Abstract
The use of heavy water (D2O) as a solvent is commonplace in many spectroscopic techniques for the study of biological macromolecules. A significant deuterium isotope effect exists where hydrogen-bonding is important, such as in protein stability, dynamics and assembly. Here we illustrate the use of D2O in additive screening for the production of reproducible diffraction-quality crystals for the Salmonella enteritidis fimbriae 14 (SEF14) putative tip adhesin, SefD.
Keywords: Deuterium oxide, crystallography, additive, pilin
1. Introduction
The rate limiting step in macromolecular X-ray crystallography is obtaining diffraction-quality crystals, where many adaptations to standard procedures have been developed to promote crystallization [1]. Traditional approaches include producing truncated versions in which flexible regions are removed and mutagenesis to alter the surface characteristics [2,3]. More recently, methods including reductive methylation of lysine side-chains [4], in situ proteolyisis [5] and ligand binding screens [6], have been deployed to increase success rates in protein crystallization. New additives have also been discovered for use with membrane proteins, which are particular difficult to study due to a limited range of suitable detergents [7].
In the absence of suitable crystals, solution-state NMR spectroscopy can be used to determine high resolution structures of small to medium sized proteins. Before the introduction of isotope labelling and multidimensional heteronuclear NMR, it was necessary to exchange samples into deuterated water (D2O) to eliminate signal interference from the solvent and exchangeable protons. Deuterium exchange is also commonly used in Infra-red spectroscopy, Raman spectroscopy and neutron scattering studies to reduce unwanted signal associated with protons. The use of D2O remains a useful strategy for improving the outcome of modern NMR experiments, particularly in studies of methyl-labelled samples of very large systems [8].
The importance of hydration has been highlighted in studies quantifying the effects of D2O on protein structure, stability and dynamics [9,10,11]. Isotope fractionation measurements have revealed that weak hydrogen bonds tend to accumulate deuterium rather than protium [12] and subsequent changes in bond lengths alter the geometry of multiple hydrogen bond networks [13]. The kinetics of protein assembly also displays marked D/H isotope effects [14,15,16]. Despite this work, the usefulness of D2O for promoting protein crystallisation in sparse matrix screening has not been described.
Salmonella enteritidis fimbriae 14 (SEF14) [17] is exported via the chaperone-usher fibre assembly pathway [18] and the putative tip adhesin, SefD, is essential for efficient uptake or survival of S. enteritidis in macrophages [19]. We originally obtained a single crystal of recombinant SefD in standard H20-based commercial screens, but this hit could not be readily reproduced. Here, we report the usefulness of D2O in overcoming this issue for SefD and describe the effects on crystal lattice formation. We propose that this D2O would be a useful additive for the use in optimising standard commercial crystallization screens.
2. Materials and Methods
2.1. Cloning, expression and purification
S. enteritidis sefD (residues 3-143, lacking the N-terminal signal sequence) was isolated from a synthetic gene (Invitrogen) with a reverse primer containing a DNKQ tetrapeptide linker, followed by the N-terminal donor strand (residues 2-18) of SefA. This produced a donor-stand complemented gene product herein referred to as SefDdscA. This was cloned into a pQE-30 plasmid (Qiagen) containing a vector encoded N-terminal His6 tag. This was transformed into E. coli BL21 (DE3) strain and grown at 37°C in LB. Expression was induced with 0.5 mM IPTG at OD600nm=0.6 followed by incubation overnight at 18°C. After harvesting the cells, they were lysed in the presence of 8 M urea and SefDdscA purified with Ni2+ affinity chromatography under denaturing conditions. After elution, SefDdscA was dialysed against 50 mM Na acetate pH 5.0, 200 mM NaCl, 1.0 M urea, followed by dialysis against 50 mM Na acetate pH 5.0, 200 mM NaCl and gel filtered with a Superdex-200 column (GE healthcare). SefDdscA was finally concentrated to 10 mg/ml (for crystallization in 0% D2O) or exchanged into buffer made up in 100% D2O and then concentrated to 10 mg/ml (for crystallization in 50% D2O).
2.2. Crystallization
Conditions for crystallization were initially screened by the sitting-drop method of vapour diffusion at 293 K using sparsematrix crystallization kits (Hampton Research, USA; Emerald BioSciences, USA; Molecular Dimensions Ltd, USA) in 96-well MRC plates with 100 nl protein solution and 100 nl reservoir solution using a Mosquito nanolitre high-throughput robot (TTP Labtech). Crystals of SefDdscA grown in 4 M ammonium acetate, 0.1 M sodium acetate pH 4.6, 0% or 100% D2O (SefDdscA exchanged into 100% D2O buffer and all well reagents reconstituted in 100% D2O) were washed in well solution plus additional 30% (v/v) glycerol and then flash frozen in liquid N2. Diffraction data were collected at 100K on beamline I24 of the Diamond Light Source (DLS), UK.
2.3. Crystal structure determination and refinement
0% D2O crystal form
Data were processed using XIA2 [20] and scaled with SCALA [21] to 3.1 Å. Molecular replacement was performed in PHASER [22] using the structure of DraD (pdb: 2AXW; residues 1-118) [23] as the search model. PARROT [24] was used to remove model bias and initial automated model building was performed with BUCANEER [25]. Refinement was carried out with REFMAC [26] using jelly body restraints, map sharpening, with 5% of the reflections omitted for cross-validation, and model building was carried out in COOT [27]. TLS B-factor refinement [28,29] is usually not carried out at low resolution, although in this case it was implemented as it reduced the Rfree by 3% and improved the map quality. Processing and refinement statistics for the final model can be found in Table 1.
Table 1.
Data collection statistics.
| 0% D2O | 100% D2O | |
|---|---|---|
| Crystal parameters | ||
| Space group | P6522 | P212121 |
| Cell dimensions | a=b=52.48, c=218.03 | a=52.66, b=87.96, c=211.75 |
|
| ||
| Data collection | ||
| Beamline | DLS I24 | DLS I24 |
| Wavelength (Å) | 0.9778 | 1.5498 |
| Resolution (Å) | 3.1-45.45 3.1-47.15 |
(3.1-3.18) (3.1-3.18) |
| Unique observations | 3659 (250) | 18325 (1290) |
| R sym | 0.072 (0.443) | 0.076 (0.45) |
| <I>/σI | 15.2 (3.2) | 14.2 (3.0) |
| Completeness (%) | 98.7 (99.5) | 98.7 (97.3) |
| Redundancy | 4.2 (4.6) | 4.9 (4.9) |
|
| ||
| Refinement | ||
| Rwork/Rfree (%) | 26.8/33.2 | 25.9/30.2 |
| Number of protein residues rmsd stereochemistry |
131 | 777 |
| Bond lengths (Å) | 0.010 | 0.009 |
| Bond angles (°) | 1.811 | 1.609 |
| Ramachandran analysis | ||
| Residues in outlier regions | 0 | 0 |
| Residues in favoured regions | 93.7% | 90.9% |
| Residues in allowed regions | 100% | 100% |
Numbers in parentheses refer to the outermost resolution shell.
Rsym = ∑ |I -<I>| / ∑I where I is the integrated intensity of a given reflection and <I> is the mean intensity of multiple corresponding symmetry-related reflections.
Rwork = ∑ ∥Fo| - |Fc∥ / ∑ Fo where Fo and Fc are the observed and calculated structure factors respectively.
Rfree = Rwork calculated using ~10% random data excluded from the refinement. rmsd stereochemistry is the deviation from ideal values.
Ramachandran analysis was carried out using Molprobity [38].
100% D2O crystal form
Data were processed using XIA2 [20] and scaled with SCALA [21] to 3.1 Å. Molecular replacement was next performed with the 100% D2O data using the previous refined structure as the search model with MOLREP [30] using the ‘use pseudo-translational symmetry’ option. Refinement was carried out in REFMAC [26] using automated NCS restraints, jelly body restraints, map sharpening, with 5% of the reflections were omitted for cross-validation, and model building was carried out in COOT [27]. Again TLS B-factor refinement was implemented [28,29] as it had a very positive effect on the final map quality and R-factors. Processing and refinement statistics for the final models can be found in Table 1.
2.4. Protein Data Bank accession number
The coordinates and structure factors have been deposited in the PDB under accession number 3UIY (0% D2O) and 3UIZ (100% D2O).
3. Results and discussion
3.1. Crystallization of SefDdscA
In the chaperone-usher family of pili, fibre polymerization occurs through a process termed donor strand exchange (DSE) [31,32], concluding with the N-terminal extension (NTE) of one pilin domain complementing the incomplete immunoglobulin-like fold of an adjacent subunit. To avoid polymerisation we created a donor-strand complemented (DSC) [33] construct of SefD (SefDdscA), in which the NTE from the SefA subunit was cloned onto the C-terminus of SefD, separated by a tetrapeptide (DNKQ) linker.
An initial crystallization screen of SefDdscA against more than 1000 standard conditions identified only microcrystals or precipitates which could not be optimized. After several months a solitary crystal developed in one condition (4 M ammonium acetate, 0.1 M sodium acetate pH 4.6), however, despite exhaustive efforts, it was not possible to reproduce this crystal with subsequent optimization in H2O. However, when the protein sample was buffer exchanged into 100% D2O (creating a 50% D2O solution in the final crystallization drop) this led to immediate reproducibility of the initial hit with crystal clusters appearing after a few days (Fig 1). Complete exchange of both precipitant mixture and protein solution into 100% D2O yielded significant improvements in crystal morphology, with large single crystals developing in the majority of the optimisation drops (Fig 1).
Figure 1.
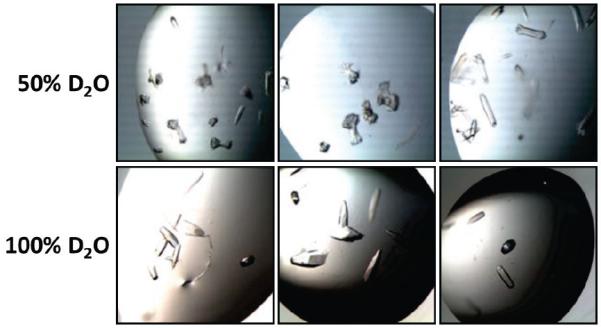
Representative images of SefDdscA crystals grown in both 50% and 100% D2O.
Diffraction data were collected from the single crystal grown in 0% D2O and several crystals from 100% D2O. The structures were solved by molecular replacement and both refined to 3.1 Å (Table 1 and Supplementary Fig. 1). Surprisingly, whilst SefDdscA in 0% D2O crystallized in space group P6522 (a=b=52.5, c=218.0), in 100% D2O SefDdscA crystallized in P212121 (a=52.7, b=88.0, c=211.8) with additional pseudotranslational non-crystallographic symmetry (Supplementary Fig. 2).
3.2. Overall structure of SefDdscA in 0% D2O
There is one molecule of SefDdscA (154 residues) in the asymmetric unit of the 0% D2O crystal form, corresponding to 54% solvent content (Vm=2.67) and all residues could be built except for the flexible N-terminal His6 tag, the flexible C-terminus of SefD (Glu118-Leu119), and the synthetic loop between SefD and the SefA NTE [34] (G-strand; Asp120-Phe125). Furthermore, the following solvent exposed side chains were not visible in the electron density map: Lys4, lys8, Glu11, Arg17, Lys22, Asn31, Arg32, Lys35, Lys36, Lys45, Lys47, Asn48, Glu72, Asp73, Asp88, Phe89, Glu92, Asn100, Asp102, Glu116, Ile117, Glu118, Lys129, Lys141 and Asn143.
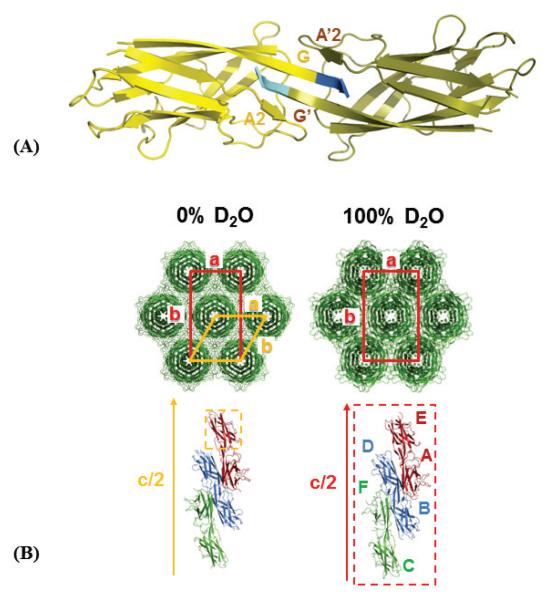
SefDdscA is composed of 9 β-strands (Supplementary Fig. 3) and its structure is similar to that of other minor pilin domains of the Aaf/Dra family [23,35]. SefDdscA purifies as a stable dimer and the structure shows that the two monomers pack against a hydrophobic face which is stabilized through domain-swapping (Fig. 2A) via the A2 and G β-strands (which include non-native vector encoded residues: Lys141, Leu142, Asn143).
Figure 2.
Global arrangement of SefDdscA molecules within crystals grown in 0% and 100% D2O. (A) Domain-swapped dimer of SefDdscA in 0% D2O. The single molecule of the asymmetric unit is coloured yellow whilst its symmetry mate is coloured olive. The dimer face is composed of a hydrophobic surface and solely backbone hydrogen bonding within the A2-G′-G-A2′ sheets. (B) Crystal packing in the 0% and 100% D2O crystal forms. Top panel: SefDdscA molecules (green) form ‘rod-like’ structures and the unit cells are viewed down the crystallographic c-axis. The spacegroup C2221 (0% D2O: red box) is a subgroup of P6522 (orange box) and is very similar to the P212121 (100% D2O: red box) packing. Bottom panel: SefDdscA domain-swapped dimers (red, blue and green) viewed parallel to half the unit cell c-axis. The asymmetric units are represented as dashed boxes.
3.3. Overall structure of SefDdscA in 100% D2O
In the 100% D2O form, there are six molecules of SefDdscA in the asymmetric unit, which corresponds to 49% solvent content (Vm=2.41) and all residues could be built except for the flexible N-terminal His6 tags, the flexible C-terminus of SefD (chains A,D: Glu118; A-D: Leu119) and the synthetic loop (chains A-D: Asp120-Lys122; A-C: Gln123-Phe125; B: Val126). Furthermore, chains E and F show considerable disorder in the loop regions which could not be modelled (chain E: Asn31-Lys36, Ile117-Val126; chain F: Ser20-Lys22, Asn78-Ile81, Phe89-Asn91, Gly99-Asn100, Glu118-Lys129) and it was not possible to build a substantial number of solvent exposed side chains, namely: Ser2 (chain B), Leu3 (chain C), Lys4 (chains A-E), Met6 (chains A,D,F), Lys8 (Chains B-F), Glu11 (chains A-F), Asp16 (chains A,B,D,E), Arg17 (chains A-F), Asn21 (chain E), Lys22 (chains A-E), His28 (chain F), Leu29 (chain F), Phe30 (chain F), Arg32 (chains B,F), Glu33 (chains A,D), Lys35 (chains A-D,F), Lys36 (chains A-D,F), Glu44 (chain E), Lys45 (chains A-F), Lys47 (chains A-C,E,F), Asn48 (chains A-F), Lys64 (chain E,F), Arg66 (chain E), Arg68 (chain F), Glu72 (chains A-F), Asp73 (chains A-D,F), Gln75 (chain E,F), Asn78 (chain E), Asp88 (chains A,B,D-F), Phe89 (chains A,B,C,D,E), Asn91 (chains A,B,C,D,E), Glu92 (chains A,C,E,F), Phe97 (chain F), Asn100 (chains A,B,C,D), Glu116 (chains A-F), Ile117 (chains A,C,D,F), Glu118 (chains B,C), Gln123 (chain D), Phe125 (chain D), Val126 (chains C,D), Asn128 (chains C,D), Lys129 (chains A,E), Lys141 (chains A-F) and Asn143 (chains A-F).
The structure of SefDdscA in 100% D2O is in essence identical to the 0% D2O form (rmsd between 0.37-0.56 Å) (Supplementary Fig. 4). The only deviations are seen in the A2 and G strands which are involved in domain-swapping within the dimer.
3.4. Comparisons between SefDdscA in 0% and 100% D2O
Crystals of SefDdscA are formed from rod-like structures packing side-by-side that are themselves composed of domain-swapped dimers, which rotate about a central crystallographic c-axis with either 65 (0% D2O) or 21 (100% D2O) screw-symmetry (Fig. 2B). The spacegroup C2221 (a=52.5, b=90.7, c=218.0) is a subgroup of P6522 and the two crystal forms are clearly similar, where the centred orthorombic has become a primitive orthorhombic system related by non-crystallographic translational symmetry in the 100% D2O form.
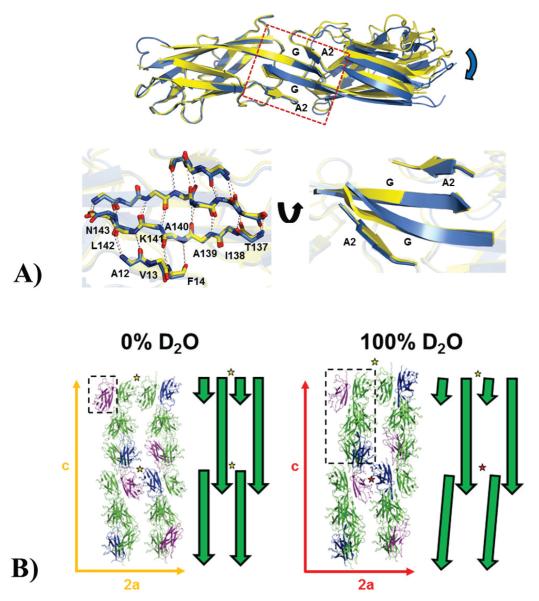
Whilst the overall structures of SefDdscA from both crystal forms are highly similar, substantial variations are evident in the orientation of the monomers within the chain A-E and B-D domain-swapped dimers where one subunit is twisted and translated in relation to the other (Fig. 3A; Supplementary Fig. 5). This inter-dimer interface is formed in part by a hydrophobic surface and also significant backbone hydrogen-bonding within the A2 and G β-strands. In addition to this domain-swapped dimer, two other lattice dimers are also present. The ‘back-to-back’ dimer is identical in both crystal forms (Supplementary Fig. 6), whilst the ‘face-to-face’ dimer exhibits some deviations (Supplementary Fig. 7). This latter dimer is responsible for the non-crystallographic pseudo-translational symmetry, where chains A and D, and chains B and C are related by the vector (0.5,0.5,0.031), and superimposes well with the 0% D2O structure (Supplementary Fig. 2; Supplementary Fig. 7). However, chains E and F do not form a dimer and as such they show substantial disorder in these ‘face-to-face’ loop regions.
Figure 3.
(A) Effects of deuterium on the inter-domain domain-swapped dimer interface. Top panel: A SefDdscA dimer from a 100% D2O crystal (blue) is shown superimposed onto a single chain of a 0% D2O dimer (yellow) and clearly demonstrates a twisting and displacement of the other subunit of the dimer in the different solvents (blue arrow). The dimer interface which orchestrates these changes is boxed and the secondary structure is labeled. Bottom panel: (left) the box has been blown up and backbone residues in the A2 and G strands are shown as sticks with inter-sheet hydrogen bonds depicted as dashed lines. (Right) This region has been rotated to highlight the distortion in the sheet structure (B) Effect of deuterium on the overall packing in crystals of SefDdscA. Packing of SefDdscA in both crystal forms are shown along the a and c unit cell axes with the E (blue) and F (purple) chains in the 100% D2O crystal and equivalent positions in the 0% D2O crystal highlighted. The asymmetric units are depicted as dashed boxes. To the right of each is a schematic representation shown as green arrows. In the 0% D2O crystals, two ‘face-to-face’ dimers are shown (yellow star), however in the 100% D2O crystal, due to changes within the domain-swapped dimer interface (Fig. 2; Fig. 3; Supplementary Fig. 5; Supplementary Fig. 7) which are propagated down the c-axis, only one of these dimers is seen (yellow star), whilst to maintain lattice integrity, chains E and F do not form the usual dimer (red star).
3.5. Interpreting the increased propensity of SefDdscA crystallization in D2O
Whilst studies have been published describing only subtle effects of D2O on crystal formation [36,37], these have only been conducted on highly compact, single domain proteins that readily crystalize in many conditions and generally contain low solvent levels. It is therefore not possible to extrapolate these conclusions to biological macromolecules that do not readily crystallise or of those crystallization conditions which cannot be reproduced. In the case of SefDdscA, the improvements in crystal reproducibility are attributed to the altered interactions in the 100% D2O crystal form. These changes are largely mediated by the interface within the domain-swapped dimer which does not contribute to the structured core. It is clear that this region is more malleable and that deuterium has altered the lengths and geometry of the hydrogen bond networks. The overall effects of these changes are propagated down the c-axis of the 100% D2O crystal form. In these crystals, the chain E-F dimer does not form, increasing the entropy of SefDdscA, whilst the chain C-F domain-swapped dimer is identical to the 0% D2O crystal form which allows it to stay in register and maintain lattice formation (Fig. 3B; Supplementary Fig. 5; Supplementary Fig. 7).
3. 4 Conclusions
Panaceas for crystallization problems do not exist and whilst replacing H2O with D2O in the crystallization medium is unlikely to be a miracle additive, our study reveals that this approach can offer genuine promise in lost causes. The ease at which any benefits of D2O can be tested, by merely buffer exchanging the protein sample into 100% D2O prior to crystallization, should enable it to become routine for sparse-matrix screening in macromolecular crystallography. Furthermore, the D/H isotope effect may have more pronounced effects with targets such as membrane proteins, large RNAs or macromolecules with substantially dynamic properties.
Supplementary Material
Acknowledgment
This work was supported by the Wellcome Trust (programme grant 079819; equipment grant 085464), BBSRC (G004668 and F007566) and MRC (G1001664). We thank the beamline scientists at I24 of the Diamond Light Source (DLS) and also members of the Centre for Structural Biology at Imperial College London for comments.
References
- [1].Chayen NE, Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal. Nat Meth. 2008;5:147–153. doi: 10.1038/nmeth.f.203. [DOI] [PubMed] [Google Scholar]
- [2].Banatao DR, Cascio D, Crowley CS, Fleissner MR, Tienson HL, Yeates TO. An approach to crystallizing proteins by synthetic symmetrization. Proceedings of the National Academy of Sciences. 2006;103:16230–16235. doi: 10.1073/pnas.0607674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallographica Section D. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- [4].Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, Fan Y, Bigelow L, Maltseva N, Wu R, Borovilos M, Duggan E, Zhou M, Binkowski TA, Zhang R.-g., Joachimiak A. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Meth. 2008;5:853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dong A, Xu X, Edwards AM. In situ proteolysis for protein crystallization and structure determination. Nat Meth. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proceedings of the National Academy of Sciences. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J-L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Meth. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-Correlated Relaxation Enhanced 1H–13C NMR Spectroscopy of Methyl Groups in Very High Molecular Weight Proteins and Protein Complexes. Journal of the American Chemical Society. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- [9].Makhatadze GI, Clore GM, Gronenborn AM. Solvent isotope effects and protein stability. Nature Structural Biology. 1995;2:852–855. doi: 10.1038/nsb1095-852. [DOI] [PubMed] [Google Scholar]
- [10].Sheu S-Y, Schlag EW, Selzle HL, Yang D-Y. Molecular Dynamics of Hydrogen Bonds in Protein–D2O: The Solvent Isotope Effect. The Journal of Physical Chemistry A. 2008;112:797–802. doi: 10.1021/jp0771668. [DOI] [PubMed] [Google Scholar]
- [11].Fu L, Villette S, Petoud S.p., Fernandez-Alonso F, Saboungi M-L. H/D Isotope Effects in Protein Thermal Denaturation: The Case of Bovine Serum Albumin. The Journal of Physical Chemistry B. 2011;115:1881–1888. doi: 10.1021/jp104769v. [DOI] [PubMed] [Google Scholar]
- [12].Loh SN, Markley JL. Hydrogen Bonding in Proteins As Studied by Amide Hydrogen D/H Fractionation Factors: Application to Staphylococcal Nuclease. Biochemistry. 1994;33:1029–1036. doi: 10.1021/bi00170a023. [DOI] [PubMed] [Google Scholar]
- [13].Sigala PA, Tsuchida MA, Herschlag D. Hydrogen bond dynamics in the active site of photoactive yellow protein. Proceedings of the National Academy of Sciences. 2009;106:9232–9237. doi: 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Panda D, Chakrabarti G, Hudson J, Pigg K, Miller HP, Wilson L, Himes RH. Suppression of microtubule dynamic instability and treadmilling by deuterium oxide. Biochemistry. 2000;39:5075–5081. doi: 10.1021/bi992217f. [DOI] [PubMed] [Google Scholar]
- [15].Santra MK, Dasgupta D, Panda D. Deuterium oxide promotes assembly and bundling of FtsZ protofilaments. Proteins-Structure Function and Bioinformatics. 2005;61:1101–1110. doi: 10.1002/prot.20671. [DOI] [PubMed] [Google Scholar]
- [16].Gripon C, Legrand L, Rosenman I, Vidal O, Robert MC, Boué F. Lysozyme-lysozyme interactions in under- and super-saturated solutions: a simple relation between the second virial coefficients in H2O and D2O. Journal of Crystal Growth. 1997;178:575–584. [Google Scholar]
- [17].Clouthier SC, Muller KH, Doran JL, Collinson SK, Kay WW. Characterization of three fimbrial genes, sefABC, of Salmonella enteritidis. Journal of Bacteriology. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis, Nature reviews. Microbiology. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Edwards RA, Schifferli DM, Maloy SR. A role for Salmonella fimbriae in intraperitoneal infections. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:1258–1262. doi: 10.1073/pnas.97.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Winter G. xia2: an expert system for macromolecular crystallography data reduction. Journal of Applied Crystallography. 2010;43:186–190. [Google Scholar]
- [21].Evans P. Scaling and assessment of data quality. Acta Crystallographica Section D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- [22].McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- [23].Jedrzejczak R, Dauter Z, Dauter M, Piatek R, Zalewska B, Mroz M, Bury K, Nowicki B, Kur J. Structure of DraD invasin from uropathogenic Escherichia coli: a dimer with swapped [beta]-tails. Acta Crystallographica Section D. 2006;62:157–164. doi: 10.1107/S0907444905036747. [DOI] [PubMed] [Google Scholar]
- [24].Cowtan K. Recent developments in classical density modification. Acta Crystallographica Section D. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallographica Section D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- [26].Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- [27].Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- [28].Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta crystallographica. Section D, Biological crystallography. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- [29].Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta crystallographica. Section D, Biological crystallography. 2005;61:465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- [30].Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallographica Section D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- [31].Barnhart MM, Sauer FG, Pinkner JS, Hultgren SJ. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. Journal of Bacteriology. 2003;185:2723–2730. doi: 10.1128/JB.185.9.2723-2730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Molecular cell. 2006;22:831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]
- [33].Sauer FG, Futterer K, Pinkner JS, Dodson KW, Hultgren SJ, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- [34].Barnhart MM, Sauer FG, Pinkner JS, Hultgren SJ. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. Journal of Bacteriology. 2003;185:2723–2730. doi: 10.1128/JB.185.9.2723-2730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cota E, Jones C, Simpson P, Altroff H, Anderson KL, Du Merle L, Guignot J, Servin A, Le Bouguénec C, Mardon H, Matthews S. The solution structure of the invasive tip complex from Afa/Dr fibrils. Molecular Microbiology. 2006;62:356–366. doi: 10.1111/j.1365-2958.2006.05375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chatake T, Ishikawa T, Yanagisawa Y, Yamada T, Tanaka I, Fujiwara S, Morimoro Y. High-resolution X-ray study of the effects of deuteration on crystal growth and the crystal structure of proteinase K. Acta Crystallographica Section F. 2011;67:1334–1338. doi: 10.1107/S1744309111031903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu XQ, Sano Y. Kinetic studies on the initial crystallization process of lysozyme in the presence of D2O and H2O. Journal of Protein Chemistry. 1998;17:9–14. doi: 10.1023/a:1022530328883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.