Background: The TCR forms nanoclusters in the plasma membrane independent of ligand binding.
Results: Membrane cholesterol and sphingomyelin facilitate TCR nanoclustering, thereby enhancing the avidity toward the ligand.
Conclusion: The membrane lipid composition regulates the degree of TCR nanoclustering and thus T-cell sensitivity.
Significance: This work contributes to the understanding of the consequences of specific lipid-membrane protein interactions.
Keywords: Lipids, Membrane Reconstitution, Protein Complexes, Synthetic Biology, T-cell Receptor, Liposomes, Protein Island
Abstract
The T-cell antigen receptor (TCR) exists in monomeric and nanoclustered forms independently of antigen binding. Although the clustering is involved in the regulation of T-cell sensitivity, it is unknown how the TCR nanoclusters form. We show that cholesterol is required for TCR nanoclustering in T cells and that this clustering enhances the avidity but not the affinity of the TCR-antigen interaction. Investigating the mechanism of the nanoclustering, we found that radioactive photocholesterol specifically binds to the TCRβ chain in vivo. In order to reduce the complexity of cellular membranes, we used a synthetic biology approach and reconstituted the TCR in liposomes of defined lipid composition. Both cholesterol and sphingomyelin were required for the formation of TCR dimers in phosphatidylcholine-containing large unilamellar vesicles. Further, the TCR was localized in the liquid disordered phase in giant unilamellar vesicles. We propose a model in which cholesterol and sphingomyelin binding to the TCRβ chain causes TCR dimerization. The lipid-induced TCR nanoclustering enhances the avidity to antigen and thus might be involved in enhanced sensitivity of memory compared with naive T cells. Our work contributes to the understanding of the function of specific nonannular lipid-membrane protein interactions.
Introduction
The T-cell antigen receptor (TCR)4 is a multisubunit transmembrane protein complex responsible for the triggering of a T-cell-mediated adaptive immune response. It consists of the antigen-recognizing TCRαβ (or TCRγδ) heterodimer and the signal-transducing CD3 dimers: CD3ϵγ, CD3ϵδ, and ζζ (1, 2). The basic functional unit of the TCR, defined as the monomeric TCR, has a TCRαβCD3δϵγϵζζ stoichiometry (3–6).
TCR nanoclusters have been detected by Blue Native (BN)-PAGE analysis, gel filtration, and co-immunopurification of two different TCRs co-expressed on the same cell (4, 7, 8) as well as by immunogold electron microscopy (4, 9, 10), high speed photoactivated localization microscopy, and dual-color fluorescence cross-correlation spectroscopy (10). TCR nanoclusters have a size of 2–30 TCRs and are co-expressed with TCR monomers (4, 10). Using bioluminescence resonance energy transfer and two-color coincidence detection microscopy, it was shown that up to 7–10% of the TCRs could be nanoclustered (11, 12). Here we use the term “nanocluster” to distinguish TCR nanoclustering, which is independent of MHC-peptide (MHCp) binding, from activation-induced microcluster formation of 20–70 TCRs (13).
TCR nanoclusters increase the sensitivity of T cells to antigenic stimulation (9), possibly because TCR nanoclusters can be stimulated at lower antigen concentrations than monomeric TCRs (4, 14). However, the molecular mechanism by which T cells regulate the extent of TCR nanoclustering is unknown.
The mechanisms that contribute to the lateral segregation of proteins and lipids are the subject of intense research. It was hypothesized that in cells, at least two distinct membrane microdomains exist: the cholesterol- and sphingomyelin (SM)-rich lipid rafts and the phospholipid-enriched non-rafts (15). Due to post-translational lipid modifications, a number of proteins partition into the raft domain (16), where they might form clusters (17). In artificial membranes containing cholesterol, SM, and phospholipids, the formation of lateral liquid-ordered (lo) and liquid-disordered (ld) phases occurs (18), which might correspond to the raft and non-raft domains. This phase separation is facilitated by the interaction of cholesterol with SM (19). The actin cytoskeleton (20, 21) and protein-protein interactions (22) can also be involved in the clustering of membrane proteins.
In this report, we study the role of the lipid environment in the formation of TCR nanoclusters and the physiological relevance of TCR nanoclustering (i.e. the avidity of the TCR-antigen interaction).
EXPERIMENTAL PROCEDURES
Reagents
The following antibodies were used: rabbit anti-ζ antiserum 448, anti-CD3 (145-2C11 from J. Bluestone), anti-hTCRβ (βF1, Endogen), anti-CD3ϵ (M20, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), anti-transferrin receptor (TfR) (7F8, Abcam), and anti-mTCRβ (H57-598, Abcam). Secondary antibodies for Western blot and anti-mouse IgG-PE were purchased from Southern Biotech. All chemicals and reagents were purchased from Sigma if not stated otherwise.
Generation of Expression Plasmids and Cell Lines
To generate the expression vector pcDNA3_mζ-SBP, the DNA fragment coding for the mouse ζ chain C-terminally linked to the streptavidin-binding peptide (SBP) purification tag (23) was amplified by PCR and cloned into the EcoRI/XhoI site of the pcDNA3 vector (Invitrogen). pcDNA3_mζ-SBP was transfected into the mouse 2B4-derived ζ-deficient line MA5.8 to yield M.mζ-SBP. The cDNA of the human TfR C-terminally linked to the SBP tag was amplified by PCR and inserted into the BglII/XhoI site of the pMIG-based expression vector, pMItom (provided by R. Y. Tsien). pMItomTfR-SBP was transfected into MA5.8 cells, to yield the M.hTfR-SBP cell line. To obtain the expression vectors for the bifluorescence complementation (BiFC) assay, cDNA coding for the mouse ζ chain was C-terminally linked to enhanced GFP, the N-terminal part (residues 1–172; YN) of a yellow fluorescent protein (Venus), and the C-terminal part (residues 155–238; CC) of enhanced cyan fluorescent protein (both from Clontech), amplified by PCR, and cloned into the BglII/XhoI site of pMItom. The vectors were transfected into M.mζ-SBP cells, yielding the M.mζ-SBP/mζ-GFP, M.mζ-SBP/mζ-YN, and M.mζ-SBP/mζ-CC cell lines. The human T cell line 31-13.scTCRβ has been described (24). All cells were cultured in complete RPMI 1640 medium supplemented with 5% fetal calf serum.
Treatments, Cell Lysis, Immunoprecipitation, and Immunoblotting
For actin depolymerization, 1 or 5 μg/ml latrunculin A was used at 37 °C for 30 min. For cholesterol depletion and loading, treatments with 2 mm methyl-β-cyclodextrin (mβCD) for 2 min or 20 μg/ml cholesterol complexed to mβCD for 3 h (both at 37 °C) were performed. The cholesterol concentration in lysates was measured using the Amplex-Red cholesterol assay kit (Invitrogen). Serial lysis was performed by resolubilizing the cellular and membrane material after each 15-min lysis and 15-min centrifugation step (14,000 × g) three times by 1% saponin and subsequently by 0.5% Brij96 in the lysis buffer containing 20 mm Tris-HCl (pH 8), 137 mm NaCl, 2 mm EDTA, 10% glycerol, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mm PMSF. Pooled saponin lysates represented the nanoclustered TCR, whereas the rest of the TCR solubilized in Brij96 was monomeric (Fig. 1A). Anti-TCR IPs were performed with 4 μg of anti-CD3ϵ (145-2C11), anti-TCRβ (H57), or anti-TCRζ (448) antibodies at 4 °C for 4 h. Native, purified TCR preparations were separated on BN-PAGE as described (25). SDS-PAGE and immunoblotting were performed by conventional methods. Western blot quantifications were done with the LI-COR Odyssey infrared imager system or with the ImageJ program after chemoluminescence detection.
FIGURE 1.
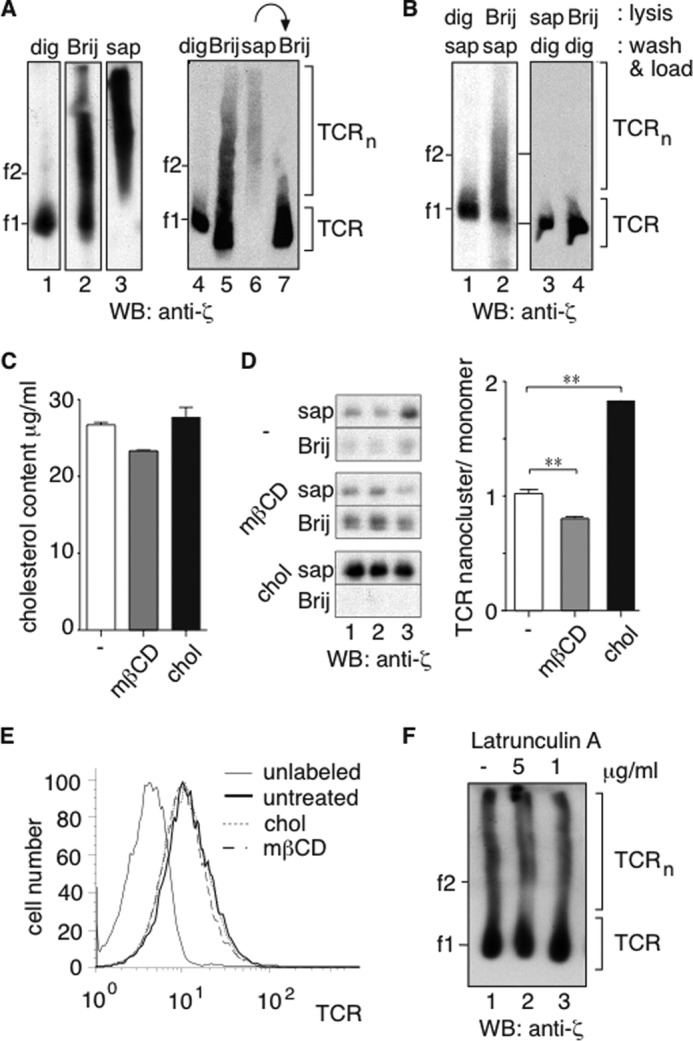
Membrane cholesterol levels control the amount of TCR nanoclustering. A, different detergents extract distinct TCR forms. M.mζ-SBP (lanes 1–3) and 31-13.scTCRβ (lanes 4–7) cells were solubilized with 1% digitonin (dig), 0.5% Brij96 (Brij), or 1% saponin (sap). In lane 7, the saponin-insoluble membranes were further lysed in 0.5% Brij96. Purified TCRs were analyzed by BN-PAGE and anti-ζ WB. The ferritin markers f1 (440 kDa) and f2 (880 kDa) are shown. B, the TCR of 31-13.scTCRβ cells was purified. The washing and the elution steps were performed with a detergent different from the one used for lysis. The TCR was analyzed as in A. C, the cholesterol content of total cell lysates of the cells treated with mβCD and cholesterol was measured with the Amplex-Red cholesterol assay kit. D, the cholesterol level controls TCR nanoclustering. M.mζ-SBP cells were left untreated or treated with mβCD or chol. Cells were lysed in 1% saponin, and the insoluble material was subsequently extracted in 0.5% Brij96. After TCR IP with anti-CD3ϵ and separation on reducing SDS-PAGE, the amount of fully assembled TCR in the nanoclustered (saponin) and monomeric (Brij96) pools was determined by anti-ζ WB. Triplicates are shown and the error bars represent the standard deviation. Band intensities were quantified with the LI-COR Odyssey infrared imager, and the ratio of the intensity of the saponin to the Brij96 sample is shown. An unpaired t test was performed; **, p < 0.01. E, the amount of surface TCR on cells treated in D was determined with anti-TCRβ staining and flow cytometry. F, actin is dispensable for the maintenance of TCR nanoclusters. M.mζ-SBP cells were treated with latrunculin A and lysed in 0.5% Brij96. The purified TCR was analyzed as in A.
MHCp-binding Experiments
To quantify the binding of MHCp tetramers and MHCp monomers to the different TCR forms, we used PbCS peptide (ABA)-H2-Kd as the MHC molecule and T1.4 hybridoma T cells that do not contain the co-receptor CD8 (26). T1.4 cells were treated with 2 mm mβCD for 30 min at 37 °C or with cholesterol complexed to mβCD as above and then serially lysed in first saponin and then digitonin-containing buffer in order to test for the disassembly of the TCR nanoclusters. TCR IP was performed with anti-CD3ϵ, and samples were subjected to SDS-PAGE, anti-ζ WB, and detection with a LI-COR Odyssey infrared imager. In parallel, treated and non-treated T1.4 cells were coupled with 250 nm PE-labeled PbCS peptide (ABA)-H2-Kd tetramers or PbCS peptide (ABA)-H2-Kd monomers for 2 h on ice. Subsequently, bound MHCp molecules were covalently cross-linked to the TCR by UV irradiation. In case of the MHCp monomer, the cells were stained with streptavidin-PE. Surface TCR expression was measured by staining with FITC-labeled anti-TCRβ. Fluorescence was measured by flow cytometry with a Calibur flow cytometer (BD Biosciences). Statistical analysis was done with the Prism4 software.
Protein Purification
Proteins were purified from T- or B-cells expressing the appropriate construct. 5 × 108 cells were lysed, and affinity purifications were performed using streptavidin-conjugated agarose (GE Healthcare) in the case of SBP-linked constructs or nitrophenol (NP)-conjugated agarose (Biosearch Technologies) in the case of the scTCRβ-containing TCR and the NP-specific B-cell antigen receptor (BCR). Elution was performed by incubation for 30 min at 4 °C with 2 mm free biotin or 2 mm nitro-iodo phenol, respectively, in BN lysis buffer containing 20 mm BisTris, pH 7.0, 500 mm ϵ-aminocaproic acid, 20 mm NaCl, 2 mm EDTA, 10% glycerol, and detergent as indicated.
Thin Layer Chromatography
A mixture of proteoliposomes (200 nmol) and 2.5% OptiPrep were added to a 3:1 mixture of CHCl3/methanol (1:1) and dried on room temperature. The pellets were resuspended in 20 μl of CHCl3 and loaded on a thin layer chromatography (TLC) plate (Silica gel 60, Merck). The TLC was run in CHCl3/methanol/NH4+/NH3 (9:7:2), and developed with Molybdenum Blue spray reagent.
TCR Reconstitution in Large Unilamellar Vesicles (LUVs)
Liposomes with different membrane compositions using soybean phosphatidylcholine, egg sphingomyelin (Lipoid), and cholesterol (Sigma-Aldrich) were prepared with the thin film method (27). The lipid film was prepared by mixing phosphatidylcholine (PC), SM, and cholesterol chloroform solutions at defined molar ratios in a round-bottom flask, removing solvent using a rotary evaporator, and drying under vacuum. The film was resuspended in phosphate-buffered salt solution (3 mm Na2HPO4, 2 mm KH2PO4, 50 mm NaCl, pH 7.0) to a lipid concentration of 20 mm. Subsequent extrusion through 200-nm (21 times) and 80-nm (51 times) polycarbonate membranes (Nuclepore, Whatman) resulted in LUVs. Lipid content was determined using a phosphorus assay and Cholesterol FS assay (DiaSys). The diameters of the vesicles fell between 100 and 200 nm, as determined by dynamic light scattering (Zetamaster S, Malvern Instruments) and electron microscopy. Approximately 0.1 μg of the purified TCR in 100 μl of 0.02% Triton X-100-containing buffer was mixed with 100 μl of 2 mm LUV preparation, and 40 μg of Triton X-100 was added. Samples were agitated for 30 min at 4 °C, and the detergent was removed by adsorption to 3 mg of BioBeads SM-2 (Bio-Rad) per sample at 4 °C overnight. The same procedure was used for the generation of TfR- or BCR-containing proteoliposomes.
Flow Cytometry and BiFC
Flow cytometry for analyzing TCR expression was performed by conventional methods with a Calibur flow cytometer. For BiFC experiments, proteoliposomes were formed by separately purifying TCRs bearing the C- and the N-terminal part of the fluorophore and adding them to the performed vesicles. As controls, TCR linked to a GFP was used, and TCR linked one half fluorophore was reconstituted in itself. After TCR reconstitution, the liposomes were lysed in lysis buffer containing 1% digitonin, and an IP was performed with anti-CD3ϵ (145-2C11) coupled to carboxylate-modified latex beads (Invitrogen). Fluorescence was measured by flow cytometry and analyzed with the FowJo 8.2 software. Statistical analysis was done with the Prism4 software.
Preparation of Giant Unilamellar Vesicles (GUVs)
GUVs were made by the electroformation technique. Proteoliposomes (3 g/liter) in droplets of 2 μl were deposited on indium tin oxide-covered glass slides. The film was partially dried overnight in a desiccator under saturated vapor pressure of a saturated NaCl solution. The indium tin oxide coverslip was assembled together with a second indium tin oxide coverslip into a flow chamber of homemade design, which was filled with a buffer containing 1 mm HEPES, 1 mm NaCl, pH 7.4. Alternating electric field (400 V/m, 10 Hz) was applied for 4 h at room temperature. GUVs were observed in the same chamber. Images were collected with a Zeiss 510 ConfoCor3 microscope using a water C-Apochromat ×40, numerical aperture 1.2 objective and avalanche photodiodes as detectors.
Photocholesterol Labeling Experiments
[3α-3H]6-Azi-5α-cholestan-3β-ol (photocholesterol) was synthesized, and the experiments were performed as described earlier (28). Jurkat cells were incubated for 16 h in lipid-free medium containing the photocholesterol-mβCD complex (5 μCi/ml) and UV-irradiated for 5 min. After cell lysis, IP and N-glycosidase F (Roche Applied Science) treatments were performed where indicated. The samples were subjected to SDS-PAGE and autoradiography.
RESULTS
Cholesterol Stabilizes TCR Nanoclusters
We established an efficient affinity purification procedure for the isolation of the native TCR from cellular lysates. We used mouse M.mζ-SBP cells derived from the 2B4 T cell hybridoma, expressing a SBP purification tag C-terminally fused to the ζ chain, and human 31-13.scTCRβ cells derived from the Jurkat T cell line, which contain a single chain variable fragment of an anti-NP antibody linked to the TCRβ chain (scTCRβ) (4, 24). We analyzed the effect of different detergents on the integrity of TCR nanoclusters using BN-PAGE (4, 25) in the case of both human and mouse T cells (Fig. 1A). 1% digitonin extracted the mouse TCR in monomeric form (lane 1), whereas 0.5% Brij96 extracted TCR monomers and nanoclusters (lane 2) as reported before (4, 5). In contrast, 1% saponin extracted only the nanoclusters (lane 3). The same held true for the human TCR (lanes 4–6). Brij96 extracted the remaining monomeric TCR from the saponin-insoluble membranes (lane 7). As controls, digitonin- or Brij96-extracted TCRs did not aggregate when kept in a saponin-containing buffer (Fig. 1B, lanes 1 and 2), whereas saponin- and Brij96-solubilized TCR nanoclusters broke down to monomeric TCR when digitonin was added (lanes 3 and 4). The detergent dependence of the TCR nanocluster stability suggested that membrane lipids play a role in the formation of the nanoclusters.
Previously, using harsh conditions to extract cholesterol from cells, we suggested that cholesterol might stabilize the TCR nanoclusters (4). To test whether cholesterol is involved in TCR nanoclustering, we reduced the amount of cholesterol using a short term low dose mβCD treatment (2 mm for 2 min at 37 °C), which does not extract membrane proteins (29). To increase the cholesterol content of the membranes, mβCD-complexed cholesterol was used. The change of the cholesterol content of total cell lysates was measured with the Amplex-Red cholesterol assay kit (Fig. 1C). To quantify the ratio of nanoclustered to monomeric TCRs, we lysed the cells serially in 1% saponin and 0.5% Brij96 (as in Fig. 1A). Anti-CD3ϵ immunopurification and SDS-PAGE were performed on the two fractions, and the amount of assembled TCR was quantified by anti-ζ WB (Fig. 1D). The nanocluster/monomer ratio decreased upon cholesterol depletion and increased upon cholesterol loading. The treatments did not extract TCRs from the membrane (Fig. 1E). These results show that TCR nanoclustering is reversible and dependent on the cholesterol content of the plasma membrane. Disrupting actin filaments by latrunculin A treatment did not disassemble TCR nanoclusters (Fig. 1F), indicating that the actin cytoskeleton is dispensable for the maintenance of the TCR nanoclusters.
The TCRβ Subunit Binds to Cholesterol in Living T Cells
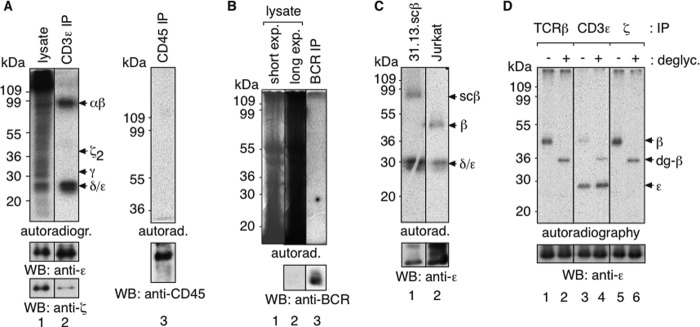
Because we found that TCR nanoclusters depend on the presence of cholesterol, we used a biochemical approach to test whether the TCR can bind to cholesterol in vivo. To this end, we used a photoactivatable radioactive analog of cholesterol (28). Photocholesterol mimics cholesterol in several assays and has been used to identify proteins that specifically interact with cholesterol (30, 31). Here, Jurkat cells were grown in the presence of photocholesterol, and upon activation by UV light, photocholesterol cross-linked to molecules in close proximity. The TCR and non-assembled CD3 dimers were immunoprecipitated with anti-CD3 antibodies from cellular lysates and analyzed by non-reducing SDS-PAGE and autoradiography (Fig. 2A). A large number of proteins were cross-linked to cholesterol in the lysate, but only two bands were detected in case of the purified TCR, corresponding to the disulfide-linked TCRαβ dimer and CD3δ and/or CD3ϵ. When isolated from the same cellular lysates, the highly abundant cell surface protein CD45 did not bind cholesterol (lane 3). Using the same method, we studied the interaction between cholesterol and the antigen receptor of A20 B cells, and we found that cholesterol did not cross-link to the BCR (Fig. 2B).
FIGURE 2.
Cross-linking of radioactive cholesterol to the TCR in live T cells. A, TCRαβ and CD3ϵ or CD3δ cross-link to photocholesterol. Jurkat cells were cultured for 16 h with 5 μCi/ml photocholesterol, and the diazirin group was activated by UV light. The Brij96 lysate and anti-CD3ϵ and anti-CD45 IPs were separated on non-reducing SDS-PAGE. The autoradiograms (top) and WBs are shown. Exposure times in lanes 1 and 2 are the same. B, the BCR of A20 B cells does not cross-link to the photocholesterol. A20 B cells were treated as in A and lysed in Brij96. BCR IP and non-reducing SDS-PAGE were performed. Autoradiography and anti-light chain WB of the lysate and the purified BCR are shown. A short exposure (exp) of the lysate shows discrete bands, and the black signal on lane 2 (lysate) resulted from the same exposure time as lane 3 (BCR IP). C, TCRβ binds to photocholesterol. 31-13.scTCRβ (lane 1) and Jurkat cells (lane 2) were treated as in A. After cell lysis and anti-CD3ϵ IP, samples were analyzed on reducing SDS-PAGE. The autoradiogram and anti-CD3ϵ WB are shown. D, non-assembled CD3ϵ cross-links to photocholesterol. Jurkat cells were treated as in A, and anti-TCRβ, anti-CD3ϵ, and anti-ζ IPs were performed. The purified proteins were left untreated (lanes 1, 3, and 5) or subjected to N-glycosidase F treatment (lanes 2, 4, and 6). The autoradiogram and the anti-CD3ϵ WB are shown.
To identify which subunits of the TCR bound cholesterol, we had to distinguish between the TCRα and TCRβ and the CD3δ and CD3ϵ chains. First, we used 31-13.scTCRβ cells, which bear the larger scTCRβ chain (4, 24) and Jurkat T cells expressing a WT TCR. After labeling with photocholesterol, a TCR IP was performed with anti-CD3ϵ, and samples were subjected to reducing SDS-PAGE and autoradiography. In addition to the signal from CD3δ and/or CD3ϵ, a protein with an apparent molecular mass of 45 kDa (in the case of Jurkat) and 80 kDa (in the case of the 31-13.scTCRβ cells; corresponding to the scTCRβ) was detected (Fig. 2C). No additional band corresponding to the size of TCRα was detected, indicating that it was TCRβ that bound to photocholesterol. CD3ϵ, in contrast to CD3δ, is not glycosylated. Therefore, we used a deglycosylation assay to distinguish between CD3ϵ and CD3δ. An IP was performed from photocholesterol-labeled lysates of Jurkat cells using anti-TCRβ, anti-CD3ϵ, or anti-ζ antibodies (Fig. 2D). Each sample was left untreated or deglycosylated using N-glycosidase F. The increased mobility of the deglycosylated TCRβ in the autoradiogram indicates efficient deglycosylation. The mobility of the low molecular weight radioactive band did not change, showing that it represents CD3ϵ. However, cholesterol-labeled CD3ϵ was only present in the anti-CD3ϵ IPs (lanes 3 and 4), although CD3ϵ was present in all IPs, as seen in the anti-CD3ϵ WB. This suggests that CD3ϵ bound to cholesterol was not part of the TCR complex and was probably located intracellularly. Therefore, within the TCR complex, TCRβ is the only subunit that associates with cholesterol in living T cells.
TCR Nanoclusters Increase the Avidity toward Multivalent MHCp
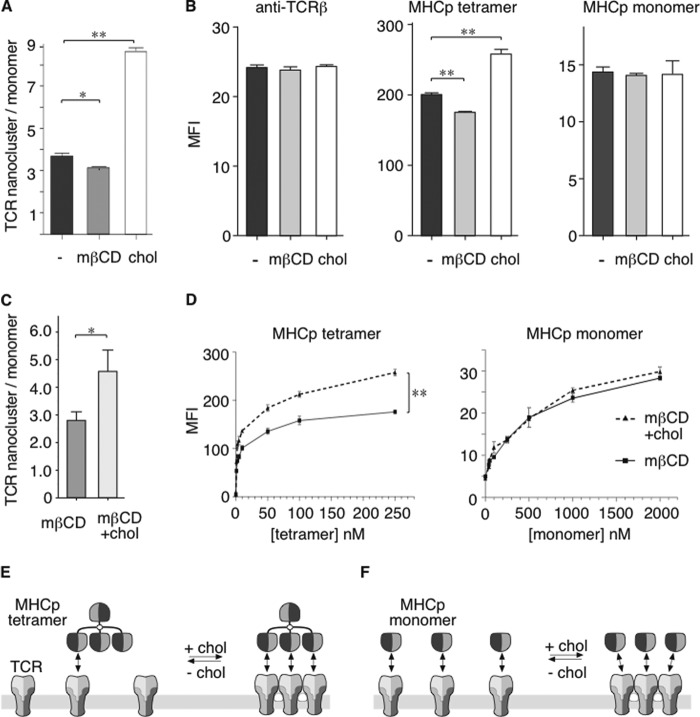
Next, we investigated whether TCR nanoclustering influences MHCp binding to the TCR. We measured the binding of fluorescent PbCS peptide (ABA)-H2-Kd (MHCp) to CD8-negative T1.4 T cells, because this system allows exact quantifications due to the fact that the MHCp can be covalently cross-linked to the TCR (24, 26). As above, we altered the TCR nanocluster/monomer ratio by extracting or loading cholesterol (Fig. 3A). The total amount of TCR in each treatment remained unchanged (Fig. 3B, left). However, there was a significant reduction in MHCp tetramer binding to cholesterol-depleted cells and an increased binding to cells where cholesterol was added (middle). Importantly, the binding of MHCp monomers was unaffected (right).
FIGURE 3.
Membrane cholesterol levels influence the binding of MHC-peptide tetramers to the TCR. A, T1.4 T cells were left untreated (−) or treated with mβCD or chol, and after serial lysis, TCR nanoclusters (saponin-extractable) and monomers (resistant to saponin lysis) were quantified as in Fig. 1D. B, T1.4 cells were treated as in A and stained with an FITC-labeled anti-TCRβ antibody (left), PE-labeled PbCS peptide (ABA)-H2-Kd tetramers (middle), or PbCS peptide(ABA)-H2-Kd monomers followed by streptavidin-PE (right). Fluorescence was measured by flow cytometry. The mean fluorescence intensity and S.E. of triplicates are shown. C, the re-addition of cholesterol after mβCD treatment restores TCR nanoclustering. The nanocluster/monomer ratio was quantified as described in the legend to Fig. 1D after mβCD treatment and mβCD treatment followed by the re-addition of cholesterol. Mean and S.E. (error bars) of triplicates are shown. D, binding curves of MHCp tetramers and monomers to T1.4 cells were taken after mβCD treatment and cholesterol replenishment following mβCD treatment. Mean fluorescence intensity and S.E. of triplicates are shown. Unpaired t tests were performed (*, p < 0.05; **, p < 0.01). E, cholesterol-driven nanoclustering of TCRs results in enhanced avidity to MHCp tetramers, due to a multiplicity in the number of binding sites. F, on the other hand, the TCR-MHCp affinity stays unaltered, as detected by MHCp monomer binding.
Moreover, the re-addition of cholesterol after previous extraction reversed the effect of mβCD treatment on MHCp tetramer binding. When, after treating the T1.4 cells with mβCD, exogenous cholesterol was added, TCR nanoclusters were restored in cells reloaded with cholesterol (Fig. 3C), whereas the amount of surface TCR remained unchanged (data not shown). A dose-response curve of MHCp tetramer binding to the cells shows that cholesterol treatment increased the tetramer binding capacity 4–5-fold (Fig. 3D, left). Again, the binding of MHCp monomers was unchanged (right). In conclusion, TCR nanoclustering significantly enhanced the TCR-MHCp avidity, represented by the MHCp tetramer binding (Fig. 3E), but did not change the TCR-MHCp affinity, as measured by MHCp monomer binding (Fig. 3F).
Reconstitution of the TCR in Large Unilamellar Vesicles
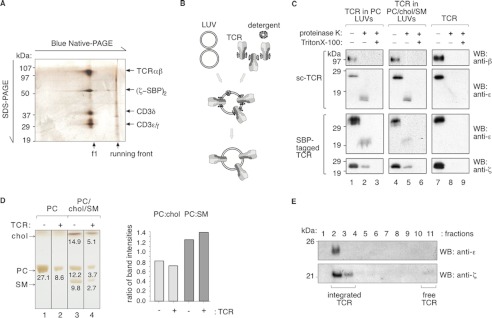
To learn more about the role of lipids in TCR nanoclustering, we reduced the complexity of cellular membranes using a synthetic biology approach. The native digitonin-solubilized TCR from M.mζ-SBP or 31-13.scTCRβ cells was purified (Fig. 4A), and it was reconstituted in LUVs with different compositions of PC, cholesterol, and SM (Fig. 4B). The orientation of the integrated TCR was determined by proteinase K digestion. For WB analysis, we used an anti-TCRβ antibody, which binds to the extracellular part of TCRβ, and anti-CD3ϵ and anti-ζ antibodies to detect the intracellular domains of CD3ϵ and -ζ. Proteinase K treatment abolished the TCRβ signal (Fig. 4C), indicating that the extracellular part of the TCR is located extraliposomally. The size of CD3ϵ decreased upon proteinase K treatment due to the loss of the extracellular domain, and a substantial fraction of ζ was resistant to digestion. Without integration into LUVs (lane 8) and after integration and subsequent lysis of the LUVs (lanes 3 and 6), the TCR was completely digested. These data show that most of the TCR was integrated into the liposomes in the same orientation as on the cell surface (i.e. the extracellular parts are extraliposomal) (Fig. 4B). Furthermore, we found that the majority of the TCR was integrated in the lipid bilayer, and the reconstitution process did not alter the lipid content of the LUVs (Fig. 4, D and E).
FIGURE 4.
Integration of the TCR into LUVs. A, purity of the SBP-tagged purified TCR. The digitonin-solubilized TCR from M.mζ-SBP cells was purified with streptavidin-coupled beads, eluted with biotin, and analyzed by two-dimensional BN-/SDS-PAGE and silver staining. B, experimental procedure. LUVs were prepared, and purified native monomeric TCR was reconstituted using a sublytic concentration of Triton X-100 and subsequent detergent adsorption to polystyrene beads. C, TCRs integrate unidirectionally into the liposomes. The purified TCR from 31-13.scTCRβ (sc-TCR) and M.mζ-SBP cells (SBP-tagged TCR) (lanes 7–9) and TCR-containing proteoliposomes comprising PC (lanes 1–3) or PC/chol/SM (40:30:30 mol %) (lanes 4–6) were treated with 100 μg/ml proteinase K in the presence or absence of 1% Triton X-100 on 37 °C for 1 h, as indicated. Reducing SDS-PAGE and WB using ectodomain-specific anti-TCRβ and cytoplasmic tail-specific anti-CD3ϵ and anti-ζ antibodies were performed. D, reconstitution of the TCR in preformed LUVs did not change their lipid composition. Sucrose gradient centrifugation was performed with TCR-containing proteoliposomes of pure PC (lane 2) and of PC/chol/SM (4:3:3 weight %); lane 4) and with LUVs of the same composition (lanes 1 and 3). Fraction 2, containing the LUVs (see E), was analyzed by thin layer chromatography. The bands corresponding to PC, cholesterol, and SM along with relative band intensity are indicated. The quantification demonstrates that the ratios of PC to SM and PC to cholesterol are unchanged after the TCR reconstitution. E, sucrose gradient centrifugation was performed with TCR-containing proteoliposomes of pure PC, and the fractions were subjected to SDS-PAGE followed by an anti-ϵ and anti-ζ WB. Fractions 2 and 3 contain the LUVs and the TCR, indicating that the TCR was integrated into the LUVs.
The TCR Forms Dimers in LUVs Composed of PC, Cholesterol, and Sphingomyelin
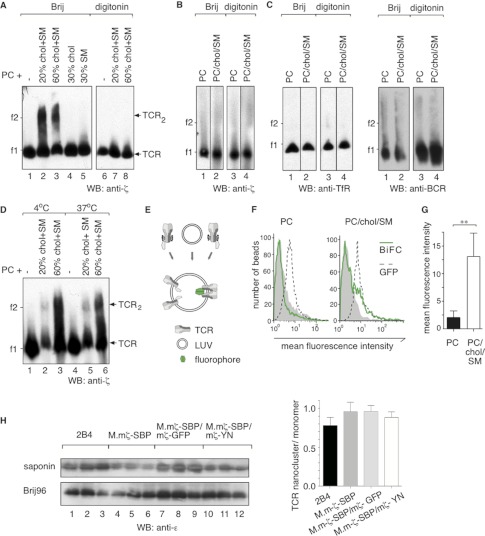
To study TCR nanoclustering, we chose the SBP-tagged TCR for its high purity and yield (Fig. 4A). The proteoliposomes of different lipid composition were sedimented, lysed in 1% saponin supplemented with 0.5% Brij96 to maintain or with 1% digitonin to disrupt the TCR nanoclusters, and analyzed by BN-PAGE. In liposomes containing a natural mixture of PC, the TCR remained monomeric (Fig. 5A, lane 1), whereas in PC/chol/SM liposomes (in either 80:10:10 or 40:30:30 mol % ratio) it formed dimers (lanes 2 and 3). The TCR remained monomeric in PC liposomes containing either 30 mol % cholesterol (lane 4) or 30 mol % SM (lane 5), showing that formation of the TCR dimers requires both cholesterol and SM. TCR dimers disassembled to monomers upon digitonin treatment (lanes 7 and 8). To exclude the possibility that the TCR dimers were the result of TCR aggregation after the lysis of the proteoliposomes, we mixed the TCR with LUVs but did not allow integration before lysing the vesicles and performing BN-PAGE. As expected, TCR dimers did not form without integration of the TCR into the bilayer of the PC/chol/SM-LUVs (Fig. 5B).
FIGURE 5.
Nanocluster analysis of liposome-reconstituted TCR, TfR, and BCR. A, The TCR forms dimers in PC/chol/SM liposomes. M.mζ-SBP cells were lysed in digitonin, and the purified TCR was reconstituted in liposomes of the lipid compositions indicated (mol %). The proteoliposomes were lysed in 1% saponin supplemented with 0.5% Brij96 or 1% digitonin and subjected to BN-PAGE and anti-ζ WB. B, liposomes of the indicated composition were mixed with the purified TCR without integration of the TCR. The liposomes and the TCR were lysed in 1% saponin supplemented with 0.5% Brij96 or 1% digitonin and analyzed by BN-PAGE and anti-ζ WB. An increased amount of TCR and long exposure times were used to detect the TCR. C and D, the SBP-tagged TfR (C) and NP-specific BCR (D) were purified, and the native proteins were reconstituted in liposomes of the indicated composition. After the lysis of the proteoliposomes in 1% saponin supplemented with 0.5% Brij96 or 1% digitonin, BN-PAGE and anti-TfR or anti-BCR WB were performed. D, proteoliposomes as in A were kept for 2 h at either 4 °C or at 37 °C, lysed in 1% saponin supplemented with 0.5% Brij96, and subjected to BN-PAGE and anti-ζ WB. E, experimental procedure. Purified TCRs bearing a ζ chain fused either to the N-terminal part of Venus or to the C-terminal part of enhanced cyan fluorescent protein were reconstituted together in liposomes. The amount of BiFC refers to the amount of dimerized TCRs in the vesicles. F, the TCR dimerizes in the presence of PC, cholesterol, and SM. Proteoliposomes containing both TCR constructs linked to the half-fluorophores (E) in a pure PC (left) or a PC/chol/SM (40:30:30 mol %) liposome (right) were lysed in 1% digitonin. The TCR was captured on anti-CD3ϵ coupled latex beads, and the fluorescence was measured by flow cytometry (green line). Proteoliposomes containing a TCR with GFP (dashed line) or a TCR with the N-terminal part of Venus only (gray) were used as positive and negative controls. G, the average of three experiments as in F is shown. A paired t test was performed (**, p < 0.01). H, cells with TCRs bearing the different tags were lysed in 1% saponin, and the insoluble material was subsequently extracted in 0.5% Brij96. After TCR-IP with anti-CD3ϵ and separation on reducing SDS-PAGE, the ratio of the nanoclustered (saponin) and monomeric (Brij96) TCR pools was determined by anti-CD3ϵ WB. Triplicates are shown, and the quantifications are given, and the error bars represent standard deviation.
To assess whether dimerization in PC/chol/SM LUVs observed for the TCR is a general feature of TM proteins, we reconstituted the native purified TfR purified from the parental cell line of the M.mζ-SBP cells and the purified BCR in PC or PC/chol/SM (40:30:30 mol %) LUVs. Neither TfR nor BCR multimers were detected (Fig. 5C). We also studied the effect of temperature on TCR clustering. Although we usually performed the reconstitution at 4 °C, we obtained the same results at 37 °C (Fig. 5D). In this particular experiment, the LUVs composed of PC contained more TCR than those composed of PC/chol/SM. The fact that TCR dimers formed only in the case of the ternary mixture indicated that TCR dimerization did not occur due to the congestion of a higher amount of proteins in the ld phase.
Furthermore, we applied BiFC as a detergent-independent read-out of TCR dimerization (32, 33). To the C terminus of the ζ chain, we fused either the N-terminal part of Venus or the C-terminal part of enhanced cyan fluorescent protein. The fusion proteins were individually expressed in M.mζ-SBP cells, which already expressed SBP-tagged ζ. Due to ζ dimer formation in the TCR complex, a portion of TCRs contained a half of the fluorophore after TCR purification via the SBP tag. Monomeric TCRs were purified from cellular lysates and mixed at equimolar concentrations for reconstitution into LUVs. The formation of TCR dimers in the LUVs allows the assembly of the fluorescent domain (Fig. 5E). Significantly higher BiFC fluorescence was detected from PC/chol/SM LUVs (40:30:30 mol %) as compared with PC LUVs (Fig. 5, F and G). These results confirm that the presence of cholesterol and SM in PC liposomes is sufficient for the formation of TCR dimers. As a control, we show that the tags appended to ζ do not influence the nanoclustering of the respective TCRs (Fig. 5H).
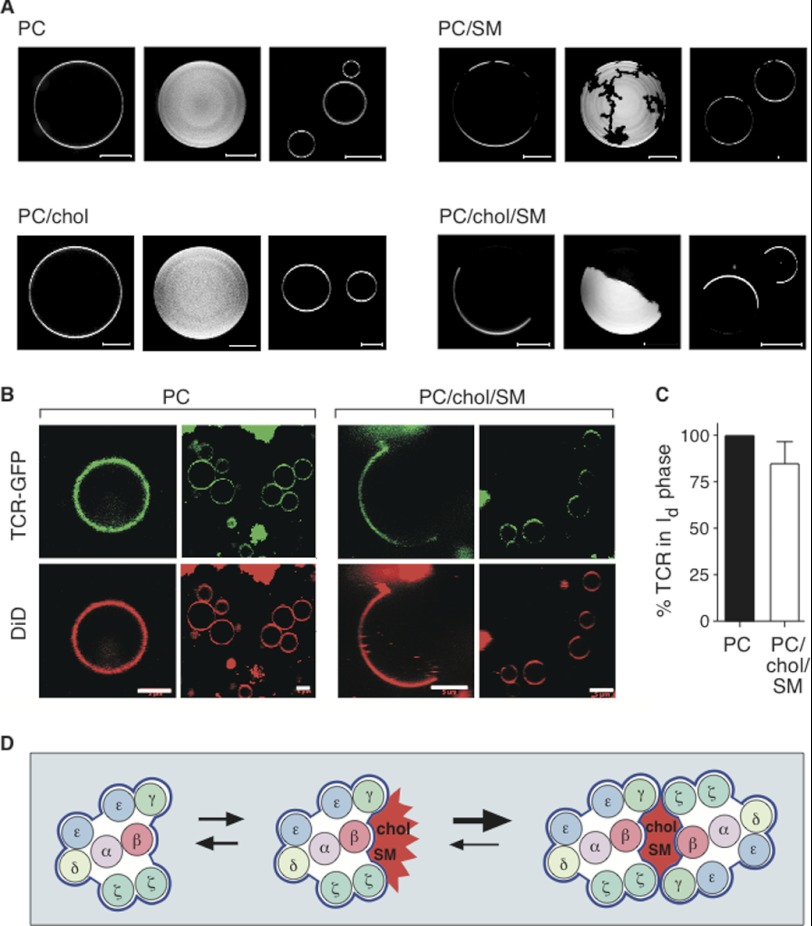
The TCR Localizes in the Liquid-disordered Phase of GUVs
The PC and SM used in this study consisted of a spectrum of acyl chain lengths. We determined the phase behavior of the bilayers in GUVs by confocal imaging. No phase separation occurred in the case of PC or PC and cholesterol (70:30 mol %), whereas in the mixture of PC, cholesterol, and SM (40:30:30 mol %) we found an lo and an ld phase (Fig. 6A). To determine TCR localization in the artificial membranes, we reconstituted the purified GFP-coupled TCR in PC or PC/chol/SM (40:30:30 mol %) LUVs supplemented with 0.05 mol % DiD, a dye staining the ld domain, and grew GUVs from the proteoliposomes. Confocal imaging revealed that the distribution of the TCR was homogenous in PC liposomes, whereas in PC/chol/SM liposomes, the TCR colocalized with the ld domain (Fig. 6B). A quantification of the distribution of the GFP-coupled TCR showed that 83 ± 11% of the TCRs was present in the ld domain in PC/chol/SM GUVs (Fig. 6C).
FIGURE 6.
The TCR localizes in the ld domain of GUVs. A, confocal images of GUVs grown from different lipid mixtures of PC, cholesterol, and SM are displayed. Fluorescent DiI-C18 was used as a marker for ld domains. Confocal slices of the mixtures are shown in the first and third column. Multiple slices of the GUVs in the first column make up the projections shown in the second column. As expected, no phase separation occurred in PC or PC/chol (70:30 mol %) vesicles. In the case of PC/SM (70:30 mol %) dark finger-shaped structures were observed, representing a solid ordered domain. In PC/chol/SM (40:30:30 mol %) domains occupied half of the GUV surface, indicating the coexistence of an lo and an ld phase. The diffusion of the DiI-C18 lipid marker was measured by fluorescence correlation spectroscopy. Diffusion was 3 times faster in bright (ld) domains as compared with the dark (lo) domains (data not shown). B, the GFP-labeled TCR was reconstituted in LUVs composed of PC or PC/chol/SM (40:30:30 mol %). GUVs were generated from these proteoliposomes, and confocal images were taken. The ld domain was identified by DiD labeling. Scale bars, 5 μm. C, the peak fluorescence intensities of the GFP-tagged TCR in the ld and lo phases (F_ld and F_lo) was quantified using line scans through the GUVs. The percentage of partitioning (%ld = F_ld/(F_ld + F_lo) was calculated from 10 GUVs as described (51), the error bars represent standard deviation. D, our model of TCR dimerization. The arrangement of the transmembrane (TM) domains of the TCR subunits within the TCR complex is shown, in line with a recent publication (47). The monomeric TCR localizes in the non-raft phase (blue, left). Cholesterol specifically binds to the TCRβ subunit, which results in the recruitment of SM (middle). The presence of a cholesterol and SM islet (red) at the TM surface in the ld phase is energetically unfavorable. If the TCR forms dimers, cholesterol and SM are shielded from the ld phase, stabilizing the TCR dimer (right). The black arrows indicate the transient nature of these interactions.
DISCUSSION
The molecular mechanism of TCR nanoclustering is poorly understood. In this study, we used T cells and a synthetic biology approach to investigate the role of lipids in antigen-independent TCR dimerization. We established a procedure to purify the complete TCR complex in native form and to reconstitute it in LUVs of different lipid composition. We found that TCR dimers formed in PC/chol/SM liposomes but not in binary mixtures or in PC alone. The effect was specific to the TCR, because the TfR and BCR remained monomeric under all conditions. Because the proteoliposomes did not contain proteins other than the TCR, we concluded that the lipid environment induced dimer formation.
A number of specific lipid-protein interactions have been revealed by x-ray crystallography (34, 35), radioactive photolipids (28, 36), and mutagenesis analyses (37, 38). Ordered cholesterol molecules were shown in the structure of metarhodopsin (39) and of the β2-adrenergic G protein-coupled receptor (40, 41). Therefore, we considered that a direct interaction with cholesterol might cause TCR dimerization. Indeed, in live T cells, photoactivatable cholesterol (28) cross-linked to the TCRβ chain but not to any other subunits of the assembled TCR. It also did not cross-link to the BCR or to CD45. This suggests that the TCR-cholesterol interaction is remarkably site-specific and that cholesterol is a nonannular lipid-binding partner of the TCR (i.e. cholesterol might bind stably to the TCR).
Annular lipids were suggested to mediate intra- and intermolecular interactions between the TM regions involved (35, 42). In analogy, we propose a mechanism of TCR dimerization, in which cholesterol and SM serve as structural components of a TCR dimer (Fig. 6D). In most (43, 44) but not all (45) reports, the TCR was found in the non-raft phase in resting T cells. Likewise, we show that the TCR is localized in the ld phase in our liposomes. Thus, we suggest that cholesterol binds to TCRβ in the ld phase of the plasma membrane. Because SM preferentially interacts with cholesterol (46), cholesterol recruits SM to the TCR TM surface (Fig. 6D). The subsequent formation of TCR dimers is energetically favored, because it leads to the shielding of cholesterol-SM from the ld phase. In addition, protein-protein interactions between TCR subunits might stabilize the TCR-TCR association (9, 47).
This model is supported by our findings that cholesterol is required for the maintenance of TCR nanoclusters. Earlier, we studied the effect of a high dose (4) and here of a mild mβCD treatment and cholesterol loading of T cells, and we found a correlation between cholesterol concentration in the membranes and TCR nanoclustering. Furthermore, disrupting the TCR-cholesterol interaction with digitonin led to disassembly of the TCR nanoclusters.
Cholesterol amounts were measured in total cell lysates; thus, the relevant concentrations of cholesterol in the plasma membrane are unknown. However, we measured an effect of cholesterol addition or removal on the ligand-binding activity of the surface TCR (Fig. 3), demonstrating that cholesterol levels were changed in the plasma membrane.
The TCR in LUVs formed dimers but not multimers. In contrast, on the surface of T cells, larger nanoclusters are present (4, 10). Because both dimers (in LUVs) and nanoclusters (in cells) are cholesterol-dependent, we suggest that the nanoclusters derive from TCR dimers. It is unclear if the lack of nanoclusters in LUVs is due to the experimental settings (e.g. not more than two TCRs are present in one LUV) or if TCR dimers and nanoclusters form along a different mechanism.
We propose that TCR dimerization is a dynamic process in which the equilibrium between the monomers and dimers is regulated by the concentration of cholesterol and SM (Fig. 6D). The cholesterol and sphingolipid content in activated T cells is higher than in naive T cells (48). This might contribute to increased TCR nanoclustering in activated T cells as compared with naive T cells (9).
Activated T cells possess enhanced avidity, but not affinity, to MHCp tetramers when compared with naive T cells (49). Increased avidity was dependent on cholesterol (45, 49), but the underlying mechanism was unknown. Using MHCp tetramers and monomers, we show that cholesterol-mediated TCR nanoclustering translates into a higher antigen-TCR avidity. This suggests that upon activation and differentiation, naive T cells up-regulate their cholesterol and SM content, thereby expressing more nanoclustered TCRs. As a result, activated and memory T cells show enhanced avidity to MHCp (avidity maturation). In fact, activated and memory T cells posses increased sensitivity to low antigen levels as compared with naive T cells (50). Our data and the new model for TCR dimerization presented here contribute to the understanding of TM protein clustering as well as to the consequences of specific binding of lipids to TM proteins.
Acknowledgments
We thank Thomas Benzing, Bernard Schermer, and Norbert Blank for help in establishing lipid assays and Ünal Coscun and Balbino Alarcon for carefully reading the manuscript. We thank Sabine Barnert and Melanie Ficht for technical help and Lipoid GmbH (Ludwigshafen, Germany) for the generous gift of lipids.
This work was supported by Deutsche Forschungsgemeinschaft Grants EXC294, SFB620, and SCHA 976/2-1 and by the European Union through Grant FP7/2007–2013 (SYBILLA).
- TCR
- T-cell antigen receptor
- GUV
- giant unilamellar vesicle
- ld
- liquid-disordered
- lo
- liquid-ordered
- LUV
- large unilamellar vesicle
- mβCD
- methyl-β-cyclodextrin
- MHCp
- MHC-peptide
- PC
- phosphatidylcholine
- SM
- sphingomyelin
- BN
- blue native
- SBP
- streptavidin-binding peptide
- TfR
- transferrin receptor
- BiFC
- bifluorescence complementation
- IP
- immunoprecipitation
- WB
- Western blot
- PE
- phosphatidylethanolamine
- NP
- nitrophenol
- chol
- cholesterol
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Davis M. M., Boniface J. J., Reich Z., Lyons D., Hampl J., Arden B., Chien Y. (1998) Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 16, 523–544 [DOI] [PubMed] [Google Scholar]
- 2. Malissen B. (2003) An evolutionary and structural perspective on T cell antigen receptor function. Immunol. Rev. 191, 7–27 [DOI] [PubMed] [Google Scholar]
- 3. Call M. E., Pyrdol J., Wiedmann M., Wucherpfennig K. W. (2002) The organizing principle in the formation of the T cell receptor-CD3 complex. Cell 111, 967–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schamel W. W., Arechaga I., Risueño R. M., van Santen H. M., Cabezas P., Risco C., Valpuesta J. M., Alarcón B. (2005) Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 202, 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Swamy M., Minguet S., Siegers G. M., Alarcón B., Schamel W. W. (2007) A native antibody-based mobility-shift technique (NAMOS-assay) to determine the stoichiometry of multiprotein complexes. J. Immunol. Methods 324, 74–83 [DOI] [PubMed] [Google Scholar]
- 6. Swamy M., Siegers G. M., Fiala G. J., Molnar E., Dopfer E. P., Fisch P., Schraven B., Schamel W. W. (2010) Stoichiometry and intracellular fate of TRIM-containing TCR complexes. Cell Commun. Signal. 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernández-Miguel G., Alarcón B., Iglesias A., Bluethmann H., Alvarez-Mon M., Sanz E., de la Hera A. (1999) Multivalent structure of an αβT cell receptor. Proc. Natl. Acad. Sci. U.S.A. 96, 1547–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Exley M., Wileman T., Mueller B., Terhorst C. (1995) Evidence for multivalent structure of T-cell antigen receptor complex. Mol. Immunol. 32, 829–839 [DOI] [PubMed] [Google Scholar]
- 9. Kumar R., Ferez M., Swamy M., Arechaga I., Rejas M. T., Valpuesta J. M., Schamel W. W., Alarcon B., van Santen H. M. (2011) Increased sensitivity of antigen-experienced T cells through the enrichment of oligomeric T cell receptor complexes. Immunity 35, 375–387 [DOI] [PubMed] [Google Scholar]
- 10. Lillemeier B. F., Mörtelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M. (2010) TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. James J. R., McColl J., Oliveira M. I., Dunne P. D., Huang E., Jansson A., Nilsson P., Sleep D. L., Gonçalves C. M., Morgan S. H., Felce J. H., Mahen R., Fernandes R. A., Carmo A. M., Klenerman D., Davis S. J. (2011) The T cell receptor triggering apparatus is composed of monovalent or monomeric proteins. J. Biol. Chem. 286, 31993–32001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. James J. R., White S. S., Clarke R. W., Johansen A. M., Dunne P. D., Sleep D. L., Fitzgerald W. J., Davis S. J., Klenerman D. (2007) Single-molecule level analysis of the subunit composition of the T cell receptor on live T cells. Proc. Natl. Acad. Sci. U.S.A. 104, 17662–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A., Tokunaga M., Dustin M. L., Saito T. (2005) Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat. Immunol. 6, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 14. Alarcón B., Swamy M., van Santen H. M., Schamel W. W. (2006) T-cell antigen-receptor stoichiometry. Preclustering for sensitivity. EMBO Rep. 7, 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Simons K., Ikonen E. (1997) Functional rafts in cell membranes. Nature 387, 569–572 [DOI] [PubMed] [Google Scholar]
- 16. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 17. Sharma P., Varma R., Sarasij R. C., Ira, Gousset K., Krishnamoorthy G., Rao M., Mayor S. (2004) Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell 116, 577–589 [DOI] [PubMed] [Google Scholar]
- 18. Veiga M. P., Arrondo J. L., Goñi F. M., Alonso A., Marsh D. (2001) Interaction of cholesterol with sphingomyelin in mixed membranes containing phosphatidylcholine, studied by spin-label ESR and IR spectroscopies. A possible stabilization of gel-phase sphingolipid domains by cholesterol. Biochemistry 40, 2614–2622 [DOI] [PubMed] [Google Scholar]
- 19. Björkbom A., Róg T., Kankaanpää P., Lindroos D., Kaszuba K., Kurita M., Yamaguchi S., Yamamoto T., Jaikishan S., Paavolainen L., Päivärinne J., Nyholm T. K., Katsumura S., Vattulainen I., Slotte J. P. (2011) N- and O-methylation of sphingomyelin markedly affects its membrane properties and interactions with cholesterol. Biochim. Biophys. Acta 1808, 1179–1186 [DOI] [PubMed] [Google Scholar]
- 20. Lillemeier B. F., Pfeiffer J. R., Surviladze Z., Wilson B. S., Davis M. M. (2006) Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 103, 18992–18997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R. S., Kondo J., Fujiwara T. (2005) Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid. High-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 34, 351–378 [DOI] [PubMed] [Google Scholar]
- 22. Sieber J. J., Willig K. I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G., Rammner B., Eggeling C., Hell S. W., Grübmuller H., Lang T. (2007) Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076 [DOI] [PubMed] [Google Scholar]
- 23. Keefe A. D., Wilson D. S., Seelig B., Szostak J. W. (2001) One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 23, 440–446 [DOI] [PubMed] [Google Scholar]
- 24. Minguet S., Swamy M., Alarcón B., Luescher I. F., Schamel W. W. (2007) Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity 26, 43–54 [DOI] [PubMed] [Google Scholar]
- 25. Swamy M., Siegers G. M., Minguet S., Wollscheid B., Schamel W. W. (2006) Blue native polyacrylamide gel electrophoresis (BN-PAGE) for the identification and analysis of multiprotein complexes. Sci. STKE 2006, pl4. [DOI] [PubMed] [Google Scholar]
- 26. Luescher I. F., Vivier E., Layer A., Mahiou J., Godeau F., Malissen B., Romero P. (1995) CD8 modulation of T-cell antigen receptor-ligand interactions on living cytotoxic T lymphocytes. Nature 373, 353–356 [DOI] [PubMed] [Google Scholar]
- 27. Torchilin V. P., Weissig V. (2003) Liposomes: A Practical Approach, Oxford University Press, Oxford [Google Scholar]
- 28. Thiele C., Hannah M. J., Fahrenholz F., Huttner W. B. (2000) Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat. Cell Biol. 2, 42–49 [DOI] [PubMed] [Google Scholar]
- 29. Rouquette-Jazdanian A. K., Pelassy C., Breittmayer J. P., Aussel C. (2006) Revaluation of the role of cholesterol in stabilizing rafts implicated in T cell receptor signaling. Cell. Signal. 18, 105–122 [DOI] [PubMed] [Google Scholar]
- 30. Charrin S., Manié S., Thiele C., Billard M., Gerlier D., Boucheix C., Rubinstein E. (2003) A physical and functional link between cholesterol and tetraspanins. Eur. J. Immunol. 33, 2479–2489 [DOI] [PubMed] [Google Scholar]
- 31. Huber T. B., Schermer B., Müller R. U., Höhne M., Bartram M., Calixto A., Hagmann H., Reinhardt C., Koos F., Kunzelmann K., Shirokova E., Krautwurst D., Harteneck C., Simons M., Pavenstädt H., Kerjaschki D., Thiele C., Walz G., Chalfie M., Benzing T. (2006) Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. U.S.A. 103, 17079–17086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J., Reth M. (2010) Oligomeric organization of the B-cell antigen receptor on resting cells. Nature 467, 465–469 [DOI] [PubMed] [Google Scholar]
- 33. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 34. Essen L., Siegert R., Lehmann W. D., Oesterhelt D. (1998) Lipid patches in membrane protein oligomers. Crystal structure of the bacteriorhodopsin-lipid complex. Proc. Natl. Acad. Sci. U.S.A. 95, 11673–11678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hunte C. (2005) Specific protein-lipid interactions in membrane proteins. Biochem. Soc. Trans. 33, 938–942 [DOI] [PubMed] [Google Scholar]
- 36. Contreras F. X., Ernst A. M., Haberkant P., Björkholm P., Lindahl E., Gonen B., Tischer C., Elofsson A., von Heijne G., Thiele C., Pepperkok R., Wieland F., Brügger B. (2012) Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature 481, 525–529 [DOI] [PubMed] [Google Scholar]
- 37. Coskun Ü., Grzybek M., Drechsel D., Simons K. (2011) Regulation of human EGF receptor by lipids. Proc. Natl. Acad. Sci. U.S.A. 108, 9044–9048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H., Papadopoulos V. (1998) Peripheral-type benzodiazepine receptor function in cholesterol transport. Identification of a putative cholesterol recognition/interaction amino acid sequence and consensus pattern. Endocrinology 139, 4991–4997 [DOI] [PubMed] [Google Scholar]
- 39. Ruprecht J. J., Mielke T., Vogel R., Villa C., Schertler G. F. (2004) Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 23, 3609–3620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hanson M. A., Cherezov V., Griffith M. T., Roth C. B., Jaakola V. P., Chien E. Y., Velasquez J., Kuhn P., Stevens R. C. (2008) A specific cholesterol binding site is established by the 2.8 Å structure of the human β2-adrenergic receptor. Structure 16, 897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Contreras F. X., Ernst A. M., Wieland F., Brugger B. (2011) Cold Spring Harb. Perspect. Biol. 3, a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Montixi C., Langlet C., Bernard A. M., Thimonier J., Dubois C., Wurbel M. A., Chauvin J. P., Pierres M., He H. T. (1998) Engagement of T cell receptor triggers its recruitment to low-density detergent-insoluble membrane domains. EMBO J. 17, 5334–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horejsí V. (2003) The roles of membrane microdomains (rafts) in T cell activation. Immunol. Rev. 191, 148–164 [DOI] [PubMed] [Google Scholar]
- 45. Drake D. R., 3rd, Braciale T. J. (2001) Cutting edge. Lipid raft integrity affects the efficiency of MHC class I tetramer binding and cell surface TCR arrangement on CD8+ T cells. J. Immunol. 166, 7009–7013 [DOI] [PubMed] [Google Scholar]
- 46. Demel R. A., Jansen J. W., van Dijck P. W., van Deenen L. L. (1977) The preferential interaction of cholesterol with different classes of phospholipids. Biochim. Biophys. Acta 465, 1–10 [DOI] [PubMed] [Google Scholar]
- 47. Kuhns M. S., Girvin A. T., Klein L. O., Chen R., Jensen K. D., Newell E. W., Huppa J. B., Lillemeier B. F., Huse M., Chien Y. H., Garcia K. C., Davis M. M. (2010) Evidence for a functional sidedness to the αβTCR. Proc. Natl. Acad. Sci. U.S.A. 107, 5094–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kaech S. M., Hemby S., Kersh E., Ahmed R. (2002) Molecular and functional profiling of memory CD8 T cell differentiation. Cell 111, 837–851 [DOI] [PubMed] [Google Scholar]
- 49. Fahmy T. M., Bieler J. G., Edidin M., Schneck J. P. (2001) Increased TCR avidity after T cell activation. A mechanism for sensing low-density antigen. Immunity 14, 135–143 [PubMed] [Google Scholar]
- 50. Dubey C., Croft M., Swain S. L. (1996) Naive and effector CD4 T cells differ in their requirements for T cell receptor versus costimulatory signals. J. Immunol. 157, 3280–3289 [PubMed] [Google Scholar]
- 51. Sezgin E., Levental I., Grzybek M., Schwarzmann G., Mueller V., Honigmann A., Belov V. N., Eggeling C., Coskun U., Simons K., Schwille P. (2012) Partitioning, diffusion, and ligand binding of raft lipid analogs in model and cellular plasma membranes. Biochim. Biophys. Acta 1818, 1777–1784 [DOI] [PubMed] [Google Scholar]