Abstract
Postmitotic neurons are subject to a vast array of environmental influences that require the nuclear integration of intracellular signaling events to promote a wide variety of neuroplastic states associated with synaptic function, circuit formation, and behavioral memory. Over the last decade, much attention has been paid to the roles of transcription and chromatin regulation in guiding fundamental aspects of neuronal function. A great deal of this work has centered on neurodevelopmental and adulthood plasticity, with increased focus in the areas of neuropharmacology and molecular psychiatry. Here, we attempt to provide a broad overview of chromatin regulation, as it relates to central nervous system (CNS) function, with specific emphasis on the modes of histone posttranslational modifications, chromatin remodeling, and histone variant exchange. Understanding the functions of chromatin in the context of the CNS will aid in the future development of pharmacological therapeutics aimed at alleviating devastating neurological disorders.
Keywords: histone PTMs, chromatin remodeling, histone variants, neuronal plasticity
INTRODUCTION
Transcriptional Control of Plasticity in the Central Nervous System (CNS)
Neuroplasticity is a multifaceted and dynamic process involving gene–environment interactions that result in both short- and long-term changes in gene expression, cellular function, circuit formation, neuronal morphology, and behavior. Elevated plasticity marks the early stages of mammalian neurodevelopment as the brain rapidly increases in size with the expansion of neuronal subtypes. Following terminal differentiation, neurons undergo periods of axonal and dendritic branching that allow for fine-tuning of synaptic contacts and the generation of elaborate circuits, many of which can persist throughout the lifetime of an individual. As the brain matures, such connections remain malleable permitting alterations in the synaptic strength of specific circuits required for various forms of experience-dependent plasticity. Throughout adulthood, environmental stimuli are continuously encoded at the level of the synapse in a process unparalleled in other tissues.
A substantial body of literature indicates that environmental stimuli experienced during early stages of neurodevelopment result in altered patterns of transcription in the brain that are essential for the establishment and maintenance of synaptic connections (Greer and Greenberg, 2008). The mammalian brain depends on numerous complex and highly regulated mechanisms to appropriately activate or silence gene programs in response to environmental input and developmental cues. At the molecular level, these events are controlled by activity-dependent signaling pathways that mediate gene expression by modifying the activity, localization, and/or expression of transcriptional-regulatory enzymes in combination with alterations in chromatin structure in the nucleus (McClung and Nestler, 2008). A large body of literature indicates that alterations in chromatin state and transcriptional programs in adult neurons are important for mediating various aspects of experience-dependent plasticity, such as learning and memory, stress responsivity, and cognition.
Chromatin Overview
Chromatin, the intimate association of genomic DNA with histone proteins, is the physiological form of our genome and the substrate for processes that regulate cellular gene expression. The fundamental repeating unit of chromatin is the nucleosome, which consists of approximately 147 bp of superhelical DNA wrapped around the radial surface of an octamer of highly conserved core histone proteins (two copies each of H2A, H2B, H3, and H4). Epigenetic mechanisms mediate distinct cellular gene expression profiles in vivo without directly affecting DNA sequence, thereby influencing transcription with far-reaching implications for human biology, health, and disease (Egger et al, 2004; Feinberg, 2007; Borrelli et al, 2008). As disruptions in chromatin are increasingly linked to human cancer, the next frontier of epigenetics, as well as human health, is to identify the role that epigenetic mechanisms might have in the potentiation of non-neoplastic diseases, including neuronal disorders.
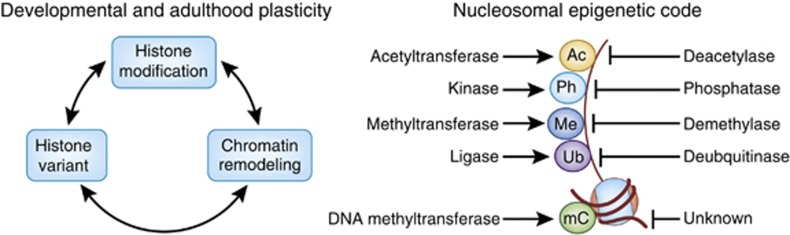
Epigenetic mechanisms regulate normal brain function, and histone modifications, along with other forms of chromatin remodeling, have been linked to neural plasticity and multiple forms of behavioral memory (Levenson and Sweatt, 2005; Borrelli et al, 2008; Dulac, 2010; Nestler, 2011). Therefore, it is not surprising that several neurological disorders involve mutations in genes that encode chromatin-binding and/or -modifying enzymes. Examples include mutations in the gene encoding the chromatin-remodeling factor, ATRX, that causes α-thalassemia and X-linked mental retardation syndrome, as well as in genes encoding the histone H3K4me3-demethylase, JARID1C, that causes epilepsy X-linked mental retardation, the histone H3K9me1/2-methyltransferase complex, G9a/GLP (EHMT2/1), that results in a human mental retardation syndrome, and the histone acetyltransferase, CREB-binding protein, that causes Rubinstein-Taybi syndrome (Gibbons et al, 1995; Alarcon et al, 2004; Kleefstra et al, 2006; Tahiliani et al, 2007; Schaefer et al, 2009). A common theme of these disorders is that mutations in epigenetic regulators can alter chromatin structure and induce a broad spectrum of neurological and behavioral deficits. Furthermore, environmentally induced alterations in chromatin structure in the absence of mutations have been shown to be critically important for a variety of neuronal functions, including those related to synaptic activity, cognition, and reward (Day and Sweatt, 2011; Robison and Nestler, 2011; West and Greenberg, 2011). In this review, we will attempt to describe the general principals of chromatin regulation as they relate to histone biology (ie, chemical modifications, chromatin remodeling, and histone variant exchange), with specific emphasis on the role that such processes may have in guiding various aspects of neural plasticity (Figure 1, left panel).
Figure 1.
Neuronal plasticity involves three major mechanisms of chromatin regulation (histone posttranslational modifications, histone variant exchange, and nucleosomal remodeling) (left panel). Nucleosomes are subject to a diverse array of covalent modifications including, but not limited to, acetylation, phosphorylation, methylation, ubiquitylation, as well as direct methylation of DNA (right panel). Note that although a bona fide DNA demethylase has yet to be described, the growth arrest and DNA damage enzyme, Gadd45, as well as oxidation reactions of 5-methylcytosine to 5-hydroxymethylcytosine, have been implicated in DNA demethylase-like activities in cells.
HISTONE MODIFICATIONS
Basic Principles of a Combinatorial Code
Histone proteins are subject to a diverse array of covalent modifications that occur primarily, but not exclusively, at amino (N-) and carboxy (C-) termini (ie, tails). The tail regions of core histones contain flexible and highly basic amino-acid sequences that are generally conserved across eukaryotic organisms, and it is well established that histone tails act as substrates for several types of posttranslational modifications, including acetylation, methylation, ADP-ribosylation, ubiquitylation, and phosphorylation (Figure 1, right panel). Such modifications have long been correlated with various nuclear functions including replication, chromatin assembly, and transcription.
Our laboratory, along with others, originally articulated the concept that distinct patterns of histone modifications act in concert with DNA methylation, noncoding RNAs (ncRNAs), and transcription factors to generate ‘histone-epigenetic codes' that are read by effector proteins (ie, regulatory molecules that bind to unmodified or modified histones, as well as specific DNA-binding complexes, to modulate chromatin function and transcriptional outputs) to elicit specific downstream transcriptional events (Strahl and Allis, 2000; Turner, 2000; Jenuwein and Allis, 2001; Jaenisch and Bird, 2003; Spitale et al, 2011). In particular, histone modifications have been demonstrated to function in a combinatorial manner, thereby increasing their indexing potentials and capacities for information storage. Roles for histone posttranslational modifications in the regulation of chromatin structure have now been extensively studied, and many groups have clearly demonstrated the validity of mapping these modifications across mammalian epigenomes. Studies of histone modifications have led to the discovery of a large number of ‘marks' (ie, histone modifications), ‘writers' (eg, enzymes such as kinases, histone acetyltransferases (HATs), and histone methyltransferases (HMTs) that modify specific substrates by adding phosphate, acetyl, or methyl groups, respectively), ‘erasers' (enzymes such as histone deacetylases (HDACs) and histone demethylases (HDMs) that catalyze the removal of specific histone modifications), and ‘readers' (these include a large number of regulatory proteins that contain unique domains that recognize acetyl, methyl, phosphate groups, etc), some of which have been linked to the aspects of neural plasticity and behavioral adaptation (Borrelli et al, 2008). Over the last decade, these concepts have gained increased attention in the field of neuroscience, as researchers attempt to identify how molecular signaling cascades encode phenotypic outputs in the nucleus to form long-lasting environmentally influenced neural circuits.
‘Active' vs ‘Repressive' Histone Modifications
Considerable research points to the critical involvement of histone modifications in transcriptional output. Among histone modifications, acetylation is by far the most extensively studied, including in the nervous system, and has been shown to directly modulate gene transcription (Brownell et al, 1996; Kundu et al, 2000). Histone methylation has also received significant attention with respect to its indexing potential, as it can exist in multiple valence states (eg, mono- (me1), di- (me2), and trimethylated (me3) forms) and exhibits slow turnover kinetics under normal cellular conditions, potentially allowing for the long-term propagation of information throughout the lifetime of an animal (ie, ‘epigenetic memory'), or even to their progeny (see Rice and Allis (2001) for a review of general differences between histone acetylation and methylation). Although evidence exists to support such propagation events throughout periods of cell division (ie, during mitosis), it is difficult to imagine how histone-based mechanisms could contribute to transgenerational inheritance of acquired parental characteristics, especially as most behavioral traits are exclusively neuronally encoded (see Future Outlooks: Histone Regulation as a Means of Generating Heritable Plasticity).
Acetylation at several lysine residues throughout the N-terminal tails of core histone proteins is generally associated with gene activation and is regarded as an epigenetic mark associated with dynamic chromatin. Acetylation acts in cis to effectively neutralize the positive charge of histone proteins, thereby decreasing the electrostatic affinity between histone tails and negatively charged DNA (Allis et al, 2007). These events ultimately function to relax chromatin structure allowing for the recruitment of various effector proteins, transcriptional coactivators (some of which contain specific acetyl–lysine recognition motifs, referred to as bromodomains), and members of the general transcriptional machinery. The enzymatic systems responsible for the addition and removal of acetyl groups have been fully characterized and consist of multiple classes of relatively promiscuous HATs, which catalyze the direct transfer of acetyl groups to histone lysine residues, and HDACs that function to remove these modifications.
Methylation of lysine and arginine residues, some of which overlap with sites of histone acetylation, can be associated with either gene activation or repression depending on the residues being modified (see Kouzarides, 2007). For example, methylation of lysine 4 or 36 on H3 is associated with transcriptional initiation/pausing and transcriptional elongation, respectively, whereas methylation of lysine 9 or 27 on H3 is more strongly correlated with transcriptional repression/silencing. As previously alluded to, categorizing methylation states as active vs repressive is further complicated because multiple methylation valences are possible, with each state being controlled by distinct ‘writers,' ‘erasers,' and ‘readers.' For example, methylation of H3K9 occurs in a seemingly non-processive manner, with the euchromatic heteromeric G9a/GLP HMT complex contributing to H3K9me1 and H3K9me2, and the heterochromatic HMT, Suv39H1, catalyzing H3K9me3. These different valence states are similarly demethylated by distinct HDMs, mainly consisting of Jumonji C (JmjC) domain-containing enzymes (eg, Jmjd2a) and are ‘read' by specific effector proteins that determine transcriptional and physiological outputs (Shinkai and Tachibana, 2011). Similar to that of acetylation, the enzymes responsible for adding methyl groups to histone tails (HMTs) have been extensively characterized. Interestingly, histone methylation was once thought to represent a stable chromatin ‘mark' that might act to control chromatin structure and the potentially related patterns of gene expression, indefinitely; however, much data now exists to refute this assumption, as numerous site and valence state-specific HDMs have been discovered (Tsukada et al, 2006).
Both histone acetylation and methylation, occurring at multiple sites throughout different histone proteins, have been the focus of an ever-growing number of studies in the field of neuroscience and appear to be especially relevant to investigations of neuropsychiatric disease (many of which will likely be discussed in subsequent reviews). It is important to consider, however, that many studies have attempted to examine the functions of histone marks in isolation, without regard to the multivalent nucleosomal landscape of the cells being examined. Such approaches require caution, as it is the combination of intra- and internucleosomal modifications that likely dictate functional outputs of chromatin. For example, a number of chromatin-associated proteins contain multiple ‘reader' domain-binding modules that are able to engage in multivalent interactions with numerous histone marks simultaneously (Ruthenburg et al, 2011). Furthermore, specific histone modifications are known to directly influence the deposition of others, as observed with H2B ubiquitination and H3K4me3 (Sun and Allis, 2002). Such interactions likely promote distinct functional outputs that cannot be predicted by a focused analysis of a single modification in isolation. Therefore, in the future, it will be necessary to employ more modern proteomic techniques (eg, large scale mass spectrometry analysis of histone modifications) to better address this issue of stimulus-driven encoding of combinatorial histone marks in neurons. Given the vast amount of literature currently available pertaining to the regulation of histone acetylation and methylation in the nervous system, we will now focus our attention on histone phosphorylation, a modification that is also likely sensitive to patterns of activity-dependent nuclear signaling and yet has received limited attention from the neuroscience community. Although not further discussed here, recent findings linking alterations in histone acetylation and methylation, as well as associated chromatin modifiers, to synaptic function, learning and memory, and psychiatric disease should inspire future studies in the field of neuroplasticity, as roles for chromatin regulation in these processes are becoming increasingly clear (Schaefer et al, 2009; Gupta et al, 2010; Maze and Nestler, 2011; Peter and Akbarian, 2011).
Histone Phosphorylation and Cross Talk Interactions in the CNS
Much less is known concerning the function of histone phosphorylation in mammalian cells. Despite its long-standing history (Stevely and Stocken, 1966; Gutierrez and Hnilica, 1967) and the presence of numerous serine, threonine, and tyrosine residues speckled throughout the primary sequences of histone proteins. Early work in this area focused primarily on the phosphorylation of linker histones (eg, H1, Sweet and Allis, 1993) and of the core histone H3 (eg, H3S10 phosphorylation (H3S10p)) as universal markers of mitotic and meiotic chromatin condensation (Wei et al, 1998, 1999). More recently, studies have begun to examine the role of phosphorylation on other core histone proteins, such as H2B, which was only recently demonstrated to function during apoptotic chromatin compaction (Ajiro, 2000; Cheung et al, 2003). To date, genome-wide analyses of histone phosphorylation marks have not been reported, making it difficult to fully appreciate the actions of these modifications in vivo.
Although H3S10p was first characterized as a mitotic signature in proliferating cells (potentially indicating a repressive function, as gene expression is significantly reduced during cell division), it is now known to exist at gene promoters in postmitotic cells and has been linked to transcriptional activation. Specifically, stimulation of growth factors and neurotransmitter-mediated kinase pathways induce rapid and transient phosphorylation of H3S10 at promoters and throughout coding regions of immediate-early genes (IEGs, Thomson et al, 1999; Clayton et al, 2000; Crosio et al, 2003). In cultured striatal neurons, for example, cAMP or glutamate stimulations have been shown to increase H3S10p enrichment at the c-fos promoter (Li et al, 2004; Brami-Cherrier et al, 2007). In the adult brain, neuronal stimulation resulting in the activated release of neurotransmitters, such as dopamine, acetylcholine, and glutamate, induce H3S10p in the dentate gyrus of the mouse hippocampus (Crosio et al, 2003). Furthermore, numerous other environmental stimuli have been found to induce H3S10p in brain, including administration of drugs of abuse (Brami-Cherrier et al, 2005) and kainic acid-induced seizures (Sng et al, 2006). At IEG promoters, H3S10p is often associated with acetylation at H3K14 (Lo et al, 2000), a modification that has been shown to be regulated by a variety of stimuli in the adult brain (eg, social stress, Covington et al, 2009) and is coupled to transcriptional activation. Combinations of specific modifications functioning together to activate gene transcription suggest the existence of common protein binders and/or concerted protein complexes that recognize multivalent histone marks.
Numerous nuclear-localized protein phosphatases and kinases have been shown to regulate histone phosphorylation across various cell types and tissues. For example, Aurora kinases A and B have been demonstrated to catalyze the addition of phosphate groups at H3S10, whereas mitogen-activated protein (MAP) kinase phosphatase-1 has previously been shown to promote the enzymatic removal of phosphate groups at this residue (Pascreau et al, 2003; Kinney et al, 2009). In brain, the dopamine and cAMP regulated protein phosphatase inhibitor, DARPP-32, as well as the MAP kinase, MSK1, provide excellent examples of enzymes regulating H3S10p in response to environmental stimuli, indicating histone phosphorylation as an important regulator of adult neuronal function (Brami-Cherrier et al, 2005; Stipanovich et al, 2008).
Similar to H3S10p, H3S28 phosphorylation has been implicated in mitotic regulation and IEG activation, with recent data indicating a direct relationship between H3S28p and transcription. Specifically, increased H3S28p, in the absence of H3S10p, was found to be sufficient to trigger the transcriptional activity of a subset of IEGs and does not appear to engage in multivalency with H3S10p within the same nucleosome (Dunn and Davie, 2005). H3S28p is believed to disrupt polycomb-mediated silencing by reducing H3K27me3, therefore allowing for the increased enrichment of H3K27ac at gene promoters and the subsequent upregulation of gene expression (Lau and Cheung, 2011). To date, this phosphomodification, along with numerous others (eg, H3T3p, H3T6p, H3T11p, H3Y41p, etc), is yet to be investigated in the nervous system. Given that neurons are highly responsive to environmental cues and are capable of rapidly responding to cellular activity through the initiation of numerous phospho-dependent signaling cascades, it is likely that histone phosphorylation states will have important roles in the integration of receptor-mediated signaling and influence transcriptional outputs necessary for proper neuronal function, either by cis- or trans-acting mechanisms (see below).
Histone Readers
Another important issue in chromatin biology and epigenetics relates to the question of how patterns of histone modifications, such as phosphorylation and acetylation, are ultimately translated into meaningful biological phenomena, both at the levels of transcription and overall chromatin structure. The identification of histone ‘reader' proteins that specifically recognize and bind histone modifications via specific histone mark-recognition motifs has greatly contributed to our mechanistic understanding of chromatin biology. For example, as H3S10p appears to be involved in both mitotic chromosomal condensation and transcriptional activation, processes that are seemingly unrelated, it stands to reason that each process might require distinct chromatin ‘readers' to appropriately execute these independent functions (ie, scenarios of broad chromatin compaction vs localized decompaction, respectively). Although the specific H3S10p readers functioning during periods of mitotic condensation have yet to be identified, it is possible that an increase in the genomic prevalence of this mark during mitosis functions to promote a binary methyl-phospho switch, which leads to the loss of heterochromatic protein 1 (HP1, an H3K9me3 reader) (Bannister et al, 2001; Lachner et al, 2001) binding to allow for proper chromosomal segregation during cell division. On the other hand, during situations of transcriptional activation, specific isoforms of the chaperone 14-3-3 have been demonstrated to directly bind H3S10p and this interaction has often been shown to increase in strength with the addition of a neighboring acetyl group at K14 (Macdonald et al, 2005). This specific phospho-acetyl combination, as well as its recognition by 14-3-3, appears to be necessary for the initiation of transcriptional elongation (Karam et al, 2010). These data further illustrate the importance of synergistic readings of distinct combinations of histone modifications, and indicate the need for future research aimed at identifying these interactions in vivo in postreplicative neurons, which likely have evolved novel mechanisms to facilitate these chromatin effector functions to meet the demands of a non-regenerative and highly plastic cellular environment.
CHROMATIN REMODELING
Basic Properties
One of the most unique properties of mammalian cells is their ability to package and sufficiently organize large amounts of DNA (∼1.7 m) into extraordinarily compact nuclei (∼5 m in diameter), thereby allowing for stable patterns of replication and transcription, which can vary greatly from tissue to tissue. Along with posttranslational modifications of histones (described above), ATP-dependent chromatin remodeling appears to be essential for both the establishment and dissolution of appropriate patterns of chromatin structural organization throughout the nucleus (Ho and Crabtree, 2010). It has been recognized for many years that nucleosomes are organized as regularly spaced, nonrandom repeating arrays, with patterns of nucleosomal spacing and occupancy varying significantly between different cell types and across organisms (Van Holde, 1989). Appropriate nucleosomal positioning and spacing patterns, as well as the ability of the cell to establish proper modes of nuclear compartmentalization and to coordinate ‘long-range' intrachromosomal interactions, are essential to all aspects of nuclear function (see Sadeh and Allis (2011) for a review of nucleosome positioning/occupancy).
Families of ATP-Dependent Chromatin-Remodeling Proteins
A large number of studies have suggested that during the transition from unicellular eukaryotes to vertebrate organisms, ATP-dependent chromatin-remodeling proteins/complexes evolved to meet the demands of a dramatically altered, and seemingly more complex, chromatin landscape. These evolutionary processes have resulted in an increased number of genes (∼30) encoding these remodeling subunits (although the increased numbers of ATPase gene products likely do not explain the full extent of the complexity observed with vertebrate remodeling complexes) and the use of combinatorial assembly, which together allow for the existence of hundreds of remodeling complexes in higher order eukaryotes. Specifically, SWI-like ATP-dependent chromatin-remodeling complexes can be categorically divided into four major subfamilies based on the structure and sequence of the ATPase subunits contained within: SWI/SNF, CHD, ISWI, and INO80 complexes. Each subfamily consists of at least one to six similar ATPases, many of which have been shown to remodel nucleosomes, transfer histone octamers in trans, and generate superhelical torsion in DNA. The assembly of ATPases into large multiprotein complexes further increases the number of specific chromatin-remodeling activities in the cell, where the same ATPase can be shared within different remodeling complexes.
Although it is true that chromatin-remodeling complexes have a vital role in guiding aspects of transcriptional regulation, it seems that these ATPase subunits do not behave in a consistent manner, making it difficult to classify these proteins simply as transcriptional activators or repressors. For example, Brahma-associated factor (BAF) complexes, which belong to the SWI/SNF family of ATP-dependent remodeling proteins, have been demonstrated to function as both repressors and activators of transcription and have further been shown to alternate these functions at a single locus (Chi, 2003). These data suggest that the relationship between chromatin-remodeling activity and transcription may be more complex than originally conceived and allow dynamic patterns of regulation through differential recruitment of effector proteins, transcription factors, and components of the general transcriptional machinery. Accordingly, BAF complexes have been demonstrated to directly interact with numerous transcription factors, oftentimes in a tissue-specific manner, thereby allowing complex subunits to acquire context-dependent, and cell type-specific, functions that are ultimately dictated by their specific interacting proteins (see Trotter and Archer (2008) for a review). Such findings indicate that although these remodeling complexes function globally in their regulation of chromatin, they may also have key roles in the precise regulation of gene programs necessary to establish distinct cellular functions. With respect to CNS development, cellular functions of BAF complexes will be described in further detail throughout this review. In addition, similar to SWI/SNF, other remodeling complexes are also necessary for many aspects of gene expression and cellular function such that ISWI remodeling complexes are important for establishing and maintaining higher order X-chromosomal structural integrity (Deuring et al, 2000), and the INO80 family of proteins appear to be necessary for telomere function and DNA replication during mitosis (see Morrison and Shen (2009) for a review).
Impact of Chromatin Remodeling on Neural Development
Recently, with respect to the development and function of the CNS, it has been argued that no other tissue within the mammalian system rivals the ability of the brain to diversify transcriptional programs to allow for such complex patterns of postmitotic gene regulation (Ho and Crabtree, 2010). These transcriptional profiles are, in turn, functionally related to the aspects of neuronal cellular identity, synapse development, circuit formation, and ultimately behavior; therefore, it is not surprising that a large number of neural-related phenotypes are beginning to emerge from genetic studies of chromatin-remodeling proteins (see Yoo and Crabtree (2009) for a review). Numerous studies now have reported the BAF family of remodeling complexes, which are based on the Brm and Brg1 core ATPases, as being essential for the development of the nervous system.
In mammals, BAF complexes are thought to be polymorphic, in that distinct subunit members, each of which are encoded by homologous gene families, assume mutually exclusive occupancy in the complex (eg, all mammalian BAF complexes contain a BAF45 subunit; however, this subunit is encoded as four distinct BAF45 isoforms (A, B, C, and D), where only one isoform can be incorporated into a single multimeric BAF complex at any given time), with the core ATPase subunit encoded by Brg1 and Brm in vertebrates (Wang et al, 1996a, 1996b; Olave et al, 2002). Distinct BAF subunits are encoded by varying number of genes allowing for numerous patterns of complex assembly; however, the core constituents of mammalian BAF complexes remain consistent. Although Brm inactivation was not shown to result in any obvious neural phenotypes (Reyes et al, 1998), Brg1 did appear to be necessary for numerous aspects of neurodevelopment. In fact, mice lacking Brg1 display pre-implantation or peri-implantation lethality, most likely due to deficits in neural tube formation based on studies performed in heterozygous mutants (Bultman et al, 2000; Lessard et al, 2007). Furthermore, mutations in BAF155, a subunit that has been proposed to shield BAF complexes from proteosomal degradation and promote the nuclear localization of the complex, similarly result in peri-implantation lethality and neural tube defects (Kim et al, 2001; Sohn et al, 2007). Unlike Brg1 inactivation in Xenopus, which promotes the expansion of neural progenitors (Seo et al, 2005), Brg1 inactivation in vertebrates was shown to interfere with neural progenitor self-renewal and results in the eventual loss of neural progenitor populations (Matsumoto et al, 2006; Lessard et al, 2007). Therefore, it can be inferred that BAF complexes function during early neurodevelopment to promote maintenance and differentiation of neural progenitors, thereby allowing for proper neural tube formation. To date, genome-wide mapping studies of BAF complex subunits in neurons have not been reported, so it remains unclear as to the exact role that BAF complexes might have in guiding chromatin-regulated transcriptional programs necessary for early neurodevelopmental processes; however, studies performed in embryonic stem (ES) cells indicate that thousands (∼10 000) of BAF-binding sites exist throughout the mammalian genome, and that nearly half of these sites localize to transcriptional start sites (Ho et al, 2009). Such patterns of BAF complex enrichment in ES cells indicate that chromatin-remodeling complexes may indeed be critical for aspects of developmental transcriptional plasticity, yet the exact roles of these genome-wide chromatin associations remain unclear.
As alluded to earlier, in vertebrates, chromatin-remodeling complexes undergo a process of combinatorial assembly, whereby distinct subunit compositions are observed between different cell types, allowing for a variety of unique protein–protein interactions between the remodeling complex and associated binders (Ho and Crabtree, 2010). These unique interactions seem to be tailored to fit the needs of specific cell types, and data indicate that such patterns of combinatorial assembly are important to neurodevelopmental processes. For example, BAF family complex members Brg1, BAF57, and BAF170 have been shown to associate as a larger complex with the neural restrictive silencing factor (NRSF/REST) and its co-repressors (eg, Sin3A, CoREST, and MeCP2) in non-neuronal cells (Ooi et al, 2006). NRSF/REST expression is restricted to non-neuronal cells, and as a zinc finger domain containing transcription factor, binds to target motifs (RE1) and represses transcription of neuronal specific genes through association with its co-repressors (Chong et al, 1995; Chen et al, 1998; Lunyak et al, 2002; Ballas et al, 2005). Recent evidence demonstrates that the repressive activity of NRSF/REST requires a functional bromodomain interaction with Brg1, whereby Brg1 is recruited to RE1 sites to synergistically control the expression of neuronal gene programs in non-neuronal cells (Ooi et al, 2006). The existence of a functional BAF complex seems to be required for NRSF/REST-mediated repression, as inactivation of Brg1 results in the unsilencing of neuronal specific genes in non-neuronal tissues. Furthermore, these events appear to concomitantly enhance histone H4 deacetylation at RE1 target loci (Watanabe et al, 2006), indicating a mechanistic link between BAF-mediated chromatin remodeling, histone posttranslational modifications, transcription factor recruitment, and gene regulation.
One can imagine a situation in neuronal cells where alterations in complex composition could result in drastically different protein–protein interactions, thereby allowing for the activation/repression of distinct transcriptional programs required for the maintenance of basal and activity-dependent neuronal function (see Yoo and Crabtree (2009) for a review). In line with this reasoning, landmark discoveries from the laboratory of Gerald Crabtree demonstrated that BAF complexes undergo a process of subunit switching during neurodevelopment, which appears to be necessary for the establishment of postmitotic cellular identity. In this work, it was discovered that during neuronal differentiation, two subunits of the neural progenitor BAF complex (npBAF), BAF45a and BAF53a, were diminished in their expression and were replaced by BAF45b and BAF53b (nBAF) specifically in postmitotic neurons (Lessard et al, 2007). Whereas the npBAF complex is functionally specialized to promote the self-renewal properties of neural stem cells, the nBAF complex seems to be dedicated to functions of fully differentiated neurons. Knockout of the nBAF complex subunit BAF53b was demonstrated to promote deficits in activity-dependent dendritic outgrowth in neurons, a phenotype that could not be rescued by add back/rescue experiments with BAF53a (Wu et al, 2007). These data suggest that neurodevelopmental patterns of combinatorial assembly are required to maintain specific biological functions in the nervous system, and that these events do not simply represent a situation of molecular redundancy. It should be noted, however, that it remains unclear how these complexes differ at the level of chromatin regulation (ie, where they associate throughout the genome, what they bind to/recruit, etc), thereby making it difficult to assess the direct vs indirect consequences of disrupting these modes of combinatorial assembly in the developing nervous system. More work is needed to mechanistically link the molecular activity of these distinct subunits to the phenotypic outcomes associated with altered neuronal function.
ATP-Dependent Chromatin Remodeling: Relevance to Neurological Disease
Although a wealth of data indicate that chromatin-remodeling activities are integral to the proper development of the nervous system, the potential impact of such activities in adult brain remain unclear. Current findings suggest that specific chromatin-remodeling enzymes/subunits have important roles in the regulation of activity-dependent transcription in the nervous system, a process important for numerous aspects of synapse development, both developmentally and in adulthood.
Genetic association studies attempting to identify putative risk alleles for schizophrenia and trisomy 21 (Down syndrome) have recently identified single-nucleotide polymorphisms (SNPs) in, or near, coding regions for specific chromatin-remodeling complex subunits. Such studies aimed at associating particular genes with neurodevelopmental disorders require identification of mutations that do not result in spontaneous abortion or other severe developmental perturbations unrelated to brain development and physiology. Specifically, SNPs in the coding region of SMARCA2, the gene that encodes BRM in the SWI/SNF-remodeling complex, were identified in association with schizophrenia, whereby risk alleles of missense SMARCA2 polymorphisms resulted in lower nuclear localization efficiencies, and risk alleles of intronic polymorphisms correlated with significantly reduced expression of SMARCA2 in adult prefrontal cortex (Sengupta et al, 2006; Koga et al, 2009). Although intriguing, these studies are presented with explicit caveats, including a lack of a genome-wide significant p-value for genetic associations, as well as the absence of an observable genetic linkage between SMARCA2 polymorphisms and schizophrenia in Caucasian individuals. Independent studies aimed at investigating the molecular mechanisms contributing to cognitive deficits characteristic of Down syndrome, have uncovered potential associations with the dual specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A), whose gene is localized in the critical region of chromosome 21 affected in Down syndrome patients. In a transgenic mouse model of Down syndrome (152F7 line), it was found that a gene dosage imbalance of Dyrk1a resulted in the deregulation of chromosomal gene clusters located in proximity to NRSF/REST target binding sites (Lepagnol-Bestel et al, 2009). It was further demonstrated that Dyrk1a, under normal circumstances, binds to the SWI/SNF BAF complex associated with NRSF/REST, and that such associations are necessary for the proper transcriptional maintenance of NRSF/REST target genes. In trisomy 21, however, DYRK1A is dysregulated, thereby leading to increased expression of NRSF/REST, subsequent disturbances in NRSF/REST-SWI/SNF complex assembly, and deficits in dendritic growth/complexity in the adult brain. Taken together, these data, along with findings demonstrating the existence of brain-specific/-restricted chromatin-remodeling proteins (eg, CHD5, a remodeling ATPase known to interact with HDAC2, which has been linked to gene expression patterns associated with aging and Alzheimer's disease) (Potts et al, 2011), demonstrate that regulatory control over chromatin-remodeling activities/enzymes in the adult CNS is important to numerous aspects of adult neurological function; however, dissecting the developmental vs adulthood contributions of such complexes to neuropsychiatric disease remains difficult.
ATRX: A Critical Regulator of Chromatin State and Histone Dynamics
Another member of the SWI/SNF family of DNA-dependent ATPases is ATRX, a protein encoded by a large X-chromosomal gene spanning greater than 300 kb of genomic DNA (Picketts et al, 1996). ATRX was originally identified because of mutations located within its coding sequence that lead to a rare mental retardation syndrome characterized by severe cognitive deficits, as well as other developmentally linked disabilities and α-thalassemia (Gibbons et al, 1995). The ATRX protein consists of a number of conserved domains that are important for its function, including a N-terminal globular domain (Argentaro et al, 2007), which is homologous to the DNMT3 family of DNA methyltransferases, known as the ATRX-DNMT3-DNMT3L (ADD) domain (the ADD domain is further composed of subdomains including GATA-1-like and plant homeodomain (PHD) zinc fingers) (Xie et al, 1999; Aapola et al, 2000). The C-terminus of ATRX contains seven conserved collinear domains conferring its ATPase activity, and other regions are important for mediating its protein–protein interactions and subnuclear localization (Bérubé, 2011) (see Figure 2 for detailed ATRX domain structure). ATRX is generally associated with repressive chromatin and has been shown to directly interact with the polycomb group protein EZH2 (Cardoso et al, 1998), the methyl-CpG-binding protein MeCP2 (Nan et al, 2007), and the heterochromatin-associated protein HP1alpha (Bérubé et al, 2000; Lechner et al, 2005). In most cell types, ATRX exclusively resides within the nucleus and tends to localize to highly repetitive sequences throughout the genome (eg, pericentromeric satellite sequences, ribosomal DNA, telomeres, etc) (McDowell et al, 1999; Law et al, 2010). In recent years, we have begun to gain better insight into the specific mechanisms controlling the tethering of ATRX to heterochromatic loci. Specifically, evidence now exists that ATRX might target chromatin directly through GATA-1-like domain interactions with the DNA template (Cardoso et al, 2000; Law et al, 2010). Alternatively, ATRX might associate with target chromatin through binding interactions with core/canonical and/or variant histone proteins, or through interactions with specific histone posttranslational modifications, either directly, or in combination with its interacting proteins HP1 (Kourmouli et al, 2005) or MeCP2 (Nan et al, 2007). It was recently demonstrated that the ADD domain of ATRX is able to efficiently bind H3K9me3 (a known heterochromatic mark) specific peptides in the absence of H3K4me3/2, due to an inability of the ATRX PHD finger domain to recognize and bind H3K4me3 (Dhayalan et al, 2011; Eustermann et al, 2011; Iwase et al, 2011). Currently, it is unclear if other adjacent chemical modifications of nucleosomes, such as H3S10p, a mark that is dramatically induced in the adult CNS in response to environmental stimulation, can impact ATRX localization in chromatin; however, given the combinatorial nature of histone modifications in vivo, it is likely that other mechanisms of ATRX recruitment/displacement will be identified.
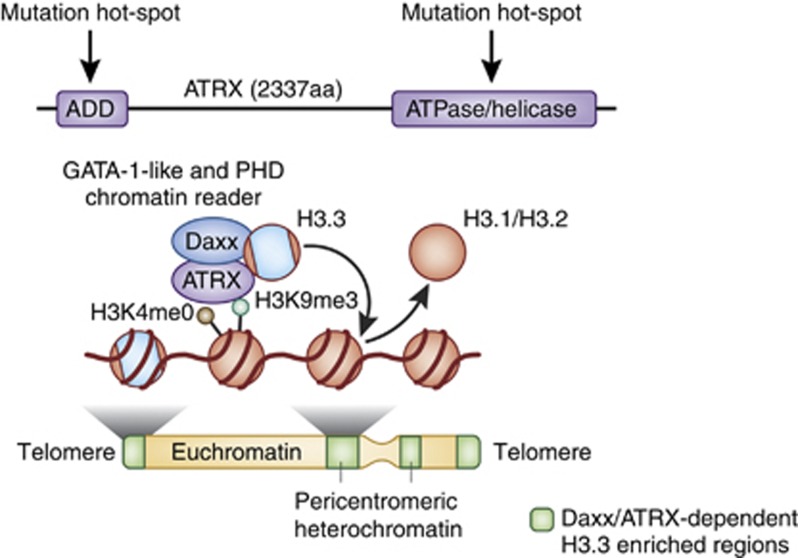
Figure 2.
The chromatin remodeler ATRX recognizes histone-modification states and modulates chromatin dynamics in vivo. The ATRX protein contains a N-terminal ADD and C-terminal ATPase domain, which function in the regulation of its binding and enzymatic activities, respectively. The ATRX ADD recognizes multiple histone marks, including H3K4me0 and H3K9me3. Daxx, an H3.3 specificity factor, and ATRX complex together as a chaperone system specifically deposit the replication-independent histone variant H3.3 at telomeric and pericentromeric heterochromatic regions of the genome. Note: mutations in ATR-X patients map to both the ATRX-DNMT3-DNMT3L (ADD) and catalytic ATPase/helicase domains.
Detailed examinations of the ATRX protein domain structure have also suggested an alternative function for this ATP-dependent remodeling protein at heterochromatin. In addition to its interactions with MeCP2 and HP1, ATRX has also been shown to directly interact with the death-associated domain protein Daxx (Xue et al, 2003; Tang et al, 2004), which is known to have an important role in apoptotic signaling in a variety of cell types, including neurons (Raoul et al, 2002; Junn et al, 2005). ATRX and Daxx are known to co-localize to pericentromeric heterochromatin and at promyelocytic leukemia bodies in vivo (Xue et al, 2003), and ATRX/Daxx complex associations have been demonstrated to exhibit ATPase-dependent mononucleosomal remodeling activity (Tang et al, 2004). Nucleosomal dynamics, specifically histone deposition and eviction, are tightly regulated by the actions of specific chaperone proteins and via the enzymatic activity of ATP-dependent remodelers. Recently, Daxx was identified as a chaperone protein for the replication-independent deposition of the histone H3 variant, H3.3, a paralog of the canonical H3 proteins, H3.1 and H3.2 (Drane et al, 2010; Lewis et al, 2010). Daxx was demonstrated to directly bind to the core AAIG motif of H3.3 (the AAIG motif partly distinguishes H3.3 from H3.1 and H3.2, as the canonical H3 variants contain a SAVM motif at amino acids 87–90), and the ATRX/Daxx complex was shown to be required for the assembly of H3.3-containing nucleosomes, specifically at pericentromeric heterochromatin, rDNA repeats, and telomeres in mouse ES cells and in mouse embryonic fibroblasts (Drane et al, 2010; Goldberg et al, 2010; Lewis et al, 2010; Wong et al, 2010) (Figure 2). Although it appears that the formation of functional ATRX/Daxx complexes is necessary for the proper establishment of specialized chromatin domains, the specific role of ATRX in this process remains unclear. ATRX seems to function primarily to maintain heterochromatic stability, in that loss of ATRX expression, at least in the case of mouse ES cells, results in the increased expression of telomeric transcripts, consistent with its role in chromatin silencing (Goldberg et al, 2010).
Alterations in the expression and/or activity of ATRX, both in rodents and humans, result in severe developmental defects, characterized by a number of negative clinical outcomes including psychomotor deficits, cognitive disability, congenital abnormalities, and α-thalassemia (Weatherall et al, 1981). Recent studies have also identified ATRX mutations in a number of cancers, including pancreatic neuroendocrine tumors and glioblastoma multiforme, suggesting that ATRX function is critically important to numerous aspect of cellular homeostasis (Heaphy et al, 2011; Schwartzentruber et al, 2012). Interestingly, Rett syndrome, a neurodevelopmental autism spectrum disorder caused by mutations in the MECP2 gene (Amir et al, 1999), has been indirectly linked to ATRX syndrome, although the direct functional consequences of disrupting ATRX/MeCP2 interactions in brain remain unclear. It seems plausible, however, that these two proteins function synergistically during neurodevelopment to regulate heterochromatic/euchromatic gene expression, as discussed above. Consistent with the notion that disruptions in heterochromatic stability are the result of ATRX dysfunction in ATR-X patients, it was discovered that many of the mutations resulting in disease phenotypes mapped to the ADD and ATPase/helicase domains of ATRX. Such mutations result in reduced expression of the protein and loss of its enzymatic activity (total null mutations in the gene encoding ATRX result in embryonic lethality) (Gibbons et al, 1995; Picketts et al, 1996). Loss-of-function and gain-of-function studies of ATRX have provided valuable insight into the consequences of ATRX dysfunction during neurodevelopment. For example, mice lacking exon 2 of ATRX, which mimics human patient mutations associated with milder forms of mental retardation, exhibit memory impairments and loss of cognitive functions, as determined through novel object recognition and contextual fear-conditioning tasks (Shioda et al, 2011). Such impairments were demonstrated to result from alterations in dendritic spine formation in the prefrontal cortex, an outcome that could be traced back to aberrant increases in the expression of CamKII, and the subsequent promulgation of CamKII-dependent phosphorylation events affecting downstream GTPase signaling in this brain region.
Broad Implications for Environmentally Influenced Chromatin Remodeling in Adult Brain
At this point, it should be apparent that neurodevelopmental disruptions in ATP-dependent remodeling activity, ATPase subunit expression and modes of complex combinatorial assembly often result in severe neurological phenotypes. However, open questions remain concerning the roles of these enzymes in the adult CNS, in the absence of mutations. For example, an expansive body of literature now indicates that a large variety of environmental stimuli (eg, stress, exposure to drugs of abuse, social interactions, sensory learning, etc) can promote alterations in the expression and activity of histone-modifying enzymes and the re-distribution of nucleosomal histone modifications throughout the genome. Given that histone-modification states and the subsequent recruitment of effector proteins are known to heavily impact chromatin-remodeling events, it seems plausible that environmentally induced alterations in the nucleosomal landscape of adult animals will result in, or will be the result of, changes in the activity of chromatin-remodeling complexes in brain. These events might occur at the level of expression or through attenuation/potentiation of distinct modes of combinatorial assembly. For example, recent studies exploring rodent models of drug abuse and stress have demonstrated dynamic alterations in the expression of H3K9me3, the heterochromatic mark recognized by the remodeling protein ATRX, throughout the brain (Hunter et al, 2009; Maze et al, 2011). These changes, although surprising, were later examined using chromatin immunoprecipitation coupled to next generation massively parallel DNA sequencing (ChIP-seq; this technique is used to map global protein/histone modification binding sites throughout the genome) to address the function of these dynamics in the adult CNS. In the case of chronic exposure to drugs of abuse, H3K9me3 was found to be significantly depleted throughout highly repetitive regions of the genome, specifically at loci encoding retrotransposable elements (eg, LINEs, SINEs) and endogenous retroviruses (Maze et al, 2011). Previously, it was demonstrated that loss of H3K9me3 enrichment at these loci correlated with increased expression of LINE elements in brain, an event that has recently been suggested to contribute to epigenetic and behavioral heterogeneity in animal populations (Singer et al, 2010). Although currently unexamined, it will be interesting to further investigate the impact of dynamic patterns of environmentally regulated H3K9me3 depletion on ATRX function in adult brain.
Given the heterogeneity of cell types throughout the CNS and the level of circuit specificity required to generate complex behaviors, it will also be necessary to further examine patterns of combinatorial assembly in adult animals exposed to a wide variety stimuli. This work will require a more detailed biochemical characterization of associations between chromatin-remodeling proteins, histone-modifying enzymes, and the histone marks, themselves. As remodeling events intrinsically result in dynamic patterns of histone exchange (ie, deposition vs eviction), more attention will need to be paid to such events, as recent studies suggest that histone turnover, under non-replicating conditions, can occur very rapidly within cells (ie, on the order of hours) (Deal et al, 2010), potentially negating the role for histone-modification dynamics at specific loci. Taken together, appropriate maintenance of neural plasticity likely requires a combination of many of these mechanistic scenarios, and caution needs to be taken in the interpretation of these events in isolation. At this point, we are arguably in the infancy of our understanding of chromatin remodeling in the adult CNS, yet to fully comprehend the contribution of histone regulation to neuropsychiatric phenomena, a more complete understanding of these processes will be required.
HISTONE VARIANTS
Background
Exciting new research indicates that in addition to nucleosomal remodeling and covalent histone modifications, eukaryotic cells generate variation in chromatin structure through the introduction of variant histone proteins. Histone variants (Allis et al, 1980) provide a means for introducing primary sequence differences that might function, at least in part, by altering the covalent modification status of these variants independently of canonical histones to expand the regulatory repertoire of chromatin (Rando and Ahmad, 2007; Henikoff, 2008). Distinct, specialized core histone proteins have been shown to differentially reside throughout specific genomic loci, including at centromeres (eg, CENP-A, a variant of H3) (Smith, 2002), within regions of ‘active' chromatin (eg, H3.3) (Ahmad and Henikoff, 2002), throughout inactivated X chromosomes in females (eg, macroH2A, a variant of H2A) (Costanzi and Pehrson, 1998), and at damage foci in chromatin following DNA double-stranded breaks (eg, γ-H2A.X) (Redon et al, 2002).
The majority of highly conserved canonical core histones (H3, H4, H2A, and H2B) are transcribed and translated in a replication/cell cycle-dependent manner in dividing metazoan cells. Many of these histone proteins are encoded from multi-copy, intronless gene clusters and are deposited in chromatin in a replication-coupled manner to allow for appropriate packaging of genomic DNA (Albig and Doenecke, 1997). All core histone proteins in mammals, with the exception of H4, have numerous sequence variants, and although much research has focused on the role of histone posttranslational modifications in the process of epigenetic inheritance (eg, genetic imprinting, where one of the two parental (maternal or paternal) alleles for a given gene is silenced in the offspring), it is becoming clear that incorporation of histone variants may provide an alternative means to encode and transmit information from one generation to the next (see Banaszynski et al (2010) for a review). Histone variants often contain minor sequence variations (eg, H3.1 vs H3.2 vs H3.3) or exhibit significantly dissimilar structures (eg, macroH2A, CENP-A) from their canonical counterparts (Rogakou and Sekeri-Pataryas, 1999). Furthermore, histone variants have been suggested to exhibit cell type-specific expression patterns, which, given the heterogeneous nature of the brain, may prove to be important for their respective functions in the nervous system.
It appears that metazoan cells have evolved histone variants, some of which are highly conserved, to accommodate the dynamic structure of the chromatin template, which is continuously subjected to factors that assemble, disrupt, and remodel nucleosomes to allow access to underlying DNA for transcriptional regulation (Ho and Crabtree, 2010). The major histone variants in mammals are deposited in chromatin in a replication-independent manner during times in which newly synthesized canonical histones are not available. For example, in terminally differentiated postmitotic neurons, which are no longer undergoing DNA replication, it can be assumed that these cells continue to have access to replication-independent histone variants, often encoded by one or two genes that are synthesized throughout the cell cycle (Frank et al, 2003). This process not only allows cells to provide continuous pools of histones for nucleosome replacement outside of S phase but also enables the generation of biochemically distinct nucleosomes that promote different patterns of chromatin regulation in cell type-specific and temporally precise manners.
H3 Variants: Basic Mechanisms
Mammalian cells express multiple distinct genetically encoded variants of histone H3 proteins (H3.1, H3.2, and H3.3/primate-specific H3.X and H3.Y (Wiedemann et al, 2010)). H3.1 and H3.2 represent canonical histones and are found throughout the genome as clustered repeat arrays, with their transcription and deposition tightly coupled to DNA replication, and their stability controlled by 3′ stem looped structures (Pandey and Marzluff, 1987). H3.3, however, is a non-canonical histone variant that is encoded by two isolated genes (H3F3A, H3F3B) and is constitutively expressed in non-dividing cells in a polyadenylated and promoter-dependent manner (Wellman et al, 1987). H3.3 differs from canonical H3 species at one amino-acid residue in the histone tail (serine 31) and at a cluster of three residues in the core histone fold (alanine 87, isoleucine 89, and glycine 90). The three amino-acid variations in the histone fold have been shown to be necessary for H3.3 replication-independent incorporation in chromatin (Ahmad and Henikoff, 2002). Although the variant histone H3.3 differs from H3.2 and H3.1 at only four and five amino acids, respectively, H3.3 is specifically enriched at transcriptionally active genes, within gene promoters, at specific genomic repeats, such as telomeres and at regulatory elements in mammalian ES cells and in neuronal precursors (Goldberg et al, 2010) (Figure 3).
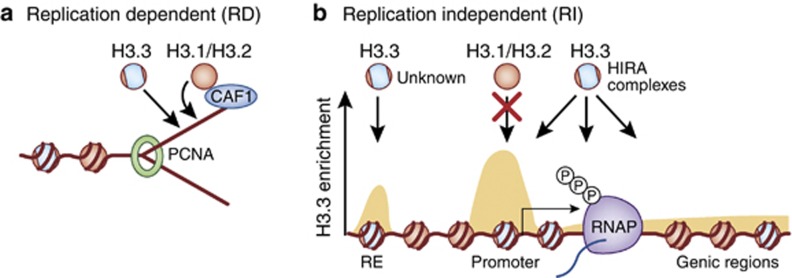
Figure 3.
The deposition of distinct H3 variant proteins involves the activity of multiple distinct chaperone complexes. (a) The replication-coupled deposition of H3.1/H3.2 is mediated by the chromatin-assembly factor 1 (CAF1) complex in association with the proliferating cell nuclear antigen (PCNA-a processivity factor for DNA polymerase in eukaryotic cells). (b) Replication-independent deposition of H3.3, however, is mediated by several HIRA complexes acting throughout genic regions of the genome, as well as by other unidentified complexes associated with H3.3 deposition at specific regulatory element (RE)-binding sites. Pericentromeric and telomeric deposition of H3.3 (not shown here, but see Figure 2) are carried out by ATRX/Daxx complexes.
As mentioned earlier, higher eukaryotes have evolved separate chaperones and deposition pathways for the different histone H3 variants, with two major deposition pathways identified through previous work; replication-coupled deposition of H3.1/H3.2 by the chromatin assembly factor 1 (CAF1) complex, and replication-independent deposition of H3.3 by the HIRA complex (see Probst et al (2009) for a review; Figure 3). HIRA (homolog of yeast Hir1) has been demonstrated to interact with ASF1a/b as a multisubunit complex specific for H3.3 chromatin deposition and has been shown to be required for H3.3 incorporation at genic loci in ES cells (Ray-Gallet et al, 2002; Tagami et al, 2004). Interestingly, H3.3 has also been shown to enrich throughout specific repetitive sequences, such as telomeres, and at most regulatory elements in a HIRA-independent manner. It is now known that the SWI/SNF chromatin remodeler ATRX, in complex with Daxx, functions to promote H3.3 localization at telomeres (Goldberg et al, 2010; Wong et al, 2010); however, the exact mechanisms controlling H3.3 enrichment at most regulatory sites throughout the genome remain unclear. Recently, HIRA was demonstrated to further interact with the SWI/SNF-remodeling protein CHD1, which has been implicated in the in vivo deposition of H3.3, and might contribute to a regulatory site-specific deposition pathway; however, H3.3 continues to be deposited in chromatin in the absence of either CHD1 or HIRA (Bonnefoy et al, 2007; Konev et al, 2007). Finally, HIRA has been shown to directly interact with Mef2 in vivo, resulting in the activation of Mef2 target genes during skeletal muscle differentiation (Yang et al, 2011). This process requires Asf1 complex association, and was demonstrated to be negatively mediated by Cabin1 binding to HIRA. Although it is unknown whether the transcriptional effects of these associations necessitate, or are responsible for, H3.3 deposition at putative target loci, one could postulate that in neurons, given the role of Mef2 in the mediation of numerous aspects of synaptic function (Flavell et al, 2006), H3.3 deposition might be tightly linked to transcriptional programs required for neural plasticity. To date, H3.3-specific deposition complexes have not been examined in postmitotic cells. Given the probable importance of the replication-independent chromatin-regulatory machinery in non-dividing cells, it is likely that future investigations in neurons will yield valuable insights into these processes.
H3 Variant Exchange: Markers of Dynamic vs Static Chromatin?
Following nucleosomal assembly and deposition, H3/H4 tetramers have been observed to display stable expression in chromatin in comparison to H2A/H2B dimers, as measured by levels of displacement during replication and transcription (Kimura, 2005). Recent cytological studies in Drosophila have demonstrated more rapid rates of H3/H4 exchange than originally predicted, indicating that nucleosomal turnover may be accelerated at specific euchromatic loci (Ahmad and Henikoff, 2002). Later studies using metabolic labeling of histones coupled to tiling arrays (CATCH-IT) provided evidence for extraordinarily rapid nucleosomal turnover kinetics and indicated H3.3 as a principal component of these dynamics (Deal et al, 2010). Genome-wide mapping studies also found that another histone variant, H2A.Z, which is similar to H3.3 in that it is encoded and deposited throughout the entirety of the cell cycle, co-localizes with H3.3 at specific promoters and regulatory elements that, due to active modes of transcriptional regulation, are oftentimes unstable and are found to be depleted of nucleosomes (termed ‘nucleosome-free regions') (Henikoff, 2009). These findings further indicate a role for H3.3 in marking/coordinating dynamic chromatin at highly active genomic loci. Consistent with these studies, ChIP-seq data from ES cells identified distinct patterns of H3.3 vs H3.2 enrichment throughout genic loci, specifically within gene bodies and at regulatory sites (Goldberg et al, 2010). Such enrichment profiles were tightly correlated with highly expressed genes, corresponding ‘active' histone modifications (eg, H3K4me1) and components of the transcriptional machinery (eg, RNA Pol II) (Ray-Gallet et al, 2011); however, enrichment at inactive genes was also observed, perhaps accounting for a ‘poised' chromatin state at these loci (Mito et al, 2007; Tamura et al, 2009). H3.3 incorporation profiles were further demonstrated to be dynamic in response to differentiation from ES cells to neuronal progenitors (Goldberg et al, 2010). Although similar studies have yet to be carried out in neurons, it will be interesting to further investigate the role of H3.3-dependent nucleosomal dynamics in the regulation of activity-dependent transcription, as plasticity in the nervous system likely necessitates rapidly re-organized and remodeled chromatin landscapes. Furthermore, if alterations in H3.3 genomic enrichment represent states of dynamic chromatin, then what are we to assume concerning the functions of H3.1 and H3.2 in the regulation of chromatin architecture? Although a new study using heavy isotope metabolic labeling of proteins in rats suggests that both H3.3 and canonical H3 proteins, as well as other variants including H2A.Z, exhibit extreme stability in rodent brain (ie, stable heavy peptides were observed up to 12 months following metabolic labeling) (Savas et al, 2012), it remains unclear as to what role, if any, H3.1 and H3.2 proteins might have in the adult CNS. On the basis of previous mechanistic studies (outlined above), one would assume that following terminal neuronal differentiation, canonical H3 proteins would no longer be able to be actively incorporated in chromatin. Therefore, if these proteins remain stable for long periods of time in brain, keeping in mind that an obvious caveat of this study was their inability to distinguish between cell types (ie, samples most likely contained a mixture of neurons, glia, etc), then one could assume that H3.1 and H3.2 act as markers of ‘static' or ‘inert' chromatin, potentially enriching throughout heterochromatic loci that are not prone to rapid turnover, unlike those expected to be occupied by H3.3. To date, these questions remain unanswered; however, data are beginning to be generated to support such a hypothesis. A quantitative mass spectrometry study aimed at examining global posttranslational-modification states between human H3.1, H3.2 and H3.3 identified differences in modification profiles, suggesting that they exhibit unique biological functions in vivo (Hake et al, 2006). Specifically, H3.3 was shown to enrich for marks associated with active chromatin (eg, lysine 9/14 acetylation, lysine 36 trimethylation, etc), whereas H3.2 contained mainly repression-associated modifications (eg, lysine 27 trimethylation). Interestingly, H3.1 was observed to enrich for both active and repressive modifications, although the repressive marks appeared to differ from those observed with H3.2. At this time, it is impossible to directly extrapolate these findings to brain, but these data indicate that H3 variants might function independently to maintain distinct chromatin states throughout the lifetime of an animal.
As described above, one interesting feature of H3.3 vs H3.1/H3.2 is the existence of a serine residue at position 31 in its N-terminal tail region (H3.3S31) that replaces an alanine found in canonical H3 proteins. H3.3S31 was demonstrated to be phosphorylated during mitosis in mammalian cells with temporal and spatial specificity in contrast to H3S10 and H3S28 phosphorylation (Hake et al, 2005). Although H3.3S31p likely contributes to the aspects of chromatin signaling during mitosis, a single replacement of the serine with an alanine at position 31 did not influence the deposition pathway of H3.3, suggesting that this phosphorylation event is not critical for H3.3 incorporation in chromatin. At this time, it is unknown whether this modification exists in neurons; however, given the potential importance of H3.3 in postmitotic cells, future attention should be paid to this possibility. Furthermore, another H3 mark, H3 threonine 32 phosphorylation (H3T32p), exists in all H3 variants; however, only in the H3.3 variant is H3T32 sequence identical to H3T11, phosphorylation of which, like H3S10p, is important for meiosis in proliferating cells (ie, only in this variant is H3T32 preceded by a serine residue at position 31, similar to Thr11, which is preceded by Serine 10) (Govin et al, 2010). Therefore, it is possible that this mark, in combination with H3.3S31p, represents a novel chromatin signature involved in cell-type-specific gene-regulatory patterns. It is becoming increasingly clear that more thorough analyses of histone-modification states in brain need to be performed. Mass spectrometric analysis has begun to identify potentially novel modification sites, as well as novel combinations of histone modifications in brain. To this effect, it will be important for future proteomic investigations to carefully examine H3-variant modification states specifically in neurons, both basally and in response to a variety of environmental stimuli.
Histone Variants in Neuroscience
Proteins involved in chromatin remodeling and regulation of histone modifications have been shown to have critical roles in processes contributing to neurodevelopment, neuronal physiology, synaptic plasticity, and behavioral memory (Borrelli et al, 2008). As chromatin maintenance in non-dividing cells relies on the incorporation of replication-independent histone variants, it is likely that histone variants will greatly impact neuronal biology. In fact, several lines of evidence support this hypothesis, with numerous histone variants (eg, H2AX, H3.3, macroH2A) likely contributing to the basic aspects of neuronal function. In line with H3.3's prominent role in postreplicative nucleosomal exchange, H3f3a RNA and H3.3 protein levels have been shown to increase in numerous models of cell differentiation (Wu and Bonner, 1982; Lord et al, 1990; Krimer et al, 1993), and high levels of H3.3 protein have been observed in rat brain (Pina and Suau, 1987). Specifically, the latter study observed a progressive increase in H3.3 protein expression, with concomitant reductions in H3.1/2, in rat cortex with age, suggesting that H3.3 is the dominant H3 protein in adult brain. However, as with the metabolic labeling study demonstrating the existence of long-lived H3.3 and H3.2 peptides in brain, no distinctions were made between cell types, so it is difficult to know whether such changes are neuronal specific. Furthermore, this study similarly demonstrated the presence of H3.1/2 expression in aged adult brain, albeit at lower levels than H3.3; therefore, future studies will be needed to fully dissect these expression pattern differences, as well as the genomic distributions and functions of H3 variants in the developing and adult CNS.
To date, very little is known concerning the biological functions of histone variants. In fact, only recently have patient mutations been identified to suggest that these variants, themselves, may be important for biological homeostasis (eg, mutations in H3F3A, specifically at lysine 27 and glycine 34, have been implicated in pediatric glioblastoma) (Schwartzentruber et al, 2012). Interestingly, in Drosophila, deletion of both H3.3 variant gene copies (A and B) results in partial but incomplete lethality, with ∼42% viability accompanied by transcriptional defects and complete sterility (Hodl and Basler, 2009; Sakai et al, 2009). Hypomorphic H3.3 mice, generated by retroviral gene trap insertion into the H3f3a gene, similarly exhibit partial neonatal lethality and deficits in fertility (Couldrey et al, 1999). In addition, H3.3 hypomorphs also display neuromuscular defects, suggesting an essential role for H3.3 in postmitotic neurons; however, the precise function of H3.3 in adult brain remains unknown, as no true knockout studies, conditional or otherwise, have been performed. Although a functional role for H3.3 in neurodevelopment has yet to be truly established, one can speculate that alterations in H3.3 deposition pathways may contribute to these processes, as mutations in ATRX, a bona fide H3.3 chaperone, result in an X-linked mental retardation phenotype. Additionally, a recent study found that calcineurin-mediated dephosphorylation of Daxx during periods of cellular activity results in increased loading of H3.3 at IEG regulatory elements in neurons (Michod et al, 2012). Although compelling, the precise mechanisms mediating Daxx localization at euchromatic loci, as well as the functional consequences of H3.3 deposition in neurons, remain unclear. Other histone variants, such as H2AX, H2A.Z, and macroH2A, are highly expressed in adult neurons (with H2AX also displaying progressive increases in expression with age in rat cortex) (Bosch and Suau, 1995; Akbarian et al, 2001); however, beyond a very basic understanding of histone variant expression in brain, little is known concerning the functions of these variants in the developing and adult nervous systems. When one considers the formal possibility that variants of one particular family can ‘mix' with other histone variants from other families, both within single mononucleosomal units and throughout localized nucleosomal arrays, a staggering potential exists in distinct biological readouts. Compositional variation at the nucleosomal level has been described as a ‘nuclesome code' (Bernstein and Hake, 2006), whose potential in neuronal functions has yet to be fully appreciated or explored.
FUTURE OUTLOOKS
Reconciling the Dangers of Hyper-Enthusiasm with Cautious Interpretations
Over the past decade, neuroscientists have made great strides in advancing our understanding of the role of chromatin biology/histone regulation throughout numerous aspects of neurodevelopment, neuroplasticity, neurophysiology, and behavior. During this time, the field has experienced an explosively renewed interest in the molecular mechanisms controlling complex behaviors associated with neuropsychiatric disease, and data are beginning to suggest that histone regulation/dysregulation might have an integral role in mediating these processes. Although these important discoveries certainly deserve celebration, it is our view that the field has only begun to scratch the surface of these complicated patterns of cellular/organismal regulation. Future attempts to define the precise chromatin-associated mechanisms involved in complex behavioral states will require a combination of more detailed biochemical and molecular approaches to appropriately integrate mechanistic analyses with descriptive output. Until such time that more mechanistic insights are ascertained, it is important that researchers approach their findings with a level of cautious optimism and interpret their data accordingly.
A growing trend in the field of ‘neuroepigenetics' is the desire to assign chromatin-related functions to alterations in the expression and/or activity of known chromatin-modifying enzymes and remodeling proteins. This current focus is logical, as a system does not currently exist to directly manipulate histone marks in mammalian cells due to the large number of redundant/clustered histone genes encoded in vertebrates. Although many chromatin-modifying enzymes (eg, HATs, HDACs, HMTs, HDMs, etc) have been demonstrated to display dynamic, and sometimes persistent, patterns of regulation in brain, events that are often correlated with global changes in histone-modification states, it is becoming increasingly clear that many, if not all, of these enzymes are capable of modifying non-histone substrates (both nuclear and cytosolic), thereby complicating functional interpretations of knockout/knockdown and overexpression studies. Although it is likely that endogenous alterations in enzyme activity contribute to changing chromatin landscapes, such assertions are difficult to causally define. Numerous groups have begun to address this issue through mutational studies, where it is possible to directly control/manipulate the catalytic activity and/or binding properties of chromatin-modifying/-remodeling enzymes (eg, acetyl bromodomains, methyl chromodomains, etc) to assess chromatin-related enzymatic and recruitment functions in vivo; however, until all chromatin-associated enzymes, many of which are likely still to be identified, are fully biochemically characterized, it will be very difficult to assign direct chromatin functions to stimulus-dependent alterations in enzymatic activity.
Similarly, it is tempting to assume that environmentally induced alterations in global histone-modification states are directly involved in the regulation of activity-dependent gene transcription in brain; however, to date, such contributions have been difficult to demonstrate outside of indirect correlations. Although it has been known for years that specific histone modifications (keeping in mind that only select modifications have been examined) are capable of influencing basal transcription (eg, H3K4me3), it remains unclear as to the exact role that stimulus-induced changes in histone modifications might have in the regulation of neuronal transcription following periods of cellular activity. Also, caution should be taken in broadly assigning functionality to specific histone modifications (eg, H3K4me3 is activating vs H3K27me3 is repressive), as rarely do these modifications exist in isolation, but rather in combination with other marks to allow for a large variety of intra- and internucleosomal associations through recruitment of effector proteins. Moreover, individual lysine residues are well known to be acetylated, methylated, or most recently crotonylated (eg, H3K9, H3K27, etc), leading to distinct biological readouts due to highly regulated ‘switching' events that are only beginning to be appreciated. We wonder if every exposed lysine residue has multiple options depending upon its physiological context, and whether the brain might draw heavily upon an array of posttranslational modifications and variant choices to guide phenotypic outcomes.
Additionally, in eukaryotic genetic model organisms, like Saccharomyces cerevisiae, a model that allows for direct mutations of histones in vivo, the majority of histone N-terminal tail point mutants have been demonstrated to be completely viable despite apparent evolutionary pressures to conserve near-invariant histone amino-acid sequences (Hyland et al, 2005; Dai et al, 2008). Although specific core amino acids located at the H3/H4 histone-fold motif have been shown to be important for aspects of transcriptional silencing in yeast (this core is centered around lysine 79 of H3 and may be important to nucleosomal stability) (Park et al, 2002), these data indicate a high level of functional redundancy in histone modifications that must be considered. With the advent of ChIP-seq, we are now beginning to gain a clearer picture of the relative distribution of histone marks throughout the mammalian epigenome, both basally and following periods of cellular activity. These data will ultimately allow us to compile comprehensive maps of histone modifications, transcription factors, remodeling complexes, etc to accurately describe associations between transcriptional outputs and histone regulation in the nervous system. It is important to keep in mind, however, that these approaches, along with global analyses of histone marks, rely heavily on the use of commercially available modification-specific antibodies, which, according to recent reports, vary greatly in their efficiencies and epitope specificities (Egelhofer et al, 2011). In some cases, the existence of one mark can greatly influence the epitope accessibility of another, especially when modifications occur in close proximity. For example, a phosphorylation event at one residue might greatly impact the ability of an antibody to recognize a methylation state at an adjacent residue (eg, H3S10p vs H3K9me3) (Duan et al, 2008), even if these marks occur in combination as identified by mass spectrometry. As a consequence, great care should be taken to appropriately validate modification-specific antibodies, as the quality of an antibody can greatly affect one's results and subsequent interpretations. As novel modifications (eg, histone–lysine crotonylation) (Tan et al, 2011), or combinations thereof, are discovered (some of which may be brain specific), it will be important to integrate these findings into the existing neuroepigenetic framework to allow for a more comprehensive understanding of chromatin biology in the context of neurological function and disease. Furthermore, viewing patterns of histone regulation in relation to other important regulatory events in the nucleus (eg, DNA methylation, ncRNA function, etc) will be essential to future investigations.
Finally, due to the cellular heterogeneity of neuronal tissues, one of the major challenges of investigating histone regulation in the CNS is our current difficulty in defining individual chromatin states and remodeling activities within specific functional regions of the brain, in neuronal subtypes (eg, dopaminergic, cholinergic, glutamatergic, GABAergic, etc) and between neurons and other cell types of the CNS (eg, glia), all of which likely vary significantly in global chromatin profiles, which support unique transcriptional outputs and cellular functions. Recent advances in the field of neuroscience have made it possible to isolate pure neuronal cell populations using fluorescence-activated cell sorting, which relies on endogenous cellular markers or transgenic tags (eg, green fluorescent protein (GFP) expressed from neuronal subtype specific promoters), to appropriately distinguish and separate distinct cell types in adult brain (Jiang et al, 2008). One potential limitation of this approach, however, lies in the processing and dissociation of cells required to sort individual cell populations for future chromatin analysis. It remains unclear if such processing can influence the chromatin state of cells, especially in the absence of fixation, and whether this technique is ultimately amenable to analysis of subtle changes in chromatin landscapes following different environmental perturbations. Nonetheless, such approaches have greatly advanced the field by allowing neuronal cell type-specific analysis in vivo and should inspire confidence in future studies aimed at tackling these difficult questions.
Histone Regulation as a Means of Generating Heritable Plasticity
One question that still remains in the field of neuroepigentics is whether environmentally mediated alterations in chromatin modifications/structure can contribute to the transgenerational transmission of complex behaviors. Although it is tempting to think that such scenarios are possible, it is difficult to conceptualize how this would occur at the level of histones. Limited examples exist to indicate that this form of epigenetic inheritance occurs during mitosis, in which ‘information' contained within focal nuclear microenvironments of parental cells can be accurately inherited by postreplicative daughter cells to allow for physiologically relevant gene expression profiles in progeny (see Zaidi et al (2010) for a review). Such sustained patterns of transcription have been shown to be accompanied by the retention of specific transcription factors, as well as associated histone modifications and patterns of DNA methylation, at promoters in a process known as mitotic bookmarking. Although these events have been suggested to represent a novel mechanism of inheritable epigenetic control, thereby allowing for the sustainability of cellular identities following mitosis, it remains unclear as to whether histone modifications themselves are important for this process, or if they simply exist as a result of the preservation of transcription factor binding at these sites following cell division.
It should be noted that reliable transmission of physiological and behavioral traits have now been observed through the paternal lineage (eg, (Ng et al, 2010; Dietz et al, 2011)), suggesting that mechanisms may indeed exist to allow for such instances of transgenerational inheritance. Some early evidence indicates a role for DNA methylation in this process; however, the exact mechanisms controlling this transmission remain unknown. During spermatogenesis, the majority of canonical histones are exchanged for protamines, which are small, highly basic proteins that promote the formation of tightly packed DNA structures important for normal sperm function. Recent high-throughput sequencing experiments have demonstrated that only ∼4% of the haploid genome is occupied by nucleosomes consisting of both canonical and variant histone proteins, some of which are testes specific (Hammoud et al, 2009). Although a number of histone modifications have been identified in sperm (eg, H3K4me2/3, H3K27me3), they tend to localize to very specific developmental loci, including HOX gene clusters, certain ncRNAs, imprinted genes, and bivalent promoters. Therefore, given that the majority of changes in environmentally regulated histone modifications in brain have been found to affect neuronal specific/enriched genes, it seems unlikely that such changes would be observed in, or would be transmitted by, sperm. Also, the heavily condensed state of sperm would appear to act as a barrier for the transmission of ‘active' chromatin, which would likely be required to transmit many of the aberrant transcriptional states described in brain following periods of plasticity. As far as maternal germ-line transmission of behaviors is concerned, much more work is needed to appropriately control for gestational influences, as in utero experiences likely supersede mechanisms of strict epigenetic inheritance and are difficult to avoid in the absence of in vitro fertilization, which itself can affect phenotypic outcomes.
Alternatively, it seems plausible that the establishment or dissolution of histone marks in brain following periods of behavioral adaptation act to initially alter chromatin structure through modulation of chromatin-remodeling events, which may subsequently lead to persistent changes in chromatin architecture and transcriptional activity. Such events ultimately result in what can be referred to as a state of chromatin metaplasticity, and it is our view that these global changes in chromatin function, even in the absence of long-lived modification changes, likely contribute to life-long patterns of behavioral plasticity. Our review has focused primarily on chromatin-remodeling pathways with an emphasis on histone-centric events. However, we acknowledge that a complete understanding of brain functions will require integration of what we have described here with other well-established epigenetic events, such as DNA methylation (see Deaton and Bird (2011) for a review), targeting of these machineries by ncRNAs (see Wang and Chang (2011) for a review) and, most certainly, other activities that have yet to be described that collectively influence the complexities of a chromatin environment. Finally, we note that non-histone proteins themselves are known to undergo extensive posttranslational modifications, sometimes reflecting closely what is seen in histone proteins. The existence of ‘histone mimicry' reminds us that the general paradigm of using this covalent ‘language' of marks, writers, readers, and erasers points to a protein ‘code' that is not limited to histones alone (Sampath et al, 2007). A future challenge will be to determine the likely key target substrates of these activities, many of which may lie beyond histone proteins.
The Holy Grail of Neuroepigenetic Research: Identification of Druggable Targets for Future Neuropsychiatric Pharmacological Intervention
It comes as no surprise that such rapid advances in neuropsychiatric/neuroepigenetic research has led to heightened enthusiasm for the development and clinical implementation of pharmacotherapies aimed at targeting chromatin-related enzymes. For example, HDAC inhibitors (HDACi) are now being used routinely in the clinic to treat a variety of human illnesses, including numerous cancers, parasitic infections, and inflammatory diseases. Although much less is known concerning the actions of HDACi in the alleviation of neuropsychiatric and neurodegenerative disorders, some deacetylase inhibitors, such as valproic acid, are commonly used to treat symptoms related to bipolar disorder (Emrich et al, 1981), and rodent work has indicated that other HDACi may prove effective in treating a variety of neurological maladies including depression-related illnesses (Covington et al, 2009), drug addiction (Romieu et al, 2008), Huntington's disease, and Parkinson's disease (see Abel and Zukin (2008) for review). These inhibitor therapies range significantly in terms of their specificity and efficacy, with numerous drugs now designed to target individual chromatin-modifying enzymes (HDACs, HMTs, etc) with extremely high-binding efficiencies. Although potentially promising as future treatments for psychiatric disorders, to date, all inhibitors are designed to target chromatin-modifying enzymes, as opposed to attributes of the histone modifications themselves. As indicated previously, chromatin-regulatory proteins are highly promiscuous in function and substrate specificity. Therefore, by inhibiting the activity of these enzymes, we are indirectly affecting more global aspects of nuclear function (ie, off-target substrate effects of histone and non-histone proteins), which may ultimately result in aberrant clinical outcomes. Indeed, the use of HDACi to treat cancer can result in severe side effects and variable efficacies (see Venugopal and Evans (2011) for review), many of which cannot be rationalized in the treatment of neuropsychiatric disorders. Another point of interest in attempting to identify druggable targets concerns the question of where we should be focusing the majority of our attention? For a number of years, it has been the goal of molecular neurobiology to identify specific genes/proteins that are dysregulated in disease, and then to use this information to develop pharmacological therapeutics; however, it is becoming clear that neuropsychiatric disorders result from, or can be characterized by, more global alterations in chromatin structure, as numerous associated remodeling and/or modifying enzymes have been demonstrated to be important for phenotypic outcomes. Therefore, it is likely that treating neurological diseases will require large-scale manipulations of chromatin regulation; however, it will be important to allow for aspects of targeted genomic specificity, a property that currently available inhibitors do not offer. Recently, inhibitors have been generated to specifically target chromatin-binding modules in regulatory proteins, which do not directly affect catalytic activity and often display higher substrate specificity, providing potentially more suitable alternatives for future neuropharmacological interventions. In 2010, a cell-permeable small molecule (JQ1) that competitively binds to acetyl–lysine recognition motifs (a.k.a. bromodomains) was characterized and shown to act as a highly potent and specific inhibitor of the bromodomain and extra-terminal (BET) family member BRD4 (Filippakopoulos et al, 2010). Such inhibition by JQ1 was later demonstrated to promote antiproliferative effects in a model of human squamous carcinoma, validating the possibility that such drugs can be used in a biologically relevant manner. Although very few of these binding-module inhibitors currently exist, it seems highly probable that similar targeting strategies may prove beneficial to the alleviation of numerous neurological diseases. Much more work is needed, however, to identify chromatin-binding proteins that may serve as suitable targets.
Conclusion
In conclusion, chromatin/histone regulation appears to have important roles in both the developing and adult CNS, and this regulation seems to be critical to many aspects of neural plasticity that directly influence the establishment of complex behavioral phenotypes. Although alterations in chromatin stability seem to contribute greatly to disease, including neuropsychiatric phenomena, the precise mechanisms underlying these clinically relevant outcomes remain unclear. As we learn more about these processes, we are beginning to truly appreciate the biological complexities of human psychiatric illness and are starting to gain a better understanding of how to clinically treat these disorders based upon their molecular and biochemical attributes. Although much more work is needed, advances in neuroepigenetics research are now occurring at a rapid pace and promise to greatly contribute to the future of molecular medicine.
Acknowledgments
We express our apologies to colleagues whose work could not be included in this review. We thank Drs Rosemary Bagot and Herbert E Covington III, as well as anonymous reviewers for critical reading of this manuscript and the members of the Allis laboratory for their helpful discussions. K-MN is supported by a Women and Science postdoctoral fellowship. IM, K-MN, and CDA are supported, in part, by grants from the NIMH (RO1 MH094698-01, P50 MH096890-01).
The authors declare no conflict of interest.
References
- Aapola U, Shibuya K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K, Henikoff S.2002The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly Mol Cell 91191–1200.This work was the first to demonstrate that H3.3 is deposited in chromatin in a replication-independent manner, and further provided evidence for a role of H3.3 in marking active chromatin. [DOI] [PubMed] [Google Scholar]
- Ajiro K. Histone H2B phosphorylation in mammalian apoptotic cells. An association with DNA fragmentation. J Biol Chem. 2000;275:439–443. doi: 10.1074/jbc.275.1.439. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Chen RZ, Gribnau J, Rasmussen TP, Fong H, Jaenisch R, et al. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol Disease. 2001;8:784–791. doi: 10.1006/nbdi.2001.0420. [DOI] [PubMed] [Google Scholar]
- Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, et al. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Albig W, Doenecke D. The human histone gene cluster at the D6S105 locus. Hum Genet. 1997;101:284–294. doi: 10.1007/s004390050630. [DOI] [PubMed] [Google Scholar]
- Allis CD, Glover CV, Bowen JK, Gorovsky MA. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980;20:609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T, Reinberg D.2007Epigenetics CSHL Press; Cold Spring Harbor, NY. [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Argentaro A, Yang JC, Chapman L, Kowalczyk MS, Gibbons RJ, Higgs DR, et al. Structural consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L (ADD) domain of the chromatin-associated protein ATRX. Proc Natl Acad Sci USA. 2007;104:11939–11944. doi: 10.1073/pnas.0704057104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Allis CD, Lewis PW. Histone variants in metazoan development. Dev Cell. 2010;19:662–674. doi: 10.1016/j.devcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Hake SB. The nucleosome: a little variation goes a long way. Int J Biochem Cell Biol. 2006;84:505–517. doi: 10.1139/o06-085. [DOI] [PubMed] [Google Scholar]
- Bérubé NG. ATRX in chromatin assembly and genome architecture during development and disease. Biochem Cell Biol. 2011;89:435–444. doi: 10.1139/o11-038. [DOI] [PubMed] [Google Scholar]
- Bérubé NG, Smeenk CA, Picketts DJ. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum Mol Genet. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- Bonnefoy E, Orsi GA, Couble P, Loppin B. The essential role of Drosophila HIRA for de novo assembly of paternal chromatin at fertilization. PLoS Genet. 2007;3:1991–2006. doi: 10.1371/journal.pgen.0030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron. 2008;60:961–974. doi: 10.1016/j.neuron.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch A, Suau P. Changes in core histone variant composition in differentiating neurons: the roles of differential turnover and synthesis rates. Eur J Cell Biol. 1995;68:220–225. [PubMed] [Google Scholar]
- Brami-Cherrier K, Lavaur J, Pages C, Arthur JS, Caboche J. Glutamate induces histone H3 phosphorylation but not acetylation in striatal neurons: role of mitogen- and stress-activated kinase-1. J Neurochem. 2007;101:697–708. doi: 10.1111/j.1471-4159.2006.04352.x. [DOI] [PubMed] [Google Scholar]
- Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, et al. 1996Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation Cell 84843–851.This pioneering manuscript was the first to provide evidence directly linking Gcn5 mediated histone acetylation to active gene regulation. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cardoso C, Lutz Y, Mignon C, Compe E, Depetris D, Mattei MG, et al. ATR-X mutations cause impaired nuclear location and altered DNA binding properties of the XNP/ATR-X protein. J Med Genet. 2000;37:746–751. doi: 10.1136/jmg.37.10.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso C, Timsit S, Villard L, Khrestchatisky M, Fontes M, Colleaux L. Specific interaction between the XNP/ATR-X gene product and the SET domain of the human EZH2 protein. Hum Mol Genet. 1998;7:679–684. doi: 10.1093/hmg/7.4.679. [DOI] [PubMed] [Google Scholar]
- Chen ZF, Paquette AJ, Anderson DJ. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–517. doi: 10.1016/s0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- Chi T. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Chong JA, Tapia-Ramirez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, et al. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Clayton AL, Rose S, Barratt MJ, Mahadevan LC. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 2000;19:3714–3726. doi: 10.1093/emboj/19.14.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Couldrey C, Carlton MBL, Nolan PM, Colledge WH, Evans MJ. A retroviral gene trap insertion into the histone 3.3A gene causes partial neonatal lethality, stunted growth, neuromuscular deficits and male sub-fertility in transgenic mice. Hum Mol Genet. 1999;8:2489–2495. doi: 10.1093/hmg/8.13.2489. [DOI] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, LaPlant QC, Vialou VF, Ohnishi YN, Berton O, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29:11451–11460. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P.2003Chromatin remodeling and neuronal response: multiple signaling pathways induce specific histone H3 modifications and early gene expression in hippocampal neurons J Cell Sci 1164905–4914.First work to indicate that numerous plasticity related signaling cascades in the CNS are linked to H3S10 phosphorylation and H3K14 acetylation, marks associated with dynamic chromatin remodeling and gene activation. [DOI] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70:813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S.2010Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones Science 3281161–1164.Using a newly developed CATCH-IT technique to metabolically label and monitor histone incorporation ratesin vivo, this manuscript demonstrated rapid histone turnover kinetics, suggesting that variant histone proteins (eg, H3.3) may display significantly lower stability in chromatin in comparison to canonical histones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuring R, Fanti L, Armstrong JA, Sarte M, Papoulas O, Prestel M, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- Dhayalan A, Tamas R, Bock I, Tattermusch A, Dimitrova E, Kudithipudi S, et al. The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9. Hum Mol Genet. 2011;20:2195–2203. doi: 10.1093/hmg/ddr107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz DM, LaPlant Q, Watts EL, Hodes GE, Russo SJ, Feng J, et al. Paternal transmission of stress-induced pathologies. Biol Psychiatry. 2011;70:408–414. doi: 10.1016/j.biopsych.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q, Chen H, Costa M, Dai W. Phosphorylation of H3S10 blocks the access of H3K9 by specific antibodies and histone methyltransferase. Implication in regulating chromatin dynamics and epigenetic inheritance during mitosis. J Biol Chem. 2008;283:33585–33590. doi: 10.1074/jbc.M803312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KL, Davie JR. Stimulation of the Ras-MAPK pathway leads to independent phosphorylation of histone H3 on serine 10 and 28. Oncogene. 2005;24:3492–3502. doi: 10.1038/sj.onc.1208521. [DOI] [PubMed] [Google Scholar]
- Egelhofer TA, Minoda A, Klugman S, Lee K, Kolasinska-Zwierz P, Alekseyenko AA, et al. An assessment of histone-modification antibody quality. Nat Struct Mol Biol. 2011;18:91–93. doi: 10.1038/nsmb.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Emrich HM, von Zerssen D, Kissling W, Moller HJ. Therapeutic effect of valproate in mania. Am J Psychiatry. 1981;138:256. doi: 10.1176/ajp.138.2.256. [DOI] [PubMed] [Google Scholar]
- Eustermann S, Yang JC, Law MJ, Amos R, Chapman LM, Jelinska C, et al. Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin. Nat Struct Mol Biol. 2011;18:777–782. doi: 10.1038/nsmb.2070. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. 2010Selective inhibition of BET bromodomains Nature 4681067–1073.Using high-througput biochemical screening, this work identified the first known example of a selective and highly potent inhibitor of BET family bromodomains, and indicated its usefulness as a novel antiproliferative cancer treatment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Frank D, Doenecke D, Albig W. Differential expression of human replacement and cell cycle dependent H3 histone genes. Gene. 2003;312:135–143. doi: 10.1016/s0378-1119(03)00609-7. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Picketts DJ, Villard L, Higgs DR.1995Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell 80837–845.Provided the first evidence that mutations in the X-chromosomally encoded gene ATRX result in a X-linked mental retardation syndrome accompanied by alpha-thalassemia. [DOI] [PubMed] [Google Scholar]
- Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, Elsaesser SJ, Stadler S, et al. 2010Distinct factors control histone variant H3.3 localization at specific genomic regions Cell 140678–691.This manuscript provided the first epigenome-wide ChIP-seq map of H3.3 localization in mouse ES cells and in neuronal progenitors, and further identified Atrx and Daxx as alternative chaperones for H3.3 at heterochromatic loci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Dorsey J, Gaucher J, Rousseaux S, Khochbin S, Berger SL. Systematic screen reveals new functional dynamics of histones H3 and H4 during gametogenesis. Genes Dev. 2010;24:1772–1786. doi: 10.1101/gad.1954910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Gupta S, Kim SY, Artis S, Molfese DL, Schumacher A, Sweatt JD, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30:3589–3599. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RM, Hnilica LS. Tissue specificity of histone phosphorylation. Science. 1967;157:1324–1325. doi: 10.1126/science.157.3794.1324. [DOI] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, et al. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- Hake SB, Garcia BA, Kauer M, Baker SP, Shabanowitz J, Hunt DF, et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc Natl Acad Sci USA. 2005;102:6344–6349. doi: 10.1073/pnas.0502413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Labile H3.3+H2A.Z nucleosomes mark ‘nucleosome-free regions. Nat Genet. 2009;41:865–866. doi: 10.1038/ng0809-865. [DOI] [PubMed] [Google Scholar]
- Ho L, Crabtree GR.2010Chromatin remodelling during development Nature 463474–484.A comprehensive review of chromatin remodeling during development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodl M, Basler K. Transcription in the absence of histone H3.3. Curr Biol. 2009;19:1221–1226. doi: 10.1016/j.cub.2009.05.048. [DOI] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyland EM, Cosgrove MS, Molina H, Wang D, Pandey A, Cottee RJ, et al. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol Cell Biol. 2005;25:10060–10070. doi: 10.1128/MCB.25.22.10060-10070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis PW, Cochrane JC, et al. 2011ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome Nat Struct Mol Biol 18769–776.A groundbreaking structural study demonstrating the importance of the ATRX ADD domain, which is often mutated in ATR-X syndrome, in H3K9me3 binding and subsequent localization at pericentromeric heterochromatin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 (Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang T, Matevossian A, Huang HS, Straubhaar J, Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junn E, Taniguchi H, Jeong BS, Zhao X, Ichijo H, Mouradian MM. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci USA. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam CS, Kellner WA, Takenaka N, Clemmons AW, Corces VG. 14-3-3 mediates histone cross-talk during transcription elongation in Drosophila. PLoS Genet. 2010;6:e1000975. doi: 10.1371/journal.pgen.1000975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA Repair. 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Kinney CM, Chandrasekharan UM, Yang L, Shen J, Kinter M, McDermott MS, et al. Histone H3 as a novel substrate for MAP kinase phosphatase-1. Am J Physiol Cell Physiol. 2009;296:C242–C249. doi: 10.1152/ajpcell.00492.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleefstra T, Brunner HG, Amiel J, Oudakker AR, Nillesen WM, Magee A, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79:370–377. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, et al. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum Mol Genet. 2009;18:2483–2494. doi: 10.1093/hmg/ddp166. [DOI] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Sung YP, Podhraski V, Chin YL, Emelyanov AV, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–1090. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourmouli N, Sun YM, Van Der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochem Biophys Res Commun. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Krimer DB, Cheng G, Skoultchi AI. Induction of H3.3 replacement histone mRNAs during the precommitment period of murine erythroleukemia cell differentiation. Nucleic Acids Res. 1993;21:2873–2879. doi: 10.1093/nar/21.12.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu TK, Palhan VB, Wang Z, An W, Cole PA, Roeder RG. Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lau PN, Cheung P.2011Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing Proc Natl Acad Sci USA 1082801–2806.First identification of a role for crosstalk between H3S28 phosphorylation and H3K27 acetylation in polycomb antagonism and subsequent regulation of transcriptional activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HPJ, Hughes JR, Garrick D, Viprakasit V, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Lechner MS, Schultz DC, Negorev D, Maul GG, Rauscher Iii FJ. The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun. 2005;331:929–937. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Lepagnol-Bestel A-M, Zvara A, Maussion G, Quignon F, Ngimbous B, Ramoz N, et al. DYRK1A interacts with the REST/NRSF-SWI/SNF chromatin remodelling complex to deregulate gene clusters involved in the neuronal phenotypic traits of Down syndrome. Hum Mol Genet. 2009;18:1405–1414. doi: 10.1093/hmg/ddp047. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, et al. 2007An essential switch in subunit composition of a chromatin remodeling complex during neural development Neuron 55201–215.The first evidence of a role for combinatorial assembly of chromatin remodeling complexes during neurodevelopment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo Y, Schroeder FA, Youngs RM, Schmidt TW, Ferris C, et al. Dopamine D2-like antagonists induce chromatin remodeling in striatal neurons through cyclic AMP-protein kinase A and NMDA receptor signaling. J Neurochem. 2004;90:1117–1131. doi: 10.1111/j.1471-4159.2004.02569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu JY, Allis CD, et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- Lord KA, Abdollahi A, Hoffman-Liebermann B, Liebermann DA. Dissection of the immediate early response of myeloid leukemia cells to terminal differentiation and growth inhibitory stimuli. Cell Growth Differ. 1990;1:637–645. [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Macdonald N, Welburn JP, Noble ME, Nguyen A, Yaffe MB, Clynes D, et al. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol Cell. 2005;20:199–211. doi: 10.1016/j.molcel.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Matsumoto S, Banine F, Struve J, Xing R, Adams C, Liu Y, et al. Brg1 is required for murine neural stem cell maintenance and gliogenesis. Dev Biol. 2006;289:372–383. doi: 10.1016/j.ydbio.2005.10.044. [DOI] [PubMed] [Google Scholar]
- Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA. 2011;108:3035–3040. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maze I, Nestler EJ. The epigenetic landscape of addiction. Ann N Y Acad Sci. 2011;1216:99–113. doi: 10.1111/j.1749-6632.2010.05893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O'Rourke DM, Bickmore WA, Pombo A, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci USA. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod D, Bartesaghi S, Khelifi A, Bellodi C, Berliocchi L, Nicotera P, et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74:122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- Morrison AJ, Shen X. Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat Rev Mol Cell Biol. 2009;10:373–384. doi: 10.1038/nrm2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Hou J, Maclean A, Nasir J, Lafuente MJ, Shu X, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci USA. 2007;104:2709–2714. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Hidden switches in the mind. Sci Am. 2011;305:76–83. doi: 10.1038/scientificamerican1211-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ.2010Chronic high-fat diet in fathers programs beta-cell dysfunction in female rat offspring Nature 467963–966.This work provided the first evidence for transgenerational inheritance of a paternally-linked phenotype (ie, obesity resulting from exposure to a chronic high-fat diet) to female offspring, potentially through an epigenetic-based mechanism (eg, DNA methylation). [DOI] [PubMed] [Google Scholar]
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- Ooi L, Belyaev ND, Miyake K, Wood IC, Buckley NJ. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J Biol Chem. 2006;281:38974–38980. doi: 10.1074/jbc.M605370200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF. The stem-loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. A core nucleosome surface crucial for transcriptional silencing. Nat Genet. 2002;32:273–279. doi: 10.1038/ng982. [DOI] [PubMed] [Google Scholar]
- Pascreau G, Arlot-Bonnemains Y, Prigent C. Phosphorylation of histone and histone-like proteins by aurora kinases during mitosis. Prog Cell Cycle Res. 2003;5:369–374. [PubMed] [Google Scholar]
- Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med. 2011;17:372–379. doi: 10.1016/j.molmed.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OWJ, Gibbons RJ. ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet. 1996;5:1899–1907. doi: 10.1093/hmg/5.12.1899. [DOI] [PubMed] [Google Scholar]
- Pina B, Suau P.1987Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons Dev Biol 12351–58.This manuscript was the first to indicate that the histone variant H3.3 becomes the predominant H3 in brain with age, consistent with what we now know regarding its replication-independent deposition. [DOI] [PubMed] [Google Scholar]
- Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH, III, et al. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Raoul C, Estevez AG, Nishimune H, Cleveland DW, deLapeyriere O, Henderson CE, et al. Motoneuron death triggered by a specific pathway downstream of Fas. potentiation by ALS-linked SOD1 mutations. Neuron. 2002;35:1067–1083. doi: 10.1016/s0896-6273(02)00905-4. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Quivy JP, Scamps C, Martini EMD, Lipinski M, Almouzni G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- Ray-Gallet D, Woolfe A, Vassias I, Pellentz Cl, Lacoste N, Puri A, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–941. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–169. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha) EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Allis CD. Histone methylation vs histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Sekeri-Pataryas KE. Histone variants of H2A and H3 families are regulated during in vitro aging in the same manner as during differentiation. Exp Gerontol. 1999;34:741–754. doi: 10.1016/s0531-5565(99)00046-7. [DOI] [PubMed] [Google Scholar]
- Romieu P, Host L, Gobaille S, Sandner G, Aunis D, Zwiller J. Histone deacetylase inhibitors decrease cocaine but not sucrose self-administration in rats. J Neurosci. 2008;28:9342–9348. doi: 10.1523/JNEUROSCI.0379-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, et al. 2011Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions Cell 145692–706.This work provided the first direct biochemical evidence for multivalent engagement of histone modifications by distinct reader modules within a single protein, further indicating the importance of combinatorial readouts of histone marks at the level of mononucleosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh R, Allis CD. Genome-wide ‘re'-modeling of nucleosome positions. Cell. 2011;147:263–266. doi: 10.1016/j.cell.2011.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Schwartz BE, Goldstein S, Ahmad K. Transcriptional and developmental functions of the H3.3 histone variant in drosophila. Curr Biol. 2009;19:1816–1820. doi: 10.1016/j.cub.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath SC, Marazzi I, Yap KL, Sampath SC, Krutchinsky AN, Mecklenbrauker I, et al. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer A, Sampath SC, Intrator A, Min A, Surmeier DJ, Tarakhovsky A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64:678–691. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzentruber J, Korshunov A, Liu X-Y, Jones DTW, Pfaff E, Jacob K, et al. 2012Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma Nature 482226–231.These data provide the first evidence of a role for dysregulation of the histone variant H3.3 in human disease, whereby driver mutations in H3F3A (K27 M, G34R/G34V) result in pediatric glioblastomas. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Xiong L, Fathalli F, Benkelfat C, Tabbane K, Danics Z, et al. Association study of the trinucleotide repeat polymorphism within SMARCA2 and schizophrenia. BMC Genet. 2006;7:34. doi: 10.1186/1471-2156-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132:105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- Shinkai Y, Tachibana M. H3K9 methyltransferase G9a and the related molecule GLP. Genes Dev. 2011;25:781–788. doi: 10.1101/gad.2027411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda N, Beppu H, Fukuda T, Li E, Kitajima I, Fukunaga K. Aberrant calcium/calmodulin-dependent protein kinase II (CaMKII) activity is associated with abnormal dendritic spine morphology in the ATRX mutant mouse brain. J Neurosci. 2011;31:346–358. doi: 10.1523/JNEUROSCI.4816-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes. Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM. Centromeres and variant histones: what, where, when and why. Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- Sng JC, Taniura H, Yoneda Y. Histone modifications in kainate-induced status epilepticus. Eur J Neurosci. 2006;23:1269–1282. doi: 10.1111/j.1460-9568.2006.04641.x. [DOI] [PubMed] [Google Scholar]
- Sohn DH, Lee KY, Lee C, Oh J, Chung H, Jeon SH, et al. SRG3 interacts directly with the major components of the SWI/SNF chromatin remodeling complex and protects them from proteasomal degradation. J Biol Chem. 2007;282:10614–10624. doi: 10.1074/jbc.M610563200. [DOI] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics. 2011;6:539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevely WS, Stocken LA. Phosphorylation of rat-thymus histone. Biochem J. 1966;100:20C–21C. doi: 10.1042/bj1000020c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453:879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD.2000The language of covalent histone modifications Nature 40341–45.This landmark paper first articulated the histone code hypothesis, which suggests that combinations of histone posttranslational modifications, along with specific recognition reader/effector proteins act in concert to promote downstream transcriptional events. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Sweet MT, Allis CD. Phosphorylation of linker histones by cAMP-dependent protein kinase in mitotic micronuclei of Tetrahymena. Chromosoma. 1993;102:637–647. doi: 10.1007/BF00352312. [DOI] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tamura T, Smith M, Kanno T, Dasenbrock H, Nishiyama A, Ozato K. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J Biol Chem. 2009;284:12217–12225. doi: 10.1074/jbc.M805651200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu S, Liu H, Stratt R, Barak OG, Shiekhattar R, et al. A novel transcription regulatory complex containing death domain-associated protein and the ATR-X syndrome protein. J Biol Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 1999;18:4779–4793. doi: 10.1093/emboj/18.17.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Van Holde KE.1989Chromatin Springer-Verlag; New York. [Google Scholar]
- Venugopal B, Evans TR. Developing histone deacetylase inhibitors as anti-cancer therapeutics. Curr Med Chem. 2011;18:1658–1671. doi: 10.2174/092986711795471284. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Coté J, Xue Y, Zhou S, Khavari PA, Biggar SR, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996b;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996a;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Mizutani T, Haraguchi T, Yamamichi N, Minoguchi S, Yamamichi-Nishina M, et al. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nonsmall cell lung carcinoma cell lines. Oncogene. 2006;25:470–479. doi: 10.1038/sj.onc.1209068. [DOI] [PubMed] [Google Scholar]
- Weatherall DJ, Higgs DR, Bunch C. Hemoglobin H disease and mental retardation. A new syndrome or a remarkable coincidence. N Engl J Med. 1981;305:607–612. doi: 10.1056/NEJM198109103051103. [DOI] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD.1998Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena Proc Natl Acad Sci USA 957480–7484.These data were the first to demonstrate a role for H3S10p in the regulation of chromosomal condensation during mitosis and meiosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Wellman SE, Casano PJ, Pilch DR, Marzluff WF, Sittman DB. Characterization of mouse H3.3-like histone genes. Gene. 1987;59:29–39. doi: 10.1016/0378-1119(87)90263-0. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME.2011Neuronal activity-regulated gene transcription in synapse development and cognitive function Cold Spring Harb Perspect Biol 3a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann SM, Mildner SN, Bonisch C, Israel L, Maiser A, Matheisl S, et al. Identification and characterization of two novel primate-specific histone H3 variants, H3.X and H3.Y. J Cell Biol. 2010;190:777–791. doi: 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, McGhie JD, Sim M, Anderson MA, Ahn S, Hannan RD, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–360. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, Olave IA, Qiu Z, Ghosh A, Graef IA, et al. 2007Regulation of dendritic development by neuron-specific chromatin remodeling complexes Neuron 5694–108.This work was the first to describe a direct relationship between neurodevelopmental chromatin remodeling and functional plasticity, whereby disruptions in proper modes of complex combinatorial assembly resulted in dendritic abnormalities. [DOI] [PubMed] [Google Scholar]
- Wu RS, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–374. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- Xie S, Wang Z, Okano M, Nogami M, Li Y, He WW, et al. Cloning, expression and chromosome locations of the human DNMT3 gene family. Gene. 1999;236:87–95. doi: 10.1016/s0378-1119(99)00252-8. [DOI] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Choi JH, Jang H, Park JY, Han JW, Youn HD, et al. Histone chaperones cooperate to mediate Mef2-targeted transcriptional regulation during skeletal myogenesis. Biochem Biophys Res Commun. 2011;407:541–547. doi: 10.1016/j.bbrc.2011.03.055. [DOI] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, et al. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]