Abstract
The piggyBac transposon system is a promising nonviral method to genetically modifiy T cells for immunotherapeutic applications. To evaluate the regulation and stability of transgene expression in human T cells modified with piggyBac transposons, peripheral blood mononuclear cells (PBMCs) were nucleofected with transposase and an eGFP-expressing transposon. Single cell clones that were subsequently stimulated and expanded exhibited homogeneous eGFP expression for >26 weeks in culture. CD3 stimulation of the T cell receptor together with CD28-mediated costimulation resulted in an approximate ten-fold transient increase in eGFP expression, but immunomodulatory cytokines, including interferon-γ, interleukin-12, interleukin-4, and transforming growth factor-β, did not alter transgene expression in actively dividing, activated, or resting T cells. Epigenetic modification with 5-azacytidine or Trichostatin-A increased transgene expression indicating that piggyBac-mediated transgene expression could be modulated by methylation or histone acetylation, respectively. We performed transposon copy number analysis of populations of stably transfected T cells, comparing transposon plasmids of 5.6kb and 3.5kb. The smaller vector achieved an average of 22 transposon copies per cell whereas the larger vector achieved 1.6 copies per cell, implying that transposon copy number can be engineered to be low or high depending on the vector employed. Our results provide important insight into the ability of piggyBac to achieve stable genetic modification of T cells for immunotherapy applications as well as how transgene expression might be regulated by TCR activation, cytokines, and epigenetic mechanisms.
Keywords: T lymphocyte, transposon, piggyBac, immunotherapy
INTRODUCTION
Transposon systems are an emerging non-viral methodology for stable genetic modification of human cells for therapeutic applications. Human T lymphocytes are a promising cell population for adoptive immunotherapies, but their ultimate success may depend on genetic modification to ensure their ability to recognize tumor cells and to function in the immunosuppressive tumor environment. Sleeping Beauty was the first transposon system used for modification of human T lymphocytes and produced stable expression first of reporter genes1 and subsequently with chimeric antigen receptors (CARs), demonstrating targeted killing of cancer cells in vitro and in animal models.2,3 Recently, the Sleeping Beauty system has been approved for a human clinical trial involving immunotherapy for CD19 positive malignancies.4
PiggyBac is a highly active transposon derived from the cabbage looper moth,5,6 that can also provide sustained transgene expression in up to ~40% of human T lymphocytes without selection, enabling several logs of expansion of primary transgenic T-cells in culture, and after magnetic bead-selection for a transgenic surface marker (truncated CD19) up to 85% of transduced T-cells could be obtained and maintained for over 9 weeks.7 Thus far, piggyBac has been used to gene-modify human T cells with reporter genes,7 a non-immunogenic suicide gene,7 a chimeric antigen receptor for CD198 or HER2,9 and a large rapamycin-resistant mTor molecule.10 Gene modified cells have shown directed tumor cell killing both in vitro and in vivo.8,9 Outside of the realm of adoptive immunotherapy, piggyBac has been used to move large transgenes,11 simultaneously co-deliver multiple (i.e. more than 5) transgenes,12 and to achieve inducible transgene expression both in cultured cells and in vivo.13,14 More recently, piggyBac has been manipulated to achieve site-directed integration in human cells.15,16
These findings suggest that the piggyBac transposon system represents an effective and adaptable tool for genetic modification of T cells for cancer immunotherapy, particularly because of its capacity for large or multiple transgenes. However, little information is known about the long-term stability of expression and regulation of transgenes in piggyBac-transgenic T-cells. Post-integrative genome silencing of transgene expression has been observed when using Sleeping Beauty in human cells, although this silencing could be partially reversed using inhibitors of DNA methylation and histone deacetylation.17
The advance of the piggyBac transposon system into clinical use requires further characterization of piggyBac-modified T cells. We therefore studied long-term regulation of transgene expression in piggyBac-modified human T-cell clones. We also evaluated whether transgene expression is influenced by immunomodulatory cytokines that T cells may encounter in vivo. As the expression of transgenes could be affected by epigenetic modification, we evaluated the expression of piggyBac transgenes in T cells in the presence of 5-aza-2′-deoxycytidine and trichostatin A, agents that are increasingly used as components of cancer therapy. We also determined the average transposon copy number per cell using transposon vectors of varying size.
MATERIALS AND METHODS
Plasmid construction
The pCMV-piggyBac (transposase) and pIRII-eGFP (transposon) plasmids have been described previously.7,18 pIRII-eGFP encodes an internal ribosome entry site (IRES) followed by the enhanced green fluorescence protein (eGFP). Both vectors are transcriptionally regulated by the cytomegalovirus immediate early gene enhancer/promoter sequence (CMV). pT-CMV-eGFP was constructed by PCR cloning a CMV-eGFP fragment into a transposon vector engineered to contain the piggyBac IR elements with the backbone from pCpGfree-MCS (Invivogen, San Diego, CA). Plasmid constructs were confirmed by restriction digestion and DNA sequencing.
Blood donors and cell lines
Peripheral blood mononuclear cells (PBMC) from healthy volunteers were obtained with informed consent from the Baylor College of Medicine Institutional Review Board. To generate activated T-cells (ATC), PBMC were cultured in complete T-cell medium (TCM) [Advanced RPMI (Gibco-BRL, Gaithersburg, MD) supplemented with 2 mM L-glutamine (GlutaMAX-I, Invitrogen, Carlsbad, CA) and 5% heat-inactivated fetal bovine serum (FBS)] in the presence of recombinant human interleukin (IL)-15 at 5 ng/mL. Artificial K562 cells (aK562, a gift from Carl June, University of Pennsylvania), engineered to express CD80, CD86, CD83 and 4-1BBL,19 were maintained in RPMI 1640 (Gibco-BRL, Gaithersburg, MD) supplemented with 10% FBS and 2mM L-glutamine after irradiation at 80 Gy and were used as feeder cells to expand gene-modified T-cells.
Gene transfer into PBMC
We previously reported an efficient gene-transfer method into human primary T-cells using piggyBac-transposons.7 In this study, the same method was used with minor modifications. Briefly, bulk PBMC (>5 × 106) were maintained in TCM with the recombinant human IL-15 (5 ng/mL) (Proleukin; Chiron, Emeryville, CA) 20 – 24 hr before nucleofection. Five million IL-15-treated PBMC were mixed with 5 μg of pIRII-eGFP and 5 μg of pCMV-piggyBac and then nucleofected using the Nucleofector Device (program U-014) in combination with the Human T-cell Nucleofector Kit according to the manufacturer (Lonza, Basel, Switzerland). The nucleofected PBMC were maintained in TCM with IL-15 (5 ng/mL) for 48 hr.
Generation of T-cell clones from piggyBac-modified T-cells
Two days after nucleofection, bulk eGFP-nucleofected cells were stained with phycoerythrin (PE)-conjugated anti-CD28 monoclonal antibody (Becton Dickinson), and ~500 cells which showed positivity for both GFP and CD28 were singly sorted into each well of five 96 well U-bottomed plates using BD Cell-Sorting system (Becton Dickinson, Franklin Lakes, NJ). Immediately, each cell was co-cultured with 1,000 irradiated (100 Gy) aK562 cells (see above), in the presence of OKT3 (50 ng/mL) (Ortho Biotech, Bridgewater, NJ) and IL-15 (5 ng/mL) in 0.2 mL TCM. IL-15 (5 ng/mL) was added to the cultures once a week. After 2 weeks, growing clones were transferred into 1 mL of TCM with IL-15 (5 ng/mL) per well of a 48-well plate. Three weeks after sorting, well-growing clones (>70% confluent in the 48-well plate) were transferred to a 12 well plate and co-cultured with 0.5 × 106 aK562 cells, OKT3 and IL-15 in 4 mL of TCM with for 3 additional weeks. At this time and every 3 weeks thereafter, 1 × 106 cells of each clone were restimulated with OKT3 in the presence of aK562 cells and IL-15 in a 12-well plate and supplemented with IL-15 twice weekly. Transgene expression was analyzed by flow cytometry just before restimulation with OKT3 and aK562 cells (i.e. every 3 weeks).
Activation of T-cell clones
Of the generated clones, 5 representative clones were used for further experiments between 9 and 15 weeks of culture. At the end of a three week stimulation period, 0.5 × 106 cells of each clone were activated on a non-tissue culture-treated 24-well plate coated with OKT3 (1 μg/mL) and anti-human CD28 monoclonal antibody (1 μg/mL) (Becton Dickinson) in TCM with IL-15 (5 ng/mL) for 4 days, and then transferred into tissue-culture treated 24-well plates. IL-15 was added to the culture twice a week. The activated clones were analyzed for GFP expression at the indicated time points.
Analysis of transgene expression after activation via T-cell receptor
To investigate influence of T-cell receptor (TCR) signaling on transgene expression, we analyzed GFP expression in 5 clones activated on anti-CD3/CD28 antibody coated plates at the indicated time points. On day 12 after activation, 0.5 × 106 cells of each activated clone were re-activated on anti-CD3/CD28 antibody coated plates for 4 days and thereafter analyzed for GFP expression at the same time points.
Analysis of influence of cytokine on transgene expression
To investigate how immunomodulatory cytokines affect transgene regulation, we activated 5 clones on anti-CD3/CD28 antibody coated plates and added cytokines (interferon-gamma (IFN-γ) (10 ng/mL), IL-12 (10 ngmL), IL-4 (1,000 IU/mL) or transforming growth factor-beta (TGF-β) (5 ng/mL) on day + 1 “actively dividing”, day + 5 “activated”, and day + 21 “resting”. Four days after treatment the clones were analyzed for GFP expression using flow cytometry.
Epigenetic analysis
To investigate the potential for epigenetic regulation of the transgene, we used the methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-AzaC) (Sigma-Aldrich, St. Louis, MO) and the histone deacetylase inhibitor, trichostatin A (TSA) (Sigma-Aldrich) for T cell treatment. To analyze the affects of demethylation, five T-cell clones were activated on anti-CD3/CD28 antibody coated plates and simultaneously treated with 5-AzaC-containing at the indicated concentration. Two days later, they received a half medium change with fresh drug. Four days after the treatment, GFP expression was analyzed. To determine the affects of histone deacetylation, we activated 5 clones on anti-CD3/CD28 antibody coated plates for 4 days, and then transferred the activated clones into IL-15-TCM in a tissue-culture treated plate. Two days later, we added the indicated concentration and further cultured for 24 hours before GFP expression analysis.
Flow cytometry
We analyzed GFP-nucleofected T cell clones for GFP expression at the indicated time points using a FACSCalibur with Cell Quest software (Becton Dickinson). Five weeks after nucleofection, we also determined CD3, CD4 and/or CD8 expression using phycoerythrin (PE)-conjugated CD4, allophycocyanin (APC)-conjugated CD8, peridinin chlorophyll protein (PerCP)-conjugated CD3 MAbs (Becton Dickinson).
Determination of transposon copy number in transgenic T-cells
PBMCs were nucleofected with 5 μg each PB-transposon plasmid (either pIRII-eGFP (5.6kb) or pT-CMV-eGFP (3.5kb)) and PB-transposase plasmid as described above. After transfection, PBMCs were stimulated on nontissue culture-treated 24-well plates coated with anti-CD3/CD28 antibodies in the presence of recombinant human IL-15 (5 ng/mL). After 3 days of stimulation, activated T cells were transferred to tissue culture-treated 24-well plates in T-cell media supplemented with IL-15 (5 ng/mL). On day 7, GFP+ cells were sorted on a FACS AriaII cell sorter (BD Biosciences, CA) and cultured in CTL media supplemented with IL-15 (5 ng/mL) and 1X Pen-Strep for 14 days after which cells were harvested for genomic DNA extraction. qPCR was performed using 1XiQ SYBR green master mix from Biorad (Hercules, CA). To measure the average number of piggyBac transposons integrated, we utilized a set of primers directed at the eGFP reporter gene: q-eGFP-F (AGAACGGCATCAAGGTGAAC) and q-eGFP-R (TGCTCAGGTAGTGGTTGTCG). Standard curves were generated with serial dilutions of the respective transposon plasmids resulting in a known copy number. In order to compare between samples, all samples were normalized to the number of genomic RNaseP copies to achieve copies query/copies RNaseP. RNaseP was chosen for normalization as it is known to be present at 2 copies/genome in human cells. A standard curve for RNaseP was generated via serial dilution of a plasmid containing the genomic RNaseP sequence using primers RPF (AGATTTGGACCTGCGAGCG) and RPR (GAGCGGCTGTCTCCACAAGT), permitting analysis of the genomic copy number of RNaseP for each cellular DNA sample assuming 6.6 pg of DNA per cellular genome.
RESULTS
Generation of piggyBac-modified T cell clones
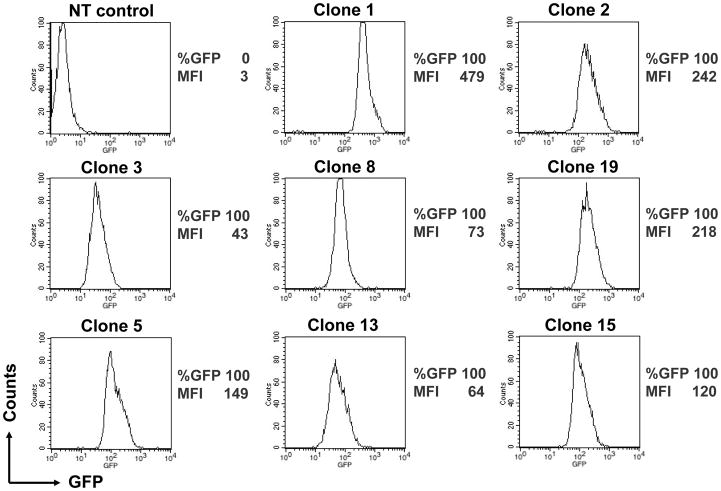
To investigate the regulation of piggyBac-inserted transgenes and their expression in T cells, we evaluated T-cell clones derived from PB-GFP-transfected, CD3/28-activated PBMCs from healthy donors. T-cell clones sorted for GFP expression were stimulated with OKT3 and aK562 cells every 3 weeks and analyzed for GFP expression. Seventeen clones expressed GFP (>99% positive) with three clones having low expression (mean fluorescence intensity [MFI] between 10 and 100), thirteen clones having medium expression (MFI between 100 and 1000), and two clones having high expression (MFI between 1,000 and 10,000). The average MFI of all clones was 473.5 with a range of 35.5 to 2211.7. Two clones did not express GFP (MFI <10). We evaluated all 17 GFP-positive clones for CD3, CD4 and CD8 expression. Twelve clones expressed CD3 and CD4, and four expressed CD3 and CD8. Over time, we obtained 8 long-lived CD4+ T cell clones while the CD8+ T cell clones did not survive past 6 weeks. All 8 of the long-lived clones expressed GFP homogeneously in >99% of cells (Figure 1).
FIGURE 1. Generation of piggyBac-modified T-cell clones.
5 × 106 IL-15-treated peripheral blood mononuclear cells (PBMCs) from a healthy donor were nucleofected with pIRII-eGFP (5 μg) and pCMV-piggyBac (5 μg). Two days later, the nucleofected cells were stained with anti-CD28 antibody, and then ~500 cells of dual GFP- and CD28-positive cells were single-sorted into five 96-well plates. Each cell was stimulated with OKT3 and co-cultured with aK562 cells in the presence of IL-15 for 3 weeks. Well-growing clones were repeatedly stimulated with OKT3 in the presence of aK562 cells in the presence of IL-15 every 3 weeks after nucleofection. GFP expression was determined by flow cytometry at 12 weeks after eGFP-gene transfer. The percentage of GFP expression and mean fluorescence intensity (MFI) are indicated in each clone.
Stable and long-term expression of piggyBac-transgenes
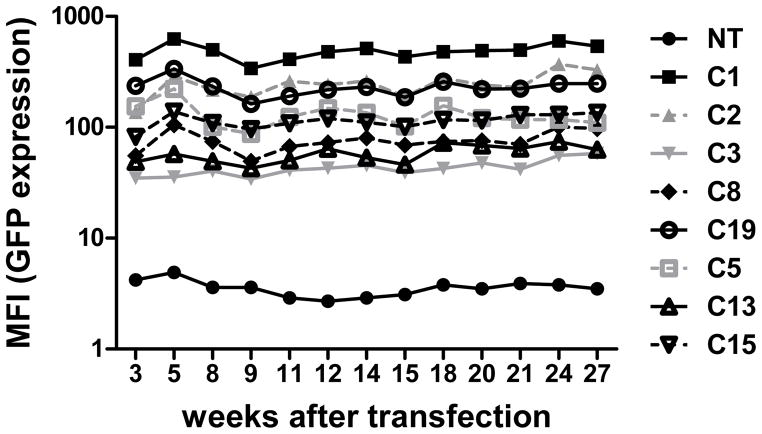
Long-term, stable expression of therapeutic genes is required for a majority of gene-modified T cell based immunotherapies. We previously found that piggyBac-modified human T cells could stably express dual transgenes for up to 9 weeks.7 To further investigate the stability and long-term transgene expression in piggyBac-modified human T cells, we cultured GFP-expressing T cell clones for up to 6 months. We found that all 8 clones continued to express GFP without significant decrease in intensity for at least 26 weeks in vitro (Figure 2). These results support and extend our previous finding that PB could mediate long-term sustained transgene expression in primary human T cells.
FIGURE 2. Stable and long-term expression of piggyBac-transgenes.
T-cell clones were stimulated with OKT3 and co-cultured with aK562 cells at a ratio of 2:1 in IL-15-containing media 3 weeks after transfection, and every 3 weeks thereafter until 24 weeks after transfection. GFP expression was determined by flow cytometry just before each stimulation (i.e. every 3 weeks). The percentage of GFP expression and mean fluorescence intensity (MFI) for each is indicated.
T cell receptor stimulation increased expression of piggyBac-transgenes
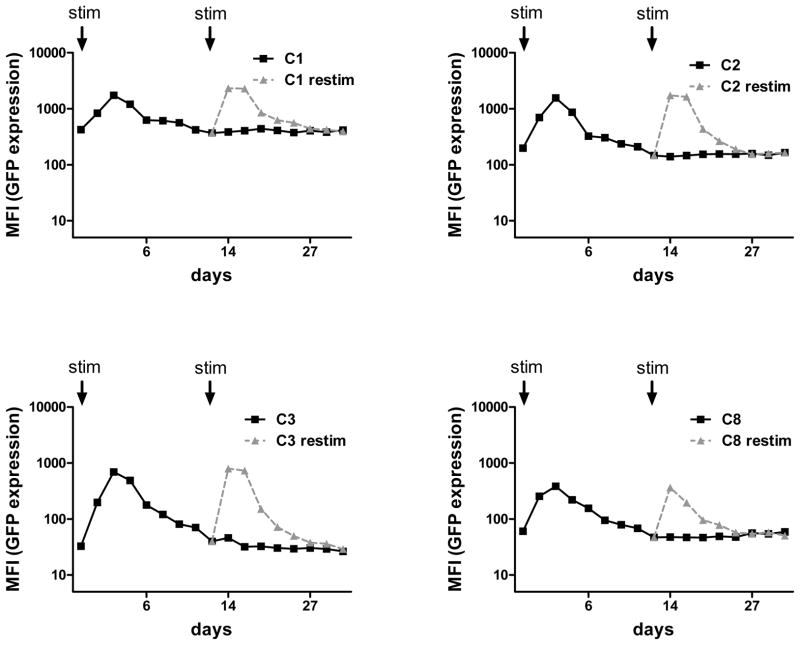
Stimulatory signals via the T cell receptor (TCR) and costimulatory receptors are critical for survival, expansion, and tumor-directed killing in vivo after adoptive transfer of T cells. To determine if TCR stimulation affected piggyBac-transgene expression, we measured the GFP expression, based on mean fluorescence index (MFI), in 5 T cell clones after stimulation with CD3 and CD28 monoclonal antibodies. On day 2 after stimulation, all 5 clones showed an increase in GFP expression (mean: 8.5 fold; range: 2.8 to 21.2 fold) compared to baseline GFP levels. However, the level of GFP returned to baseline by day 12, from which time, expression remained stable (Figure 3). To determine whether the transient increase in GFP expression occured after each TCR stimulation, we repeated the OKT3/CD28 stimulation of clones on day 12 when GFP levels had returned to baseline. Interestingly, all 5 clones showed an increase in GFP similar to that observed after the first stimulation. GFP expression on day 14 was 9.8 fold (range 3.2 to 19.8 fold) higher than that before re-stimulation on day 12 (Figure 3). These results indicate that piggyBac-modified T cells reversibly and reproducibly increase transgene expression in response to TCR stimulation and costimulation.
FIGURE 3. T cell receptor stimulation increased expression of piggyBac-transgenes.
Five representative clones were stimulated in OKT3/CD28 mAb-coated wells for 4 days and then cultured for 35 days. Activated clones were analyzed for GFP expression at the indicated time points (continuous line). On day 12 after activation, a small portion of each activated clone was reactivated with an OKT3/CD28 mAbs-coated plate for 4 days and thereafter analyzed for GFP expression as the same time points (dashed line).
Immunomodulatory cytokines do not affect transgene expression in piggyBac-modified human T cells
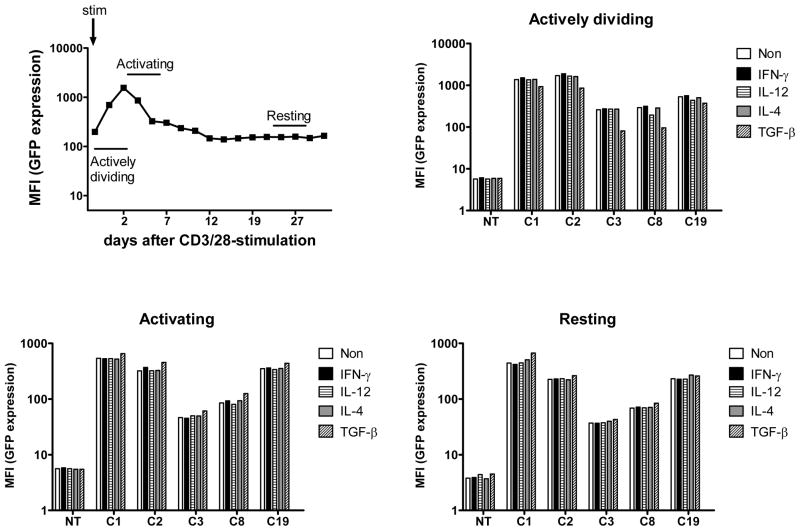
Adoptively transferred T cells for cancer immunotherapy are exposed to various immunomodulatory cytokines in vivo. Interferon (IFN)-γ is secreted by Th1 cells, CTLs, and natural killer cells thereby activating antigen presenting cells (APC) and phagocytes.20,21 IL-12 is secreted by APCs and stimulates Th1 cells.21,22 IL-4 is secreted by APCs and Th2 cells and stimulates Th2 cells.21 Transforming growth factor (TGF)-β is often produced in the tumor micro-environment and inhibits T-cell growth, activation, and cytotoxic function.23 Importantly, IFN-γ was reported to down- regulate expression of retroviral transgenes driven from the CMV promoter.24 If these cytokines negatively modulate piggyBac transgene expression, the clinical efficacy of immunotherapy might be dampened. Therefore, to elucidate whether these immunomodulatory cytokines regulate transgene expression in the T cell clones, we examined GFP expression in the presence of IFN-γ, IL-12, IL-4 or TGF-β. Because we have shown transgene expression is modulated by TCR stimulation (Figure 3), we studied 5 clones at 3 different times after OKT3/CD28-stimulation, at which we considered them to be actively dividing, activated, or resting. We found that IFN-γ, IL-4 and IL-12 did not affect transgene expression regardless of the activation state of the T cells, while TGF-β minimally inhibited the increase in transgene expression post-stimulation but only when added during the “actively dividing” phase (Figure 4). The inhibitory effect of TGF-β on transgene expression might be due to the decreased activation of clones treated with TGF-β, which proliferated poorly (data not shown). These results suggest that piggyBac-transgene expression is minimally influenced by immune stimulatory or inhibitory cytokines.
FIGURE 4. Immunomodulatory cytokines do not affect transgene expression in piggyBac-transposed human T cells.
Five representative clones (C1, C2, C3, C8, and C19) were stimulated on anti-CD3/CD28 antibody coated plates for 4 days and then cultured for 35 days. Stimulated clones were classified into 3 different groups based on the activation state such as “actively dividing”, “activated”, and “resting”. Those in “actively dividing” received cytokines on day 1, “activated” received cytokines on day 5, and “resting” received cytokines on day 21. Clones were treated with interferon-gamma (IFN-γ) (10 ng/mL), IL-12 (10 ngmL), IL-4 (1,000 IU/mL) or transforming growth factor-beta (TGF-β) (5 ng/mL) for 4 days. Four days after the treatment, the clones were analyzed for GFP expression by flow cytometry.
PiggyBac-transgenes are partially regulated by epigenetic mechanisms
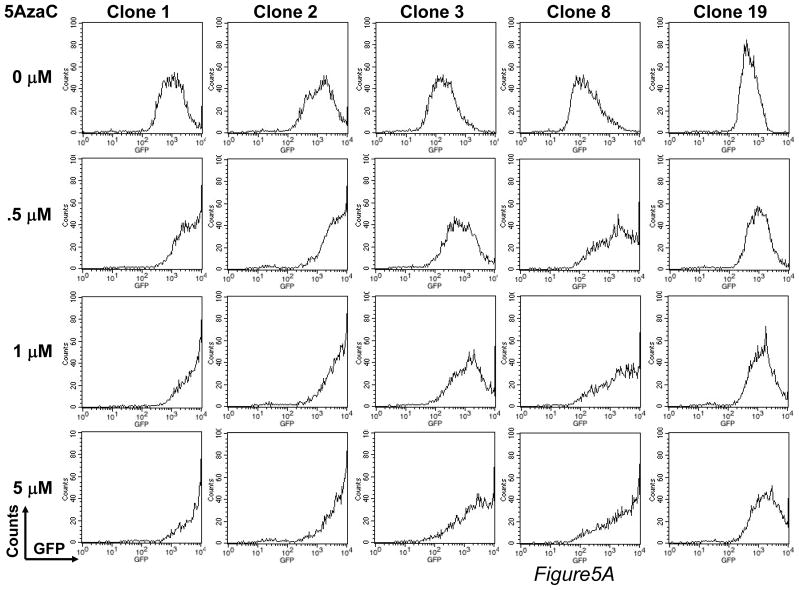
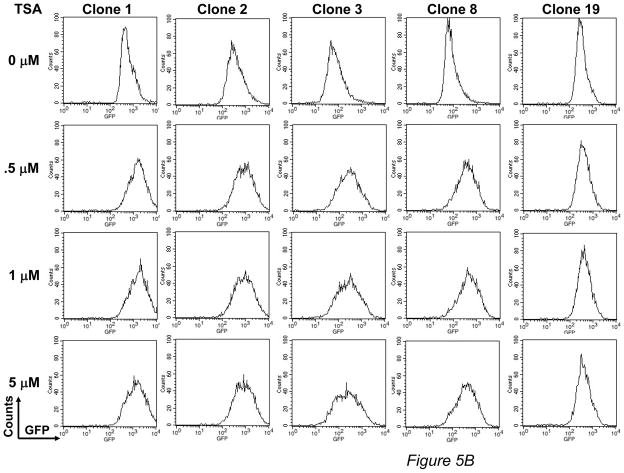
The epigenetic control of transgene expression seems to be associated with post-integrative silencing of transgenes,25,26 which could limit the use of piggyBac to modify primary human T cells for long-term immunotherapy. Currently, there is no information on epigenetic regulation of piggyBac integrated transgenes. Therefore, we analyzed GFP expression in clones treated with a methylation inhibitor, deoxymethyltransferase (5-AzaC), or a histone deacetylation inhibitor, trichostatin A (TSA). We found a dose-dependent increase in GFP expression in response to 5- AzaC (fold increase relative to untreated: 4.3 ± 2.7 at 0.5 μM; 6.1 ± 3.6 at 1 μM; 8.4 ± 5.4 at 5 μM) (Figure 5A), and to a lesser extent with TSA (fold increase relative to untreated: 3.1 ± 1.4 at 0.5 μM, 3.5 ± 1.9 at 1 μM, 3.2 ± 1.6 at 5 μM) (Figure 5B). Therefore, although piggyBac achieves stable long-term gene expression, the transgenes in piggyBac-modified primary human T cells may be regulated, in part, by epigenetic mechanisms.
FIGURE 5. PiggyBac-transgenes are partially regulated by epigenetic mechanisms.
For epigenetic analysis, two agents were used. (A) For methylation analysis, 5 representative clones were stimulated with OKT3/CD28 mAbs, in the presence of the methyltransferase inhibitor, 5-aza-2′-deoxycytidine (5-AzaC) at the indicated concentration. Four days after treatment, clones were analyzed for GFP expression by flow cytometry. (B) For histone deacetylation analysis, 5 representative clones were stimulated on OKT3/28 mAb-coated plates for 4 days. Six days after treatment, the histone deacetylase inhibitor, trichostatin A (TSA) was added into the cultures at the indicated concentration. Twenty-four hours after treatment, TSA-treated clones were analyzed for GFP expression by flow cytometry.
piggyBac transposon copy number analysis in stably transfected human T cells
To determine the transposon copy number in piggyBac-modified T cells, we transfected human PBMCs with transposase and a transposon expressing eGFP. We used transposons of two different sizes, pIRII-eGFP (5593bp in size) and pT-CMV-eGFP (3450bp in size). Because polyclonal populations of transfected T-cells rather than T cell clones would likely be used in clinical trials, we reasoned that determining copy number in a population of cells would be more clinically relevant than analysis of clones. Therefore, after 2 weeks of expansion in culture, transfected cells were sorted to isolate the population stably expressing eGFP, and we used this sorted 100% eGFP population to determine the number of piggyBac transposon integrations per cell. Copy number analysis revealed 1.6 ± 0.18 (n = 3, ± SD) transposon integrations per cell using pIRII-eGFP (MFI 201) and 22.62 ± 3.97 (n = 3, ± SD) copies per cell using the smaller transposon vector, pT-CMV-eGFP (MFI 14,074) (Figure S1). We have previously demonstrated that copy number can be titrated via altering the transposon-to-transposase ratio DNA amounts of each transfected.27 These current results also suggest that copy number can be increased when using a smaller transposon plasmid vector while transfecting the same microgram amount of DNA.
DISCUSSION
In the current study, we observed stable transgene expression for more than 6 months in piggyBac-modified human T lymphocytes in culture. Transgene expression could be increased following TCR activation and costimulation, followed by a return to previous stable baseline levels. Transgene expression appeared to be unaffected by cytokines such as IFN-γ, IL-4, IL-12, and TGF-β known to regulate T cells in vivo, but could be increased by treatment with 5-azaC or trichostatin-A. Therefore, although transgene expression remained stable for >6 months ex vivo, epigenetic mechanisms at least partially regulate transgene expression. Finally, the number of transposons that integrated varied with transposon plasmid size. Our results support the utility of the piggyBac transposon system to genetically modify T lymphocytes for human clinical adoptive transfer immunotherapy or gene therapy applications.
The long-term stability of transgene expression is encouraging for therapies that depend on prolonged gene expression. For example, when T cells are used for tumor therapy, it is important that transgene expression is stable until the tumor is eliminated and that T cells can be reactivated when tumors recur. The increase in transgene expression in response to TCR activation should also be of benefit when immunotherapeutic genes are expressed. Our previous study indicated that piggyBac had a preference for integrating into or near genes highly expressed during T cell activation,28 which may in part explain the observed increase in transgene expression following T cell reactivation.
Adoptively-transferred, genetically-modified T cells encounter both immune stimulatory and suppressive signals in tumor microenvironments. We found that transgene expression in piggyBac- modified T cells was minimally affected by IFN-γ, IL-4, IL-12 or TGFβ. Therefore, the ability of engineered T cells to express a specific transgene(s) to improve immunotherapy should not be affected by these cytokines in a tumor microenvironment. However, immunomodulatory cytokines can still affect T cell function in general as evidenced by the poor proliferation of T cell clones in the presence of TGF-β (data not shown).
Although transgene expression was stable for more than 6 months in T-cell clones, transgene expression was increased when T cells were treated with agents known to interfere with epigenetic regulatory mechanisms. Both 5-azaC and trichostatin-A treated T-cell clones showed increases in transgene expression. This could be a result of multiple transposon integrations in cells where some integrations are silenced and some are not. Treatment with these agents could increase gene expression by “re-activating” gene expression from the silenced integrations. It is also possible that treatment of the cells with these agents could affect transgene expression from unsilenced genes by global genomic affects in the cells. Both demethylating agents and HDAC inhibitors have been used as a chemotherapuetic agents.29–31 Thus, in a combined chemotherapy/immunotherapy setting, 5-azaC and/or TSA could increase transgene expression in gene-modified T cells improving function while also having an underlying chemotherapeutic affect on cancer cells.
Transposon copy number is an important consideration in evaluating their use for clinical application. This study implies that copy number can be varied by using smaller transposon plasmids while transfecting the same amount of plasmid DNA. Copy number can also be varied by altering transposon and transposase DNA amounts and the ratio of the two.27 Therefore, depending on the application, piggyBac could be used to integrate one or more transgenes for a variety of potential clinical applications.
Thus, piggyBac demonstrates characteristics in T cells that could be exploited to fill the gaps in current gene therapy and immunotherapy applications. It is non-viral and presumably less immunogenic than viral vectors. It is capable of achieving long-term stable transgene expression, which is resistant to immunomodulatory cytokines. In addition, piggyBac is capable of delivering large transgenes, such as a 14 kb rapamycine-resistant mTor10 or multiple genes to simultaneously target multiple pathways.12 Inducible expression systems13 could be used to turn genes on or off specifically at tumor sites, and although piggyBac has been used to deliver reporter genes and chimeric antigen receptors to T cells thus far,7–9 many more therapeutic transgene possibilities exist.
The current study further supports the use of the piggyBac transposon system for genetic modification of human T lymphocytes for clinical applications. Transgene expression is stable, minimally affected by immunomodulatory cytokines, and can be increased in the setting of T cell activation, and epigenetic mechanisms regulating transgene expression could be exploited in chemotherapuetic strategies.
Supplementary Material
Top, cells were modified with pIRII-eGFP (5.6kb) transposon. Shown is a representative histogram from flow cytometry (N = 3) as well as the standard curves for RNAseP (the genomic control gene) and the eGFP transgene used for copy number determination. Bottom, cells were modified with pT-CMV-eGFP (3.5kb) transposon.
Acknowledgments
Sources of support: This work was supported in parts by a Specialized Centers for Cell-based Therapy (SCCT) grant from NIH-NHLBI 1 U54 HL1081007 and an NIH-NCI lymphoma SPORE P50 CA126752. MHW was supported by a career development award from the Department of Veterans Affairs, the generous support of Dr. and Mrs. Harold M. Selzman, and in part by NIH-R01DK093660. YN was supported by JHIF scholarship awards in Herpesvirus Infections Research.
SS is a student in the Translational Biology and Molecular Medicine program supported by the HHMI Med-to-Grad initiative.
Footnotes
Financial Disclosure: All authors have declared there are no conflicts of interest in regards to this work.
References
- 1.Huang X, Wilber AC, Bao L, et al. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang X, Guo H, Kang J, et al. Sleeping Beauty transposon-mediated engineering of human primary T cells for therapy of CD19+ lymphoid malignancies. Mol Ther. 2008;16:580–589. doi: 10.1038/sj.mt.6300404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh H, Manuri PR, Olivares S, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cary LC, Goebel M, Corsaro BG, et al. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 6.Fraser MJ, Cary L, Boonvisudhi K, et al. Assay for movement of Lepidopteran transposon IFP2 in insect cells using a baculovirus genome as a target DNA. Virology. 1995;211:397–407. doi: 10.1006/viro.1995.1422. [DOI] [PubMed] [Google Scholar]
- 7.Nakazawa Y, Huye LE, Dotti G, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raja Manuri PV, Wilson MH, Maiti SN, et al. piggyBac transposon/transposase system to generate CD19-specific T cells for treatment of B-lineage malignancies. Hum Gene Ther. 2010;21:427–437. doi: 10.1089/hum.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakazawa Y, Huye LE, Salsman VS, et al. PiggyBac-Mediated Cancer Immunotherapy Using EBV-Specific Cytotoxic T-Cells Expressing HER2-Specific Chimeric Antigen Receptor. 2011;19:2133–43. doi: 10.1038/mt.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huye LE, Nakazawa Y, Patel MP, et al. Combining mTor Inhibitors With Rapamycin-resistant T Cells: A Two-pronged Approach to Tumor Elimination. Mol Ther. 2011;19:2239–48. doi: 10.1038/mt.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li MA, Turner DJ, Ning Z, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahlig KM, Saridey SK, Kaja A, et al. Multiplexed transposon-mediated stable gene transfer in human cells. Proc Natl Acad Sci U S A. 2010;107:1343–1348. doi: 10.1073/pnas.0910383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saridey SK, Liu L, Doherty JE, et al. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol Ther. 2009;17:2115–2120. doi: 10.1038/mt.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woltjen K, Michael IP, Mohseni P, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kettlun C, Galvan DL, George AL, et al. Manipulating piggyBac transposon chromosomal integration site selection in human cells. 2011;19:1636–44. doi: 10.1038/mt.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owens JB, Urschitz J, Stoytchev I, et al. Chimeric piggyBac transposases for genomic targeting in human cells. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks309. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrison BS, Yant SR, Mikkelsen JG, et al. Postintegrative gene silencing within the Sleeping Beauty transposition system. Mol Cell Biol. 2007;27:8824–8833. doi: 10.1128/MCB.00498-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson MH, Coates CJ, George AL., Jr PiggyBac Transposon-mediated Gene Transfer in Human Cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 19.Suhoski MM, Golovina TN, Aqui NA, et al. Engineering artificial antigen-presenting cells to express a diverse array of co-stimulatory molecules. Mol Ther. 2007;15:981–988. doi: 10.1038/mt.sj.6300134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehm U, Klamp T, Groot M, et al. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 21.Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- 22.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 23.Leen AM, Rooney CM, Foster AE. Improving T cell therapy for cancer. Annu Rev Immunol. 2007;25:243–265. doi: 10.1146/annurev.immunol.25.022106.141527. [DOI] [PubMed] [Google Scholar]
- 24.Qin L, Ding Y, Pahud DR, et al. Promoter attenuation in gene therapy: interferon-gamma and tumor necrosis factor-alpha inhibit transgene expression. Hum Gene Ther. 1997;8:2019–2029. doi: 10.1089/hum.1997.8.17-2019. [DOI] [PubMed] [Google Scholar]
- 25.Li CL, Emery DW. The cHS4 chromatin insulator reduces gammaretroviral vector silencing by epigenetic modifications of integrated provirus. Gene Ther. 2008;15:49–53. doi: 10.1038/sj.gt.3303009. [DOI] [PubMed] [Google Scholar]
- 26.Xia X, Zhang Y, Zieth CR, et al. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doherty JE, Huye LE, Yusa K, et al. Hyperactive piggyBac Gene Transfer in Human Cells and In Vivo. Hum Gene Ther. 2012;23:311–320. doi: 10.1089/hum.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galvan DL, Nakazawa Y, Kaja A, et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simova J, Pollakova V, Indrova M, et al. Immunotherapy augments the effect of 5-azacytidine on HPV16-associated tumours with different MHC class I-expression status. Br J Cancer. 2011;105:1533–1541. doi: 10.1038/bjc.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan O, La Thangue NB. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol Cell Biol. 2012;90:85–94. doi: 10.1038/icb.2011.100. [DOI] [PubMed] [Google Scholar]
- 31.Shakespear MR, Halili MA, Irvine KM, et al. Histone deacetylases as regulators of inflammation and immunity. Trends Immunol. 2011;32:335–343. doi: 10.1016/j.it.2011.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Top, cells were modified with pIRII-eGFP (5.6kb) transposon. Shown is a representative histogram from flow cytometry (N = 3) as well as the standard curves for RNAseP (the genomic control gene) and the eGFP transgene used for copy number determination. Bottom, cells were modified with pT-CMV-eGFP (3.5kb) transposon.