Abstract
Sirtuins are a family of NAD+-dependent protein deacetylases/deacylases that dynamically regulate transcription, metabolism, and cellular stress response. Their general positive link with improved health span in mammals, potential regulation of pathways mediated by caloric restriction, and growing links to human disease have spurred interest in therapeutics that target their functions. Here, we review the current understanding of the chemistry of catalysis, biological targets, and endogenous regulation of sirtuin activity. We discuss recent efforts to generate small-molecule regulators of sirtuin activity.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, NAD, Post-translational Modification, Sirtuins
Introduction
Accumulating data indicate that lysine acetylation is a prevalent regulatory mechanism of protein function, with thousands of acetylated proteins identified by mass spectrometry (1–3). Sir2 (silent information regulator 2 or sirtuin) protein deacetylases are a class of evolutionarily conserved enzymes that function in critical cellular processes such as transcription, DNA repair, metabolism, and stress resistance (4). Among the major classes of lysine deacetylases, the sirtuins utilize a unique catalytic mechanism that consumes NAD+, providing a direct connection between protein deacetylation and central metabolic pathways. There are seven human sirtuins (SIRT1–7), each with diverse subcellular localization and protein substrates (5). SIRT1–3 display robust deacetylation activity, whereas recent reports implicate SIRT5 as a protein desuccinylase and demalonylase (6). Thus, sirtuins can be considered deacylases. The activities of several other human sirtuins are unsettled. SIRT6 and SIRT7 display weak deacetylase activity in vitro, and SIRT4 was reported to harbor ADP-ribosyltransferase activity (7, 8). Structural analysis of the sirtuin family members reveals a conserved catalytic core composed of two subdomains, a Rossmann fold domain at one end and a smaller, more variable zinc-binding domain at the opposite end (Fig. 1). The two domains are connected by several loops that form a binding cleft for the nicotinamide and ribose moieties of NAD+ and the acyllysine substrate. Several invariant amino acids are located in the cleft and are responsible for substrate binding and catalysis. The varying hydrophobicity and charge distribution of the acyl-substrate binding cleft allow for varied substrate selectivity among the different human sirtuins (6, 9). Given their regulatory role in transcription, metabolism, and genome maintenance, sirtuins are desirable targets for therapeutic development. In this minireview, we highlight the current molecular understanding of the chemical mechanism, regulation, and substrate selectivity of sirtuins.
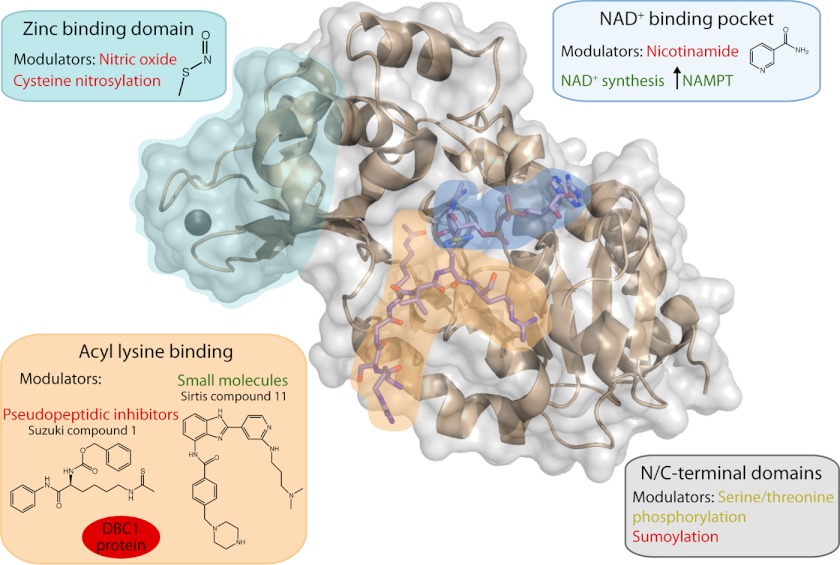
FIGURE 1.
Representative structure of a human sirtuin (Protein Data Bank code 3GLR) bound to acetylated peptide and NAD+. Key locations for sirtuin modulation are highlighted. Positive regulators of sirtuin activity are indicated in green, negative regulators are indicated in red, and regulators that can activate or inhibit depending on the sirtuin are in yellow. Proposed activators include NAD+ synthesis, Sirtris compound 11 (90) and other reported activators, and phosphorylation of SIRT1. Inhibitors include cysteine nitrosylation, DBC1 binding SIRT1, pseudopeptidic inhibitors (95) and other small molecules, nicotinamide, and sumoylation of SIRT1.
Unique Chemistry
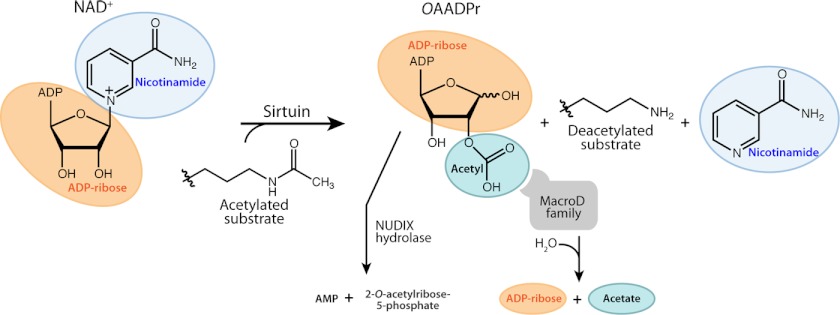
Sirtuins catalyze NAD+-dependent deacetylation of acetyllysine, resulting in the production of deacetylated lysine, nicotinamide, and 2′-O-acetyl-ADP-ribose (OAADPr)3 (Fig. 2) (10). Kinetic and biochemical studies revealed that the enzyme binds the acetyllysine substrate prior to NAD+. Nicotinamide is cleaved from NAD+ and is the first product released, followed by deacetylated lysine and OAADPr (11). In aqueous solution, non-enzymatic intramolecular transesterification yields the predominant mixture of 2′-OAADPr and 3′-OAADPr. The use of NAD+ as a co-substrate distinguishes sirtuins from other classes of protein deacetylases. Curiously, SIRT6 is the only human sirtuin capable of tightly binding NAD+ in the absence of an acetylated substrate, suggesting that SIRT6 might also function as an NAD+ sensor, possibly without active deacylation (8). Great interest lies in understanding the coupling of NAD+ consumption to the production of OAADPr, a metabolite that exhibits signaling functions but has been less studied (12). OAADPr was linked with decreased reactive oxygen species levels, gene silencing, and ion channel activation and was shown to block starfish oocyte maturation (reviewed in Ref. 12). Several OAADPr-metabolizing enzymes have been reported, including the NUDIX (nucleoside diphosphate linked to moiety x) hydrolases, ARH3 (ADP-ribosylhydrolase 3), and macrodomain proteins (12). NUDIX hydrolases cleave the pyrophosphate bond of OAADPr, forming AMP and 2- and 3-O-acetyl 5-phosphate (Fig. 2). Human macrodomain proteins are capable of hydrolyzing OAADPr, affording free acetate and ADP-ribose (13). In lower organisms, some sirtuins and macrodomain proteins are genetically coupled within the same operon or physically connected as fusion proteins, providing evidence for an as-yet-unknown pathway that involves sirtuins, macrodomain enzymes, and OAADPr (13).
FIGURE 2.
Substrates and products of the sirtuin-catalyzed reaction and potential fate of the product OAADPr. The unique use of NAD+ as a co-substrate distinguishes sirtuins from other deacetylase classes and provides a direct link to energy metabolism.
The initial chemical step of the sirtuin reaction involves nucleophilic addition of the acetyl oxygen to C1′ of the nicotinamide ribose, forming a C1′-O-alkylamidate intermediate (supplemental Fig. 1). The mechanism of nucleophilic attack has been subject to discussion, with SN1, concerted SN2, and dissociative SN2-like mechanisms proposed (reviewed in Ref. 14). A detailed study using kinetic isotope effects and computational approaches suggested that the first step of the reaction proceeds via a concerted yet highly asynchronous substitution mechanism (15). Several biochemical studies support the formation of the alkylamidate intermediate. These include nicotinamide base-exchange reactions (16) and 18O labeling studies that provide evidence for the direct transfer of the acetyl oxygen to the 1′-hydroxyl of OAADPr (17, 18). Utilization of acetyllysine analogs further demonstrated the existence of the alkylamidate intermediate. Thioacetyllysine- and acetylazalysine-containing peptides form stalled alkylamidate intermediates when used as sirtuin substrates (19, 20). Upon formation of the alkylamidate intermediate, the 2′-hydroxyl group of the NAD+ ribose is activated by a conserved active-site histidine (supplemental Fig. 1). The activated hydroxyl attacks the O-alkylamidate carbon to afford a 1′,2′-cyclic intermediate (20). Recently, the bicyclic intermediate was trapped and structurally resolved by incubating co-crystals of SIRT5 and an H3K9 thiosuccinyl-peptide with NAD+, providing direct evidence for the proposed catalytic mechanism (21). A base-activated water molecule then attacks the cyclic intermediate, affording deacetylated lysine and OAADPr (supplemental Fig. 1) (10, 17).
Although deacylation is thought to be the primary function of human sirtuins, yeast Sir2 was first implicated as having the capability to transfer ADP-ribose from NAD+ to nucleophilic amino acids on protein substrates (22). SIRT4 and SIRT6 have also been reported to catalyze ADP-ribosyl transfer to glutamate dehydrogenase and poly(ADP-ribose) polymerase 1, respectively (7, 23). This activity is not robust and has been subject to debate. Detailed kinetic characterization of the ADP-ribosyltransferase activity of a yeast and bacterial sirtuin indicated that ADP-ribosylation may be a low efficiency side reaction (∼0.1% of the deacetylation reaction) of sirtuins due to the susceptibility of active site-bound ADP-ribose to nucleophilic attack (24). Understanding the mechanistic details of sirtuin-catalyzed reactions is an important step toward a complete understanding of sirtuin function and in the development of chemical tools to probe their biology.
Substrate Recognition and Acyl Group Specificity
A number of proteomics studies have greatly enhanced our understanding of lysine acetylation as a global post-translational modification regulating diverse cellular processes (2, 3, 25). The number of reported sirtuin targets is continually increasing (Fig. 3). SIRT1 deacetylates a number of histone and non-histone proteins, including, but not limited to, histones H3 and H4 (26), p53 (27), NF-κB (28), phosphoglycerate mutase 1 (29), and peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) (30). SIRT3 is reported to deacetylate and modulate the activity of several metabolic enzymes, including ornithine transcarbamylase (31), long chain acyl-CoA dehydrogenase (32), manganese superoxide dismutase (33), acetyl-CoA synthetase 2 (34), and isocitrate dehydrogenase 2 (35, 36). With the recent expansion of the acetylome comes the challenge of identifying the physiologically relevant sites and the enzymes responsible for the addition and removal of these modifications.
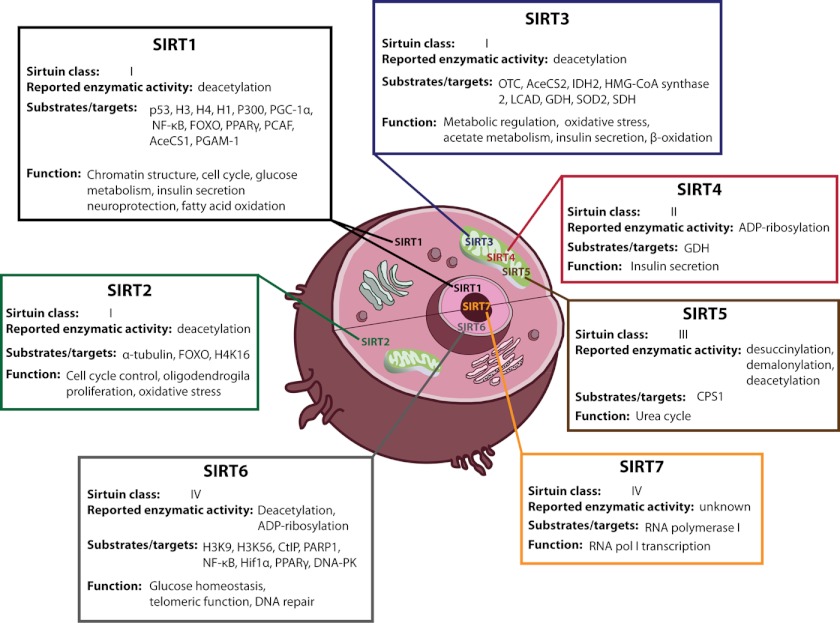
FIGURE 3.
Subcellular localization of mammalian sirtuins and reported enzymatic activities, substrates/targets, and cellular functions. PPARγ, peroxisome proliferator-activated receptor-γ; PCAF, p300/CBP-associated factor; OTC, ornithine transcarbamylase; LCAD, long chain acyl-CoA dehydrogenase; GDH, glutamate dehydrogenase; SDH, succinate dehydrogenase; pol, polymerase; CtIP, CTBP-interacting protein; DNA-PK, DNA-dependent protein kinase.
Protein Recognition
Although a full understanding of protein substrate selection is lacking, a number of reports have addressed the amino acid sequence specificity of sirtuins. Initial structural studies suggested that acetyllysine-peptide binding is largely dominated by peptide backbone hydrogen bonds, rather than through side chain interactions (37). Consistent with this idea, Blander et al. (38) used an acetyllysine-peptide library and concluded that SIRT1 displayed no substrate specificity in vitro. Different conclusions were reached when the specificity of SIRT1 was probed by a combinatorial, one-bead, one-peptide acetyl-peptide library (39) and by a mass spectrometry-based deacetylation assay of peptide substrates immobilized on self-assembled monolayers on gold slides (40). These results suggested that SIRT1 specificity is largely context-dependent, in which preference for an amino acid at a given position depends on the presence or absence of a specific amino acid at an adjacent position. Smith et al. (9) used SPOT-peptide array analysis and machine learning approaches to determine that SIRT3 displays a preference for aromatic and basic residues surrounding the acetyllysine while disfavoring negatively charged residues. Additional crystallographic studies of Thermotoga maritima Sir2 suggested that the first residue N-terminal to the acetyllysine and the second residue C-terminal to the acetyllysine play significant roles in substrate binding (41). Such unbiased library methods will be important to determine the substrate specificity for other sirtuins, including SIRT4–7, which have few known targets and possess extremely low deacetylase activity on commonly used substrates.
Sirtuin-catalyzed Protein Deacylation
In addition to acetyl-CoA, other abundant acyl-CoAs might serve as acyl donor molecules for the post-translational modification of lysine residues. Recent studies identified a series of acyl groups (propionyl, butyryl, succinyl, malonyl, and crotonyl) as post-translational modifications of lysine residues (Fig. 4) in histone and non-histone proteins (6, 42–46). Mass spectrometric and biochemical analyses identified propionyllysine and butyryllysine residues within histone H4 and on lysine 23 of histone H3 (42, 47). Several acetyltransferases, including human p300 and CBP (CREB-binding protein), Saccharomyces cerevisiae EsaI, and some bacterial acetyltransferases, can catalyze lysine propionylation and butyrylation (42, 43, 48). SIRT1–3 can catalyze depropionylation and debutyrylation, but with varying efficiencies compared with deacetylation (43, 49). Mass spectrometry-based proteomics studies recently identified succinyllysine, malonyllysine, and crotonyllysine as previously unidentified modifications of histone proteins in several eukaryotic cell types (46, 50). Crotonyllysine was shown by chromatin immunoprecipitation analysis to be associated with active promoters or enhancers in human somatic and mouse germ cell genomes, suggesting a possible role in transcriptional control (50).
FIGURE 4.
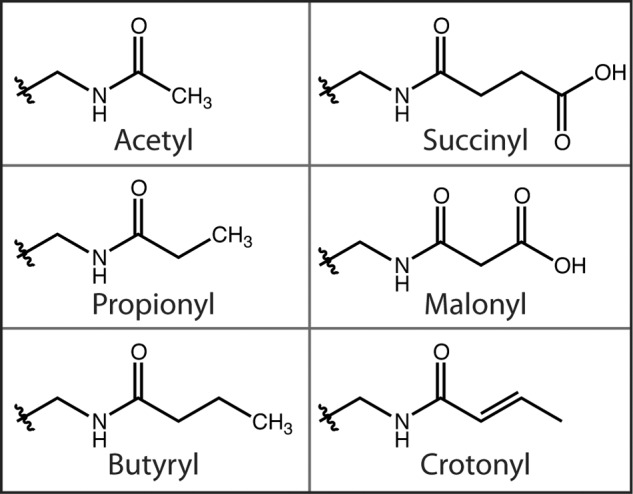
Structures of known acyl modifications found on lysine residues.
Although many of these newly described modifications were reported for histone proteins, post-translational succinylation and malonylation were identified and verified in several metabolic enzymes from mammalian cells (6, 45). Furthermore, these studies found that mitochondrially localized SIRT5 could catalyze desuccinylation and demalonylation in vitro (6, 45). Utilizing an HPLC-based assay, Du et al. (6) reported that the catalytic efficiency for demalonylation and desuccinylation for three separate peptide sequences was 29- to >1000-fold higher than that for deacetylation, suggesting that SIRT5 functions as an NAD+-dependent desuccinylase and demalonylase rather than as a deacetylase. Isolation of O-succinyl- and O-malonyl-ADPr confirmed that deacylation proceeds by the known sirtuin deacetylation mechanism. Deletion of SIRT5 appeared to increase the level of succinylation on CPS1 (carbamoyl phosphate synthase 1) (6), which was previously reported to be a target of SIRT5 (51). A crystal structure of SIRT5 bound to a succinylated peptide revealed the structural basis for this acyl group preference. The carboxyl group of the succinyllysine interacts via hydrogen bonds to Tyr-102 and Arg-105 in the active site (6). These residues are conserved among many members of the class III sirtuins, suggesting other class III sirtuins might also function as desuccinylases and demalonylases.
Cellular Regulation of Sirtuin Activity
Depending on any one particular report, sirtuins can act as either positive or negative regulators of pathways involved in disease development. For example, among published studies, SIRT1 and SIRT3 are implicated as tumor promoters or suppressors (52, 53), although the vast majority of evidence suggests that they improve health span in adult animals when their expression is induced appropriately. Because sirtuins are involved in a number of central physiological processes, endogenous signaling pathways likely control their activity in a tissue-specific, signal-dependent, and temporally programmed manner. The apparent duality of sirtuin function in disease might simply stem from an incomplete understanding of sirtuin regulation and cellular context of function. Quite surprisingly, there is relatively sparse detailed knowledge of endogenous regulatory mechanisms for sirtuins. A summary of the current understanding is discussed below.
Transcriptional Regulation
The seven sirtuins are nuclear-encoded and ubiquitously expressed in human tissues but display unique subcellular localization (5, 54). SIRT1, SIRT6, and SIRT7 localize to the nucleus; SIRT3–5 localize to the mitochondria; and SIRT2 is found primarily in the cytoplasm (Fig. 3) (5). Some evidence suggests the presence of full-length SIRT3 in the nucleus during cellular stress (55). Caloric restriction, the only confirmed treatment to extend mammalian life span (56), is known to enhance the transcription of SIRT1 and SIRT3, a result that continues to spur exploration into the role of these sirtuins in mediating the effects of caloric restriction (31, 57).
Two recent studies highlight the interplay between nutrient availability and sirtuin transcription. In response to fasting, SIRT1 expression is increased by cAMP response element-binding protein, a known inducer of gluconeogenic genes. Increased SIRT1 protein levels result in deacetylation and activation of PGC-1α, a known regulator of genes involved in mitochondrial biogenesis, thermogenesis, reactive oxygen detoxification, and gluconeogenesis. Activation of PGC-1α by SIRT1 turns on the expression of a number of catabolic proteins in metabolism. In response to refeeding, carbohydrate response element-binding protein binds to the promoter of SIRT1 and decreases its transcription, serving as a molecular switch to the anabolic state (58). Other recent studies show that the transcription of SIRT3 is induced by PGC-1α in muscle cells, brown adipose, and hepatocytes through binding to an estrogen-related receptor-binding element in the SIRT3 promoter region (59, 60). The mitochondrial metabolic reprogramming activities of PGC-1α may be mediated through increased SIRT3 protein levels. A unique cross-talk among sirtuins is suggested, as nutrient status leads to increased SIRT1 expression, which deacetylates and activates PGC-1α, ultimately leading to the induction of SIRT3 transcription.
Post-translational Modifications and Complex Formation
The highly conserved catalytic core of human sirtuins is surrounded by variable N- and C-terminal extensions, which appear to act as regulatory regions that harbor sites for post-translational modification and act as docking regions for protein complex formation.
Phosphorylation sites have been identified on all human sirtuins, but the functional impact has been investigated only for SIRT1 and SIRT2. Independent studies report multiple phosphorylation sites located in the N- and C-terminal domains of SIRT1 and implicate different kinases in regulating SIRT1 activity, including DYRK (dual specificity tyrosine phosphorylation-regulated kinase), JNK1 (c-Jun N-terminal kinase 1), cyclin B/Cdk1 (cyclin-dependent kinase 1), and PKA (61–63). These phosphorylation events are thought to activate SIRT1, perhaps through inducing allosteric conformational changes; however, the detailed mechanism is unknown. A recent study identified a cAMP-dependent phosphorylation at Ser-434 of SIRT1 that increased deacetylase activity (63). Phosphorylation of Ser-434 is thought to reduce the Km for NAD+, resulting in increased SIRT1 catalysis. SIRT2 is phosphorylated at Ser-331 and Ser-335 within the C-terminal region. Phosphorylation of Ser-331 is catalyzed by cyclin-dependent kinase and inhibits SIRT2 activity through an unknown mechanism, whereas the kinase and function of phosphorylation of Ser-335 are not known (64).
Additional post-translational modifications of SIRT1 include sumoylation, methylation, and transnitrosylation (65–67). Through an NO-dependent reaction, nitrosylation occurs at key cysteine residues in the zinc-binding domain of SIRT1 (Fig. 1). Similarly, oxidative stress and accumulation of the lipid peroxidation product 4-hydroxynonenal result in the covalent modification of SIRT3 by 4-hydroxynonenal at cysteine 280 within the zinc-binding domain (68). Proper zinc coordination is necessary for sirtuin structural integrity and catalysis (69); thus, nitrosylation and carbonylation limit zinc binding and reduce activity.
In addition to post-translational modification, protein complex formation may play an important role in regulation of sirtuin activity. Many histone-modifying proteins are commonly found in complexes that regulate their function (70). Specifically, class I and II histone deacetylases exist almost exclusively as components of large multiprotein complexes. Curiously, the formation of such regulatory complexes among mammalian sirtuins remains enigmatic. A few endogenous protein-binding partners of SIRT1 are thought to regulate its function. AROS (active regulator of SIRT1) was reported to bind to amino acids 114–217 in the N terminus of SIRT1 and stimulate deacetylation of p53 in vivo, potentially by inducing a conformational change that places SIRT1 in a more favorable catalytic conformation (71). Binding of the inhibitory protein DBC1 (deleted in breast cancer 1) to the catalytic domain of SIRT1 results in repressed deacetylation of p53 in vivo and in vitro (72, 73). The leucine zipper motif of DBC1 binds to the catalytic core of SIRT1, but not to other sirtuins, and may block substrate access to the active site. A number of other protein-binding partners of SIRT1 and SIRT3 were identified in a study using affinity purification of FLAG-tagged sirtuins followed by mass spectral identification. Whether these proteins act as regulatory factors or substrates has not been determined (74).
A recent proteomics and bioinformatics study revealed that SIRT7 interacts with several nucleolar localized chromatin-remodeling complexes, including RNA polymerase I and upstream binding factor involved in ribosomal DNA transcription (75). The results suggest that SIRT7-containing protein complexes are critical during ribosomal transcription and reveal an important role for this sirtuin, which lacks robust deacetylation activity in vitro. Understanding the function of interacting proteins might provide insight into the low deacetylase activity of some sirtuins such as SIRT4, SIRT6, and SIRT7, which might require activation or targeting to function. The development of molecular tools to capture active sirtuin complexes in cells could enable the identification of proteins involved in sirtuin regulation and activity (20, 76).
NAD+ Levels
The levels of intracellular co-substrate NAD+ and product nicotinamide are thought to influence sirtuin activity. Nicotinamide is a product inhibitor of the deacetylation reaction and is used often as a general sirtuin inhibitor. At high concentrations, nicotinamide enters the active site and reacts with the alkylamidate intermediate, reforming NAD+ and preventing the forward reaction (Fig. 2) (16). The unique catalytic consumption of NAD+ indicates that sirtuins might be sensitive to changes in intracellular NAD+ concentration. Increasing NAD+ synthesis through the NAD+ salvage pathway might be a cellular mechanism to increase sirtuin activity. Indeed, enzymes that generate NAD+ affect sirtuin activity (reviewed in Ref. 77). Nicotinamide phosphoribosyltransferase (NAMPT) catalyzes the addition of 5-phosphoribosylpyrophosphate to nicotinamide to form NMN (78). NMN adenylyltransferase then converts NMN to NAD+. There are three isoforms of NMN adenylyltransferase that localize to the mitochondria, nucleus, and cytoplasm, suggesting that there may be compartmentalized control of NAD+ synthesis and therefore subcellular control of sirtuin activity (79). Inhibitors of NAMPT have been used to decrease SIRT2 activity in the treatment of acute myeloid leukemia, providing evidence that modulating NAD+ concentration might be an effective means to regulate sirtuins (80). Furthermore, AMP-activated kinase (AMPK) is known to activate NAD+ synthesis through stimulated transcription of NAMPT. AMPK is stimulated by decreases in cellular energy status, nutrient and oxygen deprivation, and increased energy expenditure (81).
Therapeutic Potential: Small-molecule Modulation of Sirtuin Activity
Activators
Sirtuins are pharmaceutical targets due to purported roles in cell survival, fatty acid metabolism, glucose homeostasis, genomic stability, and oxidative stress reduction. Compounds that activate SIRT1 could have positive roles in regulating metabolism and influencing health span. A number of small-molecule compounds are reported to increase the deacetylase activity of SIRT1, including the naturally occurring polyphenol resveratrol, as well as a series of small-molecule compounds developed by Sirtris Pharmaceuticals, Inc. (Fig. 1) (82, 83). Although these reports have sparked great interest in the promise of sirtuin activation, other in vitro and in vivo studies have disputed the direct link to SIRT1 activation (reviewed in Ref. 84). Resveratrol is a known pleiotropic molecule, and some laboratories have reported that resveratrol activates the AMPK pathway, perhaps through direct inhibition of phosphodiesterase 4, ultimately leading to stimulated SIRT1 activity by increasing the cellular NAD+ concentration (85, 86). A recent study utilizing a tamoxifen-inducible SIRT1 knock-out in adult mice found that resveratrol improved mitochondrial function in skeletal muscle and induced a shift toward more oxidative muscle fibers in wild-type mice, but not in adult SIRT1 knock-out mice fed the same high fat diet (87). The results strengthen the physiological link connecting the positive effects of resveratrol to a SIRT1-dependent process. However, the exact molecular targets of resveratrol that influence AMPK- and SIRT1-dependent pathways remain unresolved.
The controversy surrounding resveratrol and SIRT1 originated from the observation that resveratrol could activate SIRT1 only when a fluorescently tagged peptide substrate was utilized in high throughput deacetylation assays (88, 89). More recently, isothermal titration calorimetry and tryptophan fluorescence analysis suggested that several small molecules developed by Sirtris do indeed bind directly to SIRT1 with high affinity (90). From in vitro biochemical studies, it appears that validated SIRT1 activators increase the binding affinity for acetylated peptide and that the nature of the substrate, including the amino acid sequence and/or the hydrophobic fluorescent tag, is an important factor for activation (84, 88, 90).
Inhibitors
A number of small-molecule and mechanism-based sirtuin inhibitors have been developed. Several studies have identified compounds specifically targeting either SIRT1 or SIRT2 (91–93). A structure-based approach for identifying novel isoform-specific inhibitors utilized the peptide-binding grove within the crystal structures of SIRT2, SIRT3, SIRT5, and SIRT6 (92). Characterization of several hits identified two compounds that selectively inhibited SIRT2 with low micromolar IC50 values (92). Employing a different strategy, a number of pseudopeptidic mechanistic-based inhibitors using thioacyllysine have been developed for SIRT1, SIRT2, and SIRT5 (Fig. 1) (94–97). Biochemical, kinetic, and structural analyses suggest that the thioacetyllysine act as a mechanistic inhibitor by stalling at the catalytic intermediate after nicotinamide cleavage (Fig. 2) (19, 98). Use of peptide-like inhibitors might offer added specificity and affinity. A pseudopeptidic backbone might increase bioavailability and decrease the potential for enzymatic degradation. Structure-based computational approaches to identify pseudopeptidic inhibitors provide an exciting new tool to design tight-binding bioavailable inhibitors that are isoform-specific. A detailed review of SIRT1 activators and inhibitors can be found in Ref. 99.
Concluding Remarks
Sirtuins function to regulate diverse cellular processes, and their unique consumption of NAD+ directly links sirtuin catalysis to metabolism and energy homeostasis. The expansion of the acetylome and the characterization of newly discovered acyllysine modifications, including succinylation and malonylation, broaden the cellular acylation landscape that is targeted by the sirtuins. Human sirtuins are implicated in numerous age-related diseases and, as such, have become pharmaceutical targets for small-molecule modulation. However, the molecular role of sirtuins in disease progression is not always clear. A full understanding of sirtuin function will be possible only when we have determined the complete range of their biochemical and enzymatic activities, which includes analysis of acyl group and target protein specificity. Although there has been considerable focus on developing modulators of SIRT1 and SIRT2, the importance of SIRT3 in metabolic reprogramming of mitochondria was revealed in a recent quantitative proteomics study (100). This study provided evidence that SIRT3 plays a prominent role in adaption to caloric restriction through coordinate deacetylation of proteins involved in diverse pathways of metabolism and mitochondrial maintenance. These results suggest that small-molecule modulators that promote SIRT3-dependent functions could mimic some of the positive effects on health span induced by caloric restriction (35). A deeper understanding of sirtuin catalysis and regulation will be essential to rationally design the next generation of isoform-specific therapeutics for the treatment of metabolic and age-related diseases.
Supplementary Material
J. M. D. is a consultant for Sirtris Pharmaceuticals, Inc., a GSK company. This is the first article in the Thematic Minireview Series on Sirtuins: From Biochemistry to Health and Disease.

This article contains supplemental Fig. 1.
- OAADPr
- 2′-O-acetyl-ADP-ribose
- PGC-1α
- peroxisome proliferator-activated receptor-γ coactivator 1α
- NAMPT
- nicotinamide phosphoribosyltransferase
- AMPK
- AMP-activated kinase.
REFERENCES
- 1. Norvell A., McMahon S. B. (2010) Cell biology. Rise of the rival. Science 327, 964–965 [DOI] [PubMed] [Google Scholar]
- 2. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 3. Kim S. C., Sprung R., Chen Y., Xu Y., Ball H., Pei J., Cheng T., Kho Y., Xiao H., Xiao L., Grishin N. V., White M., Yang X. J., Zhao Y. (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 23, 607–618 [DOI] [PubMed] [Google Scholar]
- 4. Frye R. A. (2000) Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 5. Michishita E., Park J. Y., Burneskis J. M., Barrett J. C., Horikawa I. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell 16, 4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 8. Pan P. W., Feldman J. L., Devries M. K., Dong A., Edwards A. M., Denu J. M. (2011) Structure and biochemical functions of SIRT6. J. Biol. Chem. 286, 14575–14587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith B. C., Settles B., Hallows W. C., Craven M. W., Denu J. M. (2011) SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem. Biol. 6, 146–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jackson M. D., Denu J. M. (2002) Structural identification of 2′- and 3′-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 277, 18535–18544 [DOI] [PubMed] [Google Scholar]
- 11. Borra M. T., Langer M. R., Slama J. T., Denu J. M. (2004) Substrate specificity and kinetic mechanism of the Sir2 family of NAD+-dependent histone/protein deacetylases. Biochemistry 43, 9877–9887 [DOI] [PubMed] [Google Scholar]
- 12. Tong L., Denu J. M. (2010) Function and metabolism of sirtuin metabolite O-acetyl-ADP-ribose. Biochim. Biophys. Acta 1804, 1617–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen D., Vollmar M., Rossi M. N., Phillips C., Kraehenbuehl R., Slade D., Mehrotra P. V., von Delft F., Crosthwaite S. K., Gileadi O., Denu J. M., Ahel I. (2011) Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J. Biol. Chem. 286, 13261–13271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sauve A. A. (2010) Sirtuin chemical mechanisms. Biochim. Biophys. Acta 1804, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cen Y., Sauve A. A. (2010) Transition state of ADP-ribosylation of acetyllysine catalyzed by Archaeoglobus fulgidus Sir2 determined by kinetic isotope effects and computational approaches. J. Am. Chem. Soc. 132, 12286–12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003) Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J. Biol. Chem. 278, 50985–50998 [DOI] [PubMed] [Google Scholar]
- 17. Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., Schramm V. L. (2001) Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry 40, 15456–15463 [DOI] [PubMed] [Google Scholar]
- 18. Smith B. C., Denu J. M. (2006) Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry 45, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith B. C., Denu J. M. (2007) Mechanism-based inhibition of Sir2 deacetylases by thioacetyl-lysine peptide. Biochemistry 46, 14478–14486 [DOI] [PubMed] [Google Scholar]
- 20. Dancy B. C., Ming S. A., Papazyan R., Jelinek C. A., Majumdar A., Sun Y., Dancy B. M., Drury W. J., 3rd, Cotter R. J., Taverna S. D., Cole P. A. (2012) Azalysine analogues as probes for protein lysine deacetylation and demethylation. J. Am. Chem. Soc. 134, 5138–5148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou Y., Zhang H., He B., Du J., Lin H., Cerione R. A., Hao Q. (2012) The bicyclic intermediate structure provides insights into the desuccinylation mechanism of human sirtuin 5 (SIRT5). J. Biol. Chem. 287, 28307–28314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tanny J. C., Dowd G. J., Huang J., Hilz H., Moazed D. (1999) An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99, 735–745 [DOI] [PubMed] [Google Scholar]
- 23. Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. (2011) SIRT6 promotes DNA repair under stress by activating PARP1. Science 332, 1443–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tanner K. G., Landry J., Sternglanz R., Denu J. M. (2000) Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 97, 14178–14182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaquero A., Scher M., Lee D., Erdjument-Bromage H., Tempst P., Reinberg D. (2004) Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16, 93–105 [DOI] [PubMed] [Google Scholar]
- 27. Vaziri H., Dessain S. K., Ng Eaton E., Imai S. I., Frye R. A., Pandita T. K., Guarente L., Weinberg R. A. (2001) hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107, 149–159 [DOI] [PubMed] [Google Scholar]
- 28. Yeung F., Hoberg J. E., Ramsey C. S., Keller M. D., Jones D. R., Frye R. A., Mayo M. W. (2004) Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 23, 2369–2380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hallows W. C., Yu W., Denu J. M. (2012) Regulation of glycolytic enzyme phosphoglycerate mutase-1 by Sirt1 protein-mediated deacetylation. J. Biol. Chem. 287, 3850–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 31. Hallows W. C., Yu W., Smith B. C., Devries M. K., Ellinger J. J., Someya S., Shortreed M. R., Prolla T., Markley J. L., Smith L. M., Zhao S., Guan K. L., Denu J. M. (2011) Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell 41, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., Farese R. V., Jr., Alt F. W., Kahn C. R., Verdin E. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hallows W. C., Lee S., Denu J. M. (2006) Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Someya S., Yu W., Hallows W. C., Xu J., Vann J. M., Leeuwenburgh C., Tanokura M., Denu J. M., Prolla T. A. (2010) Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell 143, 802–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu W., Dittenhafer-Reed K. E., Denu J. M. (2012) SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J. Biol. Chem. 287, 14078–14086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., Wolberger C. (2002) Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol. Cell 10, 523–535 [DOI] [PubMed] [Google Scholar]
- 38. Blander G., Olejnik J., Krzymanska-Olejnik E., McDonagh T., Haigis M., Yaffe M. B., Guarente L. (2005) SIRT1 shows no substrate specificity in vitro. J. Biol. Chem. 280, 9780–9785 [DOI] [PubMed] [Google Scholar]
- 39. Garske A. L., Denu J. M. (2006) SIRT1 top 40 hits: use of one-bead, one-compound acetyl-peptide libraries and quantum dots to probe deacetylase specificity. Biochemistry 45, 94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gurard-Levin Z. A., Kilian K. A., Kim J., Bähr K., Mrksich M. (2010) Peptide arrays identify isoform-selective substrates for profiling endogenous lysine deacetylase activity. ACS Chem. Biol. 5, 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cosgrove M. S., Bever K., Avalos J. L., Muhammad S., Zhang X., Wolberger C. (2006) The structural basis of sirtuin substrate affinity. Biochemistry 45, 7511–7521 [DOI] [PubMed] [Google Scholar]
- 42. Chen Y., Sprung R., Tang Y., Ball H., Sangras B., Kim S. C., Falck J. R., Peng J., Gu W., Zhao Y. (2007) Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteomics 6, 812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garrity J., Gardner J. G., Hawse W., Wolberger C., Escalante-Semerena J. C. (2007) N-Lysine propionylation controls the activity of propionyl-CoA synthetase. J. Biol. Chem. 282, 30239–30245 [DOI] [PubMed] [Google Scholar]
- 44. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Peng C., Lu Z., Xie Z., Cheng Z., Chen Y., Tan M., Luo H., Zhang Y., He W., Yang K., Zwaans B. M., Tishkoff D., Ho L., Lombard D., He T. C., Dai J., Verdin E., Ye Y., Zhao Y. (2011) Mol. Cell. Proteomics 10, M111.012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J. D., Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cel.l Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu B., Lin Y., Darwanto A., Song X., Xu G., Zhang K. (2009) Identification and characterization of propionylation at histone H3 lysine 23 in mammalian cells. J. Biol. Chem. 284, 32288–32295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Berndsen C. E., Albaugh B. N., Tan S., Denu J. M. (2007) Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Smith B. C., Denu J. M. (2007) Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J. Biol. Chem. 282, 37256–37265 [DOI] [PubMed] [Google Scholar]
- 50. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakagawa T., Lomb D. J., Haigis M. C., Guarente L. (2009) SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137, 560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang Y., Nicholl M. B. (2011) Sirtuin 1 in malignant transformation: friend or foe? Cancer Lett. 306, 10–14 [DOI] [PubMed] [Google Scholar]
- 53. Alhazzazi T. Y., Kamarajan P., Verdin E., Kapila Y. L. (2011) SIRT3 and cancer: tumor promoter or suppressor? Biochim. Biophys. Acta 1816, 80–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Frye R. A. (1999) Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 55. Scher M. B., Vaquero A., Reinberg D. (2007) SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 21, 920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anderson R. M., Weindruch R. (2012) The caloric restriction paradigm: implications for healthy human aging. Am. J. Hum. Biol. 24, 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cohen H. Y., Miller C., Bitterman K. J., Wall N. R., Hekking B., Kessler B., Howitz K. T., Gorospe M., de Cabo R., Sinclair D. A. (2004) Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science 305, 390–392 [DOI] [PubMed] [Google Scholar]
- 58. Noriega L. G., Feige J. N., Canto C., Yamamoto H., Yu J., Herman M. A., Mataki C., Kahn B. B., Auwerx J. (2011) CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 12, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kong X., Wang R., Xue Y., Liu X., Zhang H., Chen Y., Fang F., Chang Y. (2010) Sirtuin 3, a new target of PGC-1α, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE 5, e11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Giralt A., Hondares E., Villena J. A., Ribas F., Díaz-Delfín J., Giralt M., Iglesias R., Villarroya F. (2011) Peroxisome proliferator-activated receptor-γ coactivator-1α controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J. Biol. Chem. 286, 16958–16966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) Phosphorylation regulates SIRT1 function. PLoS ONE 3, e4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nasrin N., Kaushik V. K., Fortier E., Wall D., Pearson K. J., de Cabo R., Bordone L. (2009) JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE 4, e8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gerhart-Hines Z., Dominy J. E., Jr., Blättler S. M., Jedrychowski M. P., Banks A. S., Lim J. H., Chim H., Gygi S. P., Puigserver P. (2011) The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+. Mol. Cell 44, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pandithage R., Lilischkis R., Harting K., Wolf A., Jedamzik B., Lüscher-Firzlaff J., Vervoorts J., Lasonder E., Kremmer E., Knöll B., Lüscher B. (2008) The regulation of SIRT2 function by cyclin-dependent kinases affects cell motility. J. Cell Biol. 180, 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu X., Wang D., Zhao Y., Tu B., Zheng Z., Wang L., Wang H., Gu W., Roeder R. G., Zhu W. G. (2011) Methyltransferase Set7/9 regulates p53 activity by interacting with sirtuin 1 (SIRT1). Proc. Natl. Acad. Sci. U.S.A. 108, 1925–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang Y., Fu W., Chen J., Olashaw N., Zhang X., Nicosia S. V., Bhalla K., Bai W. (2007) SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 9, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kornberg M. D., Sen N., Hara M. R., Juluri K. R., Nguyen J. V., Snowman A. M., Law L., Hester L. D., Snyder S. H. (2010) GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 12, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fritz K. S., Galligan J. J., Smathers R. L., Roede J. R., Shearn C. T., Reigan P., Petersen D. R. (2011) 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem. Res. Toxicol. 24, 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Chakrabarty S. P., Balaram H. (2010) Reversible binding of zinc in Plasmodium falciparum Sir2: structure and activity of the apoenzyme. Biochim. Biophys. Acta 1804, 1743–1750 [DOI] [PubMed] [Google Scholar]
- 70. Oliver S. S., Denu J. M. (2011) Dynamic interplay between histone H3 modifications and protein interpreters: emerging evidence for a “histone language”. ChemBioChem 12, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kim E. J., Kho J. H., Kang M. R., Um S. J. (2007) Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol. Cell 28, 277–290 [DOI] [PubMed] [Google Scholar]
- 72. Kim J. E., Chen J., Lou Z. (2008) DBC1 is a negative regulator of SIRT1. Nature 451, 583–586 [DOI] [PubMed] [Google Scholar]
- 73. Zhao W., Kruse J. P., Tang Y., Jung S. Y., Qin J., Gu W. (2008) Negative regulation of the deacetylase SIRT1 by DBC1. Nature 451, 587–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Law I. K., Liu L., Xu A., Lam K. S., Vanhoutte P. M., Che C. M., Leung P. T., Wang Y. (2009) Identification and characterization of proteins interacting with SIRT1 and SIRT3: implications in the anti-aging and metabolic effects of sirtuins. Proteomics 9, 2444–2456 [DOI] [PubMed] [Google Scholar]
- 75. Tsai Y. C., Greco T. M., Boonmee A., Miteva Y., Cristea I. M. (2012) Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol. Cell. Proteomics 11, 60–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cen Y., Falco J. N., Xu P., Youn D. Y., Sauve A. A. (2011) Mechanism-based affinity capture of sirtuins. Org. Biomol. Chem. 9, 987–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Imai S., Guarente L. (2010) Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 31, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Imai S. (2009) Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 15, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nikiforov A., Dölle C., Niere M., Ziegler M. (2011) Pathways and subcellular compartmentation of NAD biosynthesis in human cells: from entry of extracellular precursors to mitochondrial NAD generation. J. Biol. Chem. 286, 21767–21778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dan L., Klimenkova O., Klimiankou M., Klusman J. H., van den Heuvel-Eibrink M. M., Reinhardt D., Welte K., Skokowa J. (2012) The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica 97, 551–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cantó C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., Elliott P. J., Puigserver P., Auwerx J. (2009) AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458, 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Milne J. C., Lambert P. D., Schenk S., Carney D. P., Smith J. J., Gagne D. J., Jin L., Boss O., Perni R. B., Vu C. B., Bemis J. E., Xie R., Disch J. S., Ng P. Y., Nunes J. J., Lynch A. V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D. A., Olefsky J. M., Jirousek M. R., Elliott P. J., Westphal C. H. (2007) Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Howitz K. T., Bitterman K. J., Cohen H. Y., Lamming D. W., Lavu S., Wood J. G., Zipkin R. E., Chung P., Kisielewski A., Zhang L. L., Scherer B., Sinclair D. A. (2003) Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 [DOI] [PubMed] [Google Scholar]
- 84. Dittenhafer-Reed K. E., Feldman J. L., Denu J. M. (2011) Catalysis and mechanistic insights into sirtuin activation. ChemBioChem 12, 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., Prabhu V. V., Allard J. S., Lopez-Lluch G., Lewis K., Pistell P. J., Poosala S., Becker K. G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K. W., Spencer R. G., Lakatta E. G., Le Couteur D., Shaw R. J., Navas P., Puigserver P., Ingram D. K., de Cabo R., Sinclair D. A. (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Park S. J., Ahmad F., Philp A., Baar K., Williams T., Luo H., Ke H., Rehmann H., Taussig R., Brown A. L., Kim M. K., Beaven M. A., Burgin A. B., Manganiello V., Chung J. H. (2012) Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell 148, 421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Price N. L., Gomes A. P., Ling A. J., Duarte F. V., Martin-Montalvo A., North B. J., Agarwal B., Ye L., Ramadori G., Teodoro J. S., Hubbard B. P., Varela A. T., Davis J. G., Varamini B., Hafner A., Moaddel R., Rolo A. P., Coppari R., Palmeira C. M., de Cabo R., Baur J. A., Sinclair D. A. (2012) SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 15, 675–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Borra M. T., Smith B. C., Denu J. M. (2005) Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 280, 17187–17195 [DOI] [PubMed] [Google Scholar]
- 89. Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E. A., Caldwell S. D., Napper A., Curtis R., DiStefano P. S., Fields S., Bedalov A., Kennedy B. K. (2005) Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280, 17038–17045 [DOI] [PubMed] [Google Scholar]
- 90. Dai H., Kustigian L., Carney D., Case A., Considine T., Hubbard B. P., Perni R. B., Riera T. V., Szczepankiewicz B., Vlasuk G. P., Stein R. L. (2010) SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J. Biol. Chem. 285, 32695–32703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Napper A. D., Hixon J., McDonagh T., Keavey K., Pons J. F., Barker J., Yau W. T., Amouzegh P., Flegg A., Hamelin E., Thomas R. J., Kates M., Jones S., Navia M. A., Saunders J. O., DiStefano P. S., Curtis R. (2005) Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 48, 8045–8054 [DOI] [PubMed] [Google Scholar]
- 92. Schlicker C., Boanca G., Lakshminarasimhan M., Steegborn C. (2011) Structure-based development of novel sirtuin inhibitors. Aging 3, 852–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sivaraman P., Mattegunta S., Subbaraju G. V., Satyanarayana C., Padmanabhan B. (2010) Design of a novel nucleoside analog as potent inhibitor of the NAD-dependent deacetylase, SIRT2. Syst. Synth. Biol. 4, 257–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Huhtiniemi T., Salo H. S., Suuronen T., Poso A., Salminen A., Leppänen J., Jarho E., Lahtela-Kakkonen M. (2011) Structure-based design of pseudopeptidic inhibitors for SIRT1 and SIRT2. J. Med. Chem. 54, 6456–6468 [DOI] [PubMed] [Google Scholar]
- 95. Suzuki T., Asaba T., Imai E., Tsumoto H., Nakagawa H., Miyata N. (2009) Identification of a cell-active non-peptide sirtuin inhibitor containing N-thioacetyl lysine. Bioorg. Med. Chem. Lett. 19, 5670–5672 [DOI] [PubMed] [Google Scholar]
- 96. Chakrabarty S. P., Ramapanicker R., Mishra R., Chandrasekaran S., Balaram H. (2009) Development and characterization of lysine-based tripeptide analogues as inhibitors of Sir2 activity. Bioorg. Med. Chem. 17, 8060–8072 [DOI] [PubMed] [Google Scholar]
- 97. He B., Du J., Lin H. (2012) Thiosuccinyl peptides as Sirt5-specific inhibitors. J. Am. Chem. Soc. 134, 1922–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hawse W. F., Hoff K. G., Fatkins D. G., Daines A., Zubkova O. V., Schramm V. L., Zheng W., Wolberger C. (2008) Structural insights into intermediate steps in the Sir2 deacetylation reaction. Structure 16, 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Blum C. A., Ellis J. L., Loh C., Ng P. Y., Perni R. B., Stein R. L. (2011) SIRT1 modulation as a novel approach to the treatment of diseases of aging. J. Med. Chem. 54, 417–432 [DOI] [PubMed] [Google Scholar]
- 100. Hebert A. S., Dittenhafer-Reed K. E., Yu W., Bailey D. J., Selen E. S., Boersma M. D., Carson J. C., Tonelli M., Balloon A., Higbee A. J., Westphall M. S., Pagliarini D. J., Prolla T. A., Assadi-Porter F., Roy S., Denu J. M., Coon J. J. (2012) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.