Abstract
The Hint1 protein, a member of the histidine triad (HIT) family, is highly conserved in diverse species and ubiquitously expressed in mammalian tissues. Previous studies in mice provided evidence that Hint1 may be haplosufficient with respect to its function as a tumor suppressor. In the present study, we investigated the aberrant methylation of Hint1 and explored possible relationships between aberrant methylation and clinicopathological features in hepatocellular carcinoma (HCC). Hypermethylation of Hint1 was evaluated by the methylation specific PCR (MSP) method in 40 patients with HCC (tumor and paired adjacent non-tumor tissues) from Taiwan, 22 cases of normal liver tissue (14 from Taiwan and 8 from the U.S.). HINT1 expression in tissues was detected by immunohistochemistry. The frequencies of hypermethylation of Hint1 in tumor, paired adjacent non-tumor and normal liver tissue were 55.0%, 37.5% and 9.1%, respectively. A statistically significant inverse association was found between Hint1 methylation status and expression of the HINT1 protein in tumor tissues (p<0.003). The relationship between Hint1 methylation status and clinical features and other, previously measured biomarkers was also analyzed. p16 hypermethylation was statistically significantly associated with Hint1 methylation status (p=0.035). There were no correlations between Hint1 methylation and HBV or HCV infection status or AFB1- and PAH-DNA adduct levels. These results suggest that promoter hypermethylation of Hint1 may play a role in hepatocarcinogenesis.
Keywords: Hint1, HCC, epigenetic changes, promoter hypermethylation, p16, environmental carcinogens
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, and a leading cause of death in many countries. The epidemiology of HCC has marked demographic and geographic variations, occurring mainly in Africa and Asia. However, the incidence is also increasing in the United States and Europe (1). The major risks for the development of HCC have been identified as chronic hepatitis B virus (HBV) and hepatitis virus C (HCV) infections and several dietary or environmental factors, including aflatoxin B1 (AFB1) and polycyclic aromatic hydrocarbons (PAHs). HBV and HCV infections and AFB1 exposure are responsible for approximately 80% of all HCCs (2;3). As with other cancers, the development of HCC is a complex, multistep process (4). The molecular pathogenesis of HCC appears to involve multiple genetic aberrations in the molecular control of hepatocyte proliferation, differentiation and death and the maintenance of genomic integrity. This process is influenced by the cumulative activation and inactivation of oncogenes, tumor suppressor genes, cell cycle control genes and other genes.
The HINT protein, a member of the histidine triad (HIT) family, is highly conserved in diverse species and ubiquitously expressed in mammalian tissues. The HIT protein superfamily consists of at least three subfamilies: Hint, Fhit and Ga1T (5). In previous studies, Hint1 deleted mice had a marked increase in susceptibility to chemical carcinogen-induced gastric tumors (6), mammary tumors and ovarian tumors (7). In addition, with aging, Hint1 deleted mice displayed an increase in the occurrence of a variety of spontaneous tumors including HCC (7). These studies in mice also provided evidence that Hint1 may be haplosufficient with respect to its function as a tumor suppressor gene. Mechanistic studies indicate that Hint1 can play a role in apoptosis and p53 expression (8) and that it can bind to and inhibit several transcription factors including MITF, USF2 and β-catenin and also inhibit AP1 activity by binding to POSH (9). It had been reported that Hint1 is transcriptionally silenced in some human non-small cell lung cancer (NSCLC) cell lines and that increased expression of Hint1 inhibits growth of the NSCLC cell lines H522 and H538 (10). Similar effects have been seen in colon cancer cells (9).
During the past decade, extensive studies in the field of epigenetics have brought an awareness that not only genetic, but also epigenetic changes, play a very important role in carcinogenesis (11;12). DNA methylation is one of the best understood epigenetic mechanisms; hypermethylation of normally unmethylated CpG islands, which are CpG dinucleotide-rich areas located mainly in the promoter regions of many genes, correlate with loss of transcription and loss of gene function (13). In HCC, a growing number of genes have been identified as undergoing aberrant promoter hypermethylation, suggesting that promoter hypermethylation is an important molecular mechanism for hepatocarcinogenesis. These include the genes p16, p15 and RASSF1A (14;15). These epigenetic changes have also been implicated as early events in the development of HCC (16–18). In recent studies, we found promoter hypermethylation of Hint1 in a subset of both colon cancer and HCC cell lines (9;19).
With the above findings as a background, in the present study, we investigated the Hint1 promoter methylation profile in HCC and paired adjacent non-tumor DNA samples from patients and also explored the correlation between Hint1 methylation status and other biomarkers and clinical parameters.
Materials and methods
Patient population and data on clinical parameters
The study samples consisted of 40 frozen dissected tumor and paired adjacent non-tumor tissues, collected in the Department of Surgery, National Taiwan University Hospital. Informed consent was obtained from patients, and the study was approved by the appropriate institutional review committees. Demographic data and clinicopathological characteristics were obtained from hospital charts, and HBV and HCV status was determined by immunoassay (see Table 1.). Fourteen normal control liver tissues were obtained from subjects affected with intrahepatic stones, liver cysts, and other non-cancerous diseases identified at the National Taiwan University Hospital. Eight U.S. normal control liver tissues were from subjects affected with heart disease identified at Columbia Presbyterian Hospital in New York City.
Table 1.
Demographics, HCV status, HBsAg status and tumor characteristics of HCC cases
| ID | Age | Gender1 | Cirrhosis in nontumorous tissue | Grade | Anti-HCV | HBsAg |
|---|---|---|---|---|---|---|
| F003 | NA2 | NA | NA | NA | NA | NA |
| F004 | 67 | M | + | II | + | + |
| F005 | 47 | M | − | II | − | + |
| F006 | 70 | F | − | I | + | − |
| F007 | NA | NA | NA | NA | NA | NA |
| F008 | NA | M | NA | NA | NA | NA |
| F009 | 56 | M | − | NA | NA | + |
| F011 | NA | NA | NA | NA | NA | NA |
| F012 | 39 | M | − | III | + | + |
| F013 | 78 | M | + | II–III | − | + |
| F014 | 42 | M | − | III | − | + |
| F015 | NA | NA | NA | NA | NA | NA |
| F017 | 70 | F | − | II | + | − |
| F018 | 51 | M | − | II | − | + |
| F019 | 20 | M | − | I–II | − | + |
| F020 | NA | NA | NA | NA | NA | NA |
| F021 | 45 | M | − | II | − | + |
| F022 | 58 | M | + | I | + | − |
| F023 | 43 | M | − | II | − | + |
| F025 | 51 | M | − | II | + | − |
| F028 | 35 | F | − | III | NA | + |
| F029 | 49 | M | + | II | − | + |
| F030 | 37 | M | − | II | − | + |
| F032 | 50 | M | − | II | − | + |
| F033 | 48 | M | − | III | + | + |
| F034 | 44 | M | − | I–II | NA | + |
| F035 | 49 | M | + | III–IV | − | + |
| F036 | 62 | F | + | III | + | + |
| F038 | 62 | M | + | II | + | + |
| F039 | 69 | M | + | III | − | − |
| F040 | 76 | M | − | III | − | + |
| F042 | 40 | M | − | II | NA | + |
| F043 | 59 | F | + | III | − | + |
| F044 | NA | NA | NA | NA | NA | NA |
| F045 | 63 | M | + | II | − | + |
| F047 | 45 | M | + | II | − | + |
| F048 | 68 | M | + | II | NA | + |
| B115 | 45 | M | + | NA | − | + |
| B169 | 53 | M | NA | NA | − | − |
| D145 | 58 | M | NA | NA | + | NA |
M, male; F, female;
NA, data not avalable
Immunohistochemical detection of Hint1 protein in paraffin-embedded sections
Detection of the HINT1 protein in 5 μm paraffin-embedded sections used a commercial polyclonal antibody (ProteinTech Group Inc. Campbell Park Dr., Chicago, IL). After deparaffinization and rehydration in graded ethanol, the slides were immersed in 10 mM citric acid (pH 6.0) and microwaved for 10 min at 400 W. Staining was carried out according to the manufacture’s instruction: the primary antibody (1:100 dilution) was added and sections were incubated overnight at 4°C. This was followed by adding the secondary antibody and ABC reagent and DAB (both ABC and DAB kits were from Vector Laboratories, Burlingame, CA). Slides were then counterstained with Harris hematoxylin (Sigma, St. Louis, MO). The following categories were used for scoring: intensity of staining, none (0), mild (1), moderate (2), strong (3); and percentage of positive staining, <5% (0), 5–25% (1), 25–50% (2), >50% (3) of cells (20). Combining intensity and percentage staining resulted in the following score 0–1; negative (−); 2–6 positive (+). Liver sections from wild type (Hint1+/+) and Hint1 knocked out (Hint1−/−) mice (7) were used as positive and negative controls.
DNA extraction
DNA was isolated from frozen tissue samples, as previous described (21). Briefly, tissue was placed in liquid nitrogen and pulverized with a blender. The tissue powder was lysed with a DNA lysing buffer (10 mM Tris, 10 mM NaCl, 0.1% sodium dodecyl sulfate at pH 7.9, and 200μg/ml proteinase K). DNA was isolated by RNase treatment, phenol/chloroform extraction and ethanol precipitation.
Analysis of Hint1 and p16 hypermethylation status: methylation–specific polymerase chain reaction (MSP)
MSP was carried out, essentially, as described previously (22) and was based on the principle that treating DNA with sodium bisulfite results in the conversion of unmethylated citosine residues into uracil. Thus, the sequence of the treated DNA will differ if the DNA is originally methylated, and is then distinguishable by sequence-specific PCR primers. Bisulfite modification of tissue DNA was conducted with the CpGnome DNA modification kit (Chemicon International, Temecula, CA). The sense and antisense primers for the methylated Hint1 promoter were 5′-TTTGCGTAGGTTTGGTTGC-3′ and 5′-AACAATCTCATCTACCATCTCGAC-3′, respectively, and the primers to detect the unmethylated Hint1 were 5′-TATTTGTGAGGTTTGGTTGTGT-3′ and 5′-AACAATCTCATCTACCATCTCAAC-3′, respectively. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining. Universal methylated DNA (Chemicon International) was used as a positive control with distilled water as a negative control.
Data on p16 methylation and AFB1- and PAH-DNA adducts on these samples were available from our previous studies (14).
Statistical analysis
Fisher’s exact tests were performed to evaluate the significance of the differences between the frequencies of Hint1 promoter hypermethylation status of the various tissue categories, comparisons with Hint1 protein expression status, and comparisons with clinical characteristics.
Results
Hint1 methylation status
Methylation of the promoter region of Hint1, determined by MSP, was frequent in the HCC tumors, with 22 of 40 (55.0%) samples positive. For the paired adjacent non-tumor tissue samples, 15 of 40 samples were positive samples (37.5%) (Table 2). Representative examples of the gel analysis of MSP are shown in Figure 1. Interestingly, unmethylated Hint1 alleles were also detected in all of samples. In the 22 normal liver controls, 14 samples were from Taiwan; 2 of 22 had promoter hypermethylation, none of the U.S. normal controls liver samples had promoter hypermethylation.
Table 2.
Hint1 Gene promoter hypermethylation status and protein expression in Taiwan HCC samples
| Hypermethylation status |
Protein expression |
||
|---|---|---|---|
| Tumor | Adjacent non-tumor | Tumor | |
| 3 | − | − | + |
| 4 | − | − | + |
| 5 | − | − | + |
| 6 | − | + | + |
| 7 | + | + | − |
| 8 | − | − | + |
| 9 | − | − | + |
| 11 | + | − | + |
| 12 | + | − | − |
| 13 | + | + | − |
| 14 | − | − | + |
| 15 | + | − | |
| 17 | − | + | − |
| 18 | − | + | − |
| 19 | − | − | + |
| 20 | + | − | − |
| 21 | − | − | + |
| 22 | + | + | − |
| 23 | + | + | − |
| 25 | + | + | − |
| 28 | + | + | + |
| 29 | − | − | − |
| 30 | + | + | + |
| 32 | + | + | + |
| 33 | + | − | − |
| 34 | + | + | − |
| 35 | + | + | + |
| 36 | + | + | + |
| 38 | + | − | + |
| 39 | − | − | + |
| 40 | + | − | + |
| 42 | − | − | + |
| 43 | + | − | − |
| 44 | − | − | + |
| 45 | − | − | + |
| 47 | + | − | − |
| 48 | − | − | + |
| B115 | − | − | + |
| B169 | + | − | − |
| D145 | + | + | − |
Figure 1.
Methylation analysis of Hint1 in DNA from HCC tissues, the HepG2 HCC cell line was used as a positive control. l. Bisulfite-treated DNA was used for PCR amplification using primers sets designed for methylated (m) and unmethylated (u) Hint1. M, molecular weight marker (100 bp); 1, DNA from the HepG2 HCC cell line; lines 2–6 DNA from 5 HCC cases; line 7, distilled water as negative control.
Expression of the Hint1 protein in HCC tissue samples
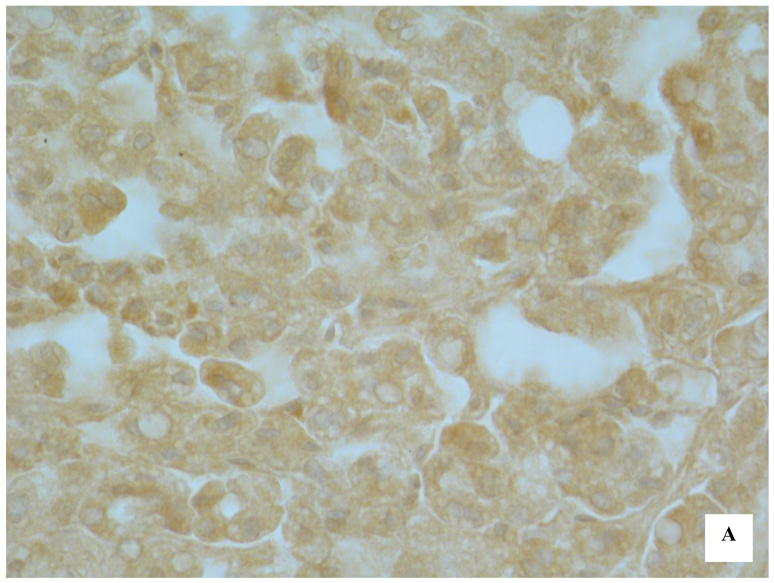
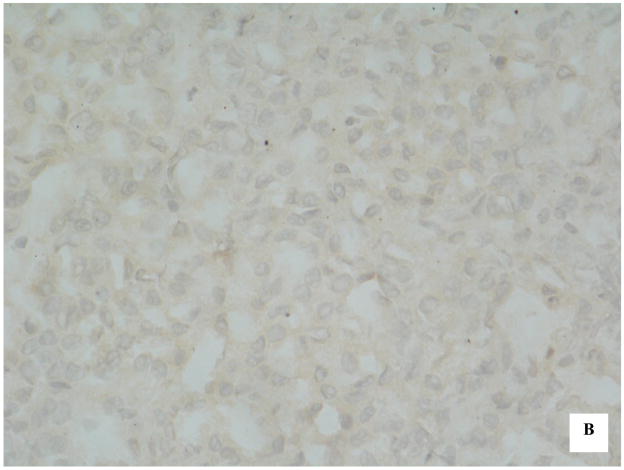
To determine whether the hypermethylation of CpG islands in the promoter region of Hint1 in HCC tissues was correlated with loss of expression of the HINT1 protein, immunohistochemical staining using an anti-HINT1 antibody was carried out on 40 HCC tissue sections. Representative examples of HINT1 protein expression are shown in Figure 2(A and B). Fifteen of the 18 unmethylated HCC samples (83%) demonstrated positive nuclear and cytoplasmic staining and 14 of 22 methylated HCCs (64%) showed loss of expression of Hint1 (Table 2). Thus, the immunostaining results were strongly correlated (p<0.003) with Hint1 methylation status (Table 3).
Figure 2.
Immunohistochemical detection of HINT1 expression in HCC samples. (A) HCC tissue with unmethylated Hint1 illustrating expression of the protein in both tumor cell nuclei and cytoplasm. X400; (B) HCC tissue with methylated Hint1 illustrating lack of expression in tumor cells. X400
Table 3.
The association of clinical characteristics with Hint1 methylation status
| Tumor tissues |
|||
|---|---|---|---|
| Variable | Non-methylated | Methylated | P |
| Adjacent non-tumor tissues | |||
| Non-methylated | 15 | 9 | 0.01 |
| Methylated | 3 | 12 | |
| Missing | 0 | 1 | |
| Hint 1 protein expression | |||
| No | 3 | 14 | 0.003 |
| Yes | 15 | 8 | |
| Grade | |||
| I orII | 11 | 8 | 0.49 |
| IIIor IV | 2 | 8 | |
| Missing | 5 | 6 | |
| Liver cirrhosis | |||
| No | 9 | 9 | 0.71 |
| Yes | 6 | 7 | |
| Missing | 3 | 6 | |
| HBsAg | |||
| Negative | 3 | 2 | 0.44 |
| Positive | 12 | 15 | |
| Missing | 3 | 5 | |
| Aflatoxin B1-adducts | |||
| Low | 11 | 7 | 0.07 |
| Medium | 3 | 11 | |
| High | 4 | 4 | |
| PAH-adducts | |||
| Low | 9 | 13 | 0.49 |
| Medium | 4 | 7 | |
| High | 4 | 2 | |
| Missing | 1 | 0 | |
| p16 methylation | |||
| No | 10 | 5 | 0.035 |
| Yes | 8 | 17 | |
Relationship between Hint1 hypermethylation and clinical parameters and other biomarkers
Possible associations between the methylation status of Hint1 and tumor stage, liver cirrhosis status, HBV infection status, p16 methylation status and levels of AFB1- and PAH-DNA adducts were investigated (Table 3). We found that p16 hypermethylation was statistically significantly associated with Hint1 methylation status (p=0.035), but there was no significant correlation with the other parameters.
Discussion
Hypermethylation of CpG islands in their promoter regions is an important mechanism for loss of function of several tumor suppressor genes, DNA repair genes and other genes in various types of human cancer (13). An increasing number of genes have been reported to undergo CpG island hypermethylation in HCC, which indicates the potential role of epigenetics in hepatocarcinogenesis (23). The promoters of ras association domain family 1A (RASSF1A) (14), p16INK4a (24), p15INK4b (25), O6-methylguanine-DNA methyltransferase (MGMT) (26), glutathione S-transferase pi (GSTP1) (16;27), suppressor of cytokine signaling 1 (SOCS-1) (28), adenomatous polyposis coli (APC) (16) and E-cadherin (E-Cad) (29) are the most frequently methylated in HCC. These findings suggest that CpG island hypermethylation is an important molecular mechanism in the development of HCC.
In previous investigations with genetically engineered mice, evidence was obtained that Hint1 is a novel hapoinsufficient tumor suppressor gene (7). However, its precise mechanism of action and relevance to specific types of human cancer is still not clear. We found a low level of expression of the HINT1 protein in the SW480 cell line when compared with four other human colon cancer cell lines and obtained evidence that this is due to methylation of the promoter region of Hint1 (9). In recent studies, we also found a low level of expression of the HINT1 protein in the human HCC cell lines HepG2 and Hep3B when compared with the human HCC cell line Huh7, by western blot analysis, and that this was also due to promoter methylation in the HepG2 and Hep3B cell lines (19). Other investigators found decreased expression of Hint1 in a subset of human NSCLC cell lines, which appeared to be due to promoter hypermethylation based on studies utilizing 5-Aza-dC (10). Thus, decreased expression of HINT1 due to hypermethylation of the promoter region of the Hint1 gene can occur in at least 3 types of human cancer cell lines.
Based on the above findings, in the present study, we investigated promoter hypermethylation of Hint1 in DNA samples from primary HCC, paired adjacent non-tumor tissues from patients with HCC, and normal liver tissue DNA. Twenty two of the 40 (55.0%) HCC samples displayed Hint1 gene promoter hypermethylation. Methylation was also observed in 37.5% of the paired adjacent nontumor tissues. This may be due to the fact that these tissue samples were not microdissected and, therefore, they may have been contaminated with a small population of HCC cells. Alternatively, since most of the adjacent nontumorous tissues are cirrhotic, promoter hypermethylation of Hint1 may be an early event in hepatocarcinogenesis (17), as is the case with other tumor suppressor genes (30). Interestingly, hypermethylation of Hint1 was also found in two normal control liver tissues, both from Taiwan and both HBV positive, but not in the 8 normal control liver tissues from the U.S. which were HBV negative. In accordance with our findings, DNA methylation of other tumor suppressor gene has been detected at a low frequency in histologically normal liver tissues (31).
In the present study, the HINT1 protein was detected in 15 of 18 (83%) tumor tissues with Hint1 promoter not methylated and 8 of 22 (36%) tumor tissues with Hint1 promoter methylated (p value for Fisher’s exact test=0.003), suggesting that methylation status correlates inversely with HINT1 expression. The discordant data may be due to the lack of tissue microdissection resulting in contamination of the tumor tissue with adjacent nontumor tissue. Immunohistochemical staining of small pieces of tissue also limits the detection of protein expression and this may also help explain the discordant data. Discrepancies between Methylation Specific PCR (MSP) and immunohistochemistry detection were reported previously (26;32).
Promoter hypermethylation of some genes is significantly linked to pathological or clinical parameters. For example, p16 hypermethylation is associated with HBV infection and expression of the HBV × protein (33;34). In our previous studies, statistically significant associations were found between RASSF1A, p16, and MGMT methylation status and the levels of AFB1-DNA adducts in Taiwan HCC samples (14;26). SOCS-1 silencing is significantly involved in the development of HCC from liver cirrhosis (28), and hypermethylation of E-Cad or GSTP1 correlates with poor survival in HCC patients (23). In the present study, the correlations between Hint1 methylation and HBV and HCV infections status and AFB1- and PAH-DNA adduct levels were also investigated, but no statistically significant correlations were found. Perhaps, HBV and HCV infections and chemical carcinogens like AFB1 and PAHs do not affect Hint1 promoter methylation status, although the relatively small sample size may limit this analysis.
Previous studies determined the frequency and chronology of methylation events of specific genes during the multistep process of hepatocarcinogenesis from cirrhosis to HCC. CpG island hypermethylation occurs in the premalignant stages and tends to accumulate during multistep hepatocarcinogenesis. The data suggest that CpG island hypermethylation of COX-2 or p16 might be potential molecular markers for the identification of patients with chronic liver disease at high risk for progression to HCC (23). Another study suggested that the most striking methylation pattern in HCC is the concurrent methylation of multiple tumor suppressor genes (TSG). Although methylation of one or two genes can be observed in nontumor and cirrhotic liver tissue, the majority of HCC cases harbored three or more methylated TSGs (16). In present study, Hint1 methylation was significantly associated with p16 hypermethylation (p=0.035). This finding is consistent with previous studies and provides further evidence that several TSGs genes accumulate methylation during progression from pre-malignant stages to HCC.
Our previous mechanistic studies suggested that Hint1 inhibits AP-1 activity by binding to a POSH-JNK2 complex, thus inhibiting the phosphorylation of c-Jun; this effect could contribute to the tumor suppressor activity of Hint1 (9). Decreased expression of the Hint1 gene through epigenetic silencing may play a role in enhancing the growth of a subset of human hepatoma cell lines by increasing the expression of genes controlled by the transcription factors β-catenin, USF2 and NFB (19).
In conclusion, to the best of our knowledge, this is the first study on detection of Hint1 promoter hypermethylation in tumor tissues. Our investigation demonstrated that epigenetic inactivation of Hint1 is a frequent event in the development of HCC and promoter hypermethylation of Hint1 can be an early event in hepatocarcinogenesis. The biologic basis and mechanisms of Hint1 inactivation by hypermethylation and the relationship between epigenetic changes in Hint1 and other risk factors for HCC is not clear at the present time. Additional studies with larger sample sizes are required to elucidate these aspects. We believe that these studies of environment-epigenetic interactions are necessary for a deeper understanding of HCC and the mechanism of action of carcinogenic exposures and could be used to identify novel opportunities for the prevention and therapy of HCC.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (ES05116 and P30ES009089). to Dr. Regina M. Santella and funds from the T.J. Martell Foundation and the National Foundation for Cancer Research to Dr. I. Bernard Weinstein
Abreviation used are
- Hint1
Hit nucleotide binding protein-1
- HCC
hepatocellular carcinoma
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- AFB1
aflatoxin B1
- PAHs
polycyclic aromatic hydrocarbons
Footnotes
Conflicts of Interest Statement
None Declared
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. New England Journal of Medicine. 1999;340:745–50. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 2.Bosch FX, Ribes J, Cleries R, Diaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191–211. v. doi: 10.1016/j.cld.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Bosch FX, Ribes J, Borras J. Epidemiology of primary liver cancer. Semin Liver Dis. 1999;19:271–85. doi: 10.1055/s-2007-1007117. [DOI] [PubMed] [Google Scholar]
- 4.Kondoh N, Wakatsuki T, Hada A, Shuda M, Tanaka K, Arai M, et al. Genetic and epigenetic events in human hepatocarcinogenesis. Int J Oncol. 2001;18:1271–8. doi: 10.3892/ijo.18.6.1271. [DOI] [PubMed] [Google Scholar]
- 5.Brenner C. Hint, Fhit, and GalT: function, structure, evolution, and mechanism of three branches of the histidine triad superfamily of nucleotide hydrolases and transferases. Biochemistry. 2002;41:9003–14. doi: 10.1021/bi025942q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su T, Suzui M, Wang L, Lin CS, Xing WQ, Weinstein IB. Deletion of histidine triad nucleotide-binding protein 1/PKC-interacting protein in mice enhances cell growth and carcinogenesis. Proc Natl Acad Sci USA. 2003;100:7824–9. doi: 10.1073/pnas.1332160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Zhang Y, Su T, Santella RM, Weinstein IB. Hint1 is a haplo-insufficient tumor suppressor in mice. Oncogene. 2006;25:713–21. doi: 10.1038/sj.onc.1209111. [DOI] [PubMed] [Google Scholar]
- 8.Weiske J, Huber O. The histidine triad protein Hint1 triggers apoptosis independent of its enzymatic activity. J Biol Chem. 2006;281:27356–66. doi: 10.1074/jbc.M513452200. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Zhang Y, Li H, Xu Z, Santella RM, Weinstein IB. Hint1 inhibits growth and activator protein-1 activity in human colon cancer cells. Cancer Res. 2007;67:4700–8. doi: 10.1158/0008-5472.CAN-06-4645. [DOI] [PubMed] [Google Scholar]
- 10.Yuan BZ, Jefferson AM, Popescu NC, Reynolds SH. Aberrant gene expression in human non small cell lung carcinoma cells exposed to demethylating agent 5-aza-2′-deoxycytidine. Neoplasia. 2004;6:412–9. doi: 10.1593/neo.03490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baylin SB, Herma JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends in Genetics. 2000;16:168–74. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 12.Yu KH, Lo YM, Tse GM, Chan KC, Chan AB, Chow KC, et al. Quantitative analysis of cell-free Epstein-Barr virus DNA in plasma of patients with nonnasopharyngeal head and neck carcinomas. Clin Cancer Res. 2004;10:1726–32. doi: 10.1158/1078-0432.ccr-0991-3. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Advances in Cancer Research. 1998;72:141–96. [PubMed] [Google Scholar]
- 14.Zhang YJ, Ahsan H, Chen Y, Lunn RM, Wang LY, Chen SY, et al. High frequency of promoter hypermethylation of the RASSF1A and p16 genes and its relationship to aflatoxin B1-DNA adducts level in human hepatocellular carcinoma. Mol Carcinogenesis. 2002;35:85–92. doi: 10.1002/mc.10076. [DOI] [PubMed] [Google Scholar]
- 15.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 16.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. American Journal of Pathology. 2003;163:1101–7. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HI, et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin Cancer Res. 2007;13:2378–84. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 18.Murata H, Tsuji S, Tsujii M, Sakaguchi Y, Fu HY, Kawano S, et al. Promoter hypermethylation silences cyclooxygenase-2 (Cox-2) and regulates growth of human hepatocellular carcinoma cells. Lab Invest. 2004;84:1050–9. doi: 10.1038/labinvest.3700118. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Li H, Zhang YJ, Harada A, Santella R, Weinstein I. Hint1 inhibits B-catenin/TCF4, USF2 and NFkB activity in human hepatoma cells. International J Cancer. 2008 doi: 10.1002/ijc.24072. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YJ, Rossner P, Chen Y, Agrawal M, Wang Q, Wang L, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. International Journal of Cancer. 2006;119:985–91. doi: 10.1002/ijc.21699. [DOI] [PubMed] [Google Scholar]
- 21.Jiang W, Zhang YJ, Kahn SM, Hollstein MC, Santella RM, Lu SH, et al. Altered expression of the cyclin D, and retinoblastoma genes in human esophageal cancer. Proc Natl Acad Sci USA. 1993;90:9026–30. doi: 10.1073/pnas.90.19.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–8. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liew CT, Li HM, Lo KW, Leow CK, Chan JY, Hin LY, et al. High frequency of p16INK4A gene alterations in hepatocellular carcinoma. Oncogene. 1999;18:789–95. doi: 10.1038/sj.onc.1202359. [DOI] [PubMed] [Google Scholar]
- 25.Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res. 2000;6:3516–21. [PubMed] [Google Scholar]
- 26.Zhang YJ, Chen Y, Ahsan H, Lunn RM, Lee PH, Chen CJ, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation and its relationship to aflatoxin B1-DNA adducts and p53 mutations in hepatocellular carcinoma. Int J Cancer. 2003;103:440–4. doi: 10.1002/ijc.10852. [DOI] [PubMed] [Google Scholar]
- 27.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–8. [PubMed] [Google Scholar]
- 28.Okochi O, Hibi K, Sakai M, Inoue S, Takeda S, Kaneko T, et al. Methylation-mediated silencing of SOCS-1 gene in hepatocellular carcinoma derived from cirrhosis. Clin Cancer Res. 2003;9:5295–8. [PubMed] [Google Scholar]
- 29.Matsumura T, Makino R, Mitamura K. Frequent down-regulation of E-cadherin by genetic and epigenetic changes in the malignant progression of hepatocellular carcinomas. Clin Cancer Res. 2001;7:594–9. [PubMed] [Google Scholar]
- 30.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95:11891–6. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–9. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 32.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–7. [PubMed] [Google Scholar]
- 33.Li X, Hui AM, Sun L, Hasegawa K, Torzilli G, Minagawa M, et al. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin Cancer Res. 2004;10:7484–9. doi: 10.1158/1078-0432.CCR-04-1715. [DOI] [PubMed] [Google Scholar]
- 34.Zhu R, Li BZ, Li H, Ling YQ, Hu XQ, Zhai WR, et al. Association of p16INK4A hypermethylation with hepatitis B virus X protein expression in the early stage of HBV-associated hepatocarcinogenesis. Pathol Int. 2007;57:328–36. doi: 10.1111/j.1440-1827.2007.02104.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.