Background: Cocaine- and amphetamine-regulated transcript (CART) plays a pivotal role in neuroprotection against stroke.
Results: Transcriptional repressor neuron-restrictive silencer factor (NRSF) represses CART through binding to two NRSEs in the CART promoter and intron.
Conclusion: NRSF represses CART transcription and antagonizes cAMP-response element-binding protein signaling through a dual NRSE mechanism.
Significance: This is the first evidence to reveal the repression mechanism of CART transcription. NRSF serves as a therapeutic target for stroke treatment.
Keywords: Corepressor Transcription, Ischemia, Neuroprotection, Stroke, Transcription Repressor, Cocaine- and Amphetamine-regulated Transcript (CART), Neuron-restrictive Silencer Factor (NRSF)
Abstract
Cocaine- and amphetamine-regulated transcript (CART) peptide plays a pivotal role in neuroprotection against stroke-related brain injury. However, the regulatory mechanism on CART transcription, especially the repression mechanism, is not fully understood. Here, we show that the transcriptional repressor neuron-restrictive silencer elements (NRSF, also known as REST) represses CART expression through direct binding to two NRSF-binding elements (NRSEs) in the CART promoter and intron 1 (named pNRSE and iNRSE, respectively). EMSA show that NRSF binds to pNRSE and iNRSE directly in vitro. ChIP assays show that NRSF recruits differential co-repressor complexes including CoREST and HDAC1 to these NRSEs. The presence of both NRSEs is required for efficient repression of CART transcription as indicated by reporter gene assays. NRSF overexpression antagonizes forskolin-mediated up-regulation of CART mRNA and protein. Ischemia insult triggered by oxygen-glucose deprivation (OGD) enhances NRSF mRNA levels and then NRSF antagonizes the CREB signaling on CART activation, leading to augmented cell death. Depletion of NRSF in combination with forskolin treatment increases neuronal survival after ischemic insult. These findings reveal a novel dual NRSE mechanism by which NRSF represses CART expression and suggest that NRSF may serve as a therapeutic target for stroke treatment.
Introduction
Cocaine- and amphetamine-regulated transcript (CART)3 is a regulated neuropeptide involved in numerous physiological processes, including reward/reinforcement circuit, feeding behavior, stress response, endocrine regulation, and sensory processing in the brain (1–6). CART is widely expressed in central nervous system and endocrine tissue, and functions as a neuropeptide to protect the brain against ischemic injury both in vivo and in vitro (7–10). Recently, it has been reported that CART peptide is increased in the blood circulation of patients with neuroendocrine malignancy and primary and metastatic breast cancer. CART has been suggested as a prognostic factor of the neuroendocrine tumor and estrogen receptor (ER)-positive, lymph node-negative breast tumors (11, 12). Therefore, CART expression must be tightly regulated. CART mRNA expression is up-regulated by psychostimulant drugs, for example, cocaine and amphetamine (13), as well as leptin (14, 15), cholecystokinin (16), estradiol (17), and glucocorticoids (3, 18). The up-regulation of CART mRNA has been well documented to be primarily achieved by PKA-CREB (cAMP response element-binding protein) signaling (9, 19–21). In addition to the CRE site, the CART promoter also contains several AP-1, STAT, and SP1 sites, and an E-box (19–23). Recently, a positive cis-acting element for the zinc-binding protein factor in pig CART promoter was reported (24). By contrast, how CART transcription is negatively regulated remains largely unclear.
We previously identified the transcriptional repressor NRSF (also known as REST) as a critical negative regulator of the CART gene (25). NRSF mediates the repression of target genes usually by direct binding to a 21–23-bp NRSE element (26). Indeed, we showed that NRSF binds to a typical NRSE site in the promoter of CART gene (pNRSE) (25). Depletion of NRSF in HeLa cells mediated by RNA interference (RNAi) results in a significant increase of CART expression and its promoter activity (25). Notably, the repression mechanism by NRSF often involves the recruitment of co-repressor complexes, such as histone deacetylases (HDACs), mSin3, CoREST (co-repressor of REST) (27–31); and the specificity of co-repressor recruitment depends on cellular context. However, whether pNRSE is the sole inhibitory site on CART and NRSF, which recruits co-repressor complexes to the CART gene, remains unknown. Moreover, how the inhibitory NRSF-NRSE mechanism concerts with the stimulatory CREB-CRE mechanism under pathological contexts also remains to be elucidated.
Here we identify an independent NRSE element in the first intron of the CART gene (iNRSE), which is functionally equivalent to the pNRSE. NRSF recruits similar but not identical complexes to pNRSE and iNRSE. The dual NRSF-NRSE mechanism exhibits an antagonizing effect on CREB-CRE signaling under ischemia insult. These findings provide novel insights on the negative regulation of CART transcription.
EXPERIMENTAL PROCEDURES
Cell Culture
Human cervical carcinoma HeLa cells and human neuroblastoma SK-N-SH cells were both cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Hyclone). Cell lines were maintained in a humidified incubator at 37 °C under an atmosphere containing 5% of CO2.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was performed as described previously (32). Briefly, nuclear extracts were prepared according to the manufacturer's instructions (Panomics, Fremont, CA), and the protein concentration was determined using bicinchoninic acid protein assay system (BCA; Pierce). Oligonucleotides containing the NRSE sequence of the CART promoter (sense, 5′-GAGTTTCAGAACGATGGAGAGCTCCCGC-3′; antisense, 5′-GCGGGAGCTCTCCATCGTTCTGAAACTC-3′) and intron I (sense, 5′-GAGTGTCAGTACCTGGGACAGCGTCCGC-3′; antisense, 5′-GCGGACGCTGTCCCAGGTACTGACACTC-3′) were synthesized and labeled with biotin at the 5′-end by AuGCT Biotechnology Synthesis Co. (Beijing, China). To perform DNA-protein binding reactions, the oligonucleotides were annealed to form double-stranded probes and incubated with the HeLa nuclear extract for 30 min in binding buffer (10 mm Tris-HCl, pH 7.5, 150 mm KCl, 5.7 mm MgCl2, 1 mm EDTA, 0.2 μg of poly(dI-dC), 5 μg of BSA, 0.67 mm DTT, 0.67 mm PMSF, 5% glycerol) in the presence or absence of unlabeled CART iNRSE probe as a competitor. For supershift assay, the NRSF antibodies (Santa Cruz Biotechnology, Santa Cruz, CA; three anti-NRSF goat and rabbit polyclonal antibodies were mixed together as a pool) were incubated with the nuclear extract and CART iNRSE probe. To ensure the protein binds to the anti-NRSF antibody, the nuclear extract and antibodies were preincubated for 20 min before the labeled probe was added. The protein/DNA binding samples were loaded onto a native polyacrylamide gel (6 and 10%, up and down) in 0.5× TBE buffer, then transferred to a nylon membrane. The membrane was blocked and incubated with stabilized streptavidin-horseradish peroxidase conjugate at room temperature for 15 min. The membrane was washed three times with washing buffer, treated with SuperSignal (Pierce Biotechnology) detection reagents, and exposed to Kodak Light films.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed according to the manufacturer's protocol (Cell Signaling Technology) and described previously (32). Briefly, 4 × 107 cells were formaldehyde cross-linked and chromatin was prepared and digested to 150–900-bp nucleosomes. One-hundred microliters of cell supernatant was diluted to a final volume of 0.5 ml in ChIP dilution buffer, and 2% of the diluted supernatant was used for input. The residual supernatant was used for overnight immunoprecipitation with 4 μg of anti-NRSF, anti-HDAC1, anti-HDAC2, anti-mSin3A, anti-mSin3B, and anti-CoREST antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at 4 °C. Normal rabbit IgG was used as negative control. Following the reverse cross-linking step, DNA was precipitated and purified by the Wizard SV Gel and PCR clean up system (Promega). Immunoprecipitated chromatin was analyzed by qPCR using SYBR Green dye with primers spanning the pNRSE and iNRSE motifs, which produce 152- and 155-bp PCR products, respectively. The primers used are as follows: CART promoter, sense, 5′-CTCCTCCCTCTTTCCTGCAC-3′, antisense, 5′-CAGAGGTAGCATCAGCAGCA-3′; and CART intron I, sense, 5′-CCCACTGCCATCCGAAGAGC-3′, antisense, 5′-CGAGGGGTGGAAACTTAGCG-3′.
Small Interfering RNA Vector Production
Small interfering RNA (siRNA) vectors containing the NRSF sequence or a scrambled sequence were prepared as previously described (33). Following our previous method, we constructed another siRNA vector that targets the human NRSF gene with the targeting sequence GTGTAATCTACAGTATCAC (34). Primers (N19)TTCAAGAGA(19N)TTTTTTC and TCGAGAAAAAA(N19)TCTCTTGAA(19N)A were designed to allow cloning into the vector pSicoR, which also expresses enhanced green fluorescent protein under control of the cytomegalovirus (CMV) promoter. Primers were annealed and cloned under control of the U6 promoter in pSicoR vector. Constructs were verified by DNA sequencing (35). The siRNA mixtures against CoREST, HDAC1, HDAC2, mSin3A, or mSin3B were purchased from Dharmacon. The siRNAs were transfected into the HeLa cells according to the manufacturer's instructions. The knockdown efficiency was determined by analysis of the mRNA level of the corresponding target molecule.
Lentivirus Generation and Infection
Lentiviruses were generated essentially as described (36). Briefly, 5 μg of pSicoR containing NRSF or scrambled control interference sequence, 4.2 μg of pLP1, 2 μg of pLP2, and 2.8 μg of pLP/VSVG were cotransfected into 293FT cells using LipofectamineTM 2000 reagent (Invitrogen). Supernatants were collected 48–72 h after transfection, filtered through a 0.45-μm filter, and used directly to infect HeLa cells in the presence of Polybrene with a final concentration of 8 μg/ml. Lentiviruses containing NRSF siRNA sequence and scrambled sequence were termed siRNA-NRSF and siRNA-Control, respectively. HeLa cells infected with siRNA-NRSF were named siRNA-NRSF #1 and siRNA-NRSF #2, and HeLa cells infected with siRNA-Control were named NC. siRNA-NRSF #1, siRNA-NRSF #2, and NC were cultured in normal culture conditions (DMEM supplemented with 10% FBS), the culture medium was changed every 3 days and passaged with 0.25% trypsin.
FACS Sorting of HeLa Cells Infected with siRNA-NRSF or siRNA-Control Lentiviruses
Cells infected with siRNA-NRSF or siRNA-Control lentiviruses were resuspended in 1× PBS, GFP-enriched cells were sorted on a FACS Vantage-SE flow cytometer (BD Biosciences) equipped with an Innova 70C-4 (488 nm) argon ion laser (Coherent, Palo Alto, CA). GFP was excited at 488 nm and fluorescence emission was detected using a 530/30 BP filter. Data from the experiments were analyzed with CellQuest software (BD Biosciences). GFP expressing cells underwent 4 rounds of GFP enrichment by FACS.
RNA Isolation and Quantitative Real-time Polymerase Chain Reaction
Total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's protocol, and 1 μg of total RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (TaKaRa) in a 10-μl reaction mixture. qPCR analysis was performed on a Bio-Rad iQ5 system using the SYBR Green PCR Master Mix (TaKaRa). The sequences of primers used for qPCR amplification were: NRSF, forward (F), 5′-ATTGAAGTTGGCTTAGTG-3′, reverse (R), 5′-TATGGGTAGATTCGTTGA-3′; CART, F, 5′-CAGCAACGACGAGTTTCAGA-3′, R, 5′-TGGGGATGTATGGAGGAGAA-3′; CoREST, F, 5′-AACTGGCAAGACGCAGTCAA-3′, R, 5′-GTTGGGCAAATCAGCCAATGA-3′; HDAC1, F, 5′-CGCCCTCACAAAGCCAATG-3′, R, 5′-CTGCTTGCTGTACTCCGACA-3′; HDAC2, F, 5′-ATGGCGTACAGTCAAGGAGG-3′, R, 5′-TGCGGATTCTATGAGGCTTCA-3′; mSin3A, F, 5′-GGTGGAGGATGCGCTATCTTA-3′, R, 5′-GGGTGTCGATGCTCTGAGATTT-3′; mSin3B, F, 5′-AGAACGAGCACGACAAGACC-3′. R, 5′-AGTCCCGTACTTCCCCACTG-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were used as control. All of the reactions were repeated at least three times.
Reporter Plasmids Construction
The plasmid pcDNA I-NRSF was generously provided by Dr. Gerward Waeber and described previously (37). The NRSF fragment was digested from the pcDNA I-NRSF construct and cloned into the pcDNA3.1 plasmid. The cloned reporter gene vector containing the proximal promoter region of the CART gene (−700 to +39 bp; P-Luc) was a kind gift from Dr. Peter Morgan (19). The reporter gene construct containing the truncated CART promoter (−700 to +12 bp) without the NRSE sequence (tP-Luc) was described previously (25). The CART intron I region fragment was generated by PCR amplification using genomic DNA from human mesenchymal stem cells as the template, and primers were as follows: sense, 5′-GGATCCTGCTGATGCTACCTCTG-3′; antisense, 5′-GTCGACGCCATACTTCTTCTCATAG-3′. The truncated NRSE-deleting CART intron I fragment (tI) was acquired by PCR using three pairs primers as follows: I sense, 5′-GGATCCTGCTGATGCTACCTCTG-3′; antisense, 5′-AGGGGTGGAAACTTAGCCGCGGGTCTCTGAGCC-3′; II sense, 5′-GGCTCAGAGACCCGCGGCTAAGTTTCCACCCCT-3′; antisense, 5′-GTCGACGCCATACTTCTTCTCATAG-3′; III sense, 5′-GGATCCTGCTGATGCTACCTCTG-3′; antisense, 5′-GTCGACGCCATACTTCTTCTCATAG-3′. Briefly, the following procedure was used. The upstream and downstream sequences of the CART intron I sequence were amplified using primers I and II, respectively. Then the PCR products were denatured, annealed, and amplified again using primer III. Last, the amplified intron I and II sequences were cloned into the BamHI/SalI sites of luciferase reporter vectors P-Luc and tP-Luc. The pGL3-control plasmid with a supplementary NdeI site was a kind gift from Dr. Xiaofei Zheng. Reporter gene constructs contain two tandem copies of pNRSE (pNRSE-Luc) and site-directed mutant pNRSE (pNRSEmut-Luc) were also described previously (25). Here we similarly synthesized two tandem copies of iNRSE, sense, 5′-CCCCGCGGTCAGTACCTGGGACAGCGTCGCTAAGTTCCCGCGGTCAGTACCTGGGACAGCGTCGCTAAGTTA-3′; antisense, 5′-GATCTAACTTAGCGACGCTGTCCCAGGTACTGACCGCGGGAACTTAGCGACGCTGTCCCAGGTACTGACCGCGGGGGTAC-3′; and site-directed mutant iNRSE (iNRSEmut) sense, (5′-CCCCGCGGTCGTTAAATGTTACGTCGTCGCTAAGTTCCCGCGGTCGTTAAATGTTACGTCGTCGCTAAGTTA-3′; antisense, 5′-GATCTAACTTAGCGACGACGTAACATTTAACGACCGCGGGAACTTAGCGACGACGTAACATTTAACGACCGCGGGGGTAC-3′. Oligonucleotide sequences containing a cleaved XbaI/NdeI cloning site were annealed and, respectively, cloned into the pGL3-control, pNRSE-Luc and pNRSEmut-Luc reporter plasmids, then reporter plasmids Luc-iNRSE, Luc-iNRSEmut, pNRSE-Luc-iNRSE, pNRSEmut-Luc-iNRSE, pNRSE-Luc-iNRSEmut, and pNRSEmut-Luc-iNRSEmut were acquired. The mutant nucleotides are underlined. All constructs were verified by DNA sequencing.
Reporter Gene Assays
For the measurement of normoxic cultured HeLa and SK-N-SH cells, cells were routinely cultured in DMEM supplemented with 10% FBS in 24-well culture dishes at a density of 1 × 105 and 2 × 105 cells/well, respectively. After an overnight adhering, the indicated luciferase reporter plasmids were transfected into cells; pRL-CMV (Promega) plasmid was co-transfected for normalization. For overexpression of NRSF or the co-repressor plasmids, the cells were incubated for 48 h and then harvested, lysed, and monitored using the dual luciferase assay system (Promega) in a luminometer (Berthold, Germany). For the depletion of CoREST or other co-repressor factors, the indicated siRNA mixture was transfected into HeLa cells and the lysates were prepared at 72 h post-transfection to ensure the high knockdown efficiency. For certain cases, the pcDNA3.1 or pcDNA3.1-NRSF and P-Luc-I plasmids were transiently co-transfected into SK-N-SH cells, and 24 h after the transfection, the cells were treated with or without 5 μm forskolin (Sigma). Luciferase activities were analyzed after a 24-h treatment. All of the reporter gene assays were repeated at least three times.
Oxygen-glucose Deprivation (OGD)
To initiate ischemia, SK-N-SH cells were subjected to combined OGD in vitro. Briefly, the normal feeding medium was removed and the cells were washed twice with glucose-free medium containing 120 mm NaCl, 25 mm Tris-HCl (pH 7.4), 5.4 mm KCl, and 1.8 mm CaCl2 and then incubated in the same volume of glucose-free medium. Cells were maintained at 37 °C in a hypoxic chamber (Billups-Rothenberg, Del Mar, CA; 95% N2 and 5% CO2) for 3 h. For reoxygenation, cells were incubated in fresh normal feeding medium at 37 °C in 5% CO2 and atmospheric air for 24 h.
Cell Death and Survival Analysis
SK-N-SH cells exposed to combined OGD were transiently transfected with NRSF or siRNA against NRSF, and treated with or without forskolin. The survival rate was measured using flow cytometry. To distinguish apoptotic from necrotic cell death, the Annexin-V-FITC apoptosis detection kit (KeyGen, China) was used according to the manufacturer's instructions. Briefly, cells were detached from the culture plate, washed twice with 1× PBS, and pellets were dissolved in 100 μl of binding buffer. Then 5 μl of Annexin-V and 5 μl of propidium iodide (PI) were added sequentially and incubated in the dark for 15 min. Apoptotic and necrotic cells were quantified by Annexin-V binding and PI uptake.
Statistical Analysis
Statistical analysis was carried out using the Student's t test.
RESULTS
Identification of a NRSE in the Intron of CART Gene
The CART gene is composed of three exons and two introns. Our careful scanning of the human CART sequence predicted a NRSE element in its first intron of the CART gene. The iNRSE is highly conserved across species, homologous to the pNRSE, and conformed to the classical NRSE sequence (Fig. 1A). No more NRSE sequences were found in the CART gene.
FIGURE 1.
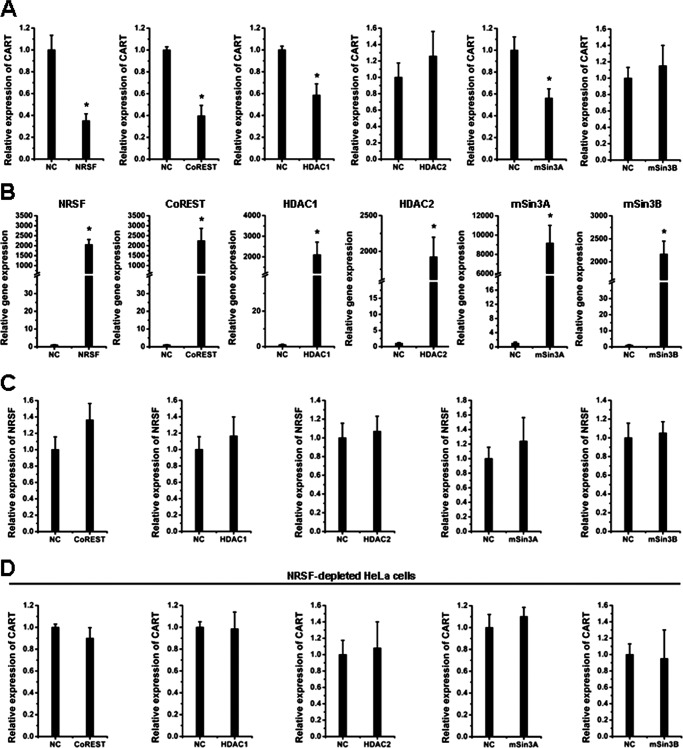
NRSF binds to pNRSE and iNRSE of the CART gene in vitro and in vivo. A, NRSE sequences in human, mouse, and rat CART genes. B, EMSA were performed with the nuclear extracts from HeLa cells and the iNRSE and pNRSE probes. C and D, ChIP assays were performed in siRNA-NRSF #1, #2, or siRNA-Control HeLa cells using antibodies against NRSF, HDAC1, HDAC2, mSin3A, mSin3B, CoREST, or control IgG. The immunoprecipitates were analyzed using quantitative real-time PCR with two sets of primers to amplify DNA including iNRSE (C) or pNRSE (D). Ct values of immunoprecipitated samples were normalized to the corresponding value for input. Bars represent mean ± S.D. *, p < 0.05. E, real-time PCR and Western blot analysis were performed to detect the expression levels of NRSF (upper) and CART (lower) in two NRSF-depleted HeLa cell lines (siRNA-NRSF #1, siRNA-NRSF #2). HeLa cells infected with siRNA-control lentivirus were termed the negative control (NC). The results displayed high knockdown efficiency by ∼85 and 86% in the levels of NRSF mRNA. Data are mean ± S.D. (n = 3). *, p < 0.05.
We first asked whether the iNRSE can bind to NRSF in vitro. Electrophoretic mobility shift assay (EMSA) showed that a clear protein-iNRSE complex formed in a dose-dependent manner when an increasing amount of HeLa nuclear extracts was incubated with the CART iNRSE probe (Fig. 1B, lanes 2–4). Further addition of a specific anti-NRSF antibody (lane 5), but not a control antibody (lane 6), resulted in a significant supershift of the complex; indicating a specific binding of iNRSE to NRSF. By contrast, addition of a large amount of unlabeled iNRSE probe abolished the iNRSE-NRSF binding (lane 7 versus lane 4). The affinity of NRSF with iNRSE was similar to that with pNRSE (lane 2 versus lane 1). These results indicate that the NRSF protein can bind to the iNRSE sequence in the CART gene in vitro.
NRSF Recruits Co-repressors to the iNRSE and pNRSE to Regulate CART Expression
Next, we carried out a series of chromatin immunoprecipitation (ChIP) assays to assess the interaction of NRSF with iNRSE in cultured mammalian cells. The cell lysates from human cervical carcinoma HeLa cells, which express a relative high level of NRSF were immunoprecipitated with the antibody against NRSF, CoREST, HDAC1, HDAC2, mSin3A, mSin3B, or a normal IgG as a negative control. We observed that NRSF and CoREST robustly, whereas HDAC1 moderately but still significantly, interacted with endogenous CART intron 1 in vivo (Fig. 1C, NC groups). By contrast, HDAC2, mSin3A, and mSin3B did not bind to iNRSE under these cellular contexts. For pNRSE, proteins NRSF, CoREST, HDAC1, and mSin3A interacted with the pNRSE element, but HDAC2 and mSin3B did not (Fig. 1D, NC groups). Notably, comparison of the quantitative ChIP data showed that the affinities of iNRSE to NRSF and CoREST proteins were more than 8-fold of those of pNRSE (Fig. 1, C and D), implicating that NRSF and CoREST might prefer to bind more tightly to the intron NRSE than to the promoter NRSE, at least in the case of HeLa cells.
The repression mechanism by NRSF often involves the recruitment of co-repressor complexes, such as HDACs, mSin3, CoREST (co-repressor of REST) (27–29); and the specificity of co-repressor recruitment depends on cellular context. To further evaluate the role of NRSF in the complex organization, endogenous NRSF was depleted by RNAi with each of two independent siRNAs. These siRNAs displayed high knockdown efficiency on NRSF expression (Fig. 1E). NRSF depletion resulted in a dramatic increase of CART levels (Fig. 1E). In the NRSF-depleted cells, the interactions of iNRSE with NRSF, CoREST, and HDAC1 were dramatically attenuated (Fig. 1C), indicating the requirement of NRSF in recruiting CoREST and HDAC1 to the iNRSE. Interestingly, the binding of iNRSE to HDAC2 and to a less extent, mSin3A and mSin3B, was enhanced after NRSF depletion (Fig. 1C). Similarly, NRSF was also required for recruitment of CoREST, HDAC1, and mSin3A to the pNRSE (Fig. 1D). Because mSin3A was specifically recruited to pNRSE but not to iNRSE, and more importantly, NRSF and CoREST displayed stronger affinity to the iNRSE than to the pNRSE, we propose that NRSF recruits a similar but not identical co-repressor complex (both the components and the amounts) to the iNRSE and the pNRSE elements.
To evaluate the role of endogenous CoREST, HDAC1, HDAC2, mSin3A, and mSin3B in the CART transcription control, each of these proteins were depleted in HeLa cells through RNAi and the CART expression levels were determined by quantitative real-time PCR. As shown in Fig. 2A, the gene knockdown efficiency of these siRNAs was ∼50–75%. Depletion of CoREST resulted in a dramatic increase of CART mRNA levels (Fig. 2B), similar to the effect of NRSF depletion (Fig. 1E). The depletion of HDAC1 or mSin3A had moderate but still significant effects on CART up-regulation, whereas the depletion of HDAC2 or mSin3B had no up-regulatory effects on CART (Fig. 2B). To evaluate whether the effects of CoREST, HDAC1, and mSin3A depletion resulted from possible alteration of NRSF mRNA levels, we monitored NRSF expression in these siRNA-transfected cells. As shown in Fig. 2C clearly, knockdown of these five genes had no significant effects on the NRSF mRNA levels. These data suggest that NRSF, CoREST, HDAC1, and mSin3A were involved in the CART regulation.
FIGURE 2.
NRSF recruits co-repressor protein factors to repress CART transcription. A–C, HeLa cells were transfected with the siRNAs against CoREST, HDAC1, HDAC2, mSin3A, or mSin3B. Seventy-two hours post-transfection, the RNA was extracted and the quantitative real-time PCR analysis was performed to measure the mRNA levels of each target molecule (A), CART (B), and NRSF (C). Data are mean ± S.D. (n = 3). *, p < 0.05. NC means the nontargeting control siRNA.
To further confirm the suppressive role of NRSF, CoREST, HDAC1, and mSin3A on CART expression, each of these proteins were overexpressed in HeLa cells and endogenous CART levels were measured. We observed that ectopic expression of NRSF, CoREST, HDAC1, and mSin3A resulted in the decrease of CART levels at various extents, whereas HDAC2 and mSin3B had no significant inhibitory effects (Fig. 3, A and B). Also, the overexpression of CoREST and others had no significant alterations on the NRSF mRNA level (Fig. 3C).
FIGURE 3.
Overexpression of NRSF and co-repressor factors repress CART transcription. A–C, HeLa cells were transfected with the indicated plasmids. Forty-eight hours later, the total RNA was extracted and the mRNA levels of CART (A), the indicated factors (B) and NRSF (C) were measured by qPCR. Data are mean ± S.D. (n = 3). *, p < 0.05. NC means the empty control vector. D, NRSF-depleted HeLa cells were prepared with the lentivirus-mediated RNAi as described under “Experimental Procedures.” These cells were transfected with the indicated plasmid or control empty vector. Forty-eight hours later, the CART mRNA levels were measured by qPCR. Data are mean ± S.D. (n = 3). *, p < 0.05.
Because HeLa cells expressed a relative high level of NRSF, we asked whether the suppressive role of CoREST, HDAC1, and mSin3A was dependent of the endogenous NRSF. In NRSF-depleted HeLa cells, overexpression of CoREST, HDAC1, or mSin3A no longer displayed the suppressive effects on CART transcription (Fig. 3D), suggesting that NRSF-mediated recruitment to NRSE elements of the CART gene was required for the regulation on CART of CoREST, HDAC1, and mSin3A. This result is consistent with the above described ChIP assay data (Fig. 1, C and D).
Both NRSEs Contribute to CART Repression
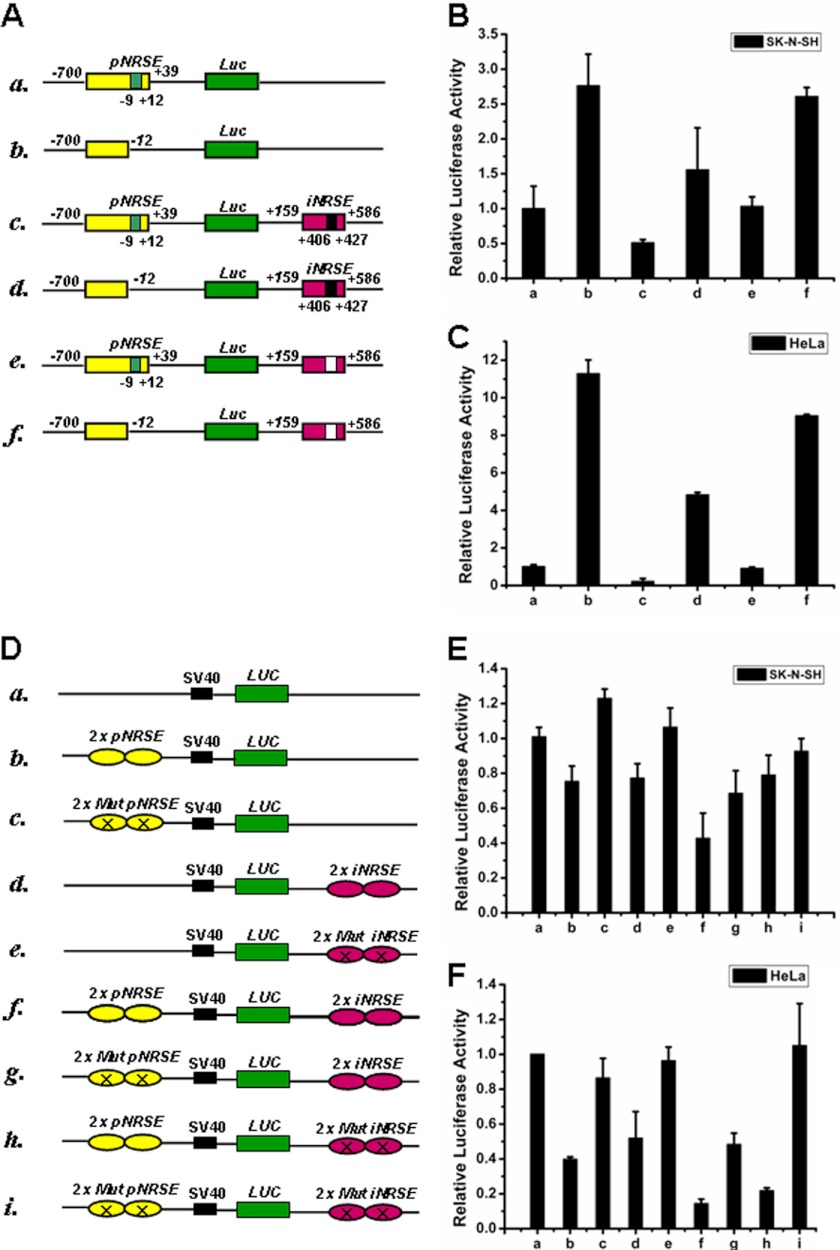
To identify the functional relevance of the two NRSE elements in CART regulation, we performed reporter gene assays with the luciferase gene fused to various combinations of CART promoter and intron sequences containing pNRSE and iNRSE (Fig. 4A). First, deletion of pNRSE resulted in dramatic increases of the reporter activity (Fig. 4, B and C, columns b, d, f versus a, c, e, respectively), indicating the suppressing role of the pNRSE. Second, the addition of intron containing iNRSE resulted in a significant inhibitory effect on the reporter activity (column c versus a, in the presence of pNRSE; d versus b, in the absence of pNRSE), and if iNRSE was deleted from the intron, such an inhibitory effect was largely attenuated (column e versus c; and f versus d), indicating the suppressing role of the iNRSE. Third, deletion of both pNRSE and iNRSE (column f) displayed an additional effect of single deletion of either pNRSE (column d) or iNRSE (column e) (compare f, d, e with c, respectively), suggesting the nonredundant roles of the pNRSE and iNRSE elements. Similar results were obtained in human neuroblastoma SK-N-SH (Fig. 4B) and human cervical carcinoma HeLa cells (Fig. 4C).
FIGURE 4.
Both pNRSE and iNRSE are required for efficient repression of CART expression. A, schematic diagram of the luciferase reporter plasmids that include the promoter and/or intron 1 of human CART gene. B and C, the indicated reporter plasmids along with the pRL-CMV vector were transfected into SK-N-SH (B) or HeLa (C) cells, which express relative lower or higher levels of endogenous NRSF, respectively. The relative luciferase activities are normalized by the activity of the pRL-CMV control. D, schematic diagram of reporter gene constructs that include pNRSE and/or iNRSE elements of the CART gene. Mutations were introduced into the NRSE elements. E and F, the indicated reporter plasmids together with pRL-CMV were transfected into SK-N-SH (E) or HeLa (F) cells and the luciferase activity was measured. Data in B and C, E and F are mean ± S.D. (n = 3).
To get more direct evidence to indicate the role of NRSE elements in CART regulation, we constructed another set of reporter plasmids consisting of pNRSE and iNRSE elements (each two copies) but without other sequences of the promoter/intron (Fig. 4D). To drive the luciferase expression, an SV40 promoter was included in these pGL3-control plasmids. Introduction of a wild-type pNRSE (column b) or iNRSE element (column d) but not a mutant one (columns c and e) resulted in a moderate reduction of the basal luciferase activity in SK-N-SH (Fig. 4E) and HeLa cells (Fig. 4F). Notably, the combination of both pNRSE and iNRSE obtained a dramatic inhibitory effect (column f). Mutation of key sites in the NRSE element reversed luciferase activity (columns g-i). Collectively, the reporter assays suggested that both pNRSE and iNRSE elements contribute to the repression of CART in cultured cells.
We have shown that NRSF, CoREST, HDAC1, and mSin3A were involved in the CART repression, possibly through recruitment to NRSE elements of CART gene (Figs. 1–3). To further evaluate the physiological role of these protein factors in the NRSE-driven CART repression, and to clarify which factors are recruited to pNRSE or iNRSE elements, reporter gene assays were performed to verify our conclusion made through ChIP assays and qPCR analysis. The luciferase reporter plasmids P-Luc-tI (the plasmid “e” described in Fig. 4A) and tP-Luc-I (the plasmid “d” described in Fig. 4A), which contains both the promoter and intron but lack either the iNRSE or pNRSE element, respectively, were used in the reporter gene assays. Each of the six protein factors (NRSF, CoREST, HDAC1, HDAC2, mSin3A, and mSin3B) were depleted from HeLa cells through siRNA transfection and P-Luc-tI and tP-Luc-I activity was measured. As shown in Fig. 5, A and B, depletion of NRSF or CoREST resulted in dramatic increases of the reporter activities of both P-Luc-tI and tP-Luc-I. Similar but lesser effects were observed with the depletion of HDAC1. Interestingly, depletion of mSin3A resulted in the specific increase of P-Luc-tI activity. By contrast, depletion of HDAC2 or mSin3B had no significant effects or even resulted in the decrease of the two luciferases (Fig. 5, A and B). These results suggest that NRSF, CoREST, HDAC1, and mSin3A are involved in pNRSE-mediated CART repression, whereas NRSF, CoREST, and HDAC1 are involved in iNRSE-mediated transcription inhibition of CART.
FIGURE 5.
The NRSF-recruited co-repressor protein factors are involved in pNRSE and iNRSE-driven CART repression. A and B, the P-Luc-tI (A) and tP-Luc-I (B) luciferase reporter plasmids described in Fig. 4A (e and d, respectively) were transfected into HeLa cells together with the pRL-CMV control luciferase plasmid, the indicated siRNA against NRSF, CoREST, HDAC1, HDAC2, mSin3A, mSin3B, or the control siRNA (NC). Seventy-two hours post-transfection, the P-Luc-tI and tP-Luc-I luciferase activities were measured and normalized. Data are mean ± S.D. (n = 3). *, p < 0.05. C and D, the P-Luc-tI (A) and tP-Luc-I (B) luciferase reporter plasmids were transfected into HeLa cells together with the pRL-CMV control luciferase plasmid, the indicated overexpression plasmids. NC means the empty vector. Forty-eight hours post-transfection, the P-Luc-tI and tP-Luc-I luciferase activities were measured and normalized. Data are mean ± S.D. (n = 3). *, p < 0.05.
To confirm the suppressive effects of these protein factors on luciferase activity, we performed reporter gene assays in HeLa cells, which were transfected, and overexpressed each of these factors. We observed that NRSF, CoREST, and HDAC1 expression significantly inhibited the activity of both luciferase reporters, whereas mSin3A only inhibited P-Luc-tI. By contrast, HDAC2 and mSin3B had no such inhibitory effects on these reporters (Fig. 5, C and D). The inhibitory effect of NRSF overexpression was not as strong as that of CoREST or HDAC1 overexpression. This might be due to the high expression level of endogenous NRSF. Taken together, the ChIP assays, the qPCR analysis, and the luciferase reporter assays suggest that NRSF functions as an organizer to recruit similar but not identical co-repressor protein factors to the promoter and the intron of CART gene. Through this manner, CART transcription was tightly repressed.
NRSF Antagonizes the CREB Activation on CART
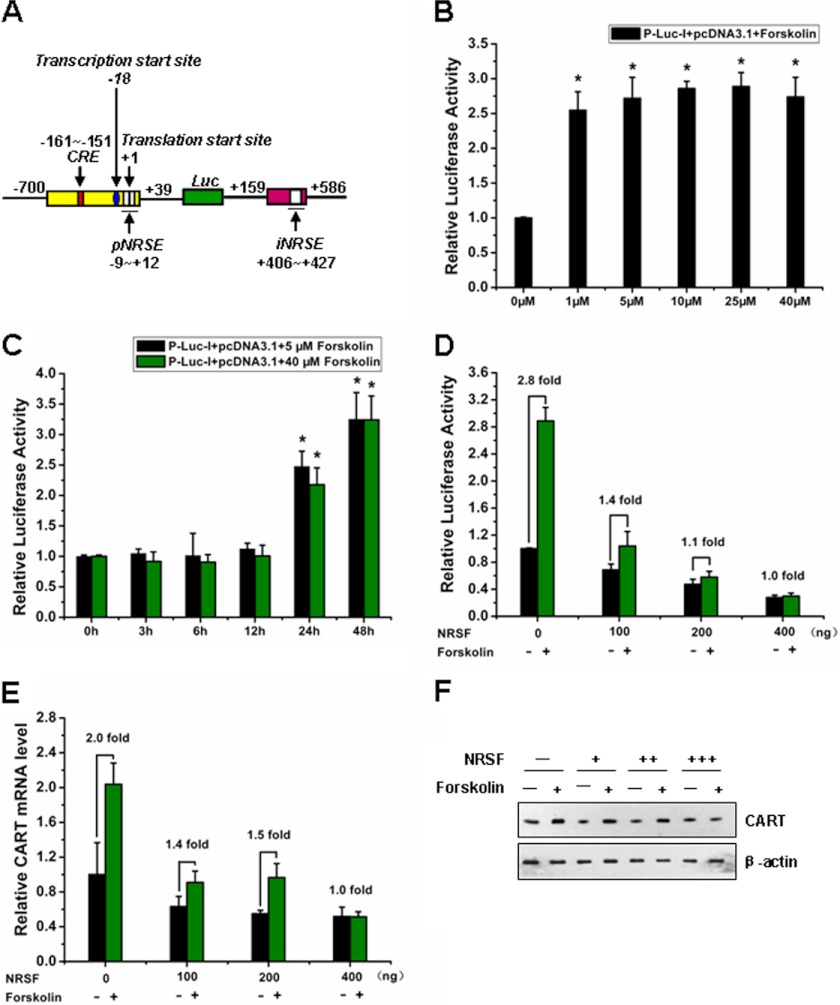
Previous studies have demonstrated that CART gene transactivation was mainly mediated by the cAMP-PKA-CREB pathway (38, 39). Forskolin is an activator of adenylate cyclase, which is the enzyme that converts ATP to cAMP (20). Through this manner, forskolin stimulates PKA activation, which in turn phosphorylates the transcriptional factor CREB. As a target gene of CREB, CART expression is up-regulated. To explore whether NRSF has any antagonizing effect on CREB signaling, we generated a luciferase reporter plasmid containing CRE, pNRSE, and iNRSE elements of the CART promoter/intron (Fig. 6A). Forskolin treatment stimulated luciferase activity (Fig. 6, B and C). NRSF overexpression significantly reduced the stimulatory effect of forskolin on CART promoter-driven luciferase (Fig. 6D). Also NRSF overexpression antagonized the up-regulation of both mRNA and protein expression of CART mediated by forskolin-CREB signaling (Fig. 6, E and F).
FIGURE 6.
Activation of the CART promoter by forskolin was repressed by NRSF. A, schematic diagram of the reporter construct P-Luc-I that includes the CRE, pNRSE, and iNRSE elements. B, forskolin stimulation promotes CART luciferase activity. The pcDNA3.1 empty vector together with P-Luc-I (shown in Fig. 4A) and pRL-CMV were co-transfected into SK-N-SH cells. Then cells were stimulated with or without forskolin at the indicated concentrations. Twenty-four hours later, the cell lysates were prepared for luciferase activity analysis. Results are expressed as mean ± S.D. (n = 3). *, p < 0.05 (compare with the un-stimulated group). C, the SK-N-SH cells transfected as described in B were stimulated with forskolin of low or high concentrations (5 μm, 40 μm) for various times as indicated. The luciferase activity was measured. Results are expressed as mean ± S.D. (n = 3). *, p < 0.05. Based on these analyses, we selected 5 μm and 24 h stimulation of forskolin for the subsequent experiments described in Figs. 8 and 9. D, the pcDNA3.1 or pcDNA3.1-NRSF together with P-Luc-I and pRL-CMV were co-transfected into SK-N-SH cells. Then cells were stimulated with or without forskolin (5 μm, 24 h), and cell lysates were prepared for luciferase activity analysis. Results are expressed as mean ± S.D. (n = 3). E, quantitative analysis for CART mRNA was performed by real-time PCR. Each experiment was performed at least three times in triplicate. Data are presented as mean ± S.D. F, Western blot analysis was performed to measure the protein levels of CART.
Ischemia Insult Up-regulates NRSF to Induce Neural Death
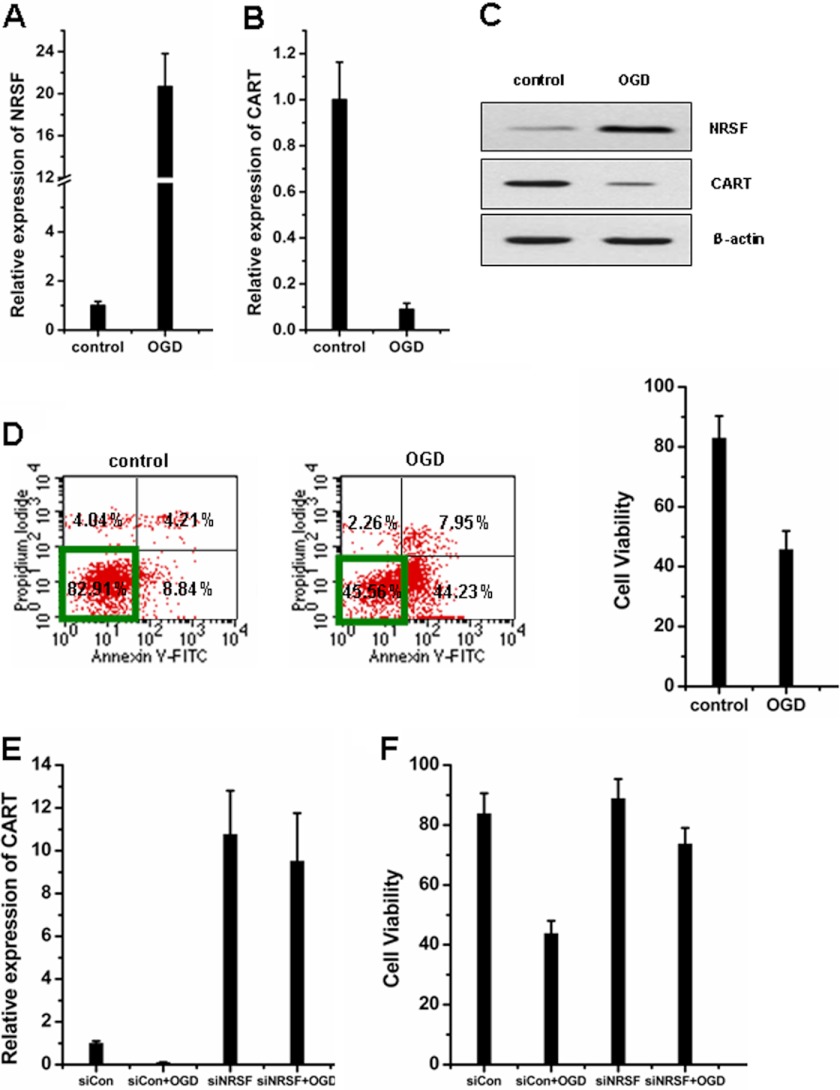
CART peptide has been defined to be protective against focal cerebral ischemia in vivo and against neuronal cell death in culture induced by OGD through preservation of mitochondrial function and prevention of energy failure (40–42). We then used the OGD model to induce neuronal cell death and evaluated the possible pathophysiological role of NRSF on CART regulation. We observed that NRSF mRNA expression was dramatically induced upon OGD treatment (Fig. 7A), whereas CART mRNA was reduced (Fig. 7B). Western blot analysis confirmed the reverse correlation of NRSF and CART expression at the protein level (Fig. 7C). OGD treatment resulted in a notable decrease in cell survival rate (Fig. 7D). Importantly, depletion of endogenous NRSF significantly attenuated the decrease of CART expression (Fig. 7E) and cell viability induced by OGD (Fig. 7F), suggesting that OGD-induced CART down-regulation and cell death were largely dependent of NRSF.
FIGURE 7.
OGD-mimicked hypoxia-ischemia up-regulates NRSF to repress CART and induce cell death. A and B, SK-N-SH cells were exposed to OGD for 3 h and then reoxygenation at 37 °C for 24 h. The expression levels of NRSF (A) and CART mRNA (B) were monitored by qPCR. C, Western blot was used to detect the protein levels of NRSF and CART. D, cell death was assessed by Annexin-V and PI labeling and flow cytometric analysis. Quadrant 1 (upper left), late necrosis; quadrant 2 (upper right), early necrosis/late apoptosis; quadrant 3 (lower left), live cells; quadrant 4 (lower right), early apoptosis. Cells unstained for Annexin V-FITC and PI are defined as viable cells. E and F, NRSF-depleted SK-N-SH cells were exposed to OGD and then reoxygenation. CART mRNA levels (E) and cell viability (F) were measured. Data in A and B and D–F are mean ± S.D. (n = 3). Each independent experiment was carried out in triplicate.
NRSF Antagonizes the Forskolin-PKA-CREB Signaling Upon Ischemia Insult and Depletion of NRSF Elevates the Cell Survival
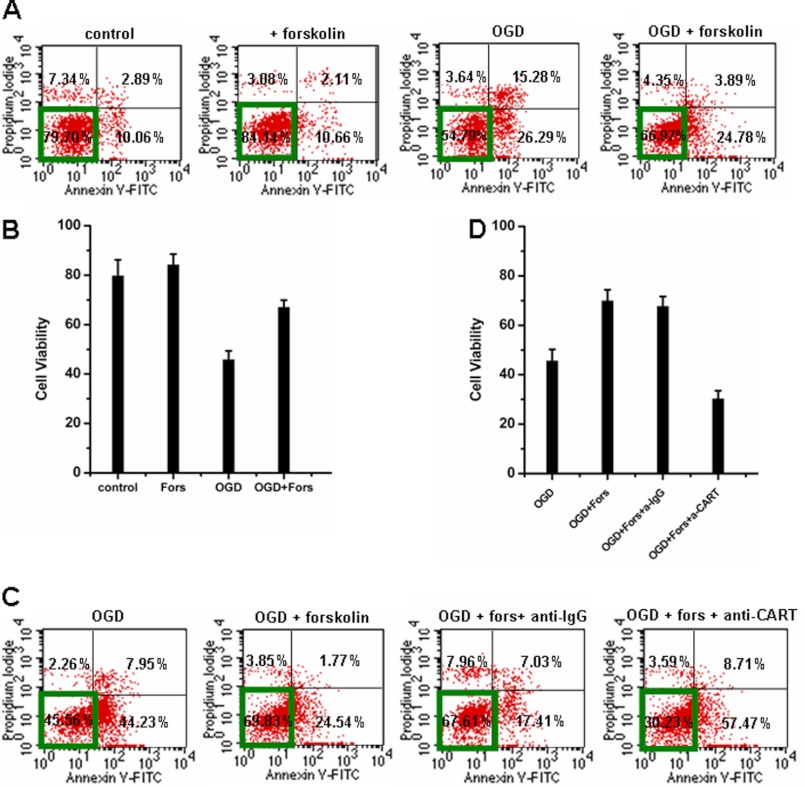
We then asked whether forskolin-stimulated CART synthesis can reduce OGD-induced cell death. Forskolin administration indeed increased the survival rate of SK-N-SH cells after OGD (Fig. 8, A and B). When a CART-blocking antibody was added into the cells, the protective effect of forskolin was remarkably attenuated (Fig. 8, C and D), suggesting that forskolin-mediated neuroprotection was dependent of CART.
FIGURE 8.
Forskolin stimulation promotes neuronal survival through up-regulation of CART. A and B, forskolin (5 μm) was added prior to OGD and maintained during OGD and throughout reoxygenation. Forskolin administration for 24 h increased the survival rate of SK-N-SH cells after OGD. C and D, addition of CART antibody attenuated the protective effect of forskolin. Data in B and D are mean ± S.D. (n = 3). Each independent experiment was carried out in triplicate. Representative FACS data of three independent experiments are shown in A and C.
Because OGD up-regulated endogenous NRSF expression, OGD treatment indeed attenuated the forskolin-stimulated CART promoter-driven luciferase activity and CART mRNA expression, which effects were further enforced by NRSF ectopic expression (Fig. 9, A and B). Moreover, NRSF also had an antagonizing role against forskolin on cell death upon OGD-induced ischemia. Ectopic expression of NRSF suppressed the protective role of forskolin (Fig. 9, C and D). By contrast, depletion of endogenous NRSF significantly elevated the cell survival both in the absence and presence of forskolin upon OGD treatment (Fig. 9, E and F).
FIGURE 9.
Ectopic expression of NRSF antagonizes, whereas depletion of NRSF synergizes, the survival-promoting effects of forskolin-CREB signaling upon OGD treatment. A, SK-N-SH cells were transfected with increasing amounts of pcDNA3.1-NRSF together with CART P-Luc-I reporter plasmid. Then cells were stimulated with or without 5 μm forskolin for 24 h, and cell lysates were prepared for luciferase activity analysis. B, quantitative analysis for CART mRNA was performed by real-time PCR. The value of SK-N-SH cells transfected with pcDNA3.1 control plasmids was arbitrarily set as 1.0. C-F, ectopic NRSF repressed (C and D), whereas siRNA against NRSF increased (E and F) forskolin-stimulated cell survival after OGD. SK-N-SH cells exposed to OGD for 3 h were transiently transfected with pcDNA3.1 or pcDNA3.1-NRSF (400 ng/well) (C and D) or siRNA against NRSF (E and F), stimulated by forskolin or not. Cell death was measured by flow cytometric analysis. Data in C and E are representative data of three independent experiments in D and F, respectively. Cells unstained for Annexin V-FITC and PI are defined as viable cells. Data are mean ± S.D. Each experiment was performed three times independently.
DISCUSSION
The main findings of the current study are: 1) NRSF binds to both pNRSE and iNRSE of the CART gene in vitro and in cultured mammalian cells; 2) NRSF recruits similar but not identical co-repressor complexes to the pNRSE and iNRSE sites to repress CART expression; 3) hypoxia-ischemia mimicked by OGD exposure results in significant NRSF up-regulation and CART down-regulation; and 4) NRSF antagonizes the forskolin-stimulated PKA-CREB activation on CART. Depletion of NRSF in combination with forskolin stimulation significantly elevates cell survival upon ischemia insult.
CART has been reported to function as an endogenous neuroprotective peptide. Increasing findings suggest that the neuroprotective mechanism of CART may be linked to activation of the ERK signaling pathway, preservation of mitochondrial function, reactive oxygen segment, and up-regulation of BDNF and NGF expression (40, 42–45). It also has been suggested that CART plays an important role in modulating post-stroke immune responses and plays a neuroprotective role in experimental stroke (45, 46). Due to the pivotal role of the CART peptide in neuroprotection, CART expression should be tightly regulated by positive and negative mechanisms. Previous studies have well defined the activation of CART, primarily PKA-CREB signaling. By contrast, little is known about the repression mechanism. This study together, with our previous findings (25), identified a dual NRSE mechanism for CART repression. As far as we know, this is the first evidence to systematically reveal the repression mechanism of CART transcription. In this regard, the current study provides novel insights into the regulation of CART expression.
Notably, we found that NRSF utilize a dual NRSE pattern to efficiently repress CART expression. Through EMSA, ChIP assays, qPCR analysis, and reporter gene assays, we provide solid data to prove that both the promoter NRSE (pNRSE) and the intron NRSE (iNRSE) are bona fide regulatory elements of CART that play equivalent roles in CART regulation. NRSF recruits co-repressor complexes to the NRSE sites to ensure the repression efficiency. The complexes include HDAC1, mSin3, and CoREST, which is the first time shown to bind to the CART gene. We noted that similar but not identical co-repressors were recruited to the pNRSE and iNRSE, i.e. (a) NRSF and CoREST displayed stronger affinity to the iNRSE than to the pNRSE (about 8-folds); (b) mSin3A was specifically recruited to pNRSE but not to iNRSE.
This study has promising implications on therapeutics of stroke-related brain injury and other CART-related diseases. We explored the dynamic role of NRSF with an in vitro ischemia neural cell death model (OGD). We observed that hypoxia-ischemia induces a dramatic NRSF up-regulation (more than 20-fold), although the detailed mechanism is yet unknown so far. Consequently, CART expression was dramatically down-regulated (less than 1/10) and cell death was increased. We further found an antagonizing role of NRSF against forskolin-CREB-CRE signaling on CART transcription and cell survival during ischemia injury. Intriguingly, a combination of forskolin with NRSF siRNA elevated the cell survival rate than the forskolin alone. These findings suggest that siRNA or a small molecular inhibitor against NRSF may be developed into new therapeutic strategy for stroke-related brain injury.
Acknowledgments
We are grateful to Xiaofei Zheng for the generous gift of pGL3-control plasmid. We also thank Changyan Li and Lingling Zhu for instrumental support.
This work was supported by National High Technology Research and Development Program of China Grant 2006AA02A107, the National Basic Research Program of China Grant 2010CB945500, Chinese National Natural Science Foundation Grant 81070618, and Beijing Natural Science Foundation Grant 5102036.
- CART
- cocaine- and amphetamine-regulated transcript
- CREB
- cAMP response element-binding protein
- CRE
- cAMP-response element
- NRSF
- neuron-restrictive silencer factor
- HDAC
- histone deacetylase
- qPCR
- quantitative PCR
- PI
- propidium iodide
- OGD
- oxygen-glucose deprivation.
REFERENCES
- 1. Kuhar M. J., Dall Vechia S. E. (1999) CART peptides. Novel addiction- and feeding-related neuropeptides. Trends Neurosci. 22, 316–320 [DOI] [PubMed] [Google Scholar]
- 2. Kuriyama G., Takekoshi S., Tojo K., Nakai Y., Kuhar M. J., Osamura R. Y. (2004) Cocaine- and amphetamine-regulated transcript peptide in the rat anterior pituitary gland is localized in gonadotrophs and suppresses prolactin secretion. Endocrinology 145, 2542–2550 [DOI] [PubMed] [Google Scholar]
- 3. Vicentic A., Dominguez G., Hunter R. G., Philpot K., Wilson M., Kuhar M. J. (2004) Cocaine- and amphetamine-regulated transcript peptide levels in blood exhibit a diurnal rhythm. Regulation by glucocorticoids. Endocrinology 145, 4119–4124 [DOI] [PubMed] [Google Scholar]
- 4. Upadhya M. A., Nakhate K. T., Kokare D. M., Singh U., Singru P. S., Subhedar N. K. (2012) CART peptide in the nucleus accumbens shell acts downstream to dopamine and mediates the reward and reinforcement actions of morphine. Neuropharmacology 62, 1823–1833 [DOI] [PubMed] [Google Scholar]
- 5. Balkan B., Keser A., Gozen O., Koylu E. O., Dagci T., Kuhar M. J., Pogun S. (2012) Forced swim stress elicits region-specific changes in CART expression in the stress axis and stress regulatory brain areas. Brain Res. 1432, 56–65 [DOI] [PubMed] [Google Scholar]
- 6. Rodrigues B. C., Cavalcante J. C., Elias C. F. (2011) Expression of cocaine- and amphetamine-regulated transcript in the rat forebrain during postnatal development. Neuroscience 195, 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Broberger C., Holmberg K., Kuhar M. J., Hökfelt T. (1999) Cocaine- and amphetamine-regulated transcript in the rat vagus nerve. A putative mediator of cholecystokinin-induced satiety. Proc. Natl. Acad. Sci. U.S.A. 96, 13506–13511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vrang N., Tang-Christensen M., Larsen P. J., Kristensen P. (1999) Recombinant CART peptide induces c-Fos expression in central areas involved in control of feeding behaviour. Brain Res. 818, 499–509 [DOI] [PubMed] [Google Scholar]
- 9. Jones D. C., Kuhar M. J. (2006) Cocaine-amphetamine-regulated transcript expression in the rat nucleus accumbens is regulated by adenylyl cyclase and the cyclic adenosine 5′-monophosphate/protein kinase a second messenger system. J. Pharmacol. Exp. Ther. 317, 454–461 [DOI] [PubMed] [Google Scholar]
- 10. Kasacka I., Janiuk I., Lewandowska A., Bekisz A., Lebkowski W. (2012) Distribution pattern of CART-containing neurons and cells in the human pancreas. Acta Histochem. 114, 695–699 [DOI] [PubMed] [Google Scholar]
- 11. Bech P., Winstanley V., Murphy K. G., Sam A. H., Meeran K., Ghatei M. A., Bloom S. R. (2008) Elevated cocaine- and amphetamine-regulated transcript immunoreactivity in the circulation of patients with neuroendocrine malignancy. J. Clin. Endocrinol. Metab. 93, 1246–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brennan D. J., O'Connor D. P., Laursen H., McGee S. F., McCarthy S., Zagozdzon R., Rexhepaj E., Culhane A. C., Martin F. M., Duffy M. J., Landberg G., Ryden L., Hewitt S. M., Kuhar M. J., Bernards R., Millikan R. C., Crown J. P., Jirström K., Gallagher W. M. (2012) The cocaine- and amphetamine-regulated transcript mediates ligand-independent activation of ERα, and is an independent prognostic factor in node-negative breast cancer. Oncogene 31, 3483–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Douglass J., McKinzie A. A., Couceyro P. (1995) PCR differential display identifies a rat brain mRNA that is transcriptionally regulated by cocaine and amphetamine. J. Neurosci. 15, 2471–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kristensen P., Judge M. E., Thim L., Ribel U., Christjansen K. N., Wulff B. S., Clausen J. T., Jensen P. B., Madsen O. D., Vrang N., Larsen P. J., Hastrup S. (1998) Hypothalamic CART is a new anorectic peptide regulated by leptin. Nature 393, 72–76 [DOI] [PubMed] [Google Scholar]
- 15. Ahima R. S., Hileman S. M. (2000) Postnatal regulation of hypothalamic neuropeptide expression by leptin. Implications for energy balance and body weight regulation. Regul. Pept. 92, 1–7 [DOI] [PubMed] [Google Scholar]
- 16. de Lartigue G., Dimaline R., Varro A., Dockray G. J. (2007) Cocaine- and amphetamine-regulated transcript. Stimulation of expression in rat vagal afferent neurons by cholecystokinin and suppression by ghrelin. J. Neurosci. 27, 2876–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sárvári M., Hrabovszky E., Kalló I., Galamb O., Solymosi N., Likó I., Molnár B., Tihanyi K., Szombathelyi Z., Liposits Z. (2010) Gene expression profiling identifies key estradiol targets in the frontal cortex of the rat. Endocrinology 151, 1161–1176 [DOI] [PubMed] [Google Scholar]
- 18. Liu L., Song Z., Sheikhahmadi A., Jiao H., Lin H. (2012) Effect of corticosterone on gene expression of feed intake regulatory peptides in laying hens. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 162, 81–87 [DOI] [PubMed] [Google Scholar]
- 19. Barrett P., Davidson J., Morgan P. (2002) CART gene promoter transcription is regulated by a cyclic adenosine monophosphate response element. Obes. Res. 10, 1291–1298 [DOI] [PubMed] [Google Scholar]
- 20. Dominguez G., Lakatos A., Kuhar M. J. (2002) Characterization of the cocaine- and amphetamine-regulated transcript (CART) peptide gene promoter and its activation by a cyclic AMP-dependent signaling pathway in GH3 cells. J. Neurochem. 80, 885–893 [DOI] [PubMed] [Google Scholar]
- 21. Lakatos A., Dominguez G., Kuhar M. J. (2002) CART promoter CRE site binds phosphorylated CREB. Brain Res. Mol. Brain Res. 104, 81–85 [DOI] [PubMed] [Google Scholar]
- 22. Jones D. C., Lakatos A., Rogge G. A., Kuhar M. J. (2009) Regulation of cocaine- and amphetamine-regulated transcript mRNA expression by calcium-mediated signaling in GH3 cells. Neuroscience 160, 339–347 [DOI] [PubMed] [Google Scholar]
- 23. Rogge G. A., Shen L. L., Kuhar M. J. (2010) Chromatin immunoprecipitation assays revealed CREB and serine 133 phospho-CREB binding to the CART gene proximal promoter. Brain Res. 1344, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ling F., Wei L., Wang T., Chen Y., Zhu X., Li J., Liu T., Du H., Wang H., Wang J. (2011) Cloning and characterization of the 5′-flanking region of the pig cocaine- and amphetamine-regulated transcript gene. DNA Cell Biol. 30, 91–97 [DOI] [PubMed] [Google Scholar]
- 25. Li Y., Liu Q., Yang Y., Lv Y., Chen L., Bai C., Nan X., Wang Y., Pei X. (2008) Regulatory role of neuron-restrictive silencing factor in the specific expression of cocaine- and amphetamine-regulated transcript gene. J. Neurochem. 106, 1314–1324 [DOI] [PubMed] [Google Scholar]
- 26. Schoenherr C. J., Paquette A. J., Anderson D. J. (1996) Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. U.S.A. 93, 9881–9886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Andrés M. E., Burger C., Peral-Rubio M. J., Battaglioli E., Anderson M. E., Grimes J., Dallman J., Ballas N., Mandel G. (1999) CoREST. A functional co-repressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. U.S.A. 96, 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grimes J. A., Nielsen S. J., Battaglioli E., Miska E. A., Speh J. C., Berry D. L., Atouf F., Holdener B. C., Mandel G., Kouzarides T. (2000) The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275, 9461–9467 [DOI] [PubMed] [Google Scholar]
- 29. Huang Y., Myers S. J., Dingledine R. (1999) Transcriptional repression by REST. Recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2, 867–872 [DOI] [PubMed] [Google Scholar]
- 30. Gao Z., Ure K., Ding P., Nashaat M., Yuan L., Ma J., Hammer R. E., Hsieh J. (2011) The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31, 9772–9786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yu H. B., Johnson R., Kunarso G., Stanton L. W. (2011) Coassembly of REST and its cofactors at sites of gene repression in embryonic stem cells. Genome Res. 21, 1284–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan L., Tian C., Wang H., Song S., Li D., Xing G., Yin Y., He F., Zhang L. (2012) Apak competes with p53 for direct binding to intron 1 of p53AIP1 to regulate apoptosis. EMBO Rep. 13, 363–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yang Y., Li Y., Lv Y., Zhang S., Chen L., Bai C., Nan X., Yue W., Pei X. (2008) NRSF silencing induces neuronal differentiation of human mesenchymal stem cells. Exp. Cell Res. 314, 2257–2265 [DOI] [PubMed] [Google Scholar]
- 34. Greco S. J., Smirnov S. V., Murthy R. G., Rameshwar P. (2007) Synergy between the RE-1 silencer of transcription and NFκB in the repression of the neurotransmitter gene TAC1 in human mesenchymal stem cells. J. Biol. Chem. 282, 30039–30050 [DOI] [PubMed] [Google Scholar]
- 35. Li B., Wang S., Liu H., Liu D., Zhang J., Zhang B., Yao H., Lv Y., Wang R., Chen L., Yue W., Li Y., Pei X. (2011) Neuronal restrictive silencing factor silencing induces human amniotic fluid-derived stem cells differentiation into insulin-producing cells. Stem Cells Dev. 20, 1223–1231 [DOI] [PubMed] [Google Scholar]
- 36. Ventura A., Meissner A., Dillon C. P., McManus M., Sharp P. A., Van Parijs L., Jaenisch R., Jacks T. (2004) Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. U.S.A. 101, 10380–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin D., Tawadros T., Meylan L., Abderrahmani A., Condorelli D. F., Waeber G., Haefliger J. A. (2003) Critical role of the transcriptional repressor neuron-restrictive silencer factor in the specific control of connexin36 in insulin-producing cell lines. J. Biol. Chem. 278, 53082–53089 [DOI] [PubMed] [Google Scholar]
- 38. Dominguez G. (2006) The CART gene. Structure and regulation. Peptides 27, 1913–1918 [DOI] [PubMed] [Google Scholar]
- 39. Hunter R. G., Jones D., Vicentic A., Hue G., Rye D., Kuhar M. J. (2006) Regulation of CART mRNA in the rat nucleus accumbens via D3 dopamine receptors. Neuropharmacology 50, 858–864 [DOI] [PubMed] [Google Scholar]
- 40. Mao P., Ardeshiri A., Jacks R., Yang S., Hurn P. D., Alkayed N. J. (2007) Mitochondrial mechanism of neuroprotection by CART. Eur. J. Neurosci. 26, 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu Y., Zhang W., Klaus J., Young J., Koerner I., Sheldahl L. C., Hurn P. D., Martínez-Murillo F., Alkayed N. J. (2006) Role of cocaine- and amphetamine-regulated transcript in estradiol-mediated neuroprotection. Proc. Natl. Acad. Sci. U.S.A. 103, 14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mao P., Meshul C. K., Thuillier P., Goldberg N. R., Reddy P. H. (2012) CART peptide is a potential endogenous antioxidant and preferentially localized in mitochondria. PLoS One 7, e29343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu B., Hu S., Yang M., Pan H., Zhu S. (2006) CART peptide promotes the survival of hippocampal neurons by up-regulating brain-derived neurotrophic factor. Biochem. Biophys. Res. Commun. 347, 656–661 [DOI] [PubMed] [Google Scholar]
- 44. Jia J., Chen X., Zhu W., Luo Y., Hua Z., Xu Y. (2008) CART protects brain from damage through ERK activation in ischemic stroke. Neuropeptides 42, 653–661 [DOI] [PubMed] [Google Scholar]
- 45. Liu Z., Huang D., Zhang M., Chen Z., Jin J., Huang S., Zhang Z., Wang Z., Chen L., Chen L., Xu Y. (2011) Cocaine- and amphetamine-regulated transcript promotes the differentiation of mouse bone marrow-derived mesenchymal stem cells into neural cells. BMC Neurosci. 12, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chang L., Chen Y., Li J., Liu Z., Wang Z., Chen J., Cao W., Xu Y. (2011) Cocaine- and amphetamine-regulated transcript modulates peripheral immunity and protects against brain injury in experimental stroke. Brain Behav. Immun. 25, 260–269 [DOI] [PubMed] [Google Scholar]