Abstract
Genome-wide association studies (GWASs) have uncovered susceptibility loci for a large number of complex traits. Functional interpretation of candidate genes identified by GWAS and confident assignment of the causal variant still remains a major challenge. Expression quantitative trait (eQTL) mapping has facilitated identification of risk loci for quantitative traits and might allow prioritization of GWAS candidate genes. One major challenge of eQTL studies is the need for larger sample numbers and replication. The aim of this study was to evaluate the robustness and reproducibility of whole-blood eQTLs in humans and test their value in the identification of putative functional variants involved in the etiology of complex traits. In the current study, we performed comphrehensive eQTL mapping from whole blood. The discovery sample included 322 Caucasians from a general population sample (KORA F3). We identified 363 cis and 8 trans eQTLs after stringent Bonferroni correction for multiple testing. Of these, 98.6% and 50% of cis and trans eQTLs, respectively, could be replicated in two independent populations (KORA F4 (n=740) and SHIP-TREND (n=653)). Furthermore, we identified evidence of regulatory variation for SNPs previously reported to be associated with disease loci (n=59) or quantitative trait loci (n=20), indicating a possible functional mechanism for these eSNPs. Our data demonstrate that eQTLs in whole blood are highly robust and reproducible across studies and highlight the relevance of whole-blood eQTL mapping in prioritization of GWAS candidate genes in humans.
Keywords: gene expression, eQTL, GWAS, whole blood
Introduction
Genome-wide association studies (GWASs) have allowed the identification of susceptibility loci influencing a wide range of complex diseases, including cardiovascular traits, diabetes and Crohn's disease, and quantitative traits such as metabolites and mRNA expression levels.1 The principal outputs of GWAS are a list of SNPs associated with the phenotype of interest. Most of the identified SNPs are intronic and do not alter protein sequence or are located in intergenic regions with unknown functionality. A major remaining challenge is the functional interpretation of GWAS results and confident assignment of the causal variant within the identified LD structure. Regulatory variation has a key role in determining human phenotypic variation and is known to influence disease susceptibility. Integration of functional data such as gene expression profiles with genotypic data allows prioritization of positional candidate genes, thereby providing a functional handle for a better understanding of the etiology of complex traits. Once genetic markers associated with the complex trait in GWAS are identified, gene expression quantitative trait (eQTL) mapping can be performed to examine if the same genetic markers are also associated with quantitative transcriptional levels of one or several transcripts. Initial eQTL studies in model organisms, such as yeast and mice, demonstrated the practicability and utility of eQTL mapping to identify susceptible loci for diseases.2, 3, 4, 5, 6 Most human eQTL studies published so far have analyzed single-cell types, such as lymphocytes or transformed lymphoblast cell lines (LCLs), while only rarely whole blood has been probed.7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 There is currently a high interest in cataloging eQTL data from a wider variety of tissues in order to uncover the tissue-specific proportion of eQTLs. This has fueled a range of eQTL studies in different tissues including brain, liver and adipose tissue.13, 18, 19, 20, 21, 22 The eQTL mapping approach facilitated the identification of several susceptibility loci for quantitative phenotypes, but the extent to which this approach can be used for mapping of disease loci remains to be explored.
The aims of the current study were threefold – (i) to identify eQTLs in humans, (ii) to analyze if these eQTLs were robust and reproducible across different studies and (iii) to elucidate whether whole-blood eQTLs allow to identify putative functional variants involved in the etiology of complex traits.
To investigate the genetic basis of natural variation in gene expression, whole-blood expression levels of 41 409 transcripts (Illumina, San Diego, CA, USA; WG-6 v2) were interrogated for their associations with 335 152 autosomal SNPs (Affymetrix, Santa Clara, CA, USA; 500K array) in 322 individuals from an explorative sample within the population-based KORA F3 discovery cohort. Significant eQTLs identified in the discovery cohort were validated in two independent replication cohorts (KORA F4 and SHIP-TREND).
Materials and methods
Biological samples
The KORA F3 and F4 data sets
The KORA (Cooperative Health research in the region Augsburg) project comprises of individuals from the general population living in the region of Augsburg in South Germany. The KORA F3 data set comprises of 2974 samples, which were collected between 2003 and 2004. The KORA F4 data set includes 6640 samples, which were collected between 2006 and 2008. The subset of individuals analyzed from the KORA F3 and KORA F4 in the current study was non-overlapping. All the participants underwent cross-sectional surveys and regular medical examination by trained staff. Informed consent was obtained from participants and all studies were approved by the local ethical committees. Study design and sampling methods have been previously described.23, 24
The SHIP-TREND data set
The study region of the SHIP-TREND data set is West Pomerania, a region located north-east of Germany. From the total West Pomerania population comprising 212 157 inhabitants, a two-stage cluster sample of adults aged 20–79 years was drawn. Baseline examinations for SHIP-TREND were performed in 2008. Because the Federal State of Mecklenburg/West Pomerania has recently established a central population registry, we used this option for SHIP-TREND to draw a stratified random sample of 8016 adults. Stratification variables are age, sex and place of residence. Subjects were sampled from the regional strata with a probability proportional to size design. Age/sex strata within counties were of equal size. Study design and sampling methods have been previously described.25
Gene expression profiling and genotyping
The expression profiling and genotyping workflow for the discovery and replication cohorts are summarized in Supplementary Figure S1.
KORA F3 discovery cohort
Gene expression profiling was performed using the Illumina Human-6 v2 Expression BeadChips as described elsewhere.23 Briefly, blood samples were collected under fasting conditions in PAXgene (PreAnalytiX, Hornbrechtikon, Switzerland) Blood RNA tubes. RNA extraction was performed using the PAXgene Blood RNA Kit (Qiagen, Hilden, Germany) and RNA was reverse transcribed and biotin-UTP-labeled into cRNA using the Illumina TotalPrep RNA Amplification Kit (Ambion, Austin, TX, USA). The cRNA was quantified using Ribogreen and the Bioanalyzer (Agilent Technologies, Boeblingen, Germany) before it was hybridized on the Illumina Human-6 v2 Expression BeadChip.
Genotyping was performed using Affymetrix 500K arrays. Hybridization of genomic DNA was done in accordance with the standard recommendations from the manufacturer. Genotypes were determined using BRLMM clustering algorithm (http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf). The genotypes were determined in batches of at least 400 chips. For quality control purposes, a positive and a negative control DNA was applied for every 48 samples.
KORA F4 replication cohort
Total RNA was extracted from whole blood under fasting conditions according to the manufacturer's instructions using the PAXgene Blood miRNA Kit (Qiagen). Purity and integrity of the RNA was assessed on the Agilent Bioanalyzer with the 6000 Nano LabChip reagent set (Agilent Technologies, Germany). Using the Illumina TotalPrep-96 RNA Amp Kit (Ambion, Darmstadt, Germany), 500 ng of RNA was reverse transcribed and biotin-UTP-labeled into cRNA. A total of 3000 ng of cRNA was hybridized to the Illumina Human HT-12 v3 Expression BeadChip, followed by washing steps as described in the Illumina protocol. The samples were genotyped on the Affymetrix 6.0 GeneChip array. The genotyping procedures are described elsewhere.26
SHIP-TREND replication cohort
RNA was prepared from whole blood under fasting conditions in PAXgene tubes (PreAnalytiX) using the PAXgene Blood miRNA Kit (Qiagen) on a QIAcube (Qiagen). DNA was isolated using the Gentra Puregene Blood Kit (Qiagen). Isolation of both RNA and DNA was performed according to the Qiagen protocols. Purity and concentration of RNA and DNA preparations were determined using a NanoDrop ND-1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Wilmington, NC, USA). To ensure a constant high quality of the RNA preparations, all the RNA samples were analyzed using RNA 6000 Nano LabChips (Agilent Technologies, Germany) on a 2100 Bioanalyzer (Agilent Technologies, Germany) according to the manufacturer's instructions. The integrity of all DNA preparations was validated by electrophoresis using 0.8% agarose-1x TBE gels stained with ethidium bromide. Processing of the RNA samples using Illumina Human HT-12 v3 Expression BeadChip arrays, as well as processing of the DNA samples using Illumina HumanOmni2.5-Quad arrays, was performed at the Helmholtz Zentrum Munich.
Data analysis
KORA F3 discovery cohort
The raw genotype and expression data were exported from the Illumina Software Genome Studio to R (http://www.R-project.org). The 500 568 SNPs from the Affymetrix 500K array were filtered using a minor allele frequency >0.05, Hardy–Weinberg P-value of <10−6 and genotyping efficiency of >95%, allowing 335 152 high-quality SNPs for further analysis. The SNP positions were updated using the hg18.SNP129 database. The average genotyping efficiency across the individuals for 335 152 SNPs was 99.11%.
The expression data was logarithmized and normalized using locally weighted smoothing algorithm (LOESS27) in R. The 50 bp probe sequences available from Illumina Human-6 v2 array annotations were mapped against the human reference sequence hg18, allowing only hits with >48 bp matches and no gaps. Of the 48 701 Illumina Human-6 v2 array probes, 7292 probes, which did not uniquely map to the human genome, were excluded from the analysis. Of the remaining 41 409 probes, 27 623 probes mapped within the annotated transcripts (25 657 probes within refSeq transcripts and 1966 probes within UCSC transcripts), whereas 13 786 probes mapped to the intergenic regions in the human genome. Stringent removal of probes for trans eQTLs was performed in accordance with cross-hybridization filtering performed by Fehrmann et al.14
Reproducibility of the microarray expression data was tested using three pairs of technical replicates and three triplets of biological replicates (Supplementary Figure S1). Linear regression algorithms implemented in the statistical analysis packages R and PLINK28 were used to test for associations between SNPs and gene expression levels. All the regressions were adjusted for gender and age. To account for multiple testing, the stringent Bonferroni correction was used. Quality checks for confounding due to population stratification was performed by calculating the genomic inflation factors for all transcripts (Supplementary Figure S2).
The power calculations were performed using QUANTO version 1.229 and assuming an additive genetic effect for continuous traits using the KORA F3 discovery cohort Bonferroni thresholds.
Previously reported SNPs significantly associated with either complex traits or quantitative traits were interrogated using the publicly available eQTL browser (http://eqtl.uchicago.edu/cgi-bin/gbrowse/eqtl/) incorporating results from 13 eQTL studies to date.
KORA F4 and SHIP-TREND replication cohorts
Genotyping was performed using the Affymetrix 6.0 arrays (KORA F4) and HumanOmni2.5-Quad arrays (SHIP-TREND). Gene expression profiling was performed on Illumina Human HT12-v3 arrays for KORA F4 and SHIP-TREND (Supplementary Figure S3).
The raw intensity data generated with the expression arrays were exported from Illumina software Genome Studio to R and processed (log transformation and quantile normalization30) using the lumi_1.12.4 package31 from the Bioconductor open source software (http://www.bioconductor.org/).
The significant SNP–probe combinations identified in the KORA F3 discovery cohort were tested in the replication cohorts. For the KORA F4 replication cohort, identical SNP–probe combinations were tested for all eQTLs. For the SHIP-TREND replication cohort, additional proxy SNPs were used in cases where the SNPs from the discovery stage analysis were not present on the genotyping array. Proxy search was carried out using the SNAP SNP Annotation and Proxy Search tool32 and only proxy SNPs with R2≥0.6 were used for testing. Concordance in allele effect directions were compared while taking into account the strand orientation and allele frequencies.
Results
KORA F3 discovery cohort
Mapping of whole-blood cis and trans eQTLs in the KORA F3 discovery cohort
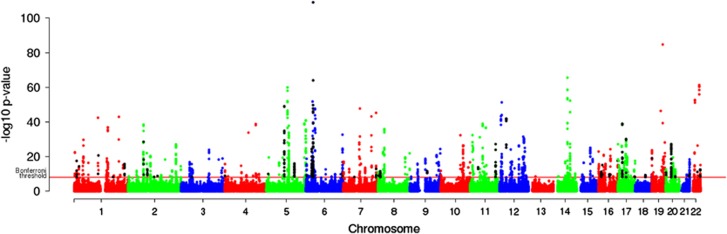
For identification of cis regulation, a cis window of +/−500 kb from the transcript coordinates was defined based on previous reports demonstrating that >90% cis SNPs were situated within 100 kb of the transcription start site.11, 18 A total of 4 802 373 SNP–probe combinations were tested for cis regulation in the KORA F3 discovery cohort. After Bonferroni correction (threshold=1.03 × 10−8), significant evidence for cis-acting regulatory variation was identified for 2149 SNP–probe pairs, comprising 363 eQTLs based on unique number of transcripts (∼1% of RefSeq genes, minimal P-value: 1.98 × 10−108)(Figures 1 and 2). For the 363 significant cis eQTLs, the cis-acting SNPs explained a substantial proportion of the total variability in gene expression: the mean expression variability explained by the SNP for each probe was 19% (median variance explained=15%) (Supplementary Figure S4). In all, 50 out of 363 cis eQTL probes contain a SNP within the transcript probe used on the Illumina expression microarray. In the current study, we assessed possible confounding of cis eQTL results due to SNP-under-probe effects and identified 10 cis eQTLs (2.8%) where the association could be caused by the SNP within the probe. For the remaining 40 eQTLs, the probe–SNPs were not associated with the eQTL SNP and among these were 20 probe–SNPs pairs representing rare variants with a minor allele frequency <1%.
Figure 1.
Manhattan plot of significant cis eQTLs in the KORA F3 discovery cohort. Manhattan plot of −log10 P-value (Y-axis) across the chromosomes (X-axis). The red line indicates the Bonferroni threshold of significance. The black dots indicate Illumina probes with annotated SNPs within their sequence. A total of 2149 SNP–probe pairs corresponding to 363 eQTLs were significant after Bonferroni corrections for multiple testing.
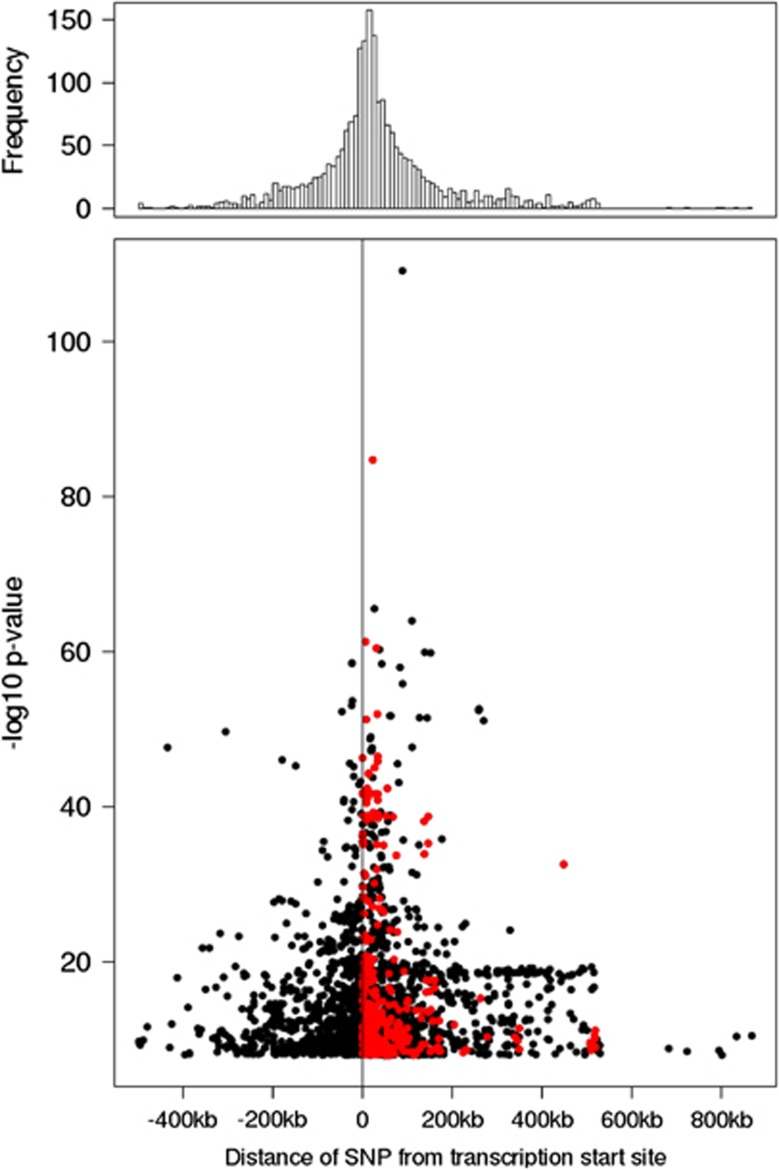
Figure 2.
Plot of distance of the SNP from the transcriptional start site (TSS) of the cis transcript in the KORA F3 discovery cohort. Histogram (10 kb window) and scatter plot of the eQTL distribution in KORA F3 discovery cohor,t depicting the distance of SNP from the TSS of the cis transcript. The red dots indicate SNPs located within the transcript.
To identify variants acting in trans, an exaustive association analysis of all the SNPs across the 41 409 transcripts outside of the defined cis window was performed. We attempted to falsify the trans eQTLs based on recent work by Fehrmann et al,14 indicating that cross-hybridization results in an inflation of the trans effects. After exclusion of possible false positives due to cross-hybridization and retaining only trans eQTLs where the SNP was on a different chromosome than the trans probe or >4 Mb from the probe, at a Bonferroni P-value threshold (threshold=3.6 × 10−12), 37 significant SNP–probe pairs corresponding to 8 trans eQTLs were identified. On average, for the eight significant trans eQTLs, the trans regulation accounted for 18.8% variance in overall expression (median variance explained=18.7%). No evidence of master regulators or possible eQTL hotspots was observed. Several identified trans eQTLs are amenable. For instance, variants in PDL5, which was found to be expressed significantly higher in a mouse model of Alzheimer's disease,33 were significantly associated with the expression levels of SLC8A2, which was found to co-localize with amyloid beta in cerebral cortex of Alzheimer's patients.34
Adjusting for possible confounders in the KORA F3 discovery cohort
To adjust for possible confounding effects of different cell counts in whole blood, the number of white and red blood cells were used as covariates in the linear regression models for the eQTL analysis. A total of 2140 cis SNP–probe pairs (352 eQTLs, 97% of all significant cis eQTLs) and all 65 trans SNP–probe pairs remained significant in KORA F3 after adjusting for the cell counts (Supplementary Table 1). Correcting for red and white blood cell counts did allow the identification of 18 additional eQTLs. In summary, only 3% of the significant cis eQTLs did not pass the significance threshold after correcting for the two cell counts, indicating that varying white and red cell counts exert a minor effect on the expression levels in this study.
Another possible confounder in the eQTL analysis is the presence of a sequence variation within the transcript probe used on the Illumina expression microarray. To check if the significant eQTLs in the discovery sample might be biased due to SNPs within the probes, the Illumina re-annotation pipeline from Barbosa-Morais et al,35 was used. Indeed we identified 10 eQTLs with a SNP in the probe, which is in high LD (R2>0.7) with the eQTL SNPs. For these eQTLs, the association could be caused by the SNP within the probe. However, several studies have previously reported allelic expression for eQTLs where Illumina probes have known SNPs within their sequence and demonstrated that SNPs-within-probes effects are not a significant problem in interpretation of eQTL results.9, 18 Although we have not excluded probes with annotated SNPs within their sequence, we provide a list of all probes containing annotated SNPs within their sequence in Supplementary Table 1.
Replication of whole-blood eQTLs in two independent cohorts
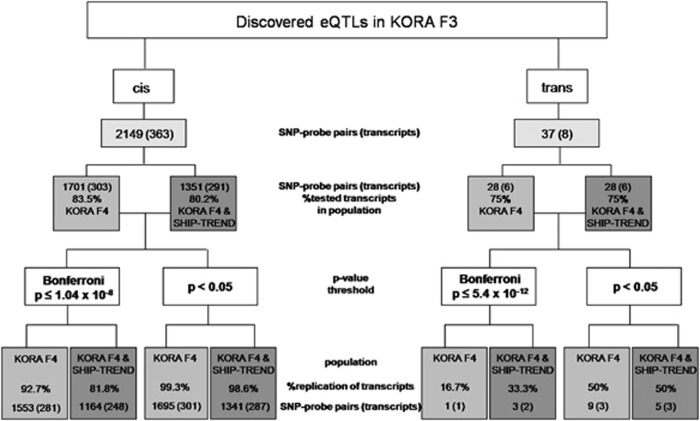
One major drawback in eQTL studies performed in humans to date is the lack of replication of eQTLs across the studies and the difficulty to compare eQTL results between different studies. To address these concerns, we tested the robustness of whole-blood eQTLs by replication in whole-blood data from two independent general population cohorts of Caucasians (KORA F4: n=740 and SHIP-TREND: n=653, Figure 3). We chose two different significance thresholds to identify eQTLs, which could be replicated in the KORA F4 and SHIP-TREND cohorts. Replication rates of 98.6% for the cis eQTLs and 40% for the trans eQTLs using P=0.05 and 81.8% for the cis eQTLs and 20% for the trans eQTLs using the discovery sample Bonferroni thresholds were observed for the eQTLs, which were tested in both data sets (Figure 4). The number of samples required to detect eQTL variance with 50, 80 and 90% power in the current study is depicted in Supplementary Figure S5. Based on these power calculations, both KORA F4 and SHIP-TREND have >90% power to detect the significant cis and trans eQTLs from the discovery cohort.
Figure 3.
Flowchart of the number of KORA F3 discovery cohort eQTLs tested and replicated in KORA F4 (n=740) and SHIP-TREND (n=653) replication cohorts. The left panel indicates replication results of the cis eQTLs, whereas the right panel indicates replication results of the trans eQTLs. Replication rates are indicated at P=0.05 and at the discovery sample Bonferroni thresholds for eQTLs, which could be successfully replicated in KORA F4 and SHIP-TREND replication cohorts.
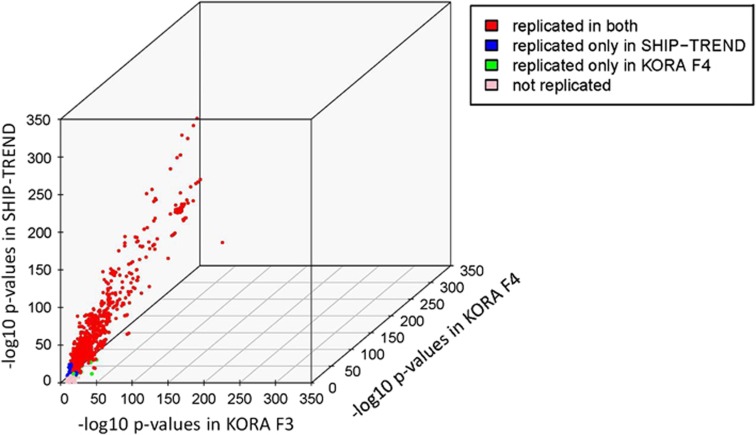
Figure 4.
3D Scatter plots of eQTL P-values in the discovery and replication cohorts. 3D scatter plots of P-values of all KORA F3 discovery cohort eQTLs versus P-values of KORA F4 and SHIP-TREND replication cohorts at the Bonferroni threshold of significance (P=1.03 × 10−8 for cis and P=3.6 × 10−12 for trans). Red dots indicate eQTLs replicated in both KORA F4 and SHIP-TREND, blue dots indicate QTLS replicated only in SHIP-TREND and green dots indicate eQTLs replicated only in KORA F4. The pink dots indicate eQTLs that could not be replicated.
Comparison of results with published peripheral blood eQTLs
We further compared our eQTL results with a recently published study, which assessed eQTLs in whole-peripheral blood from 1469 unrelated individuals.14 Fehrmann et al14 used the same array (Illumina HumanHT-12) but a different genotyping platform (Illumina HumanHap 300) resulting in an overlap of only about 50 000 shared SNPs between the Fehrmann and the KORA study. Although only 10% of the SNPs investigated in our study were available from Fehrmann et al,14 a total of 32% cis eQTLs (117/363 eQTLs) and 14% trans eQTLs (1/7 eQTLs) with identical probe–SNP combinations could be confirmed in the recently published study (Supplementary Table 1). The high replication rate provides additional support for the robustness of eQTLs in whole blood.
eQTL mapping of complex trait-associated variants
We next examined if SNPs reported to be associated with either complex traits or quantitative traits in GWAS were significantly associated with whole-blood mRNA levels. In October 2011, 1058 GWAS studies for 566 different traits had been added to the NHGRI GWAS catalog.36 A total of 7995 unique SNP–probe combinations (3699 unique SNPs and 2977 unique genes) were systematically tested using the KORA F3 discovery cohort data. Of those tested, 639 SNPs (17% of tested SNPs) were nominally associated with expression levels of the reported transcript (P=0.05) and 79 eSNPs (2% of tested SNPs) remained significant after Bonferroni correction (threshold=6.25 × 10−6) in the KORA F3 discovery cohort. Of these 79 eSNPs, 59 SNPs were previously reported to be associated with disease loci, whereas 20 were associated with quantitative trait loci.
The variance in expression explained by the 79 eSNPs ranged 5–52%, with a median variance of 11% (Table 1). For 8 of these 79 eSNPs, no significant eQTLs in any tissue have been published previously till date, whereas another 43 previosuly reported eSNPs could be validated in the current data set. The remaining 28 eSNPs were located within the HLA region. Because the LD within the HLA region is so extensive, the biological significance of these SNPs remains elusive. Although blood is not directly linked to some of these traits, several interesting candidates such as loci involved in chronic lymphocytic leukemia, beta thalassemia, hematological and biochemical traits were also present. The eight novel loci included one disease and one quantitative trait locus for which the eQTL allowed prioritization of the candidate over other transcripts in the vicinity (Supplementary Table 2).
Table 1. List of eight novel GWAS catalog eSNPs significantly associated with expression levels of the reported transcript in KORA F3.
| GWAS locus | GWAS trait | SNP | Gene | Probe ID | P-value | Adjusted R2 | Beta | Major allele | First author |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Beta thalassemia/hemoglobin E disease | rs2071348 | HBE1 | 6520176 | 0.283414 | −0.00503376 | −0.18484 | T | Nuinoon M |
| rs2071348 | HBG2 | 6400079 | 5.38E-08 | 0.09514532 | 0.52015 | T | |||
| rs2071348 | HBG2 | 6620605 | 0.030536 | 0.00723574 | −0.01432 | T | |||
| rs2071348 | HBG2 | 940181 | 0.365623 | −0.00315586 | 0.006403 | T | |||
| rs2071348 | HBG1 | 4150187 | 1.94E-06 | 0.07676854 | 0.443254 | T | |||
| rs2071348 | HBD | 6250037 | 0.195357 | −0.00360737 | −0.11191 | T | |||
| rs2071348 | HBBP1 | 7100747 | 0.881443 | −0.00768138 | −0.00186 | T | |||
| 2 | Crohn's disease | rs2058660 | IL18RAP | 6770424 | 0.154442 | −0.00297075 | −0.01054 | A | Franke A |
| rs2058660 | IL18RAP | 5130475 | 2.68E-15 | 0.18068679 | −0.44984 | A | |||
| rs2058660 | IL12RL2 | NA | NA | NA | NA | NA | |||
| rs2058660 | IL18R1 | 1500328 | 0.698045 | −0.00654905 | −0.01298 | A | |||
| rs2058660 | IL1RL1 | 670411 | 0.909656 | 0.00344823 | −0.00091 | A | |||
| rs2058660 | IL1RL1 | 3870753 | 0.349253 | −0.00614557 | −0.00746 | A | |||
| 3 | Graves' disease | rs9355610 | RNASET2 | 5310131 | 7.05E-19 | 0.21555351 | 0.27063 | G | Chu X |
| rs9355610 | FGFR1OP | 6580446 | 0.424956 | −0.00652282 | −0.00825 | G | |||
| 4 | Body mass index | rs7359397 | SH2B1 | 6620092 | 0.133159 | 0.00166276 | 0.032315 | C | Speliotes EK |
| rs7359397 | APOB48R | 2070044 | 0.927515 | 0.0020211 | 0.002928 | C | |||
| rs7359397 | SULT1A2 | 1740113 | 0.095138 | 0.01485394 | −0.01268 | C | |||
| rs7359397 | SULT1A2 | 1980554 | 0.316768 | −0.00467373 | 0.010131 | C | |||
| rs7359397 | AC138894.2 | NA | NA | NA | NA | NA | |||
| rs7359397 | ATXN2L | 990524 | 0.21315 | 0.00104259 | 0.007658 | C | |||
| rs7359397 | ATXN2L | 1300541 | 0.227558 | −0.001122 | 0.007915 | C | |||
| rs7359397 | ATXN2L | 5720435 | 0.668993 | −0.00590007 | 0.005323 | C | |||
| rs7359397 | TUFM | 6270735 | 4.89E-10 | 0.10666734 | 0.139009 | C | |||
| 5 | Systemic lupus erythematosus | rs131654 | HIC2 | 7050673 | 0.210732 | −0.00152262 | −0.01773 | T | Han JW |
| rs131654 | UBE2L3 | 770523 | 0.179421 | 0.00400821 | 0.011055 | T | |||
| rs131654 | UBE2L3 | 1050360 | 1.24E-06 | 0.06346902 | −0.15129 | T | |||
| 6 | Asthma | rs11078927 | GSDMB | 6620170 | 4.02E-18 | 0.20898095 | −0.22407 | C | Torgerson DG |
| 7 | Alzheimer's disease | rs6859 | PVRL2 | 2570544 | 7.03E-07 | 0.07723456 | −0.18595 | G | Abraham R |
| Alzheimer's disease (late onset) | rs6859 | TOMM40 | 3400747 | 0.462314 | 0.00170593 | 0.013467 | G | Naj AC | |
| rs6859 | APOE | 4150338 | 0.686794 | −0.00123172 | 0.002674 | G | |||
| 8 | Alcohol dependence | rs8062326 | SYT17 | 730725 | 5.72E-07 | 0.0710058 | 0.798277 | G | Lydall GJ |
| ITPRIPL2 | 2710551 | 0.998 | 0.003417 | 0.001 | G |
Abbreviations: NA, transcript failed alignment to genome hence removed from analysis.
These eight SNPs have not yet been reported to be associated with expression levels of the neigbouring transcripts. For several loci, the GWAS SNP was significantly associated with expression levels of one transcript (in bold), thereby allowing prioritization of this transcript compared with the other candidates in the vicinity of the SNP.
Discussion
An ongoing challenge in human genetics research is the detection of functional non-coding genetic variants. Mapping of regulatory variation in humans has the potential to facilitate the identification of susceptibility loci for complex traits. In the current study, we performed comprehensive eQTL mapping in whole blood from 322 individuals who were representative of a general Caucasian population. Given the relatively small size of our discovery sample, we had limited power to detect low-effect eQTLs. Despite this, even after stringent corrections for multiple testing, we identified a total of 363 cis and 8 trans eQTLs, numbers comparable to previous eQTL studies performed in other tissues.
Only few genome-wide replications have been performed.8, 11, 37 Owing to variant statistical methods and multiple testing corrections used, accurate comparisons of eQTLs across different studies are difficult. For instance, for the extensively investigated LCLs, genome-wide replication rates <40% for cis and 15% for trans eQTLs in humans have been previously reported.8, 11, 37 A recent study comprising of 206 discovery samples and two replication samples (n=60 and n=266) reported replication rates of 47.6% for cis and 6% for trans eQTLs in liver at P<0.05.13 In the current study, using 322 discovery samples and two independent replication cohorts (KORA F4 (n=740) and SHIP-TREND (n=653) cohorts), we demonstrated replication rates of 98.6% for the cis and 50% for the trans eQTLs at P=0.05 in whole blood.
In the current study, we assessed possible confounding of cis eQTL results due to the presence of a sequence variation within the transcript probe used on the Illumina expression microarray and identified 10 cis eQTLs (2.8%) where the association could be caused by the SNP within the probe.
One major caveat of eQTL mapping is the cell and tissue specificity of gene expression patterns. For eQTL mapping, it would be ideal to study gene expression in the affected tissue. As obtaining disease tissue samples are not feasible due to practical, ethical, legal and social issues, whole blood constitutes a surrogate tissue to assay gene expression. Peripheral blood cells have been reported to share >80% of the transcriptome with nine tissues: brain, colon, heart, kidney, liver, lung, prostate, spleen and stomach.38 Moreover, whole blood is an easily accessible resource, where RNA can be prepared at low cost and with standardized preanalytic protocols. The use of whole blood in eQTLs has been hindered by the concern of using a mixed tissue with varying proportions of cell counts. Despite possible confounding due to white and red cell counts, replication rates of 81.8% for cis and 20% for trans eQTLs in blood, even after stringent Bonferroni correction for multiple testing, far surpassed those reported for LCLs, thereby confirming the robustness and reproducibility of eQTLs in whole blood.
The value of whole-blood eQTLs was further assessed by analyzing SNPs previously reported to be associated with disease or quantitative phenotypes. Using the KORA F3 expression profiles, we uncovered 79 eSNPs (24 novel), demonstrating the utility of these eQTLs in prioritizing possible functional variants identified in GWAS. For several instances where the identity of the true causal locus from the GWAS results was not evident, expression levels of only one transcript were significantly associated with the GWAS SNP, allowing prioritization of this transcript over other candidates.
Identification of eSNPs for several non-blood related phenotypes, such as neurological diseases, provided further evidence that blood can be used as a proxy for tissue-independent eQTLs. High replication rates of whole-blood eQTLs override possible confounding factors that have hindered the use of whole blood in human eQTL mapping until now. Taken together, our results demonstrate the feasibility and robustness of eQTL mapping in whole blood and provide hypotheses for regulatory involvement of variants associated with complex traits.
Acknowledgments
We gratefully acknowledge the support of K Junghans, A Löschner, T Pham, M Borzes and K Chow in expression profiling and analyzing and P Lichtner for genotyping. We thank A Hoffmann and G Gornitzka for excellent technical assistance in the KORA F4 study. This study was supported in part by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD e.V.), the German Center for Heart Research (DZHK no. Z56010015300) and the project Systems Biology of Metabotypes (SysMBo no. 0315494A). Further support for this study was obtained from the Federal Ministry of Health (Berlin, Germany), the Ministry of Innovation, Science, Research and Technology of the state North-Rhine Westphalia (Düsseldorf, Germany) and the Federal Ministry of Education, Science, Research and Technology (NGFNplus AtheroGenomics/01GS0423; Berlin, Germany). The KORA research platform was initiated and financed by the Helmholtz Center Munich, German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Part of the financing was provided by the German National Genome Research Network (NGFN-2 and NGFNPlus: 01GS0823). KORA research was also supported within the Munich Center of Health Sciences (MC Health) as part of LMUinnovativ. SHIP-TREND is part of the Community Medicine Research net of the Ernst-Moritz-Arndt-University Greifswald, Germany, which is funded by the Federal Ministry of Education and Research, the Ministry of Cultural Affairs, and the Social Ministry of the Federal State of Mecklenburg-West Pomerania. Genome-wide SNP and expression data have been generated with support by the Federal Ministry of Education and Research (grant no. 03ZIK012).
Author contributions
Project planning was done by TM, HP, MN and UV. HP, GH and UV provided the experimental design. KORA sample collection was performed by TI, CH and MR and SHIP-TREND sample collection by MN. Genotyping was performed by CG. Expression profiling and analysis were carried out by DM, KH, MC, CS, GE and GH. Manuscript writing was carried out by DM, KH, TM and HP and critical revision of the manuscript by all the authors.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Bing N, Hoeschele I. Genetical genomics analysis of a yeast segregant population for transcription network inference. Genetics. 2005;170:533–542. doi: 10.1534/genetics.105.041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi H, Togawa K, Moritani M, Itakura M. Identification of candidate genes in the type 2 diabetes modifier locus using expression QTL. Genomics. 2005;85:591–599. doi: 10.1016/j.ygeno.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2011;27:72–79. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–282. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Murphy A, Chu JH, Xu M, et al. Mapping of numerous disease-associated expression polymorphisms in primary peripheral blood CD4+ lymphocytes. Hum Mol Genet. 2010;19:4745–4757. doi: 10.1093/hmg/ddq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Innocenti F, Cooper GM, Stanaway IB, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehrmann RS, Jansen RC, Veldink JH, et al. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet. 2011;7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Badner JA, Cheng L, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants Mol Psychiatry 2012. e-pub ahead of print 3 January 2012; doi: 10.1038/mp.2011.174 [DOI] [PMC free article] [PubMed]
- Rotival M, Zeller T, Wild PS, et al. Integrating genome-wide genetic variations and monocyte expression data reveals trans-regulated gene modules in humans. PLoS Genet. 2011;7:e1002367. doi: 10.1371/journal.pgen.1002367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wolfs MG, Deelen P, et al. Unraveling the regulatory mechanisms underlying tissue-dependent genetic variation of gene expression. PLoS Genet. 2012;8:e1002431. doi: 10.1371/journal.pgen.1002431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cheng L, Badner JA, et al. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring A, Gieger C, Mehta D, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- Holle R, Happich M, Lowel H, Wichmann HE. KORA – a research platform for population based health research. Gesundheitswesen. 2005;67 (Suppl 1:S19–S25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- Volzke H, Alte D, Schmidt CO, et al. Cohort profile: the study of health in Pomerania. Int J Epidemiol. 2010;40:294–307. doi: 10.1093/ije/dyp394. [DOI] [PubMed] [Google Scholar]
- Marzi C, Albrecht E, Hysi PG, et al. Genome-wide association study identifies two novel regions at 11p15.5-p13 and 1p31 with major impact on acute-phase serum amyloid A. PLoS Genet. 2010;6:e1001213. doi: 10.1371/journal.pgen.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland WS. Robust locally weighted regression and smoothing scatterplots. J Am Stat Assoc. 1979;74:829–836. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, MJ QUANTO.1.1: A computer program for power and sample size calculations for genetic-epidemiology studies http://hydrauscedu/gxe , 2006
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George AJ, Gordon L, Beissbarth T, et al. A serial analysis of gene expression profile of the Alzheimer's disease Tg2576 mouse model. Neurotox Res. 2009;17:360–379. doi: 10.1007/s12640-009-9112-3. [DOI] [PubMed] [Google Scholar]
- Sokolow S, Luu SH, Headley AJ, et al. High levels of synaptosomal Na(+)-Ca(2+) exchangers (NCX1, NCX2, NCX3) co-localized with amyloid-beta in human cerebral cortex affected by Alzheimer's disease. Cell Calcium. 2011;49:208–216. doi: 10.1016/j.ceca.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38:e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nica AC, Parts L, Glass D, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2010;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew CC, Ma J, Tang HC, Zheng R, Dempsey AA. The peripheral blood transcriptome dynamically reflects system wide biology: a potential diagnostic tool. J Lab Clin Med. 2006;147:126–132. doi: 10.1016/j.lab.2005.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.