Abstract
Human chromosome ends are capped by shelterin, a protein complex that protects the natural ends from being recognized as sites of DNA damage and also regulates the telomere-replicating enzyme, telomerase1–3. Shelterin includes the heterodimeric POT1-TPP1 protein, which binds the telomeric single-stranded DNA tail4–9. TPP1 has been implicated both in recruiting telomerase to telomeres and in stimulating telomerase processivity (the addition of multiple DNA repeats after a single primer-binding event)9–14. Determining the mechanisms of these activities has been difficult, especially because genetic perturbations also tend to affect the essential chromosome end-protection function of TPP115–17. Here we identify separation-of-function mutants of TPP1 that retain full telomere-capping function in vitro and in vivo, yet are defective in binding telomerase. The seven separation-of-function mutations map to a patch of amino acids on the surface of TPP1, the TEL patch, that both recruits telomerase to telomeres and promotes high-processivity DNA synthesis, indicating that these two activities are manifestations of the same molecular interaction. Given that the interaction between telomerase and TPP1 is required for telomerase function in vivo, the TEL patch of TPP1 provides a new target for anti-cancer drug development.
Genetic analysis of TPP1 regulation of telomerase is complicated by the potential of TPP1 mutations to uncap telomeres, which can give unnatural telomerase hyperextension or induce a DNA-damage response. Our search for separation-of-function mutants of TPP1 that would affect only its telomerase interactions was conducted on the TPP1 OB domain (TPP1-OB), which is separate from its POT1-ssDNA-interaction domain (Fig. 1a) and previously implicated in telomerase interaction13,14. We engineered twelve single amino-acid mutants and two double mutants of TPP1-OB, choosing residues that are both conserved among mammalian TPP1 proteins and reside on the protein surface9 (Supp. Fig. 1a and b). All mutant and wild-type (WT) proteins were expressed recombinantly in E. coli and purified (Supp. Fig. 1c; Full Methods).
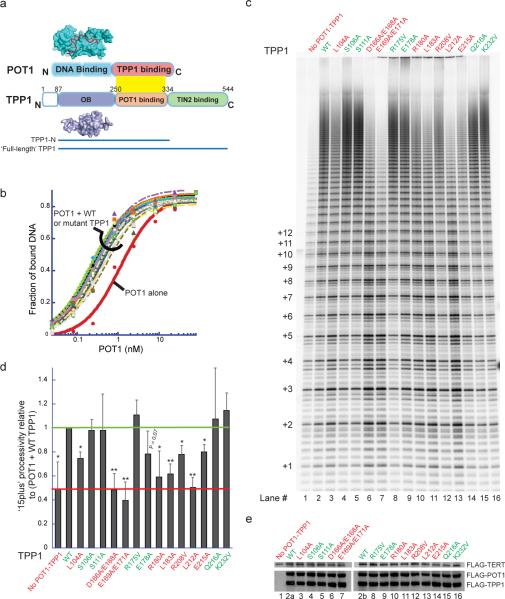
Figure 1. Separation-of-function mutants of TPP1 affect telomerase processivity without affecting telomere complex formation.
a, Domain architecture of POT1-TPP1 shown alongside the crystal structures of the DNA-binding domain of POT1 (cyan surface) bound to DNA (red ribbon; PDB: 1XJV) and the TPP1-OB domain (PDB: 2I46). b, DNA binding curves from filter-binding of mixtures containing trace amounts of 32P-labeled ssDNA (GGTTAGGGTTAG), 200 nM TPP1, and varying concentrations of POT1. See Supp. Fig. 2a for KD values. c, Direct telomerase activity assay with primer a5 of lysates from cells co-transfected with a TR plasmid and FLAG-tagged POT1, TPP1, and TERT plasmids. “No POT1-TPP1”, transfection without POT1 and TPP1. (Left), number of telomeric repeats added to primer. d, Processive extension (>15 repeats/ total) with TPP1 mutants relative to that with WT TPP1 (green line) obtained from three independent sets of experiments (as in panel c); error bars, standard deviations. Stimulation of processivity is assessed relative to the “No POT1-TPP1” negative control (red line). Two-tailed Student t-test with respect to R175V: *P<0.02, **P<0.005. Red label: significantly defective; green label: not significantly defective. e, Immunoblot of lysates used in panel c probed with anti-FLAG antibody-HRP conjugate shows uniform expression.
The POT1-TPP1 heterodimer has a higher affinity for telomeric ssDNA than does POT1 alone9, which is consistent with the role of this complex in protecting single-stranded telomeric DNA from the DNA-damage response machinery15. Quantitative DNA-binding experiments demonstrated that all the mutant proteins, like wild-type TPP1, were able to enhance POT1-DNA affinity (Fig. 1b and Supp. Fig. 2a). Furthermore, all TPP1 mutants formed stable POT1-TPP1-ssDNA ternary complexes in electrophoretic mobility shift assays (Supp. Fig. 2b). These results suggest that the TPP1-OB surface mutations do not disrupt the overall structure or the DNA-end protection function of TPP1.
Addition of purified POT1-TPP1 to telomerase enzyme activity assays enhances telomerase processivity 2–3 fold in vitro9. We developed a new direct telomerase assay in which TERT, TR, TPP1, and POT1 were recombinantly co-expressed in human cells. Lysates from these cells were used in direct telomerase activity assays involving extension of primer a5 (TTAGGGTTAGCGTTAGGG; the G-to-C mutation ensured positioning of the POT1-TPP1 complex at the 5' portion of the DNA primer, thereby providing a homogenous substrate for telomerase extension9). The fourteen TPP1 mutants varied substantially in their stimulation of processivity; some such as S106A, S111A and R175V, resulted in long extension products indicative of high processivity, similar to WT TPP1, but others, such as E169A/E171A and L212A, resulted in low-processivity patterns similar to that with no TPP1 added (Fig. 1c). Quantification of data (Fig. 1d) showed that several TPP1 mutants gave significantly decreased processivity (P < 0.005). Western blot analysis showed that TERT, POT1, and TPP1 proteins were expressed at similar levels across all transfections (Fig. 1e). Note that processivity, unlike activity, is not influenced by enzyme concentration.
As an independent test of processivity, lysates from HEK 293T cells transiently overexpressing telomerase were supplemented with purified POT1 and TPP1 proteins prior to the telomerase assay, as described previously9,11,18 (Supp. Fig. 3; see also alternative quantification of processivity in Supp. Fig. 4). This traditional assay gave results in agreement with the new co-transfection method. We also analyzed the fourteen TPP1 mutants using a primer with a purely telomeric sequence and a different sequence permutation (GGTTAGGGTTAGGGTTAG); we obtained essentially the same results as with primer a5 (Supp. Fig. 5), verifying that the TPP1 mutant phenotype was not primer-specific. All three assays identified the same separation-of-function mutants of TPP1 (L104A, D166A/E168A, E169A/E171A, R180A, L183A, and L212A), with E215A and R208V being intermediate and the other mutants having wild-type activity. Further dissection of the two double mutants into four single mutants revealed that E168, E169 and E171 but not D166 were important for telomerase stimulation (Supp. Fig. 3). Most of the TPP1 mutants that were defective in processivity also gave a statistically significant decrease in activity (Supp. Fig. 3b). In agreement with previous results9, the stimulation of activity (and its dependence on specific TPP1 residues) was observed even in the absence of added POT1 protein (Supp. Fig. 6).
The effects of TPP1 mutations might be explained by disruption of a TPP1-telomerase interaction. To test this directly, we developed a co-immunoprecipitation assay involving co-transfection of FLAG-TPP1 and untagged telomerase (TERT + TR) in HeLa-EM2-11ht cells. Inspection and quantification by immunoblot of proteins co-precipitated with anti-FLAG beads shows that TERT, but not actin, was associated with FLAG-TPP1 (Fig. 2a, compare lanes 1 and 2). More TERT protein was co-precipitated with FLAG-TPP1 when POT1, the in vivo binding partner of TPP1, was also present (compare lanes 2 and 4). Addition of primer a5 (a ligand for POT1-TPP1 and a substrate for telomerase) to lysates containing FLAG-POT1 and FLAG-TPP1 prior to immunoprecipitation increased the TPP1-telomerase interaction by ~2.5-fold compared to the basal interaction in the absence of POT1 and a5 DNA (compare lanes 2 and 6). The enhanced interaction between telomerase and POT1-TPP1 is unlikely to be occurring indirectly via bridging of TPP1 and telomerase by POT1-DNA, because FLAG-POT1 did not pull down telomerase even in the presence of a5 DNA (lane 5).
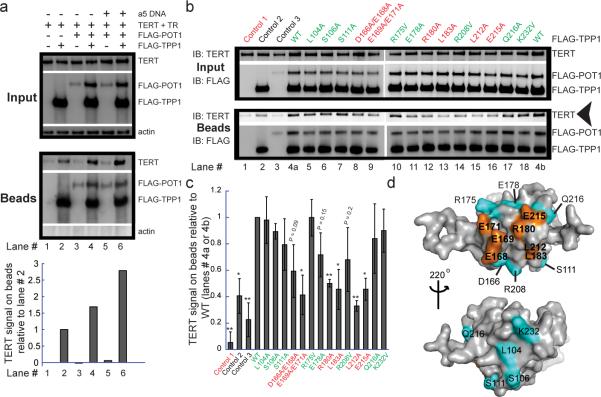
Figure 2. TPP1 mutations that disrupt telomerase stimulation also disrupt telomerase binding.
a, Pull-down of transiently expressed FLAG-TPP1, FLAG-POT1 and associated untagged TERT from HeLa-EM2-11ht lysates on anti-FLAG conjugated beads. Input, immunoblot of soluble cellular lysates prior to incubation with beads. Beads, immunoblot of proteins retained on antibody beads after 2 h incubation at 4°C and washing. The TERT signal was plotted after correction for loading differences (using the actin signal). b, Comparison of TERT pull-down by FLAG-TPP1 mutants in the presence of FLAG-POT1 and primer a5. Control 1, FLAG-POT1/TPP1 and primer a5 omitted. Control 2, FLAG-TPP1 (WT) present, FLAG-POT1 and primer omitted. Control 3, FLAG-POT1 and primer present, FLAG-TPP1 omitted. c, The mean TERT signal on beads obtained from quantification of three independent sets of experiments of which panel b is representative; error bars, standard deviations. Two-tailed Student t-test with respect to R175V: *P<0.05, **P<0.01. Red label: significantly defective; green label: not significantly defective. d, The TEL patch (amino acids in orange), the surface of the OB domain of TPP1 that mediates telomerase association and stimulation. Other amino acids mutated in this study (cyan).
The interaction between TPP1 and telomerase was reduced when either TR or TERT was omitted from co-transfections (Supp. Fig. 7a,b). These results suggest that TPP1 interacts with telomerase optimally in the context of the fully assembled RNP. The TEN (Telomerase Essential N-terminal) domain of TERT has been implicated in a functional interaction with POT1-TPP119 and in binding to TPP1-OB20. However, deletion of the TEN domain or mutating G100V19 did not abolish telomerase binding (Supp. Fig. 7c and ref 21). These data suggest that the observed immunoprecipitation of telomerase with TPP1 involves contacts both within and outside the TEN domain of TERT, which is consistent with the implication of C-terminal TERT residues in TPP1-OB-mediated recruitment to telomeres20.
When the TPP1 mutants were tested for binding telomerase (Fig. 2b,c), there was strong correlation with the telomerase stimulation results. Five of the six mutants that showed a defect in telomerase stimulation in vitro also showed defects in telomerase association in the presence of POT1 and DNA primer, and the intermediate mutant E215A was also defective in binding. An exception was mutant L104A, which showed wild-type levels of associated telomerase.
When the separation-of-function mutations that fail to stimulate processivity and also interfere with telomerase binding are mapped on the surface of the TPP1-OB structure, the seven critical amino acids (E168, E169, E171, R180, L183, L212 and E215) cluster to reveal a patch on the protein surface (orange in Fig. 2d). We name this conserved surface the TEL patch, which stands for TPP1 glutamate (E) and leucine (L)-rich patch. The fact that the TEL patch promotes both processivity and telomerase binding fulfills a prediction of a previous model for telomerase processivity11. The `back face' of TPP1-OB (bottom view of Fig. 2d) contains amino acids not involved in binding telomerase.
Previously, knockdown of TPP1 in human HTC75 cells resulted in telomere lengthening7, whereas skin keratinocytes from Tpp1D/DK5-Cre newborn mice displayed telomere shortening12. However, loss of both end-protection and telomerase-regulatory functions of TPP1 complicates interpretation of such studies. To test how the TPP1-telomerase interaction influences telomere maintenance under conditions where chromosome end-protection is unperturbed, we constructed stable cell lines containing a single-copy gene cassette with a bidirectional tetracycline-inducible promoter (Ptet-bi, Fig. 3a). The promoter expresses both an shRNA (shTPP1) to knockdown endogenous TPP1 (Fig. 3b,c and Supp. Fig. 8a–c) and a shRNA-resistant version of FLAG-TPP1 (FLAG-TPP1* or WT*; Fig. 3d and Supp. Fig. 8a,d). E169A/E171A*, R175V*, and L212A* cell lines express shRNA-resistant versions of the various mutant TPP1 proteins. (Asterisks indicate shRNA-resistant mRNAs.) We also constructed a similar set of cell lines in which various TPP1 proteins were ectopically expressed without knocking down the endogenous TPP1 (Supp. Fig. 9a,b and Supp. Fig. 10a,b).
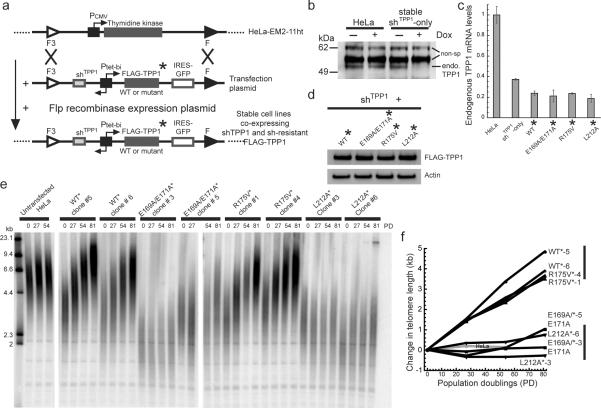
Figure 3. TPP1 TEL-patch mutants fail to stimulate telomere lengthening in human cells.
a, Engineering HeLa-EM2-11ht stable cell lines containing single-copy integration of bidirectional Tet-inducible shTPP1 and shRNA-resistant FLAG-TPP1* (WT or mutants) genes. b, A stable cell line encoding shTPP1 (no exogenous FLAG-TPP1) shows doxycycline-dependent knockdown of endogenous TPP1 protein. Non-specific bands serve as loading controls. c, Quantitative RT-PCR showing knockdown of endogenous TPP1 mRNA in the indicated cell lines ± S.D. (n =3). d, Western blot showing similar protein levels of shRNA-resistant FLAG-TPP1 in the indicated cell lines also expressing shTPP1. e, Telomeric restriction fragment (TRF) Southern blot of DNA from HeLa-EM2-11ht (Untransfected HeLa) and stable cell lines expressing shTPP1 and the indicated TPP1 constructs at the indicated population doublings (PD). Left lanes, DNA length standards. f, Change in mean telomere length for data shown in panel e was plotted against PD. The vertical bars at the right indicate the distinct ranges of telomere length attained by WT* & R175V* versus E169A/E171A* & L212A* cells after 81 PD.
Overexpression of TPP1 as described above ~25-fold; data not shown) could potentially sequester shelterin components from telomeres, leading to the appearance of Telomere-dysfunction Induced Foci (TIFs)13. However, expression of the TPP1 mutants gave the same low levels of TIFs as wild type (Supp. Fig. 11). These data are consistent with the unaltered POT1-DNA binding properties of the TPP1-OB mutants (Fig. 1b) and with a previous study showing that deletion of the entire TPP1-OB domain fails to elicit chromosome end-deprotection14.
The stable cell lines expressing wild-type and control (R175V) FLAG-TPP1 showed a robust increase in telomere length over 81 population doublings (~40 bp/PD) in a doxycycline-dependent (FLAG-TPP1 expression-dependent) manner, whereas the telomeres of the untransfected cell line were stable (Fig. 3e,f, Supp. Fig. 9c,d, and Supp. Fig. 10c,d). In stark contrast, the telomeres of cell lines expressing E169A/E171A and L212A did not change in length over 81 PDs. This result is in full agreement with our data showing that E169A/E171A and L212A have a greatly diminished ability to stimulate telomerase (Fig. 1) and to associate with telomerase (Fig. 2), but that R175V behaves like wild-type. We conclude that TPP1 increases telomerase action in vivo and that the TEL patch of TPP1 is necessary for this effect.
We tested whether the inability of the E169A/E171A and L212A mutants to stimulate telomere lengthening was due to their inability to recruit telomerase to telomeres. The cell lines stably co-expressing shTPP1 and TPP1* (WT*, E169A/E171A*, R175V*, or L212A*) were transfected with expression plasmids encoding TERT and TR and analyzed by co-immunofluorescence (co-IF) and immunofluorescence-fluorescence in situ hybridization (IF-FISH). As expected, wild-type and all mutant FLAG-TPP1 proteins localized to telomeres (Supp. Fig. 12). Telomerase was efficiently recruited to telomeres containing WT and R175V proteins, as indicated by the colocalization of TR and FLAG-TPP1 (Fig. 4a,b). By the same criterion, the E169A/E171A* and L212A* cell lines showed reduced levels of telomerase recruitment to telomeres. Instead, the TR foci in E169A/E171A* and L212A* cells resided in Cajal bodies (marked by coilin; Fig. 4c), suggesting that the corresponding TPP1 mutations prevent telomerase in Cajal bodies from being delivered to telomeres. TEL patch mutations also reduced recruitment of endogenous TR to telomeres (Supp. Fig. 13).
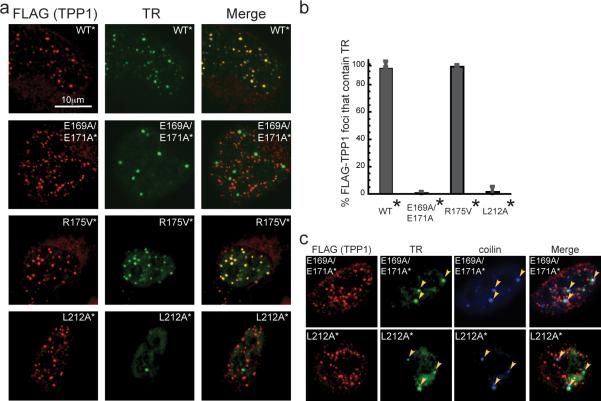
Figure 4. Failure to stimulate telomere lengthening correlates with inability to recruit telomerase to telomeres.
a, Fluorescence in situ hybridization (FISH) detects TR (green) and immunofluorescence (IF) detects the indicated FLAG-TPP1 proteins (red). Merge, Yellow spots indicate recruitment of telomerase to telomeres. b, Quantification of telomerase recruitment data of which panel a is representative. The average “% FLAG-TPP1 foci that contain TR” and standard deviations (error bars) of 15 fields of view (40–120 cells total) were plotted for the indicated stable cell lines. c, FISH-IF experiment showing that telomerase (TR in green) in E169A/E171A* and L212A* cells fails to be recruited to telomeres (FLAG-TPP1 in red) and instead remains localized in Cajal bodies (coilin in blue).
We conclude that a small patch of amino acids on the surface of the chromosome end-binding protein TPP1, the TEL patch, binds telomerase to enable both its recruitment to telomeres and its high-processivity extension of telomeric DNA. While this manuscript was under review, two groups independently reported the involvement of portions of the TEL patch in telomerase association21 and telomerase recruitment20.
In the simplest model, the TEL patch binds telomerase directly. If there were a bridging molecule, it would likely be an abundant cellular component, because TPP1-telomerase binding is robust under conditions of TPP1 and telomerase overexpression, where any telomerase-specific factors would be substoichiometric. There is precedence for such a bridging molecule in Schizosaccharomyces pombe, where Ccq1 physically connects Tpz1 (the S. pombe TPP1) to telomerase22–24. However, there is no identifiable homolog of Ccq1 in humans. (See Supp. Discussion for comparison with budding yeast recruitment.)
The substantial reduction of telomerase recruitment by subtle mutations in the TEL patch of TPP1 suggests novel strategies to inhibit telomerase for cancer therapy. By targeting the TEL patch, instead of telomerase itself, a compound could potentially inhibit telomerase action only at telomeres without interfering with other hTERT functions25,26. Furthermore, such an inhibitor should not perturb genomic stability in normal cells lacking telomerase, because the TEL patch is physically and functionally separate from the portion of TPP1 engaged in chromosome end-protection.
FULL METHODS
Oligonucleotides
Synthetic oligonucleotides used in gel-shift, filter-binding, and telomerase activity assays were purchased from Integrated DNA Technologies and resuspended in 10 mM Tris-Cl (pH 8.0) to obtain 50 μM stocks that were stored at −20°C and diluted to required concentrations immediately before use.
Plasmid constructs for cell-based assays
Mammalian cell expression plasmids encoding human TERT (pTERT-cDNA6/myc-His C18) and human TR (phTR-Bluescript II SK(+)18) were a gift from Dr. Joachim Lingner (EPFL, Lausanne, Switzerland). The pd1gfp.Ptet.miR (called pTet-BI4 in this manuscript) vector and HeLa-EM2-11ht cell line were obtained from Tet Systems Holdings GmbH & Co. KG (Germany) upon signing a materials transfer agreement. The pTet-BI4 vector, which contains a bidirectional tetracycline-inducible promoter (Ptet-bi), allows for doxycycline (dox)-dependent expression (Tet-on) of genes in the HeLa-EM2-11ht cell line, which constitutively expresses the reverse tetracycline-controlled transactivator gene28. The pIRES2-EGFP Rac2 plasmid29 that was used to PCR-amplify IRES-GFP was a gift from Gary Johnson (University of North Carolina at Chapel Hill, NC).
shRNA and shRNA-resistant constructs of TPP1
Target sequence of shRNAs shown in Supp. Fig. 8. shRNA-A: CGTTGCATCCGCTGGGTGT, shRNA-B: TGGAGTTCAAGGAGTTTGT, shRNA-C: GACTTAGATGTTCAGAAAA13. shTPP1, shown in Fig. 3, contains two repeats each of shRNA-B and -C arranged in tandem. shRNA-resistant mutations were introduced by site-directed mutagenesis using the following primers (and their reverse complements). shRNA-A resistant: GGAAACCCGGGCCCccTtCAcCCGCTcGGaGTgGCCGTGGGGATG. shRNA-B resistant: CCCCAGAAACCTAGCCTcGAaTTtAAGGAaTTcGTcGGGTTGCCCTGCAAG. shRNA-C resistant: GGGTGCCTGGTTGCAACCAAGAtcTtGATGTcCAaAAgAAGCTCTATGACTGCCTTGA GG, where lower-case indicate silent mutations introduced in the TPP1 gene to make the mRNA resistant to the indicated shRNA. In addition to possessing the duplex RNA sequence for silencing, all shRNA constructs also contained critical sequence elements derived from miRNA mR-30 for increasing the efficiency of RNA silencing30.
Immunoblotting
Standard immunoblot protocols were used with the following antibodies at specified dilutions: mouse monoclonal anti-FLAG M2-HRP conjugate (Sigma; A8592; 1:10,000), rabbit monoclonal TERT (C-term) antibody (Epitomics; 1531-1; 1:500), mouse monoclonal anti-β-actin antibody (Sigma; A5441; 1:10,000), and mouse monoclonal TPP1 antibody (Abnova; H00065057-M02; 1:1,000). Secondary horseradish peroxidase-conjugated goat antibodies against rabbit IgG (Santa Cruz Biotechnology; 1:10,000) or donkey antibodies against mouse IgG (Jackson Immunoresearch; 1:10,000) were used to reveal the primary antibodies using chemiluminescence detection by ECL plus reagents (GE Healthcare Lifesciences). The data were visualized and quantified using a FluorChem HD2 (Alpha Innotech) imaging system.
Molecular cloning
C-terminally 3X-FLAG tagged human TPP1 and C-terminally 3X-FLAG tagged human POT1 genes were amplified from hTPP1 cDNA (Open Biosystems; Thermo Scientific) and hPOT1 cDNA (pET-Smt3-POT1; unpublished results), respectively, using forward primers containing a NotI site upstream of the start codon and reverse primers that contained: a stop codon-to-serine mutation, a DNA sequence coding for a 3X-FLAG tag, and a terminal SalI site at the 5' end. For transient transfection experiments, the PCR fragments were restriction digested and cloned upstream of the IRES-GFP in the pTet-IRES-eGFP-BI4 vector (obtained by cloning a PCR-amplified and restriction digested IRES-eGFP fragment into pTet-BI4) to furnish p3X-FLAG-TPP1-BI4 and p3X-FLAG-POT1-BI4 plasmids. Using this construct, TPP1 and GFP proteins are synthesized as separate polypeptides in the cell. For stable transfection purposes, the TPP1-IRES-GFP fragment was subcloned from the p3X-FLAG-TPP1-BI4 vector to the pd1gfpPtetmiR vector (replacing the existing d1GFP gene in the vector) that contains the F/F3 Flp recombination sites flanking the multiple cloning site to yield p3X-FLAG-TPP1-F3 constructs that were subsequently used for stable transfection. p3X-FLAG-TERT-cDNA6/myc-HisC, which contains an N-terminal 3X-FLAG tag immediately upstream from the TERT start codon, was obtained via site-directed mutagenesis of pTERT-cDNA6/myc-HisC18. For stable co-transfections with shTPP1 and shTPP1-resistant TPP1, the bi-directionality of the pTet promoters of pTet-IRES-eGFP-BI4 and pd1gfpPtetmiR was exploited. Whereas the TPP1 constructs were cloned as mentioned above, the shTPP1 was cloned between the SpeI/NheI sites of the second multiple cloning site downstream of the second pTet element. The shTPP1-resistant mutations were made by site-directed mutagenesis (as detailed below).
Site-directed mutagenesis
Missense mutations were introduced into the TPP1 gene using fully complementary mutagenic primers (QuickChange® Site-Directed Mutagenesis Kit from Agilent Technologies). For the generation of TPP1-N mutants for overexpression in bacteria, the pET-Smt3-TPP1-N plasmid9 was used as the template in the PCR, whereas for expression in the HeLa-EM2-11ht cell line, p3X-FLAG-TPP1-BI4 was used as the template in the PCR. The TPP1 genes in the mutant plasmids were sequenced completely to verify the presence of the intended mutation/s and exclude the acquisition of unwanted changes during PCR amplification and cloning.
Protein purification
Purified mutant and wild-type Smt3-TPP1-N fusion proteins were obtained from soluble lysates of isopropyl β-d-thiogalactopyranoside-induced BL21(DE3) cells after nickel-agarose chromatography, treatment with Ulp1 protease to cleave the Smt3 tag31 and size-exclusion chromatography9. Purified recombinant human POT1 protein was obtained from baculovirus-infected insect cells as described previously8.
Gel-shift and filter-binding assays
Electrophoretic mobility shift assays and filter-binding assays of POT1-TPP1-oligonucleotide complexes were performed exactly as described previously32.
Cell culture
All human cells were cultured at 37°C with 5% CO2. HEK 293T cells were cultured in growth medium containing Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM Glutamax (Life Technologies), 100 units/ml penicillin, and 100 μg/ml streptomycin. HeLa-EM2-11ht and stable cell lines derived from it were propagated in growth medium containing DMEM, 10% FBS, 2 mM Glutamax, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin. Doxycycline was added to a final concentration of 200 ng/ml for induction of pTet-driven genes.
Telomerase preparations
Telomerase preparations from HEK 293T cell extracts (super-telomerase) were obtained from transient transfection of TERT and TR using a published protocol18. For telomerase preparations from HeLa-EM2-11ht cells, 300,000 cells per well of a 6-well plate were seeded and transfected 42 h later using Lipofectamine 2000 (Invitrogen) and indicated plasmid DNA using the manufacturer's recommendation. 1 μg p3X-FLAG-TERT-cDNA6/myc-HisC, and 3 μg phTR-Bluescript II SK(+)18 were added per transfection. For transfections involving POT1 or TPP1, 1 μg of p3X-FLAG-POT1-BI4 or p3X-FLAG-TPP1-BI4 was added per transfection. In control transfections where POT1 and TPP1 were omitted, pTet-BI428 (empty vector) was included. Medium was removed after 5 h and exchanged with fresh medium containing 200 ng/ml doxycycline to induce TPP1 and/or POT1 expression from the tetracycline-driven promoter. After 48 h of transfection, the cells were trypsinized, washed with phosphate buffered saline (PBS), resuspended in 100 μl CHAPS lysis buffer11 containing 1 μl of RNasin plus (Promega), mixed with rocking on a nutator at 4°C for 20 min, and centrifuged (13,600 rpm, 10 min) to remove cell debris. Aliquots of the soluble cell lysates were flash frozen in liquid nitrogen and stored at −80°C.
Telomerase activity assays
Telomerase reactions were carried out as 20 μl reactions containing: 50 mM Tris-Cl (pH 8.0), 30 mM KCl, 1 mM MgCl2, 1 mM spermidine, 5 mM β-mercaptoethanol, the indicated concentration of primer a5 or b9, 500 μM dATP, 500 μM dTTP, 2.92 μM unlabelled dGTP, 0.33 μM radiolabeled dGTP (3000 Ci/mmol), and 3 μl of super-telomerase cell extract (or telomerase from HeLa-EM2-11ht cell extracts) at 30°C for 30 min. Reactions were stopped with buffer containing 100 μl of 3.6 M ammonium acetate, 20 μg of glycogen and a 5' end-labeled (7.5 cpm/μl) 18mer oligonucleotide loading control, and precipitated with ethanol. The pellets were resuspended in 10 μl H2O and 10 μl of loading buffer (95% formamide, 5% H2O, loading dye), heated at 95°C for 10 min, and loaded onto a 10% acrylamide, 7M urea, 1X TBE sequencing gel. Gels were run at 90 W for 1.75 h, dried, exposed to an image plate and imaged on a phosphorimager (TyphoonTrio). The data were analyzed using Imagequant TL software and telomerase activities quantified as described previously11. Processivity calculations were performed as described previously11 (Supp Fig. 4) although it was not possible to include the higher molecular weight products on the gel in these calculations due to severe band overlap. To include the contributions of these upper bands (which are a result of highly processive action of telomerase; data not shown) in processivity measurements, we defined a `15plus' relative processivity term as the fraction of the total activity present in bands resulting from fifteen (number chosen arbitrarily to define the lower limit of high processivity) or more telomeric repeats added by telomerase (Fig. 1d and Supp. Fig. 3c), noting that the `15plus' method does not correct for the greater number of G nucleotides (three per repeat) in the upper bands compared to lower bands.
Co-immunoprecipitation experiments
The protocol was adapted from a previously published protocol7. HeLa-EM2-11ht cells were seeded and transfected as stated above for telomerase preparations. After 24 h of transfection, cells were washed with PBS, dislodged with a cell-scraper using 400 μl ice-cold lysis buffer [50 mM Tris-Cl (pH 7.4), 20% glycerol, 1 mM EDTA, 150 mM NaCl, 0.5% Triton X-100, 0.02% SDS, 1 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, complete protease inhibitor cocktail (Roche)] and kept on ice. After 5 min, 20 μl of 5 M NaCl was added and mixed. After another 5 min on ice, 420 μl of ice-cold water was added and mixed before immediate centrifugation (13,600 rpm, 10 min). Supernatants were collected and used directly for immunoprecipitation. Lysate (40 μl) added to 40 μl of 4X LDS sample loading buffer (Invitrogen) was kept aside for analysis of `input' samples. Anti-FLAG/M2 affinity gel beads (50 μl; Sigma) pre-incubated with 100 mg/ml of bovine serum albumin in PBS were added to lysate prepared from each well of a 6-well plate. Mixtures were mixed with rocking on a nutator for 2 h at 4°C. Beads were washed three times with 1:1 diluted lysis buffer and proteins were eluted with 180 μl 1:1 diluted 4X LDS sample loading buffer (Invitrogen) for analysis of `beads' samples. All `input' (10 μl) and `beads' (15 μl) samples were heated at 90°C for 12–14 min and analyzed by SDS-PAGE and immunoblotting.
Stable cell line generation using HeLa-EM2-11ht cells and p3X-FLAG-TPP1-F3 plasmids
Detailed protocols for stable transfection of HeLa-EM2-11ht cells are reported elsewhere33. HeLa-EM2-11ht cells were co-transfected in a 6-well format using either Lipofectamine 2000 (Life Technologies) or TransIT (Mirus) using the manufacturer's protocol with 1 μg each of the p3X-FLAG-TPP1-F3 (containing wild-type, E169A/E171A, R175V, or L212A mutants for experiments in Supp. Fig. 9, and containing shTPP1 and shTPP1-resistant wild-type, E169A/E171A, R175V, or L212A mutants for experiments in Fig. 3), and 1 μg of a plasmid expressing Flp recombinase and conferring puromycin resistance. Twelve h post-transfection, transfected cells were selected for 36 h using 5 μg/ml puromycin. Subsequently, fresh medium lacking puromycin but including 50–100 μM ganciclovir (Sigma-Aldrich) was added and negative selection was conducted for 10 days. Next, 12 individual clones were picked from each transfection and expanded. To verify the identity of the clone and distinguish it from false-positive clones that survived selection, a small aliquot of cells was induced overnight with 200 ng/ml doxycycline and observed under a fluorescence microscope (Supp. Fig. 10a). Positive clones were selected based on green fluorescence arising from their IRES-GFP locus downstream of the FLAG-TPP1 gene (Fig. 3a) and also confirmed using flow cytometry (Supp. Fig. 10b). The typical efficiency of cloning (positive clones/total survivors) using this protocol was between 8% and 50%. Positive clones were expanded until they grew to confluency in 10 cm culture dishes and induced with 200 ng/ml doxycycline for studying the effect of TPP1 mutations in cells.
Telomere length analysis
Genomic DNA was isolated from confluent 10 cm culture dishes of the original HeLa-EM2-11ht cells and FLAG-TPP1 expressing stable cell lines using the GenElute kit (Sigma). Genomic DNA (1.5 μg) was restriction digested with frequent cutters Hinf1 and Rsa1 overnight at 37°C. The DNA digests were run on a 0.8% Agarose-1X TBE gel at 50 V for a total of 1100 volt-hours. A 5' 32P-labeled (with T4 PNK; NEB) λ DNA-HindIII digest ladder (10,000 cpm) was run as a marker on a separate lane on the gel. Next morning, the gel was shaken in 0.25 M HCl for 15 min followed by two rounds (15 min each) of shaking in solution containing 0.5 M NaOH and 1.5 M NaCl. Next, the gel was shaken in solution containing 0.5 M Tris-Cl and 1.5 M NaCl (pH 7.5) for 30 min. The gel was blotted on the positively charged Hybond N+ (GE) membrane in 10X SSC buffer (1.5 M NaCl, 0.15 M sodium citrate at pH 7.2) overnight by capillary blotting. Next morning, the membrane was irradiated with UV (260 nm; set at 1200 uJ X 100 energy) and prehybridized in 15 ml Rapid-hyb (GE) for 30 min at 50°C in a hybridization oven with rotation. 5' 32P-labeled (with T4 PNK; NEB) telomeric probe of sequence (TTAGGG)4 was added (20 million cpm) and hybridization was continued for 1h at 50°C. The hybridization solution was discarded and the membrane was rinsed three times with buffer containing 0.1X SSC and 0.1% SDS. The membrane was washed three times in this buffer at 50°C, wrapped in Saran-wrap, exposed to a phosphorimager screen for 24–72 hours, and analyzed using the Imagequant TL software. The gel was calibrated using the known molecular weights of the λ DNA-HindIII ladder and the mean telomere length for each lane was plotted as a function of population doubling (PD) for each cell line. A linear regression (MS Excel) was used to calculate the rate of telomere elongation.
Quantitative RT-PCR
Total RNA was purified from the indicated cell lines using TRIzol reagent (Life Technologies). Total cDNA was prepared from total RNA using the High Capacity cDNA reverse transcription kit (Applied Biosystems). A fragment of the cDNA from endogenous TPP1 mRNA spanning a part of the 5' UTR (absent in FLAG-tagged constructs) and continuing into the ORF was amplified, and qPCR was performed with the iQ SYBR green Supermix (Biorad) using the LightCycler 480 (Roche) equipment.
Immunofluorescence (IF) and fluorescence in situ hybridization (FISH)
For IF and FISH experiments, protocols were adapted from those described previously in the literature27 and online sources (http://delangelab.rockefeller.edu/protocols.html). For TIF analysis using co-immunofluorescence (co-IF), 10,000 cells of HeLa-EM2-11ht-derived stable cell lines were seeded on coverslips in a 12 well culture plate containing growth medium adjusted to 200 ng/ml doxycycline. After 96 h, medium was removed and all subsequent steps were performed at room temperature. Cells were washed once with PBS and fixed with 3 or 4% formaldehyde in PBS for 8 min. The fixative was removed and the cells washed three times with PBS. The cells were permeablized with PBS containing 0.1% Triton X-100 (PBS-T) for 5 min and blocked in PBS-T containing nuclease-free 3% BSA for 30 min. Cells were incubated with mouse monoclonal anti-TRF2 (Imgenex; IMG-124A; 1:500) and rabbit polyclonal anti-53BP1 (Novus Biologicals; NB100–304; 1:1000 dilution) in PBS-T containing nuclease-free 3% BSA for 1 h. The cells were then washed three times in PBS (5 min each), and incubated with Alexa Fluor 568-conjugated anti-mouse IgG (Life Technologies) and Alexa Fluor 647-conjugated anti-rabbit IgG (Life Technologies) diluted 1:500 in PBS-T containing nuclease-free 3% BSA for 30 min in the dark. The cells were then washed three times in PBS and the excess PBS was removed by blotting. The coverslips were mounted on microscope slides using Vectashield mounting medium with DAPI (Vector Laboratories), sealed using transparent nail polish and stored at 4°C or −20°C until the time of imaging. Detection of FLAG-TPP1 and RAP1 by co-IF was done essentially as above, but with mouse monoclonal anti-FLAG M2 (Sigma; F1804; 1:500) and rabbit polyclonal anti-RAP1 (Novus Biologicals; NB100–292; 1:500) primary antibodies.
For combined immunofluorescence-fluorescence in situ hybridization (IF-FISH) experiments, the IF was performed before FISH. Briefly, IF was performed as described above using the appropriate primary and secondary antibodies. After the final PBS wash, the cells were fixed again in 4% formaldehyde in PBS for 10 min at room temperature. The cells were washed twice with PBS and dehydrated by successive 5 min incubations in 70%, 95%, and 100% ethanol. The ethanol was removed and the coverslips allowed to dry for 2 min. The cells were rehydrated in 50% formamide in 2× SSC for 5 min. The coverslips were placed (with cells facing down) on a drop (~40 μl) of pre-hybridization solution containing 100 mg/ml dextran sulfate, 0.125 mg/ml E. coli tRNA, 1 mg/ml nuclease-free BSA, 0.5 mg/ml salmon sperm DNA, 1 mM vanadyl ribonucleoside complexes, and 50% formamide in 2× SSC for 1 h at 37°C in a humidified chamber. Hybridization solution was made by adding a mixture of three Cy5-conjugated TR probes27 (30 ng of each probe per coverslip) to the pre-hybridization solution and the cells were hybridized in this solution overnight at 37°C in a dark humidified chamber. Next morning, the cells were washed twice in 50% formamide in 2× SSC and twice in PBS, and the coverslips were then mounted on microscope slides using ProLong Gold Antifade Reagent (Life Technologies). After 24 h at room temperature in the dark, the coverslips were sealed using transparent nail polish and stored at 4°C or −20°C until the time of imaging. In addition to antibodies mentioned above, rabbit polyclonal anti-coilin (Santa Cruz; sc-32860; 1:100), and Alexa Fluor 405-conjugated anti-mouse (and anti-rabbit) IgG antibodies (Life Technologies; 1:500) were used to stain and image FLAG-TPP1 and Cajal bodies.
Microscopy
Imaging was performed using a Nikon TE2000-U inverted fluorescence microscope equipped with Photometrics Cascade II EM-CCD camera and Yokagowa Spinning disc Confocal (CSU-Xm2) (Nikon Instruments, Inc.) and a 60× oil objective. Images were acquired using Metamorph software. Linear image adjustments were made when necessary using ImageJ and Adobe Photoshop and applied to all images in a given channel. The colors depicted in the figures do not necessarily correspond to the emission of the fluorescent labels, but are used to facilitate visualization/analysis of “merged” signals from two or more channels. Representative cells are shown in all microscopy figure panels. Quantification of colocalizations was done manually and plots of numbers of colocalizations/cell (one focal plane) show data obtained from 15 or more fields of cells (40–120 cells in total) for each sample set processed in parallel on the same day.
Supplementary Material
Acknowledgements
We thank Titia de Lange (Rockefeller University), Michael Terns (University of Georgia), and Steve Langer (University of Colorado) for suggestions and sharing protocols; Theresa Nahreini for maintenance of the departmental tissue culture facility; Jonathan Friedman and Gia Voeltz for help with confocal microscopy; and Andrea Berman, Sumit Borah, and Mai Nakashima for critical reading of the manuscript. T.R.C. is an investigator of the Howard Hughes Medical Institute (HHMI). J.N. was an HHMI fellow of the Helen Hay Whitney Foundation during a major part of this study and is supported by the National Cancer Institute of the National Institutes of Health under award number K99CA167644. This work was supported in part by US National Institutes of Health grant R01GM29090 to L.A.L. and R01GM099705 to T.R.C.
Footnotes
Supplementary information and Full Methods with associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions J.N. and T.R.C. conceived the project and designed experiments with help from I.W. and L.A.L. on biological aspects. C.F.B. with help from J.N. and A.J.Z. conducted protein purifications, DNA-binding assays and telomerase assays. J.N. and I.W. constructed the stable HeLa cell lines. J.N. performed all remaining experiments including molecular cloning, cell culture, co-IP, TRF analysis and FISH/IF. J.N. and T.R.C. wrote the paper.
Author Information The authors declare competing financial interests: T.R.C, J.N., C.F.B., and I.W. have filed a patent application relating to the identification of the TEL patch of TPP1.
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 3.Lingner J, et al. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 4.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 5.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14:1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 6.Liu D, et al. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6:673–680. doi: 10.1038/ncb1142. [DOI] [PubMed] [Google Scholar]
- 7.Ye JZ, et al. POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 2004;18:1649–1654. doi: 10.1101/gad.1215404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- 10.Lue NF. Adding to the ends: what makes telomerase processive and how important is it? Bioessays. 2004;26:955–962. doi: 10.1002/bies.20093. [DOI] [PubMed] [Google Scholar]
- 11.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tejera AM, et al. TPP1 is required for TERT recruitment, telomere elongation during nuclear reprogramming, and normal skin development in mice. Dev Cell. 2010;18:775–789. doi: 10.1016/j.devcel.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abreu E, et al. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol. 2010;30:2971–2982. doi: 10.1128/MCB.00240-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xin H, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 15.Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- 16.Guo X, et al. Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 2007;26:4709–4719. doi: 10.1038/sj.emboj.7601893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockemeyer D, et al. Telomere protection by mammalian Pot1 requires interaction with Tpp1. Nat Struct Mol Biol. 2007;14:754–761. doi: 10.1038/nsmb1270. [DOI] [PubMed] [Google Scholar]
- 18.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 2010;24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong FL, et al. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell. 2012;150:481–494. doi: 10.1016/j.cell.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sexton AN, Youmans DT, Collins K. Specificity Requirements for Human Telomere Protein Interaction with Telomerase Holoenzyme. J Biol Chem. 2012 doi: 10.1074/jbc.M112.394767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 23.Moser BA, Chang YT, Kosti J, Nakamura TM. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol. 2011;18:1408–1413. doi: 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki H, Tarumoto Y, Ishikawa F. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park JI, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majerska J, Sykorova E, Fajkus J. Non-telomeric activities of telomerase. Mol Biosyst. 2011;7:1013–1023. doi: 10.1039/c0mb00268b. [DOI] [PubMed] [Google Scholar]
- 27.Abreu E, Terns RM, Terns MP. Visualization of human telomerase localization by fluorescence microscopy techniques. Methods Mol Biol. 2011;735:125–137. doi: 10.1007/978-1-61779-092-8_12. [DOI] [PubMed] [Google Scholar]
- 28.Weidenfeld I, et al. Inducible expression of coding and inhibitory RNAs from retargetable genomic loci. Nucleic Acids Res. 2009;37:e50. doi: 10.1093/nar/gkp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abell AN, et al. Rac2D57N, a dominant inhibitory Rac2 mutant that inhibits p38 kinase signaling and prevents surface ruffling in bone-marrow-derived macrophages. J Cell Sci. 2004;117:243–255. doi: 10.1242/jcs.00853. [DOI] [PubMed] [Google Scholar]
- 30.Berger SM, et al. Quantitative analysis of conditional gene inactivation using rationally designed, tetracycline-controlled miRNAs. Nucleic Acids Res. 2010;38:e168. doi: 10.1093/nar/gkq616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 32.Nandakumar J, Podell ER, Cech TR. How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA. Proc Natl Acad Sci U S A. 2010;107:651–656. doi: 10.1073/pnas.0911099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weidenfeld I. Inducible microRNA-mediated knockdown of the endogenous human lamin A/C gene. Methods Mol Biol. 2012;815:289–305. doi: 10.1007/978-1-61779-424-7_22. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.