Abstract
Mitochondria play a central role in ageing. They are considered to be both a target of the ageing process, as well as a contributor to it1. Alterations in mitochondrial structure and function are evident during ageing in most eukaryotes2, but how this occurs is poorly understood. Here, we identify a functional link between the lysosome-like vacuole and mitochondria in S. cerevisiae, and show that mitochondrial dysfunction in replicatively-aged yeast arises from altered vacuolar pH. We found that vacuolar acidity declines during the early asymmetric divisions of a mother cell, and show that preventing this decline suppresses mitochondrial dysfunction and extends lifespan. Surprisingly, changes in vacuolar pH do not limit mitochondrial function by disrupting vacuolar protein degradation, but rather, by reducing pH-dependent amino acid storage in the vacuolar lumen. We also found that calorie restriction promotes lifespan extension at least in part by increasing vacuolar acidity via conserved nutrient sensing pathways3. Interestingly, although vacuolar acidity is reduced in aged mother cells, acidic vacuoles are regenerated in newborn daughters, coinciding with daughter cells having a renewed lifespan potential4. Overall, our results identify vacuolar pH as a critical regulator of ageing and mitochondrial function, and outline a potentially conserved mechanism by which calorie restriction delays the ageing process. Because functions of the vacuole are highly conserved throughout evolution5, we hypothesize that lysosomal pH may modulate mitochondrial function and lifespan in other eukaryotic cells.
We sought to understand the basis of age-induced mitochondrial dysfunction, using the budding yeast, S. cerevisiae. Ageing in yeast (replicative ageing) is defined as the number of times an individual cells divides4, and aged yeast cells exhibit many characteristics of age-induced mitochondrial dysfunction observed in metazoa1. These include mitochondrial fragmentation, increased levels of mitochondrial reactive oxygen species (ROS), and increased loss of mitochondrial DNA in their progeny6–9. We began our inquiry by further characterizing age-associated mitochondrial phenotypes using a genetic system in yeast that enriches for replicatively-aged mother cells, the Mother Enrichment Program (MEP)10. By monitoring a fluorescently tagged outer mitochondrial membrane protein, Tom70-GFP, we found that mitochondrial dysfunction was prevalent in aged yeast, consistent with results from earlier studies6. Normally tubular mitochondria fragmented and ultimately aggregated in aged cells (Fig. 1a). Mitochondrial fragmentation was present early in the ageing process (86% of cells at 8 divisions), and progressed to large aggregates and small fragments (93% of cells at 17 divisions) that persisted throughout ageing (median lifespan of 28 divisions). We observed similar changes using an inner mitochondrial membrane protein, Tim50-GFP (Supplementary Fig. 2a). Mitochondrial function also declined in aged cells. We measured the mitochondrial membrane potential (ΔΨ) of aged cells using DiOC6, a ΔΨ-dependent mitochondrial fluorescent dye11, and found that 80% of cells had reduced mitochondrial DiOC6 staining by 7 divisions (Supplementary Fig. 2b). At 18 divisions, ΔΨ was even lower and remained low thereafter (100% of cells by 28 divisions). Consistent with this, ΔΨ-dependent import of the mitochondrial matrix chimeric protein preCox4-mCherry9 was also reduced in aged cells (Supplementary Fig. 2c). Collectively, these results indicate that mitochondrial structure and function are progressively altered during the ageing process.
Figure 1. Age-induced mitochondrial dysfunction is suppressed by VMA1 overexpression.
Age (divisions) in lower panels of all figures represents exact age for young cells (0–5) and median age for older cells as determined by calcofluor staining of bud scars. n = 30 for all timepoints in all figures. A representative image is shown for each timepoint in all figures. a, Mitochondrial morphology in aged cells expressing the mitochondrial outer membrane protein Tom70-GFP. b, Mitochondrial membrane potential as indicated by DiOC6 staining of aged Tom70-mCherry wild-type (WT) and VMA1 overexpressing cells incubated with or without 500 nM concA for 2 h. Note that DiOC6 stains the plasma membrane of aged cells. M = mother cell.
Mitochondrial structure is regulated by ~250 genes in young cells12,13. We hypothesized that age-induced mitochondrial dysfunction might result from altered function of processes associated with one or more of these gene products. To explore this idea, we overexpressed each of the 250 genes in a MEP strain expressing Tom70-GFP, and screened for genes whose overexpression delayed the onset of altered mitochondrial morphology in aged cells. Two suppressors identified in our screen, VMA1 and VPH2, are required for activity of the Vacuolar H+-ATPase (V-ATPase). The V-ATPase is an evolutionarily conserved protein complex that acidifies the vacuole (lysosome in metazoa)5. Vma1 is the catalytic subunit of the peripheral membrane associated V1 sector of the V-ATPase14. Vph2 is an ER-localized integral membrane protein required for V-ATPase assembly15. Overexpression of VMA1 or VPH2 from a high-copy promoter suppressed age-induced mitochondrial dysfunction in 93 or 77% of aged cells, respectively (17 or 16 divisions) (Fig. 1b and Supplementary Fig. 3a). Importantly, suppression by VMA1 or VPH2 overexpression required V-ATPase activity, as treatment with the specific V-ATPase inhibitor, concanamycin A (concA)16, blocked their ability to produce normal, tubular mitochondria (Fig. 1b and Supplementary Fig. 3a). Because overexpressing proteins required for vacuolar acidity prevented age-induced mitochondrial dysfunction, we tested if vacuolar pH itself was changed in aged cells. Measurement of vacuolar acidity with four different pH-sensitive reporters revealed that the acidity of the vacuole decreased as unbudded cells became experienced mother cells (Fig. 2a, b and Supplementary Fig. 4a, b, c). Vacuoles in over 90% of young unbudded cells were acidic and stained efficiently with the fluorescent dye quinacrine, which accumulates in acidic environments17 (Fig. 2a and Supplementary Fig. 4a). However, 87% of cells which had produced daughters had decreased vacuolar acidity, as indicated by little or no quinacrine staining (Fig. 2a and Supplementary Fig. 4a). Kinetic analysis of vacuolar acidity decline using Pho8-SEP/mCherry [a dual fluorescent protein reporter system in which one copy of vacuolar Pho8 is fused to pH-sensitive super ecliptic pHluorin (SEP, that increases in fluorescence with increasing pH)18, and the other to pH-insensitive mCherry] revealed that vacuolar acidity declined progressively during the first ~4 mother cell divisions and then remained low through at least 18 divisions (Fig. 2b). However, acidity did not decline to the level observed in cells lacking V-ATPase function (Fig. 2b, ConcA), suggesting that aged cells have not completely lost vacuolar acidity. Because vacuolar acidity decreases prior to mitochondrial dysfunction in ageing cells, we asked whether overexpressing VMA1 or VPH2 suppressed mitochondrial dysfunction by preventing vacuolar acidity decline. Indeed, overexpressing VMA1 or VPH2 increased vacuolar acidity in ≥70% of aged mother cells (~18 divisions) (Fig. 2a and Supplementary Fig. 3b). Although it is unclear how VMA1 overexpression increased vacuolar acidity, overexpressing VPH2 increased levels of assembled V-ATPase at the vacuolar membrane (Supplementary Fig. 5).
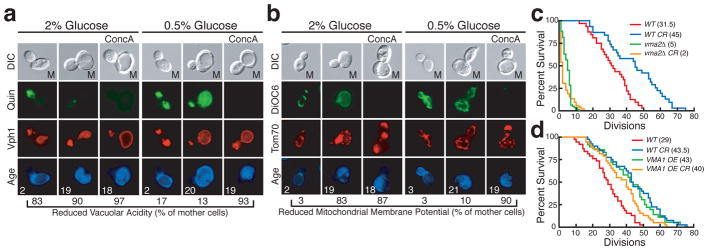
Figure 2. Vacuolar acidity is reduced in ageing cells and regulates mitochondrial function and lifespan.
Vacuolar acidity as indicated by (a) quinacrine (quin) staining of aged Vph1-mCherry wild-type (WT) and VMA1 overexpressing cells and (b) the vacuolar pH reporter Pho8-SEP in cells expressing Pho8-mCherry incubated with or without 500 nM concA for 15 minutes. c, DiOC6 staining of young cells expressing Tom70-mCherry treated with 500 nM concA for the indicated time. d, RLS of wild-type (WT) and VMA1 overexpressing cells by micromanipulation. Median lifespan is indicated. p = 0.0002, Wilcoxon rank-sum test. n = 50 for WT and 56 for VMA1 OE.
These results suggest that reduced vacuolar acidity in mother cells leads to the subsequent development of mitochondrial dysfunction. Consistent with this idea, young cells lacking the V-ATPase subunit VMA2 had reduced ΔΨ and mitochondrial morphology similar to old cells (Supplementary Fig. 6a). Additionally, treatment of young cells with the V-ATPase inhibitor concanamycin A caused mitochondrial depolarization just 30 minutes after loss of vacuolar acidity (Fig. 2c, Supplementary Fig. 6b). This loss of ΔΨ was followed by mitochondrial fragmentation and aggregation that resembled mitochondrial phenotypes present in old cells. Collectively, these results indicate that vacuolar and mitochondrial function are intimately linked, and suggest that low levels of vacuolar acidity in mother cells cause mitochondrial dysfunction by inducing mitochondrial depolarization that is followed by fragmentation and aggregation. Because mitochondrial function is a critical regulator of lifespan19, we suspected that vacuolar pH would be as well. Consistent with this idea, yeast cells lacking V-ATPase activity have drastically shortened lifespans20 (Fig. 4c). We measured the replicative lifespan (RLS) of wild-type, VMA1, and VPH2 overexpressing strains and found that overexpressing either gene significantly increased both median and maximal RLS (Fig. 2d and Supplementary Fig. 3c). These results suggest that vacuolar pH is indeed an important determinant of yeast lifespan.
Figure 4. Calorie restriction extends lifespan by regulating vacuolar acidity.
a, Quinacrine or (b) DiOC6 staining of cells expressing (a) Vph1-mCherry or (b) Tom70-mCherry aged in 2% or 0.5% glucose with or without 500 nM concA for the final 2 h of ageing. RLS of wild-type (WT), (c) vma2Δ and (d) VMA1 OE cells grown in the absence or presence of CR (0.5% glucose). Median lifespan is indicated. c, p < 0.0001 (WT vs WT CR), p = 0.0398 (vma2Δ vs vma2Δ CR), p < 0.0001 (WT vs vma2Δ), Wilcoxon rank-sum test. n = 32 for WT, 31 for WT CR, 60 for vma2Δ, and 52 for vma2Δ CR. d, p < 0.0001 (WT vs WT CR), p < 0.0001 (WT vs VMA1 OE), p = 0.0023 (WT vs VMA1 OE CR), p = 0.52 (WT CR vs VMA1 OE), p = 0.076 (VMA1 OE vs VMA1 OE CR), Wilcoxon rank-sum test. n = 38 for WT, 40 for WT CR, 36 for VMA1 OE, and 38 for VMA1 OE CR.
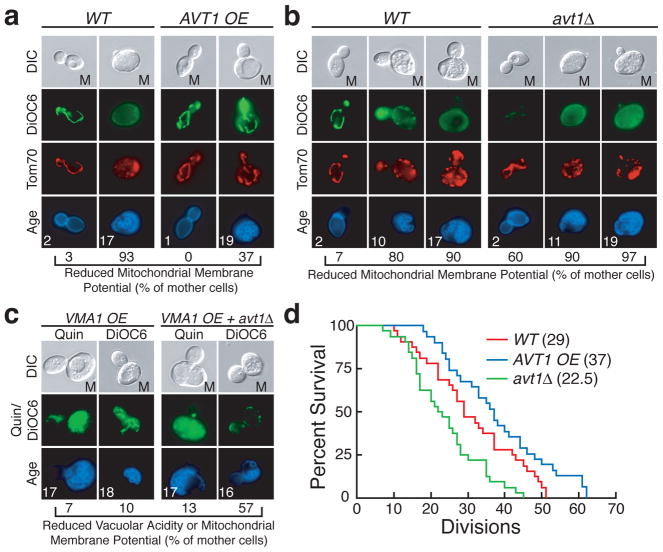
How might changes in vacuolar pH affect mitochondrial function? The vacuolar proton gradient generated by the V-ATPase is required for efficient protein degradation, and for transport of ions and basic and neutral amino acids into the vacuole for storage5. Blocking vacuolar protein degradation in a pH-independent manner by deleting the vacuolar protease PEP421 did not alter mitochondrial structure or ΔΨ (Supplementary Fig. 7). Therefore, we examined whether reduced import of vacuolar metabolites was responsible for mitochondrial dysfunction under conditions of decreased vacuolar acidity. If reduced import of a particular amino acid or ion into the vacuole led to mitochondrial dysfunction, then overexpressing the transporter required for import of that metabolite into the vacuole might suppress mitochondrial dysfunction in cells with a low vacuolar proton gradient. To test this, we reduced the vacuolar proton gradient by treating cells with low concentrations of concanamycin A, and measured mitochondrial ΔΨ with DiOC6 in cells overexpressing individual vacuolar amino acid or ion transporters. Overexpressing most characterized V-ATPase-dependent vacuolar importers22–24 had no (NHX1, VBA1, VBA3) or little (VCX1, VBA2) effect on mitochondrial function. However, overexpressing AVT1, a neutral amino acid transporter25, prevented mitochondrial dysfunction in 60% of cells (Supplementary Fig. 8a, b). This suggests that disrupting proton-dependent neutral amino acid storage in the vacuole causes mitochondrial dysfunction.
We also found that AVT1 was critical in connecting the vacuole and mitochondria during ageing. Overexpressing AVT1 prevented alterations in mitochondrial structure and ΔΨ in 63% of aged cells (19 divisions, Fig. 3a), even though vacuolar acidity was reduced in these cells (17 divisions, Supplementary Fig. 9a). Conversely, deleting AVT1 accelerated the development of age-induced mitochondrial dysfunction (Fig. 3b) without affecting the kinetics of vacuolar acidity decline (Supplementary Fig. 9b). Deleting AVT1 also prevented the suppression of mitochondrial dysfunction by VMA1 and VPH2 overexpression without affecting vacuolar acidity (Fig. 3c and Supplementary Fig. 3d). Collectively, these results suggest that reduced vacuolar acidity in aged cells might cause mitochondrial dysfunction by preventing adequate import of neutral amino acids through the vacuolar transporter Avt1. Importantly, overexpressing or deleting AVT1 was sufficient to extend or shorten replicative lifespan, respectively (Fig. 3d). This suggests that changes in vacuolar pH modulate lifespan at least in part through regulation of mitochondrial function. The differences in mitochondrial function and lifespan by AVT1 overexpression or deletion were not as dramatic as when vacuolar pH was increased or decreased (compare Figure 3 to 1 and 2), suggesting that substrates of additional V-ATPase-dependent transporters may contribute to age-induced mitochondrial dysfunction caused by reduced vacuolar acidity.
Figure 3. Reduced vacuolar acidity causes mitochondrial dysfunction by disrupting amino acid homeostasis.
DiOC6 staining of aged Tom70-mCherry WT, (a) AVT1 overexpressing and (b) avt1Δ cells. c, Quinacrine and DiOC6 staining of aged WT and avt1Δ cells overexpressing VMA1. d, RLS of wild-type (WT), avt1Δ, and AVT1 overexpressing cells by micromanipulation. Median lifespan is indicated. p = 0.0376 (WT vs AVT1 OE), p = 0.0045 (WT vs avt1Δ), Wilcoxon rank-sum test. n = 32 for WT, 32 for AVT1 OE, and 31 for avt1Δ.
Calorie restriction (CR) extends lifespan in most eukaryotes through conserved nutrient sensing pathways3. In yeast, CR is suggested to function through at least three nutrient sensing kinases: PKA, TOR, and the putative S6 kinase/AKT homolog, Sch920,26. Reduced activity of any of these kinases mimics CR and extends lifespan. We found that growth in CR carbon sources (0.5% glucose, 0.05% glucose, and 3% glycerol)26,27 increased vacuolar acidity in young cells (Supplementary Fig. 10), and prevented the decline of vacuolar acidity in ageing cells (Fig. 4a). CR also suppressed mitochondrial dysfunction in 90% of old cells (21 divisions) in a V-ATPase dependent manner (Fig. 4b).
CR appears to regulate vacuolar acidity through conserved nutrient-sensing pathways because inhibiting PKA, Sch9, or TOR by the lifespan extending mutations gpa2Δ, sch9Δ, or tor1Δ, also prevented the decline of vacuolar acidity and development of mitochondrial dysfunction in ageing cells (Supplementary Fig. 11a, b)20,26. Consistent with this, constitutively activating the PKA pathway by deleting the Ras GAP, IRA228, reduced vacuolar acidity and accelerated the development of mitochondrial dysfunction in ageing cells (Supplementary Fig. 11c, d). Deleting IRA2 also prevented CR-mediated enhancement of vacuolar acidity and suppression of mitochondrial dysfunction (Supplementary Fig. 11c, d). Furthermore, we found that lifespan extension by CR was prevented in a strain lacking V-ATPase activity (Fig. 4c), and conversely, CR did not further extend the lifespan of a strain overexpressing VMA1 (Fig. 4d). Collectively, these results indicate that vacuolar acidity is a target of CR-regulated nutrient sensing pathways, and suggest that CR suppresses age-induced mitochondrial dysfunction and extends lifespan at least in part by preventing the decline in vacuolar acidity in ageing cells.
In budding yeast, cytokinesis is asymmetric with respect to ageing. Although mother cells age, they produce daughter cells with full lifespan potential, suggesting cellular events that cause ageing are reset in daughter cells4. During our analysis, we discovered a dramatic distinction in vacuolar pH between mother and daughter cells. While vacuolar acidity was reduced in mother cells, high acidity was restored in newborn cells, even in daughters of old mothers (Fig. 2a, b and Supplementary Fig. 4a, b, c). This asymmetry required V-ATPase activity, as it was eliminated in vma2Δ cells, which lack V-ATPase activity and have constitutively reduced vacuolar acidity (Supplementary Fig. 4d). Taking all our results together, the acidity of the vacuole appears critical for re-setting lifespan in new yeast cells, and represents the earliest “rejuvenation” event observed in this organism.
Overall, our results support a model (Supplementary Fig. 1) in which a progressive decline in vacuolar acidity during early mother cell divisions limits mitochondrial function and lifespan, and provide a potential mechanism by which amino acid and glucose limitation may converge to regulate lifespan. Somewhat surprisingly, our data suggest that at least in yeast, age-induced mitochondrial dysfunction is not the result of a block in vacuolar protein degradation, but rather due to decreased uptake of neutral amino acids into the vacuolar lumen. How an apparent reduction in neutral amino acid storage leads to a decline in mitochondrial membrane potential remains to be determined, but we speculate that mitochondria-dependent catabolism of excess cytoplasmic amino acids (or their derivatives) places a large burden on proton-dependent mitochondrial carrier proteins29, and in the process, overwhelms mitochondrial ΔΨ. Other metabolites may also contribute to this vacuolar-mitochondrial cross-talk, and the nature of relevant metabolite(s) will be influenced by the cellular growth environment. Given that V-ATPase-coupled amino acid/ion import5 and lifespan extension by reduction of amino acids in food30 are conserved among many organisms, we postulate that our findings in yeast might be applicable to other eukaryotes.
METHODS
Strains
Yeast strains are listed in Supplementary Table 1. All strains are derivatives of S. cerevisiae S288c (BY) unless otherwise indicated. One step PCR-mediated gene replacement and epitope tagging were carried out using standard techniques, the previously described template plasmids pRS306, pRS400 KanMX, pKT127, and pBS3431,32, and the newly created template plasmid pKT127-SEP. pBS34 was obtained from the Yeast Resource Center at the University of Washington with permission from Roger Tsien33. Oligos for gene replacement and tagging are listed in Supplementary Table 2. Strains expressing empty vector, VMA1, VPH2, or AVT1 from a GPD promoter integrated into an empty region of chromosome I (199456-199457) were constructed by transformation of parental yeast strains with NotI-digested pAG306-GPD-empty chr I, pAG306-GPD-VMA1 chr I, pAG306-GPD-VPH2 chr I, or pAG306-GPD-AVT1 chr I, respectively. Strains expressing Vma2-GFP were derived from the yeast GFP collection34.
Plasmids
pAG306-GPD-empty chr I, pAG306-GPD-VMA1 chr I, pAG306-GPD-VPH2 chr I, and pAG306-GPD-AVT1 chr I were generated in two steps. First, we created pAG306-GPD-ccdB chr I, a plasmid for high expression of genes from the GPD promoter that can be integrated into chromosome I (199456-199457) after NotI digestion. We generated pAG306-GPD-ccdB chr I by ligation of a SmaI-digested fusion PCR product that contained two ~500 base pair regions of chromosome I flanking a NotI site into AatII-digested pAG306-GPD-ccdB (Addgene plasmid 14140)35. We generated the fusion PCR product using oligos ChrI PartB SmaI F and ChrI PartA SmaI R to amplify two templates generated by PCR of yeast genomic DNA using oligo pairs ChrI PartA NotI F and ChrI PartA SmaI R, and ChrI PartB SmaI F and ChrI PartB NotI R, respectively. Second, we inserted VMA1, VPH2, and AVT1 into pAG306-GPD-ccdB chr I from donor Gateway plasmids pDONR221-VMA1 (HIP ID ScCD00023712), pDONR221-VPH2 (HIP ID ScCD00012560), and pDONR221-AVT1 (HIP ID ScCD00012825) using LR clonase according to manufacturer’s instructions (Invitrogen)36. We created pAG306-GPD-empty chr I by digestion of pAG306-GPD-ccdB chr I with HindIII and SpeI to remove the ccdB gene, followed by blunt ended religation of the plasmid after filling in overhangs with T4 DNA polymerase (New England Biolabs).
pKT127-SEP, used for creating in frame C-terminal chromosomal fusions of a gene of interest with super ecliptic pHluorin (SEP), was created by removing GFP from pKT127 by restriction digestion with PacI/AscI and replacing it with PacI/AscI digested SEP generated by PCR of template plasmid pGM8718 using oligos SEP PacI F and SEP AscI R.
Media and cell culture
Cells were grown exponentially for 15 hours to a maximum density of 5 × 106 cells/ml prior to the initiation of all experiments. Because nutrients and growth phase affected vacuolar and mitochondrial function, extended log-phase growth was necessary to ensure vacuolar and mitochondrial uniformity across the cell population prior to the initiation of all experiments. Cells were cultured in YEPD (1% yeast extract, 2% peptone, 2% glucose) unless otherwise indicated. For CR experiments, cells were cultured in YEP plus the indicated carbon source. Yeast Complete (YC) medium used in the high copy suppressor screen was previously described37. Concanamycin A (Sigma) was added to cultures at a final concentration of 250 or 500 nM as indicated in figure legends.
Culturing and purification of aged MEP cells
MEP cells were aged and purified for downstream analysis as previously described with slight modifications10,38. Briefly, 2.5 × 107 cells from a 15 h YEPD log-phase culture were washed twice in phosphate buffered saline, pH 7.4 (PBS) and resuspended in PBS containing 3 mg/ml Sulfa-NHS-LC-Biotin (Pierce) at a final concentration of 2.5 × 107 cells/ml. Cells were incubated for 30 min at room temperature (RT) and then washed twice in PBS and once in YEPD. Biotinylated cells were resuspended in 10 ml YEPD at 2.5 × 106 cells/ml and recovered with shaking for 2 h at 30°C. These cells were used to seed cultures at a density of 2 × 104 biotinylated cells/ml in YEPD. β-estradiol (Sigma) was added to cultures at a final concentration of 1 μM to initiate the MEP and cells were cultured at 30°C for an appropriate time to obtain cells of a desired age (~1 h for median age 2, 12 h for age 8, 24 h for age 16, 48 h for age 28). Cell densities never exceeded 4 × 106 cells/ml and cells were washed and resuspended in fresh media every 24 h (12 h for CR experiments) to prevent nutrient limitation. At each ageing timepoint, 1 × 108 total cells were harvested for purification, staining, and microscopy analysis.
For live cell purification, cells were washed twice in PBS, resuspended at a density of 2 × 108 cells/ml in 500 μl of PBS, and incubated for 30 min at RT with 25 μl streptavidin-coated magnetic beads (MicroMACS, Miltenyi Biotec). After incubation, cells were washed twice in PBS, resuspended in 8 ml of PBS, and loaded onto a LS MACS column (Miltenyi Biotec) equilibrated with 5 ml of PBS. After gravity flow-through of unlabelled cells and debris, columns were washed twice with 8 ml of PBS. Columns were then removed from the magnetic field and aged cells were eluted by gravity flow with 8 ml of PBS. For cell staining experiments, purified cells were pelleted, resuspended in YEP containing 2 or 0.5% glucose at 2 × 106 cells/ml, and recovered for 1 h at 30°C prior to staining.
For purification of cells after fixation, cells were washed once in PBS and resuspended at 2 × 108 cells/ml and fixed in 500 μl of 4% paraformaldehyde in H2O for 15 minutes. Cells were then washed twice in PBS prior to incubation with streptavidin beads at 4°C instead of RT. Additionally, 2 mM EDTA was added to PBS in all fixed cell purification steps to prevent cell clumping.
Labeling and purification of aged non-MEP cells (Supplementary Fig. 2c) was conducted exactly as described for MEP cells except that labeled mother cells were purified after 12 hours of log-phase growth, reseeded, and purified a second time after another 12 hours of log-phase growth to obtain 18 division old mother cells. Cells never exceeded a density of 7 × 106 cells/ml.
Fluorescent staining and microscopy
Quinacrine (Sigma) staining was performed as previously described39. Briefly, 2 × 106 log-phase or purified aged cells were washed once in YEPD + 100 mM HEPES, pH 7.6 and resuspended in 100 μl of the same buffered media containing 200 μM quinacrine. Cells were incubated for 10 min at 30°C and then 5 min on ice. Cells were pelleted and washed twice with ice cold 100 mM HEPES, pH 7.6 + 2% glucose. Cells were resuspended in 100 mM HEPES, pH 7.6 + 2% glucose for imaging. Prior to imaging, cells were kept on ice and all images were obtained within 30 min of staining.
For staining with 5-(and-6)-carboxy-2′,7′-dichlorofluorescein diacetate11 (CDCFDA) (Invitrogen) or 2′,7′-bis (carboxyethyl)-5(6)-carboxyfluorescein40 (BCECF-AM) (Invitrogen), 2 × 106 log-phase or purified aged cells were washed once in YEPD + 100 mM HEPES, pH 7.6 and resuspended in 100 μl of the same buffered media containing 10 μM CDCFDA or 50 μM BCECF. Cells were incubated for 30 min at 30°C and then washed twice with RT 100 mM HEPES, pH 7.6 + 2% glucose. Cells were resuspended in 100 mM HEPES, pH 7.6 + 2% glucose for imaging.
3,3′-dihexyloxacarbocyanine iodide (DiOC6) (Invitrogen) staining was carried out according to manufacturer’s instructions. Briefly, 2 × 106 log-phase or purified aged cells were washed once in 10 mM HEPES, pH 7.6 + 5% glucose and resuspended in 1 ml of the same buffer containing 175 nM DiOC6. Cells were incubated for 15 min at RT and then washed twice with 10 mM HEPES, pH 7.6 + 5% glucose. Cells were resuspended in 10 mM HEPES, pH 7.6 + 5% glucose for imaging. Pho8-SEP imaging was also carried out in DiOC6 imaging buffer after incubation of cells for 20 minutes in the buffer. In all live cell experiments, calcofluor (Sigma) staining of bud scars for age determination was carried out by including 5 μg/ml calcofluor in the first post-staining wash step prior to imaging.
For fluorescence microscopy analysis, cells were visualized under 60X oil magnification using a Nikon Eclipse E800 with the appropriate filter set: UV-2E/C DAPI for calcofluor; FITC-HYQ for GFP, SEP, quinacrine, CDCFDA, BCECF, and DiOC6; and G-2E/C TRITC for mCherry. Images were acquired with a CoolSNAP HQ2 CCD camera (Photometrics) and quantified and processed using Metamorph version 7.1.1.0 imaging software. Cells that exhibited at least four-fold reduction in mean fluorescence intensity, compared to young, wild type cells, were scored as “reduced” in all figures.
Lifespan measurement by micromanipulation
RLS was measured by micromanipulation as previously described, except that cells were grown exponentially in YEPD for 15 h instead of 3 h prior to lifespan analysis10.
Membrane Protein Preparation
5×107 cells were resuspended in 50 μl B88 buffer (20 mM Hepes, pH 7.2, 150 mM KOAc, 5 mM MgOAc, and 250 mM sorbitol) plus protease inhibitors (leupeptin, pepstatin, PMSF, and aprotinin) and lysed with glass beads for 10 min in a Multi-Tube Vortexer (VWR) at 4°C. 100 μl B88 plus protease inhibitors was added to the sample and the lysate was centrifuged at 500×g for 5 min at 4°C. The supernatant of this spin was centrifuged at 20,000×g for 10 min at 4°C. The membrane pellet was resuspended in 100μl of SDS-lysis buffer (10 mM Tris-HCl, pH 6.8, 100mM NaCl, 1% SDS, 1 mM EDTA, and 1 mM EGTA) plus protease inhibitors. The supernatant of the 20,000×g spin was used as the cytosolic extract.
Imunoblotting
Membrane and cytosolic extracts were mixed with 0.25 volumes of 5× SDS loading buffer (150 mM Tris-HCl, pH 6.8, 15% SDS, 25% glycerol, 0.02% bromophenol blue, 12.5% β-mercaptoethanol) and heated to 37°C for 15 minutes. Aliquots of each sample were resolved on 7% polyacrylamide gels and transferred to nitrocellulose membranes (Invitrogen). Membranes were blocked with 5% non-fat dry milk in PBS-T (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, 0.05% Tween 20, pH 7.4) and then incubated with primary and HRP-conjugated secondary antibodies (Jackson Immunoresearch). Blots were washed in PBS-T after each antibody incubation. Bound peroxidase-conjugated antibodies were detected using the SuperSignal West Pico Chemiluminescent substrate (Pierce). Primary antibodies were anti-Vma2 13D11 (Invitrogen), anti-Pgk1 22C5 (Invitrogen), anti-Pho8 1D3 (Invitrogen), and anti-mCherry Living Colors monoclonal (Clontech).
Screen for high copy suppressors of age-induced mitochondrial dysfunction
MEP cells expressing Tom70-GFP were individually transformed in 96-well format with 250 high copy 2 μm plasmids from a tiled genomic DNA library41. Each plasmid contained a unique sequence-verified genomic DNA fragment that expressed at least one of 250 genes previously determined to affect mitochondrial morphology when deleted12,13. To identify genes that suppressed age-induced changes in mitochondrial morphology, all 250 plasmid-expressing strains were individually cultured in 5 ml of YEPD at 30°C for 6 h to a density of 2 × 104 cells/ml from overnight-saturated cultures of cells grown in YC-leucine to maintain plasmids. After 6 h in YEPD, β-estradiol was added to cultures at a final concentration of 1 μM to initiate the MEP and cells were cultured at 30°C for an additional 24 h to obtain mother cells with a median age of 16 divisions (n = 15). Cells were collected by centrifugation, fixed with 4% paraformaldehyde, stained with calcofluor, and imaged by fluorescence microscopy for mitochondrial morphology as described above. Plasmid containing strains in which 40% of aged (median age of 16, n = 15) cells exhibited young-cell like tubular mitochondrial morphology were scored as suppressors of age-induced mitochondrial dysfunction. To confirm potential suppressors, each candidate gene was transferred using LR clonase from pDONR221 Gateway entry plasmids from a previously described collection (HIP) into pAG306-GPD-ccdB chr I36. Plasmids were integrated into chromosome I of a Tom70-GFP expressing strain after NotI digestion and strains were aged, purified, and examined by fluorescence microscopy as described above. Positive integrated suppressors (such as VMA1 and VPH2) were confirmed by their ability to maintain young-cell like mitochondrial morphology in at least 70% (n = 30) of aged cells (median age of 17).
Supplementary Material
Acknowledgments
We thank L. Pallanck and members of the Gottschling lab for reviewing the manuscript; K. Henderson for helpful discussions; G. Miesenbock, D. Lindstrom, and J. Hsu for reagents; and L. Dimitrov for technical assistance. This work was supported by NIH grants AG037512 and AG023779 to D.E.G. and by fellowships from the Helen Hay Whitney Foundation and Genetic Approaches to Aging Training Grant (T32 AG000057) to A.L.H.
Footnotes
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
AUTHOR CONTRIBUTIONS
A.L.H. designed and carried out the experiments. D.E.G. provided experimental guidance and supervision. All authors discussed the results and implications of the experiments. The paper was written by A.L.H. and edited by D.E.G.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo AY, et al. New insights into the role of mitochondria in aging: mitochondrial dynamics and more. J Cell Sci. 2010;123:2533. doi: 10.1242/jcs.070490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop NA, Guarente L. Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet. 2007;8:835. doi: 10.1038/nrg2188. [DOI] [PubMed] [Google Scholar]
- 4.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: the means to the end. Annu Rev Cell Dev Biol. 2008;24:29. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li SC, Kane PM. The yeast lysosome-like vacuole: endpoint and crossroads. Biochim Biophys Acta. 2009;1793:650. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheckhuber CQ, et al. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9:99. doi: 10.1038/ncb1524. [DOI] [PubMed] [Google Scholar]
- 7.Lam YT, Aung-Htut MT, Lim YL, Yang H, Dawes IW. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radic Biol Med. 2011;50:963. doi: 10.1016/j.freeradbiomed.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Mcfaline-Figueroa JR, et al. Mitochondrial quality control during inheritance is associated with lifespan and mother-daughter age asymmetry in budding yeast. Aging Cell. 2011;10:885. doi: 10.1111/j.1474-9726.2011.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veatch JR, Mcmurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindstrom DL, Gottschling DE. The Mother Enrichment Program: A Genetic System for Facile Replicative Life Span Analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pringle JR, et al. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- 12.Dimmer KS, et al. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:847. doi: 10.1091/mbc.01-12-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altmann K, Westermann B. Role of essential genes in mitochondrial morphogenesis in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:5410. doi: 10.1091/mbc.E05-07-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirata R, et al. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:6726. [PubMed] [Google Scholar]
- 15.Jackson DD, Stevens TH. VMA12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J Biol Chem. 1997;272:25928. doi: 10.1074/jbc.272.41.25928. [DOI] [PubMed] [Google Scholar]
- 16.Drose S, et al. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993;32:3902. doi: 10.1021/bi00066a008. [DOI] [PubMed] [Google Scholar]
- 17.Weisman LS, Bacallao R, Wickner W. Multiple methods of visualizing the yeast vacuole permit evaluation of its morphology and inheritance during the cell cycle. The Journal of Cell Biology. 1987;105:1539. doi: 10.1083/jcb.105.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys J. 2000;79:2199. doi: 10.1016/S0006-3495(00)76468-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 20.Kaeberlein M, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 21.Parr CL, Keates RA, Bryksa BC, Ogawa M, Yada RY. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast. 2007;24:467. doi: 10.1002/yea.1485. [DOI] [PubMed] [Google Scholar]
- 22.Shimazu M, Sekito T, Akiyama K, Ohsumi Y, Kakinuma Y. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J Biol Chem. 2005;280:4851. doi: 10.1074/jbc.M412617200. [DOI] [PubMed] [Google Scholar]
- 23.Miseta A, Kellermayer R, Aiello DP, Fu L, Bedwell DM. The vacuolar Ca2+/H+ exchanger Vcx1p/Hum1p tightly controls cytosolic Ca2+ levels in S. cerevisiae. FEBS Lett. 1999;451:132. doi: 10.1016/s0014-5793(99)00519-0. [DOI] [PubMed] [Google Scholar]
- 24.Nass R, Cunningham KW, Rao R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J Biol Chem. 1997;272:26145. doi: 10.1074/jbc.272.42.26145. [DOI] [PubMed] [Google Scholar]
- 25.Russnak R, Konczal D, Mcintire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J Biol Chem. 2001;276:23849. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- 26.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 27.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka K, et al. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- 29.Palmieri F, et al. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta. 2006;1757:1249. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 30.Piper MD, Partridge L, Raubenheimer D, Simpson SJ. Dietary restriction and aging: a unifying perspective. Cell Metab. 2011;14:154. doi: 10.1016/j.cmet.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 33.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 34.Huh W-K, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 35.Alberti S, Gitler AD, Lindquist S. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 2007;24:913. doi: 10.1002/yea.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, et al. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Research. 2007;17:536. doi: 10.1101/gr.6037607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Leeuwen F, Gottschling DE. Assays for gene silencing in yeast. Meth Enzymol. 2002;350:165. doi: 10.1016/s0076-6879(02)50962-9. [DOI] [PubMed] [Google Scholar]
- 38.Lindstrom DL, Leverich CK, Henderson KA, Gottschling DE. Replicative age induces mitotic recombination in the ribosomal RNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2011;7:e1002015. doi: 10.1371/journal.pgen.1002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morano KA, Klionsky DJ. Differential effects of compartment deacidification on the targeting of membrane and soluble proteins to the vacuole in yeast. J Cell Sci. 1994;107:2813. doi: 10.1242/jcs.107.10.2813. [DOI] [PubMed] [Google Scholar]
- 40.Plant PJ, Manolson MF, Grinstein S, Demaurex N. Alternative mechanisms of vacuolar acidification in H(+)-ATPase-deficient yeast. J Biol Chem. 1999;274:37270. doi: 10.1074/jbc.274.52.37270. [DOI] [PubMed] [Google Scholar]
- 41.Jones GM, et al. A systematic library for comprehensive overexpression screens in Saccharomyces cerevisiae. Nat Methods. 2008;5:239. doi: 10.1038/nmeth.1181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.