Abstract
Herein, we report for the first time the design and synthesis of a novel cyclotide able to efficiently inhibit HIV-1 viral replication by selectively targeting cytokine receptor CXCR4. This was accomplished by grafting a series of topologically modified CVX15 based peptides onto the loop 6 of cyclotide MCoTI-I. The most active compound produced in this study was a potent CXCR4 antagonist (EC50 ≈ 20 nM) and an efficient HIV-1 cell-entry blocker (EC50 ≈ 2 nM). This cyclotide also showed high stability in human serum thereby providing a promising lead compound for the design of a novel type of peptide-based anti-cancer and anti-HIV-1 therapeutics.
INTRODUCTION
Chemokine receptors are G protein-coupled receptors (GPCRs) that play a key regulatory role in embryonic development and controlling leukocyte functions during inflammation and immunity.1–3 The CXCR4 receptor is one of the 19 chemokine receptors known so far. This receptor is activated exclusively by the cytokine CXCL12, also known as stromal cell-derived factor-1α (SDF1α). Activation of CXCR4 promotes chemotaxis in leukocytes,4 progenitor cell migration,5 and embryonic development of the cardiovascular, hemaotopoietic and central nervous system.6–9 CXCR4 has also been associated with multiple types of cancers where its overexpression/activation promotes metastasis, angiogenesis and tumor growth and/or survival.10, 11 Furthermore, CXCR4 is involved in HIV replication, as it is a co-receptor for viral entry into host cells.12, 13 Altogether, these features make CXCR4 a very attractive target for drug discovery.14–16 Hence, several small molecules and small peptides have been developed to antagonize CXCR4 for anti-cancer and anti-HIV activity.15 CXCR4 antagonists have also been shown to induce the mobilization of hematopoietic stem cells (HSCs) by disrupting the CXCR4-CXCL12 interaction, which is required for retaining HSCs in the bone marrow,17–19 and therefore have been used to facilitate the mobilization of HSCs to the periphery for their isolation.20
Cyclotides are small globular microproteins (ranging from 28 to 37 amino acids) with a unique head-to-tail cyclized backbone, which is stabilized by three disulfide bonds forming a cystine-knot motif 21–23 (Fig. 1A). This cyclic cystine-knot (CCK) framework provides a rigid molecular platform24, 25 with exceptional stability towards physical, chemical and biological degradation.22, 23 These micro-proteins can be considered natural combinatorial peptide libraries structurally constrained by the cystine-knot scaffold and head-to-tail cyclization, but in which hypermutation of essentially all residues is permitted with the exception of the strictly conserved cysteines that comprise the knot.26–28 Furthermore, naturally-occurring cyclotides have shown to posses various pharmacologically-relevant activities,22, 29 and have been reported to cross cell membranes.30, 31 Altogether, these features make the cyclotide scaffold an excellent molecular framework for the design of novel peptide-based therapeutics,23, 32 making them ideal substrates for molecular grafting of biological peptide epitopes.33–36
Figure 1.
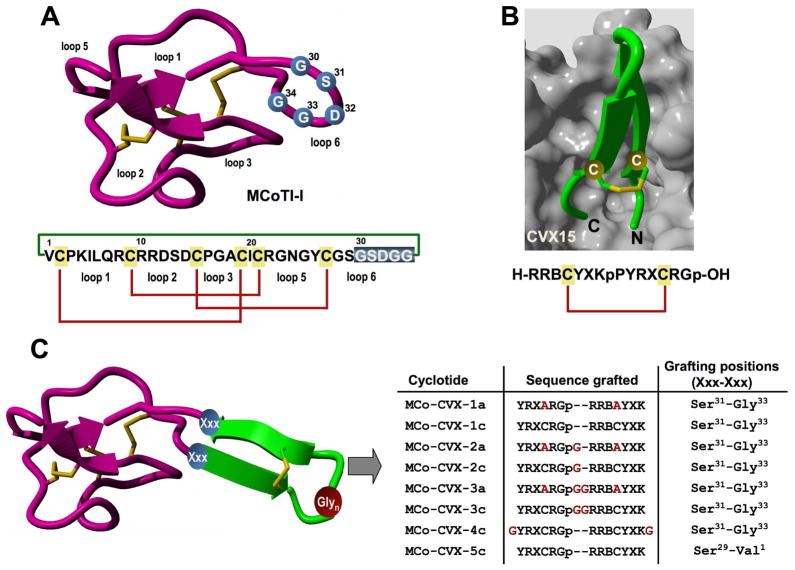
Design of MCoTI-based cyclotides to target the cytokine receptor CXCR4. A. Primary and tertiary structures of cyclotide MCoTI-I. Structure is based on a homology model using the solution structure of MCoTI-II as template (PDB: 1IB9).45 The backbone cyclized peptide (connecting bond shown in green) is stabilized by the three-disulfide bonds (shown in red). The residues used for the grafting of a CVX15-based peptide are shown in blue on the structure and sequence of MCoTI-I. B. Sequence and co-crystal structure of peptide CVX15 bound to cytokine receptor CXCR4 (PDB: 3OE0).40 Peptide CVX15 is shown as a ribbon representation in green with the side-chains of the Cys residues involved in the disulfide bond in ball-and-stick form. The solvent accessible surface of the binding site of CXCR4 is shown in grey. C. Scheme depicting the approach used to design the different MCo-CVX cyclotides. A circularly permuted version of CVX15 was grafted onto loop 6 of MCoTI-I at different residues. The CVX15-based insert was created by joining the C and N-terminus directly through a flexible Glyn linker and opening the new sequence at the D-Pro-Pro segment. Residues in red denote mutations or extra Gly residues introduced to increase flexibility. Single letter codes B, X and p represent the amino acid, 2-naphthylalanine, citruline and D-proline, respectively. Molecular graphics were built with Yasara (www.yasara.org).
Several small disulfide cyclic peptides derived from the horseshoe crab peptides polyphemusin-I/II have recently been reported to be efficient CXCR4 antagonists and effective as anti-HIV-1 and antimetastatic agents.37–39 Some of these peptides, however, have shown limited proteolytic stability and/or poor bioavailability.38 By using the crystal structure of CXCR4 bound to the polyphemusin-derived peptide CVX1540 we report here for the first time the design and synthesis of an engineered cyclotide able to effectively antagonize CXCR4 and inhibit CXCR4-tropic HIV-1 entry in human lymphocytes.
RESULTS AND DISCUSSION
To produce a novel cyclotide with CXCR4 antagonistic activity, we used MCoTI-I as a molecular scaffold (Fig. 1A). MCoTI-cyclotides have been recently isolated from the dormant seeds of Momordica cochinchinensis, a plant member of the cucurbitaceae family, and are potent trypsin inhibitors (Ki ≈ 20–30 pM).41 MCoTI-cyclotides show very low toxicity in human cells30 and represent a desirable molecular scaffold for engineering new compounds with unique biological properties.33–35
According to the X-ray crystal structure of CVX15 bound to CXCR4, the N- and C-termini of the CVX15 peptide are deeply buried into the CXCR4 binding pocket (Fig. 1B). Therefore, a circularly permuted version of the CVX15 peptide was grafted into loop 6 of the cyclotide MCoTI-I in order to preserve the biological activity of the grafted peptide. The CVX15 sequence was designed by linking the original N- and C-termini directly or through a flexible Glyn (n =1, 2) linker, removing residues D-Pro8 and Pro9 and leaving the new N- and C-terminal groups on residues Tyr10 and Lys7, respectively (Figs. 1B and 1C). Residue Gln6 was also replaced by citruline, which has been shown to increase the affinity of CVX15 for CXCR4.42 We also explored the effect of replacing the original Cys residues in the CVX15-based sequence, which are involved in a disulfide bond, by Ala residues to see the effect on the biological activity of the resulting cyclotides. The different sequences were grafted onto loop 6 by replacing residue Asp32, or the peptide segment Gly30-Gly34 (Fig. 1C). Loop 6 of MCoTI-cyclotides has been shown to be less rigid in solution24, 25 and quite tolerant to sequence grafting.33–36
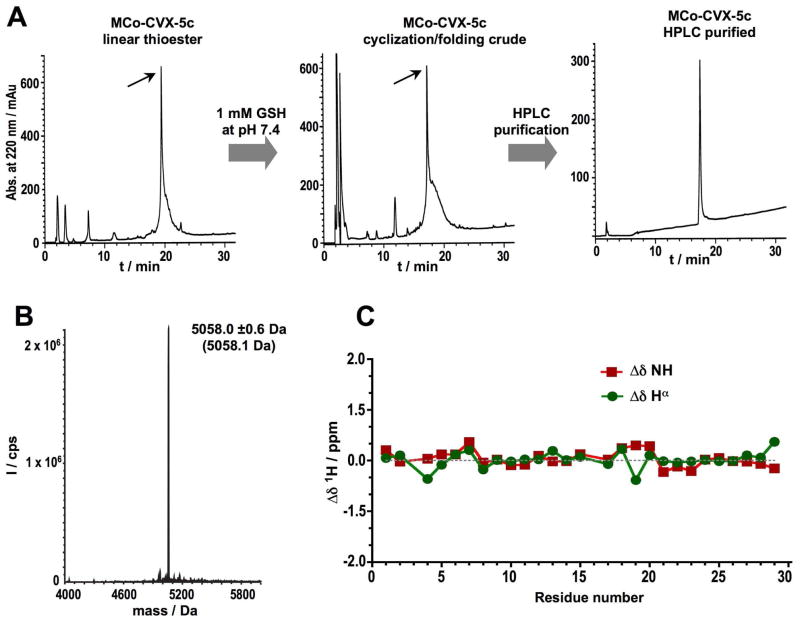
All grafted MCo-CVX cyclotides were chemically synthesized using Fmoc-based solid-phase peptide synthesis on a sulfonamide resin.30 Activation of the sulfonamide linker with iodoacetonitrile, followed by cleavage with ethyl mercaptoacetate and acidolytic deprotection, provided the fully deprotected linear peptide α-thioester (Table S1). The corresponding peptide thioester precursors were efficiently cyclized and folded in a one-pot reaction using sodium phosphate buffer at pH 7.2 in the presence of 1 mM GSH. The cyclization/folding reactions were complete in 24–96 h (Figs. 2A and S1, Table 1). The cyclization/folding yields ranged from around 20% (MCoTI-CVX-4c) to 80% (MCo-CVX-5c) (Table 1). Folded MCo-CVX cyclotides were purified by reverse-phase HPLC and characterized by ES-MS confirming ≥95% purity (Figs. 2B and S1, and Table 1). Grafted MCo-CVX-5c cyclotide was also characterized by 1H-NMR indicating that adopts a native cyclotide fold (Figs. 2C and S2).
Figure 2.
Chemical synthesis and characterization of cyclotide MCo-CVX-5c. A. Analytical HPLC traces of the linear thioester precursor, GSH-induced cyclization/folding crude after 96 h and purified cyclotide. An arrow indicates the desired peptide. B. ES-MS characterization of pure MCo-CVX-5c. The expected average molecular weight is shown in parenthesis. C. Chemical shifts differences of the backbone, NH and Hα protons between the common sequence (residues 1 through 29) of MCoTI-I24, 25 and MCo-CVX-5c (Table S2).
Table 1.
Molecular weight, cyclization/folding yield and biological activity summary for the MCo-CVX grafted cyclotides produced in this work.
| Peptide Name | Molecular weight (Da) | Cyclization/folding | EC50 (nM) | |||
|---|---|---|---|---|---|---|
| Linear thioester | Cyclized/folded | yield (%) | time (h)a | CXCR4 inhibition | HIV-1 inhibition | |
| MCo-CVX-1a | 5380.0 ± 1.0 (5380.3) | 5254.0 ± 0.4 (5252.2) | 43 | 24 | 1040 ± 45 | NDb |
| MCo-CVX-1c | 5444.4 ± 0.2 (5444.4) | 5317.1 ± 0.8 (5316.4) | 61 | 24 | 102 ±12 | NDb |
| MCo-CVX-2a | 5437.7 ± 0.5 (5437.3) | 5311.1 ± 0.5 (5311.3) | 41 | 24 | 4900 ± 600 | NDb |
| MCo-CVX-2c | 5501.4 ± 0.4 (5501.4) | 5373.2 ± 0.3 (5373.4) | 43 | 24 | 2140 ± 300 | NDb |
| MCo-CVX-3a | 5494.7 ± 0.7 (5494.3) | 5368.2 ± 0.5 (5368.3) | 26 | 24 | 23800 ± 3000 | NDb |
| MCo-CVX-3c | 5559.1± 0.5 (5558.4) | 5430.2 ± 0.6 (5430.4) | 47 | 24 | 3110 ± 480 | NDb |
| MCo-CVX-4c | 5559.2 ± 0.7 (5558.4) | 5430.2 ± 0.2 (5430.4) | 17 | 24 | 39 ± 1 | NDb |
| MCo-CVX-5c | 5185.1 ± 0.5 (5186.1) | 5058.0 ± 0.6 (5058.1) | 81 | 96 | 19 ± 3 | 2.0 ± 0.3 |
Time for efficient cyclization
Not determined
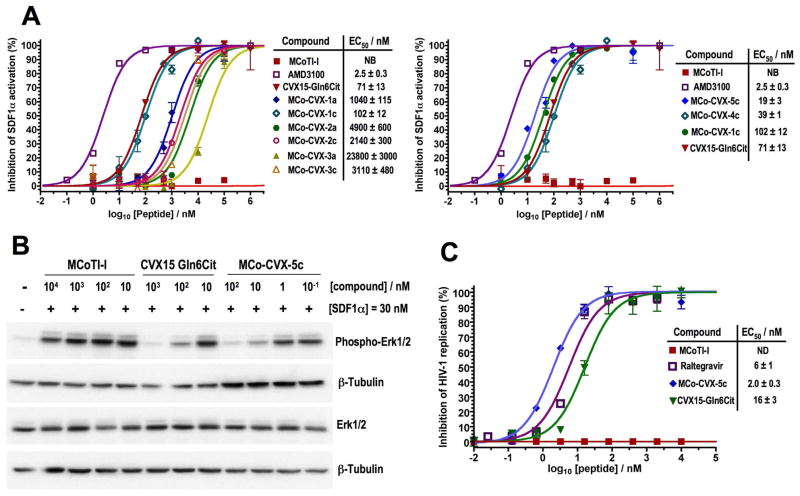
Next, we tested the ability of the CVX15-grafted cyclotides to inhibit SDF1α–mediated CXCR4 activation using a CXCR4-β-lactamase U2OS cell-based fluorescence assay (Fig 3A). All grafted cyclotides were able to block SDF1α-mediated CXCR4 activation in a dose dependent manner with EC50 values ranging from 23.8 ± 0.3 μM (MCo-CVX-3a) to 19 ± 3 nM (MCo-CVX-5c). Intriguingly, the peptide CVX15 Gln6Cit alone showed an EC50 value of 71 ± 13 nM, which is around 3 times weaker than that of the best cyclotide inhibitor (MCo-CVX-5c). As expected, the naturally-occurring cyclotide MCoTI-I did not show any inhibitory activity in this assay (Fig. 3A), indicating that the biological activity of grafted MCo-CVX cyclotides is specific and comes from the grafted sequence. The small molecule AMD3100 20 was also used as positive control. The importance of the original Cys residues in peptide CVX15 is highlighted by comparing the EC50 values of the cyclotides grafted onto Asp32. Mutation of the Cys residues to Ala significantly reduced the biological activity of the corresponding cyclotides. For example, cyclotides MCo-CVX-1c and MCo-CVX-3c were around 10-times more potent than the corresponding mutants MCo-CVX-1a and MCo-CVX-3a, respectively. The decrease in potency was less pronounced in cyclotide MCo-CVX-2a, where this mutation resulted only in a ≈ 2-fold decrease in EC50 value (Fig. 3A). The length of Gly linker used to build the CVX15-based insert, that was grafted onto the cyclotide scaffold, was also critical to the biological activity of the resulting grafted MCo-CVX cyclotides. The most active cyclotide in this series was MCo-CVX-1c (EC50 = 0.10 ± 0.01 μM), which was designed by linking directly the original N- and C-termini of the CVX15 peptide. Addition of extra Gly residues on MCo-CVX-2c and MCo-CVX-3c had a detrimental effect on their potencies yielding EC50 values around 2 μM and 3 μM, respectively (Fig. 3A and Table 1). These results are likely due to the increase in flexibility provided by the extra Gly residues, which may reduce the binding energy. Interestingly, the position on loop 6 where the CVX15-based peptide was grafted was also important for the biological activity of the resulting grafted cyclotides. The most active cyclotide was MCo-CVX-5c (EC50 = 19 ± 3 nM), where the CVX15-based peptide is grafted between residues Gly30 and Gly34. Grafting the bioactive peptide farther away from the cyclotide core resulted in less active cyclotides. Thus cyclotides MCo-CVX-1c (graft at residue Asp32) and MCo-CVX-4c (graft at residue Asp32 but with extra Gly residues at both termini of the peptide graft) showed EC50 values of 102 ± 12 nM and 39 ± 1 nM, respectively (Fig. 3A and Table 1).
Figure 3.
Biological characterization of MCo-CVX cyclotides. A. Competitive inhibition of SDF1α-mediated CXCR4 activation by different cyclotides. The peptide CVX15 Gln6Cit and the small molecule CXCR4 antagonist AMD3100 were used as controls. The assay was performed using CXCR4-bla U2OS cells. B. Inhibition of Erk phosphorylation (residues Thr202 and Tyr204) by cyclotide MCo-CVX-5c. Cyclotide MCoTI-I and peptide CVX15 Gln6Cit were used as negative and positive controls, respectively. Erk phosphorylation was visualized by Western blot using CaOV3 cells treated with increasing amounts of CXCR4 inhibitor in the presence of SDF1α. C. Dose response inhibition of HIV-1 replication in MT-4 cells by cyclotides MCoTI-I and MCo-CVX-5c. The peptide CVX15 Gln6Cit and the small molecule HIV-1 integrase inhibitor, Raltegravir, were used as positive controls. Cyclotide MCoTI-I was used as negative control. The average of standard deviation of three experiments is shown. NB and ND stand for not bound and not determined, respectively.
Cyclotide MCo-CVX-5c was also able to inhibit SDF1α-induced Erk phosphorylation and internalization of CXCR4 in a dose dependent manner, confirming that this cyclotide is an efficient CXCR4 antagonist (Figs. 3B and S3). In these experiments, cyclotide MCo-CVX-5c was around 10 times more active than the peptide CVX15 Gln6Cit. More importantly, cyclotide MCo-CVX-5c also inhibited the entry and replication of CXCR4-tropic HIV-1 in human lymphocyte MT4 cells in a dose dependent manner with an EC50 value of 2.0 ± 0.3 nM (Fig. 3C). The EC50 value for peptide CVX15 Gln6Cit was around 8-times higher (Fig. 3C), which is in agreement with the data obtained in the inhibition of Erk phosphorylation and CXCR4 internalization. More notably, cyclotide MCo-CVX-5c showed a CC50 (cytoxic concentration to reduce 50% cell viability) value in MT4 cells greater than 10 μM (data not shown), therefore providing a selectivity index of more than 4,000. It is also worth noting that cyclotide MCo-CVX-5c was 3-times more potent than Raltegravir,43 an integrase inhibitor recently approved by the FDA to treat HIV infection (Fig. 3C).
We also studied the biological stability of MCo-CVX-5c and compare it to that of the empty scaffold (MCoTI-I) and the grafted peptide (CVX15 Gln6Cit) (Fig. S4). This was accomplished by incubating the corresponding peptides in human serum at 37° C. The quantitative analysis of undigested polypeptides was performed using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Naturally occurring MCoTI-cyclotides present a very rigid structure,24, 25 which makes them extremely stable to proteolytic degradation. Remarkably, cyclotide MCo-CVX-5c showed greater stability in human serum (τ1/2 = 62 ± 3 h) than the parent cyclotide MCoTI-I (τ1/2 = 52 ± 3 h, Fig. S4). In contrast, peptide CVX15 Gln6Cit was degraded considerably faster under the same conditions (τ1/2 = 21 ± 4 h, Fig. S4). A linearized, reduced and alkylated version of MCo-CVX-5c was also rapidly degraded (τ1/2 = 21 ± 3 min) indicating the importance of the circular Cys-knot topology for proteolytic stability. We also investigated the fraction of cyclotide bound to serum proteins. Serum binding has been recently used to extend serum half-life of bioactive peptides.44 The binding, however, has to be reversible in order to be pharmacologically useful. Cyclotides MCoTI-I and MCo-CVX-5c were both found to be more than 99% bound to serum proteins under the conditions employed in the serum stability assay. The fact that these cyclotides are almost completely degraded after 120 h of treatment (Fig. S4) suggests that their binding to serum proteins may be reversible. To further explore this possibility, we studied the association and dissociation rate constants of MCo-CVX-5c to human serum proteins. This was accomplished by biolayer interferometry analysis using the commercially available platform Blitz from ForteBio. The results indicated that the cyclotide MCo-CVX-5c is able to bind serum proteins with an association and dissociation constant rates of 3.6 ± 0.7 × 103 M−1s−1 and 1.4 ± 0.2 × 10−2 s−1, respectively (Fig. S5), which provide a relatively weak dissociation constant of ≈ 4 μM when compared to the low nanomolar affinity of MCo-CVX-5c for the CXCR4. These results are in agreement with the biological activities found for MCo-CVX-5c, which were obtained in the presence of human serum, 2% and 12% for the CXCR4 translocation and HIV-1 cell entry inhibition assays, respectively.
CONCLUSIONS
In summary we report here for the first time the design and synthesis of a novel cyclotide able to efficiently inhibit the GPCR CXCR4. This was successfully accomplished by grafting a series of topologically modified CVX15 based peptides onto loop 6 of the cyclotide MCoTI-I. 1H-NMR studies also revealed that the grafting of CVX15 based peptides onto this loop did not affect the native cyclotide scaffold, indicating the tolerance of this loop for the grafting of long peptide sequences.29, 35 The most active compound produced in this study, MCo-CVX-5c, is a potent CXCR4 antagonist (EC50 = 19 ± 3 nM) and an efficient HIV-1 cell-entry blocker (EC50 = 2.0 ± 0.3 nM). Intriguingly, cyclotide MCo-CVX-5c was significantly more active than the cyclic peptide CVX15 Gln6Cit used in the design of the grafted cyclotide. Although more detailed structural studies are required to analyze the interaction between the cyclotide MCo-CVX-5c and CXCR4, altogether these results suggest that some of the residues from the neighboring loops in the cyclotide may contribute positively to the interaction with CXCR4. To further explore this possibility we built a model of MCo-CVX-5c bound to CXCR4 using the crystal structure of CVX15-CXCR4 (PDB: 3OE0)40 and the solution structure of MCoTI-II (PDB: 1IB9)45 (Fig. S6). According to this model, loops 2 and 5 may be in close proximity to the extracellular receptor surface facilitating new interactions. This should make possible the design of even more potent antagonists based on MCo-CVX-5c by the introduction of appropriate mutations in these loops to improve the molecular complementarity between the cyclotide and receptor surfaces. It is also worth noting that the cyclotide MCo-CVX-5c showed a remarkable resistance to biological degradation in human serum, with a τ1/2 value of 62 ± 3 h. This value is similar to that of the cyclotide MCoTI-I and significantly higher that the half-life of the peptide CVX15 (τ1/2 = 21 ± 4 h). In addition, the binding affinity of cyclotide MCo-CVX-5c to serum proteins was significantly weaker than for CXCR4, which should be able to decrease the renal clearance of this cyclotide without affecting its activity. Although further analysis will be required to evaluate the therapeutic value of these compounds in vivo, altogether, our results show that engineered cyclotides hold great promise for the development of a novel type of peptide-based therapeutic able to efficiently target extracellular protein/protein interactions. Our results demonstrate for the first time the design of an engineered cyclotide able to target the GPCR CXR4 with low nanomolar affinity and significant serum stability, thereby providing a promising lead compound for the design of anti-cancer and anti-HIV-1 compounds.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grants R01-GM090323 (JAC), R01-GM085006 (AS), the Southern California Clinical and Translational Science Institute Grant UL1RR031986-03 (JAC and NN), the Department of Defense Congressionally Directed Medical Research Program Grant PC09305 (JAC) and the EU-FP7 funded project CHAARM. We thank Barbara Van Remoortel for excellent technical assistance. FC is a fellow of the Industrial Research Fund (IOF).
Footnotes
Supporting Information Available: Experimental details for the synthesis, purification and characterization of MCo-CVX peptides, cell-based CXCR4 competitive binding assays, SDF1a-mediated Erk phosphorylation and CXCR4 internalization assays, HIV-1 replication and MT-4 cytotoxicity assays, NMR spectroscopy, serum stability, binding kinetics of cyclotide MCo-CVX-5c to human serum proteins and modeling of the CXCR4- MCo-CVX-5c complex. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Baggiolini M. Chemokines and leukocyte traffic. Nature. 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 2.Mackay CR. Chemokines: immunology’s high impact factors. Nat Immunol. 2001;2:95–101. doi: 10.1038/84298. [DOI] [PubMed] [Google Scholar]
- 3.Moser B, Wolf M, Walz A, Loetscher P. Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 2004;25:75–84. doi: 10.1016/j.it.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Furze RC, Rankin SM. Neutrophil mobilization and clearance in the bone marrow. Immunology. 2008;125:281–288. doi: 10.1111/j.1365-2567.2008.02950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 6.Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T, Bronson RT, Springer TA. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 8.Fulton AM. The chemokine receptors CXCR4 and CXCR3 in cancer. Curr Oncol Rep. 2009;11:125–131. doi: 10.1007/s11912-009-0019-1. [DOI] [PubMed] [Google Scholar]
- 9.Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–2931. doi: 10.1158/1078-0432.CCR-09-2329. [DOI] [PubMed] [Google Scholar]
- 10.Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 11.Balkwill F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin Cancer Biol. 2004;14:171–179. doi: 10.1016/j.semcancer.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 13.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier JL, Arenzana-Seisdedos F, Schwartz O, Heard JM, Clark-Lewis I, Legler DF, Loetscher M, Baggiolini M, Moser B. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature. 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 14.Seibert C, Sakmar TP. Small-molecule antagonists of CCR5 and CXCR4: a promising new class of anti-HIV-1 drugs. Curr Pharm Des. 2004;10:2041–2062. doi: 10.2174/1381612043384312. [DOI] [PubMed] [Google Scholar]
- 15.Choi WT, Duggineni S, Xu Y, Huang Z, An J. Drug discovery research targeting the CXC chemokine receptor 4 (CXCR4) J Med Chem. 2012;55:977–994. doi: 10.1021/jm200568c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grande F, Garofalo A, Neamati N. Small molecules anti-HIV therapeutics targeting CXCR4. Curr Pharm Des. 2008;14:385–404. doi: 10.2174/138161208783497714. [DOI] [PubMed] [Google Scholar]
- 17.Hattori K, Heissig B, Tashiro K, Honjo T, Tateno M, Shieh JH, Hackett NR, Quitoriano MS, Crystal RG, Rafii S, Moore MA. Plasma elevation of stromal cell-derived factor-1 induces mobilization of mature and immature hematopoietic progenitor and stem cells. Blood. 2001;97:3354–3360. doi: 10.1182/blood.v97.11.3354. [DOI] [PubMed] [Google Scholar]
- 18.Liles WC, Broxmeyer HE, Rodger E, Wood B, Hubel K, Cooper S, Hangoc G, Bridger GJ, Henson GW, Calandra G, Dale DC. Mobilization of hematopoietic progenitor cells in healthy volunteers by AMD3100, a CXCR4 antagonist. Blood. 2003;102:2728–2730. doi: 10.1182/blood-2003-02-0663. [DOI] [PubMed] [Google Scholar]
- 19.Tchernychev B, Ren Y, Sachdev P, Janz JM, Haggis L, O’Shea A, McBride E, Looby R, Deng Q, McMurry T, Kazmi MA, Sakmar TP, Hunt S, 3rd, Carlson KE. Discovery of a CXCR4 agonist pepducin that mobilizes bone marrow hematopoietic cells. Proc Natl Acad Sci U S A. 2010;107:22255–22259. doi: 10.1073/pnas.1009633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Clercq E. The AMD3100 story: the path to the discovery of a stem cell mobilizer (Mozobil) Biochem Pharmacol. 2009;77:1655–1664. doi: 10.1016/j.bcp.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 21.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 22.Daly NL, Rosengren KJ, Craik DJ. Discovery, structure and biological activities of cyclotides. Adv Drug Deliv Rev. 2009;61:918–930. doi: 10.1016/j.addr.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Gould A, Ji Y, Aboye TL, Camarero JA. Cyclotides, a novel ultrastable polypeptide scaffold for drug discovery. Curr Pharm Des. 2011;17:4294–4307. doi: 10.2174/138161211798999438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2010;49:7030–7034. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puttamadappa SS, Jagadish K, Shekhtman A, Camarero JA. Erratum in: Backbone Dynamics of Cyclotide MCoTI-I Free and Complexed with Trypsin. Angew Chem Int Ed Engl. 2011;50:6948–6949. doi: 10.1002/anie.201002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Austin J, Kimura RH, Woo YH, Camarero JA. In vivo biosynthesis of an Ala-scan library based on the cyclic peptide SFTI-1. Amino Acids. 2010;38:1313–1322. doi: 10.1007/s00726-009-0338-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonsen SM, Sando L, Rosengren KJ, Wang CK, Colgrave ML, Daly NL, Craik DJ. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem. 2008;283:9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 28.Huang YH, Colgrave ML, Clark RJ, Kotze AC, Craik DJ. Lysine-scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J Biol Chem. 2010;285:10797–10805. doi: 10.1074/jbc.M109.089854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia AE, Camarero JA. Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr Mol Pharmacol. 2010;3:153–163. doi: 10.2174/1874467211003030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Contreras J, Elnagar AY, Hamm-Alvarez SF, Camarero JA. Cellular uptake of cyclotide MCoTI-I follows multiple endocytic pathways. J Control Release. 2011;155:134–143. doi: 10.1016/j.jconrel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascales L, Henriques ST, Kerr MC, Huang YH, Sweet MJ, Daly NL, Craik DJ. Identification and characterization of a new family of cell-penetrating peptides: cyclic cell-penetrating peptides. J Biol Chem. 2011;286:36932–36943. doi: 10.1074/jbc.M111.264424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriques ST, Craik DJ. Cyclotides as templates in drug design. Drug Discov Today. 2010;15:57–64. doi: 10.1016/j.drudis.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 33.Gunasekera S, Foley FM, Clark RJ, Sando L, Fabri LJ, Craik DJ, Daly NL. Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J Med Chem. 2008;51:7697–7704. doi: 10.1021/jm800704e. [DOI] [PubMed] [Google Scholar]
- 34.Thongyoo P, Bonomelli C, Leatherbarrow RJ, Tate EW. Potent inhibitors of beta-tryptase and human leukocyte elastase based on the MCoTI-II scaffold. J Med Chem. 2009;52:6197–6200. doi: 10.1021/jm901233u. [DOI] [PubMed] [Google Scholar]
- 35.Chan LY, Gunasekera S, Henriques ST, Worth NF, Le SJ, Clark RJ, Campbell JH, Craik DJ, Daly NL. Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood. 2011;118:6709–6717. doi: 10.1182/blood-2011-06-359141. [DOI] [PubMed] [Google Scholar]
- 36.Sommerhoff CP, Avrutina O, Schmoldt HU, Gabrijelcic-Geiger D, Diederichsen U, Kolmar H. Engineered cystine knot miniproteins as potent inhibitors of human mast cell tryptase beta. J Mol Biol. 2010;395:167–175. doi: 10.1016/j.jmb.2009.10.028. [DOI] [PubMed] [Google Scholar]
- 37.Tamamura H, Xu Y, Hattori T, Zhang X, Arakaki R, Kanbara K, Omagari A, Otaka A, Ibuka T, Yamamoto N, Nakashima H, Fujii N. A low-molecular-weight inhibitor against the chemokine receptor CXCR4: a strong anti-HIV peptide T140. Biochem Biophys Res Commun. 1998;253:877–882. doi: 10.1006/bbrc.1998.9871. [DOI] [PubMed] [Google Scholar]
- 38.DeMarco SJ, Henze H, Lederer A, Moehle K, Mukherjee R, Romagnoli B, Robinson JA, Brianza F, Gombert FO, Lociuro S, Ludin C, Vrijbloed JW, Zumbrunn J, Obrecht JP, Obrecht D, Brondani V, Hamy F, Klimkait T. Discovery of novel, highly potent and selective beta-hairpin mimetic CXCR4 inhibitors with excellent anti-HIV activity and pharmacokinetic profiles. Bioorg Med Chem. 2006;14:8396–8404. doi: 10.1016/j.bmc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Moncunill G, Armand-Ugon M, Clotet-Codina I, Pauls E, Ballana E, Llano A, Romagnoli B, Vrijbloed JW, Gombert FO, Clotet B, De Marco S, Este JA. Anti-HIV activity and resistance profile of the CXC chemokine receptor 4 antagonist POL3026. Mol Pharmacol. 2008;73:1264–1273. doi: 10.1124/mol.107.042911. [DOI] [PubMed] [Google Scholar]
- 40.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, Trinh Hong T, Pham TT, Le Nguyen D. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry. 2000;39:5722–5730. doi: 10.1021/bi9929756. [DOI] [PubMed] [Google Scholar]
- 42.Oishi S, Fujii N. Peptide and peptidomimetic ligands for CXC chemokine receptor 4 (CXCR4) Org Biomol Chem. 2012;10:5720–5731. doi: 10.1039/c2ob25107h. [DOI] [PubMed] [Google Scholar]
- 43.Klibanov OM. Elvitegravir, an oral HIV integrase inhibitor, for the potential treatment of HIV infection. Curr Opin Investig Drugs. 2009;10:190–200. [PubMed] [Google Scholar]
- 44.McGregor DP. Discovering and improving novel peptide therapeutics. Curr Opin Pharmacol. 2008;8:616–619. doi: 10.1016/j.coph.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Felizmenio-Quimio ME, Daly NL, Craik DJ. Circular proteins in plants: solution structure of a novel macrocyclic trypsin inhibitor from Momordica cochinchinensis. J Biol Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.