Background: Peroxiredoxins are ubiquitous antioxidant enzymes reducing toxic peroxides.
Results: Two 2-Cys peroxiredoxins of Vibrio vulnificus are different in expression patterns, sensitivities to overoxidation, and kinetic properties.
Conclusion: The two peroxiredoxins are optimized for detoxifying different ranges of H2O2.
Significance: This study is the first report on the coexistence of the two peroxiredoxins along with their distinct roles in a single bacterium.
Keywords: Gene Expression, Oxidative Stress, Peroxiredoxin, Post-translational Modification, Reactive Oxygen Species (ROS), Vibrio vulnificus
Abstract
Peroxiredoxins (Prxs) are ubiquitous antioxidant enzymes reducing toxic peroxides. Two distinct 2-Cys Prxs, Prx1 and Prx2, were identified in Vibrio vulnificus, a facultative aerobic pathogen. Both Prxs have two conserved catalytic cysteines, CP and CR, but Prx2 is more homologous in amino acid sequences to eukaryotic Prx than to Prx1. Prx2 utilized thioredoxin A as a reductant, whereas Prx1 required AhpF. Prx2 contained GGIG and FL motifs similar to the motifs conserved in sensitive Prxs and exhibited sensitivity to overoxidation. MS analysis and CP-SO3H specific immunoblotting demonstrated overoxidation of CP to CP-SO2H (or CP-SO3H) in vitro and in vivo, respectively. In contrast, Prx1 was robust and CP was not overoxidized. Discrete expression of the Prxs implied that Prx2 is induced by trace amounts of H2O2 and thereby residential in cells grown aerobically. In contrast, Prx1 was occasionally expressed only in cells exposed to high levels of H2O2. A mutagenesis study indicated that lack of Prx2 accumulated sufficient H2O2 to induce Prx1. Kinetic properties indicated that Prx2 effectively scavenges low levels of peroxides because of its high affinity to H2O2, whereas Prx1 quickly degrades higher levels of peroxides because of its high turnover rate and more efficient reactivation. This study revealed that the two Prxs are differentially optimized for detoxifying distinct ranges of H2O2, and proposed that Prx2 is a residential scavenger of peroxides endogenously generated, whereas Prx1 is an occasional scavenger of peroxides exogenously encountered. Furthermore, genome sequence database search predicted widespread coexistence of the two Prxs among bacteria.
Introduction
Bacteria continually encounter toxic reactive oxygen species (ROS)3 such as superoxide anion (O2˙̄), hydrogen peroxide (H2O2), and hydroxyl radical (•OH) in their growth environments. Oxidative stress caused by increased levels of the ROS can lead to the damage of cellular components including protein, DNA, and membrane lipid. Although incomplete reduction of oxygen during respiration and aerobic metabolism is the main source of endogenous ROS for bacterial cells, exposure to metals and redox-active chemicals also cause increased exogenous ROS (1). In addition, pathogenic bacteria are inevitably exposed to ROS that are crucial for the optimal microcidal activity of neutrophils and other phagocytes of host (2). Therefore, pathogens have evolved sophisticated mechanisms to survive oxidative stress imposed by not only endogenous sources but also host defense systems, and the mechanisms are closely linked to their virulence (1).
Bacterial defense against oxidative stress relies on a variety of antioxidant defense enzymes such as superoxide dismutase, catalase, and peroxiredoxin (Prx) (2). Among these, peroxiredoxins are a family of cysteine-based peroxidases, which are also called alkyl hydroperoxidase subunit C (AhpC) in a number of bacteria (3). Typical 2-Cys Prxs that have two conserved catalytic cysteines, peroxidatic (CP) and resolving (CR) cysteines, are the largest group of Prxs. CP reacts with a peroxide and forms a cysteine sulfenic acid intermediate (CP-SOH), which is followed by the formation of an intermolecular disulfide bond with CR from another subunit. Disulfide-bonded Prxs are subsequently reduced and reactivated by thiol-containing reductants such as thioredoxin (Trx) and alkyl hydroperoxidase subunit F (AhpF) (3).
Occasionally, the intermediate CP-SOH can be overoxidized to sulfinic (CP-SO2H) or sulfonic (CP-SO3H) acids by reacting with additional peroxides, which prevent disulfide formation and hence provoke the inactivation of the enzyme (3). This overoxidation depends on the structural GGLG and YF motifs observed originally in eukaryotic 2-Cys Prxs such as human Prx1 (hPrx1) (4), which, thereby, have been termed sensitive Prxs. Bacterial 2-Cys Prxs, such as AhpC from Salmonella typhimurium (StAhpC), lack the GGLG and YF motifs, rarely undergo overoxidation, and are prototypes of robust Prxs (3). Recently, however, 2-Cys Prxs from Helicobacter pylori and cyanobacteria have been identified to be sensitive to overoxidation (5, 6). Furthermore, a computational search for the conserved GGLG (or similar) motif in the sequences of more than 3500 putative Prxs predicted a large number of sensitive 2-Cys Prxs widespread among bacteria (7).
Nevertheless, the coexistence of both robust and sensitive 2-Cys Prxs along with their distinct roles in a single bacterium has not been yet addressed. A 2-Cys Prx, which is a homologue of StAhpC from a foodborne pathogen Vibrio vulnificus, designated as Prx1 (formerly VvAhpC), was identified previously (8). Prx1, forming an NAD(P)H-dependent peroxide reductase system with AhpF, contributes to the growth and survival of the pathogen under exogenous oxidative stress. Here we present the characterization of another 2-Cys Prx of V. vulnificus, Prx2, in comparison with Prx1. Furthermore, we provide evidence that the two Prxs are different in sensitivities to overoxidation, expression patterns, and kinetic properties, and thereby play distinct roles in protecting the pathogen from oxidative stress of different levels and different sources.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Culture Conditions
The strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, the V. vulnificus strains were grown aerobically in Luria-Bertani (LB) medium supplemented with 2.0% (w/v) NaCl (LBS) at 30 °C. Anerobic conditions were obtained by using an anaerobic culture jar with AnaeroGen (Oxoid) (9).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strains or plasmids | Relevant characteristicsa | Ref. or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| MO6–24/O | Clinical isolate; virulent | Laboratory collection |
| OH0701 | MO6–24/O with prx1::nptI; Kmr | 8 |
| OH0505 | MO6–24/O with prx2::nptI; Kmr | 18 |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA 96 thi-1 relA1; plasmid replication | Laboratory collection |
| BL21(DE3) | F− ompT hsdS (rB− mB−) gal (DE3) | Laboratory collection |
| Plasmids | ||
| pGEM-T easy | PCR product cloning vector; Apr | Promega |
| pRSET A | Protein expression vector; Apr | Invitrogen |
| pHIS-parallel1 | Protein expression vector; Apr | 11 |
| pWK0704 | pRSET A with prx1; Apr | 8 |
| pWK0802 | pRSET A with ahpF; Apr | 8 |
| pOH046 | pRSET A with prx2; Apr | This study |
| pOH054 | pRSET A with trxA; Apr | This study |
| pOH053 | pRSET A with trxB; Apr | This study |
| pBANG1105 | pHIS-parallel1 with prx1; Apr | This study |
| pBANG1106 | pHIS-parallel1 with prx2; Apr | This study |
| pBANG1104 | pHIS-parallel1 with trxA; Apr | This study |
| pBANG1016 | pHIS-parallel1 with ahpFNTD; Apr | This study |
| pOH049 | pRSET A with the mutant prx2 encoding Prx2-C50S; Apr | This study |
| pOH050 | pRSET A with the mutant prx2 encoding Prx2-C171S; Apr | This study |
a Apr, ampicillin resistant; Kmr, kanamycin resistant.
Overexpression and Purification of Recombinant Proteins
Each open reading frame (ORF) of the genes encoding Prx1, AhpF, Prx2, thioredoxin A (TrxA), and thioredoxin reductase (TrR) was amplified by PCR using a pair of oligonucleotide primers as listed in Table 2. The amplified PCR products were cloned into a His6 tag expression vector, pRSET A (Invitrogen), to result in pWK0704 (for prx1), pWK0802 (for ahpF), pOH046 (for prx2), pOH054 (for trxA), and pOH053 (for trxB encoding TrR) as described in Table 1. His-tagged proteins were expressed in Escherichia coli BL21(DE3), purified by affinity chromatography according to the manufacturer's procedure (Qiagen).
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Oligonucleotide sequence (5′ → 3′)a,b | Use |
|---|---|---|
| For expression of proteins | ||
| HIS-PRX1 F | CTCGAGATGATTAACACTACTATCAAACCA | Amplification of prx1 |
| HIS-PRX1 R | AAGCTTTTAGATTTTGCCAACTAGGTC | |
| HIS-AHPF F | CTCGAGATGCTAGACCAAGCGATC | Amplification of ahpF |
| HIS-AHPF R | AAGCTTTTAGCCTTGCTTACGAATCAA | |
| HIS-PRX2 F | GGATCCATGGTACTAGTAGGTCGTC | Amplification of prx2 |
| HIS-PRX2 R | GAATTCTTATTTTTTAAGGTCAGCTG | |
| HIS-TRXA F | GAATTCTTATAGGTTCGCATCCAAAAAC | Amplification of trxA |
| HIS-TRXA R | GGATCCATGAGTGACAAGATTTTGC | |
| HIS-TRXB F | GGATCCATGAGCGATATGAAACACAG | Amplification of trxB |
| HIS-TRXB R | GAATTCTTATTTGTCGTTCAACGAGTCTAGG | |
| PRX1-pHIS F | CCATGGCTATGATTAACACTACTATCAAACCATT | Amplification of prx1 |
| PRX1-pHIS R | CTCGAGTTAGATTTTGCCAACTAGGTCTAGTG | |
| PRX2-pHIS F | AACCATGGTACTAGTAGGTCGTCAAGCC | Amplification of prx2 |
| PRX2-pHIS R | CTCGAGTTATTTTTTAAGGTCAGCTGCGTGC | |
| TRXA-pHIS F | CATATGAGTGACAAGATTTTGCAGCTGTC | Amplification of trxA |
| TRXA-pHIS R | CTCGAGTTATAGGTTCGCATCCAA | |
| HIS-AHPFNTD F | CCATGGCTATGCTAGACCAAGCGATC | Amplification of ahpFNTD |
| HIS-AHPFNTD R | AAGCTTTTAGCTTGCCGCCGCTTT | |
| For mutagenesis | ||
| PRX2-C50S F | CCACTAGACTTCACTTTCGTTAGCCCATCTGAAC | Construction of Prx2-C50S |
| PRX2-C50S R | GTTCAGATGGGCTAACGAAAGTGAAGTCTAGTGG | |
| PRX2-C171S F | CAGAAGCACGGCGAAGTAAGTCCTGCTCAATG | Construction of Prx2-C171S |
| PRX2-C171S R | CATTGAGCAGGACTTACTTCGCCGTGCTTCTG | |
| For Northern blot analysis | ||
| PRX1001 F | AACTTTGGTGTAATGCGCCC | PRX1P DNA probe |
| PRX1001 R | CTAGCGTTTGTTCGCCTTCTT | |
| PRX2002 F | ATGGTACTAGTAGGTCGTC | PRX2P DNA probe |
| PRX2002 R | TTATTTTTTAAGGTCAGCTG | |
a The oligonucleotides were designed using the V. vulnificus MO6–24/O genomic sequence (GenBank accession number CP002469 and CP002470, www.ncbi.nlm.nih.gov).
b Regions of oligonucleotides not complementary to the corresponding templates are underlined.
For purification of non-His-tagged proteins used for kinetic studies, each ORF of prx1, prx2, trxA, and the part of ORF encoding the N-terminal 206 residues of AhpF (ahpFNTD) that possess the AhpC reducing activity (10) were amplified by PCR using primers as listed in Table 2. The amplified PCR products were cloned into a His6 tag expression vector, pHIS-parallel1, containing a recombinant tobacco etch virus protease cleavage site (11), resulting in pBANG1105 (for prx1), pBANG1106 (for prx2), pBANG1104 (for trxA), and pBANG1016 (for ahpFNTD) (Table 1). Proteins were expressed, purified, and His6 tags were removed from the proteins using the recombinant tobacco etch virus protease as described elsewhere (12).
Site-specific Mutagenesis of Prx2
Cysteine 50 (Cys-50) or cysteine 171 (Cys-171) of Prx2 was replaced with serine by use of the QuikChange site-directed mutagenesis kit (Stratagene) (12). The complementary mutagenic primers listed in Table 2 were used in conjunction with the plasmid pOH046 (as a template DNA) to create pOH049 (for Prx2-C50S) and pOH050 (for Prx2-C171S) (Table 1). The mutations were confirmed by DNA sequencing, and the mutant proteins were expressed and purified as described above.
Ferrous Oxidation Xylenol Orange Assay
Peroxidase activities of Prx2 and mutant Prx2s were measured by the modified method of Jeong et al. (13). Briefly, the reaction was initiated by adding of 5 μl of 1 mm NADPH as an electron donor to 500 μl of reaction mixture containing 50 mm Hepes-NaOH (pH 7.0), 0.5 μm TrR, 1 μm TrxA, 5 μm Prx2 (or mutant Prx2s), and either 100 μm H2O2 or 100 μm tert-butyl hydroperoxide (tBHP)). At time intervals, 50-μl aliquots were removed, added to 950 μl of ferrous oxidation xylenol 1 (14) reagent, incubated at room temperature for 30 min, and then A560 was scored by Tecan Infinite M200 reader (Tecan) to quantitate the residual peroxides.
NAD(P)H Consumption Assay
Peroxidase activity of Prx2 was determined by monitoring the decrease in A340 within 13 min due to NADPH oxidation (15). The reaction was initiated by adding 200 μm NADPH to the reaction mixtures (200 μl in a final volume) including 4 μm Prx2, 0.8 μm TrxA, 0.4 μm TrR, and various concentrations of H2O2 (0.1, 1 and 5 mm) in the 50 mm potassium phosphate (pH 7.0) buffer with 1 mm EDTA and 150 mm ammonium sulfate. Similarly, 200 μm NADH, 4 μm Prx1, and 0.27 μm AhpF were employed to determine peroxidase activity of Prx1.
MALDI-TOF-MS Analysis
Each of 4 μm Prx1 and Prx2 was reacted with 1 mm H2O2 in 50 mm potassium phosphate (pH 7.0) at 30 °C for 5 min, and then with 100 mm DTT for another 10 min. Then, the Prxs bands, resolved on SDS-PAGE, were excised, reduced with 30 mm DTT, alkylated with 100 mm iodoacetamide, and digested with an endoproteinase AspN (Sigma) as described elsewhere (16). Peptides were extracted from the gel pieces with 0.1% trifluoroacetic acid in 50% acetonitrile, and MALDI-TOF-MS analyses were carried out on a Voyager-DETM STR Biospectrometry Work station (Applied Biosystems Inc.) operating in a negative-ion reflector mode. The masses of the cleavage peptides were determined using the PeptideMass software from the ExPASy proteomics server.
Western Blot Analysis
The purified His-tagged Prx1 and Prx2 were used to raise rabbit anti-Prx1 and anti-Prx2 polyclonal antibodies (AbFrontier) (17). Sulfonylated peptides DFTFVCp(SO3H)PTEL and DFTFVCp(SO3H)PSEL corresponding to the active site of Prx1 and Prx2, respectively, were synthesized and conjugated with keyhole limpet hemocyanin, and then used to raise rabbit anti-Prx1-CP-SO3H and anti-Prx2-CP-SO3H polyclonal antibodies (AbFrontier). Cultures of the wild type, prx1 (8), and prx2 (18) mutants grown to log phase (A600 of 0.5) and exposed to various levels of H2O2 for 5 min were harvested to isolate total proteins. Proteins (10 μg) were resolved on SDS-PAGE under reducing or nonreducing conditions and immunoblotted as described previously (17).
Northern Blot Analysis
Cultures of the wild type and prx2 mutant grown to log phase (A600 of 0.5) anaerobically or aerobically and exposed to various levels of H2O2 were harvested to isolate total RNAs. For Northern blot analyses, reactions were performed according to standard procedures (19) with 20 μg of RNA. The DNA probes PRX1P and PRX2P were prepared by labeling DNA fragments containing the prx1 and prx2 coding regions with [α-32P]dCTP, respectively, and used for hybridizations as previously described (20). Northern hybridization products were visualized using a phosphorimager (model BAS1500, Fuji Photo Film Co. Ltd).
Bi-substrate Kinetic Analysis of Prx1 and Prx2
To determine the kinetic parameters, Prxs, AhpFNTD, and TrxA were pre-reduced by 100 m excess of DTT and then DTT was removed as described by Parsonage et al. (15, 21). Various concentrations of H2O2 were added to the mixtures of pre-reduced Prx1 (100 nm) and AhpFNTD (5–30 μm) in the reaction buffer (50 mm Hepes-NaOH, pH 7.0), and then incubated at 25 °C. Aliquots were removed at 1-min intervals and the residual H2O2 were measured using PeroXOquantTM Quantitative Peroxide Assay kit (Thermo Scientific) to determine the initial velocities (v0) of Prx1 to reduce H2O2. Similarly, the pre-reduced Prx2 (200 nm) and TrxA (10–40 μm) were prepared and used to determine the initial velocities of Prx2 by the same procedures described above. The initial velocities were used to generate the primary and secondary Dalziel plot (22) and obtain Vmax, Km, and kcat of the Prxs. The initial velocities of H2O2 reduction and other kinetic parameters were determined using SigmaPlot Version 10.0.
Data Analyses
Mean values and mean ± S.E. of results calculated from at least three independent experiments were reported.
RESULTS
Identification and Sequence Analysis of Prx2
A database (GenBankTM CP002469 and CP002470) search for homology to the amino acid sequences deduced from Prx1, a previously identified 2-Cys Prx (8), singled out a protein, hereafter named Prx2. Sequence analysis revealed that Prx2 is identical to a thiol-specific antioxidant protein, formerly named TsaA by current authors (18). TsaA was so far poorly characterized except that it contributed to the virulence of V. vulnificus (18). As shown in Fig. 1, Prx2 is comprised of 202 amino acids with a theoretical molecular mass of 22,168 Da and a pI of 4.9. The predicted profile of the hydrophobicity (EXPASY.ch) revealed that Prx2 is a cytosolic soluble protein as observed from AhpC of other Gram-negative bacteria (23). Prx2 contains two catalytic cysteines, Cys-50 and Cys-171, in the tripeptide VCPs (Val-Cys-Pro) that are highly conserved in typical 2-Cys Prxs (24, 25). Prx1 revealed a high level of identity (78% in amino acid sequences) with StAhpC. However, the alignment of the amino acid sequences of Prx2 to those of StAhpC and Prx1 revealed only 35 and 39% identity, respectively. Prx2 is more homologous (48% identity in amino acid sequences) to eukaryotic hPrx1 than to Prx1 and StAhpC. Furthermore, Prx2 contains GGIG and FL motifs at positions corresponding to GGLG and YF motifs of eukaryotic sensitive Prxs (Fig. 1) (4).
FIGURE 1.
Sequence analysis of 2-Cys Prxs of V. vulnificus (Prx1 and Prx2), S. typhimurium (StAhpC), and human (hPrx1). The amino acid sequences retrieved from NCBI protein database (NCBI, ncbi.nlm.nih.gov) (accession numbers ADV88988 for Prx1, ADV87477 for Prx2, NP_459600 for StAhpC, and NP_859047 for hPrx1) were aligned using the CLUSTALW program. Identical sequences (black boxes), similar sequences (gray boxes), conserved sequences in the region of peroxidatic (CP) and resolving (CR) cysteines (asterisks), and missing sequences (dashes) are indicated. The motifs, GG(L/I)G and YF/FL, considered specific for sensitive Prxs are marked with triangles.
Prx2 Is a TrxA-dependent Peroxidase and Requires the Two Conserved Cysteines
As shown in Fig. 2A, Prx2 was able to reduce H2O2 and tBHP in vitro in the presence of TrxA/TrR as electron donors, indicating that Prx2 get electrons from NADPH through the TrxA/TrR system to complete a catalytic cycle as observed in other Prxs (26). In contrast, AhpF was not able to reduce oxidized Prx2 to reactivate its peroxidase activity as determined by NADH consumption assay (data not shown). These results indicated that Prx2 is a TrxA/TrR-dependent peroxidase. Thus, hereafter, the TrxA/TrR was used as a reducing system to further characterize the peroxidase activity of Prx2.
FIGURE 2.
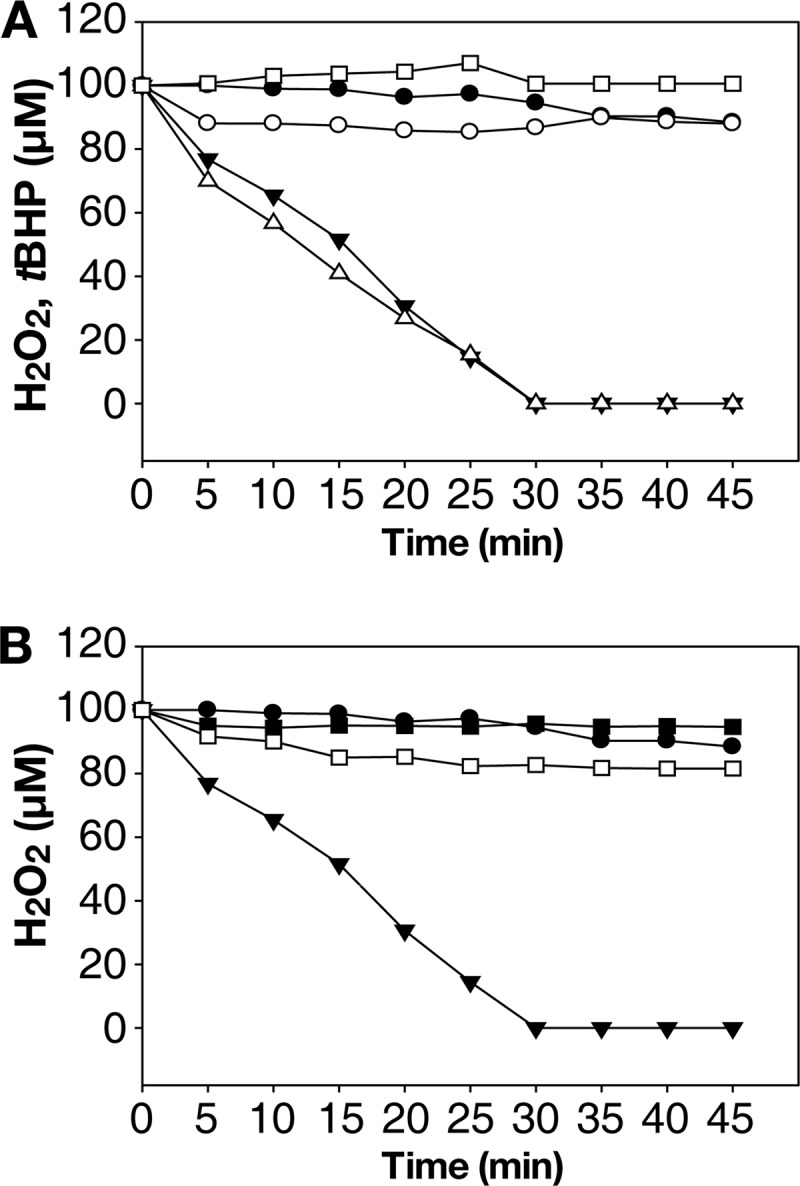
Peroxidase activities of the wild type and mutant Prx2s. To determine peroxidase activities, the residual H2O2 in the reaction mixtures were measured at the indicated times using ferrous oxidation xylenol 1 reagent. A, none (●), Prx2 (○), TrxA/TrR (□), or Prx2 and TrxA/TrR (▾) were added to the reaction mixture containing NADPH and H2O2. Residual tBHP in the reaction mixture containing Prx2, TrxA/TrR, NADPH, and tBHP in place of H2O2, were presented (▵). B, none (●), wild-type Prx2 and TrxA/TrR (▾), Prx2-C50S and TrxA/TrR (■), or Prx2-C171S and TrxA/TrR (□) were added to the reaction mixture containing NADPH and H2O2. The S.E. were too small to denote by error bars.
The mutants Prx2-C50S and Prx2-C171S, in which each of the two catalytic cysteine residues was replaced with a serine, respectively, were subjected to the ferrous oxidation xylenol assay to test their peroxidase activity. The results in Fig. 2B show that neither Prx2-C50S nor Prx2-C171S was able to decompose H2O2, indicating that both cysteines are crucial for the peroxidase activity of Prx2. These results indicated that the reaction mechanism of Prx2 as a peroxidase could be similar to that of StAhpC, in which Cys-50 directly attacks peroxides as a peroxidatic cysteine, whereas Cys-171, as a resolving cysteine, reduces the resulting sulfenic acid intermediate of the Cys-50.
Inactivation and Overoxidation of Prx2 by H2O2 in Vitro
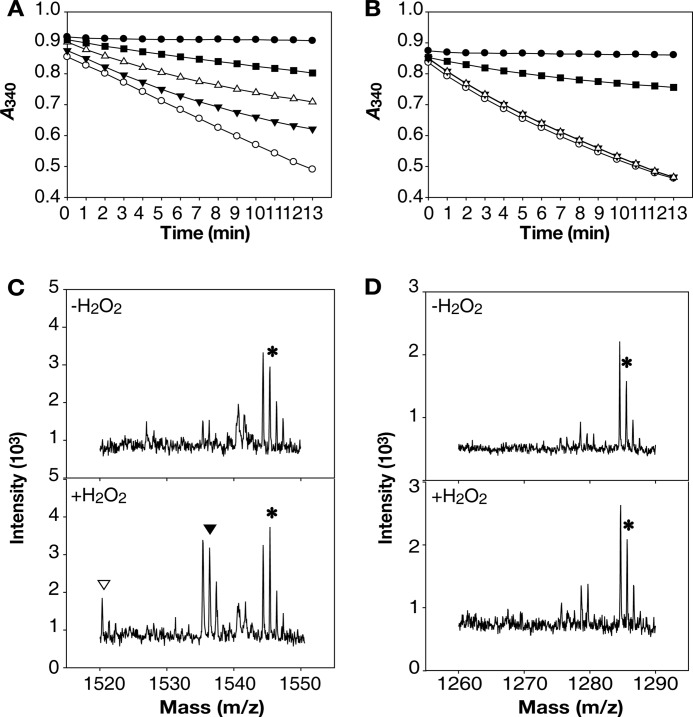
Prx2 and Prx1 activities reducing high levels of H2O2 were determined by measuring NAD(P)H consumption. When reacted with 0.1 mm H2O2, both Prx2 and Prx1 actively consumed NAD(P)H (Fig. 3, A and B). However, Prx2 consumed less NADPH when reacted with 1 and 5 mm H2O2 (Fig. 3A). The decreased consumption of NADPH reflected impaired peroxidase activity of Prx2, suggesting the inactivation of Prx2 by H2O2 exceeded 1 mm. In contrast, Prx1 still actively consumed NADH when reacted even with 1 and 5 mm H2O2 (Fig. 3B), suggesting that Prx1 was not inactivated by H2O2 at the concentrations tested. The results revealed that Prx2 is sensitive to inactivation by H2O2, whereas Prx1 is robust.
FIGURE 3.
Inactivation and overoxidation of Prxs by H2O2in vitro. A and B, the peroxidase activities of Prx2 (A) and Prx1 (B) were determined by measuring NAD(P)H consumption in the reaction mixtures. A, the reaction mixture contained none (●), TrxA/TrR (■), and TrxA/TrR and Prx2 with various concentrations of H2O2 (0.1 mm H2O2 (○), 1 mm H2O2 (▾), 5 mm H2O2 (▵)) in addition to NADPH. B, the reaction mixture contained none (●), AhpF (■), AhpF and Prx1 with various concentrations of H2O2 (0.1 mm H2O2 (○), 1 mm H2O2 (▾), 5 mm H2O2 (▵)) in addition to NADH. The S.E. were too small to denote by error bars. C and D, Prx2 and Prx1 were reacted (lower panels, +H2O2) or unreacted (upper panels, −H2O2) with 1 mm H2O2, alkylated, digested with AspN, and analyzed using MALDI-TOF-MS. C, mass spectrum of Prx2 peptides. Peptides with CP were indicated; *, m/z = 1545, CP-CH2CONH2; ▾, m/z = 1536, CP-SO3H; ▿, m/z = 1520, CP-SO2H. D, mass spectrum of Prx1 peptides. A peptide with CP was indicated; *, m/z = 1285, CP-CH2CONH2.
Both Prxs were reacted with 1 mm H2O2 and overoxidation of CP was examined. For Prx2, the theoretical monoisotopic mass of the peptide, DFTFVCPPSELIAF, in which CP is alkylated with acetamide (CP-S-CH2CONH2), is 1545. However, if the CP was overoxidized to CP-SO2H or CP-SO3H and thus not alkylated, the monoisotopic masses of the peptides would be 1520 or 1536, respectively. All three peptides were observed from the Prx2 reacted with 1 mm H2O2, whereas the overoxidized peptides with CP-SO2H or CP-SO3H were not observed from the Prx2 unreacted with H2O2 (Fig. 3C). The results indicated that 1 mm H2O2 overoxidized Prx2 at CP and resulted in the loss of the peroxidase activity.
In contrast, the peptide, DFTFVCPPTELG, containing CP alkylated with acetamide and corresponding to the monoisotopic mass of 1285, was exclusively observed from the Prx1 reacted with 1 mm H2O2. Neither overoxidized peptide with CP-SO2H nor with CP-SO3H was observed from Prx1 regardless of whether or not it reacted with H2O2 (Fig. 3D). The results indicated that 1 mm H2O2-reacted Prx1 did not undergo overoxidation at CP, which supports the result of Fig. 3B showing that Prx1 was not inactivated.
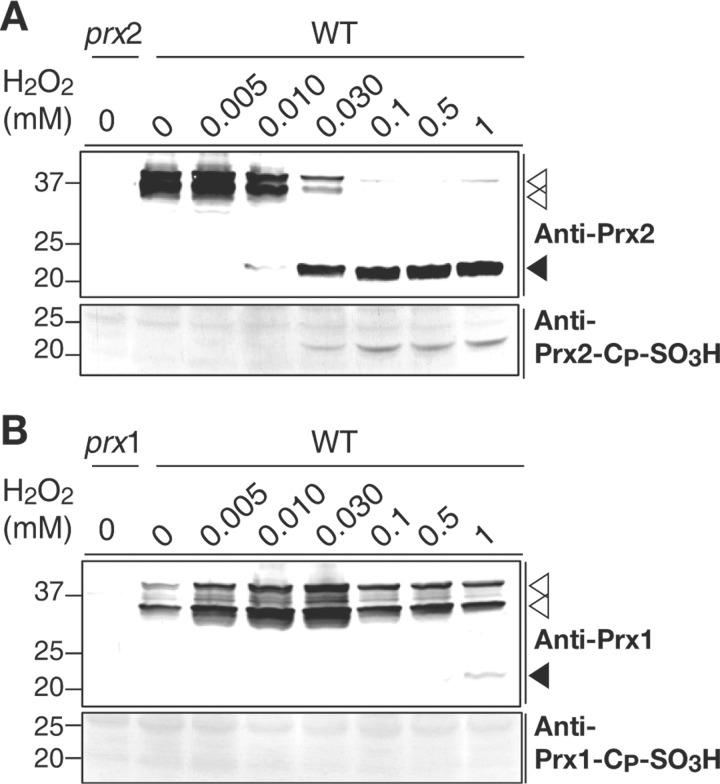
Overoxidation of Prx2 by H2O2 in Vivo
Cellular proteins on nonreducing SDS-PAGE were immunoblotted using rabbit anti-Prx2 and the molecular sizes of Prx2 were determined to examine whether or not Prx2 in cells was overoxidized (Fig. 4A, upper panel). Most Prx2 from the cells unexposed to H2O2 (control) was present as dimers, reflecting formation of the intermolecular disulfide bonds observed during the typical catalytic cycle of 2-Cys Prxs (27). Prx2 isolated from the cells exposed to H2O2 exceeding 30 μm appeared monomeric as an indication of overoxidation, accompanying gradual disappearance of the dimeric Prx2 (Fig. 4A, upper panel). In contrast, except for a minute amount of monomeric overoxidized Prx1 present in the cells exposed to 1 mm H2O2, most Prx1 were present as dimers in the cells exposed to H2O2 exceeding 30 μm (Fig. 4B, upper panel). Double bands for dimeric Prxs represent the simultaneous presence of dimers linked by one (upper band) or two (lower band) disulfide bonds as observed elsewhere (6, 28).
FIGURE 4.
Overoxidation of Prx2 and Prx1 by H2O2in vivo. Proteins extracted from the cells, after being exposed to various levels of H2O2 for 5 min as indicated, were resolved on nonreducing (upper panels) or reducing SDS-PAGE (lower panels). Immunoblotting was conducted using rabbit anti-Prx2 (A, upper panel), anti-Prx1 (B, upper panel), anti-Prx2-CP-SO3H (A, lower panel), and anti-Prx1-CP-SO3H (B, lower panel) antibodies as indicated. The protein size markers (Bio-Rad) are in kilodaltons. prx2, prx2 mutant; prx1, prx1 mutant; ▵, dimeric Prxs; ▾, overoxidized monomeric Prxs.
Overoxidized Prx2 in cells was re-examined by resolving cellular proteins on reducing SDS-PAGE and immunoblotted by the anti-Prx2-CP-SO3H antibody as described elsewhere (29). Overoxidized Prx2 was detected from the cells exposed to H2O2 exceeding 30 μm (Fig. 4A, lower panel). However, regardless of the level of exposure of the cell to H2O2, intracellular overoxidized Prx1 was not detected using the anti-Prx1-CP-SO3H antibody (Fig. 4B, lower panel). The results suggested that Prx2 is sensitive to overoxidation by H2O2 as observed in hPrx1, whereas Prx1 is robust as observed in bacterial Prxs.
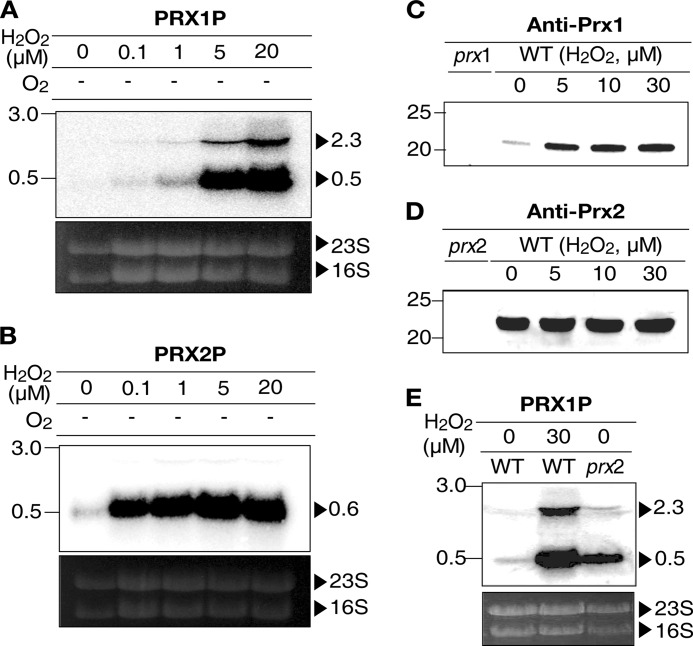
Discrete Expression Patterns of Prx1 and Prx2
Expression patterns of Prx1 and Prx2 in response to various levels of H2O2 were compared (Fig. 5). Northern blot analyses demonstrated that the transcript of prx1 was not apparent until the anaerobically growing cells were exposed to exogenous H2O2 exceeding 5 μm (Fig. 5A). In contrast, exposure to H2O2, as low as 0.1 μm, of the anaerobically growing cells was sufficient to induce the transcription of prx2 (Fig. 5B). The results indicated that prx1 and prx2 are expressed discretely in response to different levels of oxidative stress.
FIGURE 5.
Expression of prx1 and prx2 under oxidative stress. RNAs and proteins were extracted from the cultures grown anaerobically (A and B) and aerobically (C–E), after being exposed to various levels of H2O2 for 1 min (A and B) and 5 min (C-E) as indicated. For A, B, and E, RNAs were resolved and hybridized to a 32P-labeled DNA probe corresponding to the internal coding regions of prx1 (PRX1P, A and E) or prx2 (PRX2P, B). The RNA size markers (Invitrogen) and prx1 and prx2 transcripts are shown in kilobases. For C and D, total proteins were resolved on reducing SDS-PAGE, and immunoblotted using the rabbit anti-Prx1 (C) and Prx2 (D) antibody. prx2, prx2 mutant; prx1, prx1 mutant. Western blots are presented as described in the legend to Fig. 4.
Under aerobic conditions, expression of prx2, determined based on the cellular level of the Prx2 protein, was fully induced in the aerobically growing cells, even ones unexposed to exogenous H2O2 (Fig. 5D). The full expression of Prx2 and its independence on exogenous H2O2 indicated that Prx2 is induced by low levels of endogenous H2O2 and becomes a residential peroxidase in the cells of aerobic media. In contrast, Prx1 was rarely expressed in the H2O2-unexposed cells and occasionally expressed only when cells were exposed to exogenous H2O2 exceeding 5 μm (Fig. 5C).
This discrete expression pattern of the peroxidases implied that the two enzymes have distinct roles in scavenging oxidative stress of different levels and from different sources. As such, Prx2 scavenges low levels of endogenous H2O2 generated by aerobic metabolism, whereas Prx1 scavenges high levels of exogenous H2O2, which are encountered occasionally. Supporting this idea, a null mutation of prx2 increased the expression of prx1 in the H2O2-unexposed cells (Fig. 5E), reflecting that the lack of the residential Prx2 accumulated endogenous H2O2 to the level that permits the occasional expression of prx1.
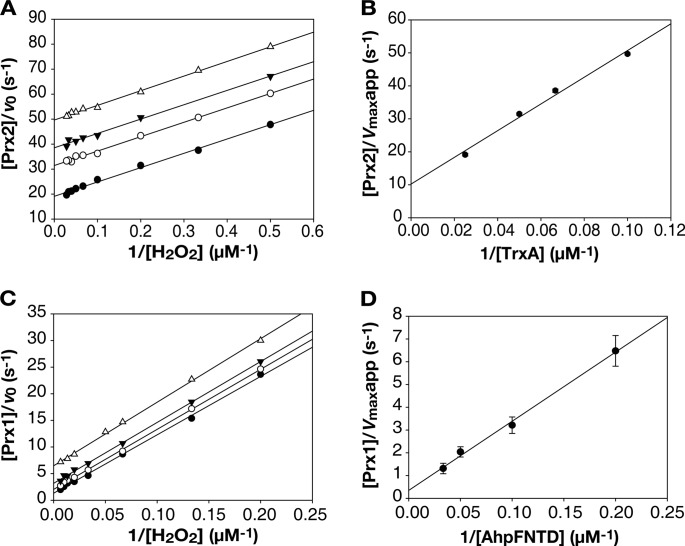
Prx2 and Prx1 Have Different Kinetic Properties to H2O2
The reciprocal initial velocities multiplied by the molarity of Prx2 ([Prx2]/v0) were plotted against the reciprocal molarities of H2O2 (1/[H2O2]) to generate the primary Dalziel plot (Fig. 6A). Parallel lines were obtained with several fixed concentrations of TrxA, indicating that Prx2 follows ping-pong kinetics. Thus, the simplified Dalziel equation, [E]/v0 = Φ0 + Φ1/[H2O2] +Φ2/[TrxA], where the ternary complex was not formed (30), was adopted to analyze other kinetic parameters of Prx2. The y intercepts from Fig. 6A ([Prx2]/Vmaxapp), where Vmaxapp were the apparent maximum velocities for infinite H2O2, were used to plot against the reciprocal molarities of TrxA (1/[TrxA]) and resulted in the secondary Dalziel plot (Fig. 6B). The real maximum velocity of Prx2 (Vmax) was obtained from the y intercept of the secondary plot, and Km for H2O2 and TrxA were obtained from the slope of the primary and secondary plots, respectively, as stated in Table 3. Using the same method, the initial velocities of Prx1 and [AhpFNTD] were also used as described above (Fig. 6, C and D), and other kinetic parameters of Prx1 were obtained (Table 3).
FIGURE 6.
Kinetic analysis of Prx2 and Prx1. Simplified systems including pre-reduced Prxs, TrxA, and AhpFNTD in the reaction buffer containing various concentrations of H2O2 were adopted to determine initial velocities of the Prxs as described under “Experimental Procedures.” In the primary Dalziel plots of Prx2 (A) and Prx1 (C), reciprocal initial velocities were multiplied by the molarity of the enzymes and plotted against the reciprocal molarities of H2O2. The y intercepts from A and C were replotted against the reciprocal molarities of TrxA and AhpFNTD, respectively, to generate the secondary Dalziel plots (B and D). A, Prx2 was mixed with 10 (▵), 15 (▾), 20 (○), and 40 μm (●) TrxA. C, Prx1 was mixed with 5 (▵), 10 (▾), 20 (○), and 30 μm (●) AhpFNTD. Error bars represent the S.E.
TABLE 3.
Kinetic parameters of Prx2 and Prx1 with hydrogen peroxide
| Enzyme | Reductant | kcat | KmHOOH | Kmreductant | kcat/KmHOOH | kcat/Kmreductant |
|---|---|---|---|---|---|---|
| s−1 | μma | m−1 s−1 | ||||
| Prx2 | TrxA | 0.098 | 1.1 ± 0.01 | 7.9 ± 0.6 | 8.8 × 104 | 1.2 × 104 |
| Prx1 | AhpFNTD | 2.9 | 32.6 ± 0.6 | 8.7 ± 0.4 | 8.8 × 104 | 3.3 × 105 |
a ± represent the S.E.
Prx2 exhibited 30-fold higher affinity to H2O2 compared with Prx1 and its Km for H2O2 was as low as 1.1 μm, whereas the turnover rate of Prx1 (kcat, Vmax/[Prx1]) was 30-fold higher than that of Prx2. Therefore, the efficiencies of Prx2 and Prx1 in reducing H2O2 appeared to not differ significantly (8.8 × 104 m−1 s−1 of kcat/Km). However, the reduction of H2O2 resulted in oxidized Prxs, which must react with their corresponding reductants to regain enzyme activity and complete a catalytic cycle. Kmreductant of Prx1 appeared similar to that of Prx2, and thus, efficiency of AhpFNTD to reduce Prx1 (3.3 × 105 m−1 s−1 of kcat/Km) was about 30-fold greater than that of TrxA to reduce Prx2 (1.2 × 104 m−1 s−1 of kcat/Km). Taken together, Prx2 seems more effective for the reduction of low levels of H2O2 because of its high affinity to H2O2. In contrast, Prx1 seems more effective at decomposing large amounts of H2O2 rapidly, as evidenced by its high turnover rate and more efficient reactivation by AhpFNTD.
DISCUSSION
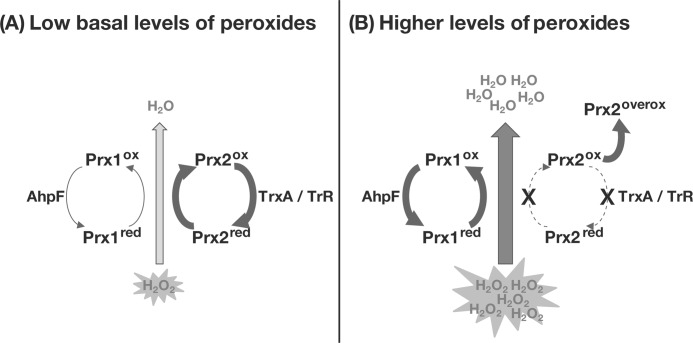
Differential roles of the two typical 2-Cys Prxs of V. vulnificus, Prx2 and Prx1, were proposed in this study and summarized in Fig. 7. The expression pattern of Prx2 that was retained during growth in aerobic media without exposure to exogenous H2O2 implied that Prx2 is a residential scavenger of low levels of H2O2 generated endogenously by aerobic metabolism, against which Prx1 is not effective (Fig. 5D). Prx1 was substantially expressed only when cells are exposed to high levels of H2O2, reflecting that Prx1 is an occasional scavenger of high levels of H2O2 provided exogenously (Fig. 5, A and C). The differential roles of Prx2 and Prx1 in scavenging H2O2 were further evidenced by their difference in sensitivity to oxidative inactivation. Sensitive inactivation by overoxidation made Prx2 difficult in reducing high levels of H2O2 (Figs. 3, A and C, and 4A). In contrast, Prx1 was resistant to overoxidation and more suitable for reducing high levels of H2O2 (Fig. 3, B and D, and 4B). Kinetic behaviors also proposed that Prx2 and Prx1 are more effective at reducing low and high levels of H2O2, respectively (Table 3). Therefore, having two Prxs in which the expression patterns, sensitivity to overoxidation, and kinetic properties are differentially optimized for the most efficient decomposition of distinct ranges of H2O2 could provide an evolutionary advantage for survival of bacteria encountering various ranges of oxidative stress.
FIGURE 7.
Proposed roles of the V. vulnificus Prx1 and Prx2 in detoxifying oxidative stress. A, in aerobically growing cells, Prx2 is induced and becomes a residential scavenger of low basal levels of peroxides, against which Prx1 is not effective because of its low level expression. B, when cells encounter high levels of exogenous peroxides, Prx1 is substantially induced and scavenges them. In contrast, Prx2 is overoxidized by high levels of peroxides, and thus unable to scavenge them. Prxred, reduced Prx; Prxox, oxidized Prx; Prx2overox, overoxidized Prx2.
Eukaryotic 2-Cys Prxs are much more sensitive to oxidative inactivation than are bacterial 2-Cys Prxs. Two amino acid motifs, the GGLG motif in the middle of the protein and YF motif in the C-terminal extension, which are uniquely present in all the sensitive 2-Cys Prxs, slow down the formation of intermolecular disulfide bonds between CP and CR, and increase the chance of overoxidation of CP to inactivate the enzymes (4). The overoxidized 2-Cys Prxs of yeast and human aggregate into the high molecular weight (HMW) complexes and exhibit chaperone activities could prevent protein misfolding under oxidative stress (31, 32). In addition to chaperone activity, eukaryotic 2-Cys Prxs regulate peroxide-mediated cellular signaling cascades (33, 34). As such, Gollnick and colleagues (34) revealed that the HMW complex of hPrx1 stimulate the secretion of proinflammatory cytokines from macrophages in a Toll-like receptor 4-dependent fashion and suggested that the hPrx1 acts as an endogenous danger signal (21).
In contrast to 2-Cys Prxs of higher organisms, bacterial 2-Cys Prxs display diversity in sensitivity to overoxidation. AhpCs of E. coli and S. typhymurium are known as robust Prx and cyanobacterial Prxs and H. pylori AhpC as sensitive (4–6). Cyanobacterial Prxs and H. pylori AhpC harbor GG(V/I)G and Y(F/L) motifs that are equivalent to the GGLG and YF motifs of hPrx1 (5, 6). Similarly, the amino acids GGIG and FL of Prx2, embedded in the regions of the GGLG and YF motifs (Fig. 1), could compose the structural features granting its sensitivity to overoxidation (Fig. 3, A and C, and 4A). The overoxidized AhpC of H. pylori can form HMW complexes characteristic of sensitive Prxs under severe oxidative stress, achieving a chaperone activity in vitro (25). In that context, it is tempting to propose that the overoxidized Prx2 would also gain new function(s), such as chaperone activity, enhancing survival of the bacteria under severe oxidative stress, and that the loss of the peroxidase activity would be not wasteful. A study is underway to address whether the overoxidized Prx2 can form HMW complexes and obtain new function(s).
To our knowledge, this is the first report on the coexistence of robust and sensitive Prxs along with distinct roles in detoxifying oxidative stress in a single bacterium. However, a search for typical 2-Cys Prxs in bacterial genomes revealed many other bacterial species that seem to be equipped with both robust and sensitive Prxs, including Enterobacter sp., Pseudomonas sp., Pseudoalteromonas sp., Salmonella sp., Shewanella sp. as well as Vibrio sp. (supplemental Table S1). Therefore, the coexistence of robust and sensitive Prxs seems one part of the bacterial nature, which is not limited to only V. vulnificus.
Supplementary Material
Acknowledgment
We thank Dr. P.-S. Chang, Seoul National University, for advice with determining kinetic parameters of the Prxs.
This work was supported by Mid-career Researcher Program Grants 2009-0092822 and 2012R1A2A1A03009679 through the National Research Foundation funded by Ministry of Education, Science and Technology and the Agriculture Research Center program of the Ministry for Food, Agriculture, Forestry and Fisheries, Republic of Korea (to S. H. C.).

This article contains supplemental Table S1.
- ROS
- reactive oxygen species
- Prx
- peroxiredoxin
- AhpC
- alkyl hydroperoxidase subunit C
- AhpF
- alkyl hydroperoxidase subunit F
- TrR
- thioredoxin reductase
- Trx
- thioredoxin
- CP
- peroxidatic cysteine
- CR
- resolving cysteine
- tBHP
- tert-butyl hydroperoxide
- Trx
- thioredoxin
- HMW
- high molecular weight.
REFERENCES
- 1. Storz G., Zheng M. (2000) in Bacterial Stress Responses (Storz Gisela, Hengge-Aronis R., eds) pp. 47–59, ASM Press, Washington, D. C [Google Scholar]
- 2. Miller R. A., Britigan B. E. (1997) Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hall A., Karplus P. A., Poole L. B. (2009) Typical 2-Cys peroxiredoxins-structures, mechanisms and functions. FEBS J. 276, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wood Z. A., Poole L. B., Karplus P. A. (2003) Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 300, 650–653 [DOI] [PubMed] [Google Scholar]
- 5. Huang C. H., Chuang M. H., Wu Y. H., Chuang W. C., Jhuang P. J., Chiou S. H. (2010) Characterization of site-specific mutants of alkylhydroperoxide reductase with dual functionality from Helicobacter pylori. J. Biochem. 147, 661–669 [DOI] [PubMed] [Google Scholar]
- 6. Pascual M. B., Mata-Cabana A., Florencio F. J., Lindahl M., Cejudo F. J. (2010) Overoxidation of 2-Cys peroxiredoxin in prokaryotes. Cyanobacterial 2-Cys peroxiredoxins sensitive to oxidative stress. J. Biol. Chem. 285, 34485–34492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson K. J., Knutson S. T., Soito L., Klomsiri C., Poole L. B., Fetrow J. S. (2011) Analysis of the peroxiredoxin family. Using active-site structure and sequence information for global classification and residue analysis. Proteins 79, 947–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baek W. K., Lee H. S., Oh M. H., Koh M. J., Kim K. S., Choi S. H. (2009) Identification of the Vibrio vulnificus ahpC1 gene and its influence on survival under oxidative stress and virulence. J. Microbiol. 47, 624–632 [DOI] [PubMed] [Google Scholar]
- 9. Miller P. H., Wiggs L. S., Miller J. M. (1995) Evaluation of AnaeroGen system for growth of anaerobic bacteria. J. Clin. Microbiol. 33, 2388–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Poole L. B., Godzik A., Nayeem A., Schmitt J. D. (2000) AhpF can be dissected into two functional units. Tandem repeats of two thioredoxin-like folds in the N terminus mediate electron transfer from the thioredoxin reductase-like C terminus to AhpC. Biochemistry 39, 6602–6615 [DOI] [PubMed] [Google Scholar]
- 11. Sheffield P., Garrard S., Derewenda Z. (1999) Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15, 34–39 [DOI] [PubMed] [Google Scholar]
- 12. Kim Y., Kim B. S., Park Y. J., Choi W. C., Hwang J., Kang B. S., Oh T. K., Choi S. H., Kim M. H. (2010) Crystal structure of SmcR, a quorum-sensing master regulator of Vibrio vulnificus, provides insight into its regulation of transcription. J. Biol. Chem. 285, 14020–14030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeong W., Cha M. K., Kim I. H. (2000) Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/alkyl hydroperoxide peroxidase C (AhpC) family. J. Biol. Chem. 275, 2924–2930 [DOI] [PubMed] [Google Scholar]
- 14. Wolff S. P. (1994) Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233, 182–189 [Google Scholar]
- 15. Parsonage D., Karplus P. A., Poole L. B. (2008) Substrate specificity and redox potential of AhpC, a bacterial peroxiredoxin. Proc. Natl. Acad. Sci. U.S.A. 105, 8209–8214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wagner E., Luche S., Penna L., Chevallet M., Van Dorsselaer A., Leize-Wagner E., Rabilloud T. (2002) A method for detection of overoxidation of cysteines. Peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochem. J. 366, 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lim M. S., Kim J. A., Lim J. G., Kim B. S., Jeong K. C., Lee K. H., Choi S. H. (2011) Identification and characterization of a novel serine protease, VvpS, that contains two functional domains and is essential for autolysis of Vibrio vulnificus. J. Bacteriol. 193, 3722–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oh M. H., Jeong H. G., Choi S. H. (2008) Proteomic identification and characterization of Vibrio vulnificus proteins induced upon exposure to INT-407 intestinal epithelial cells. J. Microbiol. Biotechnol. 18, 968–974 [PubMed] [Google Scholar]
- 19. Sambrook J., Russell D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Oh M. H., Lee S. M., Lee D. H., Choi S. H. (2009) Regulation of the Vibrio vulnificus hupA gene by temperature alteration and cyclic AMP receptor protein and evaluation of its role in virulence. Infect. Immun. 77, 1208–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsonage D., Desrosiers D. C., Hazlett K. R., Sun Y., Nelson K. J., Cox D. L., Radolf J. D., Poole L. B. (2010) Broad specificity AhpC-like peroxiredoxin and its thioredoxin reductant in the sparse antioxidant defense system of Treponema pallidum. Proc. Natl. Acad. Sci. U.S.A. 107, 6240–6245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dalziel K. (1957) Initial steady state velocities in the evaluation of enzyme-coenzyme-substrate reaction mechanisms. Acta Chem. Scand. 11, 1706–1723 [Google Scholar]
- 23. Link A. J., Robison K., Church G. M. (1997) Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis 18, 1259–1313 [DOI] [PubMed] [Google Scholar]
- 24. Poole L. B. (2007) The catalytic mechanism of peroxiredoxins. Subcell. Biochem. 44, 61–81 [DOI] [PubMed] [Google Scholar]
- 25. Chuang M. H., Wu M. S., Lo W. L., Lin J. T., Wong C. H., Chiou S. H. (2006) The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc. Natl. Acad. Sci. U.S.A. 103, 2552–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Poole L. B. (2005) Bacterial defenses against oxidants. Mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433, 240–254 [DOI] [PubMed] [Google Scholar]
- 27. Cox A. G., Winterbourn C. C., Hampton M. B. (2010) Measuring the redox state of cellular peroxiredoxins by immunoblotting. Methods Enzymol. 474, 51–66 [DOI] [PubMed] [Google Scholar]
- 28. Dietz K. J., Horling F., König J., Baier M. (2002) The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 53, 1321–1329 [PubMed] [Google Scholar]
- 29. Woo H. A., Kang S. W., Kim H. K., Yang K. S., Chae H. Z., Rhee S. G. (2003) Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J. Biol. Chem. 278, 47361–47364 [DOI] [PubMed] [Google Scholar]
- 30. Trujillo M., Ferrer-Sueta G., Thomson L., Flohé L., Radi R. (2007) Kinetics of peroxiredoxins and their role in the decomposition of peroxynitrite. Subcell. Biochem. 44, 83–113 [DOI] [PubMed] [Google Scholar]
- 31. Jang H. H., Lee K. O., Chi Y. H., Jung B. G., Park S. K., Park J. H., Lee J. R., Lee S. S., Moon J. C., Yun J. W., Choi Y. O., Kim W. Y., Kang J. S., Cheong G. W., Yun D. J., Rhee S. G., Cho M. J., Lee S. Y. (2004) Two enzymes in one two-yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117, 625–635 [DOI] [PubMed] [Google Scholar]
- 32. Moon J. C., Hah Y. S., Kim W. Y., Jung B. G., Jang H. H., Lee J. R., Kim S. Y., Lee Y. M., Jeon M. G., Kim C. W., Cho M. J., Lee S. Y. (2005) Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J. Biol. Chem. 280, 28775–28784 [DOI] [PubMed] [Google Scholar]
- 33. Rhee S. G., Woo H. A. (2011) Multiple functions of peroxiredoxins. Peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 15, 781–794 [DOI] [PubMed] [Google Scholar]
- 34. Riddell J. R., Wang X. Y., Minderman H., Gollnick S. O. (2010) Peroxiredoxin 1 stimulates secretion of proinflammatory cytokines by binding to TLR4. J. Immunol. 184, 1022–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.