Abstract
Circadian rhythms are driven by gene expression feedback loops in metazoans. Based on the success of genetic screens for circadian mutants in Drosophila melanogaster, we undertook a targeted RNAi screen to study the impact of translation control genes on circadian locomotor activity rhythms in flies. Knockdown of vital translation factors in timeless protein-positive circadian neurons caused a range of effects including lethality. Knockdown of the atypical translation factor NAT1 had the strongest effect and lengthened circadian period. It also dramatically reduced PER protein levels in pigment dispersing factor (PDF) neurons. BELLE (BEL) protein was also reduced by the NAT1 knockdown, presumably reflecting a role of NAT1 in belle mRNA translation. belle and NAT1 are also targets of the key circadian transcription factor Clock (CLK). Further evidence for a role of NAT1 is that inhibition of the target of rapamycin (TOR) kinase increased oscillator activity in cultured wings, which is absent under conditions of NAT1 knockdown. Moreover, the per 5′- and 3′-UTRs may function together to facilitate cap-independent translation under conditions of TOR inhibition. We suggest that NAT1 and cap-independent translation are important for per mRNA translation, which is also important for the circadian oscillator. A circadian translation program may be especially important in fly pacemaker cells.
Keywords: NAT1, DAP5, torin, circadian, cap-independent translation
ALL self-sustained circadian rhythms in multicellular organisms employ a transcriptional feedback loop. The clock is entrained by cues such as illumination, temperature, and nutrients and allows organisms to anticipate and accommodate daily changes in their environment. The mammalian pacemaker is driven by bHLH-PAS transcription factors CLOCK and BMAL1, which heterodimerize to drive the transcription of three Period and two Cryptochrome genes, the products of which cooperate to repress their own activation. The situation in flies is essentially identical: Clock and cycle (CLK and CYC; orthologs of CLOCK and BMAL1) activate the expression of fly period and timeless, (per and tim). PER is stabilized by TIM in the cytoplasm where both proteins accumulate post-translational modifications throughout the night (Chiu et al. 2011). The two clock proteins are eventually transported into the nucleus where they mediate repression of CLK/CYC-driven transcription. In flies, the transcriptional oscillator must be active in neurons expressing pigment dispersing factor (PDF) to stimulate rhythmic locomotor behavior (Grima et al. 2004).
Although much less well understood, translational control in flies has been suggested to stall the build-up in repressor activity and contribute to maintaining circadian oscillator function. For example, the DEAD-box helicase Lark delays circadian-gated eclosion until early morning (Newby and Jackson 1993) and influences constant darkness (DD) rhythms (Huang et al. 2009). Also, PER translation is stimulated by interactions between its 3′-UTR, TYF and PABP (Lim et al. 2011). Similar evidence is present in mammalian systems. The translation of a murine per ortholog is modulated both by mLark via the per 3′-UTR (Kojima et al. 2007) and by HNRNPq via the per 5′-UTR (Lee et al. 2011, 2012). These data suggest that translational regulation may play a role in supporting or mediating the circadian clock.
Especially in mammals but also in other organisms, there are extensive interactions between metabolic and circadian cycles (Lamia et al. 2011; Sancar et al. 2011). Because of its well-characterized sensitivity to nutrient conditions, translational control provides an attractive mechanism to explain the integration of nutrient and time-of-day information. Indeed, insulin signaling components were strongly implicated in a genome-wide screen for circadian effectors in mammalian tissue culture (Zhang et al. 2009a). Growth and nutrient signaling pathways are integrated via TOR kinase, the activity of which stimulates global cap-dependent translation initiation. Interactions between the mRNA 7mG cap and initiation factors direct small ribosomal subunits to start codons, where large ribosomal subunits are recruited and translation begins. TOR phosphorylation of eIF4B increases its stimulation of eIF4A helicase, and TOR phosphorylation of 4EBP blocks its inhibition of eIF4E; both of these events up-regulate cap-dependent translational initiation (Sonenberg and Hinnebusch 2009).
Although increases in circadian gene expression and copy number usually increase the pace of the oscillator (Baylies et al. 1987; Allada et al. 1998; Kadener et al. 2008), TOR activity was inversely correlated with the pace of rhythms in flies (Zheng and Sehgal 2010). This was surprising because increased TOR signaling increases global translation initiation. Under conditions of attenuated gene expression including mitosis and starvation, translational initiation can be carried out in a noncanonical fashion; this bypasses cap-binding requirements (Marr et al. 2007). Noncanonical translation is often promoted by paralogs of canonical translation factors (Marash et al. 2008).
To further explore the role of translation in the Drosophila circadian system, we expressed RNAi constructs targeting translation and RNA factors within two populations of brain circadian neurons and assayed locomotor activity rhythms in standard constant darkness conditions. The noncanonical translation factor NAT1 was among the strongest factors identified. Expression of its RNAi construct within adult circadian neurons slows oscillator pace, indicating a role of this protein and perhaps cap-independent translation in circadian translation. Under these knockdown conditions, PER expression is dramatically reduced in PDF cells and overexpression of PER can rescue the rhythm defect. NAT1 knockdown also decreases the amplitude of circadian reporter oscillations in cultured wings and confers sensitivity to TOR kinase inhibition upon reporter expression in both wings and S2 cells. Evidence is also shown that the per 5′- and 3′-UTRs function together to facilitate cap-independent translation. We suggest that NAT1 and cap-independent translation are important for per translation, which is important in turn for the core circadian oscillator.
Materials and Methods
Fly stocks
For all experiments, fly strains were maintained on standard cornmeal-dextrose agar media at 25° under 12-hr intervals of light:dark (LD) unless stated otherwise. RNAi lines were obtained from the Vienna Drosophila Resource Center (Dietzl et al. 2007). The primary transformant used for NAT1 was 105121. Other transformant IDs are listed in Supporting Information, Table S1. All strains have been previously described: pdf-GAL4, UAS-pdf (Renn et al. 1999), tim-GAL427 (Kaneko and Hall 2000), tim-LUC (Stanewsky et al. 2002), UAS-PER2-4 (Yang and Sehgal 2001), pdf-GAL80 (Stoleru et al. 2004), tub-GAL80TS (McGuire et al. 2003), UAS-DICER2 (Dietzl et al. 2007), and UAS-NAT1 (Yoshikane et al. 2007).
Behavior analysis:
Single flies were loaded into tubes supplemented with 5% sucrose in 2% agar solution and entrained at 25° for 3 days of 12 hr light:dark intervals followed by constant darkness for at least 6 days (Kadener et al. 2007). Data were acquired using the Trikinetic monitoring system in 1-min bins. Determination of period length and rhythmic strength was made by autocorrelation and spectral analysis Matlab tools (Levine et al. 2002). To establish a phase response curve, flies were given 10-min light pulses throughout the last night of LD and then the phase change relative to unpulsed controls was calculated after period stabilization, where an average of the phase difference was calculated for DD 2–4. Behavior experiments were all repeated at least three times with similar results.
Western blotting and RNA analysis
Expression analysis was performed in routine fashion with slight modification (Menet et al. 2010). Flies were entrained for at least 3 days and then frozen at the appropriate time points on dry ice. Flies were decapitated by vortexing and 25 heads were homogenized by electric handheld pestle twice for 30 sec in 50 μl RBS buffer supplemented with EDTA-free complete protease inhibitor cocktail (Roche). Lysates were cleared by spinning 10 min at 20,000 × g, supplemented with 5× SDS loading buffer and separated by SDS–PAGE on 3–8% acrylamide Tris-acetate Novex gels (Invitrogen). Blotting was performed with relevant antibodies (anti-PER 1:200; anti-TIM 1:1000; anti-PABP 1:1000, N. Sonenberg; anti-BEL 1:10,000, P. Lasko; HRP-conjugated antirabbit 1:3000, GE Healthcare) and developed with ECL Plus HRP system (Amersham). RNA isolation was performed with Trizol (Invitrogen) followed by DNase treatment (Turbo, Ambion) and cDNA synthesis with 3 μg of total RNA (Superscript II). Samples were treated with RNaseH (Invitrogen) prior to qPCR analysis using a SYBR Green Master mix (Qiagen). Quantification was performed in triplicate with the following primers: perF-ATAAGCACAACGACGAGATGGA, perR-GAGCCTCCTCTTTTTATCCCGT; timF-CACACCATCTTCGAGCTGAATAA, timR-AGTTGTTGATTTGATCGCATTTG; gapdh2F-CTACCTGTTCAAGTTCGATTCGAC, gapdh2R-AGTGGACTCCACGATGTATTCG; belF-CGAAAAGAACCGCAACATTT, belR-AACTGGGGATCTCCTGCTTC; and NAT1F-TGTTTAGCGGACTGAGTGTTACGG, NAT1R-TCCTTACTACCCAGCAGCTGATTG.
Confocal microscopy
Fly heads were dissected at the times indicated following 3 days of entrainment. Heads were removed under carbon dioxide anesthesia and fixed in PBS supplemented with 0.008% Triton X-100 (Sigma) and 4% paraformaldehyde (Electron Microscopy Sciences) for 45 min on ice. All washes were repeated three times, 20 min each, with gentle agitation at room temperature (RT) in PBS, supplemented here with 0.1% Triton X-100. Heads were then dissected in PBS in Sylgard-coated dishes. Brains were washed three times in PBS 0.5% Triton and then blocked in wash solution with 10% normal goat serum (NGS, Jackson Immunological Research) in PBS 0.5% Triton for 1-hr shaking at RT. Brains were incubated with primary antibodies diluted in NGS (rabbit anti-PER 1:50, rabbit anti-TIM 1:200, mouse anti-PDF 1:10, guinea pig anti-CLK 1:5000, guinea pig anti-VRI 1:500, guinea pig anti-PDP1 1:300) for 48 hr at 4° with gentle agitation. Brains were then washed three times and incubated overnight at 4° with secondary antibodies at 1:500 (Alexa Fluor Molecular Probes at 633 nm or 488 nm, Invitrogen). Brains were washed three times and mounted in Vectashield (Vector Laboratories) and imaged in 1-μm sections sequentially at ×63 in oil on a Leica SP5 confocal. Quantification was performed using National Institutes of Health Image software, where 9.3 μm2 regions of PDF cell nuclei were selected (in the section that showed the largest nuclear cross-section) with the brush selection tool and intensity measured for each.
Ex vivo luciferase monitoring
Virgin flies harboring transgenes for timeless-GAL4 (tim-GAL4) and timeless-Luciferase (tim-LUC) were crossed to UAS-NAT1RNA#105121 or UAS-GFP and the offspring entrained in a light:dark cycle for 3 days. Wings were then dissected during the light period from bodies under carbon dioxide anesthesia and placed in white 96-well Microfluor 2 plates (Fisher) with 100 μl wing media. Media were composed of M3 Shields supplemented with 12% HI-FBS, 1% penicillin-streptomycin, 0.03 mg/ml bovine insulin (Sigma), and 1.5 mM D-luciferin potassium salt (GoldBio). Monitoring was performed in a Packard Topcount multiplate scintillation counter for at least 3 days. Data were analyzed in MATLAB and Microsoft Excel. All samples with an average luciferase activity <200 Hz were eliminated, as were those for which wings did not cycle. Raw data curves were averaged for each drug or genotype condition. Individual wing profiles were fitted by a linear regression, which was then used to calculate a subtraction term to remove effects of dampening over time. These detrended data were used to calculate amplitudes for individual wings, which were assessed on the second or third day of culturing. Experiments were repeated three times with similar results.
S2 cell luciferase assays
Luciferase assays were carried out in 96-well plates (Corning). Cells were plated at 100 μl per well 1e6 cells/ml in HFX insect media supplemented with 10% HI-FBS, and 1% penicillin-streptomycin. After adhering for 3 hr, the media were replaced with serum-free media and transformations were carried out using Celfectin II reagent (Invitrogen). After 5 hr, the media were replaced with serum-containing media. The following day, torin (Tocris) in DMSO was applied in regular media. The following day the cells were harvested and assayed using the Dual-Luc system (Promega) using a Tecan plate reader. For dsRNA treatments, transcription was performed with a Megascript T7 kit (Ambion) following PCR amplification of plasmid or genomic templates with the following primers: GFP F-TTAATACGACTCACTATAGGGAGAATGGTGAGCAAGGGCGAGGAG, R-TTAATACGACTCACTATAGGGAGACTTGTACAGCTCGTCCATGCC; NAT1 (CG3845) F-TAATACGACTCACTATAGGGATCACTCAAACTCCGATGGC, R-TAATACGACTCACTATAGGGTTTCGCTCTAGGCTGTTGGT; and BEL (CG9748) F-TAATACGACTCACTATAGGGCAACAGCCTAGAAGGGATCG, R-TAATACGACTCACTATAGGGGTTCTTTTCGTTCCTCGCAC. Three hours after plating, the media were replaced with 100 μl serum-free media with 15 ng/μl dsRNA. The following day, 100 μl of HFX with 20% FBS and 2% penicillin-streptomycin was added. After 48 hr, transfection and drug treatment were performed as outlined above. The per UTRs were cloned into pAc-Luc (Kadener et al. 2008) and pAc-Renilla was a kind gift from M. Marr.
Results
RNAi screen reveals long-period rhythms in circadian-cell NAT1 knockdown
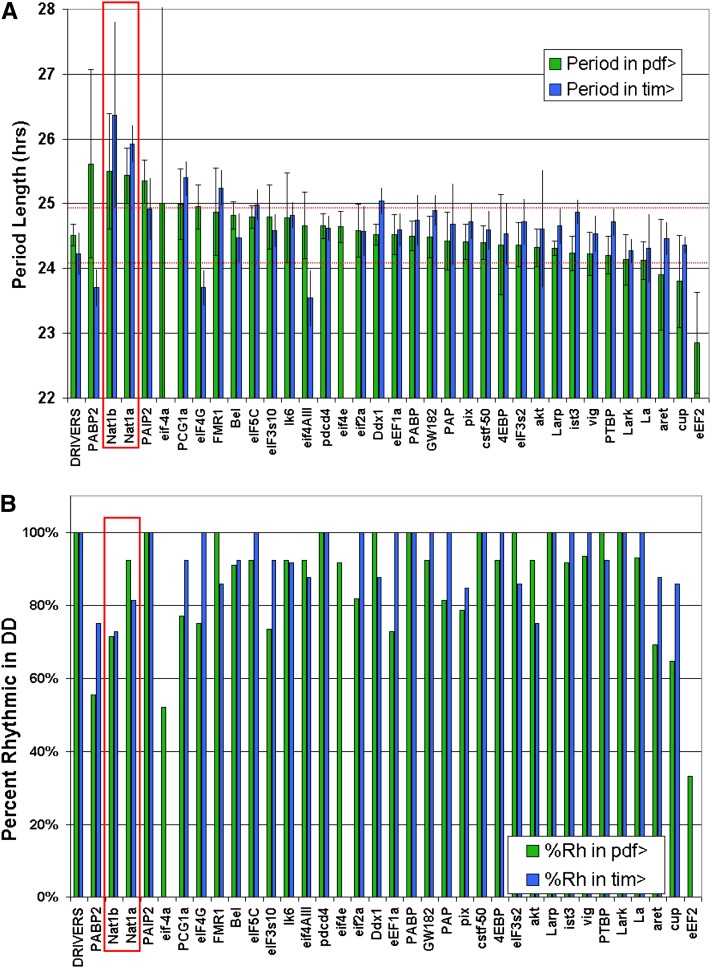
Genetic screens that assay locomotor activity rhythms in constant darkness have identified many clock genes in Drosophila. To address translational control genes, we used 70 UAS-RNAi constructs targeting 34 genes impacting translation and RNA metabolism in combination with the pdf-GAL4 driver (pdf > RNAi) as well as the tim-GAL4 driver (tim > RNAi). The former targets PDF cells, which is the major neuron type governing the period of locomotor activity rhythms in constant darkness. The latter targets a much larger number of clock neurons, probably all clock neurons including PDF cells, as well as nonclock cells. Flies were entrained in a light:dark cycle of 12 hr each and then assayed during 7 subsequent days in constant darkness. The period length of the endogenous pacemaker was then determined by standard autocorrelation analysis. For all but NAT1, the focus of this report, only the strongest result among the RNAi lines targeting a particular gene was plotted (Figure 1A, Table S1). Period lengths more than two standard deviations from the mean period length of the control lines were considered significant.
Figure 1 .
Locomotor behavior period lengths following RNAi in clock cells. Thirty-four translation and mRNA metabolism genes were knocked down in TIM+ or PDF+ cells and the DD behavior analyzed by autocorrelation. (A) Targeted genes are ranked by their period length when driven in PDF cells. Period lengths at least two standard deviations from the mean driver control were considered significant, apart from eIF-4a, which caused highly variable rhythms. When multiple targeting vectors were used, only the one providing a greater effect was plotted, except for NAT1, which is the subject of this report. (B) Percentage of highly rhythmic flies (RI > 0.2) is plotted for each knockdown.
When ranked by their effect on period length in combination with a pdf-GAL4 driver, it was apparent that most lines caused a slight increase in period length compared to control lines. The only exception was the elongation factor eEF2, which had consistently shorter periods when knocked down in PDF cells. This factor, along with two initiation factors, eIF-4a and eIF4E, also caused lethality when knocked down in TIM cells. In response to pdf-driven knockdown, eIF-4a was unusual in causing highly variable length rhythms, including slower, longer, and unstable periods in individual flies.
RNAi against the eIF4G paralog NAT1/DAP5/p97 had significant effects with both drivers and the strongest effect of all genes targeted with tim-GAL4. It also showed a strikingly similar phenotype for both tested RNAi lines. Other significant period-lengthening effects were found upon knockdown of PABP2, PAIP2, FMR1, DDX1, eIF5C, and PGC1a (Figure 1A). PGC1a (Spargel) was included in the screen because it contains an RNA-binding motif, but it is primarily associated with transcription and has already been implicated in mouse circadian rhythms (Liu et al. 2007). In addition to variation in period length, effective RNAi constructs also decreased the percentage of highly rhythmic flies. This was particularly obvious upon knockdown of NAT1, eIF-4a, and eEF2 (Figure 1B, Table S1).
To increase the power of the UAS-RNAi effects, we employed drivers recombined with a UAS-DICER2 transgene (Dietzl et al. 2007). Rescreening a subset of interesting RNAi lines with the tim- and pdf-GAL4 drivers produced similar effects to those observed without UAS-DICER2 (Figure S1). Notable differences, however, were long-period phenotypes for AKT and LK6, both of which had no effect without UAS-DICER2. Additionally, NAT1 knockdown had more severe effects on rhythmicity, with a lower percentage of rhythmic flies and longer periods for several days followed by arrhythmicity. In summary, RNAi-mediated depletion of NAT1 produced the most substantial alterations in overall period length and both UAS-RNAi constructs had significant effects. In addition, NAT1 was recently described by our lab to be a CLK direct target gene (see below), so we focused the rest of this study on NAT1.
NAT1 functions in adults to support oscillator function
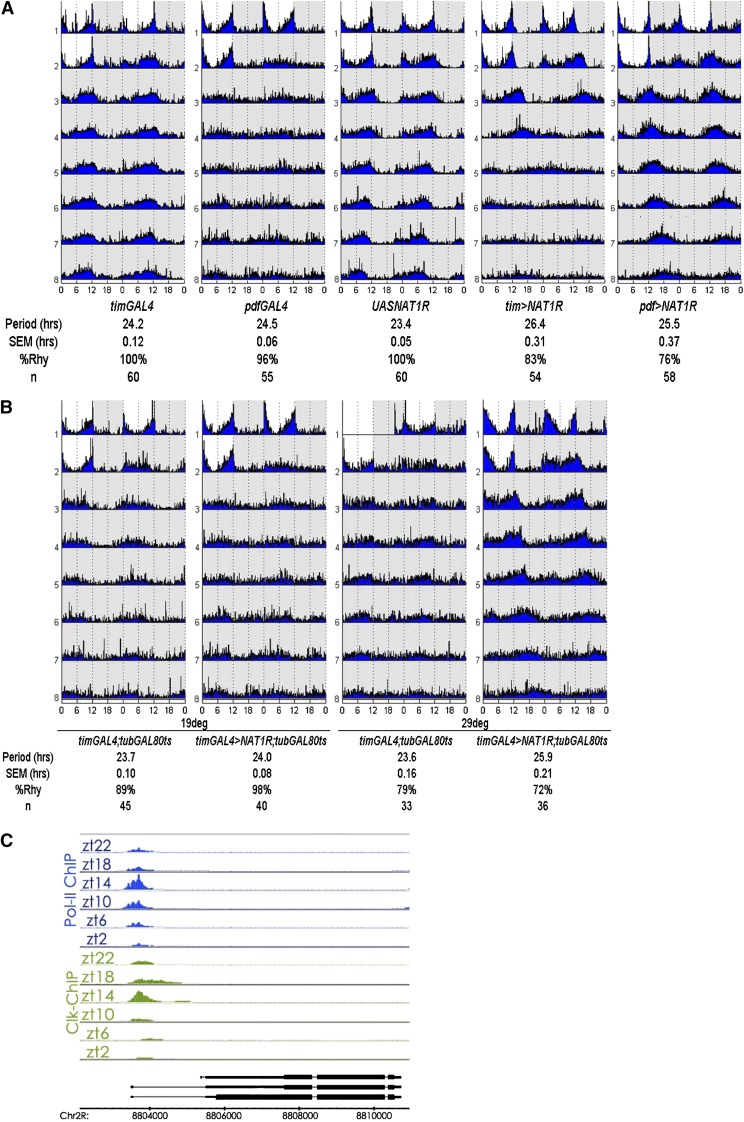
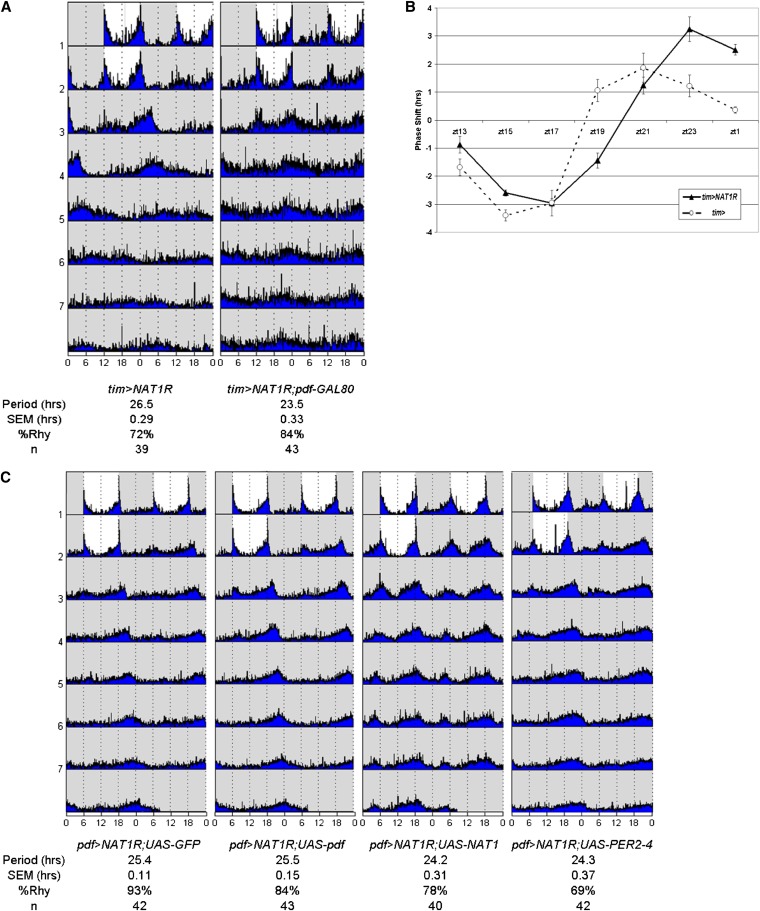
RNAi against NAT1 in PDF or TIM cells lengthened the period to 25.5 hr or 26.6 hr, respectively (Figure 2A). Although the effect was weaker with the pdf-GAL4 driver than with the tim-GAL4 driver, this may simply mean that the tim-GAL4 driver is stronger in PDF cells. It may also reflect the fact that other clock neurons, in addition to PDF cells, can affect circadian period in constant darkness. Nonetheless, effects with both drivers were highly significant compared to the slightly more than 24-hr period routinely observed for wild-type, driver, and UAS-RNAi control flies. The long-period phenotypes found with both drivers were equally stable for 1 week in constant darkness.
Figure 2 .
NAT1 supports circadian locomotor rhythms. (A) Actograms for NAT1 knockdown with tim- and pdf-GAL4 drivers demonstrate stable long-period rhythms over 7 days in constant darkness. SEM denotes standard error of the mean and %Rhy indicates the percentage of rhythmic flies. (B) Coexpression of temperature-sensitive GAL80 with NAT1RNAi can block the phenotype unless the flies are subject to 3 days of 29°, ruling out a developmental origin for the rhythm defect. (C) Schematic view of chromatin IP performed with antibodies against CLK and RNAPolII shows cyclic transcriptional activation at the NAT1 locus, with the same temporal peak as canonical CLK targets.
Because some circadian genes are required during development for proper circadian rhythms in adults (Huang et al. 2009; Goda et al. 2011) and because NAT1 alleles have been reported to cause developmental lethality (Takahashi et al. 2005), we asked whether a comparable period effect would result if we restricted the knockdown to adult circadian cells. This was done by coexpressing a temperature-sensitive GAL80 transgene driven by the ubiquitous tubulin promoter along with tim > NAT1RNAi; the GAL80 represses GAL4 activity at 21° and is inactivated at 29°, which allows GAL4 function. Shifting adult flies to 29° for 3 days resulted in the longer period characteristic of the tim > NAT1RNAi strain (Figure 2B). This indicates that the effect of NAT1 activity on circadian period is not developmental and that knockdown in adult neurons is sufficient to affect circadian rhythms.
Clock (Clk) encodes a transcription factor at the top of the circadian gene expression hierarchy in animals, and our lab has recently described its genome-wide DNA binding across the circadian cycle (Abruzzi et al. 2011). NAT1 is a direct CLK target in fly heads, consistent with the notion that oscillating levels of CLK and RNA Pol II drive its transcription with a peak at ZT14 (Figure 2C). This binding profile is similar to those of other well-described direct clock targets per, tim, vri, and Pdp1, further suggesting involvement of NAT1 in rhythms.
Dramatic BELLE reduction in the NAT1 knockdown
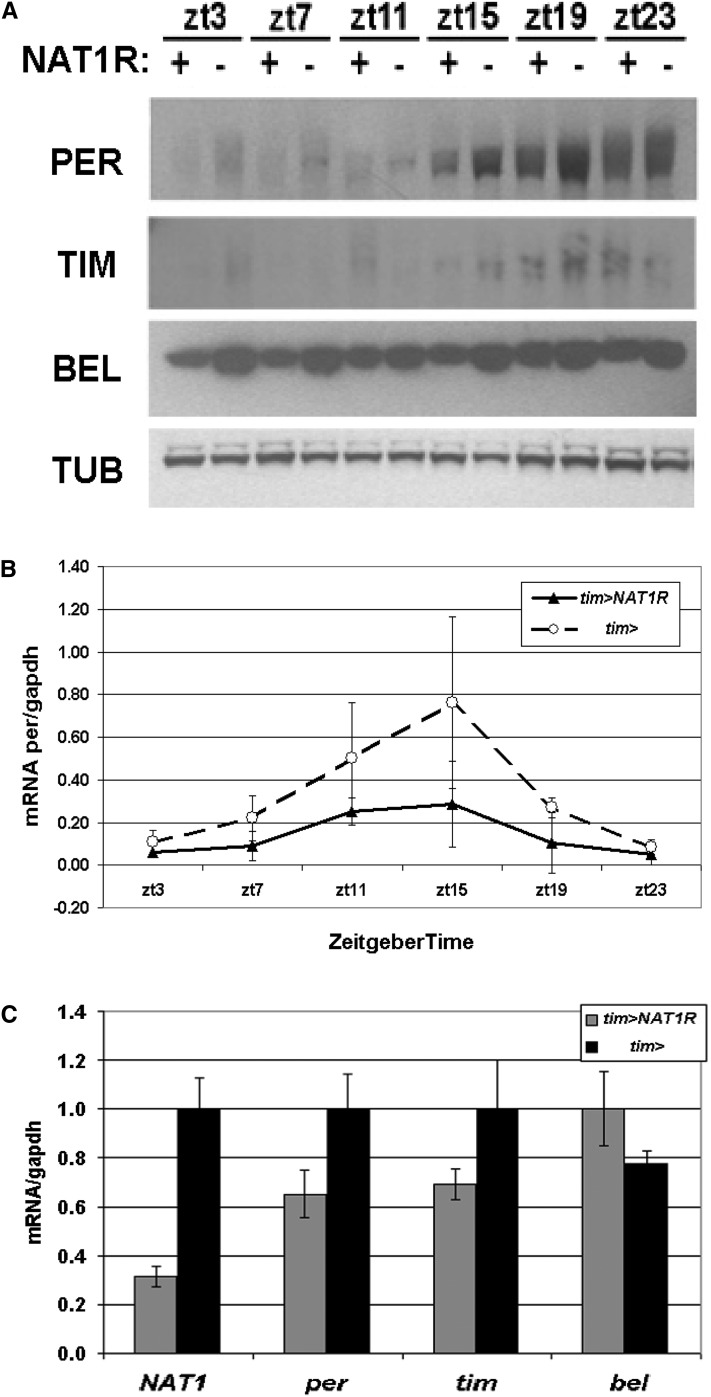
To address the mechanism by which NAT1 knockdown affects period length, we examined the expression of key circadian genes in heads. Analysis of PER and TIM by Western blotting over a 24-hr span revealed only mild effects, with protein levels slightly but consistently reduced compared to WT. In the course of examining the levels of other translation factors, we noticed a striking reduction in BELLE protein (BEL) levels in the tim > NAT1RNAi strain (Figure 3A). This translational helicase is homologous to eIF-4a and is also a direct target of CLK in fly heads (Abruzzi et al. 2011). Although unexpected and interesting (see below), the effect of the NAT1 knockdown on BEL levels may also reflect the fact that the tim-GAL4 driver expresses in most of head tissues, including the fat body, eyes, and glial tissue.
Figure 3 .
Whole head biochemistry under NAT1 knockdown. (A) Flies expressing UAS-NAT1RNAi under the control of tim-GAL4 have reduced PER, TIM, and BEL protein levels in LD conditions. (B) per mRNA cycles with reduced amplitude in tim > NAT1RNAi consistent with impaired oscillator function. (C) Assessment of mRNA levels for NAT1, per, tim, and bel at ZT12, which are all normalized to driver controls.
Examination of per mRNA levels in the tim > NAT1RNAi strain around the clock revealed a normal peak phase at ZT15, but maximum levels and amplitude of cycling were reduced to about half those of wild type (Figure 3B). This difference is greater than the protein level reduction (Figure 3A), suggesting that mRNA levels are not limiting for PER production. NAT1 mRNA levels in these flies were reduced to one-third of WT, indicating an effective knockdown (Figure 3C). The mRNA for bel is not reduced compared to wild type, suggesting a translational deficit accounts for the much larger decrease in BEL.
Staining of brains reveals reduced PER expression in small PDF cells, particularly in DD conditions
Because whole head biochemistry showed only modest decreases in circadian gene expression, we examined circadian neurons for the functional impact of NAT1 knockdown on clock gene expression. Because PDF cells provide the minimal circadian circuitry required for locomotor rhythms in constant darkness (Grima et al. 2004), we focused our staining efforts on the relevant lateral ventral regions; the PDF cells are also easily identified by co-staining for this peptide. tim-GAL4 was used as a driver for NAT1 knockdown instead of pdf-GAL4 because of its stronger behavioral phenotype.
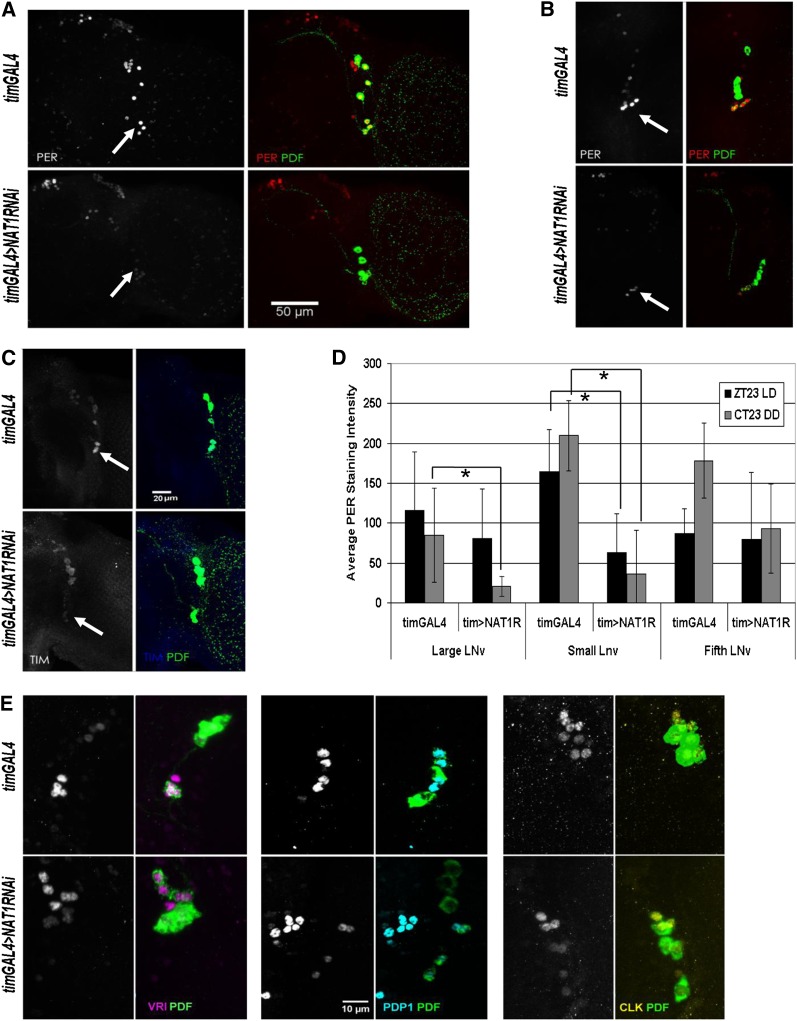
Initial inspection of NAT1 knockdown brains revealed no obvious abnormalities: PDF levels and PDF cell arborization patterns were both normal. PER staining was performed around the time of peak expression in PDF cells, at ZT23. Although PDF neurons displayed a decrease in PER staining relative to dorsal cells during LD conditions (Figure 4A), the effect was much more robust in DD, the conditions under which circadian period was assayed: PER staining was dramatically reduced at CT23 in the tim > NAT1RNAi strain compared to a wild-type strain (Figure 4B). A qualitatively identical effect was observed with TIM staining at CT23, suggesting that effects of the NAT1 RNAi knockdown are not restricted to per expression (Figure 4C). Quantification of DD1 PER levels indicates that the greatest effects occur in the small PDF cells, which normally have the highest PER expression and also dominate the behavioral phenotype under DD conditions (Grima et al. 2004). However, the large PDF cells are also substantially affected, and the fifth small lateral ventral neurons (s-LNv) (PDF− cell) is the least affected (Figure 4D).
Figure 4 .
NAT1 knockdown reduces PER expression in PDF+ cells. (A) In LD conditions, control brains (top of each panel) have the highest levels of PER signal at ZT23 in small lateral ventral neurons (s-LNv, arrowheads), but in tim > NAT1RNAi knockdown (bottom of each panel) staining is significantly reduced relative to that in dorsal cells. (B) This effect is more pronounced in DD, where cycling in whole heads rapidly damps and high amplitude PER cycling in small cells drives locomotor rhythms. (C) TIM levels are also reduced in LNv cells in DD conditions. (D) Quantification of seven hemispheres per genotype shows significant PER reductions in DD conditions for small and large PDF cells. The fifth, PDF− LNv cell is not significantly affected (*P < 0.0001, two-sample t-test; error bars represent SD). (E) Control staining for VRI at CT15, PDP1, at CT19 and CLK at CT3 in PDF cells shows only minor signal reductions in tim > NAT1RNAi brains.
To exclude the possibility that the long-period phenotype of these flies was not simply delaying the peak of expression, we checked PER staining in PDF cells 4 hr later, at CT3 of DD2. Whereas robust PER staining persisted in control flies, tim > NAT1RNAi exhibited no detectable PER at this timepoint in PDF cells (Figure S2). Although the levels of PDF staining indicated that general protein expression in these cells was normal, we wondered whether other core clock protein levels were affected by NAT1 deficits. Staining for VRI, PDP1, and CLK revealed only slight reductions under the same conditions (Figure 4E), consistent with a weaker core transcriptional oscillator.
NAT1 functions within PDF cells and supports light-induced phase responses
Because of the dramatic effect of NAT1 knockdown on per expression in PDF cells, we sought to determine the contribution that non-PDF cells made to the stronger phenotype in the TIM cell knockdown. We were also motivated by the important role of PDF cells within the circadian brain circuit. To this end, we assayed tim > NAT1RNAi flies containing an additional pdf-GAL80 transgene to block GAL4 activation only in PDF cells. This strain had a completely normal circadian period, i.e., knockdown of NAT1 in Tim+, PDF− cells was without effect. Taken together with results from the other drivers, we conclude that NAT1 knockdown in PDF cells is necessary and sufficient for the behavioral phenotype (Figure 5A).
Figure 5 .
NAT1 is primarily active in PDF+ cells. (A) Period lengthening requires knockdown in PDF cells, shown by expressing NAT1RNAi in TIM+ cells but blocking GAL4 activity with pdf > GAL80. (B) The phase response curve for tim > NAT1RNAi shows increased phase shifts and a delayed profile relative to controls in the late night, supporting a role in PDF+ morning cells. (C) Coexpression of UAS-NAT1 or UAS-PER2-4 rescues the pdf > NAT1RNAi phenotype, but UAS-GFP or UAS-pdf does not, the latter showing that the deficit in period length is not due to a shortage of this peptide. Pronounced morning peaks are evident in the UAS-NAT1 rescue only.
Especially in LD conditions, PDF cells constitute the morning oscillator and make a major contribution to late-night phase shift effects (Shang et al. 2008). To further characterize the behavioral effect of NAT1 knockdown, tim > NAT1RNAi flies were subjected to light pulses throughout the night to generate a standard phase response curve (PRC). Wild-type flies manifest a characteristic response, with an ∼4-hr delay to a light pulse administered in the early night and an ∼2-hr advance to a late night light pulse.
The phase response curve for tim > NAT1RNAi flies was both delayed (shifted to later times) and also had a substantially greater magnitude specifically in the late night-advance zone (Figure 5B). The delay probably reflects the longer period length characteristic of this strain. However, the enhanced advance zone response was unexpected. Perhaps it reflects a more prominent role for this gene in the morning oscillator (PDF) cells and/or a more prominent role of these cells in the advance zone of the PRC (see Discussion).
Because of the important role adult PDF cells play in the functional contribution of NAT1 to rhythms, we sought to affect rhythms by overexpression of other genes in PDF cells of pdf > NAT1RNAi flies. Generation of a strain carrying two copies of both pdf-GAL4 and UAS-NAT1RNAi allowed for the addition of other transgenes to this cell-specific knockdown background. Crosses to UAS-GFP produced 25.5-hr rhythms as expected for a single copy of the RNAi construct (Figure 5C). A cross to a UAS-pdf transgene-containing strain also had 25.5-hr rhythms. This suggests that this neuropeptide is not limiting for period determination under NAT1 knockdown conditions, consistent with the PDF staining results.
In contrast, addition of a UAS-NAT1 construct had a normal period length of 24.3 hr. Although the rhythm rescue was not surprising, it was accompanied by greater and more persistent levels of morning activity during constant darkness. Similar effects were found for flies homozygous for this transgene, although not for a version containing a 5′ truncation (Figure S3). This unexpectedly connects NAT1 expression levels within PDF morning cells to locomotor activity (see Discussion). Notably, addition of a UAS-PER construct also rescued the long-period rhythms of the NAT1 knockdown (Figure 5C). This suggests that low PER levels are indeed responsible for the long-period phenotype and that NAT1 somehow potentiates per mRNA translation.
To see whether viable mutants for NAT1 might offer additional clues regarding the function of this gene, we combined them with pdf > NAT1RNAi flies, expecting a more potent behavioral phenotype when only one functional copy of the NAT1 locus was present. However, the behavioral phenotypes were not different from flies with one copy each of pdf-GAL4 and UAS-NAT1RNAi transgenes, indicating that under these knockdown conditions, one NAT1 gene copy is sufficient to support stable long-period rhythms. Combining other RNAi lines with the pdf > NAT1RNAi background also did not give rise to significant period alterations (Figure S4).
Circadian oscillations in wings are stimulated by TOR inhibition in a NAT1-dependent manner
Luciferase reporter genes have been useful for studying circadian oscillator function (Stanewsky et al. 1998; Glaser and Stanewsky 2005). For example, a luciferase (Luc) transgene preceded by the promoter of the timeless gene (tim-LUC) produces luminescence driven by CLK/CYC transcriptional activity in explanted wings without the network complications present within fly brains. Importantly, the wing assay is also amenable to pharmacological manipulation. To exploit this system, flies harboring tim-GAL4 and tim-LUC transgenes were crossed to lines expressing UAS-NAT1RNAi to assess the knockdown effect on these peripheral oscillators. Wings were cultured and monitored by photoluminescence over several days under standard conditions.
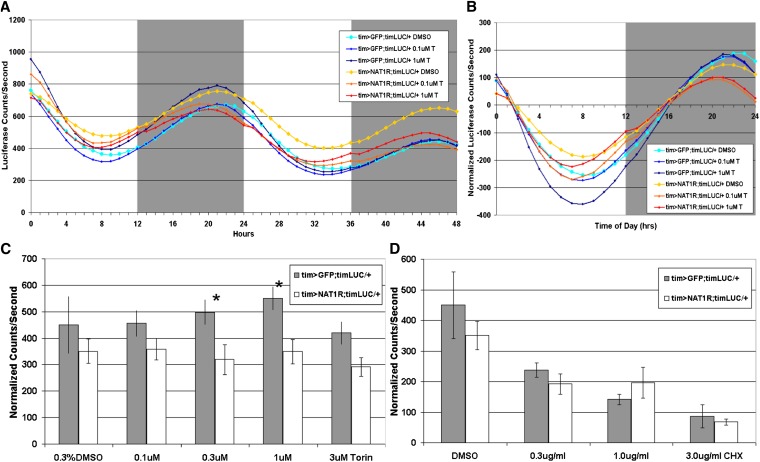
Expression of luciferase under tim promoter control was entrained by the second day of wing culture, and oscillations peaked 2–3 hr before lights on (Figure 6A). Raw values of luciferase expression in cultured wings, i.e., luminescence counts per second, were variable, making comparisons between treatments or genotypes difficult. However, amplitude measurements were reproducible and typically made on the second or third day, because of the dampening that occurs upon culturing. After baseline subtraction, normalized photoluminescence values oscillated around zero (Figure 6B). Amplitudes were obtained by subtracting trough from peak luciferase values for individual wings. Flies expressing UAS-NAT1RNAi in TIM cells had somewhat reduced amplitudes relative to control wings expressing UAS-GFP. This effect echoes the effects observed by Western blotting of fly head extracts in this same knockdown and suggests an oscillator driving lower amplitude transcription cycles.
Figure 6 .
NAT1 supports oscillator function in the peripheral circadian cells of cultured wings. (A) Average luciferase activity originating from a circadian transcriptional reporter over days 2 and 3 for control and tim > NAT1RNAi wings in culture (torin, T). (B) Detrended activity averages reveal amplitude differences. (C) Torin increases amplitude in control cultures but not in tim > NAT1RNAi. (n > 15 each condition, *P < 0.05, Mann–Whitney test; error bars are SEM). (D) Addition of cycloheximide showed similar reductions in amplitude for both genotypes, showing that global translation is not impaired in tim > NAT1RNAi.
Because of hints from the literature connecting NAT1 with cap-independent translation (Hundsdoerfer et al. 2005), we attempted to assess the relative contribution of cap-dependent and cap-independent translation on circadian oscillations and more specifically on the role of NAT1. To this end, we tested the effects of the TOR inhibitors torin and rapamycin on these wing cultures. Both chemicals block TOR kinase activation of cap-dependent translation (Thoreen et al. 2009). Their addition to cycling wings might therefore shift translation toward cap-independent activity.
Addition of torin to WT (GFP expressing) wings caused a slight increase in cycling amplitude, suggesting that cap-independent translation potentiates the amplitude of circadian oscillations. Addition of torin had the opposite effect on NAT1 knockdown wings, namely, it caused a significant reduction in cycling amplitude (Figure 6C). An interpretation is that the NAT1 knockdown made circadian translation amplitude more reliant on cap-dependent translation and therefore vulnerable to torin inhibition in a dose-dependent manner. Rapamycin caused similar but less robust effects, consistent with the fact that it is a weaker inhibitor of TOR (data not shown). An implication perhaps is that NAT1 supports cap-independent translation, which is promoted by TOR inhibition.
We also added the translation elongation inhibitor cycloheximide (CHX), which contrasts with torin by inhibiting all translation. Cycloheximide reduced the amplitude of oscillations for both genotypes in a similar dose-dependent manner (Figure 6D). Oscillations also dampened more rapidly in the presence of cycloheximide, but multiple days of cycling still occurred, sufficient to measure the amplitude of oscillations in the presence of the drug.
Untranslated regions of Period confer resistance to TOR inhibition in S2 cells
The above results suggest that NAT1 and oscillator amplitude might both be related to cap-independent translation. To connect these phenomena to per mRNA translation, we generated a number of S2 cell reporter gene constructs containing per 5′- and 3′-UTRs as well as control constructs and addressed their relationship to NAT1 and torin. Although RNAi of NAT1 in S2 tissue culture cells caused slower growth than GFP control dsRNA, growth continued sufficiently to assay the control and per reporter genes.
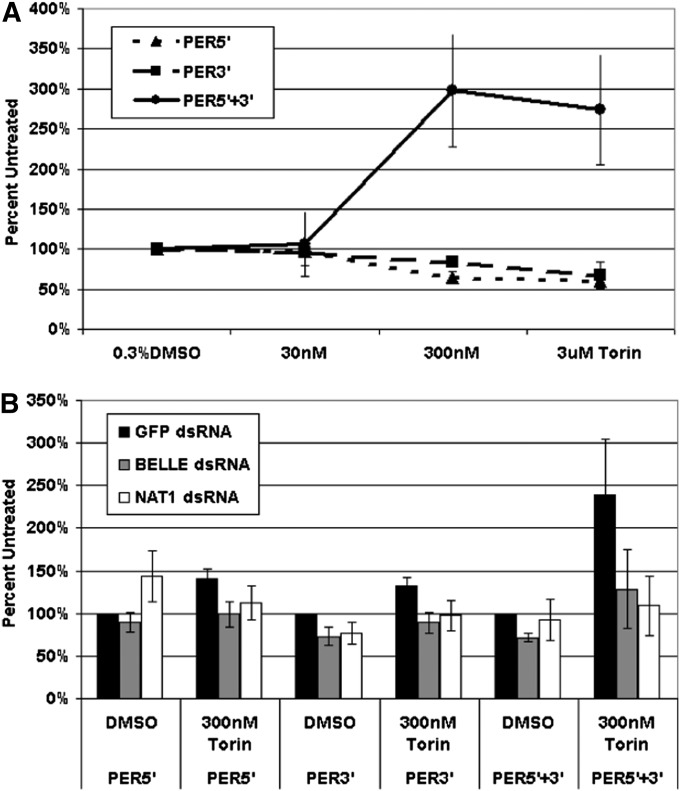
Because inhibition of TOR by torin strengthened circadian oscillations in wings, we guessed that torin addition may show an effect on circadian gene reporters in cell culture. Indeed, a construct bearing per 3′- and 5′-UTRs demonstrated resistance to 300 nM torin. Interestingly, constructs with only one per UTR, either the 5′-UTR or the 3′-UTR, were not resistant (Figure 7A). This suggests that both UTRs collaborate to promote more efficient cap-independent translation.
Figure 7 .
per mRNA untranslated regions (UTRs) act synergistically to support reporter expression under conditions of TOR inhibition. (A) In S2 cell culture, a construct bearing the luciferase coding sequence flanked by per 5′- and 3′-UTRs is relatively immune to repression of cap-dependent translation by torin. (B) RNAi knockdown of NAT1 or bel blocks the protective effects of the per UTR combination in response to torin treatment.
To assay the role of NAT1 and bel in this phenomenon, we combined torin treatment with dsRNA knockdowns against these two genes. The mRNA for each target was reduced at least fivefold (Figure S5). The results were straightforward: the relative immunity to TOR inhibition conferred by the per UTRs was dependent on both NAT1 and BEL (Figure 7B). A simple interpretation is that that both of these proteins as well as both per UTRs collaborate to promote cap-independent translation, which is important for circadian amplitude and perhaps even circadian period.
Discussion
We show that the eIF4G paralog NAT1 supports pacemaker function in flies. It was among the most potent effectors of circadian period changes in an RNAi screen of translation and RNA factors, and depletion of NAT1 decreased oscillator amplitude in both fly head Western blots and a wing-biochemical assay. The effect on circadian period in adult flies can be at least partially explained by reduced PER expression in adult PDF neurons. TOR inhibition in explanted wings caused increased oscillator amplitude, and this effect was dependent on NAT1. In S2 cells, reporter constructs bearing 5′- and 3′-UTRs of per were relatively immune to TOR inhibition. We therefore conclude that PER expression, and perhaps circadian protein expression more broadly, relies in part upon cap-independent translation and the translation factor NAT1. This is especially relevant for adult PDF neurons but also for peripheral clock cells in wings.
NAT1 was discovered by four groups in 1997 (Imataka et al. 1997; Levy-Strumpf et al. 1997; Shaughnessy et al. 1997; Yamanaka et al. 1997) and its coding sequence is highly similar to C-terminal portions of eIF4G. In mouse, it is translated from a GUG codon in a cap-independent fashion and is cleaved under conditions of cellular stress similarly to eIF4G, the protein that normally bridges eIF4E and PABP during translation. Cleavage products of both eIF4G and NAT1 are still able to bind eIF-4A, eIF3, and LK6 (MNK) (Pyronnet et al. 1999), presumably to promote cap-independent translation. Indeed, both cleavage products as well as full-length NAT1 support translation from several cellular IRES elements, including that of NAT1 itself (Henis-Korenblit et al. 2002; Nevins et al. 2003; Hundsdoerfer et al. 2005). Cap-independent translation is often upregulated when cap-dependent translation is reduced. However, full-length NAT1 is present on active polysomes in unstressed cells (Nousch et al. 2007), indicating a role for this factor under normal conditions. Indeed, RNAi against NAT1 during mitosis results in apoptosis, due to translation deficits in BCL-2 and CDK1 production (Marash et al. 2008). Also consistent with a vital role is the fact that NAT1 deficits cause developmental failure, particularly at the formation of mesoderm in several species (Yamanaka et al. 2000; Nousch et al. 2007; Yoshikane et al. 2007).
Adult PDF cells appear particularly dependent on NAT1 function, and translational regulation is more generally important for high-amplitude circadian gene expression oscillations. Within this cluster of neurons, the small PDF cells (s-LNv) have the major impact on circadian period in DD (Grima et al. 2004). Consistent with this important role in maintaining rhythms in DD, the s-LNvs maintain the most robust cycling of core clock proteins under these conditions. Dorsal neurons and large PDF cells maintain lower amplitude clock gene cycling while expression elsewhere in fly heads dampens rapidly during incubation in free-running conditions. The large PDF cells (l-LNv) have a different function: they contribute to light resetting in the late night and probably also play a role in morning locomotor activity (Shang et al. 2008; Zhang et al. 2009b; Sheeba et al. 2010).
Several findings indicate that NAT1 plays a particularly important role in PDF cells: (1) NAT1 knockdown limited to these neurons is sufficient to cause a long-period phenotype. (2) PER staining in PDF cells is dramatically reduced at peak expression times in the NAT1 knockdown, almost certainly more so than in heads by Western blotting. (3) Increased and persistent morning peak activity in DD results from NAT1 overexpression in PDF cells. (4) Overexpression of PER within PDF cells rescues the behavioral effect of the NAT1 knockdown, suggesting that NAT1 potentiates PER translation within these neurons.
We expected that NAT1 overexpression would shorten the circadian period in contrast to the RNAi effect, but the full-length or a short form of UAS-NAT1 in combination with pdf- or tim-GAL4 drivers yielded only weaker rhythms. This is similar to the result reported for TYF, which also supports PER translation in PDF cells (Lim et al. 2011). Flies homozygous for the full-length UAS-NAT1 (four gene copies total), however, did have significantly faster rhythms (23.2 hr) and a persistent morning peak (Figure S3); a truncated version of this transgene missing 5′-UTR regions was without effect. We suggest that NAT1 overexpression is buffered at the level of initiation, perhaps because of its atypical translation as mentioned above. Although most viable NAT1 mutant lines were wild type, one had a short period (23.3 hr) and a slightly pronounced morning peak (Figure S4). This line had normal levels of NAT1 mRNA. Perhaps the P-element insertion in the first intron alters the IRES-containing 5′-untranslated region and thereby increases translation initiation and NAT1 protein levels.
An additional effect of NAT1 knockdown is that late night phase responses are delayed and increased. This is probably related to the light-resetting role of PDF cells mentioned above and could also be the result of a weaker oscillator; low amplitude cycling transcription was shown to produce larger phase shifts in the mouse (Vitaterna et al. 2006). This NAT1 RNAi finding echoes other mouse results, where advance zone phase shifts are augmented by infusion of rapamycin to the SCN (Cao et al. 2010), further supporting a role for translation in late-night advance zone phase shifts. Overexpression of the TOR as well as one of its substrates, S6 kinase, lengthens behavioral period in flies (Zheng and Sehgal 2010). The mechanism by which increased TOR and S6K activity affected circadian behavior was described as via increased phosphorylation of the important circadian kinase shaggy (GSK3), which resulted in delayed TIM phosphorylation and s-LNv nuclear entry. Our results suggest another mechanism, perhaps working in parallel: increased cap-dependent translation resulting from stronger TOR activity inhibits the accumulation of PER and TIM and thereby slows the pace of the oscillator.
This view is also supported by our cultured wings luciferase experiments, where pharmacological inhibition of TOR produced higher-amplitude oscillations originating from the tim promoter. This construct offers a readout of circadian transcriptional activity, where increased amplitude should result from increased expression or activity of core clock genes and correlate with faster behavioral rhythms (Kadener et al. 2008). NAT1 RNAi driven by tim-GAL4 slightly reduced oscillator amplitude in wings. More significantly, it also blocked the torin-induced increase in amplitude (Figure 6C). Torin treatment could increase cap-independent translation of NAT1, and/or NAT1 could be more active when TOR activity is low. In either case, we speculate that PER is the link between TOR, NAT1 activity, and the luciferase reporter. TIM signal was also reduced in PDF cells (Figure 3C), indicating that tim mRNA may also be subject to atypical translation. This notion is consistent with the finding that mRNA for both genes binds to TYF (Lim et al. 2011). Although TIM is required for PER stability (Price et al. 1995), our rescue experiments in PDF cells indicate that PER is probably the NAT1 target most relevant for the behavioral phenotype.
PER appears primarily responsible for circadian transcriptional repression (Menet et al. 2010; Abruzzi et al. 2011), and modulation of per, Clk, or Pdp1 gene copy number has inverse effects on the rate of DD rhythmicity. This indicates that increases in their cycling protein levels cause shorter periods (Baylies et al. 1987; Cyran et al. 2003; Kadener et al. 2008). Recent findings also indicate that deficits in PER expression downstream of PDF cells contribute to slower and weaker behavioral rhythms in older flies (Luo et al. 2012). Our results are similar to those reported for hypomorphic tyf and indicate that reduced rather than delayed PER expression is responsible for the long periods resulting from NAT1 knockdown.
This interpretation was aided by using the untranslated regions of per to examine luciferase reporter expression. Our S2 cell transfection experiments highlight a synergistic effect of the per 5-′ and 3′-UTRs under conditions of TOR inhibition, suggesting that they work together to promote cap-independent translation. Moreover, this effect was abolished by RNAi knockdown of either NAT1 or the translational helicase bel. BEL is suggested to regulate the translation of specific targets during oogenesis (Johnstone et al. 2005; Ambrus and Frolov 2010; Yarunin et al. 2011). Decreased BEL levels in the NAT1 RNAi flies (Figure 3A) could therefore directly cause the PER deficits and the subsequent behavioral phenotype. The absence of a behavioral effect with bel RNAi argues weakly against this possibility.
Although we lack evidence for direct interaction, it is possible that NAT1 is present within initiation complexes containing per or tim mRNA and TYF, which was shown to bind both mRNAs (Lim et al. 2011). Proof of cap-independent activity for either mRNA would have been striking, but the results were negative for both 5′-UTRs in common cell-culture and in vitro assays (data not shown). It is possible that both assays were unable to recapitulate the atypical translation events that occur in vivo due, for example, to the absence of other key circadian factors such as TYF. NAT1 might also act indirectly, for example, by reducing general cap-dependent translation as a dominant-negative initiation factor.
Notable in this context is the dramatic enrichment of mRNA encoding the translational repressor 4EBP in circadian cells (Kula-Eversole et al. 2010; Nagoshi et al. 2010). Considering its role as a major substrate for TOR activity, it may play an important role in sensitizing these cells to a nutrient signaling pathway. NAT1 and 4EBP could indirectly support TYF-assisted translation of PER and TIM by attenuating global cap-dependent translation. This view is consistent with the knockdown effects of the translation inhibition genes PAIP2 (Yanagiya et al. 2010), FMR1 (Deshpande et al. 2006), and eIF5C (Lee et al. 2007), which all increase period length. In contrast, knockdown of the positive elongation factor ef2 speeds up the clock (Figure 1A, Table S1). The other translation inhibitory factors may compensate for the insufficient NAT1 activity in other parts of the fly head, which would explain the only modest reduction in protein levels observed with Western blots in tim > NAT1RNAi flies (Figure 3A).
Although we lack evidence for a direct effect of this translation factor on per or tim mRNA, as was provided recently for TYF (Lim et al. 2011), our work supports a model in which PER is translated in a noncanonical fashion due in part to NAT1 activity, especially in the PDF clock neurons. Utilization of an atypical translation pathway may be related to the fact that these cells are light sensitive and that light drives intracellular chemical changes that may affect translation (Morse et al. 1989; Powley et al. 2009). Cap-independent translation may also permit PDF neurons to remain relatively immune to conditions that downregulate cap-dependent translation, such as nutrient deprivation. This could serve to maintain normal central pacemaker activity during stress conditions. A definitive role for NAT1 within the circadian feedback loop will require identification of its direct mRNA targets, a pursuit that is currently underway.
Supplementary Material
Acknowledgments
We thank Michael Marr for extensive advice and cell culture reagents, members of the Rosbash and Marr labs for thoughtful discussion and experimental support, Ed Dougherty for imaging assistance, and Kristyna Palm-Danish for administrative assistance. FlyBase and the Vienna Drosophila Resource Center also provided invaluable resources. This work was supported by National Institutes of Health T32GM007122 (S.B.) and P01 NS44232 (M.R.).
Footnotes
Communicating editor: R. Anholt
Literature Cited
- Abruzzi K. C., Rodriguez J., Menet J. S., Desrochers J., Zadina A., et al. , 2011. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev. 25: 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allada R., White N. E., So W. V., Hall J. C., Rosbash M., 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93: 791–804. [DOI] [PubMed] [Google Scholar]
- Ambrus A. M., Frolov M. V., 2010. Mutation of the DEAD-box helicase belle downregulates the cyclin-dependent kinase inhibitor Dacapo. Cell Cycle 9: 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylies M. K., Bargiello T. A., Jackson F. R., Young M. W., 1987. Changes in abundance and structure of the per gene product can alter periodicity of the Drosophila clock. Nature 326: 390–392. [DOI] [PubMed] [Google Scholar]
- Cao R., Li A., Cho H. Y., Lee B., Obrietan K., 2010. Mammalian target of rapamycin signaling modulates photic entrainment of the suprachiasmatic circadian clock. J. Neurosci. 30: 6302–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J. C., Ko H. W., Edery I., 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyran S. A., Buchsbaum A. M., Reddy K. L., Lin M. C., Glossop N. R., et al. , 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341. [DOI] [PubMed] [Google Scholar]
- Deshpande G., Calhoun G., Schedl P., 2006. The Drosophila Fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics 174: 1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., et al. , 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156. [DOI] [PubMed] [Google Scholar]
- Glaser F. T., Stanewsky R., 2005. Temperature synchronization of the Drosophila circadian clock. Curr. Biol. 15: 1352–1363. [DOI] [PubMed] [Google Scholar]
- Goda T., Mirowska K., Currie J., Kim M. H., Rao N. V., et al. , 2011. Adult circadian behavior in Drosophila requires developmental expression of cycle, but not period. PLoS Genet. 7: e1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Chelot E., Xia R., Rouyer F., 2004. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431: 869–873. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S., Shani G., Sines T., Marash L., Shohat G., et al. , 2002. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl. Acad. Sci. USA 99: 5400–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Howlett E., Stern M., Jackson F. R., 2009. Altered LARK expression perturbs development and physiology of the Drosophila PDF clock neurons. Mol. Cell. Neurosci. 41: 196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundsdoerfer P., Thoma C., Hentze M. W., 2005. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 102: 13421–13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka H., Olsen H. S., Sonenberg N., 1997. A new translational regulator with homology to eukaryotic translation initiation factor 4G. EMBO J. 16: 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O., Deuring R., Bock R., Linder P., Fuller M. T., et al. , 2005. Belle is a Drosophila DEAD-box protein required for viability and in the germ line. Dev. Biol. 277: 92–101. [DOI] [PubMed] [Google Scholar]
- Kadener S., Stoleru D., McDonald M., Nawathean P., Rosbash M., 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev. 21: 1675–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S., Menet J. S., Schoer R., Rosbash M., 2008. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 6: e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Hall J. C., 2000. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 422: 66–94. [DOI] [PubMed] [Google Scholar]
- Kojima S., Matsumoto K., Hirose M., Shimada M., Nagano M., et al. , 2007. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 104: 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kula-Eversole E., Nagoshi E., Shang Y., Rodriguez J., Allada R., et al. , 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc. Natl. Acad. Sci. USA 107: 13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia K. A., Papp S. J., Yu R. T., Barish G. D., Uhlenhaut N. H., et al. , 2011. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480: 552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Woo K. C., Kim D. Y., Kim T. D., Shin J., et al. , 2011. Rhythmic interaction between Period1 mRNA and HnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol 32: 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H., Kim S. H., Kim D. Y., Kim S., Kim K. T., 2012. Internal ribosomal entry site-mediated translation is important for rhythmic PERIOD1 expression. PLoS ONE 7: e37936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Nahm M., Lee M., Kwon M., Kim E., et al. , 2007. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development 134: 1767–1777. [DOI] [PubMed] [Google Scholar]
- Levine J., Funes P., Dowse H., Hall J., 2002. Advanced analysis of a cryptochrome mutation’s effects on the robustness and phase of molecular cycles in isolated peripheral tissues of Drosophila. BMC Neurosci. 3: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy-Strumpf N., Deiss L. P., Berissi H., Kimchi A., 1997. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol. Cell. Biol. 17: 1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C., Lee J., Choi C., Kilman V. L., Kim J., et al. , 2011. The novel gene twenty-four defines a critical translational step in the Drosophila clock. Nature 470: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li S., Liu T., Borjigin J., Lin J. D., 2007. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature 447: 477–481. [DOI] [PubMed] [Google Scholar]
- Luo W., Chen W. F., Yue Z., Chen D., Sowcik M., et al. , 2012. Old flies have a robust central oscillator but weaker behavioral rhythms that can be improved by genetic and environmental manipulations. Aging Cell 11: 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L., Liberman N., Henis-Korenblit S., Sivan G., Reem E., et al. , 2008. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell 30: 447–459. [DOI] [PubMed] [Google Scholar]
- Marr M. T., 2nd, D’Alessio J. A., Puig O., Tjian R., 2007. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 21: 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K., Davis R. L., 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. [DOI] [PubMed] [Google Scholar]
- Menet J. S., Abruzzi K. C., Desrochers J., Rodriguez J., Rosbash M., 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev. 24: 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D., Milos P. M., Roux E., Hastings J. W., 1989. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc. Natl. Acad. Sci. USA 86: 172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E., Sugino K., Kula E., Okazaki E., Tachibana T., et al. , 2010. Dissecting differential gene expression within the circadian neuronal circuit of Drosophila. Nat. Neurosci. 13: 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins T. A., Harder Z. M., Korneluk R. G., Holcik M., 2003. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 278: 3572–3579. [DOI] [PubMed] [Google Scholar]
- Newby L. M., Jackson F. R., 1993. A new biological rhythm mutant of Drosophila melanogaster that identifies a gene with an essential embryonic function. Genetics 135: 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Reed V., Bryson-Richardson R. J., Currie P. D., Preiss T., 2007. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA 13: 374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley I. R., Kondrashov A., Young L. A., Dobbyn H. C., Hill K., et al. , 2009. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 23: 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J. L., Dembinska M. E., Young M. W., Rosbash M., 1995. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 14: 4044–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S., Imataka H., Gingras A. C., Fukunaga R., Hunter T., et al. , 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18: 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn S. C., Park J. H., Rosbash M., Hall J. C., Taghert P. H., 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- Sancar G., Sancar C., Brügger B., Ha N., Sachsenheimer T., et al. , 2011. A global circadian repressor controls antiphasic expression of metabolic genes in Neurospora. Mol. Cell 44: 687–697. [DOI] [PubMed] [Google Scholar]
- Shang Y., Griffith L. C., Rosbash M., 2008. Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. USA 105: 19587–19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy J. D., Jr, Jenkins N. A., Copeland N. G., 1997. cDNA cloning, expression analysis, and chromosomal localization of a gene with high homology to wheat eIF-(iso)4F and mammalian eIF-4G. Genomics 39: 192–197. [DOI] [PubMed] [Google Scholar]
- Sheeba V., Fogle K. J., Holmes T. C., 2010. Persistence of morning anticipation behavior and high amplitude morning startle response following functional loss of small ventral lateral neurons in Drosophila. PLoS ONE 5: e11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N., Hinnebusch A. G., 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., et al. , 1998. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95: 681–692. [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Lynch K. S., Brandes C., Hall J. C., 2002. Mapping of elements involved in regulating normal temporal period and timeless RNA expression patterns in Drosophila melanogaster. J. Biol. Rhythms 17: 293–306. [DOI] [PubMed] [Google Scholar]
- Stoleru D., Peng Y., Agosto J., Rosbash M., 2004. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431: 862–868. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Maruyama M., Tokuzawa Y., Murakami M., Oda Y., et al. , 2005. Evolutionarily conserved non-AUG translation initiation in NAT1/p97/DAP5 (EIF4G2). Genomics 85: 360–371. [DOI] [PubMed] [Google Scholar]
- Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., et al. , 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284: 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna M. H., Ko C. H., Chang A. M., Buhr E. D., Fruechte E. M., et al. , 2006. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc. Natl. Acad. Sci. USA 103: 9327–9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S., Poksay K. S., Arnold K. S., Innerarity T. L., 1997. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev. 11: 321–333. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Zhang X. Y., Maeda M., Miura K., Wang S., et al. , 2000. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 19: 5533–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagiya A., Delbes G., Svitkin Y. V., Robaire B., Sonenberg N., 2010. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J. Clin. Invest. 120: 3389–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Sehgal A., 2001. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron 29: 453–467. [DOI] [PubMed] [Google Scholar]
- Yarunin A., Harris R. E., Ashe M. P., Ashe H. L., 2011. Patterning of the Drosophila oocyte by a sequential translation repression program involving the d4EHP and Belle translational repressors. RNA Biol. 8: 904–912. [DOI] [PubMed] [Google Scholar]
- Yoshikane N., Nakamura N., Ueda R., Ueno N., Yamanaka S., et al. , 2007. Drosophila NAT1, a homolog of the vertebrate translational regulator NAT1/DAP5/p97, is required for embryonic germband extension and metamorphosis. Dev. Growth Differ. 49: 623–634. [DOI] [PubMed] [Google Scholar]
- Zhang E. E., Liu A. C., Hirota T., Miraglia L. J., Welch G., et al. , 2009a A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139: 199–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Lear B. C., Seluzicki A., Allada R., 2009b The CRYPTOCHROME photoreceptor gates PDF neuropeptide signaling to set circadian network hierarchy in Drosophila. Curr. Biol. 19: 2050–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Sehgal A., 2010. AKT and TOR signaling set the pace of the circadian pacemaker. Curr. Biol. 20: 1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.