Abstract
Purpose
To determine whether single nucleotide polymorphisms (SNPs) in genes associated with DNA repair, cell cycle, transforming growth factor beta, tumor necrosis factor and receptor, folic acid metabolism, and angiogenesis can significantly improve the fit of the Lyman-Kutcher-Burman (LKB) normal-tissue complication probability (NTCP) model of radiation pneumonitis (RP) risk among patients with non-small cell lung cancer (NSCLC).
Methods and Materials
Sixteen SNPs from 10 different genes (XRCC1, XRCC3, APEX1, MDM2, TGFβ, TNFα, TNFR, MTHFR, MTRR, and VEGF) were genotyped in 141 NSCLC patients treated with definitive radiotherapy, with or without chemotherapy. The LKB model was used to estimate the risk of severe (Grade ≥3) RP as a function of mean lung dose (MLD), with SNPs and patient smoking status incorporated into the model as dose-modifying factors. Multivariate (MV) analyses were performed by adding significant factors to the MLD model in a forward stepwise procedure, with significance assessed using the likelihood-ratio test. Bootstrap analyses were used to assess the reproducibility of results under variations in the data.
Results
Five SNPs were selected for inclusion in the multivariate NTCP model based on MLD alone. SNPs associated with an increased risk of severe RP were in genes for TGFβ, VEGF, TNFα, XRCC1 and APEX1. With smoking status included in the MV model, the SNPs significantly associated with increased risk of RP were in genes for TGFβ, VEGF, and XRCC3. Bootstrap analyses selected a median of 4 SNPs per model fit, with the 6 genes listed above selected most often.
Conclusions
This study provides evidence that SNPs can significantly improve the predictive ability of the Lyman MLD model. With a small number of SNPs, it was possible to distinguish cohorts with >50% risk versus <10% risk of RP when exposed to high MLDs.
Keywords: SNP, NTCP, biomarker, non-small cell lung cancer
INTRODUCTION
Severe radiation pneumonitis (RP) is a major dose-limiting complication of radiotherapy (RT) for non-small cell lung cancer (NSCLC). RP risk is known to depend on the radiation dose distribution to lung, and various normal-tissue complication probability (NTCP) models, including the Lyman-Kutcher-Burman (LKB) model (1,2), have been proposed to quantify risk based on the lung dose-volume histogram (DVH). One goal of such models is to enable individualization of treatment. Patients estimated to have unacceptably high RP risk might be considered for alternative treatments, such as proton therapy, which might result in lower doses to normal lung without compromising tumor coverage. Patients with very low estimated risk might be candidates for dose escalation.
Despite the success in estimating RP risk using the LKB model, there remains substantial room for improvement. RP risk is most often modeled as a function of mean lung dose (MLD), but plots showing the relationship between MLD and RP incidence Fig 2a of [3]; Fig 3 of [4]) suggest that nearly all patients have an estimated RP risk <50% with current treatments. It is likely, however, that some patients are actually at much higher than 50% risk because of biological or clinical factors not yet identified. If such factors were incorporated into NTCP models, the process of selecting the best treatment option per patient could be improved.
Clinical evidence has long suggested that some patients are intrinsically more susceptible to radiation-induced injury than others. In the 1980s-1990s there were extensive efforts to develop radiosensitivity assays to identify such patients (5). In recent years, progress has been made in identifying genetic factors contributing to differences in radiation response. A study from our group, for example, found that patients with the TT genotype of the single nucleotide polymorphism (SNP) rs1982073:869T>C of the TGFβ gene had significantly higher RP risk than patients with the CC or CT genotype, after adjusting for differences in MLD (6). Subsequently, we found that SNPs in the base excision repair genes XRCC1 and APEX1 were also significantly associated with differences in RP incidence (7).
The goal of the present study was to investigate the extent to which estimates of RP risk based on MLD could potentially be improved by incorporating SNP information into the LKB model. Rather than performing a wide screen, a limited number of SNPs was selected for study based on mechanistic considerations. The intention of the study was not to propose a specific RP risk model with SNPs included, but to present a preliminary assessment of the degree to which genetic information might improve risk estimation and to suggest genes that might be involved.
METHODS AND MATERIALS
Patient population
The cohort for the present study consisted of patients with NSCLC who underwent definitive RT with or without concurrent chemotherapy, were included in our previous analyses of clinical and dosimetric risk factors for severe RP (4,8), and had genomic DNA samples analyzed for SNPs as part of an ongoing research project (6,7,9-11). This retrospective analysis was approved by our Institutional Review Board and carried out in compliance with the Health Insurance Portability and Accountability Act.
Scoring of radiation pneumonitis
As described previously (8), RP was scored retrospectively and graded according to the National Cancer Institute’s Common Terminology Criteria for Adverse Events (CTCAE), version 3.0. Symptomatic RP that interfered with daily activities or required administration of oxygen was scored as Grade 3; the need for assisted ventilation was scored as Grade 4; and fatal RP was scored as Grade 5.
Lung DVHs
The calculation of lung DVHs is described in detail elsewhere (12). Briefly, the DVH was computed for normal lung, defined as total lung minus gross tumor volume.
Genotyping
We genotyped 16 potentially functional SNPs in genes related to DNA repair, cell cycle, transforming growth factor beta, tumor necrosis factor and receptor, folic acid metabolism, and angiogenesis. These SNPs were selected because they cause nonsynonymous amino acid changes with previous reports of an association with cancer risk, and therefore might be expected to occur with non-zero frequency in the study population. Blood collection and sample preparation were as described previously (6). Briefly, patients were genotyped using the polymerase chain reaction restriction fragment-length polymorphism (PCR-RFLP) method.
Statistical analysis
The endpoint for analysis was severe (Grade ≥3) RP. Time to RP was measured from the start of RT. Patients not experiencing severe RP were censored at last follow-up or at recurrence, if any, to avoid potential confounding effects of retreatment on the scoring of RP.
Data were analyzed using a mixture-model version of the LKB model (12,13). In the standard LKB model, the effective dose to an organ at risk is defined as
| (1) |
where n is the volume parameter, Di is the dose to relative subvolume vi, and the sum extends over all dose bins in the DVH. Complication probability is modeled as a probit function of effective dose using parameters TD50 (the dose corresponding to a 50% incidence of the endpoint) and m (a parameter inversely related to the slope of the dose-response curve) as follows:
| (2) |
where
| (3) |
The mixture LKB model is a variation of the standard LKB model that takes duration of follow-up into account (12,13). It requires specification of the distribution of latent times at which toxicity occurs, modeled here using a lognormal function with parameters μ and σ:
| (4) |
The mixture LKB model was fitted to data using maximum-likelihood analysis; the contribution to the likelihood was NTCP×f (τ) for a patient experiencing toxicity at time τ and 1-NTCP×F(τ) for a patient followed to time τ without experiencing toxicity, where F(τ) is the cumulative distribution function corresponding to f(τ). In the present study, the volume parameter was fixed at n=1 (corresponding to the MLD model) and the latency parameters were fixed at their values estimated previously (μ=1.31 and σ=0.41 [4]).
The impact of covariates, including SNP genotypes, was modeled using dose-modifying factors (DMFs) on the effective dose. Specifically, equation (3) was replaced by
| (5) |
where DMF1 through DMFk represent the DMFs corresponding to covariates Y1 through Yk. This is mathematically equivalent to modeling their effects as DMFs on TD50, as suggested previously (12), but is conceptually different in that the DMF values obtained using these two approaches are reciprocals of one another.
Genotypes were dichotomized in two different ways, with heterozygous patients grouped once with each of the homozygous subsets, e.g., once as CC versus CT or TT and again as CC or CT versus TT. The significance of improvement in the model fit with addition of a covariate was assessed using the likelihood-ratio test. A multivariate analysis was performed by adding covariates to the mixture MLD model in a forward stepwise fashion, with the factor having the smallest P-value for improvement added to the model at each step. The stepwise procedure was terminated when no additional factors improved the model further at a significance level of P<0.05. In a second multivariate analysis, SNPs were added to the MLD model with patient smoking status incorporated using DMFs in Equation 5. Compared to current smokers, the DMFs for former smokers and nonsmokers were assumed to be1.38 and 1.77, respectively, as estimated previously (4).
Bootstrap analysis was used to investigate the stability of factors selected by the multivariate model under variations in the data. Bootstrap samples were obtained by selecting patients randomly from the study cohort, with replacement, to produce datasets having the same number of patients as the original cohort. The mixture MLD model was fitted to the data from each bootstrap sample, with SNPs added to the model using the forward stepwise procedure described above. Two bootstrap analyses were performed with 100 iterations each, one in which SNPs were added to a model including MLD alone, and another in which smoking status was also included.
RESULTS
Study cohort
Complete data (clinical outcomes, lung DVH, and SNP genotypes) were available for 141 patients. The crude incidence of Grade ≥3 RP in this cohort was 28/141 (20%). Two patients had Grade 4 RP; none had Grade 5. Clinical and treatment characteristics are listed in Table 1; in previous studies of the larger cohort from which the current study population was drawn, smoking status was the only one of these factors significantly associated with risk of severe RP after correcting for differences in MLD (8). A detailed description of the SNPs investigated in this study, the frequency of each genotype, and the crude incidence of Grade ≥3 RP per genotype are shown in Table 2.
Table 1.
Patient characteristics
| Factor | Number of patients (%) | |
|---|---|---|
| Sex | ||
| Female | 60 (43) | |
| Male | 81 (57) | |
| Age (years) | ||
| Median 62 (range 35-84) | ||
| Smoking status | ||
| Current smoker | 33 (23) | |
| Former smoker | 94 (67) | |
| Nonsmoker | 14 (10) | |
| KPS | ||
| 90 | 35 (25) | |
| 80 | 86 (61) | |
| 70 | 18 (13) | |
| 60 | 2 (1) | |
| COPD | ||
| No | 112 (79) | |
| Yes | 29 (21) | |
| Histology | ||
| Adenocarcinoma | 51 (36) | |
| Squamous cell carcinoma | 45 (32) | |
| NSC NOS | 45 (32) | |
| Clinical stage | ||
| IA | 8 (6) | |
| IB | 6 (4) | |
| IIA | 2 (1) | |
| IIB | 6 (4) | |
| IIIA | 46 (33) | |
| IIIB | 67 (48) | |
| IV | 6 (4) | |
| GTV (cm3) | ||
| Median 124 (range 1.5 – 960) | ||
| RT modality | ||
| 3D-CRT | 130 (92) | |
| IMRT | 11 (8) | |
| Delivered dose (Gy) | ||
| Median 63 (range 50.4 – 72) | ||
| Fractions per day | ||
| 1 | 94 (67) | |
| 2 | 47 (33) | |
| Fraction size (Gy) | ||
| 1.2 | 47 (33) | |
| 1.8 | 53 (38) | |
| 1.85 | 1 (1) | |
| 1.9 | 1 (1) | |
| 2 | 39 (28) | |
| Induction chemotherapy | ||
| No | 87 (62) | |
| Yes | 54 (38) | |
| Concurrent chemotherapy | ||
| No | 28 (20) | |
| Yes | 113 (80) |
Abbreviations: KPS, Karnofsky performance score; COPD, chronic obstructive pulmonary disease; NSC NOS, non-small cell lung cancer not otherwise specified; GTV, gross tumor volume; 3D-CRT, three-dimensional conformal radiotherapy; IMRT, intensity-modulated radiotherapy
Table 2.
Frequencies of SNP genotypes (most frequently occurring homozygous genotype listed first in each case) and crude incidence of Grade ≥3 radiation pneumonitis for each genotype
| SNP (rs#) | Genotype | Number of patients (%) |
Crude incidence of Grade ≥3 RP (%) |
|---|---|---|---|
| TGFβ+869 T>C (rs1982073) | |||
| TT | 46 (33) | 16 (35) ** | |
| CT | 79 (56) | 10 (13) | |
| CC | 16 (11) | 2 (13) | |
| TGFβ -509 C>T (rs1800469) | |||
| CC | 87 (62) | 21 (24) ** | |
| CT | 50 (35) | 7 (14) | |
| TT | 4 (3) | 0 (0) | |
| TGFβ+915 G>C (rs1800471) | |||
| GG | 124 (88) | 26 (21) ** | |
| CG | 14 (10) | 2 (14) | |
| CC | 3 (2) | 0 (0) | |
| VEGF-1498 T>C (rs833061) | |||
| TT | 43 (31) | 9 (21) * | |
| CT | 75 (53) | 15 (20) * | |
| CC | 23 (16) | 4 (17) | |
| VEGF-634 G>C (rs2010963) | |||
| GG | 67 (48) | 10 (15) * | |
| CG | 45 (32) | 9 (20) * | |
| CC | 29 (22) | 9 (31) | |
| VEGF+936 C>T (rs3025039) | |||
| CC | 103 (73) | 20 (19) ** | |
| CT | 36 (26) | 7 (14) | |
| TT | 2 (1) | 1 (50) | |
| TNFα-308 G>A (rs1800629) | |||
| GG | 103 (73) | 18 (17) ** | |
| AG | 34 (24) | 7 (21) | |
| AA | 4 (3) | 3 (75) | |
| TNFα-1031 T>C (rs1799964) | |||
| TT | 88 (62) | 19 (22) * | |
| CT | 44 (31) | 7 (16) * | |
| CC | 9 (6) | 2 (22) | |
| TNFRSF1B+676 T>G (rs1061622) | |||
| TT | 79 (56) | 12 (15) | |
| GT | 55 (39) | 14 (25) ** | |
| GG | 7 (7) | 2 (29) | |
| TNFRSF1B-1709 A>T (rs652625) | |||
| AA | 127 (90) | 26 (20) ** | |
| AT | 12 (9) | 1 (8) | |
| TT | 2 (1) | 1 (50) | |
| MTHFR-677 C>T, Ala222Val (rs1801133) | |||
| CC | 72 (51) | 14 (19) * | |
| CT | 51 (36) | 10 (20) * | |
| TT | 18 (13) | 4 (22) | |
| MDM2 SNP309 T > G (rs2279744) | |||
| TT | 61 (43) | 10 (16) ** | |
| GT | 62 (44) | 14 (23) | |
| GG | 18 (13) | 4 (22) | |
| MTRR 66 A>G (rs1801394) | |||
| AA | 35 (25) | 10 (29) | |
| AG | 73 (52) | 15 (21) ** | |
| GG | 33 (23) | 3 (9) | |
| XRCC1 G>A, Q399R (rs25487) | |||
| GG | 40 (28) | 5 (13) | |
| AG | 63 (45) | 11 (17) * | |
| AA | 38 (27) | 12 (32) * | |
| XRCC3 C>T, T241M (rs861539) | |||
| CC | 42 (30) | 5 (12) | |
| CT | 59 (42) | 10 (17) | |
| TT | 40 (28) | 13 (33) ** | |
| APEX1 G>T, D148E (rs1130409) | |||
| TT | 39 (28) | 5 (13) | |
| GT | 70 (50) | 15 (21) * | |
| GG | 32 (23) | 8 (25) * |
Abbreviations: SNP, single nucleotide polymorphism; RP, radiation pneumonitis
Astericks (2 per SNP) indicate the genotypes of the two patients who experienced Grade 4 RP.
Multivariate SNP analyses
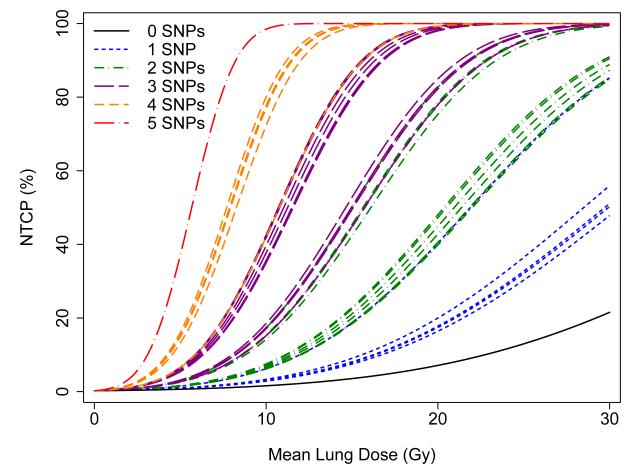
The fit of the LKB model based on MLD alone is illustrated in Figure 1. A multivariate analysis was performed in which SNPs were added to this model in a forward stepwise procedure, continuing until no additional SNPs provided a statistically significant improvement to the model fit. Five SNPs were included in the final model, in the TGFβ, TNFα, VEGF, XRCC1, and APEX1 genes; the specific SNPs selected and the model parameters are shown in Table 3. The RP risk estimates from this model fit are illustrated in Figure 2. Each curve shows the predicted incidence of RP versus MLD for patients having one of the 32 possible combinations of the 5 SNPs included in the model. Because the estimated DMFs are similar for 4 of the 5 SNPs (DMF~1.4; c.f. Table 3), the curves cluster in groups corresponding roughly to the number of adverse SNPs per patient. There are too few patients in the present cohort to illustrate the model fit for patients with specific sets of SNPs, so patients were grouped according to number of SNPs, as shown in Figure 3.
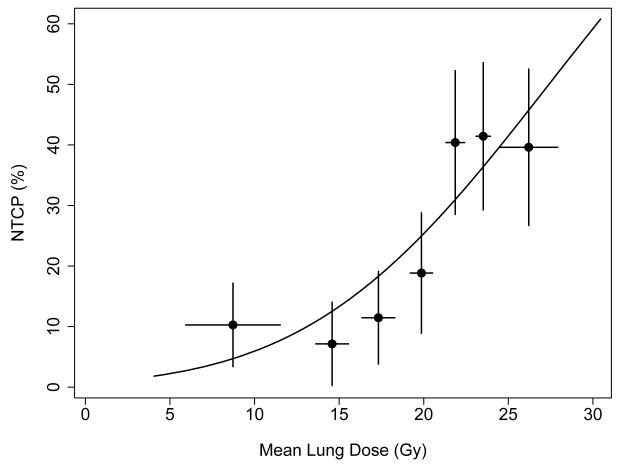
Figure 1.
Solid curve: fit of the Lyman-Kutcher-Burman model based on mean lung dose (MLD) to data from the study cohort. Parameter estimates were TD50=27.4 Gy and m=0.41. Patients were divided into 7 groups of 20-21 patients each, sorted according to MLD. Points show Kaplan-Meier estimates of Grade ≥3 radiation pneumonitis (RP) incidence at 1 year in each group, plotted at the average MLD value per group. Horizontal error bars indicate +1 standard deviation in MLD per group; vertical error bars indicate +1 standard error in RP incidence, estimated using the method of Greenwood (14).
Table 3.
Results of multivariate analyses
| Model | Parameter | Estimate |
|---|---|---|
| MLD + SNPs only | TD50 (Gy) | 41.6 |
| m | 0.354 | |
| DMF, TT allele, TGFβ+869 T>C, rs1982073 | 1.40 | |
| DMF, CC allele, VEGF-634 G>C, rs2010963 | 1.39 | |
| DMF, AA allele, TNFα-308 G>A, rs1800629 | 1.90 | |
| DMF, AA allele, XRCC1 G>A, Q399R, rs25487 | 1.46 | |
| DMF, GG allele, APEX1 G>T, D148E, rs1130409 | 1.36 | |
| MLD + SNPs + | TD50 (Gy) | 112.6 |
| smoking variables | m | 0.394 |
| DMF, Former smokers | 1.38 | |
| DMF, Nonsmokers | 1.77 | |
| DMF, TT allele, TGFβ+869 T>C, rs1982073 | 1.58 | |
| DMF, CC allele, VEGF-634 G>C, rs2010963 | 1.89 | |
| DMF, TT allele, XRCC3 C>T, T241M, rs861539 | 1.43 |
Abbreviations: SNP, single nucleotide polymorphism; MLD, mean lung dose; DMF, dose-modifying factor
Figure 2.
Predicted fit of the Lyman-Kutcher-Burman model including mean lung dose (MLD) and 5 single nucleotide polymorphisms (SNPs) in the genes listed in Table 4. Curves represent each of the 32 possible combinations of the 5 SNPs.
Figure 3.
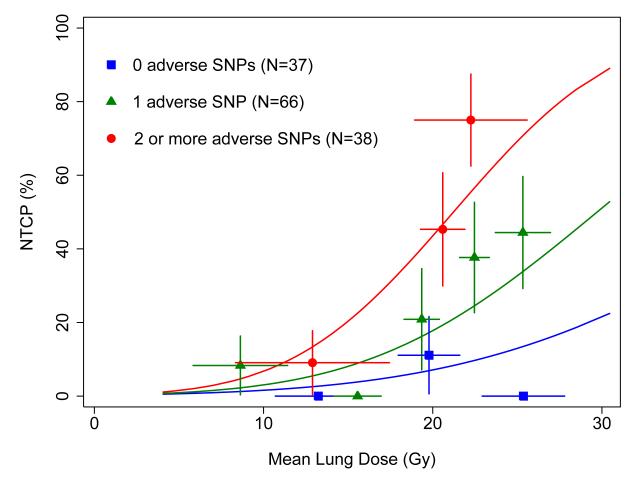
Solid curves: fit of the Lyman-Kutcher-Burman model based on mean lung dose (MLD) and number of adverse genotypes of single nucleotide polymorphisms (SNPs) in the genes listed in Table 4 (top). Patients were divided into 10 groups of 12-14 patients each according to MLD and number of SNPs. Points and error bars are as in Figure 1.
When smoking status was included in the model, three SNPs in the TGFβ VEGF, and XRCC3 genes were selected for inclusion by the multivariate analysis, as detailed in Table 3. A plot similar to that shown in Figure 2 can be obtained using these 3 SNPs, resulting in 8 curves for each of the 3 smoking categories (smokers, former smokers and nonsmokers).
Bootstrap analyses
Bootstrap analyses were performed to assess the reproducibility of variable selection in the multivariate models under variations in the data. When SNPs alone were considered as candidate factors, the median number of SNPs included in the final multivariate model per bootstrap iterate was 4 (range 1-7). The most commonly selected SNP was in the XRCC1 gene; the AA genotype was selected in 39% of iterates, with the non-GG (AA or AG) genotype selected in another 27% of cases, for a total of 66% having at least one A allele. Other SNPs selected in roughly a third or more of the bootstrap models were in genes for VEGF (VEGF-634 G>C, rs2010963; >1 C allele in 56% of models), TGFβ TGFβ+869 T>C, rs1982073; ≥1 T allele in 55%), XRCC3 (≥1 T allele in 44%), TNFα (TNFα-308 G>A, rs1800629; ≥1 A allele in 42%), APEX1 (≥1 G allele in 33%), and MTRR (≥1 G allele in 31%).
With smoking status included in the model, the median number of SNPs selected in bootstrap analyses was again 4 (range 1-7). SNPs selected most often were in genes for TGF (TGFβ+869 T>C, rs1982073; ≥1 T allele in 60% of models), TNFα (TNFα-308 G>A, rs1800629; ≥1 A allele in 53%), XRCC1 (≥1 A allele in 52%), VEGF (VEGF-634 G>C, rs2010963; ≥1 C allele in 41% of models), XRCC3 (≥1 T allele in 35%), MTRR (≥1 G allele in in 30%), and APEX1 (≥1 G allele in 25%).
DISCUSSION
This study provides evidence that SNPs in the genes for TGFβ, TNFα, VEGF, XRCC1, XRCC3, APEX1, and perhaps also MTRR, are significantly associated with differences in the risk of severe RP, after taking radiation exposure to lung (MLD) into account, with or without patient smoking status also considered. With the exception of a slight association between the genotypes of TGFβ+869 T>C, rs1982073 and TNFα-308 G>A, rs1800629 (P=0.049, chi-squared test), there were no correlations among the SNPs in these genes, indicating that the selected SNPs provide independent information about patient susceptibility to severe RP. The current study demonstrates how incorporation of genetic patient information in the form of SNPs from a relatively small set of genes can markedly improve the ability of the LKB MLD model to predict RP risk. In particular, information from SNPs can be used to stratify patients into distinct subgroups manifesting a different relationship between MLD and incidence of severe RP.
The results presented here must of course be validated, and these studies are underway. Not only do the specific SNPs identified here require confirmation for their association with an increased risk of RP, but the ways in which combinations of individual SNPs affect the relationship between MLD and complication risk also require further investigation.
A surprising finding of the present study was the large number of SNPs selected as significant by the multivariate modeling. In the model including MLD and SNPs alone, 5 SNPs were found to significantly and independently improve the fit of the model compared to use of MLD alone (Table 3, top), and a median of 4 SNPs were selected in bootstrap analyses testing the stability of variable selection. Based on many years of experience in data analysis by one of us (SLT), this is an exceptionally high number of factors selected for a data set of this size (N=141), especially since only 20% of the patients experienced the study endpoint (N=28). This finding likely reflects a strong underlying biological role played by SNPs in determining radiation response. Further evidence for this hypothesis comes from comparison of the RP incidence rates based on MLD alone versus MLD and SNPs. The incidence of severe RP in the subgroups identified using MLD alone (Figure 1) ranged from a low of ~7% to high of ~45%. When SNPs were added to the model, subgroups were identified with incidence rates over a much wider range, from 0% to nearly 80% (Figure 3).
A recent report from the inaugural meeting of a radiogenomics consortium created to establish an infrastructure for collecting data and samples from radiotherapy patients has advocated the use of genome wide association studies (GWAS) including many thousands of patients to investigate SNPs associated with the development of radiation injury to normal tissue (15). The rationale is that many GWAS have identified genes that were not previously anticipated to have an association with the phenotype under investigation. While this is indeed a worthwhile goal, the present study indicates that it will likely be possible, in the short term, to identify a relatively small number of SNPs, occurring with relatively high frequency, that have a dramatic effect on complication risk. In particular, it may be possible to begin stratifying patients by risk before the complete picture of complication susceptibility emerges from GWAS and related studies, such as those involving gene copy number or methylation status. The results of the present study also serve as a reminder that dose-volume effects should be properly taken into account when GWAS of radiosensitivity are performed using clinical data.
If confirmed, the results of the present study illustrate how genetic information might be used to guide treatment. Figure 3 shows that in the subgroup of patients with 2 or more adverse alleles, the incidence of RP is 10% when MLD is about 10 Gy. By comparison, the MLD corresponding to this same RP incidence rate is ~15 Gy in the subgroup with one adverse allele, and is ~25 Gy for the subgroup with none of the adverse alleles. Therefore, if the decision were made to treat each patient so the risk of severe RP was no more than 10%, patients in the latter group could potentially receive treatments in which the MLD values were >15 Gy higher than in the most sensitive patient group. Of course, the clinical implementation of such a scheme would need to be performed in the context of a carefully defined dose modification trial.
It is also important to note that the genes that best improve the predictive ability of the LKB model will be endpoint dependent. Some of the genes selected in the current modeling study, such as those associated with DNA repair (XRCC1, XRCC3, and APEX1), may prove to be important for many radiation-induced normal tissue endpoints. Others, such as TGFβ and TNFα, may be more strongly associated with inflammatory tissue responses, like RP, than to certain other endpoints.
CONCLUSION
The results of this study represent a proof of principle that biomarkers such as SNPs have the potential to significantly improve the predictive ability of NTCP models based on dosimetric factors alone. We are currently validating these findings in an independent cohort of patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: None
REFERENCES
- 1.Lyman JT. Complication probability as assessed from dose-volume histograms. Radiat Res Suppl. 1985;8:S13–19. [PubMed] [Google Scholar]
- 2.Kutcher GJ, Burman C. Calculation of complication probability factors for non-uniform normal tissue irradiation: the effective volume method. Int J Radiat Oncol Biol Phys. 1989;16:1623–1630. doi: 10.1016/0360-3016(89)90972-3. [DOI] [PubMed] [Google Scholar]
- 3.Marks LB, Bentzen SM, Deasy JO, et al. Radiation dose-volume effects in the lung. Int J Radiat Oncol Biol Phys. 2010;76(3):S70–S76. doi: 10.1016/j.ijrobp.2009.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tucker SL, Liu HH, Liao Z, et al. Erratum: Analysis of radiation pneumonitis risk using a generalized Lyman model. Int J Radiat Oncol Biol Phys. 2010;78:316–317. doi: 10.1016/j.ijrobp.2008.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnet NG, Johansen J, Turesson I, et al. Describing patients’ normal tissue reactions: concerning the possibility of individualizing radiotherapy dose prescriptions based on potential predictive assays of normal tissue radiosensitivity. Int. J. Cancer. 1998;79:606–613. doi: 10.1002/(sici)1097-0215(19981218)79:6<606::aid-ijc9>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Yuan X, Liao Z, Liu Z, et al. Single nucleotide polymorphism at rs1982073:T869C of the TGFβ1 gene is associated with the risk of radiation pneumonitis in patients with non-small-cell lung cancer treated with definitive radiotherapy. J Clin Oncol. 2009;27(20):3370–3378. doi: 10.1200/JCO.2008.20.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin M, Liao Z, Liu Z, et al. Functional polymorphisms of base excision repair genes XRCC1 and APEX1 predict risk of radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiotherapy. Int J Radiat Oncol Biol Phys. doi: 10.1016/j.ijrobp.2010.11.079. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jin H, Tucker SL, Liu HH, et al. Dose–volume thresholds and smoking status for the risk of treatment-related pneumonitis in inoperable non-small cell lung cancer treated with definitive radiotherapy. Radiother Oncol. 2009;91:427–432. doi: 10.1016/j.radonc.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin M, Liao Z, Huang YJ, et al. Polymorphisms of homologous recombination genes and clinical outcomes of non-small cell lung cancer patients treated with definitive radiotherapy. PLoS One. 2011;6(5):e20055. doi: 10.1371/journal.pone.0020055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan X, Yin M, Wei Q, et al. Genotypes and haplotypes of the VEGF gene and survival in locally advanced non-small cell lung cancer patients treated with chemoradiotherapy. BMC Cancer. 2010;10:431. doi: 10.1186/1471-2407-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan X, Liao Z, Ma H, et al. TNFRSF1B +676 T>G polymorphism predicts survival of non-Small cell lung cancer patients treated with chemoradiotherapy. BMC Cancer. doi: 10.1186/1471-2407-11-447. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker SL, Liu HH, Liao Z, et al. Analysis of radiation pneumonitis risk using a generalized Lyman model. Int J Radiat Oncol Biol Phys. 2008;72(2):568–74. doi: 10.1016/j.ijrobp.2008.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tucker SL, Dong L, Bosch WR, et al. Late rectal toxicity on RTOG 94-06: Analysis using a mixture Lyman model. Int J Radiat Oncol Biol Phys. 2010;78:1253–1260. doi: 10.1016/j.ijrobp.2010.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenwood M. The natural duration of cancer. Reports on Public Health and Medical Subjects. 1926;33:1–26. [Google Scholar]
- 15.West C, Rosenstein BS, et al. Establishment of a radiogenomics consortium. Int J Radiat Oncol Biol Phys. 2010;76:1295–1296. doi: 10.1016/j.ijrobp.2009.12.017. [DOI] [PubMed] [Google Scholar]