Abstract
Gene expression is controlled primarily by transcription factors, whose DNA binding sites are typically 10 nt long. We develop a population-genetic model to understand how the length and information content of such binding sites evolve. Our analysis is based on an inherent trade-off between specificity, which is greater in long binding sites, and robustness to mutation, which is greater in short binding sites. The evolutionary stable distribution of binding site lengths predicted by the model agrees with the empirical distribution (5–31 nt, with mean 9.9 nt for eukaryotes), and it is remarkably robust to variation in the underlying parameters of population size, mutation rate, number of transcription factor targets, and strength of selection for proper binding and selection against improper binding. In a systematic data set of eukaryotic and prokaryotic transcription factors we also uncover strong relationships between the length of a binding site and its information content per nucleotide, as well as between the number of targets a transcription factor regulates and the information content in its binding sites. Our analysis explains these features as well as the remarkable conservation of binding site characteristics across diverse taxa.
Keywords: population genetics, transcription regulation
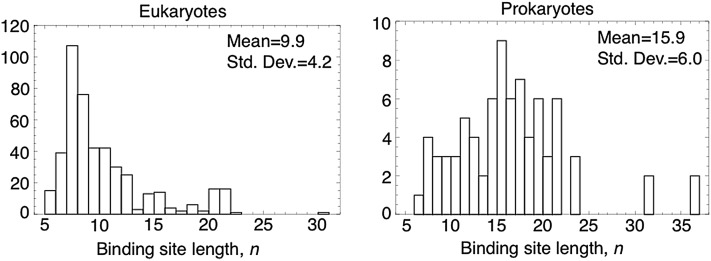
MUCH of the phenotypic divergence between species is driven by changes in transcriptional regulation, and especially by point mutations at transcription factor binding sites (Stern 2000; Carroll 2005; Ihmels et al. 2005; Prud’homme et al. 2006, 2007; Tsong et al. 2006; Wray 2007; Lemos et al. 2008; Tuch et al. 2008a,b). Such mutations can increase or decrease the affinity of a transcription factor protein to its binding sites, which in turn modifies the expression of regulated genes. Binding sites are typically ∼10 nt in length, in both eukaryotes and prokaryotes, although this number varies from as few as 5 to >30 nt (Figure 1). Binding sites are also characterized by their information content (D’haeseleer 2006), which is determined by the number of different bases that can occur at each nucleotide and still produce functional binding. The average information content varies from a maximum 2 bits per nucleotide (each nucleotide must assume a specific base to produce functional binding) to <0.25 bits (each nucleotide can assume one of several bases and still produce functional binding).
Figure 1.
The lengths of binding sites range from 5 nt to ∼30 nt, in both eukaryotes (left, 454 curated transcription factor motifs) and prokaryotes (right, 79 motifs). The information content per nucleotide ranges from ∼0.25 bits to 2 bits (see Figure S1).
What determines the length and information content of a transcription factor binding site? Biophysical factors provide some constraints, and numerous studies have explored their effects on the function of individual transcription factor binding sites (Gerland et al. 2002; Berg et al. 2004; Bintu et al. 2005; Shultzaberger et al. 2007; Mustonen et al. 2008; Gerland and Hwa 2009). Natural selection also plays an important role because, to produce the correct patterns of gene expression, transcription factors must correctly bind to some sites in the genome and avoid binding elsewhere (Sengupta et al. 2002; Shultzaberger et al. 2012; Gerland and Hwa 2002). If binding sites are too short, transcription factors bind too readily to nondesirable genomic locations, which disrupts gene expression and depletes the pool of transcription factors available to bind where they are needed. If binding sites are too long, on the other hand, sites where binding is favored are too easily disrupted by mutation. This inherent trade-off between specificity and robustness is the focus of our analysis. We adopt an evolutionary perspective and explore the effects of mutations that alter the length or information content of binding sites.
Much previous work has focused on biophysical aspects of transcription factor binding, such as how the binding energy changes with individual nucleotide substitutions and with the concentration of free transcription factor proteins in a cell. By specifying binding site fitness in terms of binding energy, these models are able to reconstruct quantitative fitness landscapes that can be used to study the evolution of transcription regulation. This has led to remarkable success in explaining such important features of transcription factor binding as the total binding energy associated with a functional binding site (Gerland et al. 2002; Berg et al. 2004) and the average change in binding energy with individual mutations (Gerland et al. 2002; Gerland and Hwa 2002) as well as some features of position weight matrices (PWMs) (Gerland et al. 2002; Sengupta et al. 2002; Moses et al. 2003; Lusk and Eisen 2008; Shultzaberger et al. 2012). It has also led to estimates for the strength of selection on functional binding sites (Hahn et al. 2003; Mustonen et al. 2008; He et al. 2011). We make use of much of this information to construct our model. However, we are principally concerned with the population genetics of transcription factor binding across a large number of sites. Therefore we specify our model in terms of aggregate properties of transcription factor binding sites derived from biophysical models, without specifying the mechanistic biophysical details that give rise to the selection coefficients we use. Nonetheless, we also verify the results of our simplified evolutionary models by comparison with simulations of detailed, biophysical models of transcription factor binding.
Materials and Methods
Biophysical model
We begin by introducing a biophysical model of transcription factor (TF)–DNA binding, derived from the literature. Detailed models of this type have been used extensively to study the properties and evolution of small numbers of binding sites (Gerland et al. 2002; Moses et al. 2003; Berg et al. 2004; Bintu et al. 2005; Sella and Hirsh 2005; Lässig 2007). These models focus on the probability that a binding site is bound by a transcription factor, which is given in terms of the properties of the binding site and in terms of the number of free transcription factor proteins available to bind to it in the cell. A binding site is assumed to consist of n contiguous nucleotides. Indexing the nucleotide positions by i ∈ {1, 2, . . . , n} and the bases by α ∈ {A, C, G, T}, the contribution to the binding energy of the site by a base α at nucleotide i is , and the total binding energy of the site is (units of kBT) (Gerland et al. 2002; Lässig 2007). Typically there are “preferred” bases at each position i that contribute 0 to binding energy (and hence make it more likely that the site is bound by a transcription factor). The number of different preferred bases, ri, is called the degeneracy at position i. All other, nonpreferred bases increase binding energy by an amount ε between 1 and 3 kBT (Gerland et al. 2002; Lässig 2007). The total binding energy of a binding site that contains m preferred bases is given to good approximation by E = ε(n − m).
A combination of empirical studies and basic thermodynamic principles has shown that, in a cell containing P free transaction factors, the probability that a binding site containing m preferred bases is bound, πm, is given by
| (1) |
Using Equation 1, the evolution of a single binding site can be studied by making the natural assumption that the fitness of a site is a linear function of the probability that it is bound. At a genome position where selection favors transcription factor binding, the fitness, wm, of a site containing m preferred bases is given by wm = 1 − s+(1 − πm), where s+ is the fitness penalty associated with the site being unbound. Similarly, at a genome position where selection disfavors transcription factor binding, the fitness of a site containing m preferred bases is given by wm = 1 − s−πm, where s− is the fitness penalty associated with the site being bound.
A simplified model
In this article we are interested in the whole set of genome positions associated with a given transcription factor that are under selection either to promote binding (true targets) or to prevent it (false targets). Such a view takes in hundreds of thousands of genome positions at once, and this makes keeping count of the number of correctly matched, preferred nucleotides at each site impractical. Therefore, to gain insight, we make a number of simplifying assumptions to reduce the complexity of the detailed biophysical model, while still retaining its essential properties.
The probability that a site with m preferred nucleotides is bound, given by Equation 1, is a sigmoidal function with threshold-like behavior, where the threshold occurs at . This means that binding sites with m > mth tend to have high binding probability (i.e., close to 1) and binding sites with m < mth tend to have low binding probability (i.e., close to 0). Our first simplifying assumption is to approximate the sigmoidal function given in Equation 1 by a step function with threshold at mth. For parameter values in the empirical range P = 100−103 and taking ε ≈ 2 (Table 1), this gives an approximate range for of 0−3. Given this, and to facilitate ease of comprehension in what follows, we take in our simplified model, which corresponds to the case in which a single mismatched nucleotide results in a nonfunctional binding site. The results of our simplified analysis are later confirmed by detailed simulations using the full biophysical model outlined in this previous section.
Table 1. Parameters and range of empirical estimates for our population-genetic model of binding site evolution.
| Parameter | Meaning | Value range |
|---|---|---|
| μ | Per-nucleotide per-generation mutation rate | 10−7−10−9 per generation (Drake et al. 1998; Lynch 2010) |
| K | Number of true targets: genomic positions at which selection favors a functional binding site | 100−103 binding sites (Lee et al. 2002; Münch et al. 2003; Harbison et al. 2004; Teixeira et al. 2006; Gama-Castro et al. 2011) |
| L | Number of false targets: genomic positions at which selection disfavors a functional binding site | 105−107 binding sites (for a genome with 103−105 genes) (Lynch and Conery 2003) |
| n | Binding-site length | 5−40 nt (Münch et al. 2003; Harbison et al. 2004; Bryne et al. 2008; Gama-Castro et al. 2011) |
| r | Binding degeneracy: the average number of different bases at each nucleotide that produce functional binding | 1.0−4.0 bases (empirical mean = 1.6 bases) (Münch et al. 2003; Harbison et al. 2004; Bryne et al. 2008; Gama-Castro et al. 2011) |
| I | Average information content per nucleotide in a binding site, | 0.25−2 bits (empirical mean = 1.3 bits) (Münch et al. 2003; Harbison et al. 2004; Bryne et al. 2008; Gama-Castro et al. 2011) |
| N | Effective population size | 104−107 individuals (Lynch 2010) |
| Ns+ | Strength of selection on true targets | ∼10 (Mustonen et al. 2008; He et al. 2011) |
| Ns− | Strength of selection on false targets | ≲1 (Hahn et al. 2003) |
| P | Number of TF proteins in a cell | 100−103 (Thattai and Van Oudenaarden 2001; Gerland et al. 2002) |
| ε | Mismatched nucleotide binding energy contribution | 1−3 (units of kBT) (Gerland et al. 2002; Lässig 2007) |
Mutation rates
In the detailed biophysical model outlined above, mutations are simple to describe. For a per-nucleotide mutation rate μ, the rate of mutations from a preferred base to a nonpreferred base at a nucleotide i is , where ri is the degeneracy at position i. Similarly, the rate of mutations from a nonpreferred base to a preferred base is . In our simplified model, we treat binding sites as either functional or nonfunctional. As described above, a functional binding site is rendered nonfunctional when one of its n nucleotides mutates to one of the disallowed bases. At true targets, where binding is favored, functional sites are rendered nonfunctional at a rate u−, given by . Writing this in terms of the average degeneracy , the rate at which a functional true target experiences a deleterious mutation, which results in loss of binding, can be written as
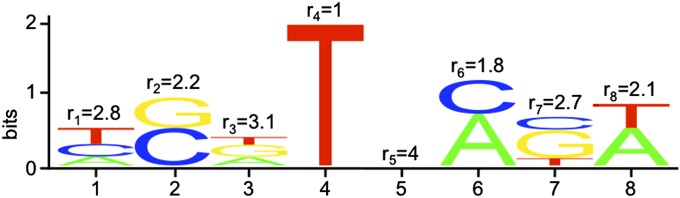
The relationship between this rate and the PWM for a given transcription factor binding site is described in Figure 2. Next we compute the rate of mutations that render nonfunctional binding sites functional at true targets, u+. The rate at which nonfunctional true targets experience gain-of-function mutations is proportional to the probability that a nonfunctional sequence is one mutation away from functional. Assuming Ns+ > 1, this probability is ∼1 for true targets, and the average rate of mutations from a nonpreferred base to a preferred base at such sequences is . Thus
Figure 2.
The rate of deleterious mutations at a binding site can be calculated directly from a PWM. An arbitrary PWM logo is shown, for a binding site of length n = 8. The x-axis shows nucleotide position, and the y-axis shows the total information contained in that nucleotide. The relative heights of the different letters (A, C, G, or T, corresponding to the different DNA bases) correspond to the relative frequency with which they occur in functional binding sites. The value of ri at each position gives the degeneracy, i.e., the average number of different bases that occur at that nucleotide in a functional binding site, calculated from the relative frequency with which each base is found in a functional site at that position. A deleterious mutation occurs when a disallowed nucleotide occurs at one of these positions. The total rate of deleterious mutations for the PWM, u−, can be determined by counting the total number of deleterious mutations that can occur at each nucleotide; i.e., , where is the average degeneracy across the PWM.
For false targets, where binding is disfavored, functional sites are rendered nonfunctional at a rate v− that is identical to that for true targets; i.e.,
However, the rate at which nonfunctional sites are rendered functional at false targets, v+, differs from the rate at true targets. Because they are typically under weak selection, Ns− ≤ 1, evolution at false targets is nearly neutral. Under neutral evolution the probability that a nonfunctional sequence is one mutation away from functional is approximately given by , while the rate of mutations at such sequences that result in a functional binding site is . Thus
Note that v+ depends on the geometric mean of the degeneracy ri at each nucleotide i across the binding site. For simplicity we have assumed that all nucleotides have the same degeneracy r, so that the geometric and arithmetic means are identical. This assumption is relaxed in simulations of the full biophysical model below. Using these rates, we construct a population-genetic model for the evolution of a large number of true and false targets, in terms of the properties of a transcription factor binding site.
Parameters of the population genetic model
We describe the evolution of binding-site properties, for a given transcription factor, using the Wright–Fisher model from population genetics. We consider a population of N individuals with a per-nucleotide mutation rate μ. We characterize the transcription factor by the number of its “true targets” (genomic positions at which binding by the factors is selectively favored), denoted K, and the number of its “false targets” (genomic positions at which binding is selectively disfavored), denoted L. There is some fitness detriment to an individual, s+, that results from nonfunctional binding at a true target, and there is another fitness detriment, s− that arises from functional binding at a false target. The binding motif of the transcription factor is described by its length n and the average degeneracy per nucleotide, r (see Table 1). Functional binding occurs when each one of n consecutive nucleotides assumes one of the r allowed bases. Thus if r > 1, more than one base is allowed at some nucleotide positions, corresponding to a PWM for which multiple sequences give rise to functional binding (Figure 2). The associated information content per nucleotide, , provides an intuitive way to quantify the specificity of each nucleotide: greater degeneracy corresponds to lower information per nucleotide.
In addition to our analytical results for this model (presented below), we performed Monte Carlo simulations of the full Wright–Fisher model for populations of N individuals. Each individual is described by the number of functional true targets i and false targets j it harbors. Code for all simulations was written in C or C++. Parameter values were chosen from the empirical ranges given in Table 1. Functional true targets mutated to nonfunctional at rate u− and nonfunctional true targets mutated to functional at rate u+. Similarly, functional false targets mutated to nonfunctional at rate v− and nonfunctional false targets mutated to functional at rate v+. The number of mutations assigned to an individual each generation, at its true and false targets, is Poisson distributed with the mutation rates described above.
Our analytical and simulation results are compared to systematic data sets on the properties of transcription factor binding sites, primarily in yeast and Escherichia coli, but also including data from a number of multicellular eukaryotes, from Drosophila to humans.
Data sets
To investigate the relationships between binding-site length and per-nucleotide information content we employ systematic data sets of transcription factors and their associated motifs from a wide range of eukaryotic and prokaryotic species. For binding-site lengths and information content in eukaryotes we used the JASPAR CORE database, which contains binding motifs from 454 eukaryotic transcription factors (Bryne et al. 2008). The JASPAR CORE database is a curated, nonredundant set of binding profiles from published articles. Binding motifs are derived from published collections of experimentally defined transcription factor binding sites for multicellular eukaryotes. Binding sites were determined either in SELEX experiments or by the collection of data from the experimentally determined binding regions of actual regulatory regions. Some of the data also arise from high-throughput techniques, such as ChIP-seq. We used data from all eukaryotes together, as well as from vertebrates and fungi separately (as shown in Figure 3).
Figure 3.
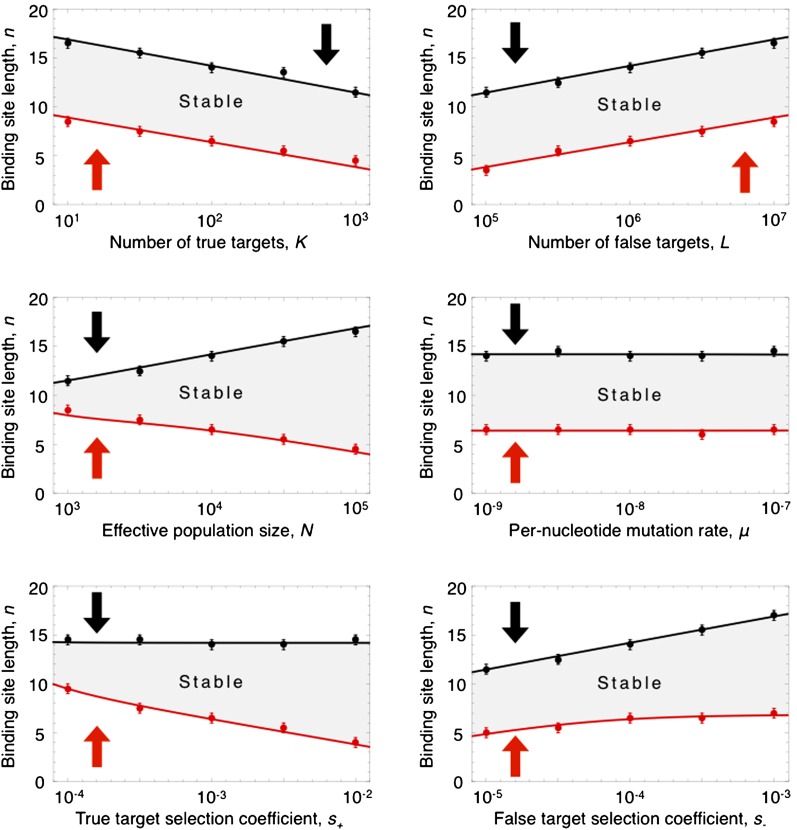
A simple population-genetic model predicts an evolutionary stable region of binding-site lengths, ranging from ∼5 nt to ∼20 nt. The stable region is largely insensitive to varying model parameters (number of true targets, number of false targets, population size, mutation rate, and selection coefficients) by three orders of magnitude. Default parameter values were derived from the literature (Table 1) and correspond to the midpoint of each x-axis. Solid circles denote the stable region determined by Monte Carlo simulations, and lines denote analytical approximations (Equations 4 and 5); arrows indicate the direction of selection outside the stable region.
For the binding site lengths and information content in prokaryotes we used data from the binding motifs of 79 prokaryotic transcription factors contained in the PRODORIC database (Münch et al. 2003). PRODORIC contains information on >2500 transcription factor binding sites identified from direct experimental evidence extracted from the scientific literature. We computed binding motifs based on multiple instances of a given transcription factor binding site, including only transcription factors with >10 known binding sites in the data set, to provide a meaningful estimate of the information content per nucleotide. The distributions of binding-site lengths for both prokaryotes and eukaryotes are shown in Figure 1 and the distributions of information content per nucleotide are shown in Supporting Information, Figure S1.
For two species we also studied relationships involving the number of true targets of each transcription factor. In this case, for Saccharomyces cerevisiae we derived the number of true targets associated with individual transcription factors from a genome-wide study of regulatory interactions (Lee et al. 2002; Harbison et al. 2004) (in Lee et al. 2002, using a cutoff P < 0.001). For E. coli true targets were assigned based on regulatory interactions taken from the curated database RegulonDB (Gama-Castro et al. 2011), which pools published data on transcription regulation in E. coli.
Results
Equilibrium distributions for true and false targets
We analyze the evolution of binding-site length and information content by considering the fate of mutations to the transcription factor that alter n or r, introduced into a population at equilibrium. Using the mutation rates u+, u−, v+, and v− for true and false targets, we can calculate the equilibrium number of functional binding sites, for a transcription factor of given length n and degeneracy per nucleotide r.
Several previous studies have looked at the population genetics of individual transcription factor binding sites (Moses et al. 2003; Berg et al. 2004; Sella and Hirsh 2005). Because binding sites are typically short (n ∼ 10), and per-nucleotide mutation rates are typically low (μ ∼ 10−8), the evolution of a single binding site in isolation can be analyzed under the weak mutation limit; i.e., Nu− ≪ 1. In this limit, a population is generally monomorphic, and evolution proceeds by a series of selective sweeps, with each mutation going to fixation with a probability given by Kimura’s famous equation (Kimura 1962). However, in this study we are concerned with the evolution of a large ensemble of transcription factor binding sites, and as a result the weak selection limit does not typically hold (i.e., the conditions Nu−K ≪ 1 and Nv+L ≪ 1 do not hold in general).
To calculate the equilibrium distribution of functional binding sites we assume that fitness is multiplicative across binding sites, and we apply standard methods (Woodcock and Higgs 1996; Krakauer and Plotkin 2002) to derive the equilibrium number of functional true targets, denoted a+K, and the equilibrium number of functional false targets, denoted a−L. These results are derived, following previous studies (Woodcock and Higgs 1996), using a finite N approximation to a quasi-species–type model. This allows us to consider the regimes Nu−K > 1 and Nv+L > 1, in which a large number of genome positions are under selection.
Under the assumption of independent fitness effects across sites, the fitness of an individual with i functional true targets and j functional false targets is . By generalizing standard techniques for multiplicative fitness landscapes we can compute the equilibrium proportion of true targets that have functional binding:
| (2) |
The first term in Equation 2 can be understood as being due to genetic drift and the second term as due to selection. In the limit N → ∞, the frequency of functional binding sites at true targets is , which agrees with the standard mutation–selection balance (Ewens 2004). A similar computation, valid in the regime Ns− ≤ 1 (see Hahn et al. 2003), provides the equilibrium proportion of false targets that are functional:
| (3) |
The mean population fitness is then (see Appendix for full derivations and complete expressions in other regimes), where the first term reflects the fitness detriment due to true targets that lack functional binding and the second term reflects the fitness detriment due to false targets that have functional binding.
Invasibility of mutants with an altered PWM
We determined whether a mutation that changes binding-site length or information content can invade a population in equilibrium. A new mutant that alters n or r can invade a population only if its fitness exceeds the mean population fitness . Because n and r are properties of the transcription factor protein itself, mutations that alter them have the potential to effect transcription factor binding at all true and false targets simultaneously. We consider the fitness associated with each of the following types of mutations: mutations that increase binding-site length, n; mutations that decrease binding-site length, n; mutations that increase the degeneracy per site, r; and mutations that decrease the degeneracy per site, r.
Mutations that increase n or decrease r will destroy a functional binding site with some probability, p. Therefore, the expected fitness of such a mutant is given by
| (4) |
Likewise, mutations that decrease n or increase r will convert a nonfunctional binding site into functional with some probability, q. Therefore the expected fitness of such a mutant, w*−, is given by
| (5) |
Invasibility criteria for the different types of mutations are determined by finding values of n and r such that and . Note that the invasibility criteria are independent of p and q. The result is a stable region in which mutations that change n or r are deleterious. Outside this region selection favors mutations that bring n and r closer to the stable region (Figure S2).
Region of stable binding site lengths
Figure 3 shows the invisibility criteria for mutations that alter binding-site length, for fixed information content. We find that mutations that increase binding-site length can invade only when n is small, whereas mutations that decrease n can invade only when n is large. Thus there is an intermediate, evolutionarily stable region in which mutations that neither increase nor decrease n can invade.
Remarkably, the size and position of the stable region depends very weakly on the number of true and false targets, the mutation rate, the strength of selection on each site, and the population size. In fact, Figure 3 shows that varying each of these parameters over three orders of magnitude consistently produces a stable region centered at ∼10 nt and ranging from a minimum of 5 nt to a maximum of 20 nt—in close agreement with the range of motif lengths observed in a systematic data set of 454 eukaryotic transcription factors and 79 prokaryotic transcription factors (Figure 1). Thus, our simple population-genetic model for the trade-off between specificity and promiscuity explains the range of binding-site lengths observed in nature, largely independent of the underlying genomic and population parameters.
We performed simulations to confirm that our analytical results on the invasibility criteria are accurate. The fitness effects for mutations that increase or decrease binding-site length in a population at equilibrium were determined for a range of n through detailed simulations of the Wright–Fisher model. Populations were evolved according to the Wright–Fisher model for a fixed binding-site length, n, and degeneracy, r, until they reached equilibrium (typically 106−108 generations). The evolved populations were then subjected to mutations that altered their binding-site length or degeneracy, and the fitness of the mutants was determined and compared to the equilibrium mean fitness of the population. The mean and variance of fitness effects of such mutations were determined for a range of values of n and r and used to numerically determine the invasibility criteria for different parameter choices. The resulting numerically determined stable regimes were compared to our analytical results (Figure 3).
Skew toward long binding sites
The empirical distribution of binding-site lengths (Figure 1) is further characterized by a long tail of transcription factors with large binding sites (several factors have motifs of length >20 nt), but a sharp cutoff at the minimum size of 5 nt. To understand this empirical observation we investigated the strength of selection on binding sites that are too short (i.e., below the stable region) and on binding-site lengths that are too long (i.e., above the stable region), finding that selection against overly short binding sites is much stronger (typical selective coefficient <0.1) than selection against overly long binding sites (typical selective coefficient ∼0.9, see Figure S2). Figure S2 shows the values of n at which mutations that increase n, or decrease n, become deleterious. Figure S2 also shows the magnitude of the fitness effects of mutations that alter binding-site lengths: changes to binding-site length have a much greater impact on fitness when binding sites are too short than when they are too long. Thus, both the shape of the evolutionary stable region under our model and the selection pressures on motifs outside this region help to explain the skew toward long binding motifs observed in eukaryotes and prokaryotes.
Information content and binding-site length
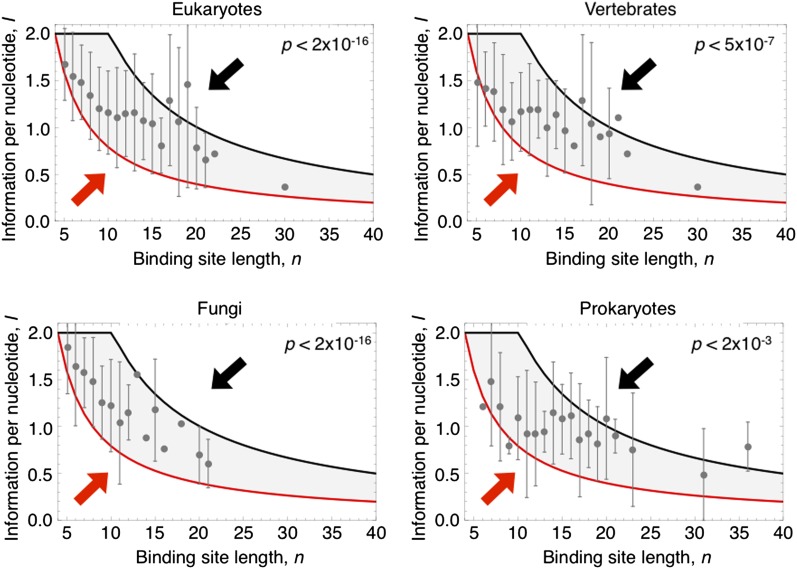
We also discovered other important relationships in the data set of empirical binding-site motifs, including a strong negative correlation between the length of a binding site and its information content per nucleotide (Figure 4, P < 2 × 10−16 in eukaryotes and P < 2 × 10−3 in prokaryotes). To understand this trend, we used our model to compute the region of stability when both binding-site length, n, and degeneracy, r, are allowed to mutate and coevolve. The resulting stable region agrees closely with the empirical data (Figure 4). In other words, even though both n and r are independent variables, they coevolve such that the predicted region of stability and empirical data both exhibit an inverse relationship between binding-site length and information per nucleotide. This result can be understood intuitively: for any binding site, the same degree of overall specificity can be achieved by simultaneously increasing binding-site length n and decreasing information content per nucleotide I. Hence there is a negative correlation between information content per nucleotide and binding-site length, although the predicted relationship is not simply linear (Figure 4).
Figure 4.
Binding motifs that are longer tend to exhibit less information content per nucleotide in a systematic empirical data set (solid circles show the mean ± 2 SD of information in motifs binned by length, for 454 eukaryotic transcription factors, 130 vertebrate factors, 177 fungal factors, and 79 prokaryotic factors). The empirical relationship between length and information content agrees with the evolutionary stable region predicted by our model. Lines denote analytic equations (Equations 4 and 5), and arrows indicate the direction of selection on length and information content outside the stable region.
Our results on the stable region of n and r are remarkably insensitive to parameter choice, as shown in Figure 3. We estimated parameters from empirical data (Table 1) and then allowed each parameter to vary by an order of magnitude to either side of the empirically estimated value. Here we show the behavior of the stable region under further variation in these parameters. Figure S3 shows the stable region when we alter the ratio of false to true targets, L/K, from those otherwise assumed. This is expected to alter the invasibility criteria of the system as described above. Although it does alter the position of the stable region, there is still good agreement with the empirical data on binding-site length and information content. Figure S3 also shows the stable region when we vary the strength of selection on true and false targets. Even when both true and false targets are under weak selection or when they are both under strong selection, the predicted stable region is still in good agreement with the empirical data.
Transcription factor specificity and number of true targets
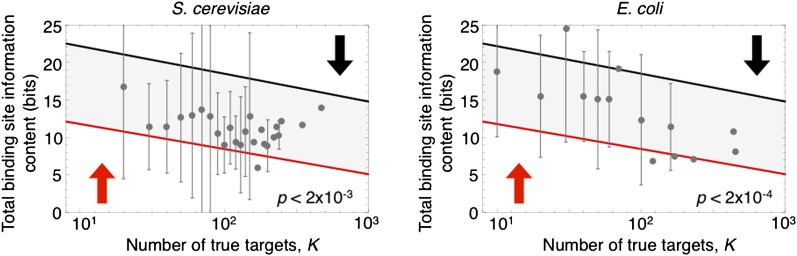
We also analyzed systematic data on the transcription networks of S. cerevisiae and E. coli and discovered that transcription factors with a larger number of true targets tend to have less total information content (I ⋅ n) in their binding sites (Figure 4, P < 2 × 10−3 for S. cerevisiae and P < 2 × 10−4 for E. coli). In other words, factors that serve as important master regulators of many genes achieve this by increased promiscuity of binding. We compared these empirical trends to the region of stability predicted by our model, for a range of values K, and once again found good agreement (Figure 5). This result can also be understood intuitively in the context of our population-genetic analysis: when there are more genomic positions at which selection favors transcription factor binding (i.e., more true targets), there are a greater number of nonfunctional true targets present in the equilibrium population. As a result, the mutational load due to nonfunctional true targets is increased relative to the load arising from functional false targets (Equations 4 and 5). This in turn favors binding sites with lower total information content.
Figure 5.
Transcription factors that regulate a greater number of target genes exhibit less total information content in their binding motifs, in both S. cerevisiae (116 factors) and E. coli (60 factors). This empirical relationship is predicted by the stable region under our model, as we vary the number of true targets. Lines denote analytic equations (Equations 4 and 5), and arrows indicate the direction of selection on I × n outside the stable region.
Evolutionary simulations
We performed evolutionary simulations to confirm our analytical results on the stable region of binding-site lengths and information content. We performed Wright–Fisher simulations in which binding-site length n and degeneracy r were themselves allowed to evolve and reach equilibrium: i.e., both quantities were heritable and subject to mutation. We first performed simulations with fixed r and allowed n to evolve. We chose values of p and q for mutations that alter n as outlined above. A typical set of simulation results is shown in Figure S4. Figure S4 shows that populations that start with binding-site lengths either above or below the stable region do indeed evolve binding-site lengths inside the stable region. These results also show that, as expected from Figure S2, short binding sites increase in length much more rapidly than long binding sites decrease in length, due to stronger selection toward the stable region. The results also show that populations evolve binding sites inside the stable region, rather than at the edge, as might be expected. This is because the invasibility criteria are derived for the effects of mutations in a population at equilibrium; however, in simulations populations experience deviations from equilibrium as their binding-site lengths evolve. Nonetheless the stable region determined by the invasibility criteria provides a good estimate of the binding-site length that will eventually evolve.
We also performed simulations in which both n and degeneracy r were allowed to evolve simultaneously. We chose values of p and q for the invisibility criteria as outlined above. The results of these simulations are shown in Figure S5. These results show that, for a range of initial conditions, populations outside the stable region will evolve toward a binding-site length and information content per nucleotide inside the predicted stable region. Figure S5A shows sample trajectories (averaged over 102 simulations) for a range of different starting conditions. Figure S5B shows the resulting distribution of binding-site length and information content per nucleotide for 102 simulations each with an initial condition chosen uniformly from the range 1 ≤ n ≤ 20 and 1 ≤ r < 4. The distribution qualitatively matches that seen in empirical data and matches the predicted stable region well.
Sexual reproduction
We performed modified Wright–Fisher simulations with limited sexual reproduction. A new offspring was produced by sexual reproduction with probability ρ and as the result of asexual reproduction with probability 1 − ρ. When an offspring was produced through sexual reproduction, a pair of parent individuals were chosen according to their fitness. The offspring then inherited a binding site from either parent with equal probability. Thus the expected Hamming class of the offspring was the average of the Hamming classes of its two parents. We determined the invasibility criteria for different values of ρ (Figure S6). These show that increasing the rate of sexual reproduction tends to increase the range of stable binding-site lengths, compared to the asexual case.
Simulations of the biophysical model
Our results up to this point have been presented for a simplified model based on aggregate properties of an ensemble of transcription factor binding sites across a genome. However, to test the validity of our results, it is important to verify them in the context of the more detailed biophysical model of transcription factor binding, from which our model is derived (Gerland et al. 2002; Sengupta et al. 2002; Berg et al. 2004; Bintu et al. 2005; Mustonen et al. 2008; Gerland and Hwa 2009). Because it is not possible to determine analytically the equilibrium distribution of mutations for this complicated model, we determined the invasibility criteria from Monte Carlo simulations. Populations of N individuals were evolved according to a Wright–Fisher process with fitness function and mutation process as described above. Populations were evolved until they reached equilibrium (typically 106−108 generations) with fixed n and r. The populations were then subjected to mutations that alter n and r, as in the simple model, to determine their fitness effect compared to the equilibrium mean fitness of the population. The resulting numerically determined invasibility criteria for n (with fixed r) are shown in Figure S7. Figure S7 shows good qualitative agreement with the results of the simple model (Figure 3). The detailed TF–DNA binding model with parameter choices within the empirically observed range continues to produce a stable region in good agreement with the observed distribution of binding-site lengths in prokaryotes and eukaryotes (Figure 1). This suggests that our results are largely insensitive to the details of the fitness landscape associated with a particular binding site, and it justifies our use of an aggregate model to describe the evolution of an ensemble of transcription factor binding sites.
To further test the validity of our results under the full biophysical binding model we ran Wright–Fisher simulations in which n and r were allowed to evolve. The fitness effects of such mutations were determined as follows. Mutations that increased n were assumed to add a random nucleotide to each binding site, such that each binding site gained a mismatched nucleotide with probability . The expected number of true targets that suffered a decrease in fitness was Kp and the expected number of false targets that suffered an increase in fitness was Lp. Mutations that decreased n were assumed to delete a random nucleotide from each binding site, such that a binding site with i matched nucleotides lost a mismatched nucleotide with probability . The expected number of true targets that suffered an increase in fitness was Kq and the expected number of false targets that suffered a decrease in fitness was Lq. Similarly mutations that decreased r at a given nucleotide by 1 caused a binding site with i correctly matched nucleotides to gain a mismatched nucleotide with probability and mutations that increased r at a given nucleotide by 1 caused a binding site with i correctly matched nucleotides to lose a mismatched nucleotide with probability . Figure S8 shows the fitness effects of mutations that increase or decrease n for fixed r and shows that, just as for the simple model, such mutations have a greater impact on the fitness of short binding sites than on that of long ones. Figure S9 shows the results of 102 replicate simulations each with an initial condition chosen uniformly from the range 1 ≤ n ≤ 20 and 1 ≤ r < 4. The distribution qualitatively matches that seen in empirical data and matches the stable region predicted by the simple model well.
Discussion
This study reveals that a simple trade-off—the need for transcription factors to be neither too specific nor too promiscuous—can explain the distribution of transcription factor binding-site lengths observed in both eukaryotes and prokaryotes. Our analysis also explains correlations observed among the characteristic features of transcription factors, such as the number of desired target genes, binding-site length, and information content. Our model considers the evolution of binding motifs through mutations that affect transcription factor proteins and change their binding-site length or information content. Other kinds of mutations at the level of the transcription factor can also occur, for example changes to the binding energy per nucleotide, ε, or to the number of proteins per cell, P (Gerland et al. 2002). However, such mutations are not subject to the same trade-off that is the focus of this article.
Our results are derived for an aggregate model, in which binding sites mutate between functional and nonfunctional and selection favors functional sites at true targets and nonfunctional sites at false targets. Although this approach neglects many of the fine details of protein–DNA binding, simulations using a more detailed biophysical model confirm our results, perhaps further reflecting the insensitivity of our results to underlying model parameters (Figure 3).
Among other parameters, our model was phrased in terms of a single fitness detriment incurred when a transcription factor binds to a false target in the genome. There are, in reality, two distinct sources of such a fitness deficit. On the one hand, binding at false targets can be deleterious because it disrupts gene expression at that target gene. On the other hand such binding can also deplete the pool of free transcription factor proteins available to bind at true targets and thereby disrupt the expression at other target genes (Lässig 2007). Although these two mechanisms of deleterious transcription factor binding are clearly distinct, the ultimate effects are similar: some target genes have their expression disrupted. Nonetheless, these two sources of fitness detriment might be studied separately in more complicated models that account for the cellular abundance of the transcription factor protein.
In reality different binding sites have a wide range of occupancies across the genome. In our model binding sites with intermediate binding strength are assumed to have intermediate fitness, since maximum fitness occurs for maximum binding strength at true targets and for minimum binding strength at false targets. Depending on the strength of selection and population size, our model predicts binding sites with a range of occupancies will coexist across the genome. However, it may be that in many cases the optimal binding strength of a true target (which gives maximum fitness) is less than the maximum achievable binding strength. Our model does not account for such a case. It is reasonable to think that when intermediate binding strengths are favored, the central trade-off studied in this article (between specificity and robustness) will still be important. However, the size and positions of the stable regions predicted by our model (Figure 3) will likely be different, if selection for intermediate binding strengths were included.
Our model does a reasonably good job of explaining the ranges of binding-site lengths and specificities found in both prokaryotes and eukaryotes. However, there are noticeable differences in these distributions between prokaryotes and higher eukaryotes; in particular, prokaryotic binding sites tend to be longer on average (Figure 1). The reason for this difference is likely due to the frequent use of complex regulatory modules and cooperative binding in eukaryotes, which tend to favor shorter transcription factor binding sites (Lässig 2007). Although we consider the ensemble of binding sites associated with a particular transcription factor, we do not make any attempt to treat the coevolution of sets of binding sites for different transcription factors that occurs in a functional module. The results of our model therefore relate most directly to the simple, unicellular organisms such as yeast and E. coli, where binding sites tend to act more independently. These are also the species for which we have the most complete data. However, it is interesting that, despite the major differences in the regulatory structure of higher eukaryotes, the trade-off between specificity and robustness still appears to constrain the distribution of transcription factor binding sites in these species, as they still fall within our predicted stable regions. This may be a further reflection of the insensitivity of our results to the underlying details of the fitness landscape associated with individual binding sites. However, the underlying fitness landscape still has important effects not accounted for by our model, which is likely reflected in the tendency of eukaryotes to have shorter, lower information content binding sites than prokaryotes have.
Higher eukaryotes also undergo sexual reproduction and recombination, which were excluded from most of our simulations. Sexual reproduction may be expected to have an impact on our results, since it acts to purge deleterious mutations from the genome and hence mitigate the effects of both low specificity and low mutational robustness, the trade-off that is the focus of this article. As shown in Figure S6, we ran limited simulations in which organisms reproduced asexually with probability 1 − ρ and outcrossed, to reproduce sexually, with probability ρ. The impact of frequent outcrossing was to broaden the range of stable binding-site lengths, compared to the case of asexual reproduction. This was even true for the low rates of outcrossing, ρ ≈ 10−3 associated with yeast (Tsai et al. 2008). It is particularly interesting that sexual reproduction allows for shorter binding-site lengths. Although our model does not predict that sexual reproduction directly favors shorter binding-site lengths, it may be that recombination is a necessary condition for the evolution of the shorter binding sites used in the complex regulatory modules of higher eukaryotes.
Higher eukaryotes enjoy a rich array of mechanisms for transcriptional and post-transcriptional control that single-cellular organisms typically lack. Nonetheless, the basic characteristics of transcription factor binding sites are remarkably conserved across life (Figure 1 and Figure S1), despite vast differences in population size, genome size, regulatory complexity, and selective regimes. Our analysis helps to explain this conservation, because the predicted characteristics of binding sites are robust to the underlying mechanistic parameters. As this explanation illustrates, natural selection acts not only on the loss or gain of individual binding sites in the genome, but also on the statistical properties of the binding motif across the genome.
Supplementary Material
Acknowledgments
J.B.P. acknowledges funding from the Burroughs Wellcome Fund, the David and Lucile Packard Foundation, the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, and grant D12AP00025 from the U.S. Department of the Interior and Defense Advanced Research Projects Agency. S. H. acknowledges NIH grant R01GM085226.
Appendix
Calculation of Equilibrium Mean Fitness and Invasibility Criteria
We focus on a transcription factor with K true targets and L false targets. Each binding site is n nucleotides in length and has an average degeneracy r per nucleotide. Each nonfunctional true target harbored by an individual reduces its fitness by an amount s+. Likewise, each functional false target harbored by an individual reduces its fitness by an amount s−. We use the Wright–Fisher model from population genetics, in which a population of N individuals reproduces according to its fitness. The fitness of an individual is assumed to be multiplicative across all the K + L true and false targets. Thus if an individual has i true targets that are functional and j false targets that are functional, it has fitness . The number of mutations experienced by each individual in each successive discrete generation is Poisson distributed, with the probability that a given nucleotide suffers a mutation given by the per-nucleotide mutation rate μ. We derive analytic equations to describe under what conditions a mutation that changes n or r can invade a population initially at equilibrium. To do this we must calculate the equilibrium mean fitness of the population.
Equilibrium mean fitness
Given the mutation rates u+, u−, v+, and v−, we apply standard results (Higgs and Woodcock 1995; Woodcock and Higgs 1996; Krakauer and Plotkin 2002) for the Wright–Fisher model on a multiplicative fitness landscape to determine the equilibrium number of functional true targets, a+K, and equilibrium number of functional false targets, a−L. The proportions a+ and a− are given, to good approximation, by Higgs and Woodcock (1995; Woodcock and Higgs 1996):
and
We can further simplify the expression for a+ by making two empirical observations. The strength of selection on true targets is typically strong (Mustonen and Lässig 2005; Mustonen et al. 2008; He et al. 2011), e.g., Ns+ ∼ 10, whereas the population-scaled per-nucleotide mutation rate is less than one for virtually all organisms, Nμ < 1. As a result, in realistic regimes, s+ must exceed u−. We can therefore expand the expression for a+ to first order in (Ns+)−1 to obtain
which is the approximation given in Results. We can likewise approximate the expression for a− by noting that false targets experience weak selection (Hahn et al. 2003), Ns− ≤ 1. Expanding the expression for a− to first order in Ns− gives
The equilibrium mean fitness of the population is then , which we can now calculate in terms of the model parameters N, s+, s−, μ, K, L, n, and r. Because s+, s− ≪ 1, the equilibrium mean fitness is well approximated by
which is the approximation given in Results.
Invasibility criteria
To determine whether a mutation that changes n or r can invade a population at equilibrium we calculate the fitness of such a mutant relative to the population as a whole. This is simple to do. A mutant that changes n or r will destroy a functional binding site with some probability p. Such a mutation is deleterious for true targets but advantageous for false targets. The expected number of functional true targets following such a mutation is a+K(1 − p) and the expected number of false targets following such a mutation is a−L(1 − p). Thus the fitness of such a mutant, w*, is
Such a mutation can invade the population only if . This occurs when a−Ls− > a+Ks+, which is our invasibility criterion. What determines the probability p? In our model we assume that mutations that increase binding-site length n or decrease degeneracy r tend to destroy functional binding sites. Therefore p is the probability that, following such a mutation, a transcription factor no longer binds to a site that was functional prior to the mutation. This will in general depend on biophysical factors relating to how the binding affinity of the protein for DNA is altered by mutation, and it may be very difficult to determine. However, as noted above, the invasibility criteria do not depend on the specific value of p. Therefore the equation above provides the invasibility criteria for mutations that increase biding-site length, n, or decrease degeneracy, r. In our evolutionary simulations we set for mutations that increase binding-site length n. This choice reflects the probability that the new nucleotide added to the binding site does not match any of the r bases that promote functional binding. For mutations that decrease average degeneracy, r, we set . This choice reflects the probability that the nucleotide at which r is decreased no longer contributes to functional binding following mutation.
Similar to the argument above, there is also some chance q that mutations that change n or r will render a nonfunctional binding site functional. Such mutations are advantageous for true targets but deleterious for false targets. The expected number of functional true targets following such a mutation is qK(1 − a+) + a+K and the expected number of functional false targets following such a mutation is , where is the number of false targets that are one mutation away from functional. Thus the fitness of such a mutant, w*, is
Such a mutation can invade the population only if . This occurs when , which is our invasibility criterion. In our model we assume that mutations that decrease binding-site length n or increase degeneracy r tend to destroy functional binding sites. This means that q is the probability that, following a mutation that decreases binding-site length or increases degeneracy, a transcription factor binds to a site that was nonfunctional prior to the mutation. Just as for p, the probability q will in general depend on biophysical factors but again the invasibility criteria do not depend on the value of q and the equation above gives the invasibility criteria for mutations that decrease n or increase r. In our evolutionary simulations we set for mutations that decrease binding-site length n. This choice reflects the probability that a binding site with only one mismatched nucleotide has zero mismatched nucleotides (and therefore becomes functional) following mutation. For mutations that increase average degeneracy, r, we set . This choice reflects the probability that a binding site with only one mismatched nucleotide has zero mismatched nucleotides (and therefore becomes functional) following a mutation that increases r.
Footnotes
Communicating editor: L. M. Wahl
Literature Cited
- Berg J., Willmann S., Lässig M., 2004. Adaptive evolution of transcription factor binding sites. BMC Evol. Biol. 4: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu L., Buchler N., Garcia H., Gerland U., Hwa T., et al. , 2005. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 15(2): 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne J. C., Valen E., Tang M.-H. E., Marstrand T., Winther O., et al. , 2008. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36(Database issue): D102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S. B., 2005. Evolution at two levels: on genes and form. PLoS Biol. 3(7): e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’haeseleer P., 2006. What are DNA sequence motifs? Nat. Biotechnol. 24(4): 423–425. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Charlesworth B., Charlesworth D., Crow J. F., 1998. Rates of spontaneous mutation. Genetics 148: 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens, W. J., 2004 Mathematical Population Genetics, Vol. 27, Ed. 2. Springer-Verlag, New York. [Google Scholar]
- Gama-Castro S., Salgado H., Peralta-Gil M., Santos-Zavaleta A., Muñiz-Rascado L., et al. , 2011. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor units). Nucleic Acids Res. 39(Database issue): D98–D105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerland U., Hwa T., 2002. On the selection and evolution of regulatory DNA motifs. J. Mol. Evol. 55: 386–400. [DOI] [PubMed] [Google Scholar]
- Gerland U., Hwa T., 2009. Evolutionary selection between alternative modes of gene regulation. Proc. Natl. Acad. Sci. USA 106(22): 8841–8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerland U., Moroz J. D., Hwa T., 2002. Physical constraints and functional characteristics of transcription factor-DNA interaction. Proc. Natl. Acad. Sci. USA 99(19): 12015–12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. W., Stajich J. E., Wray G. A., 2003. The effects of selection against spurious transcription factor binding sites. Mol. Biol. Evol. 20(6): 901–906. [DOI] [PubMed] [Google Scholar]
- Harbison C. T., Gordon D. B., Lee T. I., Rinaldi N. J., Macisaac K. D., et al. , 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431(7004): 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. Z., Holloway A. K., Maerkl S. J., Kreitman M., 2011. Does positive selection drive transcription factor binding site turnover? A test with Drosophila cis-regulatory modules. PLoS Genet. 7(4): e1002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs P. G., Woodcock G., 1995. The accumulation of mutations in asexual populations and the structure of genealogical trees in the presence of selection. J. Math. Biol. 33(7): 677–702. [Google Scholar]
- Ihmels J., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., et al. , 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309(5736): 938–940. [DOI] [PubMed] [Google Scholar]
- Kimura M., 1962. On the probability of fixation of mutant genes in a population. Genetics 47: 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer D. C., Plotkin J. B., 2002. Redundancy, antiredundancy, and the robustness of genomes. Proc. Natl. Acad. Sci. USA 99(3): 1405–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lässig M., 2007. From biophysics to evolutionary genetics: statistical aspects of gene regulation. BMC Bioinformatics 8(Suppl. 6): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Rinaldi N. J., Robert F., Odom D. T., Bar-Joseph Z., et al. , 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298(5594): 799–804. [DOI] [PubMed] [Google Scholar]
- Lemos B., Araripe L. O., Fontanillas P., Hartl D. L., 2008. Dominance and the evolutionary accumulation of cis- and trans-effects on gene expression. Proc. Natl. Acad. Sci. USA 105(38): 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk R. W., Eisen M. B., 2008. Use of an evolutionary model to provide evidence for a wide heterogeneity of required affinities between transcription factors and their binding sites in yeast. Pac. Symp. Biocomput. 2008: 489–500. [PubMed] [Google Scholar]
- Lynch M., 2010. Evolution of the mutation rate. Trends Genet. 26(8): 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2003. The origins of genome complexity. Science 302(5649): 1401–1404. [DOI] [PubMed] [Google Scholar]
- Moses A. M., Chiang D. Y., Kellis M., Lander E. S., Eisen M. B., 2003. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 3: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R., Hiller K., Barg H., Heldt D., Linz S., et al. , 2003. PRODORIC: prokaryotic database of gene regulation. Nucleic Acids Res. 31(1): 266–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Lässig M., 2005. Evolutionary population genetics of promoters: predicting binding sites and functional phylogenies. Proc. Natl. Acad. Sci. USA 102(44): 15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V., Kinney J., Callan C. G., Jr, Lässig M., 2008. Energy-dependent fitness: a quantitative model for the evolution of yeast transcription factor binding sites. Proc. Natl. Acad. Sci. USA 105(34): 12376–12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Rokas A., Kassner V. A., Williams T. M., et al. , 2006. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature 440(7087): 1050–1053. [DOI] [PubMed] [Google Scholar]
- Prud’homme B., Gompel N., Carroll S. B., 2007. Emerging principles of regulatory evolution. Proc. Natl. Acad. Sci. USA 104(Suppl. 1): 8605–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G., Hirsh A. E., 2005. The application of statistical physics to evolutionary biology. Proc. Natl. Acad. Sci. USA 102(27): 9541–9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta A. M., Djordjevic M., Shraiman B. I., 2002. Specificity and robustness in transcription control networks. Proc. Natl. Acad. Sci. USA 99(4): 2072–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger R. K., Roberts L. R., Lyakhov I. G., Sidorov I. A., Stephen A. G., et al. , 2007. Correlation between binding rate constants and individual information of E. coli Fis binding sites. Nucleic Acids Res. 35(16): 5275–5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultzaberger R. K., Maerkl S. J., Kirsch J. F., Eisen M. B., 2012. Probing the informational and regulatory plasticity of a transcription factor DNA-binding domain. PLoS Genet. 8(3): e1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. L., 2000. Evolutionary developmental biology and the problem of variation. Evolution 54(4): 1079–1091. [DOI] [PubMed] [Google Scholar]
- Teixeira M. C., Monteiro P., Jain P., Tenreiro S., Fernandes A. R., et al. , 2006. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 34(Suppl. 1): D446–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M., van Oudenaarden A., 2001. Intrinsic noise in gene regulatory networks. Proc. Natl. Acad. Sci. USA 98(15): 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai I. J., Bensasson D., Burt A., Koufopanou V., 2008. Population genomics of the wild yeast Saccharomyces paradoxus: quantifying the life cycle. Proc. Natl. Acad. Sci. USA 105(12): 4957–4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A. E., Tuch B. B., Li H., Johnson A. D., 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443(7110): 415–420. [DOI] [PubMed] [Google Scholar]
- Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D., 2008a The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6(2): e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuch B. B., Li H., Johnson A. D., 2008b Evolution of eukaryotic transcription circuits. Science 319(5871): 1797–1799. [DOI] [PubMed] [Google Scholar]
- Woodcock G., Higgs P. G., 1996. Population evolution on a multiplicative single-peak fitness landscape. J. Theor. Biol. 179(1): 61–73. [DOI] [PubMed] [Google Scholar]
- Wray G. A., 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 8(3): 206–216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.