Abstract
In the early mammalian embryo, X chromosome inactivation (XCI) achieves dosage parity between males and females for X-linked genes. During mouse development, imprinted paternal XCI is observed first and switches to random XCI in the epiblast but not placental lineages. The mechanism by which this epigenetic switch occurs is currently unknown. Here, we establish an ex vivo model for imprinting and identify a novel trans-acting regulatory factor for imprinted XCI. Using an induced trophoblast stem cell (iTS) model, we show that embryonic stem (ES) cells transdifferentiated into trophoblasts retain partial memory of the XCI imprint. Cdx2, a stem cell factor that determines commitment to the extraembryonic lineage, directly binds Xist and activates expression of Xist RNA in extrembryonic cells. Cdx2 competes with Oct4, a stem cell factor that determines commitment to the embryonic lineage, for overlapping binding sites within Xist. We propose that mutually exclusive binding between Cdx2 and Oct4 in Xist underlies the switch between imprinted and random XCI in the early mouse embryo.
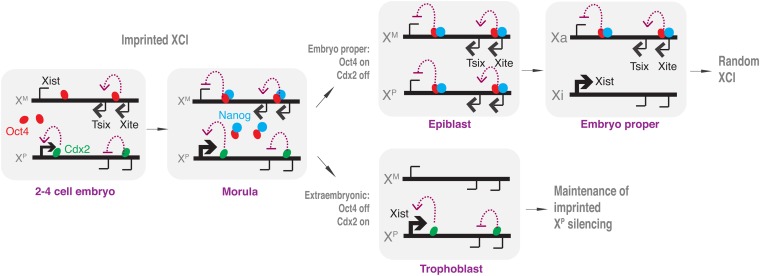
X chromosome inactivation (XCI) is an essential developmental program that achieves gene dosage parity in mammals between the XX female and the XY male (Wutz and Gribnau 2007; Payer and Lee 2008; Starmer and Magnuson 2009). XCI is epigenetically regulated and is tightly linked to changes in pluripotency and cell fate decisions in the early mouse embryo (Monk and Harper 1979). Between embryonic days 0.5 and 5.5 (E0.5–E5.5), two forms of XCI occur sequentially in the mouse. “Imprinted XCI” is a germline-determined process during which silencing occurs exclusively on the paternal X chromosome (XP) (Takagi and Sasaki 1975; Takagi 2003). Evidence of imprinted XCI is observed by the two-cell stage, where repetitive elements on XP are transcriptionally suppressed relative to those on the maternal X (XM) (Huynh and Lee 2003; Namekawa et al. 2010). Silencing progressively encompasses genic elements on XP during the next several divisions until the early mouse blastocyst stage (Okamoto et al. 2005; Kalantry et al. 2009; Namekawa et al. 2010).
In the later mouse blastocyst, embryonic (epiblast) and extraembryonic lineages (trophectoderm, primitive endoderm) become evident for the first time and it is during this time that evidence of “random XCI” is first detected. Whereas the extraembryonic cells retain imprinted XCI, the embryonic lineage reactivates XP at E4.5 (Mak et al. 2004; Okamoto et al. 2004) and undergoes a second round of XCI (Harper et al. 1982; Tan et al. 1993), this time in a “random” way such that XP and XM have an equal chance of becoming the inactive X (Xi). Random XCI is essential for in vivo differentiation of the epiblast to the three germ lineages (ectoderm, mesoderm, and endoderm) and for the ex vivo differentiation of epiblast-derived embryonic stem (ES) cells. Recent work in stem cell engineering shows that mouse XCI is also intimately linked to the reprogramming process ex vivo in the derivation of mouse induced pluripotent stem (iPS) cells (Maherali et al. 2007).
How and why the early mouse embryo switches from imprinted to random XCI present two intriguing questions. Still unknown are specific factors that dictate the decision to switch XCI pathways. Also unclear is whether the switch from imprinted to random XCI necessitates erasure of the original germline imprint, which would then allow a zygotic counting/choice mechanism to initiate random XCI. An alternative is that the zygotic counting/choice mechanism merely overwrites a parental imprint that is never erased in the epiblast. The switch in XCI pathways during early development is especially puzzling, given that the two forms are regulated by overlapping sets of factors, many based in the X-inactivation center (Xic).
The Xic includes a number of crucial long noncoding RNAs (lncRNAs). Xist RNA is a 17-kb transcript that is expressed only in female cells, coats the Xi in cis, and initiates global silencing on that chromosome (Brockdorff et al. 1992; Brown et al. 1992; Marahrens et al. 1997; Wutz and Jaenisch 2000). In the early embryo, Xist RNA is paternally expressed, required for imprinted XCI (Marahrens et al. 1997; Kalantry et al. 2009), and necessary for genic but not repetitive element silencing (Namekawa et al. 2010). Murine Xist is positively regulated by Jpx lncRNA (Tian et al. 2010) and negatively regulated by the antisense Tsix transcript (Lee and Lu 1999; Lee et al. 1999; Lee 2000; Luikenhuis et al. 2001; Sado et al. 2001; Stavropoulos et al. 2001). In cells that undergo random XCI (e.g., epiblast, ES cells), Tsix determines XCI choice and deleting Tsix results in skewed XCI to favor inactivation of the mutated X chromosome. In cells that undergo imprinted XCI (extraembryonic cells), Tsix suppresses expression of Xist on XM, and deleting Tsix on XM leads to ectopic XM-inactivation and early loss of both XX and XY embryos. Tsix is in turn controlled by Xite, a positive regulator that sustains expression of Tsix on the future active X (Ogawa and Lee 2003).
In searching for pathway-specific factors, we reasoned that, because imprinted and random XCI are tied to trophectoderm and epiblast cell fates, regulatory factors are likely to be those involved in determining lineage commitment. For random XCI, two recent studies have implicated the pluripotency factor, Oct4 (Nichols et al. 1998), in the regulation of Xite, Tsix, and Xist (Navarro et al. 2008; Donohoe et al. 2009). By binding Tsix and Xite, Oct4 upregulates Tsix and Xite expression and controls the initiation of X chromosome counting and pairing in ES cells, an established ex vivo model to study random XCI. In addition, Oct4 sites can be found in the first intron of Xist, where Oct4 is thought to directly block expression of the linked Xist allele (Navarro et al. 2008). While Oct4 is a strong candidate for the initiation of random XCI, its expression alone cannot regulate the decision to undergo imprinted or random XCI. Indeed, Oct4 is expressed ubiquitously in 2-, 4-, 8-, and 16-cell stages (Palmieri et al. 1994; Nichols et al. 1998) at a time when imprinted, but not random, XCI is observed.
During these stages, Cdx2—a protein with well-defined roles in directing commitment to the trophectoderm lineage (Strumpf et al. 2005)—is known to be expressed. Genome-wide analysis has shown that Cdx2 opposes the action of pluripotency factors by globally downregulating pluripotency factor target genes and thereby switching from embryonic to extraembryonic cell fates (Nishiyama et al. 2009). Moreover, ectopic expression of Cdx2 in ES cells leads to transdifferentiation of ES to trophoblast stem (TS) cells (Niwa et al. 2005), and one significant report has utilized the this system to study imprinted XCI (Murakami et al. 2011). Given these observations, here we test whether Cdx2 may be a regulator for the epigenetic decision to switch XCI pathways. We use the induced trophoblast stem cell (iTS) model to study imprinted XCI and show that mutually exclusive binding of Cdx2 and Oct4 to Xist underlies the switch between imprinted and random XCI.
Materials and Methods
Allele-specific RT-PCR
Allele-specific RT-PCR was performed as described in Stavropoulos et al. (2001). Briefly, total RNA was isolated with Trizol (Invitrogen, Carlsbad, CA) from cells and 2 μg was DNased with turbo DNAse (Ambion) and reverse transcribed with random hexamer primers and SuperScriptIII (Invitrogen). The following primer and restriction digest combinations were performed to amplify polymorphic Mus castaneus and M. musculus fragments:
Xist: NS33 (5′-CAGAGTAGCGAGGACTTGAAGAG-3′) and
NS66 (5′-GCTGGTTCGTCTATCTTGTGGG-3′), Scrfl digestion.
Mecp2: NS43 (5′-ATGGTAGCTGGGATGTTAGGG-3′) and
NS44 (5′-GAGCGAAAAGCTTTTCCCTGG-3′), DdeI digestion.
PGK: KHP72 (5′-CGTGATGAGGGTGGACTTCAAC-3′) and
KHP73 (5′-TAGTTTGGACAGTGAGGCTCGG-3′), MseI digestion
These fragments were fractionated on an agarose gel, Southern blotted, and probed with the 32P-end–labeled probe Xist: NS67 (5′-CCAGAGTCTGATGTAACGGAGG-3′), Mecp2: NS65 (5′-CCACTACAACCTTCAGCCCACCATTGAAGGCAAGCATGAG-3′) PGK: KHP106 (5′-TGAACTCAAATCTCTGCTGGGC-3′). Relative allelic amounts were quantitated by phorphorimage, using the software ImageQuant.
Immunofluorescence analysis
Colonies grown in ES media, TS media, or TS media + 1 μg/ml of 4-hydroxy tamoxifen (Tx; Sigma, St. Louis) were fixed using 4% paraformaldehyde and rinsed with 20 mM Tris-HCl, 0.15 M NaCl, and 0.05% Tween-20. Cells were permeabilized in 1X phosphate buffered saline with 0.05% Triton X (blocking buffer) and blocked with 4% goat serum at room temperature. Monoclonal antibodies against mouse anti-Oct3/4 monoclonal Ab (C-10; Santa Cruz, 1:500 dilution) and mouse anti-Cdx2 monoclonal Ab (Cdx2-88; BioGenex, 1:500 dilution) rabbit anti-NANOG (ab21603; Abcam) were incubated for 1 hr at room temperature. Cells were rinsed twice with blocking buffer and hybridized with the Cy3-conjugated goat anti-mouse and FITC-conjugated goat anti-rabbit secondary antibody (1:1000 dilution; Chemicon) for 1 hr. Cells were rinsed twice in blocking buffer and visualized.
Cell culture and generation of transgenic ES lines
Male J1 (40XY) and female 16.7 (40XX) ES cell lines and the TS line (TS3.5) have been described in Huynh and Lee (2003). For iTS generation, 40 μg of SalI linearized pCAG-Cdx2ER-IP (Niwa et al. 2005) was electroporated into EL16.7, J1, 3F1 (ΔTsix female line), and CG7 (ΔTsix male line) (Lee and Lu 1999) at 800 V and 3 μF in a 0.4-cm cuvette, using Gene Pulser (Bio-Rad, Hercules, CA) followed by selection with 1 μg/ml of puromycin for 8 days. Clones in which pCAG-Cdx2ER-IP were randomly integrated into genomic DNA were selected by the ability to undergo differentiation to TE by addition of 1 μg/ml of Tx (Sigma) in the standard trophoplast stem cell media (Tanaka et al. 1998), resulting in establishment of ELCDX, J1CDX, and ΔTsixCDX cells. For allele-specific RT-PCR analysis and RNA/DNA fluorescence in situ hybridization (FISH) analysis, cells were induced with 1 μg of 4-hydroxy tamoxifen and grown in standard TS media for 4 or 8 days as indicated. For chromatin immunoprecipitation (ChIP) analysis, cells were grown in ES cell media plus 1 μg of 4-hydroxy tamoxifen.
RNA/DNA FISH
FISH experiments were carried out as previously described (Lee et al. 1999). Xist was detected by Sx9 probe labeled by a nick translation kit (Roche), using FITC-dUTP (Stratagene, La Jolla, CA). A telomeric BAC probe (RP23-461E16) and a probe detecting the wild-type X (pCC3) in TsixΔCpG/+ cells was labeled by nick translation (Roche), using Cy3-dUTP (Amersham, Piscataway, NJ).
Allele-specific ChIP
ChIP analyses were carried out as described in Lee et al. (2006) and the results were averaged for three independent biological replicates. ChIP antibodies included anti-Oct4 (8628; Santa Cruz), anti-CDX2 (Cdx2-88; BioGenex), and normal rabbit IgG (2729; Cell Signaling). qPCR was performed using an iCycler iQ real-time PCR detection system (Bio-Rad) by amplifying with primer pairs listed as described in Donohoe et al. (2009): Xist intron1 (p65, ATGTTTCCTTTTGAAGCAGTTACTTGTAC; p66, CATTGTCTGGCTCTCTAGGTGATAATAC), Xite (p77, CAAGGTTGGGAACAAGGTATATCAGG; p78, GGACAAGGGACAGAAGTGCTTATTTTAC), and Lmb2 negative control (p89, ACTCCCATAATTTATCCTCTGATGTC; p90, GACAGGCTAAGTAAAGGATGTCTATTTT). PCR was performed using Amplitaq Gold (Applied Biosystems, Foster City, CA) with the following primer/restriction enzyme combinations (Stavropoulos et al. 2001): Xist intron1, XinAS-F, ACCTGCTGCGAAGGAATTTA, and XinAS-R, TGCCAGCACTTTTCAACAGA followed by BsaAI, which digests the musculus allele (63 bp and 129 bp) but not the castaneus allele (192 bp); Xite, NGP50, AGATAGGGTTTCTCTGTGTAG, and NGP51, GACTAGTTTCTGTTCGTGCAG followed by AciI digest, which does not digest the musculus allele (229 bp) and digests the castaneus allele (168 bp and 61 bp). PCR cycle number used was three cycles past the threshold cycle (Ct) from qPCR analysis. These fragments were fractionated on an agarose gel, Southern blotted, and probed with the 32P-end–labeled probe Xist-intron 1 (XINAS-F), and Xite (NGP50). Relative allelic amounts were quantitated by phorphorimage, using the software ImageQuant.
RT-PCR analysis
Two micrograms of RNA were isolated with Trizol (Invitrogen) and DNA was removed by incubation with turbo DNAse (Ambion) for 30 minutes. cDNA was synthesized by reverse transcription with random hexamer primers and SuperScriptIII (Invitrogen). Quantitative PCR was performed using an iCycler iQ real-time PCR detection system (Bio-Rad) by amplifying with the following primer pairs: Xist (NS33, CAGAGTAGCGAGGACTTGAAGAG; NS66, GCTGGTTCGTCTATCTTGTGGG), Oct4 (octF, CAGAAGGAGCTAGAACAGTTTGCC; octR, AGATGGTGGTCTGGCTGAACA), Nanog (nanogF, ACCTGAGCTATAAGCAGGTTAAGAC; nanogR, GTGCTGAGCCCTTCTGAATCAGAC), Beta-actin (actin1, agccatgtacgtagccatcc; actin2, ctctcagctgtggtggtgaa), and Cdx2 as described in Niwa et al. (2005).
Electrophoretic mobility shift assay
Reactions were incubated 20 min on ice and contained either 120 ng of recombinant Cdx2 protein (Novus Biologicals) or 140 ng of bacterially expressed rOct4 purified via a nickel column [Oct4 cDNA from J1 cells was cloned into pGEX4T1; GST-Oct4 was then purified on a glutathione resin and correct folding was verified by an electrophoretic mobility shift assay (EMSA) using an FGF4 enhancer oligo] and 0.2 pmol 32P-labeled oligonucleotides in 10 mM Tris (pH 7.5), 50 mM KC1, 2.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 μg poly(dIdC), and 4% glycerol. Cold competitors were added at 100× molar excess. The oligonucleotides were as follows: Xite-F, CCAGGTCTGCATTGATATGTAAAATAAGCACTTCTGTC, and Xite-R, GACAGAAGTGCTTATTTTACATATCAATGCAGACCTGG; Cdx2 mutated site, Xite-CdxF, CCAGGTCTGCATTGATATGGGGAATAAGCACTTCTGTC, and Xite-CdxR, GACAGAAGTGCTTATTCCCCATATCAATGCAGACCTGG; Xist intron 1 XINF, AATAGTTCTAGATGTATTATCACCTAGAG, and XINR, CTCTAGGTGATAATACATCTAGAACTATT; Xist intron 1 Cdx2 site mutated, XIN-CdxF, AATAGTTCTAGATGTGGGGTCACCTAGAG, and XIN-CdxR, CTCTAGGTGACCCCACATCTAGAACTAT; and Xist intron 1-Oct4 and Cdx sites mutated, O-C-XIN-F, AATAGTTCTAGCCCTGGGGTCACCTAGAG, and O-C-XIN-R, CTCTAGGTGACCCCAGGGCTAGAACTATT. For antibody neutralization, anti-CDX2 (Cdx2-88; BioGenex) or normal rabbit IgG (2729; Cell Signaling) was co-incubated with the protein complex for 20 min on ice before an additional 20-min incubation on ice of labeled oligonucleotide. For competitive/cobinding experiments, equal molar ratios (16 pmol each) of recombinant protein were first incubated in the above binding buffer for 20 min on ice, and then the complexes were incubated for 20 min on ice with ds-labeled oligos. Protein–DNA complexes were resolved on a 4% PAGE gel run at 300 V for 1.5 hr at 4° under native conditions.
Results
An iTS model demonstrates partial retention of the XCI imprint in ES cells
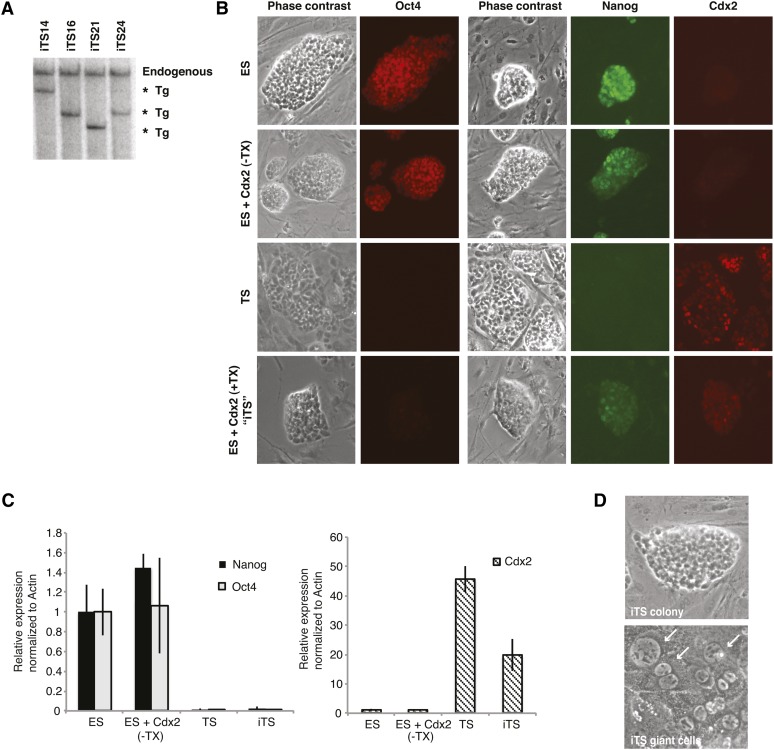
Whereas the study of random XCI has benefited hugely from ES cells as an ex vivo model, our understanding of imprinted XCI has been hampered by the absence of one. Here we took advantage of Cdx2’s known capacity to transdifferentiate ES to TS cells (Niwa et al. 2005) to develop an ex vivo model to study the transition between random and imprinted XCI. To generate iTS cells, we introduced a tamoxifen-inducible Cdx2 transgene into the hybrid female ES cell line, 16.7, which contains a paternally inherited X chromosome from M. castaneus and a maternally inherited X chromosome from M. musculus. Transgenic male ES cells were generated in parallel. Clones with low Cdx2 transgene copy numbers were selected for further analysis (Figure 1A). Southern blot analysis with an exon 1 probe for Cdx2 indicated that only one to two copies were randomly integrated into the genome.
Figure 1 .
Generation of iTS cells via ES cells carrying an inducible Cdx2 transgene. (A) Southern blot analysis of ES clones carrying an inducible Cdx2 transgene, as detected by Cdx2-exon 1 probe. Four independent clones were tested. (B) Immunofluorescence analysis for Oct4, Nanog, and Cdx2. Tx, tamoxifen. (C) Quantitative RT-PCR analysis for Oc4, Nanog, and Cdx2 mRNA. Levels are normalized to β-actin levels, with ES level set to 1. Mean ± 1 SD is shown. n = 3 biological replicates. (D) iTS cells form TS-like colonies and giant cells (arrows) typically seen in the trophoblast lineage.
Prior to Tx induction of Cdx2, the ES cells grown in standard LIF-containing media expressed the pluripotency factors, Oct4 and Nanog, comparably to the parental cell lines (Figure 1, B and C). To induced transdifferentiation into iTS cells, we grew transgenic ES cells with tamoxifen in Fgf4- and heparin-containing media for 6 days and observed high levels of Cdx2 protein in the nucleus (Figure 1, B and C). In this inducible system, tamoxifen addition promotes translocation of Cdx2 into the nucleus and transcriptionally upregulates endogenous Cdx2 via a Cdx2 auto-feedback loop (Nishiyama et al. 2009). Immunofluorescence showed that the levels and localization patterns for Cdx2 were similar to those observed in wild-type TS cells. At the same time, Oct4 and Nanog levels dramatically decreased. Furthermore, upon withdrawal of tamoxifen and growth for a further 7 days, the iTS cells differentiated into the multinucleated giant cells typically observed in differentiating TS cells, suggesting commitment to a placental lineage fate. These results are consistent with observations reported previously (Niwa et al. 2005) and were observed in both XX and XY backgrounds (Figure 1D).
We then asked whether the iTS cells would retain the random XCI pattern expected of differentiating ES cells or whether they would newly adopt the imprinted pathway ordinarily observed in TS cells. To address this question, we carried out SNP-based allele-specific RT-PCR for the X-linked genes, Pgk1, Mecp2, and Xist (Figure 2A). By inducing XCI through differentiation of wild-type female ES cells, we observed the expected allelic ratios of ∼65:35 favoring inactivation of the M. castaneus chromosome. In differentiated iTS cells, we observed a similarly skewed inactivation of the M. castaneus chromosome. For both differentiated ES and iTS cells, the X-linked genes, Pgk1 and Mecp2, have slightly skewed preferential expression from the M. musculus allele and Xist expression has slightly skewed preferential expression from the M. castaneus allele. The mild allelic skewing in ES cells is consistent with the strain-specific X-linked modifier, Xce, which favors genic expression from the X of M. castaneus origin (Cattanach and Isaacson 1967; Chadwick and Willard 2005). By contrast, in wild-type TS cells (Huynh and Lee 2004), expression of Xist was almost exclusively of paternal origin (M. castaneus), while Pgk1 and Mecp2 expression was of maternal origin (M. musculus), consistent with the occurrence of imprinted XCI in wild-type TS lines. Thus, iTS cells showed an allelic pattern that was more similar to that of the parental ES line.
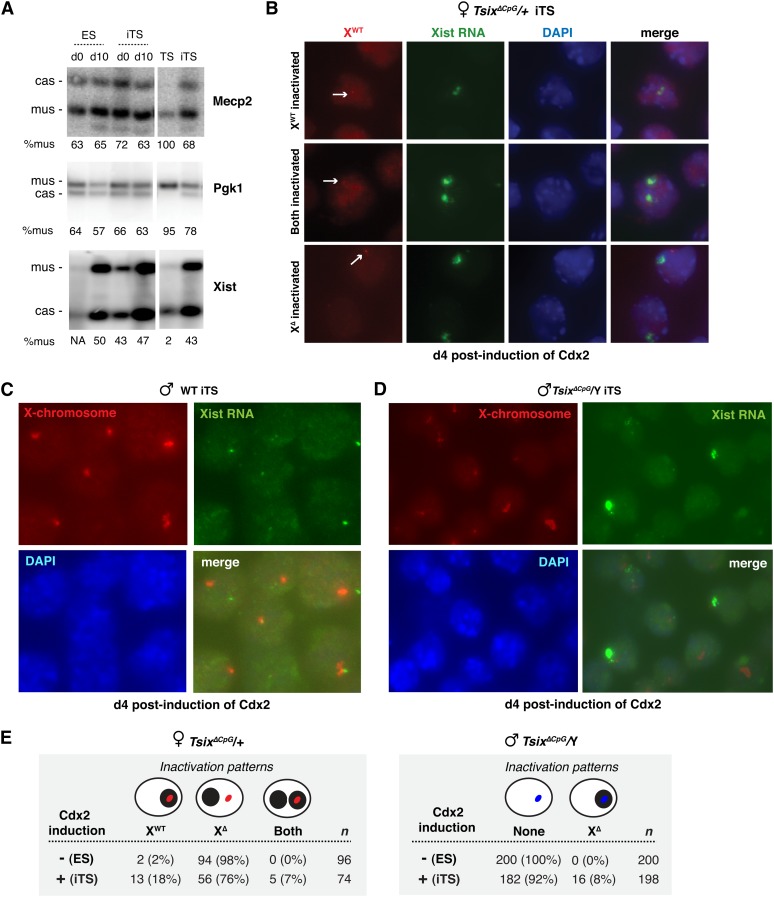
Figure 2 .
Evidence for partial memory of the XCI imprint in ES cells. (A) Allele-specific RT-PCR analysis for Pgk1, Mecp2, and Xist. d0, day 0; d10, day 10 of EB differentiation. % mus, percentage of RNA from the M. musculus allele. (B) RNA/DNA FISH of TsixΔCpG/+ female iTS clones differentiated for 4 days with tamoxifen (Cdx2). Arrow indicates wild-type allele. (C) RNA/DNA FISH of male iTS cells in a wild-type background after 4 days of Tx induction of Cdx2. (D) RNA/DNA FISH of TsixΔCpG/+ male iTS clones after 4 days of Tx induction of Cdx2. (E) Xist expression patterns observed in TsixΔCpG/+ female and male Cdx2-transgenic clones differentiated in the absence or presence of tamoxifen (ES and iTS, respectively) for 8 days. XWT, wild-type X chromosome; XΔ, Tsix-mutant X chromosome. Solid black circle, Xist RNA cloud. Red oval indicates the wild type X in females; blue oval indicates mutated X in males. n, sample size.
Although these observations seemed to indicate that iTS cells undergo random XCI, as recently observed (Murakami et al. 2011), allele-specific RT-PCR examines population-wide expression patterns and could potentially mask differences at the single-cell level. We therefore performed single-cell analysis by FISH to determine the allelic origin of Xist RNA. We used a genetically marked cell line carrying the TsixΔCpG mutation on XM, the M. musculus X chromosome (Lee and Lu 1999). TsixΔCpG carries a promoter deletion of Tsix and, because Tsix regulates the choice step of random X chromosome inactivation, the chromosome deficient for Tsix is inactivated exclusively in ES cells and also in all somatic cells of the mouse, thereby enabling us to achieve a homogeneous population that would normally inactivate XM during ES cell differentiation. However, when the deletion is maternally inherited in mice, placental tissues inactivate XM as well as XP, resulting in loss of cells carrying two Xi (Lee 2000; Sado et al. 2001).
We asked whether transdifferentiation of Cdx2-transgenic TsixΔCpG/+ female ES cells would give rise to iTS cells that retained the ES program in keeping with its original identity or adopted the imprinted program of its newly acquired TS identity. If the transdifferentiated TsixΔCpG/+ cells upregulated Xist exclusively on the maternal TsixΔCpG allele, they would recapitulate the pattern expected of the embryo proper (also ES cells). If Xist were instead expressed from the wild-type XP allele, the transdifferentiated cells would recapitulate the pattern expected of imprinted XCI in the trophectoderm. Multiple independent iTS clones were examined and each demonstrated similar results. Interestingly, serial RNA-DNA FISH showed that 18% of iTS cells expressed Xist RNA from the wild-type M. castaneus chromosome (Figure 2B). This indicated unexpected inactivation of wild-type XP in spite of the strong Tsix mutation on XM. This contrasted sharply with normal ES differentiation, in which essentially 100% of cells inactivated the Tsix-disabled XM. Notably, without TX induction of Cdx2, transgenic cells almost exclusively inactivated the mutated X (XM), as expected for ES cells. Furthermore, 7% of cells displayed two foci of Xist expression, indicating inactivation of both XM and XP in the same cell, similar to what is observed in placental tissues of Tsix-mutant mice (Lee 2000; Sado et al. 2001) but is almost never observed in normal ES differentiation (Lee and Lu 1999) and in wild-type ES female cells when induced for Cdx2 expression. This provided further evidence for memory retention, as in spite of strong pressure against inactivation of two X’s, the iTS cells could not suppress Xist from the previously imprinted XP. All female lines were confirmed for diploidy and X chromosome number (XX) by DNA FISH. Thus, transdifferentiation into iTS cells induced imprinted XCI in a subset of cells, superseding the Tsix deletion and its strong effect on XCI choice. The relatively small percentages explain why allele-specific RT-PCR measured only a small shift in allelic ratios from ES to iTS cells and indicate that effects at the single-cell level were indeed masked by population-wide measurements (Figure 2A). This imprint retention was observed in multiple independent clonal lines. Taken together, these results argue that, although random XCI can be observed in iTS cells as reported in a recent publication (Murakami et al. 2011), the germline imprint to inactivate XP may not be fully erased during XP reactivation in the epiblast in our experimental system.
To investigate further, we performed single-cell analysis in male ES and iTS cells carrying the TsixΔCpG mutation. In male ES cells, the mutation ordinarily has no effect on Xist expression during cell differentiation into embryoid bodies (EB) and also has no effect on the epiblast lineage or male soma in the mouse (Lee and Lu 1999). However, in placental tissues, maternal inheritance of the mutation results in ectopic Xist expression and inactivation of the single male X (Lee 2000). We asked whether transdifferentiation of ES into iTS cells altered the epigenetic program on X. When Cdx2 was induced for 4 days and the ES cells were transdifferentiated into iTS cells, 8% of the male cells consistently upregulated Xist (Figure 2, C–E), similar to that seen in placental tissues of mutant male embryos (Lee 2000; Sado et al. 2001) and almost never observed in normal ES differentiation (Lee and Lu 1999). Ectopic expression also did not occur in Cdx2-transgenic TsixΔCpG/Y ES cells without tamoxifen induction of Cdx2 (Figure 2E). Multiple independent clonal male lines demonstrated the same behavior. Collectively, these findings argue that ES cells retain at least partial memory of the germline imprint, which can be unmasked when ES cells are transdifferentiated into iTS cells by Cdx2. Memory retention is independent of sex and clonal derivation.
Cdx2 binds the Xite and Xist loci
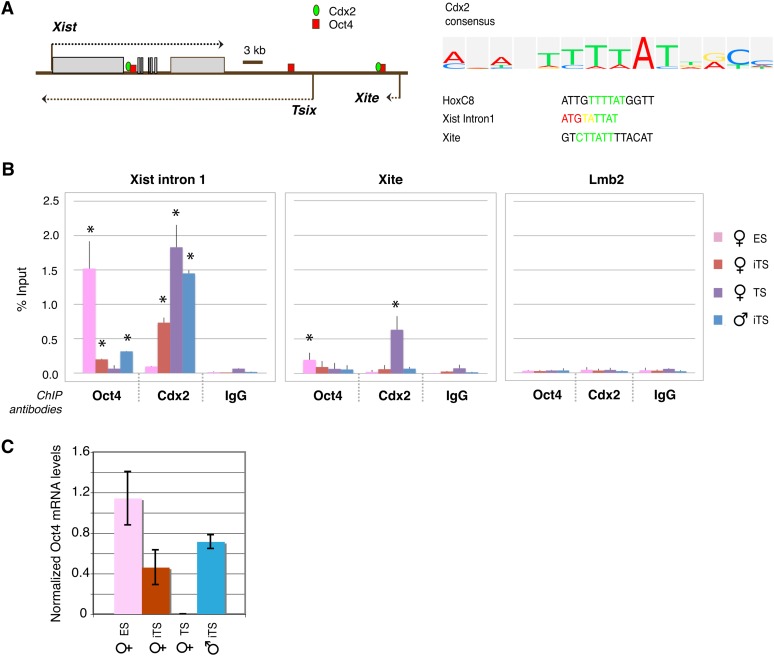
We next investigated Cdx2’s mechanism of action and asked whether Cdx2’s effect on XCI could be mediated through the Xic. We bioinformatically searched for potential Cdx2 binding sites, using the known Cdx2 consensus motif, and identified several sites within the Xic that were notable for their proximity to Oct4-binding sites (Figure 3A). Specifically, within Xite and the first intron of Xist, Cdx2 motifs colocalize with Oct4 motifs. To determine whether Cdx2 occupies these sites in vivo, we performed ChIP and observed statistically significant reciprocal binding patterns for Cdx2 and Oct4 that are dependent on cell type (P < 0.05; Figure 3B). Oct4 associated with Xist intron 1 and Xite in both male and female ES cells, consistent with previous findings (Navarro et al. 2008; Donohoe et al. 2009; Nishiyama et al. 2009) (Figure 3B).
Figure 3 .
Cdx2 and Oct4 bind to Xist and Xite in vivo. (A) Map of Oct4- and Cdx2-binding motifs within Xist and Xite. The Cdx2 consensus and binding motifs within Xist intron 1, Xite, and HoxC8 (a known Cdx2 target gene) are shown. (B) qChIP analysis of Oct4 and Cdx2 binding at designated sites in the indicated cell lines. ES, Cdx2-transgenic ES clones without Tx induction of Cdx2. iTS, Cdx2-transgenic clones transdifferentiated to TS cells by Tx induction of Cdx2. TS, trophoblast stem cells. Mean ± SEM of three independent biological replicates is shown. The statistical significance of each result is calculated compared to the control IgG ChIP (background), using a paired, one-tailed Student’s t-test. *P < 0.05. (C) qRT-PCR for Oct4 RNA in the indicated cell lines. Levels are normalized to β-actin levels. Mean ± 1 SD is shown.
Induction of Cdx2 for 48 hr and transdifferentiation into iTS cells resulted in loss of binding of Oct4 to both sites in male and female iTS cells. ChIP for Cdx2 showed the opposite pattern. Cdx2 binding was not seen in ES cells, as expected since ES cells do not ordinarily express Cdx2. Forced expression of Cdx2 and transdifferentiation into iTS cells led to de novo binding of Cdx2 protein to Xist intron 1 in iTS cells (Figure 3B). Binding to this locus was also observed in wild-type TS cells. Although Cdx2 was detected at Xite in wild-type TS cells, it did not associate significantly with Xite in iTS cells at 48 hr. In subsequent analyses, we therefore focused primarily on Xist intron 1. Quantitative RT-PCR showed that, during transdifferentiation to iTS cells, the loss of Oct4 binding occurred in advance of Oct4 depletion at 48 hr after Cdx2 induction (Figure 3C). Combined, these experiments demonstrate that the Cdx2 pathway intersects with XCI through the Xic and raise the possibility that Cdx2 binding to Xist intron 1 may be a key determinant of imprinted XCI.
Cdx2 competes with Oct4 for Xist intron 1 binding and activates the paternal Xist allele
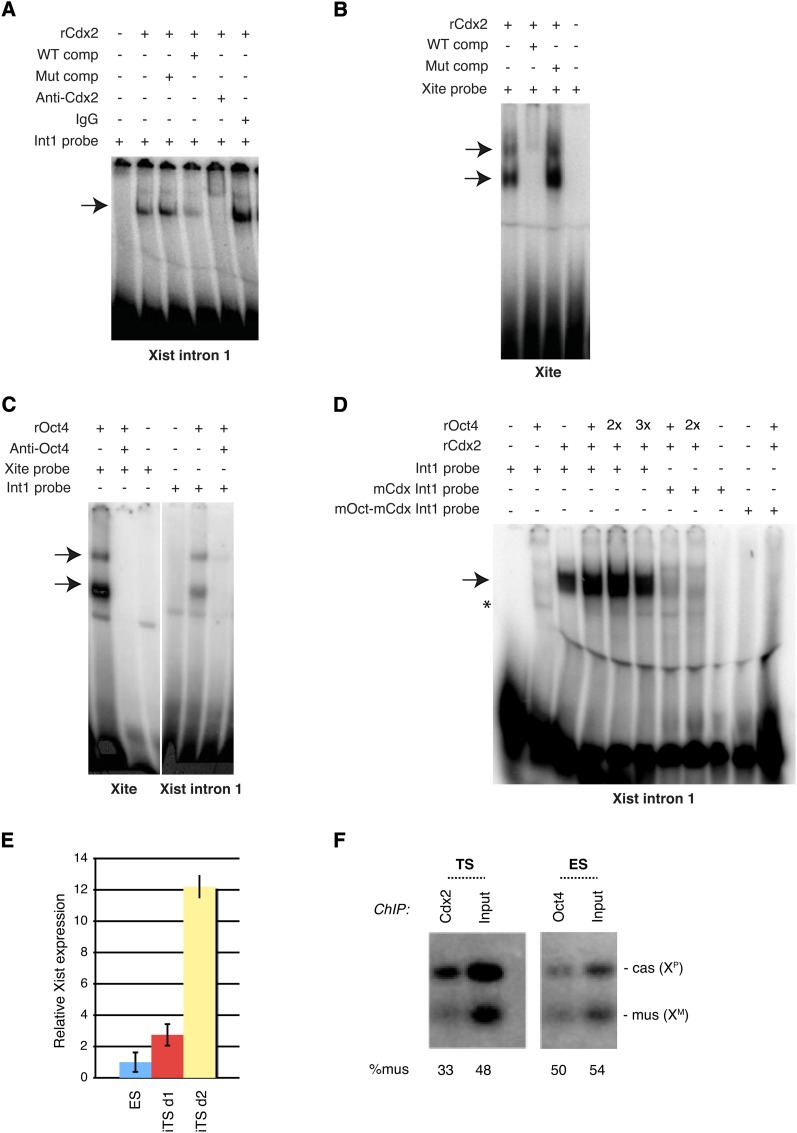
The fact that Oct4 binding to intron 1 occurred before a large increase in Oct4 levels (Figure 3C) hinted that expression of Cdx2 may displace Oct4 from the Xic. The possibility that Cdx2 and Oct4 may compete for binding to Xic was intriguing, considering that Cdx2 has been shown to interact with Oct4 (Niwa et al. 2005) and also regulate the same target genes (Nishiyama et al. 2009). Furthermore, Cdx2 sites at Xite and Xist intron 1 overlap Oct4 sites (Figure 3A). To explore this idea, we first asked whether Cdx2 directly binds the Xite and Xist motifs by performing EMSAs. Using recombinant Cdx2 protein (rCdx2), we tested its ability to shift a 32P-labeled Xist intron1 probe (Figure 4A). Indeed, Cdx2 protein shifted the intronic probe, and this interaction was significantly competed away by excess cold wild-type but not mutant oligos. Addition of α-Cdx2 antibodies neutralized and/or supershifted the protein–DNA interaction, whereas addition of normal IgG did not. Cdx2 also strongly shifted the Xite probe and this interaction was competed away by excess cold wild-type but not mutant oligos (Figure 4B). Together, these results demonstrated a direct and specific interaction of Cdx2 protein with both Xite and intron 1 of Xist.
Figure 4 .
Cdx2 and Oct4 bind directly to Xist intron 1 and Xite in vitro. (A) Gel shift of Xist intron 1 (Int1), using recombinant Cdx2 (rCdx2). Arrow, protein–DNA shift. Comp, cold competitor added at 100-fold molar excess; Mut, mutation. (B) Gel shift of the Xite motif using rCdx2. (C) Gel shift of Xist intron 1 and Xite, using recombinant Oct4 (rOct4). (D) Gel shift of Xist intron 1 motif using rOct4 and rCdx2. Two times (2×) and three times (3×),molar excess of rOct4 are shown. mCdx, mutated Cdx2-binding site; mOct-mCdx, mutated Cdx2-Oct4–binding sites. (E) qRT-PCR for Xist RNA, with levels normalized to β-actin levels. Mean ± SD is shown. ES, Cdx2-transgenic ES clones without TX induction of Cdx2. iTS, Cdx2-transgenic clones transdifferentiated to TS cells by TX induction of Cdx2 for 1 (d1) or 2 (d2) days. (F) Allele-specific gChIP analysis for Cdx2 and Oct4 binding from indicated samples.
Although the direct binding of Oct4 protein to Xite was previously demonstrated (Donohoe et al. 2009), whether Oct4 can directly bind Xist intron 1 has not been addressed, as ChIP results leave open the possibility of either direct or indirect binding (Navarro et al. 2008). To address this point, we performed EMSA using purified recombinant Oct4 protein (rOct4). Consistent with previous findings (Donohoe et al. 2009), we observed a strong rOct4-Xite complex that is neutralized by addition of α-Oct4 antibodies (Figure 4C). rOct4 also shifted Xist intron 1, although to a lesser extent than Xite. In both cases, the shifted complex was neutralized by addition of α-Oct4 antibodies. These data demonstrate that binding of Oct4 to the Xite and Xist intron 1 sequences is direct.
Collectively, these findings indicate that Oct4 and Cdx2 bind overlapping sites at the Xic and make possible the idea that they may bind mutually exclusively. Under the same in vitro conditions, Cdx2 appeared to bind the intron 1 probe more strongly than Oct4 (Figure 4D). In support of this idea, mixing equimolar amounts of Cdx2 and Oct4 with 32P-labeled Xist intron 1 resulted predominantly in formation of the Cdx2:DNA complex, suggesting that Cdx2 outcompetes Oct4 for binding when protein concentrations are equal. Increasing the amount of Oct4 protein led to increased formation of an Oct4:DNA complex. When the Cdx2-binding site was mutated (mCdx Int1), Cdx2 binding was dramatically decreased at the same time that Oct4 binding became more apparent. When both Cdx2 and Oct4 sites were mutated (mOct-mCdx Int1), binding to both sites was abolished. Our results support the notion that Oct4 and Cdx2 compete for binding to Xist intron 1. The more intense Cdx2 shifts relative to Oct4 shifts (Figure 4D) may suggest that Cdx2 has higher affinity than Oct4 for the intronic site, raising the intriguing possibility that Cdx2 expression in the early embryo may displace Oct4 from the Xic and trigger imprinted XCI.
Our data suggest that competition between Cdx2 and Oct4 for binding to Xist intron 1 may be a crucial factor in switching from random to imprinted XCI in iTS cells. We asked how Cdx2 binding to the Xic regulates Xist expression. By qRT-PCR, we observed that Xist expression increased 3- to 12-fold after 24–48 hr of Cdx2 induction (Figure 4E). This finding implies that Cdx2 may be an activator of Xist. To determine which allele binds Cdx2, we carried out allele-specific ChIP in wild-type TS cells carrying XP of M. castaneus origin and XM of M. musculus origin. Indeed, at Xist intron 1, Cdx2 preferentially bound XP (Figure 4F), the chromosome expressing Xist. By contrast, Oct4 was observed to be biallelically bound to ES cells, consistent with Oct4’s proposed repressive effect on Xist and with the presence of two active X (Xa) chromosomes in undifferentiated ES cells. We conclude that Cdx2 preferentially associates with XP in extraembryonic cells, demonstrating imprinted XP inactivation. We propose that Cdx2 is an activator of Xist and that its competitive binding with Oct4 in Xist intron 1 is a key determinant of the transition between imprinted and random XCI.
Discussion
Here we have established an ex vivo model to study imprinted XCI and identified a novel trans-acting factor that regulates imprinted XCI. Based on a Cdx2-iTS model, we show that mouse ES cells retain at least partial memory of the germline imprints that govern imprinted XP silencing. In this iTS model, a subset of cells demonstrates ectopic Xist upregulation, with female cells displaying two Xist RNA foci and male cells displaying one. Thus, although the XP has reactivated in ES cells, it still demonstrates a predisposition toward inactivation and this predisposition can be unmasked when the ES cells are transdifferentiated into trophoblast stem cells—a lineage that normally undergoes imprinted XP silencing.
It should be noted that our findings largely agree with those of a recent report using a similar iTS model (Murakami et al. 2011), in that a majority of iTS cells do overcome the XP imprint. Several experimental differences may explain why our iTS model retained the XP imprint in a subset of cells. First, there may be strain-specific differences in imprint retention, as indeed our ES cells are a M. musculus × castaneus hybrid, whereas the ES cells of Murakami et al. are of CBA × MsM background. Second, the F1(CBA × MsM) female cell line carried a puromycin-marked XP and a neomycin-marked XM and was maintained in the ES state under dual drug selection to prevent loss of the X chromosomes (there is a strong tendency for female mouse ES cells to become XO in culture). By requiring that XP remains continuously active, the system might have disrupted potential asymmetry between XM and XP, thereby selecting for ES cells that have a weakened or lost imprint. Finally and perhaps most likely, our ES cells carried a deletion of Tsix on XM, a key regulator of imprinted XCI that enables XM to resist inactivation (Lee 2000; Sado et al. 2001). It is therefore possible that the retained XP imprint may be revealed only when the maternal block to imprinted XCI is disabled. Importantly, this revelation occurs only when ES cells are transdifferentiated to iTS and is not evident when ES cells are normally differentiated into embryoid bodies.
While not all of our iTS cells revert to imprinted XCI, we observe reproducible XP skewing, strongly arguing for imprint memory. Our work is consistent with earlier studies implicating XP memory in mouse ES cells that differentiate into visceral endoderm and mesoderm of the yolk sac, an inner cell mass (ICM) lineage that also demonstrates imprinted XCI (Tada et al. 1993). In our work, remnants of the XP imprint can be observed over many ES passages, are independent of sex (both male and female ES cells demonstrate the same remnant), and are not clonally unique (multiple independent iTS clones showed similar behavior). This observed memory of an XCI imprint in ES cells supports the hypothesis that cell-type–specific factors regulate imprinted XCI in the early embryo and that the imprint can be truly erased only during gametogenesis, as is the case for autosomal imprinting (Edwards and Ferguson-Smith 2007; Wan and Bartolomei 2008; Barlow 2011). Several factors within any given ES/iTS culture could contribute to differences in reversion to imprinted XCI. For example, there could be variability at the single-cell level. Recent work suggests that a small transient population within mouse ES cultures lacks Oct4, Sox2, and Nanog expression and can contribute to either embryonic or extraembryonic tissues (Macfarlan et al. 2012). Such a subpopulation could be more permissive for imprinted XCI upon Cdx2 induction. Variable Cdx2 expression between cells could also contribute to variable recapitulation of imprinted XCI.
Using the iTS model, we identify Cdx2—a stem cell factor that defines commitment to the extraembryonic lineage—as a regulator of imprinted XCI. Cdx2 directly binds intron 1 of Xist. Its binding site overlaps with that for Oct4, the stem cell factor that defines commitment to the embryonic lineage and random XCI. Using both in vitro and in vivo techniques, we show that Cdx2 and Oct4 binding is mutually exclusive. Given that Cdx2 and Oct4 proteins interact with each other (Niwa et al. 2005), we propose that Cdx2 physically competes against Oct4 for binding in Xist intron 1 and that in Cdx2 expression in the early embryo effectively ensures that Xist is paternally expressed (Figure 5). We suggest that Cdx2 does not bind to XM because Tsix RNA—a known cis-acting maternal factor responsible for protecting XM from inactivation (Lee 2000; Sado et al. 2001)—is expressed from XM in the early embryo. At the blastocyst stage, Cdx2 becomes sequestered to the trophectoderm, maintaining paternal Xist expression in this extraembryonic lineage. In the epiblast lineage, Cdx2 is repressed, and Nanog—newly expressed in the morula—collaborates with Oct4 to repress Xist expression from both alleles. Our work shows that epiblast-derived cells (ES cells) do not fully lose memory of the paternal imprint and most likely merely override the imprint on XP to enable binding of Oct4-Nanog to the Xist promoter. Indeed, the imprint is at least partially unmasked when ES cells are transdifferentiated into TS cells and Cdx2 can once again bind to the paternal Xist allele. Thus, expression and competition between two lineage-commitment factors, Cdx2 and Oct4, underlie the switch between imprinted and random XCI in the early mouse embryo.
Figure 5 .
Opposing stem cell commitment factors, Cdx2 and Oct4, regulate the switch between imprinted and random XCI in the early mouse embryo. See Discussion for details.
Transcriptional networks of pluripotency transcription factors (pTFs) maintain expression of ES-specific genes and repression of cell-type–specific genes through a positive feedback loop. Previous work identified Cdx2 as a potent transcription factor that causes widespread downregulation of pTF-target genes by interfering with the binding of pTFs to the regulatory region of pTF-target genes on a genome-wide level (Nishiyama et al. 2009). In demonstrating Cdx2 inhibition of Oct4 binding to the Xist-intron 1 region, our results are consistent with this notion. Also consistent with our results, Xist was identified in a ChIPseq analysis as one of 337 Cdx2 targets in ES cells (Nishiyama et al. 2009).
Our data raise the possibility that Cdx2 may be involved in reading specific imprints. The nature of such imprints remains elusive. Genetic analysis suggests that DNA methylation maintains random XCI but plays a lesser role in imprinted XCI (Sado 2000, 2005; Takagi 2003; Sado et al. 2004). Strong evidence supports a maternally transmitted imprint, which acts to maintain an active XM by preventing Xist upregulation (Goto and Takagi 2000; Lee 2000; Okamoto et al. 2000; Tada et al. 2000). This maternal imprint is established during oocyte growth, can be set onto maternally transmitted autosomal transgenes, and may lie within the Tsix gene. The presence of a specific paternal imprint remains under debate. Recent work suggests that preinactivated intergenic repeats on XP may underlie paternal XCI in the early mouse embryo (Namekawa et al. 2006, 2010). If there are biparental contributions to imprinting, it is formally possible that a maternal imprint prevents Cdx2 from binding to Xist intron 1 (e.g., Tsix RNA) and/or a paternal imprint attracts Cdx2 binding to Xist intron 1. Our work provides a basis for future investigation into the molecular underpinnings of imprinting and offers a tractable ex vivo system in which to do so.
Acknowledgments
We thank H. Niwa for his generous gift of cdx2-ER plasmid. We also thank the Lee laboratory for general support and many stimulating discussions. This work was funded by a National Science Foundation Fellowship (to J.A.E.), a Human Frontiers Science Program Fellowship (to B.P.), and a National Institutes of Health grant (GM58839) (to J.T.L.). J.T.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Communicating editor: J. C. Schimenti
Literature Cited
- Barlow D. P., 2011. Genomic imprinting: A mammalian epigenetic discovery model. Annu. Rev. Genet. 45: 379–403. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., McCabe V. M., Norris D. P., et al. , 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Hendrich B. D., Rupert J. L., Lafreniere R. G., Xing Y., et al. , 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Isaacson J. H., 1967. Controlling elements in the mouse X chromosome. Genetics 57: 331–346.
- Chadwick L. H., Willard H. F., 2005. Genetic and parent-of-origin influences on X chromosome choice in Xce heterozygous mice. Mamm. Genome 16: 691–699. [DOI] [PubMed] [Google Scholar]
- Donohoe M. E., Silva S. S., Pinter S. F., Xu N., Lee J. T., 2009. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature 460: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C. A., Ferguson-Smith A. C., 2007. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 19: 281–289. [DOI] [PubMed] [Google Scholar]
- Goto Y., Takagi N., 2000. Maternally inherited X chromosome is not inactivated in mouse blastocysts due to parental imprinting. Chromosome Res. 8: 101–109. [DOI] [PubMed] [Google Scholar]
- Harper M. F., Monk M., 1982. Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J. Embryol. Exp. Morphol. 67: 127–135. [PubMed] [Google Scholar]
- Huynh K. D., Lee J. T., 2003. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426: 857–862. [DOI] [PubMed] [Google Scholar]
- Huynh K. D., Lee J. T., 2004. A continuity of X-chromosome silence from gamete to zygote. Cold Spring Harb. Symp. Quant. Biol. 69: 103–112.
- Kalantry S., Purushothaman S., Bowen R. B., Starmer J., Magnuson T., 2009. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T., 2000. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103: 17–27. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Lu N., 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Davidow L. S., Warshawsky D., 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21: 400–404. [DOI] [PubMed] [Google Scholar]
- Lee T. L., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S, et al. , 2006. Control of developmental regulators by polycomb in human embryonic stem cells. Cell 125: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikenhuis S., Wutz A., Jaenisch R., 2001. Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21: 8512–8520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., et al. , 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N., Sridharan R., Xie W., Utikal J., Eminli S., et al. , 2007. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1: 55–70. [DOI] [PubMed] [Google Scholar]
- Mak W., Nesterova T. B., de Napoles M., Appanah R., Yamanaka S., et al. , 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Panning B., Dausman J., Strauss W., Jaenisch R., 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Monk M., Harper M. I., 1979. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature 281: 311–313. [DOI] [PubMed] [Google Scholar]
- Murakami K., Araki K., Ohtsuka S., Wakayama T., Niwa H., 2011. Choice of random rather than imprinted X inactivation in female embryonic stem cell-derived extra-embryonic cells. Development 138: 197–202. [DOI] [PubMed] [Google Scholar]
- Namekawa S. H., Park P. J., Zhang L. F., Shima J. E., McCarrey J. R., et al. , 2006. Postmeiotic sex chromatin in the male germline of mice. Curr. Biol. 16: 660–667. [DOI] [PubMed] [Google Scholar]
- Namekawa S. H., Payer B., Huynh K. D., Jaenisch R., Lee J. T., 2010. Two-step imprinted X inactivation: repeat vs. genic silencing in the mouse. Mol. Cell. Biol. 30: 3187–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro P., Chambers I., Karwacki-Neisius V., Chureau C., Morey C., et al. , 2008. Molecular coupling of Xist regulation and pluripotency. Science 321: 1693–1695. [DOI] [PubMed] [Google Scholar]
- Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., et al. , 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391. [DOI] [PubMed] [Google Scholar]
- Nishiyama A., Xin L., Sharov Thomas A. AM., Mowrer G., et al. , 2009. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 5: 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., et al. , 2005. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123: 917–929. [DOI] [PubMed] [Google Scholar]
- Ogawa Y., Lee J. T., 2003. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell 11: 731–743. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Tan S., Takagi N., 2000. X-chromosome inactivation in XX androgenetic mouse embryos surviving implantation. Development 127: 4137–4145. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte A. P., Allis C. D., Reinberg D., Heard E., 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Arnaud D., Le Baccon P., Otte A. P., Disteche C. M., et al. , 2005. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438: 369–373. [DOI] [PubMed] [Google Scholar]
- Palmieri S. L., Peter W., Hess H., Scholer H. R., 1994. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev. Biol. 166: 259–267. [DOI] [PubMed] [Google Scholar]
- Payer B., Lee J. T., 2008. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 42: 733–772.
- Sado T., 2000. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 225: 294–303.
- Sado T., 2005. Imprinted X inactivation and reprogramming in the preimplantation mouse embryo. Hum. Mol. Genet. 14: R59–R64.
- Sado T., Wang Z., Sasaki H., Li E., 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Sado T., Okano M., Li E., Sasaki H., 2004. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131: 975–982. [DOI] [PubMed] [Google Scholar]
- Starmer J., Magnuson T., 2009. A new model for random X chromosome inactivation. Development 136: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulos N., Lu N., Lee J. T., 2001. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. USA 98: 10232–10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D., Mao C. A., Yamanaka Y., Ralston A., Chawengsaksophak K., et al. , 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132: 2093–2102. [DOI] [PubMed] [Google Scholar]
- Tada T., Tada M., Takagi N., 1993. X chromosome retains the memory of its parental origin in murine embryonic stem cells. Development 119: 813–821. [DOI] [PubMed] [Google Scholar]
- Tada T., Obata Y., Tada M., Goto Y., Nakatsuji N., et al. , 2000. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127: 3101–3105. [DOI] [PubMed] [Google Scholar]
- Takagi N., 2003. Imprinted X-chromosome inactivation: enlightenment from embryos in vivo. Semin. Cell Dev. Biol. 14: 319–329. [DOI] [PubMed] [Google Scholar]
- Takagi N., Sasaki H., 1975. Preferential inactivation of the paternally derived X-chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Tan S.-S., Williams E. A., Tam P. P. L., 1993. X-chromosome inactivation occurs at different times in different tissues of the post-implantation mouse embryo. Nat. Genet. 3: 170–174.
- Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J., 1998. Promotion of trophoblast stem cell proliferation by FGF4. Science 282: 2072–2075. [DOI] [PubMed] [Google Scholar]
- Tian D., Sun S., Lee J. T., 2010. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L. B., Bartolomei M. S., 2008. Regulation of imprinting in clusters: noncoding RNAs vs. insulators. Adv. Genet. 61: 207–223. [DOI] [PubMed] [Google Scholar]
- Wutz A., Gribnau J., 2007. X inactivation Xplained. Curr. Opin. Genet. Dev. 17: 387–393. [DOI] [PubMed] [Google Scholar]
- Wutz A., Jaenisch R., 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5: 695–705. [DOI] [PubMed] [Google Scholar]