Abstract
Major psychiatric disorders such as schizophrenia (SZ) and bipolar disorder (BP) with psychosis (BP+) express a complex symptomatology characterized by positive symptoms, negative symptoms, and cognitive impairment. Postmortem studies of human SZ and BP+ brains show considerable alterations in the transcriptome of a variety of cortical structures, including multiple mRNAs that are downregulated in both inhibitory GABAergic and excitatory pyramidal neurons compared with non-psychiatric subjects (NPS). Several reports show increased expression of DNA methyltransferases in telencephalic GABAergic neurons. Accumulating evidence suggests a critical role for altered DNA methylation processes in the pathogenesis of SZ and related psychiatric disorders. The establishment and maintenance of CpG site methylation is essential during central nervous system differentiation and this methylation has been implicated in synaptic plasticity, learning, and memory. Atypical hypermethylation of candidate gene promoters expressed in GABAergic neurons is associated with transcriptional downregulation of the corresponding mRNAs, including glutamic acid decarboxylase 67 (GAD67) and reelin (RELN). Recent reports indicate that the methylation status of promoter proximal CpG dinucleotides is in a dynamic balance between DNA methylation and DNA hydroxymethylation. Hydroxymethylation and subsequent DNA demethylation is more complex and involves additional proteins downstream of 5-hydroxymethylcytosine, including members of the base excision repair (BER) pathway. Recent advances in our understanding of altered CpG methylation, hydroxymethylation, and active DNA demethylation provide a framework for the identification of new targets, which may be exploited for the pharmacological intervention of the psychosis associated with SZ and possibly BP+.
Keywords: DNA methylation, hydroxymethylation, chromatin, nicotine, antipsychotics, animal models
INTRODUCTION
Patients diagnosed with schizophrenia (SZ) are characterized by clusters of symptoms exemplified by positive or psychotic symptoms, such as delusions and hallucinations, cognitive impairment, and negative symptoms. Bipolar disorder (BP) is a condition in which the patient suffers from mood swings, which consist of recurring bouts of mania and depression. Each of these psychiatric disorders is distinct and characterized by additional symptoms. The positive symptoms and cognitive impairment associated with SZ and BP disorder with psychosis (BP+) show considerable overlap in clinical presentation (Potash and Bienvenu, 2009; Keshavan et al, 2011; Ivleva et al, 2012). The origins of the psychotic symptoms of BP+ are most likely the consequence of the underlying mood disorder (mania) depending on the nature of the delusions and hallucinations described by the patient at clinical presentation. Collectively, these psychiatric disorders are complex diseases of brain function and accumulating evidence supports an overlap between the biological and genetic data associated with SZ and BP+ (Keshavan et al, 2011). Moreover, we suggest that altered DNA methylation dynamics likely underlie the pathogenesis of psychotic symptoms.
The genetics of SZ and BP+ are perhaps the most studied facet of the disorders but an emphasis on multiple linkage and association analyses over the last 60 years have provided only limited insight into the underlying etiological factors that characterize these diseases (Harrison and Weinberger, 2005). Research into the genetics of SZ and BP+ has been hindered by both the characteristic non-Mendelian inheritance and a lack of disease-specific diagnostic biomarkers. While linkage and association studies, including genome-wide association studies, are informative, the interpretation of complex genetics makes an appreciation of the role of susceptibility genes in psychotic disorders difficult. These problems include a combination of allelic, locus, and phenotypic heterogeneity (Rodriguez-Murillo et al, 2012). Even when considering recently identified copy number variants, a major source of genomic variation, the role of susceptibility genes in predicting clinical presentation is still not clear (Tam et al, 2009). The identification of de novo mutations in SHANK3 (Gauthier et al, 2010) and additional genes (Xu et al, 2011a; Girard et al, 2011) has led to a new paradigm linking de novo mutations to the genetics of SZ (Girard et al, 2012) and BP (Malhotra et al, 2011). However, of those studies reported thus far, none have identified mutations in existing susceptibility genes or copy number intervals (Mulle, 2012). While the discovery of new de novo mutations will likely continue in the forseeable future, it is unclear what is responsible for the high heritability rates of SZ.
It seems likely that the combination of susceptibility genes together with de novo mutations and the environmental/epigenetic perturbations incurred during early nervous system development increase risk for SZ or BP+. Moreover, the cognitive impairment associated with pre- and postmorbid SZ (Sørensen et al, 2006; Goff et al, 2011) is a clinical feature that has been associated with psychosis duration (Simonsen et al, 2011) as well as negative symptoms (Lewandowski et al, 2011). A recent evaluation of cognitive performance between SZ and schizoaffective disorder indicate that schizoaffective disorder patients have severe cognitive impairments, but these impairments are milder than those in SZ patients (Torniainen et al, 2012). At the same time, a recent assessment of SZ and BP+ probands showed little, if any, differences in altered cognitive performance between groups using standardized clinical and neuropsychological measures (Ivleva et al, 2012). The above studies indicate that different patient populations and different components associated with cognitive testing often lead to different conclusions. A key challenge to understanding the etiology of the psychosis associated with SZ or BP+ is identifying individual differences within each diagnostic group and the phenotypes that collectively contribute to each symptom. In other words, hallucinations are characteristic of the positive symptoms of SZ and BP+. It remains unclear as to whether visual, olfactory, and auditory hallucinations represent distinctly different symptoms and hence different endophenotypes that need to be considered as separate entities. Once a clear picture emerges in terms of what is responsible for each symptom, it may then be possible to search for the particular neuroanatomical systems responsible. This will provide a path towards an understanding of the gene expression patterns responsible, the neurons in which these are operative and how these neurons interact in a complex physiological network as a prerequisite to a better understanding of the symptomatological complexities of these psychiatric disorders. Ultimately, the specific gene regulatory cascades responsible for the collection of intermediate phenotypes evident upon clinical presentation are likely connected.
The relationship between gene copy number intensity and gene expression has recently been explored in SZ and BP patients with the goal of understanding correlations between these two measures in specific hippocampal neuron populations (Sheng et al, 2012). Previous work has shown that a network of 28 genes is associated with the regulation of glutamic acid decarboxylase 67 (GAD67) in the hippocampus and that this network interacts with other gene networks in maintaining genome integrity in specific neurons (Benes et al, 2009). GAD67, a marker for GABAergic neurons, is downregulated in the cortex and hippocampus of SZ and BP+ patients (Akbarian and Huang, 2006; Volk et al, 2000; Guidotti et al, 2000). This gene cluster was uncovered during a network association analysis of cDNAs exhibiting compromised expression in CA3/2 hippocampal GABAergic neurons in SZ and BP disorder patients (Benes et al, 2009). The more recent study (Sheng et al, 2012) shows that there are highly significant changes in the magnitude and direction of individual gene copy number intensities in this GAD67 regulatory network. Moreover, the corresponding mRNA levels of members of this network significantly correlate in CA3/2 but not in hippocampal CA1 neurons (Sheng et al, 2012). The copy number intensities and corresponding gene expression levels vary proportionately in a diagnosis-dependent manner, confirming distinct differences between SZ and BP disorders. No correlation between copy number intensity and mRNA levels is evident in CA1 neurons of the hippocampus. This suggests the possibility that genome integrity might be compromised by local environmental cues that are likely neuron- and circuitry-specific. It seems likely that the origins of SZ and BP+ may not reside strictly in DNA sequence variations (whether related to susceptibility genes, de novo mutations, or variations in copy number intensity) but rather may be coupled to epigenetic mechanisms acting as key etiopathogenic factors (Petronis, 2004; Costa et al, 2006).
SZ is a neurodevelopmental disease that may be the consequence of early life events that introduce perturbations affecting gene expression. Environmental variables acting during fetal and perinatal life, including maternal stress, obstetric complications, maternal infections, gestational nutrition, etc., impact the regulation of transcription during early development (Brown, 2011; Brown and Patterson, 2011; Markham and Koenig, 2011). While the molecular mechanisms by which these environmental (epigenetic) stressors impact brain function have yet to be elucidated, recent studies support the view that early life stresses alter processes associated with chromatin remodeling such that transcriptional regulation is abnormal in distinct anatomical structures of the brain (Matrisciano et al, 2012a; McGowan et al, 2011). Mice exposed to stress in utero exhibit altered biochemical features and behaviors reminiscent of morbidities encountered in SZ and BP+ patients after reaching adulthood (Matrisciano et al, 2011, 2012a). As more and more single (and even multiple) gene knockouts of susceptibility genes are generated and characterized, it is becoming increasingly clear that the consequences of any pathogenic mutation must be evaluated in combination with environmental manipulations that introduce stress during prenatal or perinatal development as one context for the interpretation of the targeted genes (Oliver, 2011).
Studies of postmortem human telencephalic structures from SZ and BP+ patients often fail to elucidate differences in the molecular mechanisms underlying psychosis because they provide a static view of events that are the consequence of trajectories that originated as many as several decades earlier during development. It is impossible to appreciate the dynamics of changes in DNA methylation/demethylation, histone modifications, or transcription factor availability from this static perspective because it represents the summation of numerous regulatory events that have modified the neuronal transcriptome both in response to stress during neurodevelopment and following the adaptation of the brain to this stress over long periods of time. Studies of early life stress in animals are particularly valuable in the context of understanding the impact of these stressors on gene expression once these animals become adults.
EPIGENETIC REGULATION IN NEURONS
Histone Modifications
Chromatin biology has been studied experimentally since the early 1880's, a time during which the material in cell nuclei stained by basophillic stains was first termed chromatin by the cytologist Walther Flemming. In 1884, Albrecht Kossel discovered histones and determined that these basic proteins were associated with DNA for which he won the Nobel Prize in Physiology and Medicine in 1910. In more recent times, biochemical studies on the structure and function of histone proteins began to appear in the literature during the 1950's. It soon became clear that deoxyribonucleoprotein is complex and exists in different forms as probed by deoxyribonuclease I (Mirsky, 1971). Transitions between euchromatin and heterochromatin are associated with active and inactive transcription, respectively, and are mediated by modifications in the structures of histone proteins comprising the nucleosome (Figure 1). The term ‘histone code' was coined to refer to the combinatorial nature of histone tail modifications, which represent the histone-marking system associated with chromatin-template-dependent mechanisms (Jenuwein and Allis, 2001).
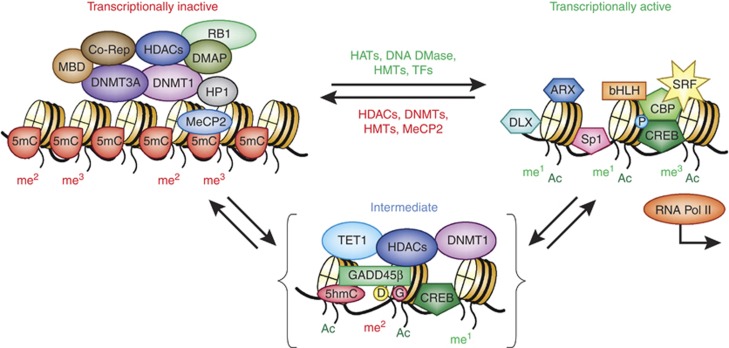
Figure 1.
Proteins bound to DNA and histones cooperate in facilitating transitions between active and inactive chromatin states. Schematic representation of the transitions between a transcriptionally inactive promoter (left) and a transcriptionally active state (right). The transcriptionally inactive state is characterized by DNA methylation and the binding of various repressor proteins, including DNA methyltransferase 1 (DNMT1) and 3A, methyl-binding domain proteins (MBDs, MeCP2), co-repressors, and modified histones associated with repressive chromatin marks (H3K9me2, H3K9me3, H3K27me2, H3K27me3, etc.). The intermediate state (shown in brackets) is stable and ‘poised' for either repression or activation. In the intermediate state, the DNA/protein complex is characterized by the binding of DNMT1 to unmethylated CpGs and ten-eleven translocase-1 (TET-1) bound to 5-methylcytosines (5mCs) and 5-hyroxymethylcytosine (5hmCs). In the transitional phase, DNMT1 is associated with histone deacetylases (HDACs) and excess DNMT3A shifts this towards the inactive state (left). The binding of TET1 to hydroxymethylated CpGs in this same intermediate state reinforces stable repression until the entry of GADD45β, which recruits proteins required for DNA demethylation (deaminases and glycosylases). DNA demethylation is accompanied by additional histone modifications (mediated by HATs and HMTs). Hydroxymethylated CpGs are further modified and removed. In this model, HDAC inhibitors facilitate a disruption of the inactive state and depending on the availability of GADD45β, DNA demethylation ensues (Kundakovic et al, 2009; Guidotti et al, 2011). In the active (open) state, various transcription factors (TFs) bind and occupy their specific DNA recognition sites enabling transcription. The specific TFs involved depend on the gene being activated and the neuronal phenotype (West and Greenberg, 2011). Some of the transcription factors are shown bound to the intermediate state (such as CREB, which upon phosphorylation (P) recruits the histone acetyltransferase CBP). Transcriptionally active promoters are represented as an open chromatin structure characterized by the presence of acetylated (H3K9ac, H3K14ac) and methylated (eg, H3K4me1, H3K4me3, H3K9me1, H3K27me1, H3K79me1, etc) histones. The model highlights repressive roles for DNMT1 and TET1, which depends upon the availability of accessory proteins (DNMT3A and GADD45β, respectively) to modify their function in postmitotic neurons. Based on localization studies of DNMT1 in GABAergic neurons (Kadriu et al, 2011) and GADD45β in pyramidal neurons (Gavin et al, 2012), these mechanisms are likely unique to specific types of neurons depending on neurotransmitter phenotype. ARX, aristaless-related homeobox; bHLH, basic helix-loop-helix transcription factors; CBP, CREB-binding protein; Co-Rep, co-repressor proteins; CREB, cyclic AMP response element-binding protein; D, deaminase; DLX, distal-less homeobox; DMAP1, DNA methyltransferase 1-associated protein; DNA DMase, DNA demethylase; G, gycosylase; HATs, Histone acetyl transferases; HMTs, histone methyl transferases; HP1, hetrochromatin protein 1; me1, monomethyl; me2, dimethyl; me3, trimethyl; MeCP2, methyl CpG-binding protein 2; P, phosphoryl group; RB1, retinoblastoma 1; SP1, promoter-specific transcription factor; SRF, serum response factor; TFs, transcription factors.
Epigenetic signaling includes a host of opposing histone modifications occurring largely at histones 3 (H3) and 4 (H4) that include phosphorylation, ubiquitination, acetylation, and deacetylation, as well as methylation and demethylation (Mersfelder and Parthun, 2006; Bannister and Kouzarides, 2011). In terms of amino acids along the histone tails, lysines (K) and arginines (R) are subject to methylation (me), while K is a site of acetylation (ac) (Jenuwein and Allis, 2001). Correlations between specific modifications on histones 3 (H3) or 4 (H4) in predicting transcription are high and dependent on the local GC content of the promoters (Karlić et al, 2010). That is, different sets of histone modifications are better predictors of gene expression driven from high GC content promoters (eg, H3K27ac and H4K20me1) compared with low GC content promoters (H3K4me3 and H3K79me1) (Karlić et al, 2010).
Recent findings regarding the negative role of histone deacetylases (HDACs) 2 (Guan et al, 2009) and 3 (McQuown et al, 2011) in synaptic plasticity and memory formation has garnered considerable interest in the identification of specific HDAC inhibitors that might prove useful in preventing the cognitive decline observed in neuropsychiatric disorders (Fischer et al, 2010; Grayson et al, 2010; Morris et al, 2010; McQuown et al, 2011; Day and Sweatt, 2012). In rats, stress induced by maternal deprivation (Qin et al, 2011) or as the consequence of attenuated maternal care (Weaver et al, 2004, 2006) has been shown to impact neurobehavioral development of the hippocampus by affecting the levels of DNA methylation of not only the glucocorticoid receptor and reelin (RELN) promoters but also a variety of additional promoters. Large numbers of mRNAs are downregulated in the hippocampus following maternal care stress and subsets of these mRNAs return to normal levels following the administration of an HDAC inhibitor (Weaver et al, 2006). This implies that not only are many of these processes reversible following treatment with drugs that modify chromatin structure but they are interconnected as well. By inhibiting histone deacetylase activity and increasing the levels of acetylated amino-terminal histone tails, HDAC inhibitors have also been shown to reverse the effects of increased DNA methylation (Weaver et al, 2006).
DNA Methylation
DNA methylation, which consists of the addition of a methyl group to the C5 position of cytosine at CpG dinucleotides (Figure 2), is an important epigenetic modification involved in the regulation of transcription in the brain. Methylation of DNA is catalyzed by a family of related DNA methyltransferases (DNMTs) that include DNMT1, DNMT3A, and DNMT3B. DNMT3L is catalytically inactive but shares homology with both DNMT3A and 3B (Aapola et al, 2001; Chédin et al, 2002). DNMT3L does not bind S-adenosylmethionine (SAM) but instead enhances the binding of SAM to DNMTs 3A and 3B and facilitates methylation in vivo (Kareta et al, 2006). SAM is the methyl donor used by DNMTs to transfer the one carbon methyl group, producing 5-methylcytosine (5mC) and S-adenosylhomocysteine (SAH) in the process (see Figure 2 and Text box 1 for additional details on DNMTs in the brain). The accumulation of homocysteine has been shown to lead to neural damage and cognitive dysfunction (Krebs et al, 2009).
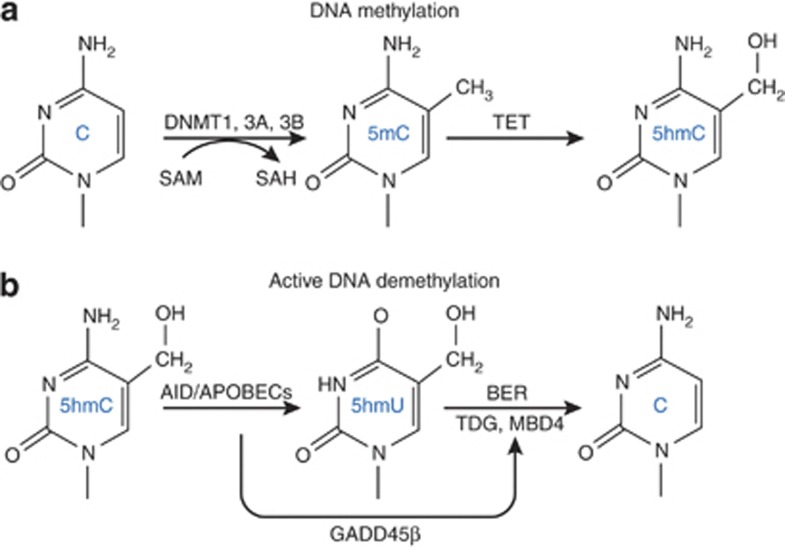
Figure 2.
DNA methylation and demethylation are in a dynamic balance in neurons. The top panel (a) shows key steps associated with DNA methylation. DNA methyltransferases (DNMTs) catalyze the methylation of the fifth position of the pyrimidine ring of cytosine in CpG dinucleotides. S-adenosylmethionine (SAM) serves as the methyl donor that is converted to S-adenosylhomocysteine (SAH) following methyl group transfer. 5-methylcytosine (5mC) can be hydroxylated in a subsequent reaction catalyzed by members of the ten-eleven translocase (TET) family of methylcytosine dioxygenases. TET1–3 are 2-oxoglutarate-Fe(II) oxygenases, which hydroxylate 5mC to 5-hydroxymethylcytosine (5hmC). TETs 1 and 3 contain a –CXXC- domain, which binds with high affinity to clustered, unmethylated CpG dinucleotides. Structural analyses of DNMT1 show that it also contains a similar –CXXC- domain (see Text Box 2). The bottom panel (b) shows steps involved with the removal of the methyl group from 5hmC. The first step is an oxidative deamination of 5hmC to produce 5-hydroxymethyluridine (5hmU) by the AID/APOBEC family of deaminases. Activation-induced cytidine deaminase (AID) is also a member of the apolipoprotein B mRNA-editing catalytic polypeptides that deaminate 5mC and 5hmC to form thymine and 5hmU, respectively. These intermediates are subsequently processed by the uracil-DNA glycosylase (UDG) family that includes thymine-DNA glycosylase (TDG, MBD4) and single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1). These latter steps are collectively part of the base excision repair glycosylases (BER) that may also generate additional reactive intermediates such as 5-formylcytosine and 5-carboxylcytosine (Wu and Zhang, 2011). GADD45β is an activity-induced neuronal immediate early gene that facilitates active DNA demethylation (Ma et al, 2009a).
DNMTs in the nervous system.
In the nervous system, DNMT1 and 3a are likely to predominate in terms of postmitotic neuronal function (Feng et al, 2005). The expression of DNMT3A and 3B were analyzed immunohistochemically during nervous system development and the results show that DNMT3B is expressed in progenitor cells during neurogenesis and that DNMT3A is present in postmitotic neurons (Watanabe et al, 2006). Using DNMT3A and DNMT3B—lacZ (+/lacZ) knock-in mice, this complementary pattern of expression was also reported (Feng et al, 2005). That is, while DNMT3B is expressed during early stages of neurogenesis, DNMT3A is present in predominantly postmitotic neurons and some oligodendrocytes. Both DNMT3A and 3B mRNAs increase in the CA1 fields of the adult rat hippocampus following contextual fear conditioning (Miller and Sweatt, 2007). More recently, it was reported that DNMT3A and 3B are induced 24 h following acute cocaine administration in the nucleus accumbens of mice implicating a role for both in the development of behavioral sensitization (Anier et al, 2010). While DNMT3A is the predominant DNMT operative in postmitotic neurons, there is increasing evidence that DNMT3B has an ancillary role in response to extraneuronal cues in the adult brain.
DNMT1 is expressed at high levels during embryogenesis and maturation of the mouse brain. Initial reports of DNMT1 mRNA levels in the brain indicated that almost all mature neurons abundantly expressed the transcript (Goto et al, 1994). Subsequent immunohistochemical analysis show DNMT1 immunostaining of postmitotic Purkinje neurons in mouse cerebellum and MAP2-positive neurons from fetal brains maintained in vitro. Interestingly, the immunohistochemical signal appears to be localized in the cytoplasm (Inano et al, 2000). Recent studies demonstrate that a small percent of DNMT1 mRNA is translated from an upstream ATG start codon that contains a mitochondrial import signal (Shock et al, 2011). While the functional implications of this protein in the context of mitochondrial DNA is not clear, both 5mC and 5hmC have been detected in neuronal mitochondrial DNA (Dzitoyeva et al, 2012). Conditional knockout of DNMT1 in neural progenitor cells results in significant global hypomethylation (Fan et al, 2001). In contrast, the conditional deletion of DNMT1 in postmitotic neurons of mice expressing cre recombinase from the neuronal calmodulin-kinase IIα (CamK) promoter does not affect global DNA methylation and has little effect on cell survival (Fan et al, 2001). These data indicate that loss of DNMT1 in neurons expressing CamK (presumably glutamatergic neurons) does not impact global DNA methylation levels. The inability of DNMT1 to catalyze de novo methylation, although still controversial, is based primarily on its preference for a hemi-methylated DNA substrate (Hermann et al, 2004; Jeltsch, 2006) and the methylation phenotype of a targeted DNMT1 deletion (Li et al, 1992). In addition, analysis of the crystal co-structure of DNMT1 with oligonucleotides containing CpGs indicates that unmethylated CpGs are not accessible to the active site unless the CpGs are hemi-methylated (Song et al, 2011b; Takeshita et al, 2011). This suggests that DNMT1 has low intrinsic de novo methylation activity. In HEK293 cells, DNMT1 overexpression leads to very small increases in gene-specific hypermethylation compared with the overexpression of variants of DNMT3A or DNMT3B (Choi et al, 2011). As HEK cells are mitotically active, this experiment warrants replication in a cell line which can be induced to differentiate and become postmitotic.
A recent immunohistochemical study of DNMT1 shows that this protein is located primarily in GABAergic neurons in telencephalic structures of the adult human (Veldic et al, 2004, 2007) and mouse brain (see Figure 4 and Kadriu et al, 2011). In adult glutamic acid decarboxylase 67 (GAD67)-eGFP knock-in mice in which eGFP is expressed from the GAD67 start codon, immunohistochemical studies co-localize DNMT1 and RELN with GAD67 immunoreactivity (Figure 4). DNMT3A immunoreactivity also coincides with GAD67 but the extent of overlap is slightly less in some brain regions such as the amygdala. Interestingly, the methyl CpG-binding protein MeCP2 is much more highly expressed in GABAergic neurons than in other cell types in the brain and mice with a selective loss of MeCP2 in GABAergic forebrain neurons display many features reminiscent of Rett Syndrome (Chao et al, 2010). The above immunohistochemical studies suggest that methylation as a mechanism for gene regulation may have a more dominant role in modulating inhibitory neuron function compared with other neurons in the brain. This concept is consistent with the hypothesis that SZ is the result of dysfunctional GABAergic transmission, which may be the consequence of an increased amount of DNMTs causing DNA hypermethylation and gene repression in telencephalic GABAergic neurons (Costa et al, 2007, 2009; Guidotti et al, 2011).
In general, DNMTs 3A and 3B function as de novo methyltransferases, whereas the main role of DNMT1 is to propagate methylation patterns during DNA replication (Hermann et al, 2004). siRNA knockdown experiments show that a loss of DNMT1 does not alter maintenance methylation patterns (Ting et al, 2004, 2006). This has led to a re-evaluation of the function of the DNMTs in dividing cells (Jin et al, 2011). Analysis of a catalytically inactive DNMT1 shows that the mutant protein has the capacity to repress an artificially introduced reporter activity (Robertson et al, 2000; Fuks et al, 2000). More recently, this observation was extended to include a large class of endogenous genes expressed in genetically modified cells in which DNMT1 expression is disrupted (Clements et al, 2012). Restoration of DNMT1 function to these cells causes the repression of many of the same transcripts and increased binding of DNMT1 to the corresponding promoters. DNMT1 both binds to the promoters and interacts with the H3K4 methylase KDM1A/LSD1 (Clements et al, 2012). KDM1A/LSD1 has previously been shown to interact with and stabilize DNMT1 (Wang et al, 2009). Moreover, the interaction of DNMT1 with KDM1A/LSD1 facilitates the depletion of H3K4 methyl groups associated with active transcription (Clements et al, 2012). The association of DNMT1 with this histone demethylase allows for the coupling of DNA methylation to the removal of active histone methyl marks. More importantly, it appears from these studies that DNMT1 functions as a transcriptional repressor independent of its methyltransferase activity (Clements et al, 2012). The above studies were carried out in mitotically active cells and confirmation of this capacity in postmitotic neurons awaits further clarification.
It has become increasingly clear that DNMT1 may be necessary for de novo methylation, while DNMT3A and 3B also contribute to maintenance methylation (Jeltsch, 2006; Jin et al, 2011). Dnmt3A and Dnmt1 cooperate functionally during de novo methylation and it may be that both enzymes work in concert (Fatemi et al, 2002). These investigators proposed a model in which DNMT3A acts to initiate de novo methylation and the newly created methyl group activates DNMT1 to methylate the opposing strand. In addition, DNMT1 antisense knockdown has been shown to induce the expression of the cell cycle inhibitor p21 and the apoptosis-inducer BCL2 by a mechanism that involves DNMT1-mediated activation of Sp1- and the related Sp3-response elements (Milutinovic et al, 2004). These findings are consistent with DNMT1 having a role in regulating the expression of GC-rich genes independent of methylation and histone acetylation. Sp1 and Sp3 transcription factors regulate multiple sets of target genes containing GC boxes, which are often present to a large extent in CpG island-containing promoters such as RELN (Chen et al, 2007).
DNA methylation appears even more complicated due to the identification of a newly recognized ATRX-DNMT3-DNMT3L (ADD) zinc-finger containing member of this family, DNMT3-like (DNMT3L, Aapola et al, 2000, 2001). While DNMT3L is catalytically inactive, it stimulates and targets the activities of DNMTs 3A and 3B (Gowher et al, 2005). For example, Dnmt3L interacts specifically with and recognizes unmethylated H3K4 (Ooi et al, 2007). Co-crystallographic analysis of human DNMT3L and DNMT3A show that the carboxyl terminal domain of DNMT3L interacts with the catalytic domain of DNMT3A. These data also demonstrate that DNMT3a and -3L form a tetrameric structure that contains two DNMT catalytic sites (Jia et al, 2007). This DNMT 3L-3A-3A-3L tetramer preferentially methylates CpGs separated by 8–10 bp, indicating a single helical turn periodicity that is often seen in both paternally and maternally imprinted genes (Glass et al, 2009). The interaction of DNMT3L with unmethylated H3K4 tails suggests a mechanism by which histone methylation and de novo methylation may be coupled (Ooi et al, 2007; Hashimoto et al, 2010). Genome-wide DNA methylation patterns indicate that DNA methylation and histone methylation are highly correlated (Meissner et al, 2008; Maunakea et al, 2010). The ADD domain of DNMT3L interacts with H3K4me0 and recruits DNMT3A to locations in which the H3K4 residue is not methylated (Hashimoto et al, 2010). There is also recent evidence that the ADD domains of DNMT3A and 3B interact with H3K4me0 directly (Zhang et al, 2010b; Otani et al, 2009). These studies provide the basis for novel mechanisms by which de novo DNA methylation and histone methylation are targeted to specific locations based on the existence of specific proteins that recognize either CpG methylation status or histone-tail modifications.
The methylation of cytosines in CpG dinucelotides allows for additional versatility in terms of genomic regulatory potential. CpG islands are regions of DNA containing a high GC content with greater than normal amounts of CpG dinucleotides. They are closely associated with ∼70% of promoters and are largely free of DNA methylation. These sequences often exist as methylation-free zones due to the abundance of GC-rich transcription factor-binding sites (Deaton and Bird, 2011). Sequencing of the human genome indicates that there are upwards of 29 000 CpG islands that are often located within approximately 1500 bp on either side of transcriptional start sites. Functionally relevant differential methylation occurs at a small proportion of these islands during development, particularly within regions that are cell-type specific (Nair et al, 2011). CpG methylation tends to be low at promoters and higher between genes (intergenic regions) and in gene bodies. Tissue-specific DNA methylation is reported to be more prevalent in the sequences flanking CpG islands, called the ‘CpG island shore regions' (Irizarry et al, 2009). CpG island shores lie up to ∼2 kb on either side of a CpG island while island shelves are further out (∼4 kb). Profiling methylated cytosines across the genome has led to an appreciation that methylation frequency is distributed bimodally (Stadler et al, 2011). That is, a large percentage of CpGs are methylated (high-methylated region) with a much smaller fraction that is under-methylated (low-methylated regions). Interestingly, low-methylated regions tend to correspond to promoter/enhancer regulatory regions based on several criteria, including the presence of the H3K4me1 histone modification. Emerging data suggest that the binding of transcription factors and related proteins do not simply protect sequences from methylation but also initiate active DNA demethylation (Stadler et al, 2011). As cells differentiate, the locations of low-methylated regions tend to migrate towards regions proximal to promoters that are transcriptionally active in the differentiated state.
DNA Hydroxymethylation
While comparing the levels of 5mC in cerebellar Purkinje and granule cells, a minor base was detected and determined to be 5-hyroxymethylcytosine (5hmC) (Kriaucionis and Heintz, 2009). In parallel, another group identified the ten-eleven translocase (TET) proteins that hydroxylate 5mC to form 5hmC using a computational search for proteins that possess oxygenase domains similar to those found in trypanosomes that oxidize the 5-methyl group of thymine (Tahiliani et al, 2009). This family of proteins (TETs 1–3) are 2-oxoglutarate and Fe(II)-dependent enzymes that are also known as the TET methylcytosine dioxygenases. Interestingly, TET-1 is a fusion protein with a histone-lysine N-methyltransferase mixed lineage leukemia gene (MLL) in acute myeloid leukemia (Tahiliani et al, 2009). There has been considerable interest in these newly described TET proteins and it was soon recognized that hydroxylation of 5mC is likely the first step in a mechanism by which cytosine methylation is reversed in the brain (see Figure 2). MLL1, a H3K4-specific methyltransferase, is also of interest as it has been shown to be involved in the cortical dysfunction associated with some cases of SZ and because it has a role in synaptic plasticity in GABAergic neurons (Huang et al, 2007).
Within a short amount of time, the identification of this new epigenetic mark (5hmC) sparked several genome-wide mapping studies of embryonic stem cells to examine both the distribution and function of 5hmC (Wu et al, 2011a, 2011b; Ficz et al, 2011; Xu et al, 2011c). In the course of these studies, it was soon realized that TET1 plays a dual role in transcriptional regulation (Wu et al, 2011b; Williams et al, 2011b). That is, in addition to catalyzing the hydroxylation of 5mC to 5hmC (Figure 1), TET1 acts to bind and repress an overlapping set of CpG-rich promoters by interacting with the polycomb repressive complex 2 (PRC2) and/or the Swi-independent3A (SIN3A) complex (Bhutani et al, 2011; Wu and Zhang, 2011). While we continue to gain new insights into genome-wide distributions of 5hmC in ES cells and in adult brain structures, the rules regarding binding and occupancy of TETs at specific 5hmCs remain unclear as does the role of this modification in facilitating transcription. This also opens the door to the possibility that the association of TET with other proteins, eg, the –CXXC- domain containing DNA-binding proteins (see Text box 2), may act cooperatively in determining whether specific CpG dinucleotides are destined to remain methylated or hydroxymethylated and also occupied by DNMT1 or TETs acting as transcriptional co-repressors. Alternatively, hydroxymethylated cytosines may be targeted by growth arrest and DNA damage-inducible (GADD45) proteins for removal by the interaction of members of the AID/APOBEC family of 5hmC deaminases and the base excision repair (BER) pathway (see Figure 2).
DNA binding by –CXXC- domain-containing proteins.
Each of the DNMTs is comprised of flexible protein loops connecting various structural and catalytic domains that mediate distinct functions (Dhe-Paganon et al, 2011; Chédin, 2011; Jurkowska et al, 2011). The amino terminal half of DNMT1, the largest of the DNMTs, contains a number of these regulatory domains. The DMAP 1 domain is involved in the interaction between DNMT1 and the transcriptional repressor DMAP (DNA methyltransferase-associated protein 1). This domain was also thought to be responsible for the binding of DNMT1 to hemimethylated CpG sites (Fatemi et al, 2001; Araujo et al, 2001). The PCNA (proliferating cell nuclear antigen)-binding domain allows the recruitment of DNMT1 to replication foci during replication and repair (Chuang et al, 1997). DNMT1 complexes with the histone methyltransferase G9a and colocalizes with dimethyl H3K9 (H3K9me2) at replication foci (Estève et al, 2006; Sharif et al, 2007). An adjacent area contains three regions that target DNMT1 to the nucleus (nuclear localization signals) and facilitates interactions with nuclear factors (Cardoso and Leonhardt, 1999). The targeting sequence is thought to be responsible for head-to-head DNMT1 dimer formation via hydrophobic interactions (Fellinger et al, 2008). There are also the BAH1 and BAH2 (bromo-adjacent homology 1 and 2) domains in the structure of DNMT1, which have been proposed to act via protein–protein interactions to silence transcription.
DNMT1 contains a –CXXC- zinc finger domain located between amino acids 645–690 of the human protein (Pradhan et al, 2008). The –CXXC- domain spans two clusters of six and two cysteines that are separated by a variable amino-acid stretch. Within the variable amino-acid stretch is a four-peptide KFGG motif that is common to additional proteins involved in binding to DNA (Allen et al, 2006). The novel fold of the –CXXC- domain allows for the tetrahedral coordination of two zinc ions, which are maintained by a repeated-CGXCXXC motif and the two cysteine cluster (CXXRXC). The –CXXC- finger domain of CFP1 (CXXC Finger Protein 1) selectively binds to unmethylated CpGs and associates with the H3K4 methyltransferase SETD1 to facilitate H3K4 methylation (Thomson et al, 2010). Interestingly, CFP1 is highly enriched at non-methylated CpG-island-containing chromatin. The CFP1 crystal structure shows that the –CXXC- domain forms a crescent-like shape, which sits in the major groove of the DNA, preferentially at CpGG tetranucleotides (Xu et al, 2011b). The association of CFP1 with the SETD1 histone (H3K4) methyltransferases provides a mechanism by which histone-modifying proteins are targeted to specific regions of the DNA. Additional proteins that facilitate histone modifications and that contain this domain include those associated with binding DNA including methyl CpG-binding domain proteins (MBD1, Jørgensen et al, 2004), histone methylation at H3K4 (MLL1, Lee and Skalnik, 2005), and histone H3K36 demethylation (JHDM1A and 1B, Tsukada et al, 2006; KDM2A, Blackledge et al, 2010). While the –CXXC- domain is absent from DNMTs 3A, 3B, and 3L, it is present in TET1, which is responsible for converting methylcytosine to hydroxylmethylcytosine (Figure 2; Wu and Zhang, 2011; Zhang et al, 2010c).
Recent in vitro binding data show that the –CXXC- domains of several of the above proteins, including CFP1, DNMT1, MLL, and TET1, exhibit robust binding to unmethylated CpGs. MLL1 is a histone methyltranserase that methylates H3K4, which is typically associated with transcriptionally active chromatin (Ayton et al, 2004). MLL1 binds to unmethylated CpGs through its –CXXC- domain with a higher affinity than for 5mC, which is consistent with its action in marking active chromatin. The –CXXC- domains of DNMT1 and TET1 are very similar with the exception that the TET1 domain lacks the conserved KFGG motif mentioned above. DNMT1 binds strongly to unmethylated and less so to methylated CG-containing oligonucleotides with a preference for unmethylated CpGs by about 48 to 1 (Zhang et al, 2010c). In contrast, TET1 prefers unmethylated CpGs but the ratio between affinities is closer to 3 to 1. Mutant TET1 proteins containing an intact –CXXC- domain but lacking catalytic hydroxylase activity fail to induce transcription of a fully methylated promoter/reporter template (Zhang et al, 2010c). Another study demonstrated by transient expression that the –CXXC- domain of TET1 fails to bind DNA and is dispensable for catalytic activity (Frauer et al, 2011). The results suggest that while the –CXXC- domain of DNMT1 is a functionally independent binding motif, the corresponding domain of TET1 likely requires additional regions of the protein for full biological function. The presence of a –CXXC- domain may not be sufficient to confer DNA binding in isolation, although controls for determining proper folding were not included. As one example, MBD1 contains three –CXXC- domains and only one of these is responsible for CpG-binding activity (Jørgensen et al, 2004). To assess the impact of 5hmC on transcription, transient transfections of 5hmC-containing promoter/reporter constructs showed that the presence of 5hmC strongly represses transcription when this modification is proximal to a promoter. Interestingly, when 5hmC is present in the body of the gene, there is only a minimal effect on transcription (Robertson et al, 2011). Correlations between the genome-wide locations of 5mC and 5hmC and transcription have not clarified this issue and do not directly address whether these DNA modifications simultaneously reside or overlap in the same genomic/promoter regions in vivo.
Available data indicate that both DNMTs and TET proteins have the ability to modify the methylation status of CpG dinucleotides and to act as transcriptional repressors. This occurs through interactions with ancillary proteins. For example, the amino terminus of DNMT1 functions as a transcriptional repressor by directly binding to HDAC2 (Rountree et al, 2000). In addition, the lymphoid-specific helicase interacts with both DNMT1 and DNMT3B, as well as HDACs 1 and 2 to facilitate the formation of repressive chromatin (Myant and Stancheva, 2008). DNMT1 has also been shown to bind to several promoters expressed in GABAergic neurons, such as reelin and GAD67, forming repressor complexes in cooperation with HDACs, and MeCP2 (Kundakovic et al, 2007, 2009). Similarly, in ES cells, TET1 associates with MBD3, HDACs 1 and 2 as part of a complex of nucleosome remodeling and deacetylase factors which bind to 5hmC and repress transcription (Yildirim et al, 2011). Brahma-related gene 1 (BRG1), which serves as a catalytic subunit of the switch/sucrose non-fermentable (SWI/SNF) ATP-dependent chromatin remodeling complexes, acts in opposition to TET1/MBD3 and serves as an antagonistic chromatin regulator in facilitating transcription of 5hmC-marked genes. Collectively, the above studies argue that both DNMT1 and TET1 act as context-dependent transcriptional repressors in addition to their normal catalytic activities.
A comparison of recently published genome-wide studies that utilized different methodological approaches to define 5hmC status maps shows some discrepancies between studies. For example, the genome-wide distribution of TET1 binding at promoters containing both H3K4me3 (permissive) and H3K27me3 (repressive) marks (so-called bivalent promoters enriched at polycomb repressive complexes) matches the presence of 5hmC at the same locations (Matarese et al, 2011). However, the binding of TET1 to CpG island-containing promoters and the near absence of 5hmC at these promoters suggests the possibility that TET1 performs different functions depending on regional contextual cues (see Text box 2). TET1 may be pleiotropic, activating transcription by converting 5mC to 5hmC but also blocking transcription by participating in a repressor complex with SIN3A or PRC2 (Matarese et al, 2011). In other words, TET proteins may act as bifunctional switches in setting up and maintaining repressive chromatin or in facilitating demethylation by hydroxylating 5mC. By performing successive oxidation reactions, TET proteins can further convert 5mC into 5-hmC, 5-formylcytosine, and 5-carboxylcytosine (Wu and Zhang, 2011). It is too early to ascertain whether these latter modifications represent intermediates in DNA demethylation or whether they represent newly identified epigenetic marks. With the advent of single molecule real time DNA sequencing, direct detection of modified nucleotides, including 5-mC and 5-hmC, may be soon at hand. This technology is still in its early stages, but will likely enable genome-wide methylation profiling and the identification of modified epigenetic marks directly (Flusberg et al, 2010; Song et al, 2011a; Booth et al, 2012).
Activity-Induced Active DNA Demethylation
Classically, DNA methylation suppresses transcription by targeting CpG islands, as documented in the silencing of transposable elements and tumor suppressor genes (Baylin and Herman, 2000; Bird, 2002). However, it is also plausible that CpG islands serve as multifunctional chromatin-remodeling centers that send signals bidirectionally to facilitate DNA and histone methylation in response to cellular signals to modify local and more distant promoters depending on local environmental cues. In light of the recent findings that a portion of 5mCs are converted to 5hmCs, the nature of these modifications is now being re-examined. Both methylated CpGs and hydroxymethylated CpGs are sites for 5mC- (MBDs, MeCP2) and 5hmC-binding proteins that facilitate transitions between open and closed states depending on additional signals or additional regulators.
Ever since the recognition that methylation proximal to CpG island-promoters is associated with the regulation of transcription, there have been reports that this methylation is likely a reversible epigenetic mark. Passive demethylation occurs following DNA replication when daughter strands fail to retain parental methylation patterns. Because DNMT1 has a very low affinity for recognizing and binding to 5hmC, this modification is thought to lead to replication-dependent active demethylation in dividing cells. For many years, it was believed that active DNA demethylation (demethylation in the absence of cell division) was energetically prohibitive in higher organisms (Ooi and Bestor, 2008). Initial reports provided evidence that demethylation is catalyzed by DNA demethylases (Gjerset and Martin, 1982). Somewhat later, attention turned to the role of MBDs such as MBD2 in catalyzing demethylation (Bhattacharya et al, 1999; Ramchandani et al, 1999) although mice deficient in MBD2 show normal genomic methylation patterns (Hendrich et al, 2001). Subsequently, it was reported that the histone deacetylase inhibitor valproic acid (VPA) induces replication-independent DNA demethylation of a cytomegalus virus promoter (Detich et al, 2003). In addition, VPA treatment demethylates neuronal promoters such as RELN and GAD67 in vitro (Chen et al, 2002, 2011) and in vivo (Tremolizzo et al, 2002, 2005; Dong et al, 2007). Studies have also provided evidence for cyclical DNA methylation/demethylation of selected promoters (trefoil factor 1(TFF1), also known as pS2) through an action of the estrogen receptor α by a mechanism involving DNMTs 3A and 3B (Kangaspeska et al, 2008; Métivier et al, 2008). However, it seems likely that cyclical methylation of these promoters involves deamination followed by steps involving BER, in which the entire base becomes substituted (Reid et al, 2009).
The use of DNMT inhibitors provides direct evidence for a role for DNA demethylation in activating transcription of genes in neurons linked to synaptic plasticity. For example, treatment of postmitotic neurons in hippocampal slice preparations with DNMT inhibitors leads to the demethylation of various synaptic plasticity genes, including RELN and brain-derived neurotrophic factor (BDNF), and this was coupled to long-term potentiation at Schaffer collateral synapses (Levenson and Sweatt, 2005; Levenson et al, 2006; Miller and Sweatt, 2007). The treatment of mice in vivo with HDAC inhibitors attenuates the hypermethylation induced at several promoters expressed in GABAergic neurons by co-administration with methionine (MET, Dong et al, 2007). More recently, it was shown that neuronal activation of mature hippocampal neurons induces the immediate early gene GADD45β, which couples neuronal excitation and DNA demethylation (see Figure 2, Ma et al, 2009a, 2009b). Electroconvulsive treatment induces GADD45β in an NMDA receptor-dependent manner with kinetics comparable to that of other immediate early genes such as cfos and cjun. Moreover, data show that the GADD45β induction correlates with the demethylation of regulatory regions within the BDNF-IX and FGF-1B promoters (Ma et al, 2009a).
Genome-wide profiling of DNA methylation following ECT demonstrates that approximately 1.4% of the CpGs measured in dentate granule neurons exhibit changes in their methylation status and that many of these are associated with brain-specific genes linked to synaptic plasticity (Guo et al, 2011b). While these data support a link between NMDA receptor activation and GADD45β induction and downstream changes in genome-wide methylation, they provide little mechanistic insight into how these changes might come about or how specific promoters are targeted.
GADD45β has properties of an immediate early gene and acts as a transcriptional coactivator that is induced in liver by compensatory regeneration or drug-induced hyperplasia (Tian et al, 2011). GADD45β is one of a family of three small molecular size proteins (GADD45 α, β, and γ) that act as regulators of DNA demethylation (Ma et al, 2009b) through interactions with a variety of proteins involved with DNA repair, cell cycling, genome stability, stress, and immune responses. GADD45 (α, β, γ) proteins contain signature motifs considered necessary for the binding of coactivators to multiple nuclear hormone receptors (Yi et al, 2000). While GADD45β is induced in the nervous system by synaptic signaling, GADD45α appears to respond preferentially to signals related to cell damage and injury. Recent data indicate that GADD45α interacts with the nucleotide excision repair pathway (see Figures 1 and 2) which includes the DNA repair endonuclease XPG (Barreto et al, 2007).
Active demethylation of oct4 in Xenopus oocytes in vitro requires recruitment of GADD45α to the site of demethylation and binding of the repair endonuclease enzyme. Data from zebrafish embryos provide evidence that GADD45 proteins promote demethylation by coupling 5mC deamination, catalyzed by the activation-induced cytidine deaminase (AID), with a G:T mismatch-specific thymine DNA glycosylase (TDG) or methyl-CpG-binding domain protein 4 (MBD4) (Rai et al, 2008; see Figure 2). MBD4 contains both an amino-terminus methyl-CpG-binding domain and a carboxyl-terminal glycoylase domain that acts on G:T and G:U mismatches (Hendrich et al, 1999). Overexpression of AID or MBD4 in vivo also facilitates genomic DNA demethylation (Rai et al, 2008). Current thinking suggests that deamination of 5mC or 5hmC by either AID or apolipoprotein B mRNA-editing, enzyme-catalytic (APOBEC) enzymes is followed by BER (Ma et al, 2009b; Fritz and Papavasiliou, 2010; Bhutani et al, 2011; Cortellino et al, 2011). The APOBEC family of cytidine deaminases is comprised of fourteen members that are linked to a series of cellular processes that involve deamination of single-stranded DNA or RNA (Bransteitter et al, 2009). In vitro, both TDG and MBD4 have been shown to recognize AID-generated 5hmU and complete the demethylation of 5hmC (Hashimoto et al, 2012). This mechanism provides a means by which GADD45 proteins couple deamination to base excision by interacting with the AID/MBD4 complexes and members of the APOBEC family of proteins (Rai et al, 2008; Figure 2).
Bidirectional DNA Methylation Dynamics
A key question regarding the role of the DNA methylating and demethylating proteins in regulating gene expression concerns the possible bifunctional nature of DNMT1 and TET1. In vitro studies show that TET1 can bind to non-methylated C (5C), 5mC and 5hmC and repress transcription (Zhang et al, 2010c). While TET1 binds to unmethylated CpGs vs methyl CpGs with a ratio of 2.8 : 1 in vitro, the extent to which this ratio changes in vivo is not known. However, of those DNA proteins that interact with DNA through a –CXXC- domain, TET1 has the highest affinity for 5hmC. Similarly, DNMT1 also binds 5C through its –CXXC- domain (see Text box 2) and similarly represses transcription in the absence of DNMT3A. The 5C to 5mC preference ratio for DNMT1 is 48 : 1 (Zhang et al, 2010c). It seems plausible that a large number of promoters may be kept in a temporarily dormant state by DNMT1 and TET1 binding to either non-methylated or methylated CpGs, respectively (see Figure 1). The subsequent recruitment of DNMT3A, HDACs, and methyl-binding domain proteins (MBDs) by DNMT1 would anchor this dormant state by facilitating de novo methylation. The recent finding that levels of 5hmC in mouse cerebellum vary inversely with MeCP2 levels indicates that the binding of MeCP2 to 5mC might serve to protect that base from conversion to 5hmC by TET1 (Szulwach et al, 2011b). In vitro binding studies show that the MBD-binding proteins (so-called methylation readers; MeCP2, MBD1, MBD2b, MBD3, and MBD4) bind 5hmC but with a lower affinity than 5mC (Hashimoto et al, 2012). Similarly, the binding of TET1 to 5hmC in the absence of GADD45β, APOBEC, and MBD4 might also be expected to facilitate the stabilization of transcriptionally repressed chromatin. The neuronal activity-induced increase of GADD45β and additional proteins that signal DNA demethylation coordinates a shift of specific promoters between dormant and active status. It seems plausible that GADD45β targets promoters that contain 5hmC and that are not simultaneously occupied by proteins acting as repressors, including DNMTs, TETs, and MBDs. This model, although consistent with the known capacities of these proteins, awaits additional experimental testing.
Epigenetic regulation of gene expression operates through a bidirectional DNA methylation/demethylation cascade that occurs through both active and passive mechanisms (Chen and Riggs, 2011; Bhutani et al, 2011; see Figure 2). The oxidation of 5mC to 5hmC by members of the TET family of proteins is associated with changes in local chromatin architecture and the modified base can be either permissive or repressive depending on the location within the gene and the availability of proteins that bind to the modified epigenetic marks (Hashimoto et al, 2010; Zhang et al, 2010c; Williams et al, 2011b; Wu and Zhang, 2011; Xu et al, 2011c). Genome-wide mapping of DNA methylation and hydroxymethylation in embryonic stem cells (Xu et al, 2011c; Ficz et al, 2011; Pastor et al, 2011; Stroud et al, 2011; Szulwach et al, 2011a; Wu et al, 2011a; Wu and Zhang, 2011), during postnatal development (Szulwach et al, 2011b) and in the adult brain (Guo et al, 2011a, 2011b) has provided a wealth of new contextual information. The identification of genome-wide methylation and hydroxymethylation marks is useful in establishing correlations in the expression of sets of related genes or gene networks. It does not provide information regarding how specific genes or CpGs are targeted by this mechanism. The recent examination of activity-dependent changes in the DNA methylation landscape confirms and extends the idea that neuronal depolarization induces the expression of GADD45, which in turn signals additional proteins associated with DNA demethylation to various targets such as BDNF (Ma et al, 2009a; Guo et al, 2011b). How these gene-specific changes are recognized as targets by the various enzymes is only beginning to be explored (Lienert et al, 2011; Bird, 2011). Ultimately however, we will better appreciate DNA methylation as a potential mechanism associated with the etiology of SZ and BP+ once we are able to superimpose the effects of pre- and postnatal stresses on the locations of these epigenetic marks both during development and in the adult brain.
EVIDENCE SUPPORTING ALTERED METHYLATION IN SZ AND BP+
One Carbon Metabolism and SZ
One carbon metabolism has been linked to the etiology of SZ from reports regarding high maternal homocysteine levels, dietary folate deficiencies, methylene-tetrahydrofolate reductase (MTHFR) polymorphisms and the involvement of one carbon substrates that bind to the glycine site of the NMDA-selective glutamate receptor (Krebs et al, 2009; Iwamoto and Kato, 2009). MET is a precursor of SAM, the universal methyl donor. Following methyl group transfer, SAM is converted to SAH, which is subsequently hydrolyzed producing homocysteine. Elevated levels of homocysteine have been reported in the plasma of adolescent male SZ patients (Levine et al, 2002; Adler Nevo et al, 2006) and in newly admitted male schizophrenic patients (Applebaum et al, 2004; Levine et al, 2005). Interestingly, in spite of the marked difference in plasma homocysteine levels, no differences were observed in cerebrospinal fluid levels between SZ patients and controls (Levine et al, 2005). In at least one report, elevated plasma homocysteine was linked to tardive movement disorders (Lerner et al, 2005). Follow-up studies confirmed the original finding that elevated homocysteine was associated with SZ and suggested that a common polymorphism (677 C>T) in the MTHFR gene might be responsible (Muntjewerff et al, 2006). Hyperhomocysteinemia did not appear to impact global DNA methylation in leukocytes of patients (Bromberg et al, 2008) or in mice with artificially elevated levels of homocysteine (Bromberg et al, 2011). Archived maternal serum samples assayed for levels of homocysteine during pregnancy indicate that an elevated homocysteine level, particularly during the third trimester, is associated with a greater than twofold increase in risk for SZ (Brown et al, 2007). Folate supplementation has been used in clinical studies of SZ patients to alleviate negative symptoms with somewhat mixed results (Levine et al, 2006; Hill et al, 2011).
Transmethylation and MET Loading in SZ Patients
The transmethylation theory came about after two decades of work that suggested a striking structural similarity between psychotomimetics and naturally occurring neurotransmitters (Matthysse and Lipinski, 1975). For example, dopamine and norepinephrine and N,N-dimethyltryptamine and serotonin differ by a single methyl group. It seemed plausible that methylation of catecholamines might produce dimethoxyphenethylamine, which is structurally related to mescaline. Analysis of urine samples from SZ patients provided mixed results. The advent of mass spectrometry showed dimethoxyphenethylamine to be present in the urine of SZ patients and also in several naturally occurring sources (Creveling and Daly, 1967; Stabenau et al, 1970). N,N-Dimethyltryptamine (DMT) was also considered as a pathogenic agent and the discovery of an enzyme in brain capable of synthesizing DMT from tryptamine fostered this notion as well (Mandell and Morgan, 1971; Saavedra and Axelrod, 1972).
A series of MET-loading studies were performed in the 1960's and 1970's (reviewed in Wyatt et al (1971); Costa et al (2003) and Grayson et al (2009)). Data from at least one of these studies indicate that MET loading causes an exacerbation of psychotic symptoms (Pollin et al, 1961). These data were subsequently replicated by several other groups (Cohen et al, 1974). Results from the MET-loading experiments appear to invalidate the transmethylation theory as no evidence was ever uncovered showing increased levels of methylated catecholamine metabolites in SZ patients. Moreover, NP (non-psychiatric) subjects showed no response to MET loading. An analysis of prefrontal cortical tissue (Brodmann's area 9) demonstrates that in patients with SZ and BP+ there is an approximate twofold increase in the levels of SAM (Guidotti et al, 2007). Based on available data, it seems plausible that SZ patients treated with MET may have suffered from the consequences of a MET-induced hypermethylation of CpG island-containing promoters (Costa et al, 2003; Grayson et al, 2009).
DNMTs in SZ and BP+
The relevance of the RELN and GAD67 promoters to the current discussion is that previous studies have shown these genes to be consistently downregulated in SZ and BP+ patients. The downregulation of GAD67 mRNA in the prefrontal cortex (PFC) of SZ patients was originally reported some years ago (Akbarian et al, 1995), while the observation that both GAD67 and RELN mRNAs and proteins are downregulated in GABA neurons of the PFC of patients with SZ and BP+ came somewhat later (Impagnatiello et al, 1998; Guidotti et al, 2000; Fatemi et al, 2000). GAD67 is one of two enzymes associated with the formation of GABA, whereas RELN is an extracellular matrix protein synthesized and secreted from GABAergic neurons in the adult brain. Neuroanatomical and neurophysiological evidence support the concept of a reciprocal interaction between GABAergic interneurons and glutamatergic pyramidal principal neurons in the cortex and hippocampus (Figure 3). These reciprocal interactions are altered in SZ and BP+ patients and this is likely due to a disruption of GABAergic neuron function which disrupts inhibitory neurotransmission and activates excitatory pyramidal neurons. An inhibitory/excitatory imbalance in various brain circuitries likely underlies the onset of positive and negative symptoms and the cognitive dysfunction seen in SZ patients.
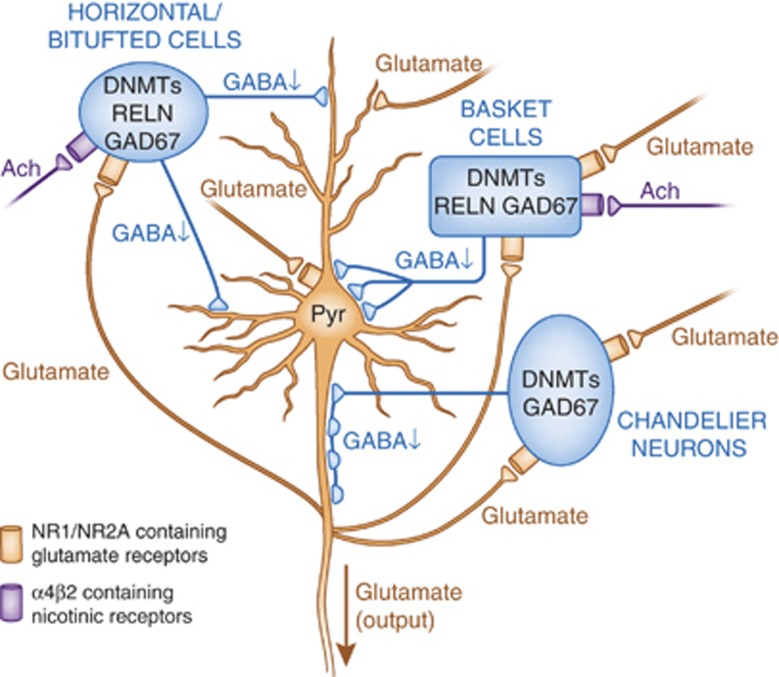
Figure 3.
DNA methyltransferase (DNMT) overexpression leads to the downregulation of mRNAs in GABAergic neurons, increased methylation, and reduced gamma-aminobutyric acid (GABA) output (hypofunction). Schematic representation of the principal neuronal circuits in the cortex showing the reciprocal interaction between GABAergic innervation of pyramidal neurons and glutamatergic innervation of horizontal/bitufted, basket, and chandelier GABAergic interneurons. The GABAergic promoter downregulation in schizophrenia (SZ) and bipolar disorder with psychosis (BP+) patients is characterized by increased DNMT1 and 3A, and reduced GAD67, RELN and a variety of interneuron markers (Fung et al, 2010). These neurons also exhibit compromised expression of additional genes associated with inhibitory neuron function including NR1/NR2-containing-NMDA selective glutamate receptors and α4β2-containing nicotinic receptors. Glutamatergic inputs (shown in red) are meant to exhibit the excitatory input that arises from proximal pyramidal neurons or additional brain regions such as the thalamus. Ach, acetylcholine; Pyr, pyramidal neuron.
The genes for both RELN and GAD67 have been studied extensively in the context of the epigenetics of psychiatric disorders (Chen et al, 2002, 2011; Abdolmaleky et al, 2005; Grayson et al, 2005; Akbarian and Huang, 2006; Zhang et al, 2010a). An analysis of the human RELN promoter indicates that it is embedded in a large CpG island and that its expression, at least in vitro, correlates with DNA methylation status (Chen et al, 2002). Various HDAC and DNA methylation inhibitors induce expression of the endogenous RELN mRNA in neuroprogenitor cells and this increase in mRNA is marked by the appearance of DNase I hypersensitive sites, which map to GC-rich regions upstream of the corresponding promoter. The locations of the hypersensitive site(s) are proximal to several Sp1 sites critical for transcription based on transient transfection studies (Chen et al, 2007). In vitro methylation of the promoter abolishes downstream transcriptional activity. An evaluation of the GAD67 gene shows that this promoter is also GC rich and is also likely regulated through changes in promoter methylation (Chen et al, 2011). These findings coupled with the MET-loading studies prompted the hypothesis that increased expression of DNMTs in human brain might at least in part be responsible for the downregulation of mRNAs in GABAergic neurons of patients with psychosis.
In situ hybridization studies show that in SZ and BP+ subjects DNMT1 mRNA is higher in BA10 cortical layers I–IV than in patients with major depressive disorder, bipolar disorder pateints without psychosis, and non-psychiatric subjects (NPS) (Veldic et al, 2004). The layer-specific increases in DNMT1 mRNA were shown to parallel the reduced levels of GAD67 and RELN mRNAs and suggest an inverse correlation between DNMT1 and these transcripts. In neurons, DNMT1 mRNA co-localizes with GAD65/67 protein and RELN mRNA (see Figure 4). DNMT1 is also preferentially expressed in GABAergic interneurons in BA9 of the McClean 66 cohort (Veldic et al, 2005). This study demonstrates that DNMT1 mRNA and protein are increased in cortical layers I, II and IV of BA9 of SZ and BP+ patients. The polyclonal antibody used to detect DNMT1 immuno-histochemically is directed against an amino-terminal synthetic peptide that does not cross-react with other antigens as determined by western blotting. The data provide evidence of a negative correlation between DNMT1 mRNA levels and the numbers of GAD67-immunopositive neurons. Previous work shows that in the adult brain RELN is co-expressed in the majority of GABAergic neurons (Pesold et al, 1999; Guidotti et al, 2000; Figures 3 and 4). A co-variance analysis of subject demographics ruled out the possibility that these data might be due to postmortem interval, brain pH, or RNA integrity number. Interestingly, DNMT1 increases fail to occur when patients with psychosis are treated with both an antipsychotic and VPA (Veldic et al, 2005).
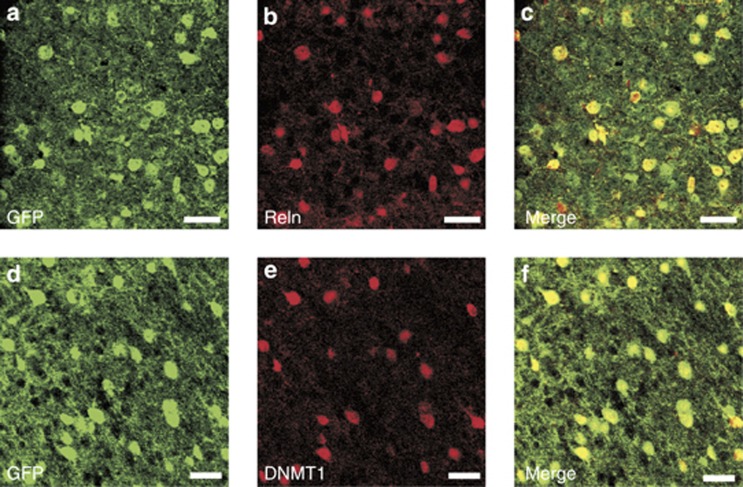
Figure 4.
Co-localization of DNA methyltransferase 1 (DNMT1) and RELN immunoreactivities with GAD67 expression in mouse cortical neurons. The enhanced green fluorescent protein (GFP) was knocked into the GAD67 start codon to create GAD67+/−mice (Tamamaki et al, 2003). In these mice, GFP is expressed from the GAD67 promoter so that GFP immunoreactivity could be used to mark GAD67-positive (GABAergic) neurons. Fixed sections were incubated with their respective primary and secondary antibodies (Kadriu et al, 2011). Top panels show the co-localization of GFP immunoreactivity (GAD67) (a) and RELN (b) immunoreactivity. The merged signal shows co-localization (c). Similarly, GFP (GAD67) immunoreactivity (d) co-localizes with DNMT1 (e) as shown in f. Scale bar=40 μm. For details regarding immunohistochemistry, see Kadriu et al (2011).
A further study of DNMTs in SZ and BP+ patients compared with NP subjects shows that both DNMTs 1 and 3A are increased in cortical GABA neurons of BA10. Unlike DNMT1, DNMT3A is overexpressed only in layers I and II (Zhubi et al, 2009). DNMT3B mRNA was examined and not detected in any cortical layer. Interestingly, both DNMT1 and DNMT3A mRNAs are overexpressed in peripheral blood lymphocytes of SZ patients. Both DNMT mRNAs are increased approximately twofold and neither increase is associated with time or dose of antipsychotic treatment (Zhubi et al, 2009). Laser capture microdissection (LCM) of layer I GABAergic neurons from the PFC of SZ patients allowed for a more robust (threefold) increase in DNMT1 mRNA compared with layer I GABA neurons of NP subjects (Ruzicka et al, 2007). The DNMT1 transcript is readily detected in these neurons and is virtually absent from layer V pyramidal neurons. Similar to previous studies, the increase in DNMT1 mRNA levels correlates with decreased levels of GAD67 and RELN mRNAs in these same interneurons (Ruzika et al, 2007). The findings provide evidence for an epigenetic disruption of GABAergic neurons that has the potential to impact the expression of numerous downstream target promoters ultimately impacting neuronal function. Postnatal ablation of the NR1 (GRIN1) subunit of the NMDA receptor in GABAergic neurons of conditional knockout mice results in SZ-like symptoms during adolescence (Belforte et al, 2010; Nakazawa et al, 2012). It remains a possibility that NR1 or other subunits of the NMDA receptor could be targets of DNMT1 action which would, in turn effectively produce a defect in GABAergic transmission.
Recent evidence suggests that DNMT1 can act as a repressor in binding to promoters and because it is over-expressed in GABAergic neurons of SZ and BP+ patients, the downregulated promoters and corresponding mRNAs are likely characteristic of inhibitory neurons. A repressor role for DNMT1 is consistent with the recent crystal structure analysis of both mouse and human DNMT1 bound to DNA-containing unmethylated CpGs (Song et al, 2011b). The binding of the –CXXC- domain of DNMT1 and its downstream BAH1 linker region to unmethylated CpGs prevents the CpGs from coming into contact with the active site. This autoinhibitory mechanism prevents de novo methylation and may serve to stabilize the binding of DNMT1 to unmethylated CpGs in the DNA (see Text box 2). To date, no structures for DNMT1 bound to hemi-methylated DNA have been reported, suggesting the possibility that this enzyme/substrate complex may be rapidly turned over. The data are consistent with the notion that gene repression may occur in GABAergic neurons of SZ and BP+ patients by a DNMT1-mediated mechanism that is independent of DNA methylation.
DNA Methylation in SZ and BP+
Candidate genes –dopamine D2 receptor (DRD2)
Initial reports that focused on examining methylation in the context of psychiatric disease included those that examined the promoters of genes associated with SZ, such as the dopamine D2 receptor (DRD2), as a means of studying differential regulation of gene expression (Popendikyte et al, 1999). For example, the methylation profiles corresponding to a defined region of the DRD2 promoter would be expected to be different in neurons from the striata (DRD2-expressing) compared with the same DNA region in lymphocytes (non-expressing) of individuals with no history of psychiatric disorders. These authors reported finding that the extent of methylation in DNA from the DRD2 promoter of lymphocytes is higher than that detected in the striatal samples. Moreover, the right striatal hemisphere showed higher levels of methylation and the amount of methylation tended to increase with the individual's age (Popendikyte et al, 1999). Two early reports on methylation in monozygotic twins evaluated methylation differences in sets of twins either concordant or discordant for SZ (Tsujita et al, 1998; Petronis et al, 2003). The earlier study found discrepancies in the methylation of global Not1 restriction cleavage sites in DNA from leukocytes and the authors argued that these differences might reflect differential gene regulation which was also consistent with the absence of SZ in one twin (Tsujita et al, 1998). The second study used bisulphite conversion of lymphocytic DNA to fine-map methylation differences in the DRD2 gene (Petronis et al, 2003). Numerous differences between MZ twins were reported with the discordant ‘affected' twin being epigenetically closer to the concordant ‘affected' twins (Petronis et al, 2003). The authors also note that while the analysis included only several hundred base pairs of the DRD2 promoter, there was a large amount of non-uniformity across clones from the same individual and from different individuals. This finding was used as an argument for the universality of intra- and inter-individual epigenetic variation (Petronis et al, 2003). While this concept is discussed in the following section, the notion that epigenetic marks (in the case methylation) are highly variable may be related to stochastic events associated with major psychosis (McGuffin et al, 1994; Woolff, 1997).
Candidate genes-RELN
During the early part of this century, there was considerable interest in the biological role of methylation and the regulation of genes such as RELN, DRD2, OPRM, HTR2A, COMT, ALOX5, and others that are downregulated in psychosis (Petronis et al, 1999; Andria and Simon, 1999; Chen et al, 2002; Zhang et al, 2004; Abdolmaleky et al, 2005, 2006; Polesskaya et al, 2006). Methylation was proposed as a mechanism for the silencing of genes expressed in cortical neurons in SZ. This hypothesis was based, in part, on a re-evaluation of contemporary concepts of genetic susceptibility genes. In addition, the MET-loading clinical trials described above were consistent with the so-called methylation hypothesis. The observations that DNMT1 and 3A were more highly expressed in GABA neurons of SZ and BP+ patients (Veldic et al, 2004, 2005) and reports that gene expression in the brain is inversely correlated with methylation at specific promoters support this hypothesis (Costa et al, 2002, 2003).
Subsequently, two studies appeared describing higher amounts of methylation in the human RELN promoter in postmortem SZ cortices compared with NP subjects (Abdolmaleky et al, 2005; Grayson et al, 2005). Each group analyzed bisulphite-modified DNA by sequencing of amplified DNA or by methylation-specific PCR. The locations of higher methylation noted in the first study, which analyzed postmortem BA9/10 from the Harvard Tissue Resource Center, were between −440 and −360 bp relative to the RNA start site (see Chen et al, 2002 for numbering) and flanking putative CRE and SP1 transcription factor-binding sites (Abdolmaleky et al, 2005). The hyper-methylated CpG sites in the second study were located closer to the RNA start site with the most common sites at −139 and −134 bp (Grayson et al, 2005, 2006). Point deletion constructs were generated and used to show that the removal of the single C at the −139 bp position abolished 50% of the promoter activity. As noted, methylation profiles varied considerably within this latter study (Grayson et al, 2005), which evaluated a small number of samples from two patient cohorts, including occipital cortices from the Stanley Medical Research Institute (ten SZ and ten NPS) and BA9/10 from the Harvard Tissue Resource Center (five SZ and five NPS). Interestingly, there was some consistency in each patient cohort, but the extent of positional overlap between different groups of patients occurs only at three positions. A third report (Tochigi et al, 2008) of methylation at the RELN promoter in postmortem psychiatric subjects, used pyrosequencing of bisulfite-modified DNA to evaluate methylation levels in DNA obtained from BA10 of the 15 SZ and 15 NP from the Stanley Medical Research Institute. This latter study found no differences between NP and SZ subjects at any of the sites reported by either previous study. Moreover, this latter group showed nearly no methylation of any of the CpG sites within the RELN CpG island (Tochigi et al, 2008). Finally, using BssHII methylation-sensitive restriction site mapping of human RELN from postmortem forebrain tissue of SZ, BP, and NP subject of the Stanley Medical Research Institute, another study reported RELN promoter methylation data between these diagnostic groups (Tamura et al, 2007). This latter study demonstrated a statistically significant correlation between age and levels of DNA methylation in healthy subjects. In contrast, no such correlations were evident in either SZ or BP patient brains. These data confirm that methylation is most likely a key element in regulating RELN expression and that aberrant methylation status is likely associated with psychiatric disorders (Tamura et al, 2007).
More recently, a temporal analysis of RELN DNA methylation in postmortem temporocortical tissue (BA 41 and 42) from pre- and postpubertal individuals was reported (Lintas and Persico, 2010). The data indicate that the promoter is only lightly methylated in pre-pubertal tissue, whereas in postpubertal DNA, RELN promoter methylation increases 25-fold, particularly in the region between −131 and −98 bp relative to the transcriptional start site (Lintas and Persico, 2010). The somewhat striking increase is the result of a substantially greater number of methylated bases and a fourfold rise in the intensity of methylation at each methylated CpG (Lintas and Persico, 2010). The authors argue that the paucity of methylation in the RELN promoter (and hence high expression of RELN mRNA and protein) in prepubertal individuals allows RELN-mediated synaptic plasticity during early childhood (Lintas and Persico, 2010). At or during puberty, various hormones and/or neurosteroids associated with adolescence could constrain synaptic plasticity, hence enhancing vulnerability to neurodevelopmental disorders. Consistent with this logic is the possibility that steroid hormones facilitate increased DNMT1 gene expression, which would boost either DNA methylation or DNMT-mediated repression at promoters expressed in GABAergic neurons. In susceptible individuals, this might contribute to the onset of SZ as adolescence is a sensitive time period for the remodeling of cortical and limbic circuits (Sisk and Zehr, 2005). Analysis of the RELN promoter in a variety of human cancers shows that RELN is often methylated and that RELN-expressing cancers are less aggressive than RELN-negative cancers (Sato et al, 2006; Stein et al, 2010; Okamura et al, 2011; Dohi et al, 2010). In each of these reports, the data show that increased methylation of the RELN promoter is consistent with transcriptional silencing as measured by RELN mRNA levels.
Candidate genes-GAD67
Analysis of GAD67 promoter methylation patterns was carried out using chromatin isolated from the PFC enriched in methyl-histone-containing mononucleosomes (Huang and Akbarian, 2007). By precipitating chromatin with antibodies against either H3K4me3 (open) or H3K27me3 (closed), these researchers were able to fractionate open and repressive chromatin from total chromatin which allowed for substantial signal amplification (Huang and Akbarian, 2007). The study showed that while GAD67 mRNA is reduced in SZ patients, the amount of methylation at the corresponding promoter in these patients is reduced significantly in the repressive chromatin fraction (Huang and Akbarian, 2007). A recent in vitro examination of the regulation of the human GAD67 promoter shows that there are key regions that exhibit differential methylation and these correlate with the ability of this promoter to drive expression (Chen et al, 2011). The differentially methylated regions lie just downstream of the first exon and were not analyzed in any of the above-mentioned studies (Mill et al, 2008; Huang and Akbarian, 2007).
Genome-wide methylation studies
A comprehensive epigenome-wide study of methylation in psychosis using postmortem human tissue detected significant methylation differences in a large number of genes between SZ and BP patients or combined (major psychosis group) vs controls (Mill et al, 2008). For these studies, the unmethylated fractions of each genome were enriched using a combination of methylation-sensitive restriction enzymes and the DNA was PCR amplified. The amplified DNA was hybridized to a CpG island microarray to detect DNA methylation (Mill et al, 2008). The microarray screen showed evidence of psychosis-associated differences at numerous loci for genes involved in neurotransmission, brain development, mitochondrial function, stress response, and additional pathways linked to disease etiology (Mill et al, 2008). Moreover, data were parsed by psychiatric disorder as well as gender (SZ and BP vs NPS for males and females, separately). Some of the observed differences in methylation were present in genes linked to disease etiology and genes relevant to both glutamatergic and GABAergic signaling. While promoter methylation levels showed differences based on gender, mRNA levels did not always correlate. For example, the vesicular glutamate transporters 1 and 2 (VGLUT1 and VGLUT2) exhibit complementary expression patterns in glutamatergic neurons of the brain (Fremeau et al, 2001). Interestingly, the promoter of VGLUT1 is hypermethylated in female SZ DNA (Mill et al, 2008), but the corresponding mRNA is downregulated in both male and female SZ postmortem human brain (Eastwood and Harrison, 2005). In contrast, VGLUT2 mRNA is upregulated in SZ patients (Smith et al, 2001) and the promoter is hypomethylated in DNA obtained from female SZ samples. KCNJ6, a G protein-coupled potassium channel linked to GABA neurotransmission, is hypermethylated in SZ (males and females) and major-psychosis males. MARLIN-1, a RNA-binding protein that regulates GABAB receptors, is hypermethylated in SZ (males and females), BP (males and females), and major psychosis (females only). In addition, there was a strong correlation between methylation of the MEK1 gene promoter (mitogen-activated protein kinase) and lifetime antipsychotic use in SZ patients (Mill et al, 2008). An analysis of methylation across a selected set of some 12 or so positive genes was performed to compare the microarray data and site-specific methylation patterns (as measured by pyrosequencing). For many of the genes that show changes in DNA-methylation, there is a good agreement with reported changes in the corresponding mRNA levels (Mill et al, 2008). Candidate gene analysis of RELN (Abdolmaleky et al, 2005; Grayson et al, 2005) and COMT (Abdolmaleky et al, 2006), two genes previously reported to be hyper- and hypo-methylated, respectively, showed no evidence of methylation changes associated with either SZ or BP (Mill et al, 2008).
Analysis of methylation in peripheral blood DNA from unique sets of twins indicates that numerous loci exhibit disease-specific methylation changes between twins discordant for SZ and BP disorder. The top psychosis-associated differentially methylated region is located in the promoter of ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 (ST6GALNAC1) (Dempster et al, 2011). The ST6GALNAC1 promoter is hypomethylated in affected individuals compared with their unaffected co-twin and the corresponding mRNA encodes a protein important for cell–cell interactions. The authors note that only five of the one hundred or so differentially methylated regions were also identified in the previous epigenome-wide analysis (Mill et al, 2008). These include gametogenetin (GGN), solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (SLC17A7), single-strand-selective monofunctional uracil-DNA glycosylase 1 (SMUG1), SRY (sex determining region Y)-box 1 (SOX1) and transcription factor 7-like 2 (T-cell specific, HMG-box) (TCF7L2), suggesting that these methylated DNA regions warrant additional study.
A temporal evaluation of DNA methylation profiles in CpG islands showed a number of genes in which DNA methylation increases consistently with age (Siegmund et al, 2007). A more recent analysis of the temporal profiles of site-specific methylation shows that overall patterns of genome-wide methylation are age dependent (Numata et al, 2012). Another report evaluated CpG methylation at some 27 000 CpGs in frontal and temporal cortices, cerebellum and pons and demonstrated a significant and consistent correlation between CpG methylation and age (Hernandez et al, 2011). Moreover, those sites, where this association is significant reside close to promoters associated with DNA binding and transcriptional regulation. In the former study, at least four types of developmental profiles were observed: (1) those genes in which methylation increases progressively with age, (2) those that exhibit methylation increases until ∼10 years of age and then remain constant, (3) those that are highly methylated at birth and decline during the first decade reaching a plateau, and (4) those genes that possess low levels of methylation until ∼50 years of age and increase thereafter. These data underscore that methylation at specific loci varies with age and also that age dependence must be a consideration when evaluating methylation in disease cohorts. GAD67 falls into the first group of genes that become progressively methylated with age. As mentioned previously, the expression of GAD67 has been shown by a number of groups to be downregulated in SZ brain.
Genome-wide analysis of DNA methylation in neuronal (NeuN)-positive neurons isolated from the cortex indicates that neuronal nuclei have a greater potential to change their methylation profiles than non-neuronal nuclei (Iwamoto et al, 2011). It seems evident that because DNA methylation is cell-type specific, the ability to discern subtle changes in methylation in bulk tissue may be difficult to establish with a high degree of certainty. That is, until the sensitivity of methyl-mapping approaches increases to the point at which single (GABA vs pyramidal) neurons or small groups of neurons are able to be assessed with confidence, the cellular heterogeniety of tissue from postmortem brain will be problematic. Moreover, in the absence of functional data defining the location of regulatory elements corresponding to promoters and enhancers, finding the locations of the methylation differences is also problematic (Grayson, 2010). A comparison of site-specific DNA methylation levels in NeuN+ nuclei compared with prefrontal cortical tissue shows that the global methylation status of neurons is lower and promoters exhibit a higher degree of interindividual variation. In contrast, the DNA methylation patterns obtained from NeuN− nuclei are identical to bulk cortex. In other words, separation of neuronal and non-neuronal nuclei from bulk cortical tissue allows for a comparison of the methylation patterns in each (Iwamoto et al, 2011). DNA methylation patterns detected from the analysis of neuronal nuclei were distinctive and correlated with promoter function as measured by transcription factor-binding site analysis. It seems likely that the methylation patterns observed when isolating DNA from postmortem brain tissue may be overshadowed by the methylation signals present in non-neuronal chromatin. In addition, neuronal methylation patterns from phenotypically distinct types of neurons contribute to the patterns observed when examining NeuN+ nuclei. This means that the patterns observed from isolated neurons represent the summation of signals arising from both excitatory (glutamatergic) and inhibitory (GABAergic) neurons. Moreover, both types of neurons can be further subdivided based on additional criteria.
A lack of correlation between gene expression and promoter proximal methylation in material dissected from the dorsolateral PFC was reported in a recent study (Numata et al, 2012). These authors showed that across all CpG sites, methylation is equally likely to positively or negatively correlate with gene expression (Numata et al, 2012). However, as noted above, the methylation signals in the total population of cells (neurons and glia) contribute to gene-specific methylation signals but only a fraction of these express the corresponding gene of interest. It would be highly unlikely that the DNA used for the methylation analysis would be enriched from only one type of neuron. Assuming that increased DNMTs leads to increased methylation, to accurately assess the consequences of DNMT1 and DNMT3A overexpression in SZ or BP+ prefrontal cortical GABAergic neurons will require the isolation of sufficiently large numbers of these neurons by LCM or by another technology. It would also be important to highlight changes proximal to transcription factor-binding sites near RNA start sites to simplify subsequent correlation analyses. While a direct link between the methylation of specific genes and psychosis has remained elusive, technological advances used in evaluating epigenomic profiling are improving (Rutten and Mill, 2009). Collectively, the data suggest that site-specific DNA methylation patterns are dynamic and are modulated by environmental, hormonal, nutritional, behavioral, and pharmacological influences. The molecular mechanisms underlying how these processes are targeted to specific CpGs is incompletely understood. More precisely, the rules governing where (promoter vs gene body vs intergenic regions) and which CpGs within clusters of CpG dinucleotides in islands, shores, and shelves have not yet been deciphered.
Variability in DNA methylation analyses
A key issue related to DNA methylation studies remains the high variability in not only the same individual but also between individuals and between collections of cohorts. While DNA methylation appears to be stable to pH changes (Ernst et al, 2008), the possibility remains that experimental differences other than pH account for different findings. This variability is most likely related to uncertainties associated with either the methodological approach used or the cellular heterogeneity of the tissue from postmortem human tissue (Pidsley and Mill, 2011; Abdolmaleky and Thiagalingam, 2011). With bisulphite-modified genomic DNA, there can be problems with incomplete conversion of non-methylated cytosines, particularly in regions where the GC content is particularly high. This phenomenon would lead to variability in the profiles and the appearance of methylated bases when the actual positions are not modified (false positives). DNA methylation is a cell-specific event and different neurons of the brain exhibit altered methylation patterns depending on neurotransmitter phenotype and neuronal function. When analyzing pieces of tissue for methylation levels, the signal obtained will represent a composite of the signals present in the specific population of neurons and glia. Another source of variability between individuals is the heterogenous nature of psychiatric disorders, discussed in the introduction. Not all SZ patients present with the same symptoms and there appears to be a wide range of differences in terms of symptom severity. Because patient clinical presentation reflects clusters of symptoms, it would be helpful to have an appreciation of which mRNAs and in which neurons these likely contribute to distinct symptom groups. Then it might be possible to understand which changes are more likely to be consistent between individuals with similar diagnoses. Recent data also show that in the cortex of patients with psychosis there is increased TET1 mRNA and protein and an increase in the amount of 5hmC levels at the GAD67 and BDNF promoters (Dong et al, 2012). Because 5hmC is the oxidation product of 5mC and is not detected using conventional bisulfite conversion, definitive evaluation of promoter methylation levels will likely require neuronal-specific measurements of both 5mC and 5hmC. As discussed below, nicotine improves cognition and attention in patients diagnosed with psychiatric disoders (Maloku et al, 2011; Wallace and Porter, 2011). In rodents, nicotine and α4β2 agonists have been shown to reduce the expression of DNMT1 and DNA methylation in the cortex of mice (Satta et al, 2008; Maloku et al, 2011). As a high percentage of SZ patients smoke cigarettes, this raises another potential source of variability among patients. Finally, various antipsychotic medications and the mood-stabilizing drug VPA have been shown to impact gene expression and DNA methylation in mice and in human neuroprogenitor cells. Therefore, the medications used in treating psychiatric patients are also likely to contribute to steady-state promoter methylation levels (Chen et al, 2002; Guidotti et al, 2009, 2011).
DNA Hydroxymethylation in SZ and BP+
In psychotic patients, abnormal cortical mRNA expression patterns may be the consequence of overexpression of DNMTs and the hypermethylation of promoters in GABAergic neurons. However, proteins of the TET enzyme pathway modify the 5mC mark by hydroxylating the methyl moiety to form 5hmC (Figure 2). It has been shown that hydroxylation of 5mC by TET promotes active DNA demethylation and increased gene transcription in the dentate gyrus of the adult mouse brain (Guo et al, 2011a). Because multiple candidate genes are downregulated in the brains of psychotic patients, one might anticipate finding reduced conversion of 5mC to 5hmC, perhaps owing to decreased amounts of the TET family of proteins. In other words, given the overall reduced mRNA expression observed in cortical and other brain areas of psychotic patients, it would be consistent that the overall levels of repressive epigenetic marks might increase. A recent analysis of the levels of TET mRNAs and proteins in the parietal cortices of psychotic patients shows a marked increase in the levels of TET-1 (and not TET2 or 3) mRNA (Dong et al, 2012). This increase is associated with an increased genome-wide level of 5hmC and also an increase of 5hmC levels at the BDNF and GAD67 promoters. However, the BDNF and GAD67 promoters are downregulated in cortical neurons of SZ and BP+ patients.
There are two plausible explanations for the increase in TET1 expression and reduced mRNA expression observed in psychotic patients. The first is that downstream cytidine-deaminating enzymes that convert 5hmC into 5hmU are also reduced in psychotic patients preventing subsequent demethylation steps and favoring an accumulation of 5hmC. In this case, 5hmC would act as an epigenetic modification that recruits repressor proteins such as MBD3 (Yildirim et al, 2011) or members of co-repressor complexes (Williams et al, 2011a) suppressing transcription (Robertson et al, 2011). This has been most extensively studied in embryonic stem cells (Williams et al, 2011a). A second possibility is that TET1 acts as a repressor independent of its enzyme activity. Because of its ability to bind CpGs via its –CXXC- domain, TET1 could act to repress transcription either directly or indirectly by associating with co-repressor proteins essential for inhibiting the transcription of subsets of genes (see Text box 2). The –CXXC- domain present in TETs 1 and 3 contains sites for two zinc atoms. These zinc-finger-like motifs allow binding to 5C, 5mC and 5hmC (Xu et al, 2011b). In either case, the presence of the –CXXC- domain in TETs 1 and 3 and other proteins provides a mechanism whereby under certain conditions 5hmC represents a repressive DNA modification.
DNA Demethylation in SZ and BP+
Members of the GADD45 immediate early gene family facilitate active DNA demethylation by recruiting cytosine deaminases and thymine glycosylases to specific promoter regions (BDNF-IX, FGF-1) in response to activity-dependent cues for rapid transcriptional activation in mouse brain (Ma et al, 2009a). The expression of several components of the DNA-demethylation pathway was measured in postmortem PFC or the inferior parietal lobule of psychotic patients (Gavin et al, 2012, Dong et al, 2012). The levels of GADD45β mRNA and protein are increased nearly threefold, while a 50–60% decrease in APOBEC 3A and 3C is evident with no changes in the levels of thymine glycosylases. GADD45β is increased particularly in cortical layers II, III, and V (Gavin et al, 2012). GADD45β immunoreactivity is detected in cortical pyramidal neurons and to a lesser extent in GABAergic neurons (Gavin et al, 2012). This suggests that the increased GADD45β mRNA levels might be due to increased basal levels of excitation, perhaps owing to the GABA hypofunction attributed to reduced levels of GAD67 in patients with psychosis (Akbarian et al, 1995; Guidotti et al, 2000; Volk et al, 2000). Based on the observed increase of GADD45β in the brains of psychotic patients, one might expect increased demethylation at specific promoter regions with an associated increased transcriptional activity. When BDNF IX (BDNF-IXabcd) is used as the readout to determine the effects of GADD45β expression in cortical tissue from psychotic patients, BDNF-IX shows reduced methylation (based on methyl DNA immunoprecipitation assays (MeDIP) and more hydroxymethylation (based on hMeDIP assays)). The amount of GADD45β binding to this same promoter region is significantly less (Gavin et al, 2012). Further, there is reduced BDNF-IX abcd mRNA expression. Given that GADD45β does not contain a DNA-binding domain in its structure, these observations suggest that the reduced amounts detected by ChIP in brains of psychotic patients are likely the consequence of reduced interactions of GADD45β with a secondary protein(s), possibly even covalently modified histone tails (see Figure 2). On balance, we suggest that DNA-methylation dynamics may be altered in psychosis, with an increase of DNMT1 binding to 5mC at promoters and transcriptional start sites owing to the DNMT overexpression, and an even larger increase of 5hmC due to the increase in TET1 protein.
DNA HYPERMETHYLATION AS A TARGET OF DRUG ACTION
Epigenetic Pharmacology of Nicotinic Acetylcholine Receptors
Tobacco smoking is significantly more common in the mentally ill than in the population at large. The prevalence of abuse and resistance to cessation is especially evident in patients with anxiety disorders, major depression, schizoaffective disorder, and SZ (Dierker and Donny, 2008; Kumari & Postma, 2005; Ziedonis et al, 2008; Matthews et al, 2011). As nicotine is the major cholinergic component inhaled in tobacco smoke and both the expression and function of nicotinic acetylcholine receptors (nAChRs) are downregulated in the brains of SZ and BP+ patients (reviewed by Albuquerque et al (2009)), it has been proposed that the inhalation of nicotine in tobacco-smoking patients is an attempt to correct a cholinergic (nicotinic) neurotransmission deficit. Indeed, protracted and repeated nicotine administration to laboratory animals and humans increases the expression of the high-affinity nAChRs in the brain (Auta et al, 1999; Breese et al, 2000). In the cortex and hippocampus, nicotine primarily targets high-affinity α4β2 and low-affinity α7 nAChR subtypes, which are abundantly expressed in GABAergic neurons and modulate the release of GABA (Albuquerque et al, 2009). However, little is known about the long-term actions of nicotine or nicotine analogues on specific nuclear mechanisms underlying alterations in the transcriptome in these neurons (Brown et al, 2000; Mansvelder et al, 2007; Levin and Rezvani, 2000)
When the postmortem brains of SZ and BP+ disorder patients are compared with that of NPS, a GABAergic neuropathology is found to be prevalent in the hippocampus and cortex along with a decrease of high- and low-affinity nAChR subtypes in the same brain areas (Maloku et al, 2011). Based on the hypothesis that the downregulation of genes specifically expressed in cortical GABAergic neurons of SZ and BP+ disorder patients is mediated by an increase in DNMT1 and DNMT3a (Costa et al, 2007) and that epigenetic disruptions are potentially reversible, a novel strategy to alleviate the neuropathology of SZ and BP+ would be to selectively reduce the increased levels of these DNMTs in GABAergic neurons. Potent DNMT inhibitors have been identified and are being used in the treatment of numerous cancers (Szyf, 2009). Some of these (doxorubicin and 5-aza-2′-deoxycytidine) have been used to treat neuroprogenitor cells in vitro and show that RELN and GAD67 mRNAs increase following administration (Kundakovic et al, 2007). Zebularine and 5-aza-2-deoxycytidine, when added to hippocampal slice preparations in vitro reduce the methylation associated with the RELN and BDNF promoters and abolish hippocampal long-term potentiation (Levenson et al, 2006). Unfortunately, these DNMT inhibitors fail to readily cross the blood–brain barrier and become toxic if administered systemically in large doses. Instead, an alternative approach is to selectively reduce the expression of DNMT1 and/or DNMT3a in GABAergic neurons by pharmacologically activating nAChRs.
Repeated injections of doses of nicotine that achieve plasma concentrations comparable to those reported by heavy cigarette smokers result in reduced levels of DNMT expression and an increased expression of GAD67 in cortical and hippocampal GABAergic neurons (Table 1, Satta et al, 2008). Furthermore, data show that the doses of nicotine that induce GAD67 expression also diminish the levels of 5mC at the GAD67 promoter (Satta et al, 2008). As the effects of nicotine are blocked by the non-competitive nAChR open-channel blocker mecamylamine, these data support a role for nAChRs (α4, β2, α7) as potential therapeutic targets to correct the dysfunction present in cortical and hippocampal GABAergic neurons of SZ patients. Varenicline is a high-affinity partial agonist at α4β2-containing receptors and a lower affinity full agonist at α7 neuronal nAChRs (Mihalak et al, 2006; Rollema et al, 2007). Repeated administration of varenicline reduces DNMT1 levels in a dose-dependent manner (Maloku et al, 2011). Cortical DNMT1 levels decreased ∼30–40% following treatment, whereas no changes were observed in striatal DNMT1 levels. While varenicline did not alter cortical DNMT3A mRNA levels, the expression of GAD67 mRNA and protein increased significantly (Maloku et al, 2011). A-85380, a high-affinity full agonist at α4β2 nAChRs (but devoid of activity at α3β4 or α7-containing receptors), also facilitates the DNMT1 mRNA decrease and GAD67 mRNA increase in cortical GABAergic neurons. In contrast, PNU-282987, a selective α7-nAChR agonist fails to decrease cortical DNMT1 mRNA or to induce GAD67 expression. These studies (summarized in Table 1) suggest that α4β2 nAChR agonists may be better suited than α7 nAChR agonists to attenuate the epigenetic alterations of GABAergic neurons in psychotic patients. Further, they support a role for varenicline as a pharmacological tool to increase GAD67 expression in patients with psychosis. In this regard, nAChR ligands are some of the more promising adjunct treatment options currently under investigation for SZ (Freedman, 2007). The molecular mechanism by which nAChR stimulation is coupled to DNMT1 expression remains to be elucidated. In the cortex and hippocampus, nAChR stimulation depolarizes selected GABAergic neurons, which results in a concomitant increase in intracellular Ca2+ levels and the activation of Ca2+-dependent kinases (Albuquerque et al, 2009). It seems likely that this might be linked to decreased transcription or increased mRNA degradation of DNMT1 mRNA observed in mice treated with nAChR ligands (Satta et al, 2008; Maloku et al, 2011). Studies in mouse cortical neuron cultures show that depolarization in vitro (high potassium, veratridine) also reduces the levels of DNMT1 mRNA and protein (Sharma et al, 2008). Thus, selectively reducing the expression of DNMT1 in GABAergic neurons by stimulating nAChRs may prove to be a useful way of reducing promoter methylation in SZ and BP+ patients.
Table 1. Effect of nAchR Agonists on DNMT1 and GAD67 Expression in Mouse Frontal Cortex.
| Drug |
nAchR subtype stimulation |
Maximal dose mg/kg (5 days) | DNMT1a mRNA % | GAD67a protein % | |
|---|---|---|---|---|---|
| α4β2 | α7 | ||||
| Vehicle | − | − | — | 100 | 100 |
| Nicotine | +++ | ++ | 3.5 | 53* | 225* |
| A-85380 | +++ | − | 2.5 | 65* | 225* |
| Varenicline | ++ | + | 5 | 48* | 250* |
| PNU-282981 | − | +++ | 5 | 105 | 119 |
Data from Satta et al (2008) and Maloku et al (2011).
*Drugs were given subcutaneously twice daily with the exception of nicotine, which was given four times per day.
*p<0.05.
Antipsychotic Drugs Reduce Promoter Methylation
VPA has been used in the United States as an anticonvulsant for the treatment of epilepsy since its approval in 1978 (Chapman et al, 1982). In psychiatry, the use of VPA as a drug to treat acute mania was approved in 1995 and it also acts to enhance GABAergic transmission (Löscher, 2002) by inducing the expression of GAD67 (Tremolizzo et al, 2002, 2005). The mode of VPA action is complex although it has been shown to inhibit class I HDACs (Phiel et al, 2001; Göttlicher et al, 2001). Initial studies suggested that the symptomatic benefits and faster onset elicited by a combination of antipsychotics and VPA in the treatment of SZ patients might prove beneficial, especially to patients with hostility or who exhibit acute psychotic symptoms (Wassef et al, 2000; Citrome, 2003; Lopez et al, 2004; Citrome et al, 2004; Kelly et al, 2006). More recent clinical studies report results that are mixed and show little benefit of adjunctive VPA administration in SZ (Casey et al, 2009; Larrison et al, 2011). A review of the literature suggests that VPA can be effective in treating some forms of SZ and aggression, ie, SZ with comorbid mood disorder (schizoaffective disorder) and treatment-resistant patients (Schwarz et al, 2010). A comprehensive clinical trial involving large numbers of psychiatric admissions determined that the efficacy of VPA depends on whether the extended release formulation (divalproex) or generic VPA is initially administered (Wassef et al, 2005). It should be pointed out that many of above studies were carried out using VPA in association with other medications including both typical and atypical antipsychotics. Medication dosing was variable as was the stage of disease during which the VPA/antipsychotic was administered. Based on arguments presented below, it seems plausible that HDAC inhibitors that are more selective or subtype-specific in conjunction with antipsychotics, such as clozapine, quetiapine, or olanzepine, might prove useful as adjunctive therapy in treating the psychosis associated with BP+ and SZ.
The possility that VPA might reverse DNA methylation was tested in mice in which DNA hypermethylation was first induced by the administration of MET for 7 days. As discussed below, MET downregulates the expression of promoters associated with GABAergic neurons. VPA, administered to mice at doses comparable to those used clinically, increases the amounts of acetylated H3K9 or H3K14 proximal to promoters and dramatically accelerates subsequent promoter demethylation (Dong et al, 2007). This finding is also observed in human NT2 cells in vitro following the administration of the HDAC inhibitor MS-275 (Kundakovic et al, 2009; Chen et al, 2011). The ability of VPA and other HDAC inhibitors to facilitate demethylation is not caused by a direct inhibitory action of the drug on DNMT activity or DNMT levels nor on an inhibitory activity of VPA on SAM biosynthesis (Guidotti et al, 2011). However, the mechanism by which VPA increases GABAergic function is by facilitating the increased expression of GAD67 and other promoters expressed in GABA neurons. This has been studied extensively and involves: (a) the inhibition of HDACs and relaxed local chromatin (Tremolizzo et al, 2002), (b) decreased recruitment of co-repressor complexes that include MeCP2, TET1, and DNMTs at the GAD67 promoter, (c) increased expression of the demethylation-targeting protein GADD45 (Matrisciano et al, 2011; Guidotti et al, 2011), and (d) decreased methylation at the GAD67 promoter (Dong et al, 2010; Guidotti et al, 2011; Chen et al, 2011).
Subsequent experiments examined the possibility that VPA might work together with antipsychotics in facilitating DNA demethylation. Studies in mice were used to evaluate the potential of clinically relevant doses of various antipsychotics, including haloperidol (a selective D2 receptor antagonist), clozapine (a 5HT2a-preferring receptor antagonist), risperidone (D2 and 5HT2A receptor antagonists), olanzapine (a 5HT2a-preferring receptor antagonist), and sulpiride (a D2/D3 receptor antagonist). GAD67 and RELN promoter methylation induced by 7 days of MET treatment (of mice) is reversed by clozapine, olanzapine, sulpiride and quetiapine but not by haloperidol or risperidone given either alone or with VPA (Table 2). The administration of clozapine but not that of haloperidol induces an increase in nuclear H3K9 acetylation (Dong et al, 2008) and also an increase of GADD45β mRNA (Matrisciano et al, 2011). The mechanisms by which these drugs induce DNA demethylation likely involves chromatin relaxation and the targeting of the DNA demethylation machinery by GADD45β.
Table 2. Effects of VPA and Various Antipsychotics on RELN Promoter Demethylation in Mouse Prefrontal Cortex.
| Druga | Effective dose (mg/kg) |
Promoter demethylationb |
|
|---|---|---|---|
| −VPA | +VPAc | ||
| Vehicle | − | − | + |
| Clozapine | 1.25 | + | ++++ |
| Olanzapine | 10 | + | +++ |
| Quetiapine | 10 | + | ++++ |
| Sulpiride | 10 | + | ++++ |
| LY379268d | 0.5 | + | +++ |
| Risperidone | 10 | − | + |
| Haloperidol | 1.5 | − | + |
Antipsychotics were given subcutaneously twice a day for 3 days.
RELN promoter demethylation was determined as described in Dong et al (2009).
VPA was administered twice daily (subcutaneously) for 3 days.
Data are modified from Guidotti et al (2009) and Matrisciano et al (2011).
The above studies suggest that the action of clozapine, olanzapine, and sulpiride on chromatin remodeling is independent of their action on catecholamine or serotonin receptors. In fact, the novel antipsychotic LY379268, which acts as an agonist at mGluR2/3 receptors and is devoid of action at monoamine receptors, elicits the demethylation of the GAD67 and RELN promoters comparable to that seen with atypical antipsychotics (Matrisciano et al, 2011). If successful, an ‘epigenetic neuroleptic treatment' will shift the emphasis in psychosis treatment from the use of drugs acting exclusively at membrane dopaminergic and other neurotransmitter receptors to drugs that directly address a chromatin remodeling defect (Guidotti et al, 2009). By activating DNA demethylation, clozapine (and its derivatives) and LY379268 together with VPA or other more potent and selective HDAC inhibitors, should be considered promising treatment strategies for normalizing the hypermethylation and the mRNA downregulation of genes expressed in GABAergic neurons detected in brains of psychotic patients.
EVALUATING THE EPIGENETIC POTENTIAL OF ANTIPSYCHOTICS
MET Mouse Model of DNA Hypermethylation
One of the adverse consequences of MET loading in rats is an increased sensitivity to convulsant drugs leading to reduced seizure thresholds (Gallagher, 1969). This could be due to reduced levels of GAD67, as a consequence of increased MET-induced promoter methylation. Acute treatment of heterozygous reeler mice (HRM) with MET elicits a dose-dependent increase in SAM and SAH in cortical tissue (Tremolizzo et al, 2002). Chronic MET administration produces a marked downregulation of RELN and GAD67 in both wild-type and HRM as compared with non-treated mice (Tremolizzo et al, 2002). The mRNAs corresponding to RELN, GAD67, VGAT (GABA vesicular transporter, Slc32a1), and GAT (neurotransmitter transporter, GABA, Slc6a1) are decreased in cortical RNA of MET-treated mice (Tremolizzo et al, 2002; Ying Chen, unpublished data). The MET-induced downregulation of RELN and GAD67 mRNAs is associated with an increase in CpG methylation of the corresponding promoters. In primary cultures of mouse cortical neurons maintained in vitro, MET treatment (2 mM) downregulates RELN and GAD67 mRNA levels and increases methylation of the RELN promoter (Noh et al, 2005). Interestingly, antisense knock down of DNMT1 prevents the MET-induced RELN and GAD67 mRNA downregulation. These data suggest that in the presence of exogenous MET, DNMT1 is capable of de novo methylation or that added MET facilitates the formation of a repressor complex that includes DNMT1 binding at specific promoters.
Wild-type mice treated with MET were also tested for behavioral changes that might be representative of an epigenetic mouse model for psychosis (Tremolizzo et al, 2005). For example, in addition to facilitating RELN promoter hypermethylation, mice given MET twice daily for 2 weeks show increased histone H3K9 dimethyl content (a repressive chromatin mark), exhibit reduced social interaction time with an intruder mouse, are less aggressive when socially isolated, and develop a deficit in prepulse inhibition to startle (Tremolizzo et al, 2005). The co-administration of VPA with MET for 2 weeks increases histone H3 acetylation in the frontal cortices, reduces the content of dimethyl H3K9, reverses the RELN promoter hypermethylation, and attenuates the observed MET-induced behavioral modifications (Tremolizzo et al, 2005). Moreover, the MET-induced increase in promoter methylation is accompanied by an increased binding of methyl domain-binding proteins (MBD, such as MeCP2) which is reversed by the co-administration of VPA (Dong et al, 2005).
More recently, two structurally unrelated HDAC inhibitors, VPA and the benzamide, MS-275 (Simonini et al, 2006), were shown to decrease RELN and GAD67 promoter methylation when given at the termination of MET treatment. This suggests that the effects of protracted MET administration are reversible (Dong et al, 2005). These data prompted the search for an HDAC inhibitor-induced DNA demethylase activity present in cortical extracts of VPA-treated mice (Dong et al, 2010). An evaluation (Tueting et al, 2010) of the effects of protracted MET administration in vivo on mouse cortical neurons shows that there are decreases in dendritic spine density in layer III pyramidal neurons similar to that reported in HRM (Liu et al, 2001) and in SZ subjects (Glantz and Lewis, 2000; Costa et al, 2001). Moreover, this response is blocked by the co-administration of VPA (Tueting et al, 2010). Collectively, these studies support the concept that the exacerbation of symptoms previously noted in SZ patients treated with MET is likely the consequence of SAM-induced DNA hypermethylation that further downregulates the expression of mRNAs in GABA neurons of susceptible individuals (Costa et al, 2003; Grayson et al, 2009).
Based on studies described thus far, it appears that MET facilitates increases in methylation at sites that are relevant to physiological cortical function. We still have no clear information regarding how much methylation is sufficient to enable a closed chromatin conformation, particularly in regions containing low levels of DNA methylation. Transient transfection experiments with the RELN promoter in vitro show that even partial methylation reduces promoter activity by as much as 50% (Chen et al, 2002). It would also be informative to determine correlations between percent methylation across defined DNA stretches and percent occupancy by methyl CpG-binding proteins such as MeCP2. Given the recently described roles of DNMT1 and TET1 as transcriptional repressors (see Text box 2), it will be important to determine which CpGs are bound by these proteins and what signals trigger subsequent hydroxymethylation. A more complete picture of MET-induced DNA methylation awaits an analysis of the genome-wide DNA methylome and hydroxymethylome in MET-treated neurons in vitro or in neurons isolated by LCM from treated and wild-type animals.
Prenatal Restraint Stress Model of Psychosis
In view of the data presented above, it is important to study the effects of nAChR agonists, VPA (and other HDAC inhibitors), and antipsychotics on DNMTs and on proteins participating in the DNA-demethylation cascade in normal (ie, wild-type) animals to establish basic mechanisms of action. This information will provide a framework for assessing the action of these compounds in animal models that mimic the downregulation of selected promoters, such as RELN and GAD67, in GABAergic neurons and that show behavioral signs reminiscent of psychosis (ie, altered prepulse inhibition of startle (PPI), exploratory behaviors, social interactions, and fear conditioning). One presumes that these types of comparisons will allow insight into the potential therapeutic actions of antipsychotic drugs on individuals with psychiatric illnesses. Early life stress has been shown to impact the methylation of selected promoters associated with stress (Weaver et al, 2004, 2005, 2006) and neuroendocrine responses (Murgatroyd et al, 2009), as well as GAD67 (Zhang et al, 2010a). Studies examining the neurochemical, behavioral, and pharmacological responses observed in the offspring of mothers exposed to prenatal restraint stress (PRS mice) have been carried out during corticogenesis to address this issue (Markham and Koenig, 2011; Matrisciano et al, 2012a).
Quantitation of DNMT mRNA levels in the cortex and hippocampus of PRS mice indicates that DNMT1 and DNMT3A are elevated at postnatal day 1 (P1) and remain elevated by twofold or more just after birth (see Figure 5, Matrisciano et al, 2012a). The DNMT levels remain elevated relative to controls until at least 70 days following birth. The increased expression occurs in GAD67-immunopositive GABAergic neurons of the frontal cortex and hippocampus and is accompanied by decreased RELN and GAD67 expression (Matrisciano et al, 2012a). Interestingly, there is increased binding of DNMT1 and MeCP2 to the corresponding promoters and increased levels of both 5mC and 5hmC (Matrisciano et al, 2012a). Importantly, the epigenetic phenotype present in cortex and hippocampus of PRS mice is reminiscent of that present in the brains of SZ and BP+ patients (Guidotti et al, 2011; Roth et al, 2009). Behaviorally, PRS mice show locomoter hyperactivity, enhanced responsiveness to the NMDA-receptor antagonist MK-801, and deficits in social interaction, paired pulse inhibition to startle, and fear conditioning (Mastricano et al, 2012a). PRS mice also exhibit reduced amounts of mRNA and protein levels of group II metabotropic glutamate receptors (Matrisciano et al, 2012a). Examination of MeCP2 and DNMT1 binding to mGluR2 and 3 receptor promoters show increased binding to both, as well as to the GAD67 and BDNF-IX promoters. Systemic treatment of PRS mice with Group II-selective mGlu2/3 receptor agonists corrects the biochemical and behavioral abnormalities associated with PRS mice. This action is likely related to the observed capacity of these compounds to increase levels of GADD45β and induce DNA demethylation (Matrisciano et al, 2011).
Figure 5.
DNA methyltransferase 1 (DNMT1) is elevated after birth and progressively declines with age in frontal cortex of control mice. Compared with controls, DNMT1 elevation is greater in offspring of prenatally stressed mice (PRS) even at 2 months of age. For the prenatal stress paradigm, pregnant mice were restrained in a transparent tube for 30 min twice daily from day 7 of pregnancy to delivery (Matrisciano et al, 2011). At the indicated times, RNA was harvested and analyzed for DNMT1 mRNA levels. *p<0.05, Student's t-test vs control values. Figure 5 is modified from Matrisciano et al (2012a).
To investigate the hypothesis that alterations in DNA methylation in GABAergic neurons of PRS mice might be responsible for the observed behavioral abnormalities, VPA and clozapine were administered in doses that are known to act on chromatin remodeling, inducing RELN and GAD67 promoter demethylation. At these doses, VPA and clozapine abolish observed changes in locomotor activities, stereotypy, social interactions, and PPI present in PRS mice. Furthermore, clozapine blocks the increased stereotypical behavior in PRS mice induced by small doses of MK-801. It is noteworthy that the doses of VPA and clozapine active on these behaviors in PRS mice fail to have a significant effect on the behaviors of control mice. This suggests that the drugs act specifically on the altered epigenetic mechanisms that underlie the behavioral pathology in PRS mice.
Brains of PRS mice exhibit increased levels of DNMT1 and DNMT3A in GABAergic neurons from birth until adulthood. The elevated expression is likely the consequence of an environment created by increased levels of stress, which when experienced prenatally induces an increased expression of DNMTs that persists until adulthood. We cannot establish at the present time if a similar time course in DNMT expression occurs in human brain, but it is conceivable that similar neurodevelopmental changes take place in response to stressful situations either in utero or during early postnatal life. We propose that early life stress prevents the reduction in DNMT expression that occurs during normal development. This hypothesis is supported by reports that the exposure of pregnant women to psychological stress, malnutrition, or viral infection during pregnancy is associated with an increased incidence of psychosis later in the life of the offspring (Mittal et al, 2008; Markham and Koenig, 2011; Howes et al, 2004). Hence, the neurochemical, behavioral, and pharmacological responses observed in adult offspring of dams exposed to restraint stress during pregnancy appear to parallel some of the responses observed in adult-onset SZ and strongly support the prenatal stress model in mice as an appropriate endophenotypic animal model of psychosis.
Collectively, the effects of prenatal stress on behaviors in mice indicate an imbalance in inhibitory/excitatory circuitry mediated by changes in the expression of DNMTs. Increased DNMT levels alter GAD67, RELN, and additional promoters expressed in GABA neurons hence altering GABA neuron function. PRS mice represent a suitable animal model to study the epigenetic mechanisms associated with psychosis. Moreover, PRS mice are useful for establishing the validity of new compounds with potential antipsychotic activity. Having said this, there are multiple rodent models that have been used to mimic the endophenotypes characteristic of SZ. These include the ventral hippocampal lesion model in rats (Lipska et al, 1993), gene knockout models (Papaleo et al, 2012), pharmacological models (Amann et al, 2010), neurodevelopmental models (Wilson and Terry, 2010) and additional stress models (Oliver, 2011). Given the large numbers of rodent models currently used to mimic the phenotypes associated with SZ, it is becoming increasingly clear that validating these models will depend on the extent to which clinicians provide pertinent behavioral information and the accuracy with which behavioral testing of mice mimics the human endophenotype (Young et al, 2010). As mentioned earlier, clinical presentation of psychiatric disorders is heterogenous and the more we understand results from behavioral testing of rodent models, the better we may be able to match animal phenotypes with clinical endophenotypes.
FUTURE DIRECTIONS AND CLINICAL IMPLICATIONS
The findings discussed here suggest that DNA-methylation levels are maintained by a dynamic process that has an important role in the regulation of gene expression in specific neuronal populations. The enzymes responsible for DNA methylation (ie, DNMTs) or DNA demethylation (ie, TET, APOBEC, GADD45) are highly expressed in mammalian brain. Initially, the overexpression of the maintenance methyltransferase DNMT1 in SZ and BP+ was somewhat surprising, as the vast majority of telencephalic neurons are postmitotic. In addition, similar to MeCP2 (Chao et al, 2010), DNMTs (1 and 3A) are abundantly expressed in GABAergic neurons compared with other neurons or glia. This suggests that the reported DNMT overexpression in GABAergic neurons of SZ and BP+ patients might be responsible for the promoter hypermethylation and repression of genes in these neurons, including GAD67, RELN, NMDA receptor subunits (GRIN1, 2A), GABA transporter (GAT1), TRKb receptors (TRKB), somatostatin (SST), cholecystokinin (CCK) and others, which occur in the absence of changes in the numbers of GABAergic neurons. DNMT1 is not only more abundant in GABAergic neurons (Kadriu et al, 2011), but can also be transported to mitochondria (Shock et al, 2011). Through the use of an upstream transcriptional initiation site, a fraction of the DNMT1 mRNA (∼1–2% in proliferating cells) contains an upstream in-frame ATG start codon that imparts a mitochondrial import signal to the translated protein (Shock et al, 2011). Recent data indicate that in addition to its actions on nuclear DNA, DNMT1 has a role in modulating mitochondrial DNA in response to hypoxia and the loss of the tumor suppressor p53 (Shock et al, 2011). Although the field of mitochondrial epigenetics is new, the finding of 5mC and 5hmC in the mitchondrial genome (Dzitoyeva et al, 2012) suggests that the rules regulating the dynamics of these proceses may be comparable to those we have outlined above.
The observation that DNMT1 is able to repress large numbers of promoters independent of methylation status indicates that DNMT1 acts to provide multiple layers of gene silencing (Clements et al, 2012). It seems likely that there are two classes of genes repressed by DNMTs, some of which are hypermethylated and others that are not. The binding of DNMT1 to these promoters is linked to the recruitment of a lysine (K)-specific histone demethylase (LSD1) that erases positive methylation marks on H3K4 (Clements et al, 2012). That is, the overexpression of DNMT1 in postmortem human brains of patients with SZ and BP+ could silence transcription by distinct mechanisms, one of which is catalytic and the other as a CpG binding repressor protein that interacts with HDACs, HMTs and co-repressors. Results from studies examining DNA methylation at specific promoters in SZ and BP+ patients have been mixed. This is not entirely surprising given that the vast majority of DNMT1 and 3A resides in GABAergic neurons. The issue may not be resolved until protocols for increasing the sensitivity of measuring genome-wide methylation profiles from single neurons are developed. Moreover, in patients with psychosis, the binding of DNMT1 and TET1 to promoters accessible in GABAergic neurons and a reduced DNA-demethylating capacity may be responsible for the downregulation of GABAergic transmission that has been reported to be operative in reducing inhibitory constraints on pyramidal neurons. The use of LCM for brain slice preparations to isolate morphologically identical neurons coupled to genome-wide ChIP-Seq and methyl DIP-Seq would provide additional relevant information. Disinhibited pyramidal neurons presumably have an important pathogenetic role in the development of cognitive impairment and psychotic symptoms in SZ patients (Gonzalez-Burgos and Lewis, 2008).
Accumulating evidence of cortical and hippocampal GABAergic deficits in SZ and BP+ patients suggest that to improve the efficacy of therapeutic treatments, future efforts should be directed at developing drugs that can normalize the functional deficits in GABAergic transmission (Guidotti et al, 2005). A logical approach would be to administer drugs that act as direct (muscimol or 4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-olhydrochloride (THIP)) or indirect (various benzodiazepines) GABAA receptor agonists. Unfortunately, unwanted side effects including sedation, amnesia, tolerance, and dependence have limited the utility of these drugs, particularly those active at α1 subunit-containing GABAA receptors. MK-0777, a selective GABAAα2/α3 receptor subunit partial agonist with reduced unwanted side effects, was reported to improve delayed memory performance and decrease reaction times on selected measures of cortical function in SZ patients (Lewis et al, 2008). However, in a more recent randomized clinical trial, MK-0777 showed little benefit in improving cognitive performance in patients with SZ (Buchanan et al, 2011). In view of these results, α2- and α3- subunit-containing GABAA receptors remain promising drug targets but more potent and selective receptor modulators need to be developed to treat the cognitive deficits associated with SZ (Guidotti et al, 2005; Buchanan et al, 2011).
An alternative approach to treating the GABAergic deficit associated with psychosis would be to target the aberrant increase in DNMTs and TET1 in GABAergic neurons. The implications of TET1 overexpression in patients with psychosis are not currently appreciated. DNMT1 overexpression in GABAergic neurons facilitates the reduced expression of numerous genes, likely due to its action as a transcriptional repressor. In contrast, DNMT3A overexpression would be expected to lead to promoter hypermethylation and the downregulation of an overlapping set of target genes. Because DNMT inhibitors that cross the blood–brain barrier and that are devoid of toxic effects are not available, a combination multi-pharmaceutical approach to bypass the epigenetic consequences of DNMT overexpression might prove efficacious. nAChR agonists or partial agonists (ie, varenicline) with selectivity at α4β2 receptor subtypes and possibly HDAC inhibitors (ie, VPA) in combination with antipsychotics (clozapine, sulpiride, olanzapine) might alleviate the promoter downregulation in GABAergic neurons, GABAergic hypofunction, and associated psychotic symptoms.
The mechanisms whereby nAChR agonists and VPA and selected antipsychotics interfere with DNA-methylation dynamics are different. For example, nAChR agonists reduce the expression of DNMTs in GABAergic neurons selectively, whereas HDAC inhibitors and selected antipsychotics facilitate the activation of DNA demethylation in different types of neurons. The molecular mechanisms by which HDAC inhibitors and antipsychotics activate DNA demethylation remain unknown. However, current data suggest that the action of these drugs on potentiating gene expression in GABAergic neurons is independent from the stimulation of monoaminergic receptors and may be mediated by a direct action on chromatin remodeling. Considering the emerging evidence supporting a prominent role of an epigenetic GABAergic dysfunction in SZ and BP+, the identification of pharmacological agents, such as nAChR agonists that target epigenetic mechanisms selectively in these neurons, might be crucial in the development of new lines of pharmacological interventions to treat SZ and BP+ morbidity.
Acknowledgments
The authors would like to thank Dr Bashkim Kadriu for his help in preparation of Figure 3a.
The authors have no financial interests to disclose.
References
- Aapola U, Kawasaki K, Scott HS, Ollila J, Vihinen M, Heino M, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65:293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- Aapola U, Lyle R, Krohn K, Antonarakis SE, Peterson P. Isolationand initial characterization of the mouse Dnmt3l gene. Cytogenet Cell Genet. 2001;92:122–126. doi: 10.1159/000056881. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Faraone SV, Wilcox M, Glatt SJ, Gao F, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum Mol Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolmaleky HM, Cheng KH, Russo A, Smith CL, Faraone SV, Wilcox M, et al. 2005Hypermethylation of the reelin (RELN). promoter in the brain of schizophrenic patients: a preliminary report Am J Med Genet B Neuropsychiatr Genet 13460–66.The first paper to provide biochemical evidence that the reelin promoter is hypermethylated in the brain of schizophrenia patients. [DOI] [PubMed] [Google Scholar]
- Abdolmaleky HM, Thiagalingam S. Can the schizophrenia epigenome provide clues for the molecular basis of pathogenesis. Epigenomics. 2011;3:679–683. doi: 10.2217/epi.11.94. [DOI] [PubMed] [Google Scholar]
- Adler Nevo G, Meged S, Sela BA, Hanoch-Levi A, Hershko R, Weizman A. Homocysteine levels in adolescent schizophrenia patients. Eur Neuropsychopharmacol. 2006;16:588–591. doi: 10.1016/j.euroneuro.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Huang H-S.2006Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders Brain Res Rev 52293–304.This paper was the first to establish that the expression of GAD67 mRNA is reduced without loss of neurons in prefrontal cortex of schizophrenia patients. [DOI] [PubMed] [Google Scholar]
- Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- Albuquerque EX, Pereira EF, Alkondon M, Rogers SW. Mammalian nicotinic acetylcholine receptors: from structure to function. Physiol Rev. 2009;89:73–120. doi: 10.1152/physrev.00015.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, et al. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. EMBO J. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann LC, Gandal MJ, Halene TB, Ehrlichman RS, White SL, McCarren HS, et al. Mouse behavioral endophenotypes for schizophrenia. Brain Res Bull. 2010;83:147–161. doi: 10.1016/j.brainresbull.2010.04.008. [DOI] [PubMed] [Google Scholar]
- Andria ML, Simon EJ. Localization of promoter elements in the human mu-opioid receptor gene and regulation by DNA methylation. Brain Res Mol Brain Res. 1999;70:54–65. doi: 10.1016/s0169-328x(99)00126-6. [DOI] [PubMed] [Google Scholar]
- Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35:2450–2461. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applebaum J, Shimon H, Sela BA, Belmaker RH, Levine J. Homocysteine levels in newly admitted schizophrenic patients. J Psychiatr Res. 2004;38:413–416. doi: 10.1016/j.jpsychires.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Araujo FD, Croteau S, Slack AD, Milutinovic S, Bigey P, Price GB, et al. The DNMT1 target recognition domain resides in the N terminus. J Biol Chem. 2001;276:6930–6936. doi: 10.1074/jbc.M009037200. [DOI] [PubMed] [Google Scholar]
- Auta J, Longone P, Guidotti A, Costa E. The regulation of hippocampal nicotinic acetylcholine receptors (nAChRs). after a protracted treatment with selective or nonselective nAChR agonists. J Mol Neurosci. 1999;13:31–45. doi: 10.1385/JMN:13:1-2:31. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto G, Schäfer A, Marhold J, Stach D, Swaminathan SK, Handa V, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Belforte JE, Zsiros V, Sklar ER, Jiang Z, Yu G, Li Y, et al. 2010Postnatal NMDA receptor ablation in corticolimbic interneurons confers schizophrenia-like phenotypes Nature Neurosci 1376–83.Created and characterized a conditional knock-in mouse in which the NR1 subunit of the NMDA receptor was selectively eliminated in cortical and hippocampal interneurons. Mice with this deletion exhibit a schizophrenia-like phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Lim B, Subburaju S. Site-specific regulation of cell cycle and DNA repair in post-mitotic GABA cells in schizophrenic versus bipolars. Proc Natl Acad Sci USA. 2009;106:11731–11736. doi: 10.1073/pnas.0903066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bird A. Putting the DNA back into DNA methylation. Nat Genet. 2011;43:1050–1051. doi: 10.1038/ng.987. [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Bransteitter R, Prochnow C, Chen XS. The current structural and functional understanding of APOBEC deaminases. Cell Mol Life Sci. 2009;66:3137–3147. doi: 10.1007/s00018-009-0070-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese CR, Lee MJ, Adams CE, Sullivan B, Logel J, Gillen KM, et al. Abnormal regulation of high affinity nicotinic receptors in subjects with schizophrenia. Neuropsychopharmacology. 2000;23:351–364. doi: 10.1016/S0893-133X(00)00121-4. [DOI] [PubMed] [Google Scholar]
- Bromberg A, Levine J, Belmaker R, Agam G. Hyperhomocysteinemia does not affect global DNA methylation and nicotinamide N-methyltransferase expression in mice. J Psychopharmacol. 2011;25:976–981. doi: 10.1177/0269881110388328. [DOI] [PubMed] [Google Scholar]
- Bromberg A, Levine J, Nemetz B, Belmaker RH, Agam G. No association between global leukocyte DNA methylation and homocysteine levels in schizophrenia patients. Schizophr Res. 2008;101:50–57. doi: 10.1016/j.schres.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Brown AS. The environment and susceptibility to schizophrenia. Prog Neurobiol. 2011;93:23–58. doi: 10.1016/j.pneurobio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Bottiglieri T, Schaefer CA, Quesenberry CP, Jr, Liu L, Bresnahan M, et al. Elevated prenatal homocysteine levels as a risk factor for schizophrenia. Arch Gen Psychiatry. 2007;64:31–39. doi: 10.1001/archpsyc.64.1.31. [DOI] [PubMed] [Google Scholar]
- Brown AS, Patterson PH. Maternal infection and schizophrenia: implications for prevention. Schizophr Bull. 2011;37:284–290. doi: 10.1093/schbul/sbq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RW, Gonzalez CL, Kolb B. Nicotine improves Morris water task performance in rats given medial frontal cortex lesions. Pharmacol Biochem Behav. 2000;67:473–478. doi: 10.1016/s0091-3057(00)00398-1. [DOI] [PubMed] [Google Scholar]
- Buchanan RW, Keefe RS, Lieberman JA, Barch DM, Csernansky JG, Goff DC.et al (2011A randomized clinical trial of MK-0777 for the treatment of cognitive impairments in people with schizophrenia Biol Psychiatry 69442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MC, Leonhardt H. DNA methyltransferase is actively retained in the cytoplasm during early development. J Cell Biol. 1999;147:25–32. doi: 10.1083/jcb.147.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey DE, Daniel DG, Tamminga C, Kane JM, Tran-Johnson T, Wozniak P, et al. Divalproex ER combined with olanzapine or risperidone for treatment of acute exacerbations of schizophrenia. Neuropsychopharmacology. 2009;34:1330–1338. doi: 10.1038/npp.2008.209. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, et al. Dysfunction in GABA signaling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–269. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Keane PE, Meldrum BS, Simiand J, Vernieres JC. Mechanism of anticonvulsant action of valproate. Prog Neurobiol. 1982;19:315–359. doi: 10.1016/0301-0082(82)90010-7. [DOI] [PubMed] [Google Scholar]
- Chédin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
- Chédin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci USA. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dong E, Grayson DR. Analysis of the GAD1 promoter: trans-acting factors and DNA methylation converge on the 5′ untranslated region. Neuropharmacology. 2011;60:1075–1087. doi: 10.1016/j.neuropharm.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, Grayson DR. Induction of the reelin promoter by retinoic acid is mediated by Sp1. J Neurochem. 2007;103:650–665. doi: 10.1111/j.1471-4159.2007.04797.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR.2002On the epigenetic regulation of human reelin promoter expression Nucleic Acids Res 302930–2939.The first data to show that human reelin expression is regulated by CpG island promoter methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286:18347–18353. doi: 10.1074/jbc.R110.205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Heo K, Byun HM, An W, Lu W, Yang AS. Identification of preferential target sites for human DNA methyltransferases. Nucleic Acids Res. 2011;39:104–118. doi: 10.1093/nar/gkq774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LS, Ian HI, Koh TW, Ng HH, Xu G, Li BF. Human DNA-(cytosine-5). methyltransferase-PCNA complex as a target for p21WAF1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- Citrome L. Schizophrenia and valproate. Psychopharmacol Bull. 2003;37 (Suppl 2:74–88. [PubMed] [Google Scholar]
- Citrome L, Tremeau F, Wynn PS, Roy B, Dinakar H. A study of the safety, efficacy, and tolerability of switching from the standard delayed release preparation of divalproex sodium to the extended release formulation in patients with schizophrenia. J Clin Psychopharmacol. 2004;24:255–259. doi: 10.1097/01.jcp.0000125687.27961.64. [DOI] [PubMed] [Google Scholar]
- Clements EG, Mohammad HP, Leadem BR, Easwaran H, Cai Y, Van Neste L, et al. 2012DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes Nucleic Acids Res 404334–4346.Data establish evidence that there are multiple genes for which DNMT1 acts as a transcriptional repressor independent from its methyltransferese activity and that the repressive function occurs through its interactions with histone-modifying enzymes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM, Nichols A, Wyatt R, Pollin W. The administration of methionine to chronic schizophrenic patients: a review of ten studies. Biol Psychiatry. 1974;8:209–225. [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Chen Y, Davis J, Dong E, Noh JS, Tremolizzo L, et al. 2002Reelin and schizophrenia: a disease at the interface of the genome and the epigenome Mol Interventions 247–57.Established an important physiological role for reelin in regulating extrasomal protein synthesis requisite for dendritic spine plasticity. Also, proposed that reelin is deficient in schizophrenia because of an epigenetic hypermethylation of its corresponding promoter. [DOI] [PubMed] [Google Scholar]
- Costa E, Chen Y, Dong E, Grayson DR, Kundakovic M, Maloku E, et al. GABAergic promoter hypermethylation as a model to study the neurochemistry of schizophrenia vulnerability. Expert Rev Neurother. 2009;9:87–98. doi: 10.1586/14737175.9.1.87. [DOI] [PubMed] [Google Scholar]
- Costa E, Davis J, Grayson DR, Guidotti A, Pappas GD, Pesold C. Dendritic spine hypoplasticity and downregulation of reelin and GABAergic tone in schizophrenia vulnerability. Neurobiol Dis. 2001;8:723–742. doi: 10.1006/nbdi.2001.0436. [DOI] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2:29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Ruzicka WB, Simonini MV, Veldic M, et al. Epigenetic targets in GABAergic neurons to treat schizophrenia. Adv Pharmacol. 2006;54:95–117. doi: 10.1016/s1054-3589(06)54005-0. [DOI] [PubMed] [Google Scholar]
- Costa E, Grayson DR, Guidotti A. Epigenetic downregulation of GABAergic function in schizophrenia vulnerability: potential for pharmacological intervention. Mol Interventions. 2003;3:220–229. doi: 10.1124/mi.3.4.220. [DOI] [PubMed] [Google Scholar]
- Creveling CR, Daly JW. Identification of 3,4-dimethoxyphenethylamine from schizophrenic urine by mass spectrometry. Nature. 1967;216:190–191. doi: 10.1038/216190a0. [DOI] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. Epigenetic treatments for cognitive impairments. Neuropsychopharmacology. 2012;37:247–260. doi: 10.1038/npp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, et al. 2011Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder Hum Mol Genet 204786–4796.Report that numerous loci demonstrate disease-associated DNA methylation differences between twins discordant for schizophrenia and bipolar disorder. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M.2003Valproate induces replication-independent active DNA demethylation J Biol Chem 27827586–27592.Reported that valproate, induces replication-independent DNA demethylation in HEK 293 kidney cells by inhibiting HDAC activity. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S, Syeda F, Park L. DNA methyl transferase 1: regulatory mechanisms and implications in health and disease. Int J Biochem Mol Biol. 2011;2:58–66. [PMC free article] [PubMed] [Google Scholar]
- Dierker L, Donny E. The role of psychiatric disorders in the relationship between cigarette smoking and DSM-IV nicotine dependence among young adults. Nicotine Tob Res. 2008;10:439–446. doi: 10.1080/14622200801901898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohi O, Takada H, Wakabayashi N, Yasui K, Sakakura C, Mitsufuji S, et al. Epigenetic silencing of RELN in gastric cancer. Int J Oncol. 2010;36:85–92. [PubMed] [Google Scholar]
- Dong E, Agis-Balboa RC, Simonini MV, Grayson DR, Costa E, Guidotti A. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci USA. 2005;102:12578–12583. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Chen Y, Gavin DP, Grayson DR, Guidotti A. Valproate induces DNA demethylation in nuclear extracts from adult mouse brain. Epigenetics. 2010;5:730–735. doi: 10.4161/epi.5.8.13053. [DOI] [PubMed] [Google Scholar]
- Dong E, Gavin DP, Chen Y, Davis J.2012Up-regulation of TET1 and down-regulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients Translational Psychiatryin press. [DOI] [PMC free article] [PubMed]
- Dong E, Grayson DR, Guidotti A, Costa E. Antipsychotic subtypes can be characterized by differences in their ability to modify GABAergic promoter methylation. Epigenomics. 2009;1:201–211. doi: 10.2217/epi.09.2. [DOI] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Grayson DR, Costa E.2007Demethylation of reelin and glutamic acid decarboxylase 67 brain promoters is induced by hyperacetylation of histones Proc Natl Acad Sci USA 1044676–4681.Provided evidence that when promoters are hypermethylated by methionine treatment in vivo, the HDAC inhibitors valproate and MS-275 facilitate the demethylation of reelin and GAD67 by a mechanism involving the hyperacetylation of histones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA-demethylase. Proc Natl Acad Sci USA. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzitoyeva S, Chen H, Manev H.2012Effect of aging on 5-hydroxymethylcytosine in brain mitochondria Neurobiol AgingEpub ahead of print. [DOI] [PMC free article] [PubMed]
- Eastwood SL, Harrison PJ. Decreased expression of vesicular glutamate transporter 1 and complexin II mRNAs in schizophrenia: further evidence for a synaptic pathology affecting glutamate neurons. Schizophr Res. 2005;73:159–172. doi: 10.1016/j.schres.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: in vitro and post-mortem brain studies. J Neurosci Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- Estève PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20:3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Beard C, Chen RZ, Csankovszki G, Sun Y, Siniaia M, et al. 2001DNA hypomethylation perturbs the function and survival of CNS neurons in postnatal animals J Neurosci 21788–797.Conditional deletion of DNMT1 in mitotic precursor cells produces daughter cells that exhibit global DNA hypomethylation. The DNA hypomethylation perturbs the function and the survival of CNS neurons postnatally. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi M, Hermann A, Gowher H, Jeltsch A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur J Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- Fatemi M, Hermann A, Pradhan S, Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J Mol Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Earle JA, McMenomy T. Reduction in reelin immunoreactivity in hippocampus of subjects with schizophrenia, bipolar disorder and major depression. Mol Psychiatry. 2000;5:654–663. doi: 10.1038/sj.mp.4000783. [DOI] [PubMed] [Google Scholar]
- Fellinger K, Rothbauer U, Felle M, Längst G, Leonhardt H. Dimerization of DNA methyltransferase 1 is mediated by its regulatory domain. J Cell Biochem. 2008;106:521–528. doi: 10.1002/jcb.22071. [DOI] [PubMed] [Google Scholar]
- Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, et al. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398–402. doi: 10.1038/nature10008. [DOI] [PubMed] [Google Scholar]
- Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C, Rottach A, Meilinger D, Bultmann S, Fellinger K, Hasenöder S, et al. Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011;6:e16627. doi: 10.1371/journal.pone.0016627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman R. Exacerbation of schizophrenia by varenicline. Am J Psychiatry. 2007;164:1269. doi: 10.1176/appi.ajp.2007.07020326. [DOI] [PubMed] [Google Scholar]
- Fremeau RT, Jr, Troyer MD, Pahner I, Nygaard GO, Tran CH, Reimer RJ, et al. 2001The expression of vesicular glutamate transporters defines two classes of excitatory synapse Neuron 31247–260.First to show that the localization of two excitatory amino-acid transporters define distinct excitatory synapses. [DOI] [PubMed] [Google Scholar]
- Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation. Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fung SJ, Webster MJ, Sivagnanasundaram S, Duncan C, Elashoff M, Weickert CS. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am J Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- Gallagher BB. Amino acids and cerebral excitability. J Neurochem. 1969;16:701–706. doi: 10.1111/j.1471-4159.1969.tb06448.x. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafrenière RG, Xiong L, Spiegelman D, Brustein E, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, Beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531–542. doi: 10.1038/npp.2011.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Dion PA, Rouleau GA. Schizophrenia genetics: putting all the pieces together. Curr Neurol Neurosci Rep. 2012;12:261–266. doi: 10.1007/s11910-012-0266-7. [DOI] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, et al. Increased exonic de novo mutation rate in individualswith schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Gjerset RA, Martin DW., Jr Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J Biol Chem. 1982;257:8581–8583. [PubMed] [Google Scholar]
- Glantz LA, Lewis DA.2000Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia Arch Gen Psychiatry 5765–73.Demonstrated a region- and disease-specific decrease in dendritic spine density on dorsolateral prefrontal cortex layer 3 pyramidal neurons of schizophrenia patients. [DOI] [PubMed] [Google Scholar]
- Glass JL, Fazzari MJ, Ferguson-Smith AC, Greally JM. CG dinucleotide periodicities recognized by the Dnmt3a-Dnmt3L complex are distinctive at retroelements and imprinted domains. Mamm Genome. 2009;20:633–643. doi: 10.1007/s00335-009-9232-3. [DOI] [PubMed] [Google Scholar]
- Goff DC, Hill M, Barch D. The treatment of cognitive impairment in schizophrenia. Pharmacol Biochem Behav. 2011;99:245–253. doi: 10.1016/j.pbb.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Lewis DA.2008GABA neurons and the mechanisms of network oscillations: implications for understanding cortical dysfunction in schizophrenia Schizophr Bull 34944–961.Discuss data suggesting that phasic synaptic inhibition mediated by specific subtypes of cortical gamma-aminobutyric acid (GABA) neurons is essential for the production of synchronized network oscillations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Göttlicher M, Minucci S, Zhu P, Krämer OH, Schimpf A, Giavara S, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H, Stockdale CJ, Goyal R, Ferreira H, Owen-Hughes T, Jeltsch A. De novo methylation of nucleosomal DNA by the mammalian Dnmt1 and Dnmt3A DNA methyltransferases. Biochemistry. 2005;44:9899–9904. doi: 10.1021/bi047634t. [DOI] [PubMed] [Google Scholar]
- Grayson DR. Schizophrenia and the epigenetic hypothesis. Epigenomics. 2010;1:201–211. doi: 10.2217/epi.10.22. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Chen Y, Costa E, Dong E, Guidotti A, Kundakovic M, et al. The human reelin gene: transcription factors (+), repressors (−) and the methylation switch (+/−) in schizophrenia. Pharmacol Ther. 2006;111:272–286. doi: 10.1016/j.pharmthera.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Chen Y, Dong E, Kundakovic M, Guidotti A. From trans-methylation to cytosine methylation: evolution of the methylation hypothesis of schizophrenia. Epigenetics. 2009;4:144–149. doi: 10.4161/epi.4.3.8534. [DOI] [PubMed] [Google Scholar]
- Grayson DR, Jia X, Chen Y, Sharma RP, Mitchell CP, Guidotti A, et al. 2005Reelin promoter hypermethylation in schizophrenia Proc Natl Acad Sci USA 1029341–9346.Provide bisulfite-sequencing data to demonstrate that the reelin promoter exhibits increased methylation at positions∼135 bp from the transcriptional start site in brains of schizophrenia patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson DR, Kundakovic M, Sharma RP. Is there a future for HDAC inhibitors in the pharmacotherapy for psychiatric disorders. Mol Pharmacol. 2010;77:135–126. doi: 10.1124/mol.109.061333. [DOI] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Chen Y, Davis JM, Dong E, Gavin DP, et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology. 2011;60:1007–1016. doi: 10.1016/j.neuropharm.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Di Giorgi-Gerevini V, Dwivedi Y, Grayson DR, et al. 2000Decrease in reelin and glutamic acid decarboxylase67 (GAD67). expression in schizophrenia and bipolar disorder: a postmortem brain study Arch Gen Psych 571061–1069.Present evidence of a GABAergic neuropathology in the prefrontal cortex of schizophrenia and bipolar disorder patients characterized by a decreased expression of reelin and GAD67. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Auta J, Davis JM, Dong E, Grayson DR, Veldic M, et al. GABAergic dysfunction in schizophrenia: new treatment strategies on the horizon. Psychopharmacology. 2005;180:191–205. doi: 10.1007/s00213-005-2212-8. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Kundakovic M, Satta R, Grayson DR.2009Characterization of the action of antipsychotic subtypes on valproate-induced chromatin remodeling TiPS 3055–60.Provided the first demonstration that clozapine and sulpiride and not haloperidol synergize with valproate in facilitating chromatin remodeling and activating DNA demethylation. [DOI] [PubMed] [Google Scholar]
- Guidotti A, Ruzicka W, Grayson DR, Veldic M, Pinna G, Davis JM, et al. S-adenosyl methionine and DNA methyltransferase-1 mRNA overexpression in psychosis. Neuroreport. 2007;18:57–60. doi: 10.1097/WNR.0b013e32800fefd7. [DOI] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011b;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011a;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, et al. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Vertino PM, Cheng X. Molecular coupling of DNA methylation and histone methylation. Epigenomics. 2010;2:657–669. doi: 10.2217/epi.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15:710–723. doi: 10.1101/gad.194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- Hermann A, Gowher H, Jeltsch A. Biochemistry and biology of mammalian DNA methyltransferases. Cell Mol Life Sci. 2004;61:2571–2587. doi: 10.1007/s00018-004-4201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill M, Shannahan K, Jasinski S, Macklin EA, Raeke L, Roffman JL, et al. Folate supplementation in schizophrenia: a possible role for MTHFR genotype. Schizophr Res. 2011;127:41–45. doi: 10.1016/j.schres.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Howes OD, McDonald C, Cannon M, Arseneault L, Boydell J, Murray RM. Pathways to schizophrenia: the impact of environmental factors. Int J Neuropsychopharmacol. 2004;7 (Suppl 1:S7–S13. doi: 10.1017/S1461145704004122. [DOI] [PubMed] [Google Scholar]
- Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2:e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HS, Matevossian A, Whittle C, Kim SY, Schumacher A, Baker SP, et al. 2007Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters J Neurosci 2711254–11262.Data from this study implicated a role for histone methylation in schizophrenia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. 1998A decrease of reelin expression as a putative vulnerability factor in schizophrenia Proc Natl Acad Sci USA 9515718–15723.First to report that reelin is highly expressed in adult human brain and is decreased in the telencephalon of psychotic patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, et al. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva EI, Morris DW, Osuji J, Moates AF, Carmody TJ, Thaker GK, et al. Cognitive endophenotypes of psychosis within dimension and diagnosis. Psychiatry Res. 2012;196:38–44. doi: 10.1016/j.psychres.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Ueda J, Oldham MC, Ukai W, Hashimoto E, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome Res. 2011;21:688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Kato T. Epigenetic profiling in schizophrenia and major mental disorders. Neuropsychobiology. 2009;60:5–11. doi: 10.1159/000234811. [DOI] [PubMed] [Google Scholar]
- Jeltsch A. On the enzymatic properties of Dnmt1: specificity, processivity, mechanism of linear diffusion and allosteric regulation of the enzyme. Epigenetics. 2006;1:63–66. doi: 10.4161/epi.1.2.2767. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD.2001Translating the histone code Science 2931074–1080.Proposed that distinct histone amino-terminal modifications function as a ‘histone code' by generating synergistic or antagonistic interactions between chromatin-associated proteins. These modifications in turn dictate dynamic transitions between transcriptionally active and transcriptionally inactive chromatin states. [DOI] [PubMed] [Google Scholar]
- Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy. Genes Cancer. 2011;2:607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen HF, Ben-Porath I, Bird AP. Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains. Mol Cell Biol. 2004;24:3387–3395. doi: 10.1128/MCB.24.8.3387-3395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chem Biochem. 2011;12:206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- Kadriu B, Guidotti A, Chen Y, Grayson DR.2011The DNA methyltransferases1 (DNMT1). and 3a (DNMT3a). co-localize with GAD67-positive neurons in the GAD67-GFP mouse brain J Comp Neurol 5201951–1964.Demonstrated that DNMTs (1 and 3a) are highly expressed in GABAergic telencephalic neurons in the GAD67-eGFP mouse brain. Only low levels of expression are found in pyramidal neurons or glial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kareta MS, Botello ZM, Ennis JJ, Chou C, Chédin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J Biol Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107:2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DL, Conley RR, Feldman S, Yu Y, McMahon RP, Richardson CM. Adjunct divalproex or lithium to clozapine in treatment-resistant schizophrenia. Psychiatr Q. 2006;77:81–95. doi: 10.1007/s11126-006-7963-9. [DOI] [PubMed] [Google Scholar]
- Keshavan MS, Morris DW, Sweeney JA, Pearlson G, Thaker G, Seidman LJ.et al (2011A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale Schizophr Res 133250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs MO, Bellon A, Mainguy G, Jay TM, Frieling H. One-carbon metabolism and schizophrenia: current challenges and future directions. Trends Mol Med. 2009;15:562–570. doi: 10.1016/j.molmed.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N.2009The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain Science 324929–930.Report that the nuclear DNA base 5-hydroxymethylcytosine is abundant in Purkinje cells and in the brain in general, suggesting a role in epigenetic control of neuronal function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari V, Postma P. Nicotine use in schizophrenia: the self-medication hypotheses. Neurosci Biobehav Rev. 2005;29:1021–1034. doi: 10.1016/j.neubiorev.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Costa E, Grayson DR. DNA methyltransferase inhibitors coordinately induce expression of the human reelin and glutamic acid decarboxylase genes. Mol Pharmacol. 2007;71:644–653. doi: 10.1124/mol.106.030635. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Chen Y, Guidotti A, Grayson DR.2009The reelin and GAD67 promoters are activated by epigenetic drugs that facilitate the disruption of local repressor complexes Mol Pharmacol 75342–354.Demonstrated that HDAC inhibitors facilitate disruption of local repressor complexes and DNA demethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrison AL, Babin SL, Xing Y, Patel SS, Wassef AA, Sereno AB. Effects of adjunct valproic acid on clinical symptoms and saccadic eye movements in schizophrenia. Hum Psychopharmacol. 2011;26:517–525. doi: 10.1002/hup.1236. [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1). is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lerner V, Miodownik C, Kaptsan A, Vishne T, Sela BA, Levine J. High serum homocysteine levels in young male schizophrenic and schizoaffective patients with tardive parkinsonism and/or tardive dyskinesia. J Clin Psychiatry. 2005;66:1558–1563. doi: 10.4088/jcp.v66n1210. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, et al. 2006Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus J Biol Chem 28115763–15773.Demonstrated that the promoters for reelin and BDNF, both implicated in synaptic plasticity, exhibit rapid and dramatic changes in cytosine methylation when DNMT activity is inhibited. Moreover, inhibition of DNMT activity with zebularine and 5-aza-2-deoxycytidine blocks the induction of LTP in hippocampus. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Sweatt JD. Epigenetic mechanisms in memory formation. Nat Rev Neurosci. 2005;6:108–118. doi: 10.1038/nrn1604. [DOI] [PubMed] [Google Scholar]
- Levin ED, Rezvani AH. Development of nicotinic drug therapy for cognitive disorders. Eur J Pharmacol. 2000;393:141–146. doi: 10.1016/s0014-2999(99)00885-7. [DOI] [PubMed] [Google Scholar]
- Levine J, Sela BA, Osher Y, Belmaker RH. High homocysteine serum levels in young male schizophrenia and bipolar patients and in an animal model. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1181–1191. doi: 10.1016/j.pnpbp.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Levine J, Stahl Z, Sela BA, Gavendo S, Ruderman V, Belmaker RH. Elevated homocysteine levels in young male patients with schizophrenia. Am J Psychiatry. 2002;159:1790–1792. doi: 10.1176/appi.ajp.159.10.1790. [DOI] [PubMed] [Google Scholar]
- Levine J, Stahl Z, Sela BA, Ruderman V, Shumaico O, Babushkin I, et al. Homocysteine-reducing strategies improve symptoms in chronic schizophrenic patients with hyperhomocysteinemia. Biol Psychiatry. 2006;60:265–269. doi: 10.1016/j.biopsych.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Lewandowski KE, Cohen BM, Ongur D. Evolution of neuropsychological dysfunction during the course of schizophrenia and bipolar disorder. Psychol Med. 2011;41:225–241. doi: 10.1017/S0033291710001042. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Cho RY, Carter CS, Eklund K, Forster S, Kelly MA, et al. Subunit-selective modulation of GABA type A receptor neurotransmission and cognition in schizophrenia. Am J Psychiatry. 2008;165:1585–1593. doi: 10.1176/appi.ajp.2008.08030395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- Lintas C, Persico AM. Neocortical RELN promoter methylation increases significantly after puberty. Neuroreport. 2010;21:114–118. doi: 10.1097/WNR.0b013e328334b343. [DOI] [PubMed] [Google Scholar]
- Lipska BK, Jaskiw GE, Weinberger DR. Postpubertal emergence of hyperresponsiveness to stress and to amphetamine after neonatal excitotoxic hippocampal damage: a potential animal model of schizophrenia. Brain Res Dev Brain Res. 1993;75:213–222. doi: 10.1038/npp.1993.44. [DOI] [PubMed] [Google Scholar]
- Liu WS, Pesold C, Rodriguez MA, Carboni G, Auta J, Lacor P, et al. Down-regulation of dendritic spine and glutamic acid decarboxylase 67 expressions in the reelin haploinsufficient heterozygous reeler mouse. Proc Natl Acad Sci USA. 2001;98:3477–3482. doi: 10.1073/pnas.051614698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez LM, Wassef AA, Molloy MS, Williams NG. Valproic acid induces manifestations of simultaneous dopamine enhancement and reduction in schizophrenia. Neuropsychopharmacology. 2004;29:1217. doi: 10.1038/sj.npp.1300461. [DOI] [PubMed] [Google Scholar]
- Löscher W. Basic pharmacology of valproate: a review after 35 years of clinical use for the treatment of epilepsy. CNS Drugs. 2002;16:669–694. doi: 10.2165/00023210-200216100-00003. [DOI] [PubMed] [Google Scholar]
- Ma DK, Guo JU, Ming G-L, Song H. DNA excision repair proteins and GADD45 as molecular players for active DNA demethylation. Cell Cycle. 2009b;8:1526–1531. doi: 10.4161/cc.8.10.8500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, et al. 2009aNeuronal activity-induced Gadd45β promotes epigenetic DNA demethylation and adult neurogenesis Science 3231074–1077.Present the first evidence that the neuronal activity-induced GADD45b promotes BDNF promoter demethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, McCarthy S, Michaelson JJ, Vacic V, Burdick KE, Yoon S, et al. High frequencies of de novo CNVs in bipolar disorder and schizophrenia. Neuron. 2011;72:951–963. doi: 10.1016/j.neuron.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloku E, Kadriu B, Zhubi A, Dong E, Pibiri F, Satta R, et al. Selective α4β2 nicotinic acetylcholine receptor agonists target epigenetic mechanisms in cortical GABAergic neurons. Neuropsychopharmacology. 2011;36:1366–1374. doi: 10.1038/npp.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell AJ, Morgan M. Indole(ethyl)amine N-methyltransferase in human brain. Nat New Biol. 1971;230:85–87. doi: 10.1038/newbio230085a0. [DOI] [PubMed] [Google Scholar]
- Mansvelder HD, Fagen ZM, Chang B, Mitchum R, McGehee DS. Bupropion inhibits the cellular effects of nicotine in the ventral tegmental area. Biochem Pharmacol. 2007;74:1283–1291. doi: 10.1016/j.bcp.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JA, Koenig JI. Prenatal stress: role in psychotic and depressive diseases. Psychopharmacology. 2011;214:89–106. doi: 10.1007/s00213-010-2035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarese F, Carrillo-de Santa Pau E, Stunnenberg HG. 5-Hydroxymethylcytosine: a new kid on the epigenetic block. Mol Syst Biol. 2011;7:562. doi: 10.1038/msb.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Dong E, Gavin DP, Nicoletti F, Guidotti A.2011Activation of group II metabotropic glutamate receptors promotes DNA demethylation in the mouse brain Mol Pharmacol 80174–182.Authors report that the systemic injection of group II metabotropic glutamate receptor agonists promotes DNA demethylation in the mouse frontal cortex and reduces defects in social interaction observed in mice pretreated with methionine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrisciano F, Tueting P, Dalal I, Kadriu B, Grayson DR, Davis JM, et al. 2012aEpigenetic modifications of GABAergic interneurons are associated with the schizophrenia-like phenotype induced by prenatal stress in mice NeuropharmacologyEpub ahead of print. [DOI] [PMC free article] [PubMed]
- Matrisciano F, Tueting P, Maccari S, Nicoletti F, Guidotti A. Pharmacological activation of group-II metabotropic glutamate receptors corrects a schizophrenia-like phenotype induced by prenatal stress in mice. Neuropsychopharmacology. 2012b;37:929–938. doi: 10.1038/npp.2011.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AM, Wilson VB, Mitchell SH. The role of antipsychotics in smoking and smoking cessation. CNS Drugs. 2011;25:299–315. doi: 10.2165/11588170-000000000-00000. [DOI] [PubMed] [Google Scholar]
- Matthysse S, Lipinski J. Biochemical aspects of schizophrenia. Annu Rev Med. 1975;26:551–565. doi: 10.1146/annurev.me.26.020175.003003. [DOI] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Suderman M, Sasaki A, Huang TC, Hallett M, Meaney MJ, et al. Broad epigenetic signature of maternal care in the brain of adult rats. PLoS One. 2011;6:e14739. doi: 10.1371/journal.pone.0014739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Asherson P, Owen M, Farmer A. The strength of the genetic effect. Is there room for an environmental influence in the aetiology of schizophrenia. Br J Psychiatry. 1994;164:593–599. doi: 10.1192/bjp.164.5.593. [DOI] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersfelder EL, Parthun MR. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res. 2006;34:2653–2662. doi: 10.1093/nar/gkl338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Mihalak KB, Carroll FI, Luetje CW. Varenicline is a partial agonist at alpha4beta2 and a full agonist at alpha7 neuronal nicotinic receptors. Mol Pharmacol. 2006;70:801–805. doi: 10.1124/mol.106.025130. [DOI] [PubMed] [Google Scholar]
- Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. 2008Epigenomic profiling reveals DNA-methylation changes associated with major psychosis Am J Hum Genet 82696–711.Document that in the frontal cortex, epigenomic profiling reveals psychosis-associated DNA methylation differences at numerous loci, including several involved in GABAergic and glutamatergic neurotransmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- Mirsky AS. The structure of chromatin. Proc Natl Acad Sci USA. 1971;12:2945–2948. doi: 10.1073/pnas.68.12.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal VA, Ellman LM, Cannon TD. Gene-environment interaction and covariation in schizophrenia: the role of obstetric complications. Schizophr Bull. 2008;34:1083–1094. doi: 10.1093/schbul/sbn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris MJ, Karra AS, Monteggia LM. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav Pharmacol. 2010;21:409–419. doi: 10.1097/FBP.0b013e32833c20c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulle JG. Schizophrenia genetics: progress, at last. Curr Opin Genet Dev. 2012;22:238–244. doi: 10.1016/j.gde.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Muntjewerff JW, Kahn RS, Blom HJ, den Heijer M. Homocysteine, methylenetetrahydrofolate reductase and risk of schizophrenia: a meta-analysis. Mol Psychiatry. 2006;11:143–149. doi: 10.1038/sj.mp.4001746. [DOI] [PubMed] [Google Scholar]
- Murgatroyd C, Patchev AV, Wu Y, Micale V, Bockmühl Y, Fischer D, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- Myant K, Stancheva I. LSH cooperates with DNA methyltransferases to repress transcription. Mol Cell Biol. 2008;28:215–226. doi: 10.1128/MCB.01073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, et al. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Zsiros V, Jiang Z, Nakao K, Kolata S, Zhang S, et al. GABAergic interneuron origin of schizophrenia pathophysiology. Neuropharmacology. 2012;62:1574–1583. doi: 10.1016/j.neuropharm.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh JS, Sharma RP, Veldic M, Salvacion AA, Jia X, Chen Y, et al. 2005DNA methyltransferase 1 regulates reelin mRNA expression in primary mouse cortical cultures Proc Natl Acad Sci USA 1021749–1754.Provide evidence that the antisense-mediated downregulation of DNMT1 leads to changes in the expression of reelin and GAD67 in mouse cortical neuronsin vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, Wininger M, et al. 2012DNA methylation signatures in development and aging of the human prefrontal cortex Am J Hum Genet 90260–272.Comprehensive analysis of genome-wide methylation levels in the prefrontal cortices across the lifespan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Nomoto S, Kanda M, Hayashi M, Nishikawa Y, Fujii T, et al. Reduced expression of reelin (RELN) gene is associated with high recurrence rate of hepatocellular carcinoma. Ann Surg Oncol. 2011;18:572–579. doi: 10.1245/s10434-010-1273-z. [DOI] [PubMed] [Google Scholar]
- Oliver PL. Challenges of analyzing gene-environment interactions in mouse models of schizophrenia. Scientific World Journal. 2011;11:1411–1420. doi: 10.1100/tsw.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otani J, Nankumo T, Arita K, Inamoto S, Ariyoshi M, Shirakawa M. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 2009;10:1235–1241. doi: 10.1038/embor.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaleo F, Lipska BK, Weinberger DR. Mouse models of genetic effects on cognition: relevance to schizophrenia. Neuropharmacology. 2012;62:1204–1220. doi: 10.1016/j.neuropharm.2011.04.025. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesold C, Liu WS, Guidotti A, Costa E, Caruncho HJ. Cortical bitufted, horizontal, and Martinotti cells preferentially express and secrete reelin into perineuronal nets, nonsynaptically modulating gene expression. Proc Natl Acad Sci USA. 1999;96:3217–3222. doi: 10.1073/pnas.96.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronis A. The origin of schizophrenia: genetic thesis, epigenetic antithesis, and resolving synthesis. Biol Psychiatry. 2004;55:965–970. doi: 10.1016/j.biopsych.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Petronis A, Gottesman II, Kan P, Kennedy JL, Basile VS, Paterson AD, et al. 2003Monozygotic twins exhibit numerous epigenetic differences: clues to twin discordance Schizophr Bull 29169–178.Authors studied D2 receptor promoter methylation in monozygotic twins discordant or concordant for schizophrenia. It was discovered that the affected twin from the pair discordant for schizophrenia is epigenetically closer to the affected concordant twins than to his unaffected MZ co-twin. [DOI] [PubMed] [Google Scholar]
- Petronis A, Paterson AD, Kennedy JL. Schizophrenia: an epigenetic puzzle. Schizophr Bull. 1999;25:639–655. doi: 10.1093/oxfordjournals.schbul.a033408. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS.2001Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen J Biol Chem 27636734–36741.Describe the actions of valproic acid as a histone deacetylase inhibitor. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Mill J. Epigenetic studies of psychosis: current findings, methodological approaches, and implications for postmortem research. Biol Psychiatry. 2011;69:146–156. doi: 10.1016/j.biopsych.2010.03.029. [DOI] [PubMed] [Google Scholar]
- Polesskaya OO, Aston C, Sokolov BP. Allele C-specific methylation of the 5-HT2A receptor gene: evidence for correlation with its expression and expression of DNA methylase DNMT1. J Neurosci Res. 2006;83:362–373. doi: 10.1002/jnr.20732. [DOI] [PubMed] [Google Scholar]
- Pollin W, Cardon P, Kety S. Effects of amino acid feedings in schizophrenic patients treated with iproniazid. Science. 1961;133:104–105. doi: 10.1126/science.133.3446.104. [DOI] [PubMed] [Google Scholar]
- Popendikyte V, Laurinavicius A, Paterson AD, Macciardi F, Kennedy JL, Petronis A. DNA methylation at the putative promoter region of the human dopamine D2 receptor gene. Neuroreport. 1999;10:1249–1255. doi: 10.1097/00001756-199904260-00018. [DOI] [PubMed] [Google Scholar]
- Potash JB, Bienvenu OJ. Neuropsychiatric disorders: Shared genetics of bipolar disorder and schizophrenia. Nat Rev Neurol. 2009;5:299–300. doi: 10.1038/nrneurol.2009.71. [DOI] [PubMed] [Google Scholar]
- Pradhan M, Estève PO, Chin HG, Samaranayke M, Kim GD, Pradhan S. CXXC domain of human DNMT1 is essential for enzymatic activity. Biochemistry. 2008;47:10000–10009. doi: 10.1021/bi8011725. [DOI] [PubMed] [Google Scholar]
- Qin L, Tu W, Sun X, Zhang J, Chen Y, Zhao H. Retardation of neurobehavioral development and reelin down-regulation regulated by further DNA methylation in the hippocampus of the rat pups are associated with maternal deprivation. Behav Brain Res. 2011;217:142–147. doi: 10.1016/j.bbr.2010.10.018. [DOI] [PubMed] [Google Scholar]
- Rai K, Huggins IJ, James SR, Karpf AR, Jones DA, Cairns BR. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135:1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci USA. 1999;96:6107–6112. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G, Gallais R, Métivier R. Marking time: the dynamic role of chromatin and covalent modification in transcription. Int J Biochem Cell Biol. 2009;41:155–163. doi: 10.1016/j.biocel.2008.08.028. [DOI] [PubMed] [Google Scholar]
- Robertson J, Robertson AB, Klungland A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochem Biophys Res Commun. 2011;411:40–43. doi: 10.1016/j.bbrc.2011.06.077. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Murillo L, Gogos JA, Karayiorgou M. The genetic architecture of schizophrenia: new mutations and emerging paradigms. Annu Rev Med. 2012;63:63–80. doi: 10.1146/annurev-med-072010-091100. [DOI] [PubMed] [Google Scholar]
- Rollema H, Chambers LK, Coe JW, Glowa J, Hurst RS, Lebel LA, et al. Pharmacological profile of the alpha4beta2 nicotinic acetylcholine receptor partial agonist varenicline, an effective smoking cessation aid. Neuropharmacology. 2007;52:985–994. doi: 10.1016/j.neuropharm.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Sodhi M, Kleinman JE. Epigenetic mechanisms in schizophrenia. Biochim Biophys Acta. 2009;1790:869–877. doi: 10.1016/j.bbagen.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Rutten BP, Mill J. Epigenetic mediation of environmental influences in major psychotic disorders. Schizophr Bull. 2009;35:1045–1056. doi: 10.1093/schbul/sbp104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka W, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A.2007Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection Mol Psychiatry 12385–397.Authors establish that DNMT1 is highly expressed in GABAergic interneurons of frontal cortical layers I and II using laser capture microdissection and that it is strikingly increased in schizophrenia patients. [DOI] [PubMed] [Google Scholar]
- Saavedra JM, Axelrod J. Psychotomimetic N-methylated tryptamines: formation in brain in vivo and in vitro. Science. 1972;175:1365–1366. doi: 10.1126/science.175.4028.1365. [DOI] [PubMed] [Google Scholar]
- Sato N, Fukushima N, Chang R, Matsubayashi H, Goggins M. Differential and epigenetic gene expression profiling identifies frequent disruption of the RELN pathway in pancreatic cancers. Gastroenterology. 2006;130:548–565. doi: 10.1053/j.gastro.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, et al. 2008Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons Proc Natl Acad Sci USA 10516356–16361.Report that activation of nicotinic acetylcholine receptors impacts the expression of cortical DNA methyltransferase 1 in GABAergic neurons. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz C, Volz A, Li C, Leucht S. Valproate for schizophrenia. Cochrane Database Syst Rev. 2010;(3):CD004028. doi: 10.1002/14651858.CD004028.pub3. [DOI] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Sharma RP, Tun N, Grayson DR. Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics. 2008;3:74–80. doi: 10.4161/epi.3.2.6103. [DOI] [PubMed] [Google Scholar]
- Sheng G, Demers M, Subburaju S, Benes FM.2012Differences in the circuitry-based association of copy numbers and gene expression between the hippocampi of patients with schizophrenia and the hippocampi of patients with bipolar disorder Arch Gen PsychiatryEpub ahead of print. Authors Show that gene copy number intensities are variable in different neurons of the hippocampus. [DOI] [PubMed]
- Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM.2011DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria Proc Natl Acad Sci USA 1083630–3635.Describe how alternative codon usage generates a mitochondrial import signal for DNMT1 that targets the protein to mitochondria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonini MV, Camargo LM, Dong E, Maloku E, Veldic M, Costa E, et al. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci USA. 2006;103:1587–1592. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen C, Sundet K, Vaskinn A, Birkenaes AB, Engh JA, Faerden A, et al. Neurocognitive dysfunction in bipolar and schizophrenia spectrum disorders depends on history of psychosis rather than diagnostic group. Schizophr Bull. 2011;37:73–83. doi: 10.1093/schbul/sbp034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisk CL, Zehr JL. Pubertal hormones organize the adolescent brain and behavior. Front Neuroendocrinol. 2005;26:163–174. doi: 10.1016/j.yfrne.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Smith RE, Haroutunian V, Davis KL, Meador-Woodruff JH. Expression of excitatory amino acid transporter transcripts in the thalamus of subjects with schizophrenia. Am J Psychiatry. 2001;158:1393–1399. doi: 10.1176/appi.ajp.158.9.1393. [DOI] [PubMed] [Google Scholar]
- Song CX, Clark TA, Lu XY, Kislyuk A, Dai Q, Turner SW, et al. Sensitive and specific single-molecule sequencing of 5-hydroxymethylcytosine. Nat Methods. 2011a;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Rechkoblit O, Bestor TH, Patel DJ.2011bStructure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation Science 3311036–1040.Authors describe the structures of the mouse and human DNMT1 protein bound to DNA-containing unmethylated CpG sites. The study identifies an autoinhibitory mechanism in which unmethylated CpG dinucleotides are occluded from the active site to ensure that only hemimethylated CpG dinucleotides undergo methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen HJ, Mortensen EL, Parnas J, Mednick SA. Premorbid neurocognitive functioning in schizophrenia spectrum disorder. Schizophr Bull. 2006;32:578–583. doi: 10.1093/schbul/sbj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabenau JR, Creveling CR, Daly J. The ‘pink spot,' 3,4-dimethoxyphenylethylamine, common tea, and schizophrenia. Am J Psychiatry. 1970;127:611–616. doi: 10.1176/ajp.127.5.611. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stein T, Cosimo E, Yu X, Smith PR, Simon R, Cottrell L, et al. Loss of reelin expression in breast cancer is epigenetically controlled and associated with poor prognosis. Am J Pathol. 2010;177:2323–2333. doi: 10.2353/ajpath.2010.100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol. 2011;12:R54. doi: 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, et al. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011a;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011b;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. Epigenetics, DNA methylation, and chromatin modifying drugs. Annu Rev Pharmacol Toxicol. 2009;49:243–263. doi: 10.1146/annurev-pharmtox-061008-103102. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. 2009Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1 Science 324930–935.Study describes that TET1, a fusion partner of the MLL gene in acute myeloid leukemia, is also a 2-oxoglutarate (2OG)- and Fe(II)-dependent enzyme that catalyzes the conversion of 5mC to 5-hydroxymethylcytosine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita K, Suetake I, Yamashita E, Suga M, Narita H, Nakagawa A, et al. Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1) Proc Natl Acad Sci USA. 2011;108:9055–9059. doi: 10.1073/pnas.1019629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GW, Redon R, Carter NP, Grant SG. The role of DNA copy number variation in schizophrenia. Biol Psychiatry. 2009;66:1005–1012. doi: 10.1016/j.biopsych.2009.07.027. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki JI, Obata K, Kaneko T. Green fluorescent protein expression and colocalization with calretinen, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 2003;467:60–79. doi: 10.1002/cne.10905. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatry. 2007;12:593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Huang H, Hoffman B, Liebermann DA, Ledda-Columbano GM, Columbano A, et al. Gadd45β is an inducible coactivator of transcription that facilitates rapid liver growth in mice. J Clin Invest. 2011;121:4491–4502. doi: 10.1172/JCI38760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting AH, Jair KW, Schuebel KE, Baylin SB. Differential requirement for DNA methyltransferase 1 in maintaining human cancer cell gene promoter hypermethylation. Cancer Res. 2006;66:729–735. doi: 10.1158/0008-5472.CAN-05-1537. [DOI] [PubMed] [Google Scholar]
- Ting AH, Jair KW, Suzuki H, Yen RW, Baylin SB, Schuebel KE. CpG island hypermethylation is maintained in human colorectal cancer cells after RNAi-mediated depletion of DNMT1. Nat Genet. 2004;36:582–584. doi: 10.1038/ng1365. [DOI] [PubMed] [Google Scholar]
- Tochigi M, Iwamoto K, Bundo M, Komori A, Sasaki T, Kato N, et al. Methylation status of the reelin promoter region in the brain of schizophrenic patients. Biol Psychiatry. 2008;63:530–533. doi: 10.1016/j.biopsych.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Torniainen M, Suvisaari J, Partonen T, Castaneda AE, Kuha A, Suokas J.et al (2012Cognitive impairments in schizophrenia and schizoaffective disorder: relationship with clinical characteristics J Nerv Ment Dis 200316–322. [DOI] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB, Mitchell CP, Sugaya I, Tueting P, et al. 2002An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability Proc Natl Acad Sci USA 9917095–17100.Study demonstrates that systemic injection of valproate in mice results in a dose-related increase of histone3 acetylation in the frontal cortex and hippocampus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremolizzo L, Doueiri MS, Dong E, Grayson DR, Pinna G, Tueting P, et al. Valproate corrects the schizophrenia-like epigenetic behavioral modifications induced by methionine in mice. Biol Psychiatry. 2005;57:500–509. doi: 10.1016/j.biopsych.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Tsujita T, Niikawa N, Yamashita H, Imamura A, Hamada A, Nakane Y, et al. Genomic discordance between monozygotic twins discordant for schizophrenia. Am J Psychiatry. 1998;155:422–424. doi: 10.1176/ajp.155.3.422. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tueting P, Davis JM, Veldic M, Pibiri F, Kadriu B, Guidotti A, et al. L-methionine decreases dendritic spine density in mouse frontal cortex. NeuroReport. 2010;21:543–548. doi: 10.1097/WNR.0b013e3283373126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Caruncho HM, Liu WS, Davis J, Satta R, Grayson DR, et al. 2004DNA-methyltransferase-1 mRNA is selectively overexpressed in telencephalic GABAergic interneurons of schizophrenia brains Proc Natl Acad Sci USA 101348–353.The first immuno-histochemical demonstration that DNMT is primarily expressed in GABAergic neurons and is selectively overexpressed in these neurons in postmortem schizophrenia brains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E, Davis JM, Costa E. In psychosis, cortical interneurons overexpress DNA-methyltransferase I. Proc Natl Acad Sci USA. 2005;102:2152–2157. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldic M, Kadriu B, Maloku E, Agis-Balboa RC, Guidotti A, Davis JM, et al. Epigenetic mechanisms expressed in basal ganglia GABAergic neurons differentiate schizophrenia from bipolar disorder. Schizo Res. 2007;91:51–61. doi: 10.1016/j.schres.2006.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- Wallace TL, Porter RH. Targeting the nicotinic alpha7 acetylcholine receptor to enhance cognition in disease. Biochem Pharmacol. 2011;82:891–903. doi: 10.1016/j.bcp.2011.06.034. [DOI] [PubMed] [Google Scholar]
- Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, et al. The lysine demethylase LSD1 (KDM1). is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- Wassef AA, Dott SG, Harris A, Brown A, O'Boyle M, Meyer WJ, III, et al. Randomized, placebo-controlled pilot study of divalproex sodium in the treatment of acute exacerbations of chronic schizophrenia. J Clin Psychopharmacol. 2000;20:357–361. doi: 10.1097/00004714-200006000-00011. [DOI] [PubMed] [Google Scholar]
- Wassef AA, Winkler DE, Roache AL, Abobo VB, Lopez LM, Averill JP, et al. Lower effectiveness of divalproex versus valproic acid in a prospective, quasi-experimental clinical trial involving 9,260 psychiatric admissions. Am J Psychiatry. 2005;162:330–339. doi: 10.1176/appi.ajp.162.2.330. [DOI] [PubMed] [Google Scholar]
- Watanabe D, Uchiyama K, Hanaoka K. Transition of mouse de novo methyltransferases expression from Dnmt3b to Dnmt3a during neural progenitor cell development. Neuroscience. 2006;142:727–737. doi: 10.1016/j.neuroscience.2006.07.053. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE, Dymov S, Sharma S, Meaney MJ, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver ICG, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR.et al (2004Epigenetic programming by maternal behavior Nat Neurosci 7847–854.Data showing that the offspring of mothers that exhibit high levels of licking and grooming behaviors have reduced glucocorticoid receptor promoter methylation in the hippocampus as compared with offspring of low licking and grooming mothers. The epigenomic state of a gene can be established through behavioral programming and is potentially reversible. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands. EMBO Rep. 2011b;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011a;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C, Terry AV., Jr Neurodevelopmental animal models of schizophrenia: role in novel drug discovery and development. Clin Schizophr Relat Psychoses. 2010;4:124–137. doi: 10.3371/CSRP.4.2.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolff CM. Does the genotype for schizophrenia often remain unexpressed because of canalization and stochastic events during development. Psychol Med. 1997;27:659–668. doi: 10.1017/s003329179700473x. [DOI] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011b;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011a;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt RJ, Benedict A, Davis J. Biochemical and sleep studies of schizophrenia: a review of the literature-1960–1970. Schiz Bull. 1971;4:10–44. [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011a;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Lam R, Dong A, Min J.2011bThe structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain Nat Commun 2227.Report the co-crystal structure of the CXXC domain of CFP1 with different CpG-containing oligonucleotides. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Wu F, Tan L, Kong L, Xiong L, Deng J, et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol Cell. 2011c;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Commun. 2000;272:193–198. doi: 10.1006/bbrc.2000.2760. [DOI] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147:1498–1510. doi: 10.1016/j.cell.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JW, Zhou X, Geyer MA. Animal models of schizophrenia. Curr Top Behav Neurosci. 2010;4:391–433. doi: 10.1007/7854_2010_62. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Chen CQ, Manev H. DNA methylation as an epigenetic regulator of neural 5-lipoxygenase expression: evidence in human NT2 and NT2-N cells. J Neurochem. 2004;88:1424–1430. doi: 10.1046/j.1471-4159.2003.02275.x. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Hellstrom IC, Bagot RC, Wen X, Diorio J, Meaney MJ.2010aMaternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus J Neurosci 3013130–13137.Report data showing that maternal behavior impacts epigenetic regulation of the GAD67 promoter through changes in promoter methylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jurkowska R, Soeroes S, Rajavelu A, Dhayalan A, Bock I, et al. Chromatin methylation activity of Dnmt3a and Dnmt3a/3L is guided by interaction of the ADD domain with the histone H3 tail. Nucleic Acids Res. 2010b;38:4246–4253. doi: 10.1093/nar/gkq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010c;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An up-regulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophrenia Res. 2009;111:115–122. doi: 10.1016/j.schres.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziedonis D, Hitsman B, Beckham JC, Zvolensky M, Adler LE, Audrain-McGovern J, et al. Tobacco use and cessation in psychiatric disorders: National Institute of Mental Health report. Nicotine Tob Res. 2008;10:1691–1715. doi: 10.1080/14622200802443569. [DOI] [PubMed] [Google Scholar]