Abstract
Quantitative trait loci (QTL) with small effects on phenotypic variation can be difficult to detect and analyze. Because of this a large fraction of the genetic architecture of many complex traits is not well understood. Here we use sporulation efficiency in Saccharomyces cerevisiae as a model complex trait to identify and study small-effect QTL. In crosses where the large-effect quantitative trait nucleotides (QTN) have been genetically fixed we identify small-effect QTL that explain approximately half of the remaining variation not explained by the major effects. We find that small-effect QTL are often physically linked to large-effect QTL and that there are extensive genetic interactions between small- and large-effect QTL. A more complete understanding of quantitative traits will require a better understanding of the numbers, effect sizes, and genetic interactions of small-effect QTL.
Keywords: yeast, quantitative trait loci (QTL), complex trait, sporulation efficiency
COMPLEX traits exhibit non-Mendelian inheritance patterns, which arise from the segregation of multiple quantitative trait loci (QTL) (Lander and Schork 1994). A QTL is a region of the genome containing an allelic difference that causes a change in phenotype. Many medically and agriculturally important traits exhibit complex genetic architecture, including phenotypes ranging from diabetes and cancer penetrance to meat quality and frost tolerance in crops (Glazier et al. 2002; Heuven et al. 2009; Dumont et al. 2009; Gaudet et al. 2010). QTL with relatively large effects are the easiest to identify and analyze, yet most QTL have small average effects on complex traits (Mackay 2001). Thus, while theory and experiment suggest that a large fraction of the variation of many phenotypes will be explained by QTL with smaller effect sizes (Fisher 1930; Lango Allen et al. 2010; Yang et al. 2011), our current understanding of complex traits is based primarily on analyses of QTL with the largest effect sizes. Because small-effect QTL are necessarily more difficult to detect and analyze, a large fraction of the genetic architecture of most complex traits is not well understood. A more complete model of complex traits should include an understanding of the numbers, effect sizes, and interactions of small-effect QTL.
To identify and study small-effect QTL, we used sporulation efficiency in the yeast Saccharomyces cerevisiae as a model complex trait (Gerke et al. 2006). This system offers several advantages for the study of QTL with relatively small-effect sizes. Sporulation efficiency is a highly heritable trait in yeast (Gerke et al. 2006). The measurements of sporulation efficiency can be performed in controlled environments that provide the statistical power to detect QTL with small-effect sizes. With this system, we previously identified four quantitative trait nucleotides (QTN) that have large effects on sporulation efficiency (Gerke et al. 2009). Here we describe crosses designed to uncover additional QTL that account for phenotypic variation not explained by the major-effect QTN. For the purposes of this study we define these additional QTL as small-effect QTL.
One previous effort in yeast uncovered additional small-effect QTL using a single targeted backcross to fix a single large-effect QTL (Sinha et al. 2008). While this method was effective at identifying some small-effect QTL, as a backcross it could only assay the subset of the variation remaining in the F2 parent. We present an alternate approach for identifying small-effect QTL in which we fix causal SNPs previously identified as large-effect QTL, allowing all other variation in the parental genomes to segregate freely. In separate crosses we fixed the four large-effect SNPs as both variants, allowing us to determine the dependence of small-effect QTL on particular alleles of large-effect QTL.
We describe the identification of small-effect QTL that together account for 40–55% of the remaining variance in sporulation efficiency when the large-effect QTL are fixed. Unlike the large-effect QTN, small-effect alleles both increase and decrease sporulation efficiency. Our ability to detect small-effect QTL was highly dependent on the allelic status of the large-effect QTL, suggesting strong genetic interactions between small- and large-effect QTL. Our mapping results also reveal that small-effect QTL are often physically linked to large-effect QTL, and in one case we show that a small-effect QTL resides within the same gene as a large-effect QTN. Our results suggest that QTL may harbor mixtures of small- and large-effect causative alleles that interact to influence complex trait phenotypes.
Materials and Methods
Strains
Parental strains BC240 (vineyard), BC248 (oak), BC713 (vineyard(oooo)), and BC728 (oak(vvvv)) were described previously (Gerke et al. 2009). All strains contain a GFP reporter fused to the SPS2 ORF and marked by an antibiotic resistance cassette. BC240 contains the natMX4 marker, conferring resistance to nourseothricin, BC248 contains the hygMX4 marker, conferring resistance to hygromycin, while BC713 and BC728 contain the kanMX4 marker, conferring resistance to G418 (Wach et al. 1994; Goldstein and McCusker 1999). BC713 and BC728 were created by single-nucleotide replacement followed by multiple rounds of intercrossing and backcrossing to ensure that phenotypes were not affected by second-site mutations(Gerke et al. 2010). The oak strain containing all four vineyard QTN (oak(vvvv), strain BC728) sporulates at 7.7%, while the vineyard strain containing the four oak QTN (vineyard(oooo), strain BC713) sporulates at 68.8% (Gerke et al. 2009). For each cross, doubled haploid offspring were collected as tetrads and are available on request. For the BC240 × BC728 cross (hereafter called the vineyard-fixed cross), 164 offspring were genotyped, while for the BC248 × BC713 cross (hereafter called the oak-fixed cross), 175 offspring were genotyped.
Growth and sporulation measurement
Strains were grown in standard yeast–peptone–dextrose (YPD, 1% yeast extract, 2% peptone, 2% dextrose) media unless otherwise indicated. Hybridization was selected for by supplementing media with either G418 (200 mg/liter, Invitrogen) and nourseothricin (100 mg/liter, Werner BioAgents) or G418 (200 mg/liter, Invitrogen) and hygromycin (300 mg/liter, Roche) and selecting for resistance to both drugs. Offspring tetrads were checked to confirm 2:2 segregation for the drug resistances.
Offspring from both crosses were phenotyped for sporulation efficiency as described previously, except that the overnight growth was extended from 14 to 15 hr (Gerke et al. 2006). Sporulation efficiency was assayed via flow cytometry and calculated as described previously (Gerke et al. 2006). Heritability was calculated as described previously (Gerke et al. 2006).
Genotyping and QTL analysis
DNA was extracted using the YeaStar Genomic DNA Kit (Zymo Research, Orange, CA). Markers were identified and assayed using a modified restriction site-associated DNA (RAD) tag approach (Baird et al. 2008). In 96 well plates, approximately 0.3 μg of DNA from each strain was digested with 5 units each of MfeI and MboI [New England Biolabs (NEB)] in NEB buffer 4 for 1 hr at 37°, then heat inactivated for 20 min at 65°. The digested DNA from each strain to be genotyped was ligated to one pair of modified Illumina sequencing adapters (IDT, sequences available in supporting information, Table S1) containing 1 of 48 different 4-bp barcodes using 1000 units of T4 ligase (NEB) in 1× ligation buffer containing dATP. Adapters were preannealed and added so that the final concentration was 100 nM for sequencing adapter (P1) and 10 µM for secondary adapter (P2). The ligation was run for 20 min at 20° and the reaction was heat inactivated for 20 min at 65°.
Fifteen microliters of each ligated sample was pooled into groups of 48 containing 44 offspring and 2 of each parent as controls and purified using the QIAquick PCR purification kit (Qiagen). Pools were gel extracted to isolate 200- to 500-bp fragments using the QIAquick gel extraction kit (Qiagen). Five to 10 ng of this product was used in a reaction to add the final 20 bp of sequencing adapter onto the fragments using Phusion mastermix (NEB) and 18 cycles of PCR. Samples were then sequenced using standard Illumina GAIIx primers and protocol for 36 cycles. Raw sequencing reads were deposited in NCBI’s Sequence Read Archive under submission numbers SRA056533 and SRA056489.
To select markers, reads were binned by barcode and any reads containing ambiguous barcodes were discarded. Barcodes were removed and reads were collapsed into unique sequences within barcode groupings. Only reads that occurred three or more times were considered for further analysis. Parental controls from each pool were separated and used to select markers. A set of repeatable markers was determined by selecting only those that occurred at least seven times in all parental control samples sequenced. Two types of markers were used: those with sequence polymorphisms between the parents within a given read and those that had a sequence polymorphism in a restriction site that caused a read to be absent in one parent but present in the other. Reads were mapped to the reference S. cerevisiae genome using Bowtie v. 0.12.7 and options suppressing all nonunique alignments, specifying raw file type, and “best” alignment method (Langmead et al. 2009). Because we specified unique alignments, we did not use a maximum coverage threshold.
From this set of usable markers, the genotyping set was selected to maximize genomic coverage with a minimum of markers; markers were selected to be at least 10 kb away from the next nearest marker. Number of usable markers varied with sequencing depth. For the vineyard-fixed cross, 369 markers were used. For the oak-fixed cross, 536 markers were used. All marker positions provided refer to the beginning of the read mapped to the reference genome (Table S2). The average total number of reads per genotyped offspring or parental control was 271,313 for the vineyard-fixed cross and 667,511 for the oak-fixed cross (Table S1). The average number of reads that mapped to genotyping markers was 18,837 for the vineyard-fixed cross and 40,619 for the oak-fixed cross (Table S1). The average reads per marker is reported in Table S2. This is mostly a concern for presence/absence markers, as lack of sequencing depth could lead to miscalling a marker that is actually present as absent. In both the oak-fixed and vineyard-fixed crosses, the lowest average number of reads for a presence marker was 22, with most being significantly higher (an average of 62 for the vineyard-fixed cross and 104 for the oak-fixed cross). Assuming a Poisson distribution of read counts, with an average number of reads of 22 the probability of a presence read being mistakenly called absent is extremely low (P = 7.4 × 10−8).
Final genotyping data used can be found in Table S3 and Table S4. A genetic map was constructed using Mapmaker/EXP v. 3.0 (Whitehead Institute, Cambridge, MA), and QTL peaks were mapped using composite interval mapping (CIM) as implemented in WinQTL Cartographer v. 2.5 (Wang, Basten, and Zeng 2011) and described previously (Gerke et al. 2009). Thresholds for significance were set using 1000 permutations of each data set.
Analysis of epistasis
Three methods were used to quantify epistasis. The data were bootstrapped by offspring to confirm robustness of QTL, a linear model was constructed for each QTL peak location in a combined cross analysis to test whether the contribution of a marker to the phenotype was cross specific, and statistical power was calculated. Bootstrapping was accomplished by using custom R-scripts to resample the progeny pool with replacement, generating 1000 data sets with the same size as the original progeny pool. For each bootstrapped data set, QTL mapping was performed with the original map distances preserved, and a significance threshold was determined using 1000 permutations of the resampled data set. For each bootstrapped data set, all QTL that rose above the threshold were collected, and from these a percentage of bootstrap repetition was calculated for each marker position across the genome.
Since our two crosses were genetically identical except for the four large-effect polymorphisms fixed as either oak or vineyard alleles, we were able to use identical markers at each of the QTL loci to analyze the crosses together. Using the marker nearest each QTL peak from both crosses the following two models were compared,
where SE is the sporulation efficiency of the offspring strain, Marker is allelic status (oak or vineyard) at the marker nearest the QTL location being tested, Large-Effect QTN indicates whether that offspring contains the vineyard or oak alleles of the large-effect QTN, and Marker:Large-Effect QTN interaction term tests whether there is a significant two-way interaction between the marker and the large-effect QTN.
Statistical power was calculated as described by Hu and Xu (2008). For each QTL detected in one fixed cross, the centimorgan distance between the nearest markers in the opposite fixed cross were used along with the QTL effect size. For all calculations cross type was doubled haploid. Number of offspring was 164 for the vineyard-fixed cross and 175 for the oak-fixed cross.
Reciprocal hemizygosity analysis
Reciprocal hemizygosity analysis of putative QTL genes in the oak(vvvv) × vineyard cross was performed as described previously (Steinmetz et al. 2002). Parent strains were made haploid by deleting the HO locus using the d-serine deaminase dsdAMX4 cassette, which confers ability to grow on d-serine as a sole nitrogen source (Vorachek-Warren and McCusker 2004). Genes to be tested were then knocked out with the MX hygromycin B cassette (Goldstein and McCusker 1999), and reciprocal hemizygotes were constructed by crossing the knockout to the other parent strain. Each gene was tested using five independent knockouts of each allele crossed to the opposite parental strain, with the exception of RAD52 (two knockouts of the vineyard allele) and IME1 (4 knockouts of the oak(vvvv) allele). All five replicates of reciprocal hemizygotes were phenotyped three times starting from frozen stocks, and results of these three technical replicates were averaged. Significance was tested by calculating a t-test on the two groups of five allele knockouts. The strain containing the complete vineyard IME1 locus replacement in the oak background was described previously (Gerke et al. 2009).
Results
To identify small-effect QTL that influence sporulation efficiency, we first eliminated the contribution of large-effect QTN. We previously identified four large-effect QTN that were responsible for 80% of the difference in sporulation efficiency between a high-sporulating oak tree strain (BC248) and a low-sporulating vineyard-derived strain (BC240) (Gerke et al. 2006, 2009). We reasoned that we could better uncover small-effect QTL in crosses in which the large-effect QTL were no longer segregating among the offspring. We genetically modified the parental strains (Materials and Methods) to replace each QTN with its alternate allele and crossed the strains in which the QTN were fixed to the unmodified parental strains.
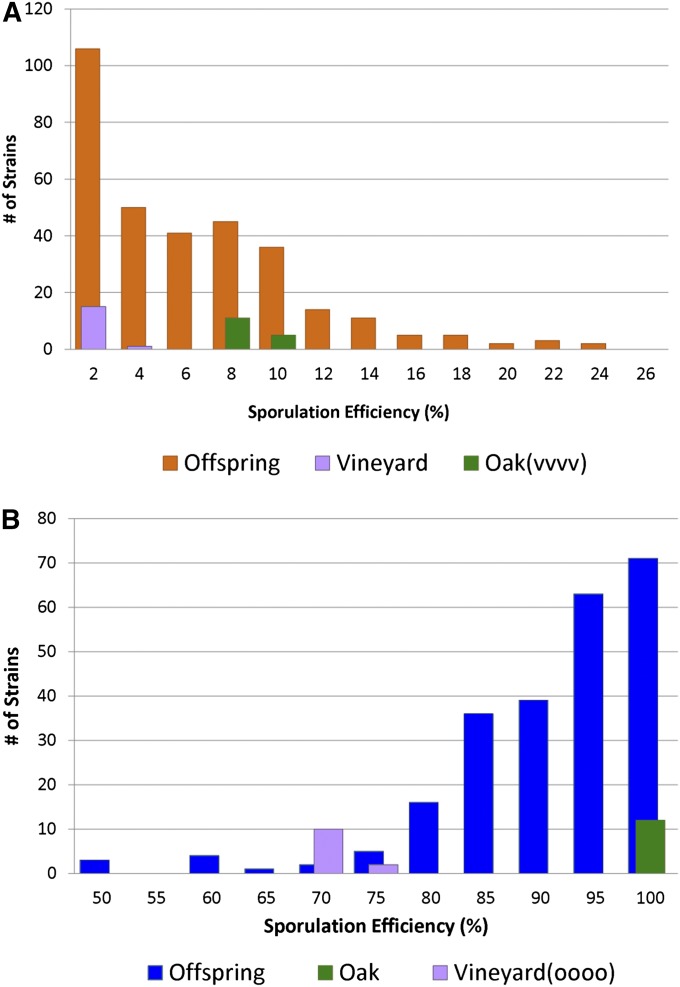
We first consider the cross in which all of the major-effect QTN were fixed as their vineyard alleles, or the “vineyard-fixed cross” (BC240 × BC728). Aside from the four fixed QTN, all of the remaining sequence variation between the parental strains was left to segregate among offspring. We saw a range of sporulation efficiencies across the 320 offspring phenotyped, confirming that there was still variation in this trait after the major effects had been fixed (Figure 1a). Given that heritability was high in the cross between the vineyard and oak parent strains in our previous study (Gerke et al. 2006, 2009), we expected most of the remaining variation to be genetic in nature. Heritability for the vineyard fixed cross was 98.0%, indicating that most of the phenotypic variation in this cross is the result of genetic differences. This gave us confidence that we had the statistical power to detect some of the remaining small-effect QTL. Additionally, some offspring exceed the phenotypic values established by the parental strains (transgressive segregation), indicating that the oak(vvvv) and vineyard strains both contain alleles that increase and decrease sporulation efficiency.
Figure 1 .
Histograms of sporulation efficiency for progeny from fixed crosses. In each fixed cross, replicates of the oak parent strain are indicated in green while replicates of the vineyard parent strain are purple. (A) The distribution of sporulation efficiencies for the offspring and parents of the vineyard-fixed cross, vineyard (BC240) × oak(vvvv) (BC728). Offspring are shown in orange. (B) The distribution of sporulation efficiencies for the offspring of the oak-fixed cross, oak (BC248) × vineyard(oooo) (BC713).
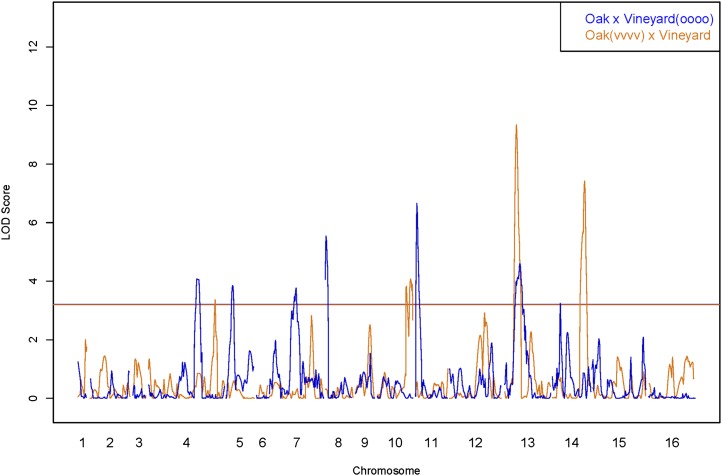
We next mapped QTL using 164 genotyped offspring (Materials and Methods). A CIM scan for QTL resulted in the orange trace in Figure 2; the threshold for significance was 3.2 LOD. Four genomic locations were significant, including regions on chromosomes 4, 10, 13, and 14. A list of nearest markers and reference genomic coordinates for the identified QTL is found in Table 1.
Figure 2 .
QTL scan for sporulation efficiency. LOD peaks from the vineyard-fixed cross (vineyard × oak(vvvv)) are shown in orange. LOD peaks from the oak-fixed cross (oak × vineyard(oooo)) are overlaid in blue. The threshold for significance was set using 1000 permutations of the data sets and corresponds to 3.2 LOD.
Table 1 . Markers nearest to QTL peak apex.
| Chromosome | Cross | Marker | Location | Oak allele effect direction |
|---|---|---|---|---|
| 4 | Oak fixed | CCA04.58 | 976738 | − |
| 4 | Vineyard fixed | CCA04.76 | 1322285 | + |
| 5 | Oak fixed | CCA05.10 | 119193 | − |
| 7 | Oak fixed | CCA07.41 | 511909 | + |
| 8 | Oak fixed | CCA08.03 | 24495 | + |
| 10 | Vineyard fixed | CCA10.31 | 654963 | − |
| 11 | Oak fixed | CCA11.04 | 46428 | − |
| 13 | Vineyard fixed | CCA13.18 | 244489 | + |
| 13 | Oak fixed | CCA13.20 | 293935 | + |
| 14 | Oak fixed | CCA14.16 | 176365 | + |
| 14 | Vineyard fixed | CCA14.43 | 651306 | − |
Cross column indicates the cross in which the marker listed was found to be significant. Location column corresponds to the start of the reference genome mapped sequencing read for that marker.
Since we observed transgressive segregation in sporulation efficiency among the offspring, we expected that each parent would have at least one QTL that increases and one QTL that decreases sporulation efficiency. As expected, the small-effect QTL we identified act in both directions, with the oak(vvvv) parent contributing two alleles that increase sporulation efficiency as well as two that decrease it. For the two QTL located on chromosome 4 and 13 the allele from the higher sporulating oak(vvvv) strain increases sporulation efficiency (Table 1). In contrast, the QTL on chromosomes 10 and 14 had oak alleles that correlated with a decrease in sporulation efficiency, indicating that these two QTL are at least partly responsible for the transgressive segregation we identified among the offspring. This is in contrast to the results from our previous study on large-effect QTL where all oak alleles increase sporulation efficiencies and all vineyard alleles decrease sporulation efficiency (Gerke et al. 2009).
The four small-effect QTL we identified explain a significant fraction of the remaining heritability that is not accounted for by the major effect QTN. Although a truly accurate model of effect size requires knowledge of the causal polymorphism, we estimated effect sizes using the marker nearest to the apex of each QTL peak. This calculation results in an underestimate of effect size as there will likely be some offspring that have a crossover event between the marker and the causal variant. Conversely, the Beavis effect could result in a slight overestimation of effect sizes (Beavis 1998). With these caveats, the linear model we obtained via stepwise regression contains additive terms for all four QTL (Table 2). The additive effect of each QTL was estimated to be between 2.0 and 5.0%, which matches our expectations for small-effect alleles. There was also evidence for interaction between the QTL on chromosome 13 and the QTL on both chromosomes 4 and 14. The R2 for this model is 0.40, suggesting that these four QTL explain at least 40% of the remaining variation in sporulation efficiency after the large effects are fixed as the vineyard QTN.
Table 2 . Coefficients for a stepwise linear regression using nearest markers for the vineyard-fixed cross (vineyard × oak(vvvv)).
| Term | Effect | Significance |
|---|---|---|
| intercept | 0.053 | 0–0.001 |
| 4 | −0.032 | 0–0.001 |
| 10 | 0.02 | 0–0.001 |
| 13 | −0.033 | 0–0.001 |
| 14 | 0.05 | 0–0.001 |
| 13:14 | −0.03 | 0.001–0.01 |
| 4:13 | 0.024 | 0.01–0.05 |
Small-effect QTL are dependent on large-effect QTN
We had previously observed that the genetic background of a strain influences the effect sizes of the four large-effect QTNs on sporulation efficiency (Gerke et al. 2009, 2010). We therefore asked whether the small-effect QTL we identified would be influenced by the allelic status of the large-effect QTN. To address this question, we performed the reciprocal fixed cross in which we fixed the four large-effect QTN as oak alleles, using the vineyard(oooo) strain (BC713) (Gerke et al. 2009). If the contributions of the small-effect QTL to sporulation efficiency were independent of the large-effect QTN, we would expect to uncover the same small-effect QTL in the reciprocal fixed cross. By mapping small-effect QTL in this reciprocal fixed cross, we sought to determine whether the large-effect QTN affect the small-effect QTL.
We collected 240 offspring from the oak × vineyard(oooo) cross (BC248 × BC713) and phenotyped them for sporulation efficiency (Figure 1b). We first confirmed that phenotypic variation in the oak-fixed cross was heritable. Heritability was 97.6%, indicating that phenotypic variation was again caused by genetic differences and that we had the power to identify small-effect alleles. We again saw transgressive segregation, with a number of offspring sporulating both lower and higher than the parental bounds, suggesting that each parent contains alleles that increase as well as decrease sporulation efficiency.
We scanned for QTL using 175 genotyped offspring (Materials and Methods). In the oak-fixed cross, we identified seven significant QTL peaks. The nearest markers and reference genome coordinates for QTL identified in the oak-fixed cross are found in Table 1. While we observed a QTL in the same region of chromosome 13 in both crosses, six of the seven QTL (on chromosomes 4, 5, 7, 8, 11, and 14) identified in the oak-fixed cross were not significant in the vineyard-fixed cross (Figure 2, blue trace). The lack of overlap between small-effect QTL regions in the two crosses suggests that most small-effect QTL are epistatic to the large-effect QTN. With the exception of the chromosome 13 QTL, our ability to detect small-effect QTL depends on the allelic status of at least one large-effect QTN (see below).
As the offspring from the oak-fixed cross showed transgressive segregation, we again expected to find QTL in which alleles from the high-sporulating oak strain decrease sporulation efficiency. We found three transgressive QTL on chromosomes 4, 5, and 11, where the oak allele acted to decrease sporulation efficiency in the offspring. The QTL located on chromosomes 7, 8, 13, and 14 all contain an oak allele that is correlated with increased sporulation efficiency among offspring. As in the vineyard-fixed cross, the small-effect QTL in the oak-fixed cross are split evenly for effect direction, with the oak strain containing four QTL that increase sporulation efficiency and three that decrease it relative to the vineyard alleles at the same locations.
The chromosome 13 QTL is the only genomic location that was found to be significant in both crosses. This locus has the same direction of effect in each cross, the 99% confidence intervals overlap, and the nearest markers for the QTL are only 50 kb apart. By contrast, while both crosses have significant peaks on chromosomes 4 and 14, the QTL on each of these chromosomes have opposite effect directions in each cross and are located much further apart, almost 350 kb on chromosome 4 and more than 450 kb on chromosome 14. These data suggest that the chromosome 4 and 14 peaks that we found in the two crosses are unlikely to be the result of the same causal variant in both crosses. The lack of overlap between small-effect QTL regions in the two crosses suggests that with the exception of the chromosome 13 QTL, our ability to detect small-effect QTL depends on the allelic status of at least one large-effect QTN.
We set up a linear model using the markers nearest to each QTL peak apex to estimate the effect size of each QTL. The linear model obtained from a stepwise regression on the oak-fixed cross data resulted in a significant additive effect for all peaks as well as a few two-way interactions, indicating that there is some epistasis between small effects (Table 3). The R2 for this model is 0.55, suggesting that these QTL explain at least half of the remaining variation in sporulation efficiency in the oak-fixed cross.
Table 3 . Coefficients for a stepwise linear regression using nearest markers for the oak-fixed cross (oak × vineyard(oooo)).
| Term | Effect | Significance |
|---|---|---|
| Intercept | 0.914 | 0–0.001 |
| 4 | 0.094 | 0–0.001 |
| 5 | 0.042 | 0.001–0.01 |
| 7 | −0.033 | 0–0.001 |
| 8 | −0.085 | 0–0.001 |
| 11 | 0.06 | 0–0.001 |
| 13 | −0.04 | 0–0.001 |
| 14 | −0.076 | 0–0.001 |
| 4:5 | −0.057 | 0.001–0.01 |
| 4:11 | −0.048 | 0.01–0.05 |
| 11:14 | 0.041 | 0.01–0.05 |
| 5:8 | 0.04 | 0.01–0.05 |
Combined cross-statistical analyses
We performed a bootstrap analysis, combined cross linear modeling, and power calculations to evaluate the epistatic QTL we observed. To test the robustness of QTL we bootstrapped our data by resampling offspring. In 1000 bootstrapped data sets, all significant QTL appeared 100% of the time for their cross except for the chromosome 4 QTL in the vineyard-fixed cross, which showed 32% replication, and the chromosome 14 QTL in the oak-fixed cross, which showed 83% replication. Notably, with the exception of the two QTL on chromosome 13, no QTL from the vineyard-fixed cross was ever significant in the oak-fixed cross bootstrap, and no QTL from the oak-fixed cross was ever significant in the vineyard-fixed cross. This supports our conclusion that the majority of the small-effect QTL are epistatic to the large-effect QTN.
To more directly test whether small-effect QTL are dependent on the large-effect QTN background, we set up a combined cross analysis using a series of linear models to analyze the combined data from both crosses for each QTL. The significance of each marker, large-effect QTN, and marker:large-effect QTN term was assayed in models with and without interaction. If the marker:large-effect QTN interaction is significant, it indicates that the small-effect QTL contribution to change in sporulation efficiency is specific to one set of large-effect QTN. For the small-effect QTL located on chromosome 14 in the vineyard-fixed cross and chromosomes 4, 5, 8, 11, and 14 in the oak-fixed cross, inclusion of a marker:large-effect QTN term contributed significantly to the fit of the model (Tables 4 and 5), indicating that these small-effect QTL act primarily through one or more large-effect QTN. In all of these models except the vineyard-fixed model for the chromosome 14 QTL, inclusion of the marker:large-effect QTN interaction in the model resulted in loss of statistical significance for the additive marker term, indicating that most loci had an effect on the phenotype of sporulation efficiency only when they co-occurred with one set of large-effect QTN. These data support our conclusion that the small-effect QTL are dependent on the presence of the alleles of the large-effect QTN.
Table 4 . Combined cross analysis linear models for each QTL locus identified in the vineyard-fixed Cross.
| Chr 4 | Chr 10 | Chr 13 | Chr 14 | |
|---|---|---|---|---|
| CCA04.76 | CCA10.31 | CCA13.18 | CCA14.43 | |
| Marker | * | NS | * | NS |
| Large-effect QTN | * | * | * | * |
| Marker | NS | NS | * | * |
| Large-effect QTN | * | * | * | * |
| Marker:large-effect QTN | NS | NS | NS | * |
Above the break, model without interaction between marker and large-effect QTN, below the break, model with interaction. *, the term was significant in that model (F-statistic, P < 0.05). NS, the term was not significant.
Table 5 . Combined cross analysis linear models for each QTL locus identified in the Oak-fixed Cross.
| Chr 4 | Chr 5 | Chr 7 | Chr 8 | Chr 11 | Chr 13 | Chr 14 | |
|---|---|---|---|---|---|---|---|
| CCA04.58 | CCA05.10 | CCA07.41 | CCA08.03 | CCA11.04 | CCA13.20 | CCA14.16 | |
| Marker | NS | * | * | * | * | * | * |
| Large-effect QTN | * | * | * | * | * | * | * |
| Marker | NS | NS | NS | NS | NS | * | NS |
| Large-effect QTN | * | * | * | * | * | * | * |
| Marker:large-effect QTN | * | * | NS | * | * | NS | * |
Above the break, model without interaction between marker and large-effect QTN; below the break, model with interaction term. *, the term was significant in that model (F-statistic, P < 0.05). NS, the term was not significant.
We also calculated the statistical power of each of our crosses to detect QTL of various sizes, to ensure that we are not seeing cross-specific peaks simply because we lack the power to detect them in the opposite cross. We adapted a method (Hu and Xu 2008) to calculate power for a proposed cross given cross type, number of offspring, interval distance in centimorgans, heritability explained by the QTL, and alpha (type I error) to calculate a power for the actual marker intervals present in the QTL region of the opposite cross, assuming that the effect size of the QTL in question would be the same. These power calculations can be found in Table 6. For the peaks on chromosomes 4, 8, 11, and 14 in the oak-fixed cross we had good (>65%) power to detect QTL in the corresponding regions in the vineyard-fixed cross, suggesting that it is unlikely that those four QTL affect sporulation efficiency in the vineyard-fixed cross. Given our power to detect the 10 QTL we identified, the cumulative probability of finding zero or one QTL in common between the two crosses is small (P = 0.0024). This calculation, along with the combined cross modeling and bootstrapping data, suggests that these QTL do depend on the particular allele present at the large-effect QTL.
Table 6 . Statistical power to detect QTL in the opposite fixed cross.
| Chromosome | Cross | Absolute effect size | Distance in vineyard-fixed cross (cM) | Distance in oak-fixed cross (cM) | Power to detect QTL in vineyard-fixed cross | Power to detect QTL in oak-fixed cross |
|---|---|---|---|---|---|---|
| 4 | Oak fixed | 0.09 | 22.1 | 0.82 | ||
| 5 | Oak fixed | 0.04 | 17.8 | 0.41 | ||
| 7 | Oak fixed | 0.03 | 3 | 0.35 | ||
| 8 | Oak fixed | 0.08 | 8.4 | 0.84 | ||
| 11 | Oak fixed | 0.06 | 11.5 | 0.67 | ||
| 13 | Oak fixed | 0.04 | 8.3 | 0.46 | ||
| 14 | Oak fixed | 0.07 | 5.6 | 0.79 | ||
| 4 | Vineyard fixed | 0.03 | 14 | 0.33 | ||
| 10 | Vineyard fixed | 0.02 | 7.3 | 0.22 | ||
| 13 | Vineyard fixed | 0.03 | 16.7 | 0.32 | ||
| 14 | Vineyard fixed | 0.05 | 15.2 | 0.57 |
Small effects are linked to large effects
The four large-effect QTN are found in three genes, with two causal SNPs in IME1 and one causal SNP each in RME1 and RSF1 (Gerke et al. 2009). Our mapping analyses revealed that all of the large-effect QTN have a small-effect QTL nearby, an observation unlikely to occur by chance (P < 0.01, multinomial distribution). One possible reason for this result was that the small-effect polymorphisms resided within the same genes as the large-effect QTN. To determine what genes were likely candidates to contain the causal variation in the new QTL region, we considered the 99% confidence interval of the QTL, which is approximated by a 2 LOD drop from the apex. In the oak-fixed cross, the QTL on chromosome 7 has a 99% confidence interval which includes the RME1 gene, implying that this particular QTL could be the result of other polymorphisms within RME1. In the vineyard-fixed cross, the 99% confidence interval for the QTL on chromosome 10 includes the IME1 gene. The 99% confidence interval for the QTL on chromosome 13 in the vineyard-fixed cross does not include RSF1, but the 99% confidence interval is wider in the oak-fixed cross and does contain the RSF1 gene. Thus, our data show that all three of the previously identified large-effect genes are adjacent to small-effect QTL, and two of the large-effect genes could contain the small-effect causal variation.
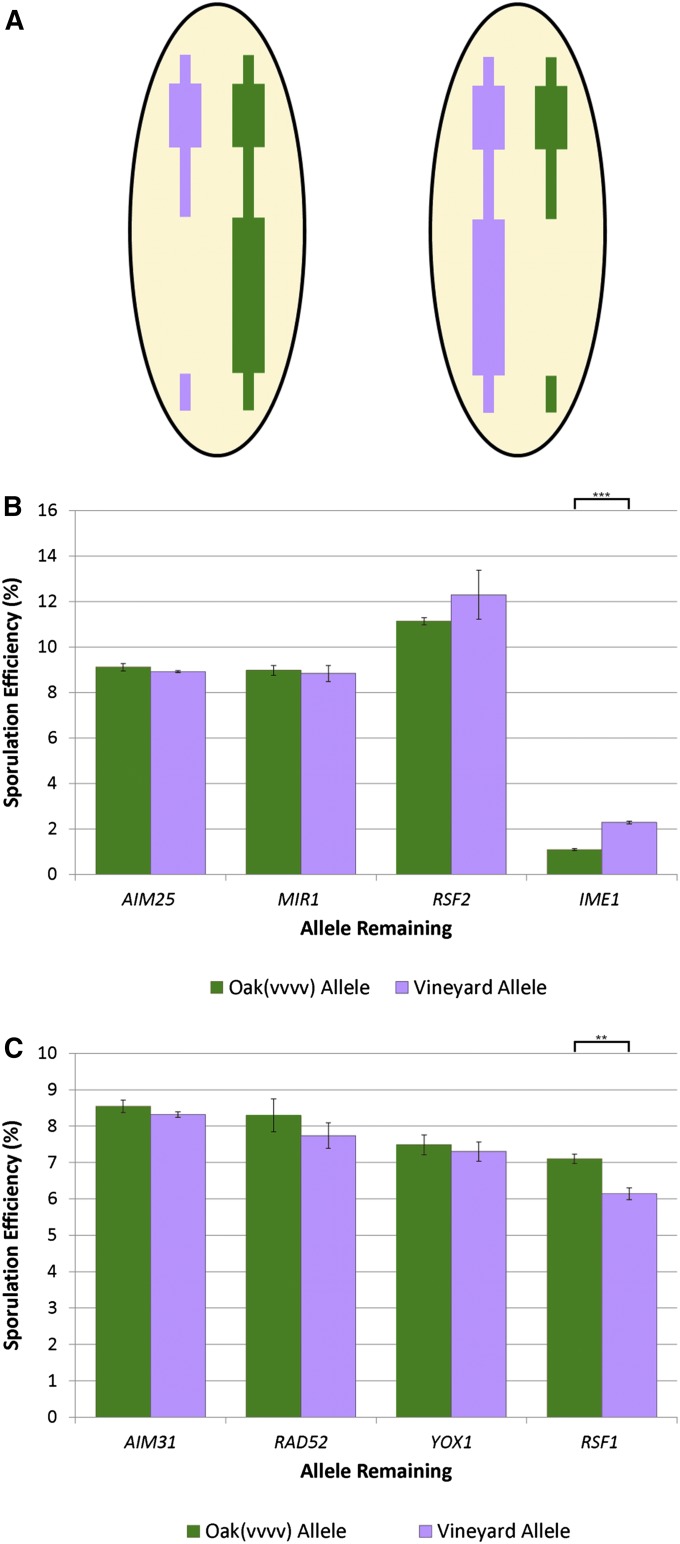
We attempted to determine whether polymorphisms in large-effect genes might underlie adjacent small-effect QTL. We performed reciprocal hemizygosity analysis (Steinmetz et al. 2002) on genes under small-effect QTL that were located near large-effect QTN (Figure 3A). In addition to the large-effect gene, three other genes were selected for analysis based on annotation for sporulation, cell cycle, or respiration processes. In the chromosome 10 region we tested IME1, RSF2, MIR1, and AIM25 (Figure 3B). Alleles of RSF2, MIR1, and AIM25 do not contribute significantly differently to sporulation efficiency in the hybrid background. Only alleles of IME1 showed a significant difference (t-test, P < 0.001), indicating that there is at least one more polymorphism in IME1, which contributes to the sporulation difference remaining between the oak(vvvv) and vineyard strains. As expected given the direction of effect of the chromosome 10 QTL, the reciprocal hemizygote containing the vineyard allele of IME1 sporulated higher than the strain containing the oak-fixed version. These data show that IME1 contains additional small-effect variation as well as the previously identified large-effect QTN.
Figure 3 .
Reciprocal hemizygosity of chromosome 10 and 13 QTL genes. (A) Schematic of reciprocal hemizygosity test comparing the effect of each parental allele on phenotype in hemizygote hybrid strains. The hybrid depicted on the left contains only the oak allele of the gene being tested; data from these strains are shown in B and C as green bars. The hybrid on the right contains only the vineyard allele of the gene being tested; data from these strains are shown as purple bars. (B) The AIM25, MIR1, RSF2, and IME1 genes from the chromosome 10 QTL region were tested. Only IME1 alleles contribute significantly differently to sporulation efficiency (t-test, *** indicates P < 0.001). Error bars indicate standard error. (C) The AIM31, RAD52, YOX1, and RSF1 genes from the chromosome 13 QTL region were tested. Only RSF1 alleles contributed significantly differently to sporulation efficiency (t-test, ** indicates P < 0.01). Error bars indicate standard error.
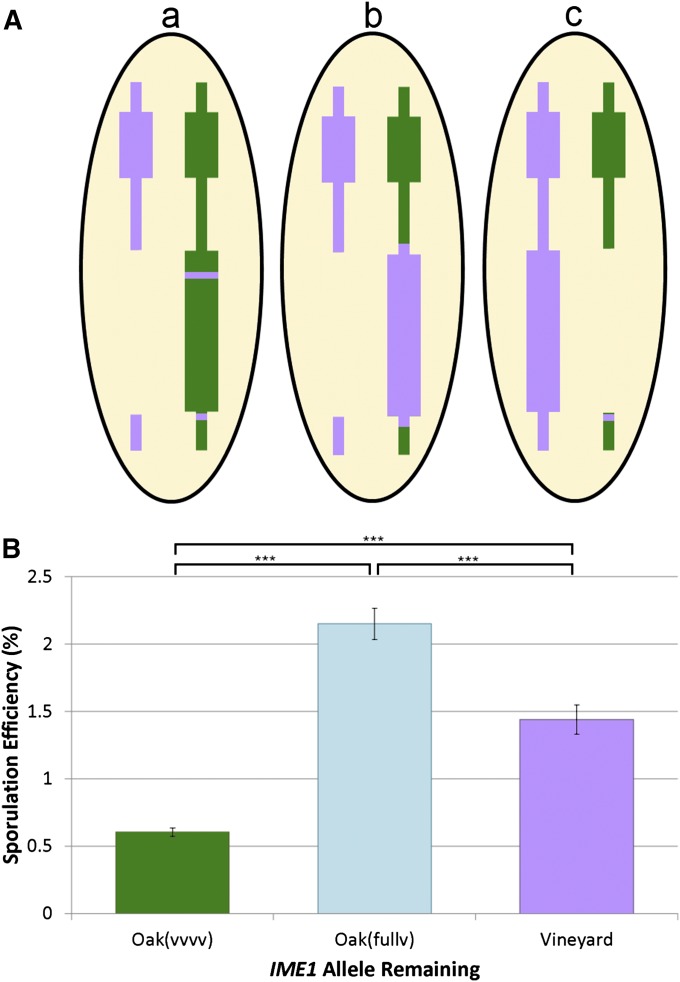
Since we had previously tested polymorphisms within the IME1 coding and noncoding sequences (Gerke et al. 2009), we suspected that a cis-acting polymorphism outside of the immediate promoter sequence might underlie the small-effect QTL in this gene. To test this, we replaced the entire IME1 locus in the oak strain with the vineyard sequence of IME1, including all SNPs within 2 kb of the upstream promoter sequence as well as all coding polymorphisms. We crossed this strain to the vineyard strain IME1 knockout and repeated the reciprocal hemizygosity test (Figure 4A, hybrid b). If the causal polymorphism was within the region replaced, the hybrid with the fully vineyard IME1 allele on the oak chromosome (hybrid b) would sporulate the same as the hybrid with the vineyard allele on the vineyard chromosome (hybrid c). If instead the polymorphism was outside of the region we replaced and independent of IME1, then we expected the fully vineyard IME1 oak hybrid (hybrid b) to sporulate with the same efficiency as the oak(vvvv) hybrid (hybrid a). Instead, the hybrid with the fully vineyard IME1 allele on the oak chromosome sporulated higher than either the hybrid with the oak(vvvv) allele or the vineyard allele on the vineyard chromosome (Figure 4B), indicating that there is a cis-acting polymorphism on the oak chromosome, which affects IME1. The oak allele of this cis-acting polymorphism increases sporulation efficiency, which suggests that this is not the polymorphism that underlies the small-effect QTL we detected in the vineyard-fixed cross. Our results suggest the presence of yet another layer of small-effect QTL beneath those that we detected in the vineyard-fixed cross.
Figure 4 .
Modified reciprocal hemizygosity analysis of the IME1 locus. (A) Hybrid strains being compared in this analysis. Strain a (left) is a hybrid that carries only the oak(vvvv) allele of IME1. This allele has all oak polymorphisms except for the two major effect QTN, which were changed to the vineyard SNPs, shown as purple lines on the chromosome; this strain is represented by a green bar in B. Strain b (center) is a hybrid that has the full complement of vineyard polymorphisms in IME1, but located on the oak chromosome. This strain is labeled Oak(fullv) and is represented by a light blue bar in the graph in B. Strain c (right) is a hybrid that contains only the vineyard allele of IME1 and is represented by a purple bar in the graph in B. (B) A comparison of the sporulation efficiency of the three IME1 hemizygote strains. All pairwise comparisons between the hybrids are significant (t-test, *** indicates P < 0.001).
We also performed reciprocal hemizygosity analysis to analyze the small-effect QTL we detected on chromosome 13 near the large-effect gene RSF1 in the vineyard-fixed cross. We tested reciprocal hemizygotes of AIM31, RAD52, YOX1, and RSF1 (Figure 3C). While RSF1 was not in the 99% confidence interval of this QTL we included it because it did contain the causal polymorphism for one of the previously identified major effect QTL. Only the alleles of the large-effect gene RSF1 showed a significant effect on sporulation efficiency (t-test, P = 0.002). Since the RSF1 gene is located ∼50 kb outside of the 99% confidence interval for the vineyard-fixed cross, it is unlikely that this is the effect responsible for the QTL peak we observed. Instead, this is likely a smaller effect in RSF1 that we did not detect in our mapping crosses. This result again suggests that there are QTL with even smaller effects than those we detected in our crosses and that these layers of small-effect QTL genetically interact with the large-effect QTL to modify phenotype.
Discussion
The two crosses we performed here differ from each other and the parent cross at only the four large-effect QTN, yet we largely detected different QTL in each cross. This means that while the small-effect alleles are present in both crosses, they are detected only as QTL in the cross containing the interacting alleles of the large-effect QTN. The degree of complexity found in this trait is similar to what has been found elsewhere (Ehrenreich et al. 2010, 2012; Parts et al. 2011), but the level of epistasis we observe has not previously been established. Our data suggest that the allelic status of large-effect QTL influence the effect sizes of additional genetic variants present in the genome of an individual.
We do not yet know which small-effect QTL interact with which large-effect QTN in our crosses. In the simplest scenario, each small-effect QTL might depend on a single large-effect QTN. However, the interactions could be much more complicated, with different small-effect QTL depending on two, three, or even all four large-effect QTN. Our results suggest another reason why complex trait analysis in outbred populations has proven difficult—if small-effect alleles affect a trait only in the presence of certain large-effect alleles, then a small-effect allele could be present at high frequency in a population but not contribute much to a trait’s variation because in many individuals it does not co-occur with the necessary large-effect allele.
We also found that all previously identified large-effect QTN had small-effect QTL physically linked to their chromosomal locations. QTL fractionation, the phenomenon in which large-effect QTL subdivide into multiple smaller effect QTL, is common in quantitative genetics (Steinmetz et al. 2002; Jordan et al. 2006; Studer and Doebley 2011). Because we were working with causative QTN, the large effects in our study could not fractionate. Instead by fixing the large-effect QTN we found tightly linked small-effect QTL. Without the high resolution afforded by having the causative large-effect QTN, this type of architecture would appear as a fractionating large-effect QTL. Our work shows that the QTN within fractionating QTL can be very closely linked; they may even reside in the same genes. This highlights the importance of identifying the causative QTN underlying quantitative traits.
The QTL on chromosome 13 near the large-effect gene RSF1 is unique among the small-effect QTL in that it is present in the same location in both fixed crosses, indicating that it is not epistatic to the large-effect QTN. However, since it is tightly linked to the RSF1 locus, it was indistinguishable from that large effect in the original cross and would likely not have been distinguishable from that QTL if we had not fixed the large-effect QTN. When we tested genes in this region in the vineyard-fixed cross, most did not have a differential effect on sporulation efficiency. RSF1 itself did have a small effect, despite not being within the expected region for the QTL. Like the evidence for an enhancer-like effect near IME1, this is likely a further layer of variation that we did not have the power to detect in this set of crosses. None of the candidates we tested in the RSF1 region were responsible for the QTL that we identified, suggesting that this effect was due to a gene not known to be involved in sporulation efficiency.
We found that a few small-effect alleles were able to explain a large fraction of the variation remaining after the large-effect QTL were fixed. For each of our fixed crosses, four to seven small-effect QTL explain around half of the remaining variation. Since this estimate is made using the nearest linked marker rather than the causal nucleotide, it is an underestimation of how much these alleles explain. Our results suggest that there are more, still smaller-effect alleles that remain to be detected in these strains.
Supplementary Material
Acknowledgments
We wish to thank Justin Gerke and members of the Cohen Lab for advice and comments on earlier versions of the manuscript. This work was funded by National Science Foundation grant MCB0948512.
Footnotes
Communicating editor: S. F. Chenoweth
Literature Cited
- Baird N., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PloS One 3(10): e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis W. D., 1998. QTL analyses: power, precision, and accuracy, pp. 145–162 in Molecular Dissection of Complex Traits, edited by A. H. Paterson. CRC Press, Cleveland, OH; Boca Raton, FL.
- Dumont E., Fontaine V., Vuylsteker C., Sellier H., Bodèle S., et al. , 2009. Association of sugar content QTL and PQL with physiological traits relevant to frost damage resistance in pea under field and controlled conditions. Theoret. Appl. Genet. 118(8): 1561–1571. [DOI] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464(7291): 1039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Bloom J., Torabi N., Wang X., Jia Y., et al. , 2012. Genetic architecture of highly complex chemical resistance traits across four yeast strains. PLoS Genet. 8(3): e1002570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. A., 1930. The Genetical Theory of Natural Selection. Oxford University Press, Oxford. [Google Scholar]
- Gaudet M. M., Kirchhoff T., Green T., Vijai J., Korn J. M., et al. , 2010. Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet. 6(10): e1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Cohen B., 2009. Genetic interactions between transcription factors cause natural variation in yeast. Science 323(5913): 498–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Lorenz K., Ramnarine S., Cohen B., 2010. Gene–environment interactions at nucleotide resolution. PLoS Genet. 6(9): e1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J. P., Chen C. T. L., Cohen B., 2006. Natural isolates of Saccharomyces cerevisiae display complex genetic variation in sporulation efficiency. Genetics 174: 985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier A. M., Nadeau J. H., Aitman T. J., 2002. Finding genes that underlie complex traits. Science 298(5602): 2345–2349. [DOI] [PubMed] [Google Scholar]
- Goldstein A. L., McCusker J. H., 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15(14): 1541–53. [DOI] [PubMed] [Google Scholar]
- Heuven H. C., van Wijk R. H., Dibbits B., van Kampen T. A., Knol E. F., et al. , 2009. Mapping carcass and meat quality QTL on Sus scrofa chromosome 2 in commercial finishing pigs. Genet. Select. Evol. 41: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Xu S., 2008. A simple method for calculating the statistical power for detecting a QTL located in a marker interval. Heredity 101(1): 48–52. [DOI] [PubMed] [Google Scholar]
- Jordan K. W., Morgan T. J., Mackay T. F. C., 2006. Quantitative trait loci for locomotor behavior in Drosophila melanogaster. Genetics 174: 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E. S., Schork N. J., 1994. Genetic dissection of complex traits. Science 265(5181): 2037–2048. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3): R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lango Allen H., Estrada K., Lettre G., Berndt S. I., Weedon M. N., et al. , 2010. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 467(7317): 832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T. F., 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35: 303–339. [DOI] [PubMed] [Google Scholar]
- Parts L., Cubillos F., Warringer J., Jain K., Salinas F., et al. , 2011. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 21(7): 1131–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H., David L., Pascon R. C., Clauder-Münster S., Krishnakumar S., et al. , 2008. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 180: 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz L. M., Sinha H., Richards D. R., Spiegelman J. I., Oefner P. J., et al. , 2002. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416: 326–330. [DOI] [PubMed] [Google Scholar]
- Studer A. J., Doebley J. F., 2011. Do large effect QTL fractionate?: a case study at the maize domestication QTL Teosinte Branched1. Genetics 188: 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorachek-Warren M. K, McCusker J. H., 2004. DsdA (d-serine deaminase): a new heterologous MX cassette for gene disruption and selection in Saccharomyces cerevisiae. Yeast 21(2): 163–171. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pöhlmann R., Philippsen P., 1994. New Heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10(13): 1793–1808. [DOI] [PubMed] [Google Scholar]
- Wang S., Basten C. J., Zeng Z.-B., 2011. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. [Google Scholar]
- Yang J., Manolio T., Pasquale L. R., Boerwinkle E., Caporaso N., et al. , 2011. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 43(6): 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.