SUMMARY
In addition to sculpting eukaryotic transcripts by removing introns, pre-mRNA splicing greatly impacts protein composition of the emerging mRNP. The exon junction complex (EJC), deposited upstream of exon-exon junctions after splicing, is a major constituent of spliced mRNPs. Here we report comprehensive analysis of the endogenous human EJC protein and RNA interactomes. We confirm that the major “canonical” EJC occupancy site in vivo lies 24 nucleotides upstream of exon junctions and that the majority of exon junctions carry an EJC. Unexpectedly, we find that endogenous EJCs multimerize with one another and with numerous SR proteins to form megadalton sized complexes in which SR proteins are super-stoichiometric to EJC core factors. This tight physical association may explain known functional parallels between EJCs and SR proteins. Further, their protection of long mRNA stretches from nuclease digestion suggests that endogenous EJCs and SR proteins cooperate to promote mRNA packaging and compaction.
Keywords: RNA processing, splicing, spliceosome, exon junction complex, SR proteins, mRNP structure, transcriptome, deep sequencing, mass spectrometry
INTRODUCTION
In cells, all RNAs exist as ribonucleoprotein particles (RNPs). The protein components of these particles both add functionality to the RNA and serve as key structural elements of the overall complex. The most compositionally diverse RNPs are those containing messenger RNAs (mRNPs), and several hundred different polypeptides have now been identified as mRNP components (Baltz et al., 2012; Castello et al., 2012; Dreyfuss et al., 2002). Among these are proteins recognizing specific structures (e.g., the cap and polyA tail binding proteins) and others associating along the RNA's length in a largely sequence independent manner (e.g., YB-1 protein) (Moore, 2005). Other abundant mRNP components are heterogeneous nuclear RNP (hnRNP) and serine/arginine-rich (SR) proteins, which recognize short sequence motifs via sequence-specific RNA binding domains (Dreyfuss et al., 2002; Singh and Valcarcel, 2005). SR proteins characteristically contain one to two N-terminal RNA recognition motifs (RRMs) and a C-terminal domain rich in RS dipeptides subject to dynamic phosphorylation (Long and Caceres, 2009). These proteins are often loaded co-transcriptionally and many accompany the fully processed mRNA to the cytoplasm. The three best-studied shuttling SR proteins are SRSF1 (ASF/SF2), SRSF3 (SRp20) and SRSF7 (9G8). All three become dephosphorylated during the course of pre-mRNA processing and, in the dephosphorylated state, promote mRNP transit through the nuclear pore by interacting with export factors (Huang and Steitz, 2005). Also facilitating mRNP export is the transcription/export (TREX) complex (UAP56, Aly/REF and THO subcomplex) loaded near the 5′ cap during transcription and pre-mRNA splicing (Cheng et al., 2006).
Another set of proteins stably associated with both nuclear and cytoplasmic mRNPs is the exon junction complex (EJC). EJCs are deposited upstream of exon-exon junctions as a consequence of pre-mRNA splicing (Le Hir et al., 2000a). Unlike hnRNP and SR proteins, the EJC has no apparent sequence dependence. Rather, it binds RNA primarily through eIF4AIII, a sequence-independent DEAD-box protein (Shibuya et al., 2004) that grasps RNA stably only when associated with its partners, Y14 and Magoh. A fourth protein, MLN51, can further stabilize eIF4AIII on RNA (Ballut et al., 2005; Bono and Gehring, 2011). This core serves as a binding platform for additional “peripheral” EJC factors. These include Pinin, Upf3, SKAR, SRm160, the “ASAP” complex (RNPS1, Acinus, SAP18), and the TREX proteins UAP56 and Aly/REF (Bono and Gehring, 2011). When assembled in in vitro splicing reactions, the EJC has been estimated to be ~350 kDa and protects 8–10 nts from nuclease digestion (Le Hir et al., 2000a).
In mammalian cells, core EJC proteins and peripheral factors interface with numerous machineries controlling mRNA export, translation and decay (Giorgi and Moore, 2007; Tange et al., 2004). The EJC is known to promote export of spliced mRNAs in vertebrates, perhaps through its close association with the TREX complex (Cheng et al., 2006). One of the best-understood EJC functions is its role in discriminating between premature and normal translation termination events. When translation terminates on a mammalian mRNA upstream of at least one EJC, Upf3 and its co-factors Upf1 and Upf2 orchestrate a series of events that destabilize the message by a process known as nonsense-mediated mRNA decay (NMD) (Rebbapragada and Lykke-Andersen, 2009). Another documented role for the EJC in mammalian cells is enhanced translation of newly synthesized mRNAs through interactions between SKAR and activated S6-kinase (Gudikote et al., 2005; Ma et al., 2008; Nott et al., 2004; Wiegand et al., 2003).
To date, all studies exploring mammalian EJC deposition, structure and composition have been carried out either in vitro, or using exogenously expressed proteins and/or reporter RNAs in vivo (Bono and Gehring, 2011; Tange et al., 2004). In this study, we report the first comprehensive analysis of the endogenous EJC interactome, including the complete EJC proteome and all tightly associated RNA fragments strongly protected from nuclease digestion. Surprisingly, endogenous EJCs multimerize both with one another and with numerous SR proteins to form megadalton sized complexes in which SR proteins are super-stoichiometric to EJC core factors. This intimate physical association may explain known functional similarities between EJCs and SR proteins. Additionally, their protection of extended mRNA regions from nuclease digestion suggests that cooperation between endogenous EJCs and SR proteins leads to higher order mRNP structures, which may facilitate overall mRNA packaging and compaction.
RESULTS
An unexpectedly long footprint for endogenous human EJCs
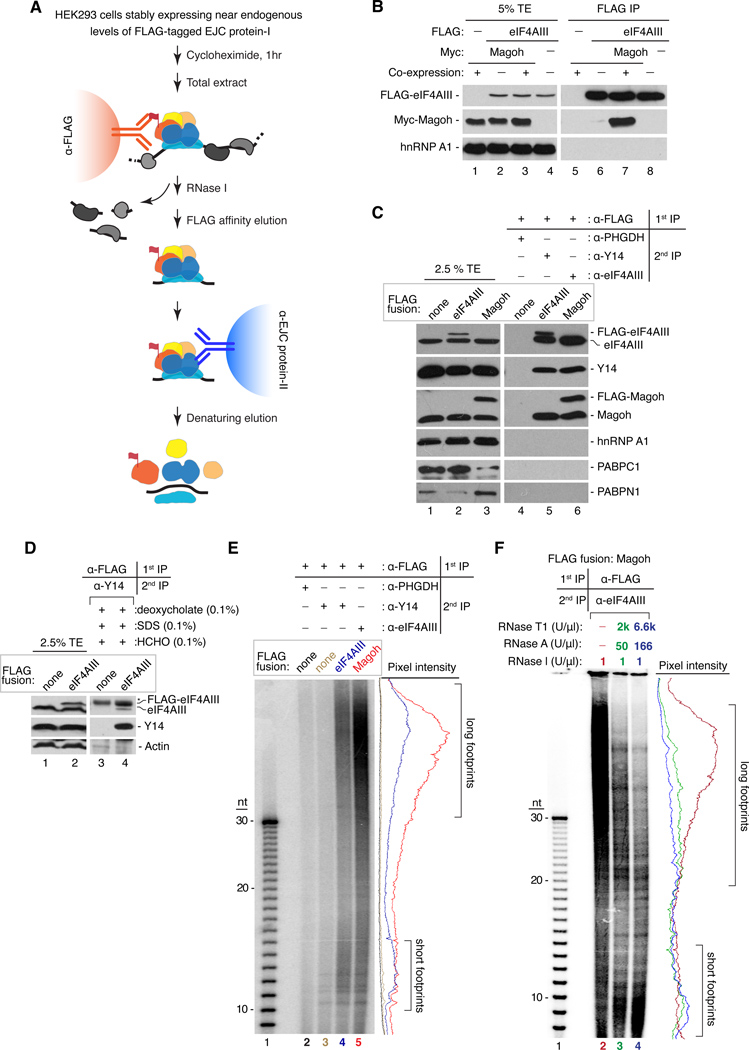
EJC deposition in vitro is strictly splicing dependent and, once formed, the complex is remarkably stable (Le Hir et al., 2000a). Therefore, we reasoned that a native RNA:protein immunoprecipitation in tandem (RIPiT) approach could be used to purify endogenous EJCs and their RNA footprints (Figures 1A and S1A). Validating this approach, Myc-Magoh only co-purified with FLAG-eIF4AIII when both were co-expressed in human embryonic kidney (HEK293) cells, but not when the two proteins were expressed separately and mixed following cell lysis (Figure 1B). Thus our strategy is specific for preexisting EJCs, because no new EJC-like complexes appear to form after cell lysis (Mili and Steitz, 2004).
Figure 1. Purification of endogenous EJCs and RNA footprints.
A. RIPiT schematic. EJC core factors, colored; non-EJC proteins, gray; RNA, black line.
B. Co-purification of EJC factors requires their co-expression. Western blot of total extract (TE, lanes 1–4) or FLAG IPs (lanes 5–8) from extracts in which FLAG-eIF4AIII and Myc-Magoh were either co-expressed (+) or expressed in separate cells (−) and mixed prior to IP.
C. Confirmation of EJC purification. Western blots of EJC or non-EJC RNA-binding proteins in total extracts (lanes 1–3), control RIPiT (lane 4) or EJC RIPiTs (lanes 5 and 6). For each lane, indicated are the FLAG-fusion protein (box above) and antibodies used for the 1st and 2nd IPs (table above lanes 4–6).
D. EJC purification under denaturing conditions after formaldehyde crosslinking. Western blots as in C showing protein levels in total extracts (lanes 1 and 2) or RIPiTs (lanes 3 and 4) from formaldehyde-crosslinked cells. SDS and deoxycholate were included during the α-FLAG IP. * indicates IgGH.
E. Length distribution of RNase I-resistant EJC footprints. Base-hydrolyzed synthetic polyU30 RNA (lane 1) or immunoprecipitated RNA fragments from RIPiTs indicated at top (lanes 2–5) were 5′-end 32P-labeled and separated by denaturing PAGE.
Autoradiograph pixel intensity profiles of lanes 2–5 are at right.
F. Same as E except samples in lanes 3 and 4 were additionally incubated with RNases A+T1 after α-eIF4AIII IP.
See also Figure S1.
Two different RIPiTs (FLAG-eIF4AIII:Y14 and FLAG-Magoh:eIF4AIII), but not two control RIPiTs [FLAG only:PHGDH (3-phosphoglycerate dehydrogenase) and FLAG-only: Y14], yielded efficient and specific enrichment of the EJC core components Y14, eIF4AIII and Magoh (Figure 1C and data not shown). Surprisingly, even under the most stringent nuclease digestion conditions, we always observed co-purification of the endogenous untagged protein with its FLAG-tagged counterpart (Figure 1C, lanes 5 and 6; Figure S1B). This co-purification was eliminated under conditions disrupting EJC core integrity (e.g., >450 mM NaCl or 0.5 M urea; Figures S1B and S1C) but persisted when cells were formaldehyde-crosslinked prior to cell lysis and subjected to RIPiT under denaturing conditions (Figure 1D). This suggests that EJC monomers physically interact with one another in vivo (see below).
Another surprising finding was that in addition to the small RNA footprints expected for monomeric EJCs (Ballut et al., 2005; Le Hir et al., 2000a; Stroupe et al., 2006), even more abundant and much longer ~30–150 nt RNase-resistant fragments were consistently observed in all of our EJC preps (Figure 1E). Both footprint sizes were observed in the absence or presence of cycloheximide, with all nucleases tested, but were lost if RNA-protein complexes were denatured with high-salt or by incubation at 70 °C prior to nuclease digestion (Figures S1D-F). The longer footprints persisted even after exposure to extreme nuclease conditions, although their size and intensity were reduced with a concomitant increase in shorter footprints (Figure 1F). The unexpectedly large RNA footprints, combined with the evidence above for EJC-EJC interactions, suggested greater complexity for endogenous EJCs than previously supposed.
The EJC proteome
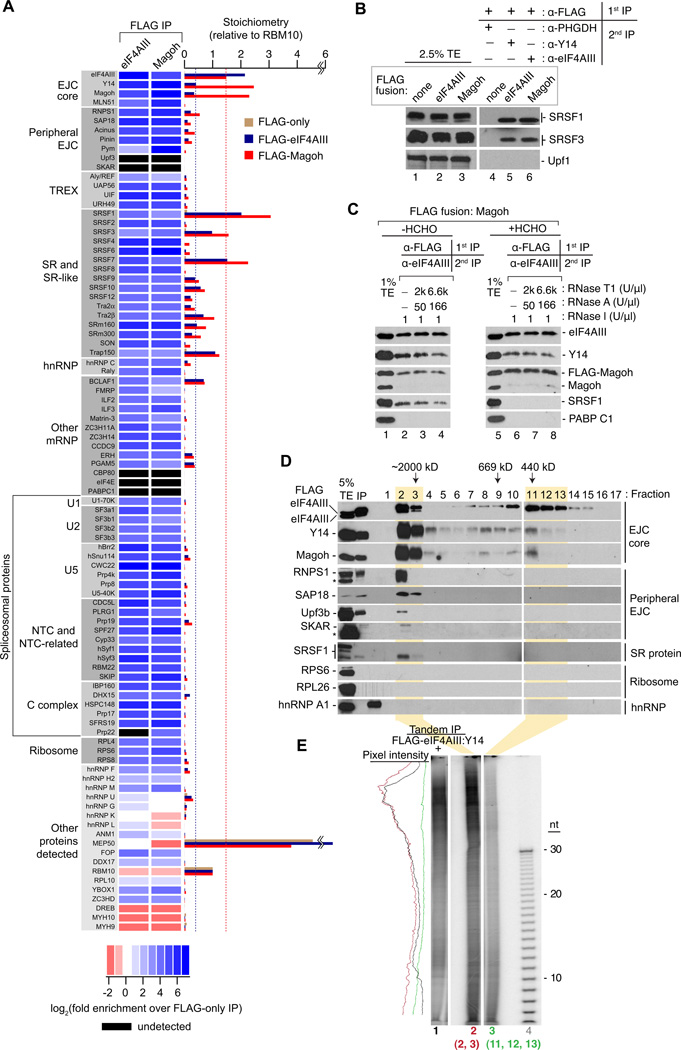
To assess the full complement of proteins stably associated with EJC core factors in vivo, we performed comprehensive mass spectrometry analyses of samples subjected to extensive RNase I-digestion and FLAG IP from cells expressing FLAG-eIF4AIII, FLAG-Magoh, or FLAG peptide only (to control for nonspecific interactions). Remarkably, ~70 different proteins were at least 10-fold enriched in both EJC core factor IPs compared to the control IP, and these enriched proteins could be organized into readily definable classes (Figures 2A and S2A, Table S1). Amongst the most abundant were the three EJC core factors, eIF4AIII, Y14 and Magoh. Also readily detectable were late-stage spliceosome components (i.e., U5 snRNP, NTC and NTC-related, and C complex proteins) consistent with reports that EJC assembly occurs concurrent with the second step of splicing and/or spliced exon release (Ideue et al., 2007; Reichert et al., 2002; Zhang and Krainer, 2007). The most highly enriched U5 snRNP component was the eIF4AIII-recruitment factor Cwc22 (Barbosa et al., 2012; Steckelberg et al., 2012). The majority of known peripheral EJC proteins/protein complexes [such as MLN51, Pinin, Pym, the ASAP complex (Acinus, SAP18, RNPS1), and TREX components (UAP56, UIF, URH49, Aly/REF)] were also detected, but they were all sub-stoichiometric to the EJC core factors. Notably absent were Upf3 and SKAR, although they were marginally detectable by western blotting (Figures S2B and 2C).
Figure 2. The EJC interactome.
A. The EJC proteome. Left: Protein names organized into indicated classes. Center: Heat map showing fold-enrichment of each protein in FLAG-eIF4AIII or FLAG-Magoh IPs over its level in a FLAG-only (control) IP (not shown). Black rectangles: protein not detected. All proteins shown were at least 10-fold enriched in FLAG-EJC protein IPs except in the “Other proteins detected” class, which includes <10-fold enriched RNA binding proteins, previously reported EJC-interacting proteins similarly enriched in our EJC and control IPs, and select proteins depleted in the EJC IPs compared to the control IP. Bottom: Heat map log2 color scale. Right: Bar graph showing protein stoichiometries relative to RBM10 in FLAG-eIF4AIII (blue), FLAG-Magoh (red) or FLAG-only (tan) IPs. Dashed blue line: level of co-purifying Y14 and Magoh in FLAG-eIF4AIII IP. Dashed red line: level of co-purifying eIF4AIII in FLAG-Magoh IP. These dashed lines assembled EJC core stoichiometries in each IP.
B. Confirmation of EJC proteomic analyses. Western blots of total extracts or RIPiTs as in Figure 1B showing levels of EJC-interacting factors either detected or not in mass spec analysis.
C. SRSF1 is tightly associated with but does not crosslink to EJC core factors. Western blots of indicated proteins in total extracts (lanes 1 and 5) or RIPiTs (lanes 2–4 and lanes 6–8). Native and denaturing α-FLAG IPs were as in Figure 1C and 1D, respectively.
D. Two distinct size complexes containing EJC core proteins. Western blots of indicated proteins (right) in total cell extract (TE), input FLAG-eIF4AIII IP sample (IP) and gel filtration fractions of RNase A-treated FLAG-eIF4AIII complexes (lanes 1–17). Peak elution fractions of known MW size markers are indicated at top. * indicates IgGH (RNPS1 panel) or a cross-reacting unknown protein (SKAR panel).
E. RNA footprints of EJC-core containing complexes. Autoradiogram as in Figure 1D showing RNA profiles from FLAG-eIF4AIII:Y14 RIPiT (lane 1) or from indicated gel filtration fractions in C (lane 2: fractions 2 and 3; lane 3: fractions 11, 12 and 13). Pixel intensity profile is on the left.
Surprisingly, the entire complement of SR proteins was robustly enriched, with numerous members being either nearly stoichiometric (SRSF9, 10, 12) or even superstoichiometric (SRSF1, 3 and 7) to the EJC core factors. Further, several other proteins containing SR domains were also enriched. These “SR-like” proteins (SRm160, SRm300, Trap150, Tra2α, Tra2β and SON) function as splicing co-activators and regulators in vivo (Ahn et al., 2011; Lee et al., 2010; Long and Caceres, 2009), and are also known mRNPs components (Baltz et al., 2012; Castello et al., 2012; Merz et al., 2007). Thus, the list of EJC-associated SR-like proteins extends significantly beyond the previously known peripheral EJC components Pinin, Acinus and RNPS1 (Le Hir et al., 2000a; Tange et al., 2005).
To assess the stability of SR protein and EJC core association, we performed western blots of RIPiT samples. Consistent with the EJC being a post-splicing complex, western blotting confirmed that only the hypo-phosphorylated forms of SRSF1 and SRSF3 co-purified with the EJC (Figures 2B, 2C, S2C and S2D). Further, even after extreme nuclease treatment SRSF1 was readily detectable in native RIPiT and IP samples (Figures 2C and S2C). In contrast, SRSF1 was not detectable in samples that had been formaldehyde-crosslinked prior to cell lysis, RNase digestion and denaturing RIPiT (Figure 2C). This indicates that nuclease-resistant EJC and hypophosphorylated SR protein-containing complexes detected under native conditions are predominantly due to residence of EJC cores and SR proteins on the same highly protected fragments of RNA.
Higher order EJC interactions
To assess the size of endogenous EJCs, we performed gel filtration analysis of RNase-treated FLAG-eIF4AIII (Figure 2D) or FLAG-Magoh (data not shown) complexes subsequent to affinity elution with FLAG peptide (Figure 1A). Western blotting revealed two peaks containing the EJC core proteins eIF4AIII, Y14 and Magoh. These same two peaks were also observed upon gel filtration of RNase A-treated cell lysates (data not shown), indicating that they were not formed as a consequence of the purification. The ~450 kDa peak contained only eIF4AIII, Y14 and Magoh and, in purified samples, only the FLAG-tagged protein used for affinity purification, but not its endogenous counterpart. This complex, which is similar in size to the in vitro-assembled EJC (Le Hir et al., 2000a), thus likely represents EJC monomers, perhaps consisting of the core and other peripheral proteins undetected here.
The larger complex [high molecular weight (HMW) EJC] contained both the FLAG-tagged protein and its untagged endogenous counterpart. This complex also contained “peripheral” EJC proteins and other EJC-associated factors identified above, such as hypophosphorylated SRSF1. The larger complex eluted in the void volume of the gel filtration column, which had an exclusion limit of >2 MDa. Sucrose gradient fractionation revealed that the larger complex co-sedimented with ribosomal subunits (Figure S2E), confirming their large size. In gel filtration experiments, the 30–150 nt RNA fragments almost exclusively fractionated with the HMW EJC, while the shorter fragments co-fractionated with both the large and small complexes (Figure 2E). Consistent with the relative abundance of the long and short RNA footprints in our RNase I digests (Figure 1E), the HMW EJC constituted ~70% of the total. Taken together, these results indicate that cells contain two EJC populations: monomeric EJCs with short footprints consisting primarily of the core factors, and HMW EJCs protecting much longer stretches of RNA.
Mapping of EJC- and nuclear mRNP-protected sites
To investigate the nature of the short and long EJC-protected regions, we generated a series of libraries for deep sequencing on the SOLiD platform (Figure S3A). Short footprint EJC libraries were generated from 12–25 nt RNA fragments copurifying in FLAG-eIF4AIII:Y14 or FLAG-Magoh:eIF4AIII RIPiTs or in a FLAG-only:Y14 control RIPiT. Two libraries were generated from long EJC footprints: one from the 70–100 nt RNA fragments from the FLAG-Magoh:eIF4AIII RIPiT, and another from 30–100 nt RNA fragments co-fractionating with the HMW EJC (gel-filtration fractions 2 and 3 in Figure 2D). To assess how the short and long EJC footprints compared to all protected regions on endogenous mRNAs, we also prepared a library of 24–36 nt fragments protected from RNase I digestion in native nuclear mRNPs. Finally, to quantify expression levels of endogenous mRNAs, we generated a polyA-selected RNA-Seq library from one of our HEK293 cell lines. Raw reads were mapped to both the human genome and a reference exon junction dataset (Figure S3B, see Supplemental Experimental Procedures).
Among all libraries, reads mapping uniquely to the human genome and/or transcriptome were ~20–37 fold enriched in exonic regions (Figure S3D). Exon mapping reads in our short footprint libraries proved highly reproducible, both between biological replicates and between different IP configurations (Figure S3E). As expected, these reads were significantly more abundant for mRNAs from intron-containing genes than mRNAs from intronless genes (Figures S3F and S3G). In these libraries, ~40% of exonic reads mapped to the expected EJC deposition site (i.e., 15–31 nts upstream of exon-exon junctions). This read distribution led to readily identifiable peaks on individual exons (Figure 3A). For quantitative purposes, we developed an algorithm that defined “EJC peaks” as regions significantly enriched with short EJC footprint reads across multiple libraries (see Supplemental Experimental Procedures). Around half of such computationally identified EJC peaks mapped within exons (Figures S3D and S4A). In comparison, reads from the long EJC footprint and mRNP protection libraries were much more broadly distributed across exons (Figure 3A), so did not readily lend themselves to peak calling. Nonetheless, the exonic read densities within transcripts were highly correlated between the long and short EJC footprint libraries (Figures S3H and S4B), indicating enrichment of similar RNA sequences in long and short footprints.
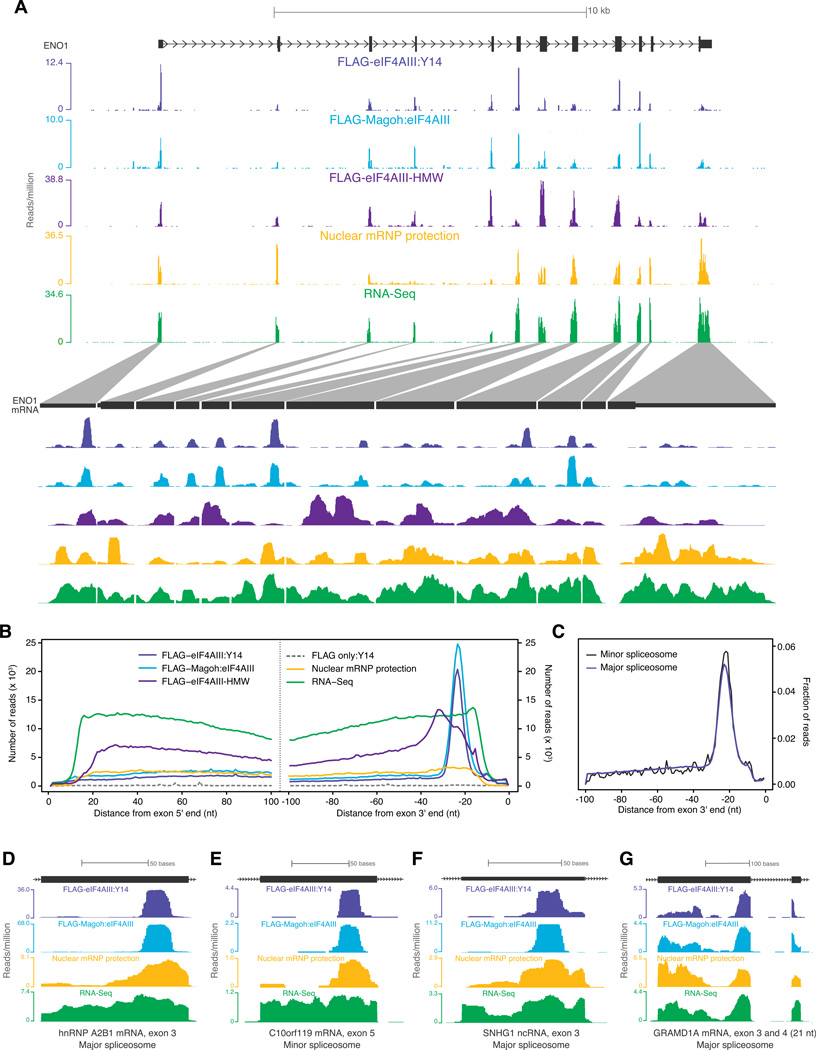
Figure 3. EJC occupancy on endogenous mRNAs.
A. Distribution of short EJC footprints (FLAG-eIF4AIII:Y14 and FLAG-Magoh:eIF4AIII), HMW EJC footprints (FLAG-eIF4AIII-HMW), nuclear mRNP footprints or RNA-Seq reads on a representative gene (top half), ENO1, and its spliced mRNA (bottom half). Scale is indicated at top; reads per million at the highest position within each library window are indicated at left.
B. Meta-exon analysis of read distribution. Composite plots of distances from the centers of all exon-mapping reads to the 5′ (left panel) or 3′ end (right panel) of each read's parent exon for the indicated libraries.
C. Meta-analysis of exons upstream of major or minor spliceosome introns. Composite plots for FLAG-Magoh:eIF4AIII library reads mapping to exons upstream of introns excised by the “major” (~1.7×105 introns) or “minor” (427 introns) spliceosome. In both plots, the fraction of reads at each nucleotide position out of all reads within 100 nt from exon 3′ end was plotted.
D-G. Profiles of short EJC footprints, nuclear mRNP footprints and RNA-Seq reads (as in A) on individual exons. Distinguishing features of each exon are indicated at bottom.
See also Figure S3.
The canonical EJC binding site is 24 nts upstream of exon-exon junctions
To map the predominant EJC deposition site in vivo, we calculated the distance from the center of each exon mapping read in all libraries to the 5′ or 3′ end of its parent exon. Whereas no preferential read positioning was detected in the RNA-Seq library, a composite plot revealed a striking accumulation of reads centered 24 nucleotides upstream of exon 3′ ends in all short EJC footprint libraries, with an apparent footprint width of ~14 nts (Figure 3B). No such accumulation was observed relative to any other mRNA feature such as exon 5′ ends, transcription start sites, or polyA sites (Figure 3B and data not shown). Reads in the long EJC footprint library were also preferentially skewed toward exon 3′ ends, but were distributed more broadly than the short EJC footprints. Although protection at the −24 position (24 nts upstream of exon junctions) was evident on some individual exons in the nuclear mRNP protection library (Figure 3A), this signal was only slightly enriched over other exonic sites in the composite plot (Figure 3B).
Preferential EJC association at the −24 position was readily apparent on individual spliced exons from both protein-coding and non-coding RNAs, as well as exons upstream of introns removed by both the "major" and "minor" (U2- and U12- dependent, respectively) spliceosomes (Figures 3C-F). Further, the strict position of EJC deposition relative to exon 3′ ends is maintained even on exons smaller than 24 nucleotides. In such cases, the EJC deposition site lies within the preceding spliced exon (Figure 3G). Thus, the major or "canonical" EJC (cEJC) deposition site is centered 24 nucleotides upstream of exon-exon junctions transcriptome-wide.
EJC occupancy is variable within mRNAs
We noticed that cEJC reads exhibited significant exon-to-exon variability within individual mRNAs (Figures 3A, S4B and S4C). To test the validity of this observation, we calculated for each mRNA the coefficient of variation (Cv) of the number of reads at cEJC sites. Consistent with profiles of individual mRNAs, the median Cv was much higher for the EJC libraries than for the RNA-Seq library (Figure 4A). One possible explanation was differential EJC epitope accessibility at different exon junctions, leading to differential IP efficiencies. However, the nuclear mRNP protection library, which should be free of any IP bias, exhibited variability similar to the EJC libraries within the −15 to −31 window (Figure 4A). High variability at the cEJC site was also observed in a library generated from formaldehyde-crosslinked, FLAG-eIF4AIII cells lysed and FLAG-IPed under denaturing conditions (Figures 4A and S4C). Therefore, variability in cEJC reads is unlikely explained entirely by differences in epitope accessibility or EJC stability during purification.
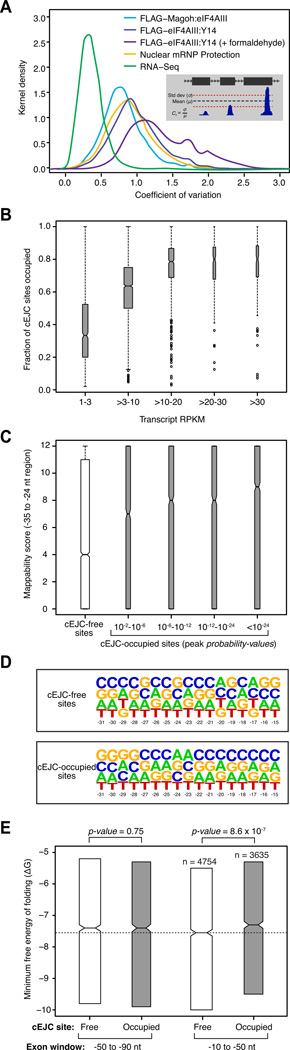
Figure 4. Variability in cEJC occupancy and contributing factors.
A. Signal variability among cEJC sites on individual mRNAs. Smoothed histograms of coefficient of variation (Cv) in the number of reads at each cEJC site (−15 to −31 nts from exon junctions) within a single transcript in a set of 4366 highly expressed transcripts. Inset: parameters used to calculate Cv.
B. Fractions of occupied cEJC sites per transcript. Included were all mRNAs with >10 introns and representative transcript RPKM >1. All cEJC sites with a significant peak (probability-value < 0.01) overlapping the −24 position in the FLAG-Magoh:eIF4AIII set were considered occupied. The box-plots show the interquartile range (IQR) of fraction occupied cEJC sites in each RPKM bin. The whiskers are drawn at 1.5 times the IQR and outliers are shown as open circles. Median (black horizontal line) and its confidence interval (notch) are also indicated within each box-plot.
C. Range of mappability scores at cEJC-free (white) and cEJC-occupied (gray) sites from all spliced transcripts with RPKM >10. Occupied sites were binned by peak significance (probability-values). Box-plots are as in B.
D. Nucleotide frequency plot at cEJC-free and -occupied sites. All sites with mappability score ≥8 from all spliced transcripts with RPKM >10 were used.
E. Interquartile range of the minimal free energy of folding for two different 40 nt exonic windows (−50 to −90 nt from exon junctions, left; −10 to −50 nt from exon junctions, right) for cEJC-free (white) and cEJC-occupied (gray) sites from D except that included cEJC-occupied sites had a peak probability-value <10−12. The total number of sites in each dataset is indicated. Box-plots are as in B except only the IQR is shown. The dashed line accentuates statistically significant (indicated by non-overlapping notches) difference in median minimal free energy of cEJC-free and -occupied sites.
See also Figure S4.
Surprisingly, numerous exons completely lacked any EJC signal at the canonical site (e.g., ENO1 exons 8 and 11, Figure 3A). We estimated the frequency of such occurrences in all mRNAs detectably expressed in our cell lines (reads-mapped per kilobase per million (RPKM) >1 in our RNA-Seq library). Within these, for each of the 4156 mRNAs with 10 or more introns we determined the fraction of cEJC sites with a detectable peak in our largest peak set (FLAG-Magoh:eIF4AIII). Among lower expressed mRNAs, the fraction of exons with a cEJC peak was a direct function of transcript abundance (Figure 4B), suggesting that apparent EJC absence was mostly due to incomplete sampling. But at RPKM ≥10, the fraction of cEJC occupied sites plateaued at a median of ~80%. Thus, even in the most highly expressed mRNAs a fraction of cEJC sites appeared unoccupied.
Some apparently cEJC-free sites in highly expressed mRNAs were undoubtedly due to poor mapping of the short EJC footprint reads. As compared to cEJC-occupied sites, cEJC-free sites exhibited a significantly lower (p-value <1.35×10−40, Wilcoxon rank sum test) median mappability score (Figure 4C). Nonetheless, many canonical sites lacking an cEJC peak within highly expressed mRNAs (RPKM ≥10) also had high mappability scores (≥8). When compared to occupied sites in the same mRNAs, these readily mappable but cEJC-free sites showed no significant difference with regard to either sequence composition (Figure 4D) or conservation (overall or wobble-position only; data not shown) in the −15 to −31 window. We also detected no difference in predicted 5′ or 3′ splice site strengths between cEJC-free and occupied sites (Figures S4D and S4E). Nonetheless, there were small but significant differences in the exon and downstream intron length distributions, with occupied sites tending to occur on slightly shorter exons (~10 nt difference in median, p-value=7.26×10−5) or upstream of somewhat longer introns (~150 nt difference in median, p-value=1.6×10−3) than cEJC-free sites (Figures S4F and S4G). But the most significant difference between the two sets was the predicted propensity of nts −10 to −50 to form a secondary structure (p-value= 8.6×10−7, Figure 4E). That no such difference was apparent for the adjacent window (−50 to −90 nt) strengthens the idea that EJC deposition is inhibited by local secondary structure (Mishler et al., 2008). In sum, our data indicate that cEJCs are deposited on the vast majority of exon-exon junctions in human mRNAs, but observed occupancy is influenced by exon and intron length and the availability of single-stranded RNA at the canonical deposition site.
mRNAs encoding RNA processing factors display higher overall cEJC occupancy
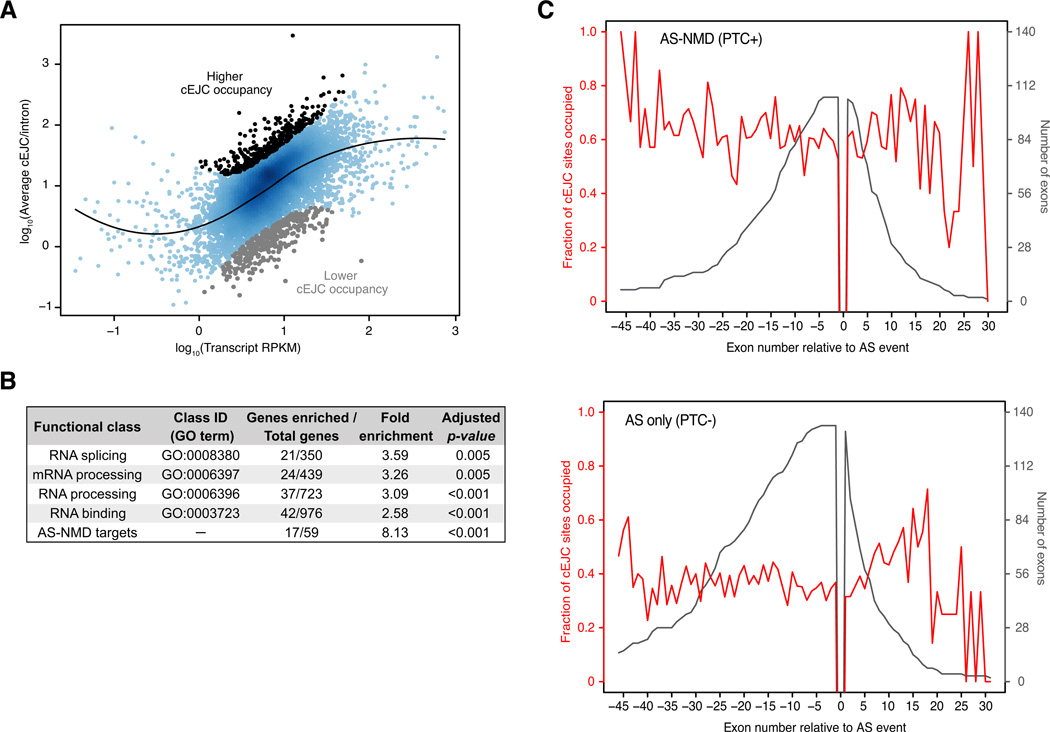
In addition to variable cEJC occupancy amongst exons within a single mRNA, we also wondered to what extent overall cEJC occupancy varied amongst mRNAs. We chose the 7536 mRNAs from genes with one or more introns that had the most reproducible cEJC peaks, determined the average cEJC peak height for each mRNA and then identified mRNAs with significantly lower or higher occupancy given their expression level (Figure 5A). Whereas no functionally related gene class (defined by Gene Ontology terms) was enriched in the low occupancy set, those pertaining to RNA metabolism were ~3-fold enriched in the higher occupancy set (Figure 5B). Notably, many mRNAs in this class regulate their own expression by alternative splicing linked to nonsense-mediated mRNA decay (AS-NMD), an EJC-dependent process (McGlincy and Smith, 2008). Consistent with this, the most enriched category (~8-fold) in the high cEJC occupancy class constituted known AS-NMD targets (Saltzman et al., 2008). This enrichment cannot be explained simply by a higher turnover rate for these mRNAs, as other classes of unstable mRNAs (e.g., transcription factors and cell-cycle proteins) were not similarly enriched (Figure S5A).
Figure 5. Functional classes enriched in cEJCs.
A. Average cEJC peak-height versus expression level (RPKM) for transcripts of 7536 mRNAs with highly reproducible cEJC peaks. Each dot represents an mRNA with 4 or more exons. Black line: locally weighted non-linear fit (LOESS). The top and bottom 5% deviating furthest from the fit are indicated.
B. Functional classes enriched in the higher cEJC occupancy set. The p-values shown were adjusted for multiple hypotheses testing.
C. Average cEJC occupancy up- and downstream of alternative spliced exons. AS-NMD (PTC+): Red line: fraction of cEJC occupied sites on successive exons flanking PTC-inducing alternative exon inclusion/exclusion events; Black line: total number of exons at each position for AS-NMD transcript set (RPKM > 1). AS only (PTC-): Same as above, except for a dataset where cassette exon inclusion/exclusion does not generate a PTC.
One possible explanation for the prevalence of AS-NMD targets in the high cEJC occupancy set could be enhanced cEJC occupancy downstream of the PTC-inducing alternative splicing event. To test this, we compared cEJC occupancies on exons flanking alternative splicing events that either did (AS-NMD mRNAs) or did not (AS-only mRNAs) introduce a PTC. For both datasets, there was no apparent preferential cEJC occupancy either 5′ or 3′ to the event (Figure 5C). What was readily apparent, however, was the higher overall cEJC occupancy on the AS-NMD mRNAs relative to the AS-only set. Thus there may well be a propensity for increased cEJC occupancy on AS-NMD transcripts, but this occupancy is not differentially distributed relative to the PTC-inducing event.
Non-canonical EJC peaks in exons
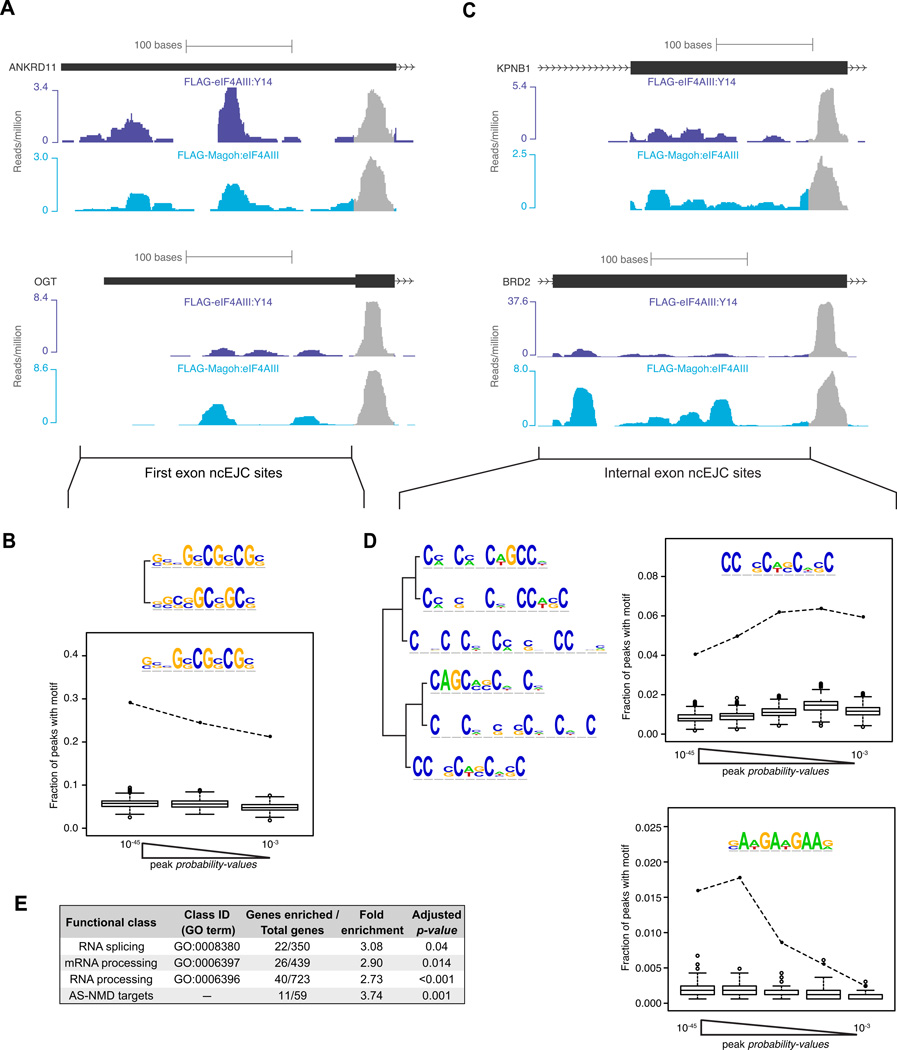
In addition to the cEJC peaks centered at the −24 position, we reproducibly observed peaks at other exonic locations ("non-canonical" or ncEJC peaks; Figures 3A and 6A). Such ncEJC peaks were also present in the library prepared from formaldehyde-crosslinked FLAG-eIF4AIII:Y14 lysates subjected to denaturing RIPiT, validating these as bona fide in vivo interactions (Figure S4C). Among our set of the most reproducible exonic peaks in the non-crosslinked sample libraries, these ncEJCs represented 40% of the total (Figure S6A). Unlike cEJCs, ncEJC peaks contained clearly identifiable sequence motifs (Figure 6). Highly enriched among first-exon ncEJC peaks was a CG-rich motif (Figures 6A, 6B and S6C) almost identical to an mRNA export promoting sequence found in mRNAs lacking 5′-UTR introns (Cenik et al., 2011). Two other motif classes were identifiable under ncEJC peaks in internal exons (Figures 6C and 6D). Several C-rich motifs (Figure 6D, top half; Figures S6B and S6D) resemble known high affinity sites for SRSF3, while an AG-rich motif (Figure 6D, bottom half; Figure S6E) closely resembles the in vivo SRSF1 binding site determined by CLIP as well as a reported binding site of the SR-like protein Tra2β (Long and Caceres, 2009 and references therein; Sanford et al., 2009). These internal exon motifs were also similarly enriched in the HMW EJC footprints (data not shown). Further, hexamers predicted to function as exonic splicing enhancers (ESEs) occurred more frequently as compared to exonic splicing silencers (ESSs) in the HMW EJC footprints (Figure S6F). ESEs are generally bound by SR proteins, whereas ESSs usually bind hnRNPs (Dreyfuss et al., 2002; Singh and Valcarcel, 2005). Thus some ncEJC peaks likely represent short footprints from the SR proteins found to copurify with EJC core factors in our proteomics analysis (Figure 2).
Figure 6. Non-canonical EJC (ncEJC) sites.
A-B. First exon ncEJCs.
A. ANKRD11 (top) and OGT exon 1 showing read distributions at canonical (gray) and non-canonical (colored) sites from indicated short footprint libraries.
B. Top: Two related sequence motifs enriched under first exon ncEJC peaks. Bottom: Frequency of occurrence of inset motif among three equal size bins of first exon ncEJC peaks (black dots connected by dashed line) or in scrambled peak sequences (box-plots: IQR and median (horizontal line with box-plot) of the frequencies from 1000 iterations; whiskers are at 1.5 times IQR and outliers are as open circles). Peaks were binned according to their probability-values.
C-D. Internal exon ncEJCs.
C. cEJC and ncEJC sites in KPNB1 exon 10 (top) and BRD2 exon 11 (bottom, color scheme as in A).
D. Left: Dendrogram showing related motifs detected in internal exon ncEJC peaks. Right: Same as B, except internal ncEJC peaks were divided into five equal size bins.
E. Functional classes enriched in ncEJC read densities.
See also Figure S6.
Plotting ncEJC read densities versus transcript RPKM revealed that, like cEJCs, ncEJC signal is most enriched on RNA processing factor and AS-NMD mRNAs (Figure 6E). Further, there is a very high correlation (r = 0.81) between average cEJC signal and ncEJC density across mRNAs (Figure S6G). Finally, as expected, these same gene classes are also enriched in the HMW EJC footprints (data not shown).
cEJCs and ncEJCs collaborate to create higher order mRNP structure
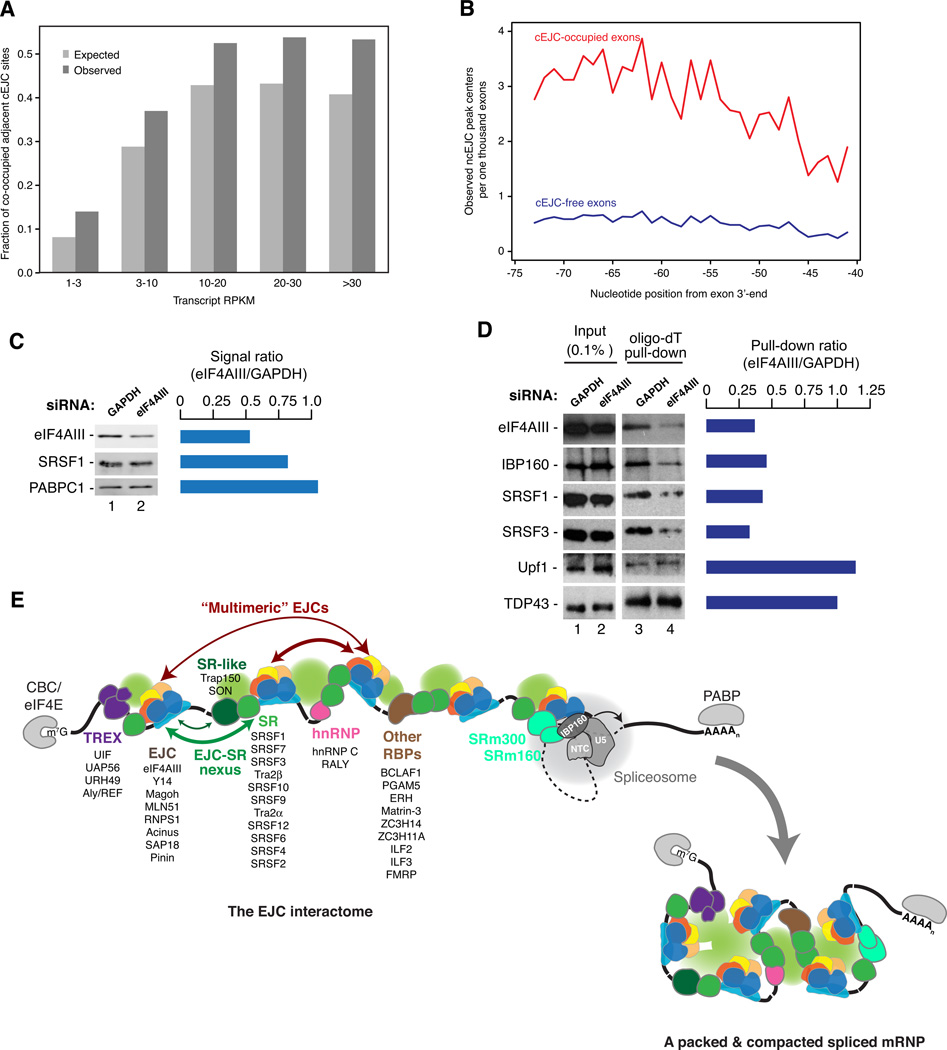
Our biochemical analysis of cellular EJCs had revealed that EJC monomers physically interact in vivo (Figure 1). Such interactions most likely occur between neighboring EJCs. Supporting this, co-occupancy of adjacent cEJC sites was significantly higher than expected (Figure 7A). This effect is strongest between proximal sites and becomes progressively reduced at more distal sites (Figure S7A). A strong correlation was also observed for ncEJC peak co-occurrence on neighboring exons (Figure S7B). Taken together, these data support the hypothesis of stable physical interactions between adjacent EJC binding sites.
Figure 7. A new view of mRNP structure incorporating EJC-EJC and EJC-SR protein collaboration.
A. Co-occupancy of adjacent cEJC sites. Bar-plots showing expected (light gray) and observed (dark gray) frequencies of co-occupied cEJC sites (mappability ≥8) at indicated transcript RPKMs. The two distributions are significantly different (p-value < 1×10−15, chi-squared test).
B. Occurrence of ncEJC peaks on cEJC-occupied (red line) and cEJC-free exons (blue line).
C. Knockdown efficiency of eIF4AIII quantified by Odyssey imaging. Bar plot: Lane 2:1 ratio for each protein.
D. mRNA binding efficiency of proteins upon eIF4AIII knockdown. Western blots showing levels of indicated proteins in total extracts (lanes 1 and 2) and in oligo-dT selected RNAs (lanes 3 and 4) from UV-crosslinked HEK293 cells. Bar plot: Lane 4:3 ratio for each protein.
E. The EJC interactome and a new view of mRNP structure: Solid black line: exonic RNA; Dashed black line: a generic intron; Color ovals: proteins enriched more >10-fold in the EJC proteome (Figure 2) listed in descending order of stoichiometry. Black ovals: undetected proteins known to bind to mRNA ends; Green spheres: bridging protein-protein interactions.
See also Figure S7.
Also striking is the correlation between ncEJC and cEJC peaks. That is, ncEJC peaks are ~2–4 fold more likely to occur adjacent to cEJC-occupied sites than unoccupied sites. This is consistent with our biochemical data showing tight physical association between EJC cores and numerous SR proteins. To determine whether this physical interaction is of any functional consequence, we measured the crosslinking efficiencies of several mRNA binding proteins to polyA+ RNA upon knockdown of eIF4AIII (Figures 7C and 7D). Whereas crosslinking of non-EJC interacting proteins was unaffected by ~50% eIF4AIII knockdown, crosslinking of SRSF1 and SRSF3 was reduced more than two-fold. Thus eIF4AIII abundance, and likely EJC core deposition (Shibuya et al., 2004), functionally impacts recruitment and/or stable association of SRSF1 and SRSF3 with spliced mRNA.
DISCUSSION
A prevailing view based largely on in vitro studies is that EJCs are deposited upstream of every exon-exon junction on mammalian mRNAs, where they exist as singular units decorating the spliced mRNA much like beads on a string. Our new analysis of spliced mRNPs derived from cells, however, shows that while EJCs are deposited on the majority of intron excision sites, this occupancy is not absolute. Further, our data indicate that EJCs exist in higher-order complexes containing multiple EJC cores in intimate contact with other mRNP components, especially SR proteins bound to their preferred target sites. These higher order complexes protect long stretches of RNA from nuclease digestion. Our results thus provide a novel view of cellular mRNPs wherein the regions adjacent to sites of intron excision are protected and compacted by proteinaceous complexes consisting largely of EJCs and SR proteins (Figure 7E). The existence of these complexes explains numerous functional parallels between EJC and SR proteins.
In vivo EJC occupancy
Our data indicate that EJCs are deposited ~24 nts upstream of the vast majority of spliced exon junctions in human mRNAs (Figures 3 and 4). Nonetheless, ~20% of cEJC sites appear unoccupied. Consistent with a previous in vitro study showing that local secondary structure can hamper EJC deposition (Mishler et al., 2008), the strongest contributor to EJC absence in vivo is the propensity of the −24 region to engage in local secondary structure (Figure 4E). cEJC occupied sites, on the other hand, are skewed toward shorter upstream exons and longer downstream introns, features that positively correlate with the presence of ESEs (Dewey et al., 2006) that generally bind SR proteins. As discussed below, this may reflect binding cooperativity between SR proteins and EJCs.
The high degree of cEJC occupancy in human cells contrasts with recent findings from Drosophila melanogaster where only a few introns tested led to EJC deposition (Sauliere et al., 2010). This apparent discrepancy could either be due to the different experimental approaches employed, or real differences in EJC deposition and stability. That mammalian NMD is highly EJC-dependent, whereas Drosophila NMD is largely EJC-independent (Conti and Izaurralde, 2005) supports the latter view. In both organisms, numerous RNA binding proteins regulate their own expression by AS-NMD. In Drosophila, it appears that AS-NMD is EJC-independent and is accomplished primarily by modulating the distance between the stop codon and the polyA tail (i.e., 3′- UTR length) (Hansen et al., 2009). In mammals, however, AS-NMD is most often accomplished by inserting a PTC upstream of at least one intron (i.e., EJC deposition site). Our data showing significant enrichment of both c- and ncEJCs on known ASNMD targets (Figures 5 and 6) supports this tight connection between EJC deposition and AS-NMD in mammalian cells. While we observed no preferential cEJC enrichment downstream of the PTC-inducing AS event, higher overall EJC occupancy could enhance targeting of these transcripts to NMD by (1) preferentially driving them into the translationally active pool (Gudikote et al., 2005; Nott et al., 2004; Wiegand et al., 2003), and then (2) enhancing PTC recognition due to higher cEJC levels downstream. Significant enrichment of ncEJC signal on AS-NMD targets is consistent with previous observations that SRSF1 overexpression stimulates NMD (Sato et al., 2008; Zhang and Krainer, 2004). One possible explanation is that increased SRSF1 occupancy on its preferred sequence motif serves to promote or stabilize adjacent EJC deposition (see below).
Extensive interactions between EJCs and SR proteins
A major surprise in this work is that the in vivo EJC proteome is much more extensive than previously imagined from in vitro studies (Figures 2 and S2). Particularly unexpected was our finding that, not only do numerous SR and SR-like proteins co-purify with the EJC core, but several (SRSF1, 3 and 7) are super-stoichiometric to the core. Consistent with this high abundance, preferred RNA binding sequences for SRSF1 and 3 were significantly enriched under ncEJC peaks in our deep sequencing libraries. Our stringent IP conditions (300mM NaCl, 0.5% Empigen-BB) and extensive nuclease digestion conditions were specifically designed to eliminate all but the most tightly bound EJC-associated factors and RNA footprints. After nuclease digestion, subsequent purification steps usually required 4–5 additional hours. Therefore, any RNA sequence present in our short and long footprint libraries had to be very tightly associated with the EJC core. We conclude that many EJCs exist within larger complexes predominated by SR proteins bound to their specific recognition sequences.
Our finding of a tight physical association between EJCs and SR proteins in vivo is consistent with our previous results showing SR protein co-purification with in vitro assembled EJCs (Le Hir et al., 2000b; Tange et al., 2005), and may help explain the many known functional overlaps between these two protein sets. In addition to promoting NMD (Sato et al., 2008; Zhang and Krainer, 2004), both the EJC and SRSF1 can stimulate translation of bound mRNAs (Gudikote et al., 2005; Ma et al., 2008; Michlewski et al., 2008; Nott et al., 2004; Wiegand et al., 2003). Further, the EJC and multiple SR proteins all promote nucleocytoplasmic mRNA export (Huang and Steitz, 2005; Le Hir et al., 2001). In yet another functional parallel, knockdown of SRSF1 or any EJC core component leads to genomic instability (Li and Manley, 2005; Silver et al., 2010). Finally, both EJC core factors and the SR-like protein RNPS1 are required for splicing of long introns in Drosophila (Ashton-Beaucage et al., 2010; Roignant and Treisman, 2010), and the EJC, RNPS1 and other ASAP complex components are required for splicing of apoptosis gene introns in human cells (Michelle et al., 2012).
Our data strongly suggest that rather than being attributable to functional redundancies, these functional parallels reflect tight physical association between EJCs and SR proteins. Reduced SR protein crosslinking to polyA+ RNA upon eIF4AIII knockdown suggests that EJC core deposition helps stabilize SR protein association with spliced mRNA (Figure 7). Evidence that sequences distal to the cEJC site can promote EJC deposition in Drosophila (Sauliere et al., 2010) further suggests that this relationship is reciprocal (i.e., SR proteins may also stabilize EJC core association). Further, this model is consistent with the cooperative mode of target recognition already proposed for the SR proteins themselves, which generally display weak binding with poor specificity in isolation (Singh and Valcarcel, 2005). The exact nature of the physical interactions between EJC core factors and SR proteins, and the degree to which their many roles in modulating mRNA metabolism are functionally dependent on these physical interactions, remain to be elucidated.
Higher order interactions within mRNPs
In comparison to other large RNPs such as the ribosome and spliceosome, rather little is known about overall mRNP architecture. Electron microscopy of purified Saccharomyces cerevisiae mRNPs has revealed them to be on average 10–15 times shorter than the calculated extended length of their resident mRNAs (Batisse et al., 2009). Even more compacted (~200-fold) are the gigantic Balbiani ring mRNPs assembled on long (~35–40 kB) Chironomus tentans salivary gland mRNAs (Skoglund et al., 1983). Extensive electron microscopy has shown these mRNPs to exist in the nucleus as 50 nm ring-like granules having a distinct coiled and folded structure containing several SR-like proteins (Bjork et al., 2009).
In human mRNPs, EJC proteins are present on the vast majority of exons and they form stable, high molecular weight multimers (Figures 1, 2 and 7). Further, we find a high degree of correlation between cEJC and ncEJC sites (Figure 7). Taken together, these results suggest that physical interactions between and among EJC cores and sequence-specific SR proteins could be major driving forces for mRNP compaction. Such compaction could be necessary for faithful and efficient execution of downstream events such as intra-nuclear mRNP mobility, mRNP export through the nuclear pore, and mRNP transport and translation in the cytoplasm. Of great interest for future studies will be the structural nature of the HMW EJC complexes, and how spliced RNA sequences are packaged within them. Intriguingly, many spliceosomal and mRNP components, including EJC core factors and SR proteins, are enriched in inherently disordered regions (Castello et al., 2012; Korneta and Bujnicki, 2012), and the disordered region in another RNA binding protein, TLS/FUS, was recently shown to mediate phase transition of RNPs into hydrogels (Han et al., 2012; Kato et al., 2012). Thus the presence of EJCs and SR proteins may give cellular mRNPs an intrinsic propensity to adopt hydrogel-like structures.
EXPERIMENTAL PROCEDURES
Extended experimental procedures are provided in the Supplemental Information section.
RNA:protein immunoprecipitation in tandem (RIPiT)
Stable HEK293 cells expressing near endogenous levels of a FLAG-tagged EJC core protein were treated with cycloheximide (100 µg/ml; 1 hr) and sonicated in a buffer containing 300 mM NaCl. RNA:protein complexes captured on FLAG-beads were washed and RNase I treated at 37°C for 10 min. Washed, FLAG-affinity eluted complexes were then subjected to a second IP with an antibody recognizing a different EJC core component.
Mass spectrometry
Stably expressed FLAG-eIF4AIII, FLAG-Magoh or FLAG-peptide were IPed and RNase I digested as above in a lysis buffer also containing 0.5% Empigen BB and 1 µg/ml FLAG peptide. A short unresolved SDS-PAGE gel slice containing IPed, reduced and alkylated proteins was subjected to in-gel trypsin digestion. Chromatographed peptides identified on a LTQ Orbitrap Velos mass spectrometer were searched against the human SwissProt index (09/21/11) using Mascot. Label-free quantitation via extracted ion chromatograms was performed using Mascot Distiller.
Gel filtration
FLAG-eIF4AIII-IPed complexes were fractionated on a 30 ml Sephacryl-S400 HR column. Proteins from 1 ml fractions were precipitated with trichloroacetic acid and western blotted. ~30–100 nt RNA fragments from HMW EJC fractions were extracted to make a deep sequencing library.
Nuclear mRNP footprints
HEK293 nuclei were lysed in a buffer containing 250 mM NaCl and 0.5% Triton X-100. PolyA-containing RNPs from cleared nuclear extract were captured on oligo-dT cellulose and released by a mild RNase I treatment in a low salt buffer. RNase-protected RNAs were extracted and size-selected for deep sequencing.
RNA-Seq and generation of deep sequencing libraries
4 µg polyA+ RNA from FLAG-eIF4AIII expressing cells was base-hydrolyzed at 90°C for 20 min. ~25–36 nt fragments were converted into a deep sequencing library. All short RNA fragment libraries were generated using the small RNA expression kit (Applied Biosystems) following manufacturer recommendations, and sequenced on the SOLiD™ platform.
Mapping and quantification of deep sequencing data
SOLiD™ colorspace reads converted into sequence space using reference sequences (human genome and exon junction sequences) were aligned to reference sequences using Bowtie and viewed on the UCSC genome browser. Uniquely mapping reads from biological replicates of short EJC footprint libraries were combined for peak calling (signal/background probability-value <10−2; peak height = number of reads at the most occupied position). The most reproducible peaks were those common between the FLAG-Magoh:eIF4AIII and FLAG-eIF4AIII:Y14 RIPiT sets. Data analysis was performed using the R statistical software package.
siRNA knockdown and oligo-dT pull-down of polyA+ RNA-crosslinked proteins
HEK293 Flp-In cells transfected (Lipofectamine RNAiMAX) with 50 nM siRNA oligos were UV-crosslinked at 800 mJ/cm2 ~50 hrs post-transfection and lysed in buffer containing 0.5%SDS and 0.5 M NaCl. PolyA+ RNA was captured on oligo-dT cellulose. RNA-crosslinked proteins were eluted by RNase A + T1 digestion, TCA-precipitated and analyzed by western blots.
Supplementary Material
HIGHLIGHTS.
-
-
EJCs reside at ~80% of exon-exon junctions in human mRNAs.
-
-
EJCs and SR proteins multimerize to form high molecular weight complexes.
-
-
EJCs stabilize SR protein association with polyA+ RNA.
-
-
This EJC/SR protein collaboration likely functions in mRNA compaction.
ACKNOWLEDGEMENTS
For valuable antibodies, we thank Elisa Izaurralde (α-Magoh), Jens Lykke-Andersen (α-Upf, α-Y14, α-eIF4AIII), Akila Mayeda (α-RNPS1), and John Blenis (α-SKAR). Frederick Roth and Hon Nian Chua generously provided a modified AlignACE algorithm. Ellie Kittler and the deep sequencing core are thanked for generating the deep sequencing data. We thank members of the Moore lab for discussions and critiques, and Hervé Le Hir for communicating results prior to publication. This work was supported by funding from HHMI and NIH RO1-GM53007 (MJM). G.S. was partially supported by Charles A. King trust post-doctoral fellowship program. MJM is an HHMI Investigator. G.S. and M.J.M. originally conceived the project, and G.S. executed all wet bench experiments. A.K. implemented all computational analyses with input from C.C. and Z.W. on computational method design and statistical analysis. J.D.L and S.A.S. performed mass spec. All authors contributed to data analysis and interpretation. With input from all authors, G.S. and M.J.M. were primarily responsible for writing the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All sequencing data is available in NCBI’s GEO database under accession number GSE41154.
References
- Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol. Cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrancois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, et al. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell. 2010;143:251–262. doi: 10.1016/j.cell.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 2012 doi: 10.1038/nsmb.2380. 10.1038/nsmb.2380. [DOI] [PubMed] [Google Scholar]
- Batisse J, Batisse C, Budd A, Bottcher B, Hurt E. Purification of nuclear poly(A)-binding protein Nab2 reveals association with the yeast transcriptome and a messenger ribonucleoprotein core structure. J. Biol. Chem. 2009;284:34911–34917. doi: 10.1074/jbc.M109.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork P, Jin S, Zhao J, Singh OP, Persson JO, Hellman U, Wieslander L. Specific combinations of SR proteins associate with single pre-messenger RNAs in vivo and contribute different functions. J. Cell Biol. 2009;184:555–568. doi: 10.1083/jcb.200806156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono F, Gehring NH. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 2011;8:24–29. doi: 10.4161/rna.8.1.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cenik C, Chua HN, Zhang H, Tarnawsky SP, Akef A, Derti A, Tasan M, Moore MJ, Palazzo AF, Roth FP. Genome analysis reveals interplay between 5'UTR introns and nuclear mRNA export for secretory and mitochondrial genes. PLoS Genet. 2011;7:e1001366. doi: 10.1371/journal.pgen.1001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5' end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 2005;17:316–325. doi: 10.1016/j.ceb.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Dewey CN, Rogozin IB, Koonin EV. Compensatory relationship between splice sites and exonic splicing signals depending on the length of vertebrate introns. BMC genomics. 2006;7:311. doi: 10.1186/1471-2164-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin. Cell Dev. Biol. 2007;18:186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- Gudikote JP, Imam JS, Garcia RF, Wilkinson MF. RNA splicing promotes translation and RNA surveillance. Nat. Struct. Mol. Biol. 2005;12:801–809. doi: 10.1038/nsmb980. [DOI] [PubMed] [Google Scholar]
- Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149:768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Lareau LF, Blanchette M, Green RE, Meng Q, Rehwinkel J, Gallusser FL, Izaurralde E, Rio DC, Dudoit S, et al. Genome-wide identification of alternative splice forms down-regulated by nonsense-mediated mRNA decay in Drosophila. PLoS Genet. 2009;5:e1000525. doi: 10.1371/journal.pgen.1000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Steitz JA. SRprises along a messenger's journey. Mol. Cell. 2005;17:613–615. doi: 10.1016/j.molcel.2005.02.020. [DOI] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 2007;21:1993–1998. doi: 10.1101/gad.1557907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneta I, Bujnicki JM. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput Biol. 2012;8:e1002641. doi: 10.1371/journal.pcbi.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsensemediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000a;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000b;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Hsu Ia W, Tarn WY. TRAP150 activates pre-mRNA splicing and promotes nuclear mRNA degradation. Nucleic Acids Res. 2010;38:3340–3350. doi: 10.1093/nar/gkq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- McGlincy NJ, Smith CW. Alternative splicing resulting in nonsensemediated mRNA decay: what is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Merz C, Urlaub H, Will CL, Luhrmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S, et al. Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol. Cell. Biol. 2012;32:954–967. doi: 10.1128/MCB.06130-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol. Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–1694. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishler DM, Christ AB, Steitz JA. Flexibility in the site of exon junction complex deposition revealed by functional group and RNA secondary structure alterations in the splicing substrate. RNA. 2008;14:2657–2670. doi: 10.1261/rna.1312808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Reichert VL, Le Hir H, Jurica MS, Moore MJ. 5' exon interactions within the human spliceosome establish a framework for exon junction complex structure and assembly. Genes Dev. 2002;16:2778–2791. doi: 10.1101/gad.1030602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell. 2010;143:238–250. doi: 10.1016/j.cell.2010.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol. Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Sauliere J, Haque N, Harms S, Barbosa I, Blanchette M, Le Hir H. The exon junction complex differentially marks spliced junctions. Nat. Struct. Mol. Biol. 2010;17:1269–1271. doi: 10.1038/nsmb.1890. [DOI] [PubMed] [Google Scholar]
- Shibuya T, Tange TO, Sonenberg N, Moore MJ. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, Liaw HJ, Myung K, Walsh CA, Gaiano N, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat. Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nat. Struct. Mol. Biol. 2005;12:645–653. doi: 10.1038/nsmb961. [DOI] [PubMed] [Google Scholar]
- Skoglund U, Andersson K, Bjorkroth B, Lamb MM, Daneholt B. Visualization of the formation and transport of a specific hnRNP particle. Cell. 1983;34:847- –855. doi: 10.1016/0092-8674(83)90542-1. [DOI] [PubMed] [Google Scholar]
- Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH. CWC22 Connects Pre-mRNA Splicing and Exon Junction Complex Assembly. Cell Rep. 2012 doi: 10.1016/j.celrep.2012.08.017. 10.1016/j.celrep.2012.1008.1017. [DOI] [PubMed] [Google Scholar]
- Stroupe ME, Tange TO, Thomas DR, Moore MJ, Grigorieff N. The three-dimensional arcitecture of the EJC core. J. Mol. Biol. 2006;360:743–749. doi: 10.1016/j.jmb.2006.05.049. [DOI] [PubMed] [Google Scholar]
- Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl. Acad. of Sci. USA. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Involvement of SR proteins in mRNA surveillance. Mol. Cell. 2004;16:597–607. doi: 10.1016/j.molcel.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Krainer AR. Splicing remodels messenger ribonucleoprotein architecture via eIF4A3-dependent and -independent recruitment of exon junction complex components. Proc. Natl. Acad. of Sci. USA. 2007;104:11574–11579. doi: 10.1073/pnas.0704946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.