Abstract
Maternal inheritance is one of the hallmarks of animal mitochondrial DNA (mtDNA) and central to its success as a molecular marker. This mode of inheritance and subsequent lack of heterologous recombination allows us to retrace evolutionary relationships unambiguously down the matriline and without the confounding effects of recombinant genetic information. Accumulating evidence of biparental inheritance of mtDNA (paternal leakage), however, challenges our current understanding of how this molecule is inherited. Here, using Drosophila simulans collected from an East African metapopulation exhibiting recurring mitochondrial heteroplasmy, we conducted single fly matings and screened F1 offspring for the presence of paternal mtDNA using allele-specific PCR assays (AS–PCR). In all, 27 out of 4092 offspring were identified as harboring paternal mtDNA, suggesting a frequency of 0.66% paternal leakage in this species. Our findings strongly suggest that recurring mtDNA heteroplasmy as observed in natural populations of Drosophila simulans is most likely caused by repeated paternal leakage. Our findings further suggest that this phenomenon to potentially be an integral part of mtDNA inheritance in these populations and consequently of significance for mtDNA as a molecular marker.
Keywords: mtDNA, paternal leakage, biparental inheritance, heteroplasmy, mitochondria
Introduction
Two main hypotheses have been put forward to explain uniparental inheritance of mtDNA: The first hypothesis argues that this system has evolved to exclude sperm mtDNA from transmission because it is more likely to carry deleterious mutations (Rand, 1994; Allen, 1996). The increased mutational load of sperm mtDNA is thought to accumulate due to its exposure to reactive oxygen species during mitochondrial activity necessary to reach and fertilize the egg (Rand, 1994; Allen, 1996). In contrast, the egg retains a quiescent metabolism with its mtDNA presumably remaining in a pristine state until after fertilization. The second hypothesis argues that the presence of genetically distinct mtDNA haplotypes within single zygotes may be unfavorable as it allows for natural selection and might occur at the expense of single individuals and subsequently of whole populations (Hastings, 1992; Hurst, 1996).
Contrary to the long prevailing, but finally discredited hypothesis that paternal mtDNA is excluded from the animal zygote by the exclusion of the sperm tail containing the mitochondria, we now know that it is rather the interplay of several stochastic and molecular mechanisms that inhibits the transmission of paternal mtDNA (Pitnick and Karr, 1998; White et al., 2008; DeLuca and O'Farrell, 2012). These mechanisms include: (i) the dilution of paternal by maternal mtDNA in the zygote, that is, paternal mtDNA is vastly outnumbered by maternal mtDNA, (ii) the selective tagging (ubiquitination) and subsequent destruction of paternal sperm mitochondria upon fertilization in mammals (Sutovsky et al., 1999, 2000) and (iii) the presence of a genetic bottleneck during early developmental stages that is likely to exclude minor allele contributions, for example, paternal mtDNA (Bergstrom and Pritchard, 1998; White et al., 2008). What is more, DeLuca and O'Farrell (2012) suggested that mtDNA is removed from Drosophila sperm mitochondria during spermatogenesis leading to gametes maintaining mitochondrial structures seemingly devoid of mtDNA.
Despite these mechanisms in place, there is increasing evidence for paternal leakage of mtDNA during recent years, covering invertebrates to vertebrates including at least one documented case in humans (Schwartz and Vissing, 2002). In some cases, only single individuals are affected whereas in others the introgression and replacement of mtDNA haplotypes affects entire populations or species (Goodman et al., 1999; Alves et al., 2008; Oliveira et al., 2008; Zakharov et al., 2009).
If paternal leakage occurs frequently, then it might have far-reaching consequences. First, mitochondrial genomes co-evolve alongside nuclear genomes and mitochondrial function relies on a concerted interplay of both genomes (Puslednik et al., 2012). Paternal leakage might lead to the introgression of new haplotypes into populations disrupting these mitonuclear interactions, reducing organismal and/or population fitness (Sackton et al., 2003; Ballard et al., 2007; Ellison and Burton, 2008). Second, due to its maternal mode of inheritance, mitochondrial genomes most often respond to selection in the female line (Wade and Brandvain, 2009). This process is assumed to lead to the accumulation of mtDNA mutations deleterious to male-specific traits (Innocenti et al., 2011). The introgression of paternal haplotypes could potentially counter-balance such sex-specific selective sieve. Third, paternal leakage introduces additional haplotypes into populations, potentially leading to erroneous reconstruction of population or species histories in mtDNA analyses, and generally also creates individuals with more than one type of mtDNA (heteroplasmy), resulting in ambiguities in sequence interpretation. Furthermore, heteroplasmy might also allow for heterologous mtDNA recombination to occur, a process that has been documented in a several animal species (White et al., 2008). Such process has the potential to significantly impact on the evolution of the molecule if newly arisen haplotypes propagate. It is hence of utmost importance that we aim to determine whether observed cases of biparental inheritance are an integrated facet of mtDNA evolution rather than only exceptions to the general rule or, alternatively, peculiarities in the mode of inheritance of single species (for example, doubly uniparental inheritance in some bivalves; Breton et al., 2007).
In this study, we aimed to reveal the source of recurring heteroplasmy in natural populations of D. simulans. This species has three distinct mitochondrial haplogroups, siI, -II and -III, that differ by 3% (Baba-Aissa et al., 1988; Ballard, 2000). The siI haplogroup occurs on Indian/Pacific islands. The siII harboring flies are globally distributed but do not occur in sympatry with siI flies. Flies with siIII mtDNA live alongside siII flies in East Africa and Reunion (Ballard et al., 2007). Despite recent findings by DeLuca and O'Farrell (2012), suggesting that drosophilid sperm mitochondria are devoid of mtDNA, genetic surveys have repeatedly revealed the presence of heteroplasmy (6–12%) for siII and siIII among individuals collected from natural populations in East Africa and Reunion island, indicating recurring paternal leakage of mtDNA (Satta et al., 1988; Matsuura et al., 1991; Dean et al., 2003). Maintenance of heteroplasmy in 2 isofemale lines (out of 17) of D. simulans from Reunion island for 6 years suggests the frequent occurrence of heteroplasmy and/or presence of large copy number of variants in these flies (Satta et al., 1988). Similarly, in East Africa, Dean et al. (2003) reported that 15% of siIII flies also harbored siII mtDNA using an allele-specific PCR assay (AS-PCR) and in 2011 we identified 9% (19 out of 203 individuals) as heteroplasmic for both haplotypes in Kenya (data unpublished).
D. simulans is an ideal model in which to investigate the mechanism of mitochondrial inheritance because of the recurrence and persistence of mtDNA heteroplasmy and because paternal leakage has previously been documented in this species (Kondo et al., 1990, 1992; Sherengul et al., 2006). The occurrence of two distinct haplotypes within East African populations facilitates the detection of paternal leakage and heteroplasmy which in other models may remain unnoticed due to missing or lesser haplotypic variety. In contrast to our experimental approach, earlier studies were greatly influenced by backcrossing and hybridization, breeding regimes hypothesized to promote paternal leakage in insects and mammals (Kaneda et al., 1995; Sutovsky et al., 2000; Sherengul et al., 2006). Conducting single fly matings and subsequent haplotyping of the F1 generation without the confounding factors of backcrossing and hybridization, we aimed to provide direct evidence for paternal leakage as potential cause for observed heteroplasmy in natural populations of D. simulans. We further aimed to determine the frequency at which this phenomenon occurs. To conduct these experiments on flies maintained in the laboratory and not on individuals collected in the wild is of significance for this approach because Drosophila are known to mate multiple times, rendering attempts to accurately type the paternal parent impossible (Scott and Richmond, 1990).
Materials and methods
Samples
The isofemale lines 2-KY0418 (GenBank JQ691660) and 3-KY0410 (GenBank JQ691661) of D. simulans were collected in Nairobi (Kenya) in November 2004. Lines are labeled according to mtDNA haplotype (2: siII, 3: siIII), locality (KY: Kenya), year of collection (2004) and the last two numbers indicate specific line numbers. A test for infection with Wolbachia revealed these lines to be free of the parasite. Virgin females were collected from both lines and mated individually to individual males from each of the two lines: 2-KY0418 females × 3-KY0410 males (mating experiment 1, hereafter ME1) and 2-KY0418 males × 3-KY0410 females (mating experiment 2, hereafter ME2). To control for haplotype- and gender-specific effects, experiments were conducted bidirectionally. A total of 200 pairs were placed in individual vials for each mating direction. Each pair was allowed 5 days for mating and egg laying followed by preservation at −20 °C. In all, 33 offspring from 62 pairs per mating experiment (ME) were collected and tested for the presence of paternal mtDNA.
Molecular methods
DNA was extracted in 96-well plate format using the Gentra Puregene Cell Kit (Qiagen Sciences, Germantown, MD, USA) as previously described (Holleley, 2007). Two AS-PCR assays were used: AS-PCR1 was specifically designed for this study (Supplementary Figure 1A) and AS-PCR2 followed methods as outlined in Dean et al. (2003) (Supplementary Figure 1B). AS-PCR1 consists of two allele-specific reverse primers and one common forward primer (Supplementary Figure 1A; Supplementary Table 1). To examine whether the primers were amplifying the mitochondrial COI region and not non-functional nuclear copies of mitochondrial genes (NUMTs), the paternal and maternal mtDNA representative sequences from intraspecific crosses were blasted against the D. simulans autosomal sequence. All of the sequences corresponded to the COI region of mtDNA and none matched with any sequence in the nuclear genome.
PCRs were carried out according to Dean et al. (2003) for AS-PCR2 and with 10 pmol of primer 1432+, 10 pmol of primer 1793−, and 3 pmol of primer 2314−, 2 mℳ MgCl2, 5 μl 5 × Crimson PCR buffer (New England BioLabs Inc., Ipswich, MA, USA), 0.2 mℳ dNTPs (Bioline, London, UK) and 0.5 U Taq DNA polymerase (New England BioLabs Inc.) for AS-PCR1. Thermal profile for AS-PCR1: initial denaturation at 95 °C for 2 min followed by 95 °C for 20 s, 57 °C for 20 s and 68 °C for 60 s for 35 cycles. Both AS-PCR1 and AS-PCR2 were carried out in 25 μl reactions and with 30 ng genomic DNA.
The sensitivity of AS-PCR1 for siIII in a mix of both haplotypes is 1:1000 (3-KY0410:2-KY0418, Supplementary Figure 1C) and for the reverse direction 1:10 (2-KY0418:3-KY0410; hereafter AS-PCR1-R; Supplementary Figure 1E). The sensitivity of AS-PCR2 for siII in a mix of both haplotypes is 1:100 (2-KY0418:3-KY0410; Supplementary Figure 1D) and for the reverse direction 1:100 (3-KY0410:2-KY0418; hereafter AS-PCR2-R; Supplementary Figure 1F).
For Sanger sequencing, PCR products were purified using ExoSAP-IT according to manufacturer's instructions (USB Amersham, Buckinghamshire, UK) and sequenced using the Big Dye terminator chemistry (Applied Biosystems, Foster City, CA, USA). The volume of the reaction was 10 μl containing 2 μl purified PCR product and 3 pmol primer. Samples were analyzed at the Ramaciotti Centre for Gene Function Analysis (University of New South Wales, Sydney, Australia) on an ABI-3730 DNA sequencer (Applied Biosystems Inc.). All samples suggestive of paternal leakage in ME1 and ME2 were sequenced using the common primer of AS-PCR2. A subset of positive samples, failing to reveal the paternal signal using the common primer, was sequenced using the respective paternal-specific primer.
The stability of mtDNA heteroplasmy induced by paternal leakage was evaluated in two single parent populations (crosses #72 and #85) maintained as mixed-generation populations after 10 months (that is, these populations have been maintained without separating generations for ∼20 generations). For each cross, 12 offspring were collected and tested for the presence of the founder parents' maternal and paternal mtDNA.
In silico analysis followed using the Sequencher version 5.0 sequence analysis software (Gene Codes Corp., Ann Arbor, MI, USA). Randomization tests (20 000 iterations per test) for both MEs using MATLAB (MathWorks, Natick, MA, USA) were conducted to evaluate whether observed paternal inheritance of mtDNA is distributed randomly among all offspring of single MEs or if the distribution is pair specific. Pair-specific distribution indicates differences between single pairs on the level of mtDNA inheritance whereas a random distribution suggests homogeneity among pairs.
Results
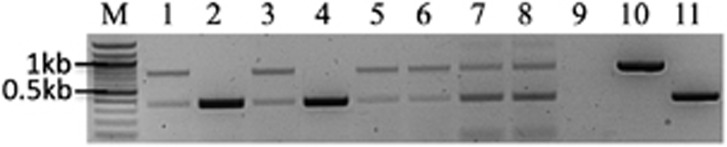
DNA from 2046 offspring (33 offspring × 62 independent mating pairs) was extracted from ME1 and screened for the presence of paternal mtDNA using AS-PCR1. This assay identified six offspring from two independent mating pairs suggestive of paternal leakage. Four of these offspring belonged to pair #4 and two to pair #21. Of these six offspring, one individual per pair revealed two bands indicative of the presence of both maternal and paternal mtDNA (that is, heteroplasmy), whereas the remainder revealed only one amplicon, indicating a potential turnover from maternal to paternal mtDNA (Figure 1). A second screen of all individuals suggestive for paternal leakage with AS-PCR2 confirmed this result (data not shown).
Figure 1.
Subset of samples revealing paternal mtDNA using AS-PCR1. M: 100 bp ladder; 1 and 2: two positive cases from pair #4; 3 and 4: two positive cases pair from #21; 5 and 6: two positive cases from pair #72; 7 and 8: two positive cases from pair #85; 9: no template control; 10: positive control for 2-KY0418; 11: positive control for 3-KY0410. Gender of offspring: 1, 2, 5–7: female; 3, 4, 8: male.
In ME2, applying AS-PCR2 during the initial screen, two independent mating pairs revealed the presence of paternal mtDNA among 2046 offspring. Two offspring of pair #85 and 19 offspring of pair #72 revealed two bands, indicative of heteroplasmy. A second screen of all individuals suggestive for paternal leakage with AS-PCR1 confirmed this result (data not shown).
To further validate this result, we sequenced the founder parents and offspring positive for paternal mtDNA to confirm respective haplotypes (Table 1; Supplementary Table 2). The region amplified using AS-PCR2 harbors a total of 29 diagnostic single-nucleotide polymorphisms (SNPs) between the two haplotypes allowing for unambiguous assignment of haplotypes to single samples (Table 1). We first sequenced both parents from pairs #4 and #21 from ME1 along with their offspring revealing paternal leakage (Table 1). As observed with both AS-PCRs, sequences of four positive cases (pair #4, offspring 1–3; pair #21, offspring 1) showed the same pattern as the founder males confirming the occurrence of paternal leakage. The lack of a heteroplasmic signal is suggestive of a replacement of the maternal 2-KY0418 haplotype by the paternal 3-KY0410 haplotype (Table 1). Though a very weak secondary signal, the remaining two positive cases of pairs #4 and #21 revealed a mixed signal of both haplotypes (heteroplasmy), reflective of the two bands observed during the AS-PCR screens (Table 1). After completing the screening of ME2, we also sequenced the parental generation of pairs #72 and #85, all offspring of pair #85 and 11 offspring of pair #72 suggestive of paternal leakage (see Supplementary Table 2 and Supplementary Figure 2). The parental haplotype and the presence of paternal mtDNA in all offspring examined were confirmed. Two offspring of pair #72 revealed a weak heteroplasmic signal (Supplementary Figure 2).
Table 1. Base identities observed at 17 diagnostic SNP positions among offspring revealing paternal leakage and their parents of ME1 (subset of SNPs obtained via unidirectional sequencing).
| Sample | Diagnostic SNP a | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
5254 |
5269 |
5278 |
5305 |
5329 |
5387 |
5524 |
5528 |
5738 |
5759 |
5762 |
5796 |
5807 |
5849 |
5915 |
5918 |
5921 |
| 2-KY0418 reference | A | C | C | C | A | T | A | G | C | T | T | T | T | C | T | T | C |
| Female parent pair #4 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Female parent pair #21 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| 3-KY0410 reference | G | T | T | T | T | C | G | A | T | A | C | C | C | T | A | C | T |
| Male parent #4 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Male parent #21 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Pair #4 offspring 1 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Pair #4 offspring 2 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Pair #4 offspring 3 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Pair #4 offspring 4 | R | Y | Y | Y | W | Y | R | R | Y | W | Y | Y | Y | Y | W | Y | Y |
| Pair #21 offspring 1 | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • |
| Pair #21 offspring 2 | R | Y | Y | Y | W | Y | R | R | Y | W | Y | Y | Y | Y | W | Y | Y |
To determine whether the observed cases of paternal leakage were random events, we conducted randomization tests for both MEs. As expected, these tests revealed the distribution of offspring carrying paternal mtDNA to be non-random (P=0 for ME1; P=0 for ME2).
The frequency of paternal leakage in this study was estimated at 12% (pair #4: 4 offspring) and 6% (pair #21, 2 offspring) within two mating pairs and at 0.29% among 2046 offspring of ME1. For the reciprocal experiment ME2, the frequency of biparental inheritance was estimated at 58% for mating pair #72 (19 offspring) and 6% for mating pair #85 (two offspring), resulting in an overall frequency of 1.03% for this mating direction. Combined, the frequency of paternal leakage was estimated at 0.66% for 4092 offspring and at 3.23% per cross (4 out of 124 crosses).
The stability of heteroplasmy induced by paternal leakage was tested after 20 generations for crosses #72 and #85. All offspring tested for cross #72 revealed a strong heteroplasmic signal whereas all offspring of cross #85 returned to homoplasmy of the maternal mtDNA.
Discussion
Despite the plenitude of mechanisms promoting maternal inheritance of mtDNA, our study revealed paternal leakage at an overall rate of 0.66%. To exclude the potential impact of contamination or technical errors that may have led to falsely identifying paternal mtDNA in offspring, we have sequenced all positive cases and we also conducted a similar screen after 20 generations to further validate the presence of leaked paternal mtDNA. While in line with previous findings documenting heteroplasmy between siII and siIII haplotypes in East Africa and Reunion Island, this study would appear to contradict a recent study suggesting that drosophilid sperm mitochondria are devoid of mtDNA (DeLuca and O'Farrell, 2012). These authors, however, conducted their work on D. melanogaster and it is possible that deviations between findings may reflect species- or line-specific differences in mtDNA inheritance. Further experiments on both species are necessary to solve this discrepancy.
Conducting our experiments bidirectionally and observing biparental inheritance in two pairs per ME, a dominant gender- or line-specific effect is unlikely the driving force of the observed occurrence of paternal leakage. Due to the limited sensitivity of the AS-PCRs the overall frequency of biparental inheritance in these experiments is likely to be higher than what we have estimated because the contribution of paternal mtDNA may lie in some cases well beyond what we can detect using our experimental approach (1:1000 for siIII; 1:100 for siII). Our estimates of 0.29 and 1.03% for the frequency of biparental inheritance of mtDNA for ME1 and ME2, respectively, may hence have to be regarded as conservative estimate of biparental inheritance within these crosses.
Overall, the three-fold higher rate of biparental inheritance in ME2 is in line with earlier studies of de Stordeur (1989) revealing a competitive advantage of siII over siIII in micro-injection studies. While we can confirm de Stordeur's (1997) findings by observing a higher rate of paternal leakage in ME2 (with siII as the introgressing haplotype), our results are not in full agreement because the only complete replacements of maternal mtDNA were found in ME1. Considering a competitive advantage of siII over siIII in heteroplasmic flies, we assumed complete replacement of maternal by paternal mtDNA, if occurring at all, more likely in ME2 than in ME1. One factor, potentially leading to the replacement of the maternal haplotype is positive selection in favor of the paternal haplotype. Increased cell respiration and adaptive advantage between the introduced haplotype and the host's nuclear background have been put forward as potential driver of this process (de Stordeur, 1997). A change in respiratory capacity would be in line with studies revealing the disruption of mitonuclear interactions to negatively impact on organismal fitness (Ellison and Burton, 2008; Sackton et al., 2003). If this concept finds general applicability, then paternal mtDNA should be selected against. A recent study comparing catalytic properties of siII and siIII lines (identical to those used in our study), however, reported that the mitochondrial respiratory potential was not impaired by introgressing either haplotype into varying nuclear backgrounds (Pichaud et al., 2012).
Alternatively, a replicative advantage or environmental factor may impact on the inheritance and propagation of paternal mtDNA. One factor with the capacity to cause selective transmission or propagation of single haplotypes in Drosophila is temperature. This mechanism depends on both temperature and nuclear background and can lead to the selective transmission of certain haplotypes at specific temperatures (Doi et al., 1999; Matsuura et al., 1993). We deem temperature unlikely a major driving force in our study because temperature was kept constant in our experiments and a temperature-dependent effect, if occurring, should have been observable in both mating directions. An uneven distribution of mtDNA haplotypes could also be caused by a bottleneck effect or by random assortment of large mitochondrial sperm derivates (assuming these contain mtDNA) during early developmental stages (Bergstrom and Pritchard, 1998; Pitnick and Karr, 1998). These are stochastic processes, however, and are thus not in agreement with the observed pair-specific distribution of paternal leakage here. Conducting a similar study including additional haplotypes and/or changing the nuclear background could support or refute the alternate hypotheses. The high penetrance and stable state of heteroplasmy as observed in cross #72 after 20 generations, however, is in disagreement with purifying selection or a clear competitive advantage of one haplotype over another. Instead, the loss of heteroplasmy in cross #85 may be indicative of a minimum proportion of heteroplasmic individuals necessary to propagate and maintain heteroplasmy within a population (in F1, cross #72 revealed 58% and cross #85 only 6% heteroplasmic offspring).
The pair specificity of observed paternal leakage in our experiments is in line with previous studies (Kondo et al., 1990; Kondo et al., 1992; Sherengul et al., 2006). These studies, however, reveal paternal leakage at frequencies as high as 0.86 per line, a finding most likely attributable to the utilization of interspecific or backcrossed fly lines, crossing regimes believed to promote paternal leakage (Kaneda et al., 1995; Sutovsky et al., 2000; White et al., 2008). We can only speculate why mechanisms to prevent paternal leakage fail in single pairs. If it was a purely stochastic phenomenon, then we would expect the distribution of affected individuals carrying paternal mtDNA to be even among all offspring and not pair specific (Bergstrom and Pritchard, 1998; Wolff et al., 2011). Pair specificity instead requires one or more mechanisms promoting maternal inheritance to fail in single pairs, allowing for the repeated occurrence among a pair's offspring. We thus argue that it is more likely to be on a molecular than on a stochastic level that the mechanistics promoting maternal inheritance are affected to agree with such a pattern. Slight differences between lines or pairs in the nuclear background have the potential to play a major role in causing this effect. As progeny of isofemale lines, all offspring analyzed in this study are in theory genetically identical but newly arisen mutations (SNPs or chromosomal rearrangements) or transposable element movement, may have manifested in individual pairings during zygotic meiosis, germ line development or early embryonic development, leading to genetic heterogeneity.
There are at least two mechanisms that have, if affected, the potential to cause pair- or line-specific patterns of paternal leakage. First, the directed sequestration of mitochondrial sperm derivatives into the midgut and subsequent defecation during early embryonic development, a mechanism put forward to exclude paternal mtDNA from inheritance in D. melanogaster and D. pachea (Pitnick and Karr, 1998). A failure of this mechanism could potentially lead to the inclusion of paternal mtDNA within the developing embryo. Second, from the mammalian system we know that paternal mitochondria are selectively tagged (ubiquitinated) during spermatogenesis followed by their degradation by the 26S proteasome when sperm mitochondria enter the egg cytoplasm (Sutovsky et al., 1999). Assuming this mechanism also applies to Drosophila and that the mitochondrial remnants in Drosophila midgut are a rudiment of sperm mitochondrion degradation in the zygote, a distinct window of opportunity presents itself during which such a mechanism could fail: a widespread proteasomal degradation of proteins left from oogenesis occurs in the Drosophila zygote at the onset of embryo development (DeRenzo and Seydoux, 2004). If this process is compromised, then some mitochondria could escape and propagate their DNA. Alternatively, a mismatch between paternal mitochondrial proteins and ooplasmic ubiquitin ligases, the enzymes responsible for a substrate-specific tagging of proteins recycled by the 26S proteasome, could be due to polymorphisms in genes encoding some such enzymes in different strains/haplogroups of the same Drosophila species.
In this study, we have not quantified heritability of heteroplasmy. However, if we follow a highly simplified inheritance model we can begin to estimate the heritability necessary to sustain heteroplasmy levels that are observed in natural populations as H=h × f+h2 × f+…+hn × f where h is the heritability and f the frequency of paternal leakage where n approaches infinity. Considering an overall frequency of 0.66% for paternal leakage and heteroplasmy levels of 6–15%, the heritability would need to lie within the range of about 0.9 and 0.96. Although this estimate finds some support in the high percentage of heteroplasmy of cross #72 after 20 generations this simple model makes a broad set of assumptions that need to be tested, and the model refined, before its accuracy is known. It is likely that the effective population size of D. simulans is very large in east Africa (Lachaise et al., 1988) and it is possible that flies harboring distinct haplotypes randomly mate in nature (Ballard et al., 2002). Bioenergetic differences conferred by the distinct haplotypes remain in an introgressed background at 24 °C (Pichaud et al., 2012) and it is assumed that the nuclear genome of D. simulans flies in East Africa is effectively randomized with respect to the mtDNA (Melvin et al., 2008). In East Africa, the frequency of haplotypes is not equal and the distinct strains of Wolbachia that infect D. simulans flies in the region may plausibly influence the rates of heteroplasmy (Turelli et al., 1992; Dean et al., 2003). In this study, flies were confirmed Wolbachia free so the symbiont did not influence the observed rates of paternal leakage. One assumption that may be violated is that mtDNA genomes have equal potential to go to fixation in a heteroplasmic fly (de Stordeur et al., 1989), although this potential may be temperature dependent (Doi et al., 1999; Matsuura et al., 1993).
The broad scale consequences of high levels of heteroplasmy on the evolutionary dynamics of mtDNA are unknown. It is reasonable to assume, however, the inheritance of paternal mtDNA to be of significance if the introgressing molecule propagates and replaces the maternal mtDNA or if heterologous recombination takes place and the resulting chimeric molecule manifests in populations (Eyre-walker, 2000). Such processes would not only impact on the evolution of the molecule but also may impact species or population fitness and may counterbalance the female selective sieve acting on mtDNA (Bromham et al., 2003; Gemmell et al., 2004; Innocenti et al., 2011). Our study shows that a replacement of maternal by paternal mtDNA is possible in single individuals but additional studies are necessary to examine whether these novel mitonuclear lineages have the capacity to propagate within entire populations in this species as reported for other species (Alves et al., 2008; Zakharov et al., 2009). Similarly, further research is needed to evaluate the potential impact of impending heterologous recombination on dynamics of mitochondrial evolution and organismal or population fitness (Bromham et al., 2003).
In conclusion, we have successfully demonstrated extensive paternal leakage in D. simulans, strongly suggesting this phenomenon as the cause for recurring heteroplasmy in natural population of this species in East Africa and Reunion Island. Paternal leakage must thus be considered as integral component of mtDNA transmission in these populations. This consideration is important, especially in light of impending heterologous recombination between the two haplotypes, as these phenomenon have the potential to introduce new haplotypes into populations, and with that a potentially distorting effect on mtDNA analyses. Accurate interpretation of mtDNA sequence data in evolutionary contexts or attempts to predict probabilities of the heritability of mitochondrial disease critically depend on a mechanistic understanding of mitochondrial inheritance. It is hence of utmost importance to gain a better understanding of mechanisms involved in shaping the evolution of mtDNA and those leading to the leakage of paternal mtDNA.
Data archiving
Data have been deposited as follows: Repository/DataBank Accession: GenBank, Accession ID: JQ691660, Databank URL: http://www.ncbi.nlm.nih.gov/genbank/ and Repository/DataBank Accession: GenBank, Accession ID: JQ691661, Databank URL: http://www.ncbi.nlm.nih.gov/genbank/.
Acknowledgments
We thank the Ballard and Wilton lab group for helpful discussion and comments during the study and on the manuscript. We are highly indebted to Daniel Masiga at ICIPE to host us, enabling the collection of D. simulans in Nairobi, Kenya, in 2011. This research was supported by a Discovery Project grant (DP110104542) of the Australian Research Council to JWOB, PS and JNW. JNW holds a postdoctoral fellowship from the Australian Research Council.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Allen JF. Separate sexes and the mitochondrial theory of ageing. J Theor Biol. 1996;180:135–140. doi: 10.1006/jtbi.1996.0089. [DOI] [PubMed] [Google Scholar]
- Alves PC, Melo-Ferreira J, Freitas H, Boursot P. The ubiquitous mountain hare mitochondria: multiple introgressive hybridization in hares, genus Lepus. Philos Trans R Soc Lond B Biol Sci. 2008;363:2831–2839. doi: 10.1098/rstb.2008.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba-Aissa F, Solignac M, Dennebouy N, David JR. Mitochondrial DNA variability in Drosophila simulans: quasi absence of polymorphism within each of the three cytoplasmic races. Heredity. 1988;61:419–426. doi: 10.1038/hdy.1988.133. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Chernoff B, James AC. Divergence of mitochondrial DNA is not corroborated by nuclear dna, morphology, or behavior in Drosophila simulans. Evolution. 2002;56:527–545. doi: 10.1111/j.0014-3820.2002.tb01364.x. [DOI] [PubMed] [Google Scholar]
- Ballard JWO. Comparative genomics of mitochondrial DNA in Drosophila simulans. J Mol Evol. 2000;51:64–75. doi: 10.1007/s002390010067. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Melvin RG, Katewa SD, Maas K. Mitochondrial DNA variation is associated with measurable differences in life-history traits and mitochondrial metabolism in Drosophila simulans. Evolution. 2007;61:1735–1747. doi: 10.1111/j.1558-5646.2007.00133.x. [DOI] [PubMed] [Google Scholar]
- Bergstrom CT, Pritchard J. Germline bottlenecks and the evolutionary maintenance of mitochondrial genomes. Genetics. 1998;149:2135–2146. doi: 10.1093/genetics/149.4.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Beaupre HD, Stewart DT, Hoeh WR, Blier PU. The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough. Trends Genet. 2007;23:465–474. doi: 10.1016/j.tig.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Bromham L, Eyre-Walker A, Smith NH, Maynard Smith J. Mitochondrial Steve: paternal inheritance of mitochondria in humans. Trends Ecol Evol. 2003;18:2–4. [Google Scholar]
- de Stordeur E. Nonrandom partition of mitochondria in heteroplasmic Drosophila. Heredity. 1997;79:615–623. doi: 10.1038/hdy.1997.207. [DOI] [PubMed] [Google Scholar]
- de Stordeur E, Solignac M, Monnerot M, Mounolou JC. The generation of transplasmic Drosophila simulans by cytoplasmic injection: effects of segregation and selection on the perpetuation of mitochondrial DNA heteroplasmy. Mol Gen Genet. 1989;220:127–132. doi: 10.1007/BF00260866. [DOI] [PubMed] [Google Scholar]
- Dean MD, Ballard KJ, Glass A, Ballard JWO. Influence of two Wolbachia strains on population structure of East African Drosophila simulans. Genetics. 2003;165:1959–1969. doi: 10.1093/genetics/165.4.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca SZ, O'Farrell PH. Barriers to male transmission of mitochondrial DNA in sperm development. Dev Cell. 2012;22:660–668. doi: 10.1016/j.devcel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Doi A, Suzuki H, Matsuura ET. Genetic analysis of temperature-dependent transmission of mitochondrial DNA in Drosophila. Heredity. 1999;82:555–560. doi: 10.1038/sj.hdy.6885080. [DOI] [PubMed] [Google Scholar]
- Ellison CK, Burton RS. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution. 2008;62:631–638. doi: 10.1111/j.1558-5646.2007.00305.x. [DOI] [PubMed] [Google Scholar]
- Eyre-walker A. Do mitochondria recombine in humans. Philos Trans R Soc Lond B Biol Sci. 2000;355:1573–1580. doi: 10.1098/rstb.2000.0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19:238–244. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Goodman SJ, Barton NH, Swanson G, Abernethy K, Pemberton JM. Introgression through rare hybridization: A genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll, Scotland. Genetics. 1999;152:355–371. doi: 10.1093/genetics/152.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings IM. Population genetic aspects of deleterious cytoplasmic genomes and their effect on the evolution of sexual reproduction. Genet Res. 1992;59:215–225. doi: 10.1017/s0016672300030500. [DOI] [PubMed] [Google Scholar]
- Holleley CE. Economical high-throughput DNA extraction procedure in 96-well format for Drosophila tissue. Dros Inf Serv. 2007;90:137–138. [Google Scholar]
- Hurst LD. Why are there only two sexes. Proc R Soc Lond B Biol. 1996;263:415–422. [Google Scholar]
- Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011;332:845–848. doi: 10.1126/science.1201157. [DOI] [PubMed] [Google Scholar]
- Kaneda H, Hayashi J, Takahama S, Taya C, Lindahl KF, Yonekawa H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:4542–4546. doi: 10.1073/pnas.92.10.4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo R, Matsuura ET, Chigusa SI. Further observation of paternal transmission of Drosophila mitochondrial DNA by PCR selective amplification method. Genet Res. 1992;59:81–84. doi: 10.1017/s0016672300030287. [DOI] [PubMed] [Google Scholar]
- Kondo R, Satta Y, Matsuura ET, Ishiwa H, Takahata N, Chigusa SI. Incomplete maternal transmission of mitochondrial DNA in Drosophila. Genetics. 1990;126:657–663. doi: 10.1093/genetics/126.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Cariou ML, David JR, Lemeunier F, Tsacas L, Ashburner M. Historical biogeography of the Drosophila melanogaster species subgroup. Evol Biol. 1988;22:159–225. [Google Scholar]
- Matsuura ET, Fukuda H, Chigusa SI. Mitochondrial DNA heteroplasmy maintained in natural populations of Drosophila simulans in Reunion. Genet Res. 1991;57:123–126. doi: 10.1017/s0016672300029189. [DOI] [PubMed] [Google Scholar]
- Matsuura ET, Niki Y, Chigusa SI. Temperature-dependent selection in the transmission of mitochondrial DNA in Drosophila. Jpn J Genet. 1993;68:127–135. doi: 10.1266/jjg.68.127. [DOI] [PubMed] [Google Scholar]
- Melvin RG, Katewa SD, Ballard JWO. A candidate complex approach to study functional mitochondrial DNA changes: sequence variation and quaternary structure modeling of Drosophila simulans cytochrome c oxidase. J Mol Evol. 2008;66:232–242. doi: 10.1007/s00239-008-9078-4. [DOI] [PubMed] [Google Scholar]
- Oliveira R, Godinho R, Randi E, Alves PC. Hybridization versus conservation: are domestic cats threatening the genetic integrity of wildcats (Felis silvestris silvestris) in Iberian Peninsula. Philos Trans R Soc Lond B Biol Sci. 2008;363:2953–2961. doi: 10.1098/rstb.2008.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichaud N, Ballard JWO, Tanguay RM, Blier PU. Natually occurring mitochondrial DNA haplotypes exhibit metabolic differences: insight into functional properties of mitochondria. Evolution. 2012;66:3189–3197. doi: 10.1111/j.1558-5646.2012.01683.x. [DOI] [PubMed] [Google Scholar]
- Pitnick S, Karr TL. Paternal products and by-products in Drosophila development. Proc Biol Sci B. 1998;265:821–826. doi: 10.1098/rspb.1998.0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puslednik L, Yeates DK, Faith DP, Ballard JWO. Protein-protein interactions and the cytochrome c oxidase DNA barcoding region. Sys Entomol. 2012;37:229–236. [Google Scholar]
- Rand DM. Thermal habit, metabolic rate and the evolution of mitochondrial DNA. Trends Ecol Evol. 1994;9:125–131. doi: 10.1016/0169-5347(94)90176-7. [DOI] [PubMed] [Google Scholar]
- Sackton TB, Haney RA, Rand DM. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003;57:2315–2325. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- Satta YN, Toyohara C, Ohtaka Y, Tatsuno TK, Watanabe ET, Matsuura SI, et al. Dubious maternal inheritance of mitochondrial DNA in D. simulans and evolution of D. mauritiana. Genet Res. 1988;52:1–6. [Google Scholar]
- Schwartz M, Vissing J. Paternal inheritance of mitochondrial DNA. N Engl J Med. 2002;347:576–580. doi: 10.1056/NEJMoa020350. [DOI] [PubMed] [Google Scholar]
- Scott D, Richmond R. Sperm loss by remating Drosophila melanogaster females. J Insect Physiol. 1990;36:451–456. [Google Scholar]
- Sherengul W, Kondo R, Matsuura ET. Analysis of paternal transmission of mitochondrial DNA in Drosophila. Genes Genet Syst. 2006;81:399–404. doi: 10.1266/ggs.81.399. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–372. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- Sutovsky P, Moreno RD, Ramalho-Santos J, Dominko T, Simerly C, Schatten G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol Reprod. 2000;63:582–590. doi: 10.1095/biolreprod63.2.582. [DOI] [PubMed] [Google Scholar]
- Turelli M, Hoffmann AA, McKechnie SW. Dynamics of cytoplasmic incompatibility and mtDNA variation in natural Drosophila simulans populations. Genetics. 1992;132:713–723. doi: 10.1093/genetics/132.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ, Brandvain Y. Reversing mother's curse: selection on male mitochondrial fitness. Evolution. 2009;63:1084–1089. doi: 10.1111/j.1558-5646.2009.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- Wolff JN, White DJ, Woodhams M, White HE, Gemmell NJ. The strength and timing of the mitochondrial bottleneck in salmon suggests a conserved mechanism in vertebrates. PLoS ONE. 2011;6:e20522. doi: 10.1371/journal.pone.0020522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharov EV, Lobo NF, Nowak C, Hellmann JJ. Introgression as a likely cause of mtDNA paraphyly in two allopatric skippers (Lepidoptera: Hesperiidae) Heredity. 2009;102:590–599. doi: 10.1038/hdy.2009.26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.