (1) | 1 | 0 | |
| K. pneumoniae | (1) | 1 | 0 | ||
| Total | (6) | 5 (86%) | 1 (14%) | ||
| Total: 1521 | (901) | 443 | 458 | ||
Positive, indeterminate, and negative results were defined according to the recommendations of the Clinical and Laboratory Standards Institute.
True-positive and inconclusive results were defined according to this study's proposal (see Figure 1).
The modified Hodge test results according to the sensitivity, specificity and predictive values for KPC detection are shown in Table 2. The modified Hodge test had a 100% negative predictive value.
Table 2.
Performance of the modified Hodge test (MHT) for KPC detection.
| MHT | N° Isolates | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
| True-positive | 386 | 100 | 98 | 98 | 100 |
| Inconclusive | 154 | 100 | 82 | 51 | 100 |
| Positivea) | 540 | 100 | 81 | 85 | 100 |
True-positive plus inconclusive results.
PPV - positive predictive value, NPV – negative predictive value.
Specimens yielding KPC-producing isolates were primarily urine samples (37%), rectal swabs (17%), blood samples (10%), and catheters (10%).
The antimicrobial susceptibility profile of the KPC-producing isolates revealed high resistance rates to all cephalosporins, aztreonam (98.9%), piperacillin/tazobactam (99.3%), and fluoroquinolones (92%). Amikacin and gentamycin had susceptibility rates of 95% and 88%, respectively. Tigecycline had an 8% susceptibility rate, and colistin had an MIC50 of 2 mg/l and MIC90 of 4 mg/l. The meropenem results for KPC isolates from the automated system indicated that 50% were resistant, but the disk method indicated that 70% of isolates were resistant (data not shown). The KPC non-producing isolates also had high rates of resistance to all cephalosporins, aztreonam, piperacillin/tazobactam, and fluoroquinolones, and the detailed information is summarized in Table 3.
Table 3.
In vitro activity of antimicrobial agents tested against 901 ENSE isolates.
| Antibiotica) | KPC producers (458) | KPC non-producers (443) | |||||||||
| %R | %I | %S | MIC50 | MIC90 | %R | %I | %S | MIC50 | MIC90 | ||
| PTZ | 99.3 | 0.7 | 0 | 128 | 128 | 93.3 | 2.1 | 4.6 | 128 | 128 | |
| Ceftazidime | 82.9 | 5.6 | 11.5 | 64 | 64 | 91 | 2 | 7 | 64 | 64 | |
| Cefotaxime | 94.7 | 3.7 | 1.5 | 64 | 64 | 95.9 | 0.7 | 3.4 | 64 | 64 | |
| Cefepime | 54.0 | 23.5 | 22.5 | 64 | 64 | 68.7 | 4.6 | 26.7 | 64 | 64 | |
| Aztreonam | 98.9 | 0.3 | 0.8 | 64 | 64 | 93.4 | 0.8 | 5.8 | 64 | 64 | |
| Ertapenem | 100 | 0 | 0 | 8 | 8 | 99.5 | 0.5 | 0 | 4 | 8 | |
| Imipenem | 69.8 | 16.4 | 13.8 | 8 | 16 | 22.5 | 8 | 69.5 | 1 | 8 | |
| Meropenem | 50 | 7 | 43 | 2 | 16 | 27.9 | 3.6 | 68.4 | 0.25 | 8 | |
| Amikacin | 3.1 | 1.5 | 95.4 | 2 | 16 | 17 | 3 | 80 | 2 | 64 | |
| Gentamicin | 11 | 1 | 88 | 1 | 16 | 48.3 | 2.3 | 49.4 | 8 | 16 | |
| Ciprofloxacin | 92 | 1 | 7 | 4 | 4 | 79.5 | 2.5 | 18 | 4 | 4 | |
| SUT | 94 | 0 | 6 | 16 | 16 | 60.4 | 0 | 39.6 | 16 | 16 | |
| Colistinb) | 32 | 0 | 68 | 0.5 | 16 | 11.7 | 0.5 | 87.8 | 0.5 | 16 | |
| Tigecyclinec) | 8 | 15 | 77 | 2 | 4 | 11 | 13 | 76 | 1 | 8 | |
Minimum inhibitory concentration (MIC) breakpoint in mg/L, as determined by the Vitek2® system.
Breakpoint for P. aeruginosa according to the Clinical and Laboratory Standards Institute.
U.S. Food and Drug Administration (FDA) breakpoints.
SUT: trimethoprim/sulfamethoxazole; PTZ: piperacillin/tazobactam; ENSE: Enterobacteriaceae not susceptible to ertapenem; KPC: Klebsiella pneumoniae carbapenemases; R: resistant; I: intermediate; S: susceptible.
The DNA sequencing of the KPC-producing isolates confirmed that they carried the KPC-2 variant.
DISCUSSION
The presence of blaKPC deserves special attention because isolates harboring this plasmid gene are usually multidrug resistant, limiting the therapeutic options available and causing a high mortality rate (17).
KPC represents an epidemiological challenge for infection control groups because it can spread very quickly, and there are limited treatment options available (18). The early detection of such isolates is critical. Screening for KPC producers by identifying Enterobacteriaceae isolates that are not susceptible to ertapenem has a low specificity but is a starting point to identify isolates harboring carbapenemases. The modified Hodge test is relatively easy to perform in standard microbiology laboratories, and the standardization of the interpretation of the results may improve the specificity of this test for KPC detection.
Infections caused by KPC-producing isolates are now a concern in our institution, and 30% of the Enterobacteriaceae isolates that were not susceptible to ertapenem were KPC positive in this study period. The majority of those isolates (37%) were from urine cultures, but the positivity rate for KPC among blood isolates was proportionally higher (data not shown).
The molecular detection of blaKPC is the gold standard for diagnosis, but the majority of standard laboratories in our country do not have the resources necessary to perform this test on a routine basis. Phenotypic tests should be made available to detect KPC early or rule it out. The modified Hodge test may detect the presence of carbapenemases, but it is not specific for KPC and may have false-positive results due to non-carbapenemase enzymes, such as AmpC and/or extended-spectrum beta-lactamases (ESBLs), combined with porin loss. Positive results could also be due to other carbapenemases. In the literature, the specificity of the modified Hodge test differs because some authors analyzed data for distinct bacteria groups, such as non-fermenters and Enterobacteriaceae, and detected different carbapenemases, such as NDM (13). In our study, the modified Hodge test was evaluated only for Enterobacteriaceae bacteria and KPC producers, which may explain the high level of agreement with the molecular results.
The proposed positive modified Hodge test category had 98% agreement with the molecular blaKPC results, highlighting the good positive predictive value of KPC detection among Enterobacteriaceae when a standardized method for interpretation is in place. Seven isolates showed true-positive results and were negative for blaKPC, but an accurate analysis of the reason for these discrepancies was limited because no tests were performed for other carbapenemases. Of the isolates in the inconclusive group, 49% were negative for blaKPC, and these negative strains most likely produced CTX-M or hyperproduced AmpC in association with porin loss. Another possibility is that these isolates produced carbapenemases other than KPC.
The indeterminate result, as proposed by the CLSI, does not have a clear microbiological or clinical interpretation in the literature and should be the focus of future studies.
Enterobacteriaceae bacteria that not susceptible to ertapenem but have a negative modified Hodge test result are not KPC producers.
Although tests other than the modified Hodge test, such as the aminophenylboronic acid and dipicolinic acid tests, may also be options for the phenotypic screening for carbapenemases (19), the supplies necessary for these tests are not routinely available in the majority of Brazilian laboratories.
The antibiograms of the 458 KPC isolates generated using the automated system revealed multidrug resistance and showed that 43% of isolates were susceptible to meropenem (MIC ≤1 µg/ml). The meropenem susceptibility rates were not reproducible by TREK microdilution panels (Cleveland, OH) or the disk diffusion method (OXOID) (data not shown), with 45% very major errors (VMEs) for this drug. Corroborating these findings, the literature contains reports of unacceptable VME levels for meropenem when KPC isolates were tested using automated systems. Microbiologists and clinicians should be aware of the current problems regarding false susceptibility results for meropenem among KPC isolates when only automated systems are used to screen isolates (20). Regarding the colistin and tigecycline Vitek results, our internal validation process did not show major discrepancies with the E-test (data not shown), and published studies have not reported major problems with these drugs (21). Every laboratory should perform its own validation of automated systems when CLSI breakpoints are in place.
The modified Hodge test results for Enterobacteriaceae using standardized readings are a useful phenotypic and epidemiological tool to identify KPC isolates while waiting for molecular results. Negative modified Hodge test results ruled out all KPC isolates in our institution in an efficient and timely manner.
Footnotes
No potential conflict of interest was reported.
REFERENCES
- 1.Deshpande LM, Rhomberg PR, Sader HS, Jones RN. Emergence of serine carbapenemases (KPC and SME) among clinical strains of Enterobacteriaceae isolated in the United States Medical Centers: report from the MYSTIC Program (1999-2005) Diagn Microbiol Infect Dis. 2006;56(4):367–72. doi: 10.1016/j.diagmicrobio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Chiang T, Mariano N, Urban C, Colon-Urban R, Grenner L, Eng RH, et al. Identification of carbapenem-resistant klebsiella pneumoniae harboring KPC enzymes in New Jersey. Microb Drug Resist. 2007 Winter. 13(4):235–39. doi: 10.1089/mdr.2007.767. [DOI] [PubMed] [Google Scholar]
- 3.Bratu S, Mooty M, Nichani S, Landman D, Gullans C, Pettinato B, et al. Emergence of KPC-possessing Klebsiella pneumoniae in Brooklyn, New York: epidemiology and recommendations for detection. Antimicrob Agents Chemother. 2005;49(7):3018–20. doi: 10.1128/AAC.49.7.3018-3020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M, et al. First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother. 2006;50(8):2880–2. doi: 10.1128/AAC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maltezou HC, Giakkoupi P, Maragos A, Bolikas M, Raftopoulos V, Papahatzaki H, et al. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece) J Infect. 2009;58(3):213–9. doi: 10.1016/j.jinf.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Navon-Venezia S, Chmelnitsky I, Leavitt A, Schwaber MJ, Schwartz D, Carmeli Y. Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob Agents Chemother. 2006;50(9):3098–101. doi: 10.1128/AAC.00438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naas T, Nordmann P, Vedel G, Poyart C. Plasmid-mediated carbapenem-hydrolyzing beta-lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob Agents Chemother. 2005;49(10):4423–4. doi: 10.1128/AAC.49.10.4423-4424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–5. doi: 10.1128/AAC.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavez M, Mamizuka EM, Lincopan N. Early dissemination of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53(6):2702. doi: 10.1128/AAC.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monteiro J, Santos AF, Asensi MD, Peirano G, Gales AC. First report of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53(1):333–4. doi: 10.1128/AAC.00736-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrade LN, Curiao T, Ferreira JC, Longo JM, Climaco EC, Martinez R, et al. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother. 2011;55(7):3579–83. doi: 10.1128/AAC.01783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. CLSI document Wayne: PA, USA; 2011. Performance Standards for antimicrobial susceptibility testing; p. M100-S21. [PubMed] [Google Scholar]
- 13.Girlich D, Poirel L, Nordmann P. Value of the modified Hodge test for detection of emerging carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2012;50(2):477–9. doi: 10.1128/JCM.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carvalhaes CG, Picao RC, Nicoletti AG, Xavier DE, Gales AC. Cloverleaf test (modified Hodge test) for detecting carbapenemase production in Klebsiella pneumoniae: be aware of false positive results. J Antimicrob Chemother. 2010;65(2):249–51. doi: 10.1093/jac/dkp431. [DOI] [PubMed] [Google Scholar]
- 15.Castanheira M, Sader HS, Jones RN. Antimicrobial susceptibility patterns of KPC-producing or CTX-M-producing Enterobacteriaceae. Microb Drug Resist. 2010;16(1):61–5. doi: 10.1089/mdr.2009.0031. [DOI] [PubMed] [Google Scholar]
- 16.Ilstrup DM. Statistical methods in microbiology. Clin Microbiol Rev. 1990;3(3):219–26. doi: 10.1128/cmr.3.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, et al. Predictors of Mortality in Bloodstream Infections Caused by KPC-Producing Klebsiella pneumoniae: Importance of Combination Therapy. Clin Infect Dis. 2012;55(7):943:50. doi: 10.1093/cid/cis588. [DOI] [PubMed] [Google Scholar]
- 18.Zarkotou O, Pournaras S, Tselioti P, Dragoumanos V, Pitiriga V, Ranellou K, et al. Predictors of mortality in patients with bloodstream infections caused by KPC-producing Klebsiella pneumoniae and impact of appropriate antimicrobial treatment. Clin Microbiol Infect. 2011;17(12):1798–803. doi: 10.1111/j.1469-0691.2011.03514.x. [DOI] [PubMed] [Google Scholar]
- 19.Pasteran F, Mendez T, Rapoport M, Guerriero L, Corso A. Controlling false-positive results obtained with the Hodge and Masuda assays for detection of class a carbapenemase in species of enterobacteriaceae by incorporating boronic Acid. J Clin Microbiol. 2010;48(4):1323–32. doi: 10.1128/JCM.01771-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulik CC, Fauntleroy KA, Jenkins SG, Abuali M, LaBombardi VJ, Nicolau DP, et al. Comparison of meropenem MICs and susceptibilities for carbapenemase-producing Klebsiella pneumoniae isolates by various testing methods. J Clin Microbiol. 2010;48(7):2402–6. doi: 10.1128/JCM.00267-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lat A, Clock SA, Wu F, Whittier S, Della-Latta P, Fauntleroy K, et al. Comparison of polymyxin B, tigecycline, cefepime, and meropenem MICs for KPC-producing Klebsiella pneumoniae by broth microdilution, Vitek 2, and Etest. J Clin Microbiol. 2011;49(5):1795–8. doi: 10.1128/JCM.02534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]


