Abstract
Constitutional mismatch repair deficiency (CMMR-D) syndrome is a rare inherited childhood cancer predisposition caused by biallelic germline mutations in one of the four mismatch repair (MMR)-genes, MLH1, MSH2, MSH6 or PMS2. Owing to a wide tumor spectrum, the lack of specific clinical features and the overlap with other cancer predisposing syndromes, diagnosis of CMMR-D is often delayed in pediatric cancer patients. Here, we report of three new CMMR-D patients all of whom developed more than one malignancy. The common finding in these three patients is agenesis of the corpus callosum (ACC). Gray matter heterotopia is present in two patients. One of the 57 previously reported CMMR-D patients with brain tumors (therefore all likely had cerebral imaging) also had ACC. With the present report the prevalence of cerebral malformations is at least 4/60 (6.6%). This number is well above the population birth prevalence of 0.09–0.36 live births with these cerebral malformations, suggesting that ACC and heterotopia are features of CMMR-D. Therefore, the presence of cerebral malformations in pediatric cancer patients should alert to the possible diagnosis of CMMR-D. ACC and gray matter heterotopia are the first congenital malformations described to occur at higher frequency in CMMR-D patients than in the general population. Further systematic evaluations of CMMR-D patients are needed to identify possible other malformations associated with this syndrome.
Keywords: constitutional mismatch repair deficiency syndrome, agenesis of corpus callosum, gray matter heterotopia, biallelic germline mutation, childhood cancer
Introduction
Constitutional mismatch repair deficiency (CMMR-D) syndrome is a rare childhood cancer syndrome caused by biallelic germline mutations in one of the four mismatch repair (MMR) genes, MLH1, MSH2, MSH6 or PMS2.1 The MMR genes are mainly involved in the correction of single base-pair mismatches and small insertion–deletion loops that arise during DNA replication. Moreover, the MMR system is involved in the cellular response to a variety of agents that damage DNA.2 Monoallelic germline mutations in these MMR genes are responsible for Lynch syndrome (LS). The first reported CMMR-D patients were descendants from consanguineous LS families.3, 4 Since then more than hundred CMMR-D patients from ∼68 families have been reported. The broad tumor spectrum deduced from all published CMMR-D patients can be divided into four main groups:5 (1) hematological malignancies;6 (2) brain/CNS tumors;7 (3) LS-associated tumors as well as multiple intestinal polyps;8, 9 and (4) other malignancies including embryonic tumors and rhabdomyosarcoma. Many CMMR-D patients show signs reminiscent of neurofibromatosis type 1 (NF1). In particular, multiple café au lait macules (CALMs) are often found as a first manifestation of the underlying cancer syndrome. Areas of skin hypopigmentation have also been reported in CMMR-D patients. It has therefore been suggested that all patients with a childhood cancer, not clearly associated with NF1, who show the skin abnormalities as seen in NF1 should be suspected of having CMMR-D syndrome. Furthermore, defects in immunoglobulin class switch recombination (indicated by decreased levels or absence of IgG2, IgG4 and IgA together with increased IgM levels) are also associated with CMMR-D syndrome.10, 11 At least one CMMR-D patient initially presented with a primary immunodeficiency.11
Despite publication of >100 CMMR-D patients, diagnosis may still be delayed in many childhood cancer patients. Here, we report three new patients confirming this, as they were suspected to suffer from CMMR-D only when they developed their second or third malignancy. The common finding in these three patients is agenesis of the corpus callosum (ACC) and cerebral gray matter heterotopia in two of them. These congenital malformations may be used as an additional feature suggestive of the diagnosis of CMMR-D in childhood cancer patients.
Patients and methods
Patients
Suspicion of CMMR-D syndrome was raised in all three patients when they developed a second or third malignancy. Parents gave written informed consent to molecular genetic testing. The main clinical findings of the patients are summarized in Table 1.
Table 1. List of three CMMR-D patients with agenesis of the corpus callosum and cerebral heterotopias.
| Patient | Gene | Mutations | Family history | Malignant tumors (age in years at diagnosis or at death of disease) | Signs of neurofibromatosis or other non-neoplastic features | Results of microsatellite instability (MSI) | Results of immunohistochemical analysis in tumor | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | PMS2 | c.(2397_2400del)+(2397_2400del) p.(Ser801GlufsX15)+(Ser801GlufsX15) | Consanguineous parents (paternal grandfather and maternal grandmother are sibs); no FH of cancer (one brother died at early age of unknown cause) | B-cell NHL (9 years), muco-epidermoïd carcinoma of the parotis (11 years), T-cell NHL (11.5 years) | >10 CALMs; MRI-brain: ACC, interhemisferic cyst, heterotopia; decreased IgA levels (total 0.18 g/l); IgG and IgM in normal range with 7.7 and 0.84 g/l, respectively | No MSI in parotis cancer and surrounding normal parotid tissue and in T-cell NHL | MLH1/MSH2/MSH6/PMS2: +/+/+/− (in parotis cancer and in T-cell NHL) | This report |
| 2 | MLH1 | c.(218T>G)+(218T>G) p.(Leu73Arg)+(Leu73Arg) | No known consanguinity (parents both from same Pacific Island population); maternal grandfather (50 years), great aunt (48 years) and great-great grandfather (79 years) CRC; paternal FH unknown | Glioblastoma multiforme (3 years) T-cell lymphoblastic lymphoma (5.5 years) | >15 CALMs, renal cysts; MRI-brain: near complete ACC, interhemispheric cyst, frontal heterotopia, intracerebral cysts (angiomas?) | NA | MLH1/MSH2/MSH6/PMS2: −/+/+/− (in glioblastoma and in T-cell lymphoblastic lymphoma) | This report |
| 3 | PMS2 | c.(538−?_803+?del)+(1121dupA) p.(Glu180HisfsX30)+(Gln375AlafsX7) | No consanguinity, no FH of CRC | Anaplastic astrocytoma of the cervicothoracic spinal cord (3 years), T-cell lymphoblastic lymphoma (5.5 years) | No CALMs, capillary haemangioma; MRI-brain: ACC | NA | MLH1/MSH2/MSH6/PMS2: −(a)/+/+/− (in anaplastic astrocytoma) | This report |
| 4 | PMS2 | c.(1840A>T)+(1840A>T) p.(Lys614X)+(Lys614X) | Consanguineous parents (paternal and maternal grandfather are sibs); identical twin anaplastic oligodendroglioma (no ACC!), mother endometrial cancer (56 years), maternal grandmother and maternal great-grandmother and uncle CRC (52, 80 and 60 years), paternal grandmother cancer of paranasal sinus (62 years) | Adenocarcinoma of sigmoid colon (14 years) astrocytoma grade III (19 years) | Multiple ‘congenital compound nevi' (CALMs); MRI-brain: ACC; hypogammaglobulinemia as infant (attributed to Transcobalamin II deficiency) | No MSI in glioma and CRC | MLH1/MSH2/MSH6/PMS2: +/+/+/− (in glioma, CRC and normal brain and colonic tissue) | Gururangan et al29 |
Abbreviations: ACC, agenesis of corpus callosum; CALMs, café-au lait macules; CMMR-D, constitutional mismatch repair deficiency; CRC, colorectal cancer; FH, family history; MSI, microsatellite instability; NA, not analyzed; NHL, non-hodgkin lymphoma; +, positive IHC staining; −, negative IHC staining.
GenBank reference sequences for PMS2 and MLH1 are NM_000535.5 and NM_000249.3, respectively.
MLH1 loss only in the tumor cells but not in the non-neoplastic cells.
Tumor analysis
Immunohistochemical (IHC) staining for the MLH1, MSH2, MSH6 and PMS2 protein expression was performed on 4-μm sections of formalin-fixed, paraffin-embedded normal and tumor tissues, from all three patients. Microsatellite instability (MSI) analysis was performed on the parotid tumor and surrounding normal tissue, as well as on the T-cell non-Hodgkin lymphoma (NHL) of patient 1 using a multiplex fluorescence-based PCR assay with a panel of six microsatellite markers (BAT25, BAT26, BAT40, D2S123, D5S346 and D17S250).12
Mutation analysis
Exon sequencing was used as primary assay to identify the PMS2 mutation in patient 1 and the MLH1 mutation in patient 2. PMS2 exons were amplified using mainly previously published PMS2-specific primers,13 some of which were redesigned to achieve higher specificity (available upon request). To confirm the mutation identified in patient 1 at transcript level, a PMS2-specific RT-PCR product containing exons 10–15 (PCR product B in Etzler et al14) was generated from RNA isolated from a cycloheximide treated short-term lymphocytes culture and sequenced with three-internal primers. Only low-quality RNA was available of patient 2. Therefore, only a 366-bp RT-PCR fragment from exons 2–5 (fwd primer: 5′-ATCCAGCGGCCAGCTAATG-3′ reverse primer: 5′-CTTGATTGCCAGCACATGG-3′) was amplified from RNA isolated from the puromycin-treated short-term lymphocytes culture of the patient to exclude a possible splicing effect of the identified MLH1 missense variant in exon 3. The previously described RNA-based mutation analysis protocol14 was used as primary assay to analyze MLH1 and PMS2 in patient 3.
PMS2 MLPA was performed with SALSA kits P008-A1 and/or P008-B1 (MRC-Holland, Amsterdam, The Netherlands)15, 16 according to the manufacturer's instructions. MLH1 MLPA was performed with the SALSA kit P003 (MRC-Holland, Amsterdam, The Netherlands).
In vitro mismatch repair MMR activity assay and in silico analyses
To assess the effect of the MLH1 variant c.218T>G identified in patient 2, the mismatch repair efficiency of the aberrant protein MLH1-L73R was tested by an in vitro MMR assay as previously described.17, 18 Briefly, aberrant protein was produced in and isolated from Spodoptera frugiperda (SF9) insect cells with its cognate heterodimerisation partner PMS2. Newly produced wild-type and variant protein were used to complement the MMR-deficient HCT116 nuclear extract (NE). NE of the MMR-proficient HeLa cells was used as a positive control. Negative controls were an uncomplemented HCT116 extract and a NE-free mock sample. NEs were incubated in the presence of 100 ng of circular heteroduplex DNA containing a G·T mismatch downstream from a 5′ single-strand nick. Successful repair converts the G·T heteroduplex into an A·T homoduplex generating a BglII-restriction site. Therefore, the repair efficiency can be measured by the cleavage efficiency of BglII using GeneTools 3.08 (SynGene Cambridge, England). The mean repair percentages from three repeated experiments as well as the SD were calculated.
In silico methods (Polyphen 219 (http://genetics.bwh.harvard.edu/pph2/), sorting intolerant from tolerant (SIFT)20 (http://sift.jcvi.org/) and the multivariate analysis of protein polymorphism (MAPP-MMR)21, 22 (http://mendel.stanford.edu/SidowLab/)) were used for prediction of the pathogenicity of the missense MLH1 variant.23
Array analysis
For patient 1 CGH-array analysis was performed using 105K microarray slides from Agilent Technologies (Santa Clara, CA, USA) following manufacturer's protocols. Data analysis was performed using DNA analytics 4.76 software from Agilent technologies using the ADM-2 algorithm. Interpretation of CNV-data was performed as described in the guidelines of Vermeesch et al.24 Array analysis for patients 2 and 3 was performed using the HumanCytoSNP-12v2.1 beadchips (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. Raw data were processed using the Genotyping Analysis Module of GenomeStudio 1.6.3 (Illumina), for further CNV analysis we used Nexus Copy Number 6.0 standard edt. (Biodiscovery, El Segundo, CA, USA).
FLNA mutation analysis
The entire coding sequence of the FLNA gene (exons 2–48 and flanking intronic sequences) was PCR amplified from gDNA and sequenced as previously published.25
Results
Analysis of patient 1
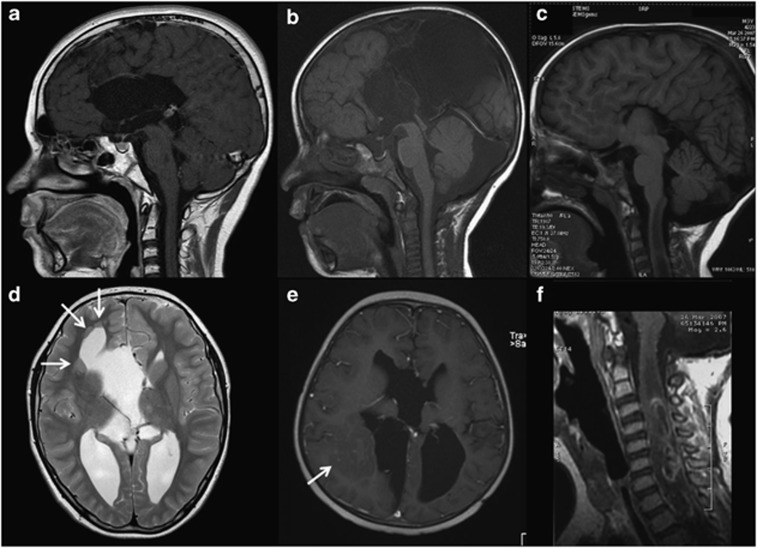
This 11-year-old boy was seen at the genetics clinic after he developed a third primary malignancy. He was born after an uneventful term pregnancy. He was the first of four children of healthy consanguineous parents (Table 1). Apart from mild behavior problems, he had a normal psychomotor development. A younger brother died at early age from unknown cause. The two sisters were healthy. There was no family history of cancer, seizures, congenital abnormalities or intellectual disabilities. At the age of 9 years, patient 1 was diagnosed with a B-cell NHL, for which he underwent chemotherapy (SNWLK-94 regimen). At 11 years of age, a grade II muco-epidermoïd carcinoma of the parotid was diagnosed, which was radically resected and locally treated with brachytherapy. MRI of the face performed for staging of the parotid tumor revealed ACC (Figure 1a), an interhemispheric cyst and several periventricular gray matter heterotopias (Figure 1d). Six months later, he was diagnosed with a peripheral T-cell NHL grade I, which was treated with chemotherapy (according the EURO-LB-02 protocol). On physical examination he had multiple (>10) CALMs without other signs of neurofibromatosis. Array-CGH study and NF1 mutation analysis showed normal results. The combination of several primary malignancies at young age, multiple CALMs and consanguineous parents was suggestive of CMMR-D.
Figure 1.
(a–c). T1-weighted midsagittal magnetic resonance (MR) images of the brain demonstrating agenesis of the corpus callosum in the first ((a) – contrast enhanced), second ((b) – postoperative) and third ((c) – showing also a tumor in the cervical cord) patient. (d) Axial T2-weighted MR image through the brain of patient 1 illustrating the periventricular gray matter heterotopias. (e) Axial contrast-enhanced T1-weighted MR image through the brain of patient 2 (postoperative) illustrating the large gray matter heterotopia (arrow) in the right occipital region. (f) Sagittal contrast-enhanced T1-weighted MR image through the cervical spine of patient 3, illustrating the intramedullary anaplastic asctrocytoma.
MSI and IHC analysis for all four MMR genes was performed on the parotid tumor as well as the T-cell NHL. No allelic shifts were observed for any of the six microsatellite markers (data not shown). Therefore, the tumors were considered microsatellite stable. All tissues stained positive for MLH1/MSH2/MSH6. However, PMS2 staining was repeatedly negative in both tumors and healthy tissue (Figures 2a and b), consistent with a constitutive biallelic PMS2 mutation. Exon sequencing from gDNA, revealed a homozygous 4 bp deletion, c.(2397_2400del)+(2397_2400del), leading to a premature stop codon, p.Ser801Glufs*15, in PMS2 exon 14. Sequencing of exon 14 demonstrated a weak normal signal underlying the homozygous deletion most likely deriving from co-amplification of the PMS2CL pseudogene. To circumvent pseudogen co-amplification and/or to exclude allelic drop-out because of the presence of a pseudogene-derived sequence within one of the PMS2 alleles,26, 27 direct cDNA sequencing was performed. This confirmed homozygosity for c.2397_2400del in the PMS2 transcripts. As the parents declined molecular testing, we could not confirm that they are heterozygous carriers of the mutation. Hemizygosity of the identified frameshift mutation owing to a larger single or multi-exon deletion was excluded by MLPA analysis.
Figure 2.
Photomicrographs showing IHC staining of the mismatch repair proteins PMS2 (left side) and MLH1 (right side) in malignancies of the three patients. The inset in panel (d) and (f) show staining for MSH2 as positive control. The muco-epidemoid carcinoma in the parotis of patient 1 (panel (a) and (b)) shows isolated PMS2 expression loss in neoplastic cells and surrounding normal tissue while MLH1 is expressed. A mediastinal mass biopsy of the T-cell lymphoblastic lymphoma of patient 2 (panel (c) and (d)) shows MLH1 and concomitant PMS2 expression loss. Weak, equivocal reactivity in background cells is of uncertain significance and may result from weak expression of the aberrant MLH1-L73R protein. The spinal cord anaplastic astrocytoma of patient 3 (panel (e) and (f)) shows PMS2 and MLH1 loss in neoplastic cells.
Analysis of patient 2
This boy was seen at the genetics department at the age of 5 years when he developed his second malignancy. He was the first born child to Polynesian parents after an uneventful term pregnancy. Early milestones were delayed (first walking at 18 months and at 3 years of age having only single words). At the age of 3 years he presented with right hemiparesis, subsequently diagnosed to be secondary to a left parieto-occipital-temporal glioblastoma multiforme. Previous neurological examination has been normal. MRI of the brain showed near complete ACC (Figure 1b), interhemispheric and intracerebral cysts, and right subcortical and periventricular heterotopia (Figure 1e). The glioblastoma was managed with surgical resection followed by intracranial irradiation (59.4Gy fractionated over 50 days) and concurrent temozolomide followed by 6 cycles of adjuvant temozolomide. He suffered from seizures after treatment but remained otherwise healthy for the next 26 months. At the age of 5 years 6 months, he presented with acute respiratory distress because of a mediastinal mass subsequently diagnosed as T-cell lymphoblastic lymphoma. He was treated with the Children's Oncology Group A5971 chemotherapy protocol, but presented acutely septic at the completion of phase IV (delayed intensification) and died of staphylococcal septicemia. On clinical examination he had normal height, weight and head circumference. Apart from multiple (>15) large CALMs he had no dysmorphic features. Parents denied known consanguinity but were from the same Pacific Island population and SNP-array analysis revealed extended regions of homozygosity, but was otherwise normal. Maternal family history was positive for colorectal cancer (see Table 1).
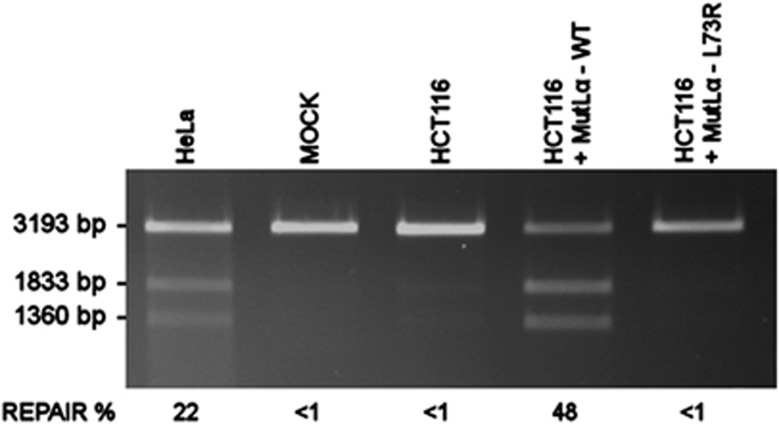
The patient's phenotype and family history suggested the diagnosis of CMMR-D syndrome. IHC staining for all four MMR genes of both tumors showed nuclear staining for MSH2 and MSH6, and absence of MLH1 and PMS2 staining in the lymphoma (Figures 2c and d), glioblastoma and in the normal tissue present on the slides. This result prompted MLH1 sequencing analysis, which revealed homozygosity for c.218T>G in exon 3, predicted to lead to amino-acid change p.Leu73Arg. This result (without any evidence for aberrant splicing of MLH1 exon 3) was also obtained by sequence of a short RT-PCR product containing exons 2–5. Hemizygosity of this alteration was largely excluded by normal MLPA results. As this amino-acid change is not listed in the InSiGHT database and has not been previously published, we aimed at confirming the pathogenicity of this missense variant. Several in silico programs (PolyPhen, SIFT and MAPP-MMR) evaluated this mutation as pathogenic. This prediction was further confirmed by an in vitro MMR assay testing for the G·T mismatch repair efficiency of the aberrant protein, MLH1-L73R. MMR-proficient HeLa NE and MMR-deficient HCT116 NE extracts complemented with wild-type protein used as positive-assay controls, showed repair efficiencies of 22% and 48%, respectively. However, HCT116 NE extracts complemented with MLH1-L73R protein repeatedly failed to produce any detectable repair (<1%) (Figure 3).
Figure 3.
Results of in vitro MMR assay of the G·T mismatch repair efficiency of MLH1-L73R. The MMR-proficient HeLa NE is used as a positive-assay control where as the MOCK contains heteroduplex DNA with no protein. The uncomplemented MMR-deficient HCT116 NE is used as a negative control. The results of the HCT116 NE complemented with wild-type MLH1 (+MutLα-WT) and mutant MLH1 (+MutLα-L73R) are compared. The top fragment (3193 bp) in each lane shows the migration of the unrepaired linearized G·T mismatch containing construct, and the two smaller fragments (1833 and 1360 bp) represent the repaired and double-digested fragments of the construct. The indicated repair percentages are calculated from three independent repeats and represent the percentage of repaired DNA from total DNA in the reaction. The MutLα-L73R variant repeatedly failed to produce any detectable repair (<1%), where as the wild-type MutLα had a mean repair efficiency of 48% (STD±9%). The mean repair efficiency of the assay control was 22% (STD±2).
Analysis of patient 3
Prenatal ultrasound at 36th week of this pregnancy (first common child of non-consanguinious parents) demonstrated bilateral dilatation of the occipital horns of the lateral ventricles. A male (second child of the mother) was subsequently delivered at term after an uncomplicated pregnancy and postnatal ultrasound revealed ACC. At birth two capillary hemangiomas of the skin were noted on the right lower leg and the left groin, both remained unchanged since. Beside hemangiomas and a small number of cutaneous nevi, there were no other congenital malformations or dysmorphic features. Psychomotor development was normal in the boy (as it was for his older half-brother). At the age of 2 years and 10 months this patient was diagnosed with an anaplastic astrocytoma of the spinal cord (C3–Th3) (Figure 1f). MRI of the brain confirmed complete ACC with hypertrophic anterior commissure (Figure 1c). Secondary to neurosurgical treatment and the tumor development, he developed spastic tetraparesis, more pronounced in the lower extremities, and partial neurogenic bladder. He subsequently underwent radiochemotherapy treatment according to HIT-GBM-D protocol, with partial response on MRI studies and mild improvement of the neurological deficits. At the age of 5 years and 6 months a mediastinal mass was detected. Cytological analysis and immunophenotyping of a pleural effusion disclosed a T-lymphoblastic lymphoma. Bone marrow cytology and immunophenotyping showed no significant infiltration. Chemotherapy according to EURO-LB-02 protocol was initiated with initially good response. The patient died 9 months later from septic infection during severe bone marrow aplasia.
Diagnosis of this second malignancy led to the suspicion of CMMR-D syndrome. Retrospective IHC staining of the spinal cord anaplastic astrocytoma tissue demonstrated MLH1 and PMS2 loss in the tumor cells, suggestive of a MLH1 mutation (Figures 2e and f). Direct cDNA sequencing revealed regular biallelic MLH1 expression as evidenced by heterozygosity of a polymorphic variant (c.655A>G; rs.1799977) and no evidence of a pathogenic alteration. As loss of MLH1 was only observed in the tumor cells, but not in the surrounding non-neoplastic cells (suggestive of a possible somatic MLH1 mutation restricted to the neoplastic cells and an underlying germline PMS2 mutation), direct PMS2 cDNA sequencing was performed. This demonstrated heterozygous skipping of the out of frame exons 6 and 7 (r.538_803del) as well as heterozygosity for c.1121dupA in exon 10. A heterozygous deletion of exons 6 and 7 (c.538−?_803+?del) was confirmed by MLPA analysis and sequencing of exon 10 from gDNA confirmed the frameshift mutation resulting in a premature stop codon, p.Gln375Alafs*7. Analysis of the parental gDNA confirmed that the two mutations were located in trans. Retrospectively performed SNP-array analysis was normal (no evidence of copy number changes).
FLNA analysis in patients 1 and 2
Mutations of the X-linked FLNA gene account for up to 24% of cases with periventricular heterotopias.28 Analysis of this gene in the two patients (patients 1 and 2; patient 3 did not show gray matter heterotopia) revealed no abnormalities.
Discussion
The common feature in the present three novel CMMR-D patients was ACC. Gray matter heterotopia and interhemispheric cysts were observed in two patients. ACC was previously reported in only one CMMR-D patient with anaplastic astrocytoma and a very early colorectal cancer with an inferred homozygous PMS2 mutation (patient 4 in Table 1).29 With the present report, there are now at least 4 out of the 115 published CMMR-D cases who show these cerebral malformations. Sixty of those had a brain tumor and therefore presumably had comprehensive imaging of the central nervous system. The prevalence of callosal anomalies varies widely among different studies. Wang et al30 found among 2309 clinically normal term neonates three with ACC (0.09%). However, a study on perinatal/neonatal autopsy series found ACC in 15/4122 (0.36%) of cases.31 These two studies may delineate the lower and upper end of ACC frequency among live births. With a frequency of at least 3.5–6.6% (4/115 or 4/60), the incidence of ACC in CMMR-D patients is higher than in the general population, suggesting that ACC is a feature of CMMR-D.
Patients 1 and 2 also had gray matter heterotopia and interhemispheric cysts. No evidence of a germline FLNA mutation was found in both patients. There might be other explanations for periventricular heterotopia (eg, insults, other rare genetic causes32) in these patients. However, on the basis of the scattered distribution of the heterotopic nodules in both patients, the most likely explanation is that this malformation as well as ACC (which is occasionally associated with gray matter heterotopia33) is related to the underlying defect in MMR.
ACC is frequently associated with neurological disorders and/or severe developmental delay. However, as in our three patients, ACC may also be entirely asymptomatic or associated with only mild developmental or behavioral deficits.34 ACC results from disruption of any one of the multiple steps of fetal callosal development, such as cellular proliferation and migration, axonal growth and guidance, or glial patterning at the midline.34 The genetics of ACC reflect the underlying complexity of callosal development. The London Dysmorphology Database lists >350 syndromes associated with ACC. In patients with callosal defects, mutations in at least 13 human genes have been identified. Equally, murine studies implicate an extensive list of genes involved in callosal development (for review see Kamnasaran35 and Richards et al36). It is conceivable that one or more of the genes implicated in the callosal development are the target of a constitutional defect in the DNA repair, as it has been shown with the NF1 gene.37 Signs of NF1 in CMMR-D patients are thought to result from somatic mutations present in a segmental or mosaic status.38, 39 Similarly, it is possible that early embryonic somatic mutations (presumed to occur more frequently in CMMR-D patients) in one or more of the genes implicated in the callosal development are responsible for the occurrence of isolated, largely asymptomatic ACC with or without gray matter heterotopia in CMMR-D patients.
As with other previously reported CMMR-D patients, our patients were suspected of having CMMR-D only when they developed their second or third malignancy. This demonstrates that diagnosis of CMMR-D may be frequently delayed in pediatric cancer patients. Apart from the lack of awareness for this rare cancer predisposition syndrome among pediatric hematologists–oncologists, other factors that may play a role in delayed diagnosis include the lack of clearly disease-specific clinical features and the overlap with other cancer predisposing syndromes, including NF1,1 familial adenomatous polyposis9 and to some extent also with Fanconi anemia.40, 41, 42 In view of the wide tumor spectrum, it has been suggested that CMMR-D should be considered in the differential diagnosis in all pediatric patients with malignancies (except clearly NF1-associated tumors), who show one or more of the following features: (1) CALMs and/or other signs of NF1 and/or hypo-pigmented skin lesions; (2) consanguineous parents; (3) family history of LS-associated tumors; (4) second malignancy; and (5) sibling with childhood cancer. We suggest including ACC with or without gray matter heterotopia as an additional feature. As the example of patient 3 shows, this could help to establish more timely diagnosis and implementation of surveillance programs for pre-symptomatic detection of malignancies43 in similar CMMR-D cases. Patient 3 lacked CALMs and was the only-child from unrelated parents without a family history for Lynch-syndrome-associated tumors. Hence, without the criterion of ACC he fulfilled only diagnostic criterion (4) when he developed a second malignancy. Of note, this patient had also two capillary haemangiomas of the skin, a feature that has been previously reported once in CMMR-D.44
Patient 1 developed a muco-epidermoïd carcinoma of the parotid, a malignancy that so far has not been described in a CMMR-D patient. Negative IHC PMS2 staining in neoplastic and surrounding normal cells from both the parotid tumor and T-cell NHL, together with the lack of MSI, was puzzling and initially interpreted as a failure of proper staining. Hence, this case further underscores that the lack of MSI, particularly in extra-intestinal tumors of CMMR-D patients,1 may represent a pitfall in molecular analysis performed to verify the suspected diagnosis of CMMR-D.44
ACC and gray matter heterotopia are the first congenital malformations described to be associated with CMMR-D. We suggest that ACC with or without gray matter heterotopia should be included in the clinical diagnostic criteria for CMMR-D. Further systematic studies may answer the question whether other malformations are also found at higher frequency in CMMR-D patients than in the general population.
Acknowledgments
We thank Dr I Kardum, Dr D Batinic and Dr K Zarkovic for their thorough respective cytological, immunophenotyping and pathohistological diagnostic workup of patient 3, ML Berenbroek for performing cDNA sequencing in patient 1, Dr T Letteboer for supervision of patient 1 and Dr M Coulibaly-Wimmer for helpful review of MRI images. AW and KW acknowledge support from the Austrian Science Fonds (FWF-Grant no P 21172-B12). MK and MN acknowledge support through the European Research Council (2008-AdG-232635), the Sigrid Juselius Foundation and the Helsinki Graduate Program in Biotechnology and Molecular Biology. SPR is supported by Curekids New Zealand.
The authors declare no conflict of interest.
References
- Wimmer K, Etzler J. Constitutional mismatch repair-deficiency syndrome: have we so far seen only the tip of an iceberg. Hum Genet. 2008;124:105–122. doi: 10.1007/s00439-008-0542-4. [DOI] [PubMed] [Google Scholar]
- Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- Ricciardone MD, Ozcelik T, Cevher B, et al. Human MLH1 deficiency predisposes to hematological malignancy and neurofibromatosis type 1. Cancer Res. 1999;59:290–293. [PubMed] [Google Scholar]
- Wang Q, Lasset C, Desseigne F, et al. Neurofibromatosis and early onset of cancers in hMLH1-deficient children. Cancer Res. 1999;59:294–297. [PubMed] [Google Scholar]
- Wimmer K, Kratz CP. Constitutional mismatch repair-deficiency syndrome. Haematologica. 2010;95:699–701. doi: 10.3324/haematol.2009.021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T, Beger C, Rahner N, et al. Constitutional mismatch repair deficiency and childhood leukemia/lymphoma - report on a novel biallelic MSH6 mutation. Haematologica. 2010;95:841–844. doi: 10.3324/haematol.2009.015503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesma P, van der Klift H, van Grieken N, et al. Childhood brain tumours due to germline bi-allelic mismatch repair gene mutations. Clin Genet. 2011;80:243–255. doi: 10.1111/j.1399-0004.2011.01635.x. [DOI] [PubMed] [Google Scholar]
- Durno CA, Holter S, Sherman PM, Gallinger S. The gastrointestinal phenotype of germline biallelic mismatch repair gene mutations. Am J Gastroenterol. 2010;105:2449–2456. doi: 10.1038/ajg.2010.215. [DOI] [PubMed] [Google Scholar]
- Herkert JC, Niessen RC, Olderode-Berends MJ, et al. Paediatric intestinal cancer and polyposis due to bi-allelic PMS2 mutations: case series, review and follow-up guidelines. Eur J Cancer. 2011;47:965–982. doi: 10.1016/j.ejca.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Gardes P, Forveille M, Alyanakian MA, et al. Human MSH6 Deficiency Is Associated with Impaired Antibody Maturation. J Immunol. 2012;188:2023–2029. doi: 10.4049/jimmunol.1102984. [DOI] [PubMed] [Google Scholar]
- Peron S, Metin A, Gardes P, et al. Human PMS2 deficiency is associated with impaired immunoglobulin class switch recombination. J Exp Med. 2008;205:2465–2472. doi: 10.1084/jem.20080789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks YM, Jagmohan-Changur S, van der Klift HM, et al. Heterozygous mutations in PMS2 cause hereditary nonpolyposis colorectal carcinoma (Lynch syndrome) Gastroenterology. 2006;130:312–322. doi: 10.1053/j.gastro.2005.10.052. [DOI] [PubMed] [Google Scholar]
- Etzler J, Peyrl A, Zatkova A, et al. RNA-based mutation analysis identifies an unusual MSH6 splicing defect and circumvents PMS2 pseudogene interference. Hum Mutat. 2008;29:299–305. doi: 10.1002/humu.20657. [DOI] [PubMed] [Google Scholar]
- Vaughn CP, Hart KJ, Samowitz WS, Swensen JJ. Avoidance of pseudogene interference in the detection of 3' deletions in PMS2. Hum Mutat. 2011;32:1063–1071. doi: 10.1002/humu.21540. [DOI] [PubMed] [Google Scholar]
- Wernstedt A, Valtorta E, Armelao F, et al. Improved MLPA analysis identifies a deleterious PMS2 allele generated by recombination with crossover between PMS2 and PMS2CL Genes Chromosomes Cancer 2012. e-pub ahead of print 14 May 2012; doi: 10.1002/gcc.21966 [DOI] [PMC free article] [PubMed]
- Kantelinen J, Kansikas M, Korhonen MK, et al. MutSbeta exceeds MutSalpha in dinucleotide loop repair. Br J cancer. 2010;102:1068–1073. doi: 10.1038/sj.bjc.6605531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom-Lahti M, Perrera C, Raschle M, et al. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33:160–167. [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao EC, Velasquez JL, Witherspoon MSL, et al. Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR) Hum Mutat. 2008;29:852–860. doi: 10.1002/humu.20735. [DOI] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kansikas M, Kariola R, Nystrom M. Verification of the three-step model in assessing the pathogenicity of mismatch repair gene variants. Hum Mutat. 2011;32:107–115. doi: 10.1002/humu.21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeesch JR, Fiegler H, de Leeuw N, et al. Guidelines for molecular karyotyping in constitutional genetic diagnosis. Eur J Hum Genet. 2007;15:1105–1114. doi: 10.1038/sj.ejhg.5201896. [DOI] [PubMed] [Google Scholar]
- Robertson SP, Twigg SR, Sutherland-Smith AJ, et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet. 2003;33:487–491. doi: 10.1038/ng1119. [DOI] [PubMed] [Google Scholar]
- Ganster C, Wernstedt A, Kehrer-Sawatzki H, et al. Functional PMS2 hybrid alleles containing a pseudogene-specific missense variant trace back to a single ancient intrachromosomal recombination event. Hum Mutat. 2010;31:552–560. doi: 10.1002/humu.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klift HM, Tops CM, Bik EC, et al. Quantification of sequence exchange events between PMS2 and PMS2CL provides a basis for improved mutation scanning of Lynch syndrome patients. Hum Mutat. 2010;31:578–587. doi: 10.1002/humu.21229. [DOI] [PubMed] [Google Scholar]
- Parrini E, Ramazzotti A, Dobyns WB, et al. Periventricular heterotopia: phenotypic heterogeneity and correlation with Filamin A mutations. Brain. 2006;129:1892–1906. doi: 10.1093/brain/awl125. [DOI] [PubMed] [Google Scholar]
- Gururangan S, Frankel W, Broaddus R, et al. Multifocal anaplastic astrocytoma in a patient with hereditary colorectal cancer, transcobalamin II deficiency, agenesis of the corpus callosum, mental retardation, and inherited PMS2 mutation. Neuro-oncology. 2008;10:93–97. doi: 10.1215/15228517-2007-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LW, Huang CC, Yeh TF. Major brain lesions detected on sonographic screening of apparently normal term neonates. Neuroradiology. 2004;46:368–373. doi: 10.1007/s00234-003-1160-4. [DOI] [PubMed] [Google Scholar]
- Pinar H, Tatevosyants N, Singer DB. Central nervous system malformations in a perinatal/neonatal autopsy series. Pediatr Dev Pathol. 1998;1:42–48. doi: 10.1007/s100249900005. [DOI] [PubMed] [Google Scholar]
- Lu J, Sheen V. Periventricular heterotopia. Epilepsy Behav. 2005;7:143–149. doi: 10.1016/j.yebeh.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Hetts SW, Sherr EH, Chao S, Gobuty S, Barkovich AJ. Anomalies of the corpus callosum:an MR analysis of the phenotypic spectrum of associated malformations. AJR Am J Roentgenol. 2006;187:1343–1348. doi: 10.2214/AJR.05.0146. [DOI] [PubMed] [Google Scholar]
- Paul LK, Brown WS, Adolphs R, et al. Agenesis of the corpus callosum: genetic, developmental and functional aspects of connectivity. Nat Rev Neurosci. 2007;8:287–299. doi: 10.1038/nrn2107. [DOI] [PubMed] [Google Scholar]
- Kamnasaran D. Agenesis of the corpus callosum: lessons from humans and mice. Clin Invest Med. 2005;28:267–282. [PubMed] [Google Scholar]
- Richards LJ, Plachez C, Ren T. Mechanisms regulating the development of the corpus callosum and its agenesis in mouse and human. Clin Genet. 2004;66:276–289. doi: 10.1111/j.1399-0004.2004.00354.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Montmain G, Ruano E, et al. Neurofibromatosis type 1 gene as a mutational target in a mismatch repair-deficient cell type. Hum Genet. 2003;112:117–123. doi: 10.1007/s00439-002-0858-4. [DOI] [PubMed] [Google Scholar]
- Alotaibi H, Ricciardone MD, Ozturk M. Homozygosity at variant MLH1 can lead to secondary mutation in NF1, neurofibromatosis type I and early onset leukemia. Mutat Res. 2008;637:209–214. doi: 10.1016/j.mrfmmm.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Auclair J, Leroux D, Desseigne F, et al. Novel biallelic mutations in MSH6 and PMS2 genes: gene conversion as a likely cause of PMS2 gene inactivation. Hum Mutat. 2007;28:1084–1090. doi: 10.1002/humu.20569. [DOI] [PubMed] [Google Scholar]
- Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet. 2007;44:1–9. doi: 10.1136/jmg.2006.043257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch B, Shimamura A, Moreau L, et al. Association of biallelic BRCA2/FANCD1 mutations with spontaneous chromosomal instability and solid tumors of childhood. Blood. 2004;103:2554–2559. doi: 10.1182/blood-2003-06-1970. [DOI] [PubMed] [Google Scholar]
- Reid S, Renwick A, Seal S, et al. Biallelic BRCA2 mutations are associated with multiple malignancies in childhood including familial Wilms tumour. J Med Genet. 2005;42:147–151. doi: 10.1136/jmg.2004.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durno CA, Aronson M, Tabori U, Malkin D, Gallinger S, Chan HS.Oncologic surveillance for subjects with biallelic mismatch repair gene mutations: 10 year follow-up of a kindred Pediatr Blood Cancer 2011. e-pub ahead of print 16 December 2011; doi: 10.1002/pbc.24019 [DOI] [PubMed]
- Leenen C, Geurts-Giele W, Dubbink H, et al. Pitfalls in molecular analysis for mismatch repair deficiency in a family with biallelic pms2 germline mutations. Clin Genet. 2010;80:558–565. doi: 10.1111/j.1399-0004.2010.01608.x. [DOI] [PubMed] [Google Scholar]