Background: Ribonucleotides in DNA are associated with genome instability.
Results: Human DNA polymerase ϵ catalyzes efficient incorporation of ribonucleotides and extension from primers terminating in multiple consecutive ribonucleotides.
Conclusion: Human DNA polymerase ϵ is able to extend ribonucleotide-terminal primers through a reduction in its proofreading activity.
Significance: Leading strand replication may have a unique relationship to ribonucleotides, RNA, and genome stability.
Keywords: DNA Enzymes, DNA Polymerase, DNA Repair, DNA Replication, Nucleotide, Nucleotide Selectivity, Ribonucleotides
Abstract
Replicative DNA polymerases (Pols) help to maintain the high fidelity of replication in large part through their strong selectivity against mispaired deoxyribonucleotides. It has recently been demonstrated that several replicative Pols from yeast have surprisingly low selectivity for deoxyribonucleotides over their analogous ribonucleotides. In human cells, ribonucleotides are found in great abundance over deoxyribonucleotides, raising the possibility that ribonucleotides are incorporated in the human genome at significant levels during normal cellular functions. To address this possibility, the ability of human DNA polymerase ϵ to incorporate ribonucleotides was tested. At physiological concentrations of nucleotides, human Pol ϵ readily inserts and extends from incorporated ribonucleotides. Almost half of inserted ribonucleotides escape proofreading by 3′ → 5′ exonuclease-proficient Pol ϵ, indicating that ribonucleotide incorporation by Pol ϵ is likely a significant event in human cells. Human Pol ϵ is also efficient at extending from primers terminating in up to five consecutive ribonucleotides. This efficient extension appears to result from reduced exonuclease activity on primers containing consecutive 3′-terminal ribonucleotides. These biochemical properties suggest that Pol ϵ is a likely source of ribonucleotides in human genomic DNA.
Introduction
Replication of the eukaryotic nuclear genome is carried out primarily by three DNA polymerases (Pols),2 α, δ, and ϵ (1, 2). These enzymes help to ensure accurate genome duplication by maintaining a high degree of selectivity for the correctly base-paired incoming dNTP over the incorrectly base-paired dNTP (3). Replicative Pols were believed to select against ribonucleotides to such a high degree that incorporation in vivo was thought to be negligible. However, recent studies have demonstrated that the yeast replicative Pols do incorporate ribonucleotides efficiently both in vitro and in vivo (4, 5). Furthermore, human Pol γ, the sole polymerase responsible for replicating the mitochondrial genome, has also been shown to efficiently incorporate ribonucleotides (6). Underscoring the differences in sugar selectivity by DNA Pols, human Pol β, a critical repair polymerase, inserts ribonucleotides 10,000 times less efficiently than deoxyribonucleotides (7). Given the three billion nucleotides of the human genome, the ability to incorporate ribonucleotides during replication could have profound effects on genome stability. Yet to date, the ribonucleotide selectivity of a human nuclear replicative DNA polymerase has not been studied.
In eukaryotic cells, the level of a particular NTP exceeds that of its cognate dNTP by anywhere from 10- to 2000-fold (8–10). The relative abundance depends on which nucleotide is being compared in which organism. In a survey of a wide variety of mammalian cell types both in cell culture and in tissues, the observed NTP/dNTP ratios ranged between 9.6 and 130 (10). A more recent analysis of cultured human cells generally agreed with these observations (8).
The sugar selectivity values of DNA polymerases (or the NTP/dNTP incorporation ratio) range over 6 orders of magnitude depending on the Pol studied and the measurement technique used (11). For example, Pol μ has a sugar selectivity value for the dT/rU pair of 101, whereas phage φ29 Pol has a sugar selectivity value of almost 107 for the same pair. Underscoring the importance of preventing the large scale incorporation of ribonucleotides into genomic DNA, Pols from all families contain residues capable of minimizing NTP binding and insertion into DNA. The primary determinant of ribonucleotide discrimination has been called the steric gate (12). Although the steric gate is a glutamate in Family A Pols, it can be either a tyrosine or phenylalanine in Family Y Pols. Sugar discrimination in Family X Pols is more complicated and involves interactions with the peptide backbone rather than steric clashes with side chains (7, 13–15).
All B family DNA polymerases contain a conserved tyrosine whose bulk causes a steric clash with the 2′-OH on the ribose. This includes Pol ϵ where substitution of the steric gate tyrosine with alanine leads to a dramatic reduction in yeast growth, although the effects of this substitution on sugar selection are unknown (16). Active site mutants in yeast Pol ϵ that displayed reduced ribonucleotide selectivity in vitro also showed an increase in alkaline-sensitive genomic DNA in vivo, indicative of ribonucleotide incorporation during cellular growth (4). These mutant alleles give rise to a mutator phenotype characterized by short 2–5-bp deletions in perfect or imperfect direct repeat sequences. This same error specificity is observed at sites of high transcription in a topoisomerase I-dependent manner (17, 18) and when topoisomerase I processes incorporated ribonucleotides (19).
The majority of RNase H activity in eukaryotes is carried out by RNase H2, a three-subunit enzyme (20). RNase H2 hydrolyzes RNA-DNA hybrids and importantly is the only enzyme known to specifically remove single ribonucleotides from RNA-DNA duplexes (21, 22). Impairing the removal of ribonucleotides from genomic DNA by inactivating RNase H2 is not lethal in yeast but causes a mutator phenotype (19, 23, 24). RNase H2 is essential for mammalian development, however, and cells lacking RNase H2 contain increased genomic ribonucleotides and DNA damage response activation (25, 26). In humans, mutation in any of the three RNase H2 subunits results in Aicardi-Goutières syndrome, a neurological disorder with phenotypic characteristics mimicking chronic viral infection (27). Some disease mutant alleles of RNase H2 have been shown to possess reduced removal of rNMPs from DNA in vitro (28, 29). Removal of ribonucleotides from genomic DNA is also critical to prevent slowing in S phase and the elevation of dNTP pools, which are indicative of replication stress (4, 30).
We report here the ability of human DNA polymerase ϵ to efficiently incorporate ribonucleotides at physiological nucleotide concentrations. With its proofreading function intact, 3′ → 5′ exonuclease-proficient Pol ϵ is still able to extend from almost half of the incorporated ribonucleotides. Exonuclease-proficient human Pol ϵ is also able to efficiently extend from multiple consecutive ribonucleotides at the 3′ primer terminus. Pol ϵ exonuclease activity is reduced by the presence of increasing numbers of ribonucleotides at the primer terminus.
EXPERIMENTAL PROCEDURES
Materials
Unmodified oligonucleotides were purchased from Invitrogen, and modified oligonucleotides from Midland Certified Reagent Co. (Midland, TX). Oligonucleotides were purified using PAGE. Deoxyribonucleotides and ribonucleotides were purchased from Roche Applied Science, and both were of the highest purity available. The catalytic fragment of wild-type and exonuclease-deficient human Pol ϵ was expressed and purified as described previously (31).
NTP Incorporation Assays
Primer oligonucleotides were 5′-radiolabeled as described (31). Briefly, the primer was incubated with T4 polynucleotide kinase (Invitrogen) and γ-32P-labeled ATP (PerkinElmer Life Sciences) for 30 min at 37 °C. Unincorporated 32P was separated by passage over an Illustra MicroSpin G-25 column (GE Healthcare), and the purified radiolabeled primer was then annealed to a complementary 45-mer DNA oligonucleotide at a final concentration of 2 μm. For paired single nucleotide incorporation assays, the following primer oligonucleotides were used: for rCTP + dA/G/TTP and for dATP/rATP, 5′-CCTCTTCGCTATTACGCC-3′; for dGTP/rGTP and dTTP/rUTP, 5′-CCTCTTCGCTATTACGCCA-3′; and for dCTP/rCTP, 5′-CCTCTTCGCTATTACGCCAG-3′. The following template oligonucleotides were used: for dATP/rATP, dCTP/rCTP, and dGTP/rGTP, 5′-TTGCAGCACATCCCCCTTTCGCCAGCTGGCGTAATAGCGAAGAGG-3′, and for dTTP/rUTP, 5′-TTGCAGCACATCCCCCTTTCGCCAGATGGCGTAATAGCGAAGAGG-3′. Reaction mixtures (30 μl) contained 50 mm Tris, pH 7.4, 8 mm MgCl2, 1 mm DTT, 10% glycerol, 100 nm DNA primer-template, and varying concentrations of the indicated dNTP or NTP (Table 1). Reactions were carried out at 37 °C and started by the addition of the indicated amounts of enzyme. Aliquots were removed at the indicated times and stopped by the addition of an equal volume of 95% formamide followed by incubation for 5 min at 95 °C, chilling on ice, and analysis by denaturing PAGE. The gels were then dried, exposed to a phosphorimaging screen, and scanned using a Typhoon Trio+ Imager (GE Healthcare). Nucleotide incorporation was quantitated using ImageQuant5.2 software (GE Healthcare). Nucleotide discrimination factors were calculated using the following equation.
 |
TABLE 1.
Physiological nucleotide concentrations used in this study
Values were taken from Traut (10).
| dNTP | Concentration | NTP | Concentration |
|---|---|---|---|
| μm | μm | ||
| dATP | 24 | ATP | 3,150 |
| dCTP | 29 | CTP | 280 |
| dGTP | 5.2 | GTP | 470 |
| dTTP | 37 | UTP | 570 |
rUMP Bypass Assays
An 18-mer oligonucleotide, 5′-CCTCTTCGCTATTACGCC-3′, was 5′-end-labeled with 32P using T4 polynucleotide kinase as described above and hybridized to a 45-mer containing rUMP at the position highlighted in parentheses, 5′-TTGCAGCACATCCCCCTTTCGCCAG(rU)TGGCGTAATAGCGAAGAGG-3′. Reaction mixtures (30 μl) contained 50 mm Tris, pH 7.4, 8 mm MgCl2, 1 mm DTT, 10% glycerol, 250 μm each dNTP, and 100 nm DNA primer-template. Reactions were started by the addition of 1 nm Pol ϵ and carried out at 37 °C. Nucleotide incorporation was quantitated by measuring band intensities using a phosphorimaging system. These values were used to determine bypass efficiencies past uracil as described (32). Briefly, bypass probabilities were calculated by dividing the sum of band intensities for products of synthesis past uracil (>N + 2) by the sum of band intensities for products that initiated synthesis (>N) where N is the unreacted primer and N + 2 is uracil or the equivalent position on the undamaged substrate. Bypass efficiency was calculated by dividing the uracil bypass probability by the bypass probability calculated for the same site on the undamaged substrate. Termination probabilities were calculated as the band intensity at any site, n, divided by the sum of band intensities at all positions ≥n.
Ribonucleotide Primer Extension and Excision Assays
The oligonucleotides listed in Table 2 were used to measure extension and excision from 3′-terminal ribonucleotides. Reaction mixtures (30 μl) to measure extension contained 50 mm Tris, pH 7.4, 8 mm MgCl2, 1 mm DTT, 10% glycerol, 25 μm each dNTP, and 100 nm DNA primer-template. Reaction mixtures to measure excision were identical except dNTPs were withheld. Reactions were started by the addition of 10 nm Pol ϵ and carried out at 37 °C.
TABLE 2.
DNA substrates used in ribonucleotide extension and excision experiments
Ribonucleotides in the indicated primer positions are noted in bold.
| DNA | Sequence |
|---|---|
| R0 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R1 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R1(U) | 5′-CCTCTTCGCTATTACGCCAU-3′ |
| 3′-GGAGAAGCGATAATGCGGTAGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R2 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R3 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R4 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ | |
| R5 | 5′-CCTCTTCGCTATTACGCCAG-3′ |
| 3′-GGAGAAGCGATAATGCGGTCGACCGCTTTCCCCCTACACGACGTT-5′ |
Single Turnover Kinetic Assays of Primer Excision
Each DNA substrate listed in Table 2 with an increasing number of primer 3′-terminal ribonucleotides was used to measure the excision rate by wild-type Pol ϵ.
A preincubated solution of 300 nm wild-type Pol ϵ and 30 nm 5′-radioactively labeled DNA in the buffer (50 mm Tris, pH 7.4, 1 mm DTT, 50 mm NaCl, 10 μm EDTA, 1× BSA, and 10% glycerol) was rapidly mixed with an equal volume of the same buffer solution containing additional 16 mm MgCl2 at 37 °C in a rapid chemical quench flow apparatus (KinTek Corp., Snow Shoe, PA), resulting in final reaction concentrations of 150 nm wild-type Pol ϵ, 15 nm DNA, and 8 mm MgCl2. Reactions were terminated by the addition of 0.37 m EDTA after various times. The reaction mixtures were separated by sequencing gel electrophoresis. The plot of product concentrations versus reaction times was fit to the following equation.
where A and kexo represent the reaction amplitude and observed DNA excision rate, respectively.
RESULTS
Insertion and Extension of Ribonucleotides by Human Pol ϵ
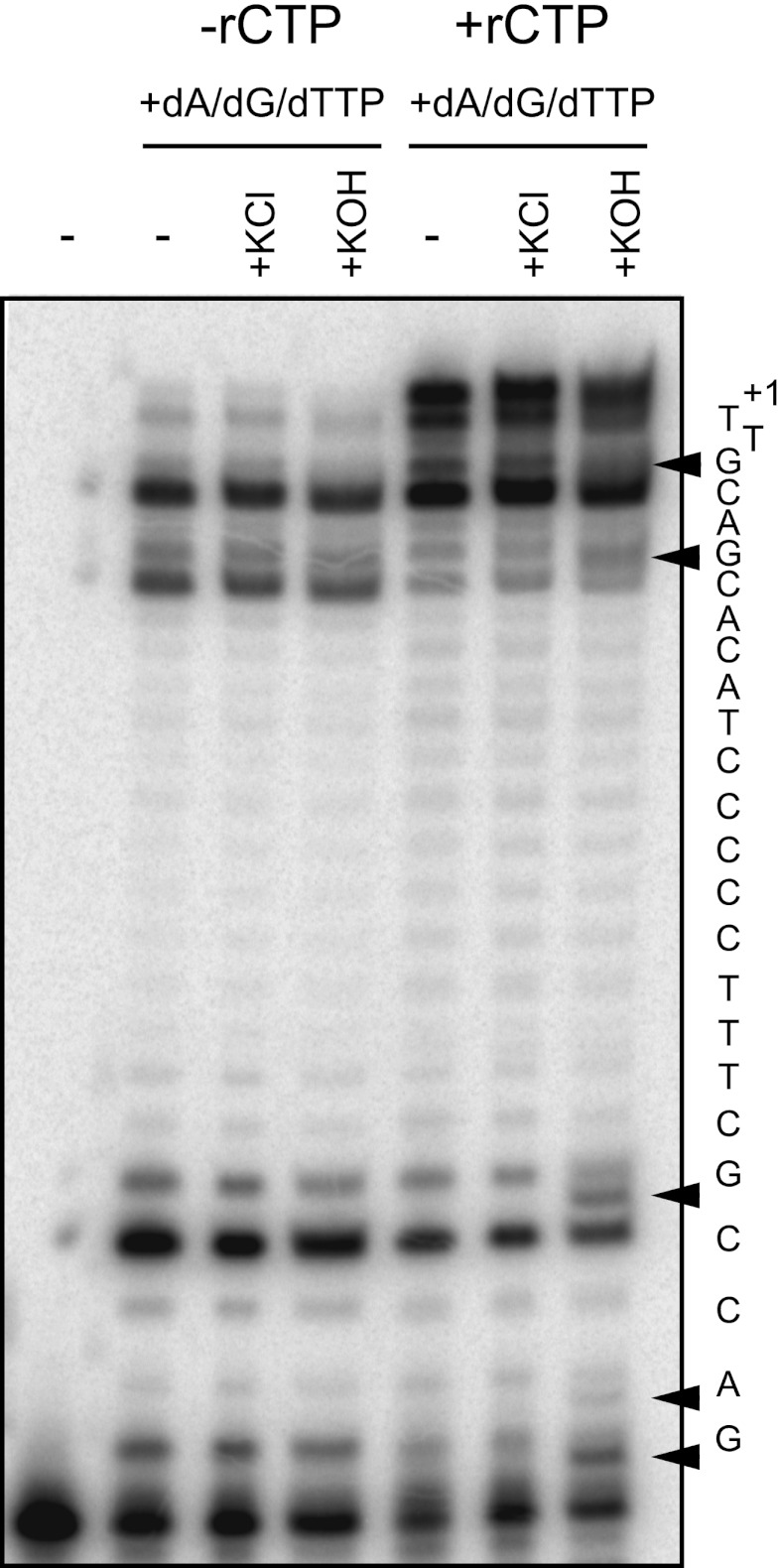
Several eukaryotic DNA polymerases have recently been shown to incorporate ribonucleotides with varying degrees of efficiency (4–6). However, little is known about the ability of any human nuclear replicative DNA polymerase to incorporate ribonucleotides. To address this lack of knowledge, we first asked whether human Pol ϵ was able to insert a ribonucleotide during processive synthesis. For human Pol ϵ, we used the N-terminal 140-kDa fragment that we characterized previously (31). We used a 32P-5′-end-labeled 18-mer DNA oligo hybridized to a 45-mer DNA oligo (Table 2, R0) containing four template G residues distributed throughout (Fig. 1). When human Pol ϵ was incubated with only three dNTPs, dATP, dGTP, and dTTP, the polymerase was unable to carry out efficient DNA synthesis to the end of the template due to the absence of dCTP. Pol ϵ was able to insert incorrectly base-paired dNTPs opposite template G, albeit inefficiently, as evidenced by products terminating with insertion opposite and past template Gs (Fig. 1, −rCTP). When rCTP was included as the sole source of cytosine in the reaction, fully extended products were observed (Fig. 1, +rCTP), indicating incorporation of rCMP by human Pol ϵ. To verify that these products were due to rCMP incorporation, we incubated the products with 0.3 m KOH at 55 °C, which cleaves ribonucleotides via alkaline hydrolysis. Products resulting from ribonucleotide hydrolysis leave a 3′-phosphate, not an entire nucleotide, and are not expected to align with the ladder of unmodified bases. We observed cleavage products specific to alkaline hydrolysis predominantly at template G sites (Fig. 1, black arrowheads). Concomitant with the appearance of the alkaline cleavage products, we saw a corresponding decrease in full-length products, suggesting that Pol ϵ was able to fully extend past the incorporated rCMP.
FIGURE 1.
Incorporation of rCTP by human Pol ϵ. Purified Exo− human Pol ϵ (1 nm) was incubated with 18-mer/45-mer substrate (100 nm) for 10 min at 37 °C in the presence of a 250 μm concentration of each of dATP, dGTP, and dTTP. The rCTP nucleotide was either withheld (−rCTP) or added at 250 μm (+rCTP). These reaction products were incubated with either 300 mm KCl or 300 mm KOH for 2 h at 55 °C. Products were resolved on a 12% denaturing gel. Black arrowheads indicate extension products containing incorporated rNMPs that were hydrolyzed by alkaline treatment. The template sequence is shown to the right.
Ribonucleotide Selectivity of Human Pol ϵ
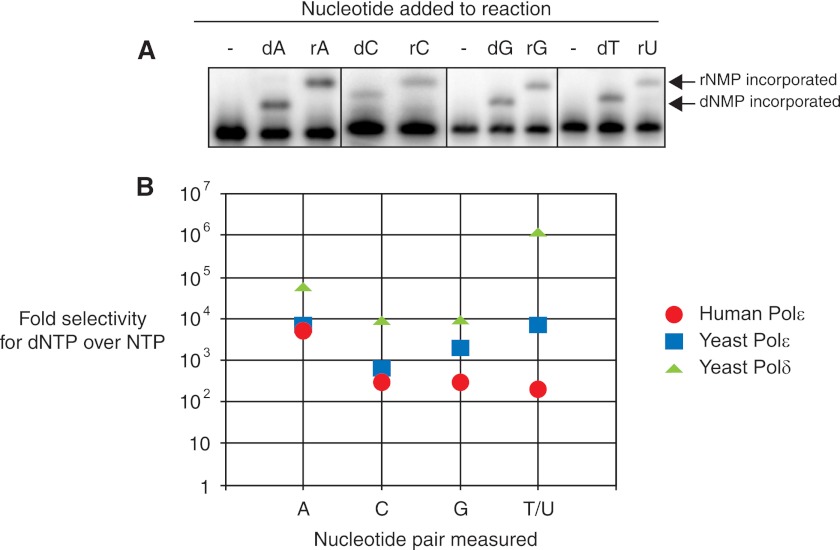
Because ribonucleotides are in 10- to 100-fold excess over their deoxyribonucleotide equivalents in vivo (8, 10) and because human Pol ϵ is able to incorporate ribonucleotides during polymerization, we next addressed the intrinsic selectivity of human Pol ϵ for deoxyribonucleotides over ribonucleotides for each pair. We used four different primer-template pairs that allow observation of a single nucleotide incorporation event with each one of the four bases separately. The ribonucleotide or deoxyribonucleotide was added at the observed physiological concentration (Table 1), whereas the enzyme concentration was varied to allow observation of nucleotide incorporation under single hit conditions (33). These are conditions in which substrate is present in excess so that DNA synthesis can be measured during a single polymerase binding event. We quantified the insertion of each nucleotide at physiological concentrations (Fig. 2A). The 2′-OH present in the ribonucleotide causes the rNMP insertion product to migrate more slowly than the equivalent dNMP insertion product on a denaturing polyacrylamide gel. Based on the amount of nucleotide incorporated, the relative nucleotide concentrations, the enzyme concentrations, and times used, we were able to calculate the intrinsic nucleotide selectivity values for each base (Fig. 2B, red circles). Human Pol ϵ preferred to insert dTTP over the ribonucleotide analog, rUTP, by only 210-fold. This result is similar to the 300-fold discrimination observed between dGTP and rGTP and between dCTP and rCTP. Incorporation of rATP was more strongly selected against with a calculated discrimination factor of 5300-fold. These discrimination values are similar to those reported for the yeast Pol ϵ (Fig. 2B, blue squares; yeast values were taken from McElhinny et al. (5)) with the exception of the dTTP-rUTP value where human Pol ϵ was 34-fold less selective against the ribonucleotide. Pol ϵ discrimination against each ribonucleotide was in the order rA > rC ≅ rG ≅ rU.
FIGURE 2.
Ribonucleotide selectivity of human Pol ϵ. A, human Pol ϵ was incubated with 100 nm primer-template substrate where the next correctly base-paired incoming nucleotide is indicated. The indicated nucleotide was added at its physiological concentration (Table 1). The amount of Pol ϵ was adjusted until less than 20% of the primer was extended. For each dNTP, 0.3 nm Pol ϵ was used. For ribonucleotide incorporation, the following concentrations of Pol ϵ were used: rATP, 12 nm; rCTP, 6 nm; rGTP, 3 nm; rUTP, 0.6 nm. Shown are individual pairs of deoxyribo- and ribonucleotide for comparison. Arrows indicate the migration of the single incorporated dNMP or rNMP product. B, ribonucleotide selectivity values from experiments in A were calculated using Equation 1 (described under “Experimental Procedures” and in Ref. 5) and then plotted for human Pol ϵ as the -fold selectivity of dNTP over NTP (red circles). Ribonucleotide selectivity values for yeast Pol ϵ (blue squares) and yeast Pol δ (green triangles) are shown for comparison and were taken from Ref. 5.
Bypass of a Ribonucleotide during DNA Synthesis
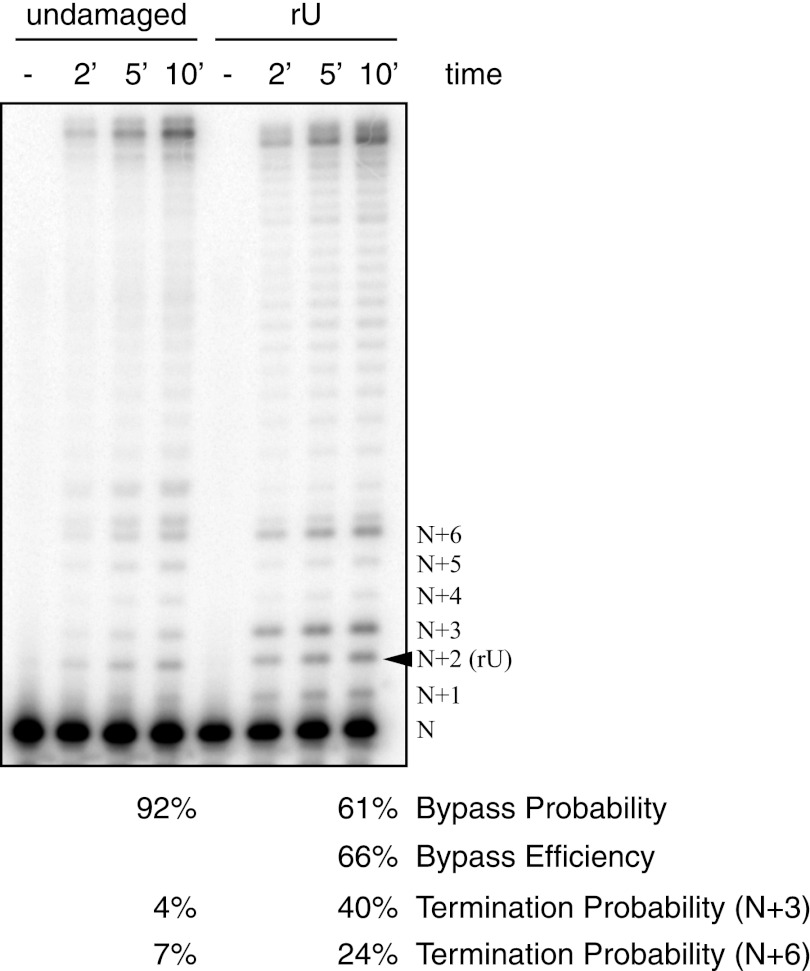
The ability of yeast replicative DNA polymerases to insert ribonucleotides in vitro correlates with their ability to do so in vivo (4, 34). If left unrepaired, these ribonucleotides would remain in the template strand during subsequent rounds of replication, potentially blocking nascent DNA synthesis. Because human Pol ϵ is able to incorporate ribonucleotides in vitro, we asked what the consequences were for DNA synthesis past a template ribonucleotide by measuring the bypass efficiency of DNA synthesis past a site-specific template ribouracil (32, 35) under conditions where each substrate molecule was estimated to encounter at most one active DNA polymerase molecule (Fig. 3). When synthesis past rU was compared with the undamaged template, the bypass efficiency was calculated to be 66%, indicating that human Pol ϵ bypassed a single rU nucleotide in the template strand under the reaction conditions used.
FIGURE 3.
Bypass of template rU by human Pol ϵ. Human Pol ϵ was incubated with all four dNTPs at 25 μm each and 100 nm substrate containing either entirely template deoxyribonucleotides (undamaged) or a single rUMP at the N + 2 position (rU) where N is the unreacted primer. Reactions were carried out as described under “Experimental Procedures.” Reactions were performed at 37 °C and started with the addition of 1 nm enzyme, and aliquots were removed at 2, 5, and 10 min (′). Control substrate with no enzyme added is shown (−). Products were resolved on a 12% denaturing acrylamide gel. Bypass probabilities, bypass efficiency, and termination probabilities were calculated as described (see “Experimental Procedures” and Refs. 31 and 32) using the following equations: Bypass probability = ΣBand intensity(>N + 2)/ΣBand intensity(>N), Bypass efficiency = (Bypass probabilityrU)/(Bypass probabilityundamaged), and Termination probability = Band intensity(n)/ΣBand intensity(≥n).
The termination probability for Pol ϵ one nucleotide after insertion opposite the uracil (N + 3) was 40%, 10-fold higher than at the same site in the undamaged substrate. The termination probability for Pol ϵ four nucleotides after insertion opposite the uracil (N + 6) was 24%, 3.4-fold higher than the same position in the undamaged substrate. Both results indicate that although Pol ϵ can efficiently bypass uracil in the template it can still detect rU at a distance from the active site.
Extension from a 3′-Terminal Ribonucleotide: Exonuclease-deficient Pol ϵ
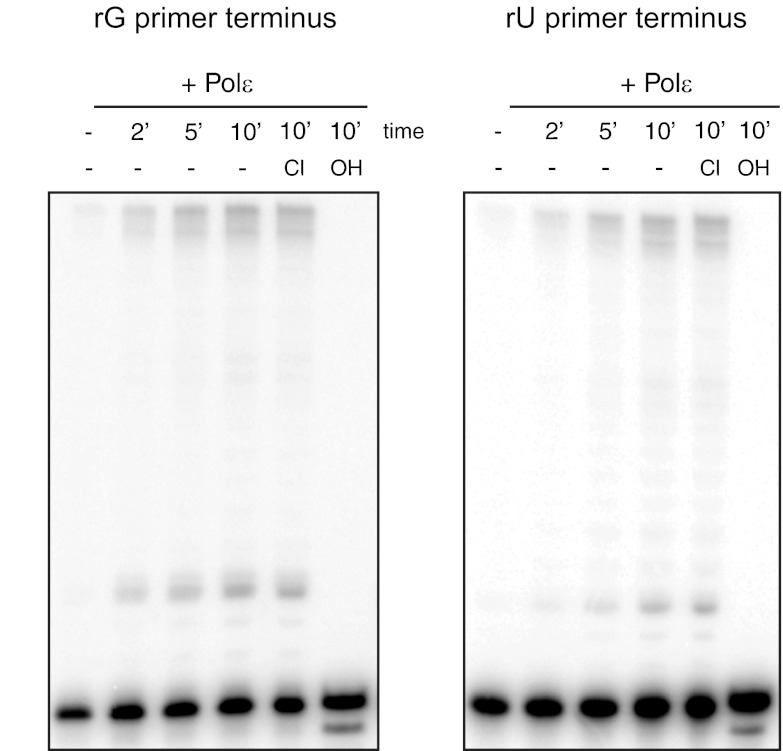
We then asked whether human Pol ϵ was able to extend from an incorporated ribonucleotide. We used a 20-mer oligonucleotide consisting of 19 deoxyribonucleotides with a single terminal ribonucleotide. Using an rGMP primer terminus, we observed highly processive synthesis during single hit conditions (Fig. 4, left panel). Although this indicates that the terminal 2′-OH group did not impede processive DNA synthesis, we observed a pause site at the n + 4 position. To ensure that the observed products were truly extended from the rNMP at the primer terminus, we incubated reaction products with KOH. We observed appearance of an alkaline cleavage product migrating slightly faster than the unreacted primer. This band is the product of alkaline hydrolysis of the ribonucleotide-containing extension product, resulting in a 20-mer with a terminal 3′-phosphate group. The appearance of this band also correlated with complete disappearance of both the fully extended and n + 4 products. This indicates that the Pol ϵ reaction products resulted from the extension of the rG-terminal oligonucleotide. To determine whether the nature of the base at the 3′ terminus affected the ability of Pol ϵ to extend, we repeated these reactions using a primer terminating in rU. We observed no differences in processive synthesis, the n + 4 pause product, or the alkaline sensitivities of either (Fig. 4, right panel).
FIGURE 4.
Extension from a 3′-terminal ribonucleotide by exonuclease-deficient human Pol ϵ. Exo− human Pol ϵ was incubated with all four dNTPs at 25 μm each and a substrate containing a deoxyribonucleotide primer with a single 3′-terminal rGMP (left panel) or rUMP (right panel) nucleotide. Reactions were carried out as described under “Experimental Procedures.” Reactions were performed at 37 °C and started with the addition of 1 nm enzyme, and aliquots were removed at 2, 5, and 10 min (′). Control substrate with no enzyme added is shown (−Pol ϵ). Products were untreated (−) or incubated at 55 °C for 2 h in either KCl (Cl) or KOH (OH). Products were resolved on a 12% denaturing acrylamide gel.
Extension from a 3′-Terminal Ribonucleotide: Exonuclease-proficient Pol ϵ
Endogenous human Pol ϵ retains a 3′ → 5′ exonuclease activity capable of proofreading incorrectly inserted nucleotides (31, 36). It remains unknown, however, how efficiently the enzyme proofreads ribonucleotides. We asked how the proofreading function of Pol ϵ affects extension from an oligonucleotide terminating in a 3′-ribonucleotide.
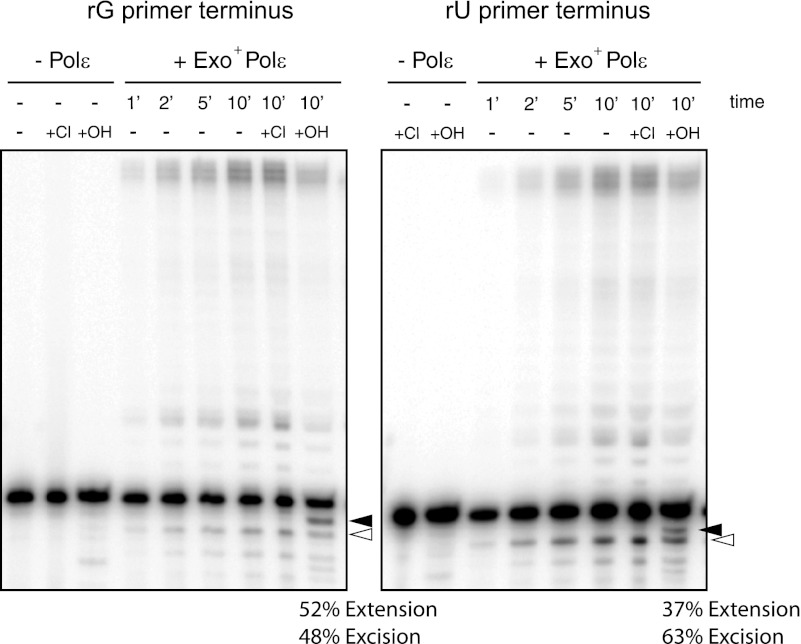
Upon binding of exonuclease-proficient Pol ϵ to the ribonucleotide-terminal substrate, one of several productive outcomes can potentially occur: excision, excision followed by DNA synthesis, or DNA synthesis directly from the rNMP primer terminus. Excision results in products of n − 1 or shorter length. Excision followed by synthesis and extension directly from the ribonucleotide both result in products greater than n in length. To distinguish between these two outcomes, we treated the reaction products with KOH. Products resulting from direct extension from the rNMP terminus retain the alkaline-sensitive ribonucleotide. Upon KOH treatment, these direct extension products are resolved to a species (Fig. 5, black arrowhead) that migrates between the unreacted rNMP-terminal primer and the n − 1 excision product (Fig. 5, white arrowhead). This product of alkaline hydrolysis is a 20-mer that contains a 3′-phosphate group, causing it to migrate slightly faster than the unhydrolyzed R1 20-mer. Products resulting from excision followed by DNA synthesis do not contain any ribonucleotides and are resistant to alkaline hydrolysis. We thus measured the disappearance of extended reaction products after KOH treatment to determine the relative ability of exonuclease-proficient human Pol ϵ to extend from primers terminating in a ribonucleotide. These measurements were all done under single hit conditions to ensure that synthesis and/or excision were measured during the course of a single polymerase binding event.
FIGURE 5.
Quantitation of extension from versus excision of a 3′-terminal ribonucleotide by exonuclease-proficient human Pol ϵ. Exonuclease-proficient human Pol ϵ was incubated with all four dNTPs at 25 μm each and 100 nm substrate containing a deoxyribonucleotide primer containing a single 3′-terminal rGMP (left panel) or rUMP (right panel) nucleotide. Reactions were carried out as described under “Experimental Procedures.” Reactions were performed at 37 °C and started with the addition of 1 nm enzyme, and aliquots were removed at 1, 2, 5, and 10 min (′). Control substrate with no enzyme added is shown (− Pol ϵ). Products were untreated (−) or incubated at 55 °C for 2 h in either KCl (Cl) or KOH (OH). Products were resolved on a 12% denaturing acrylamide gel. The cleavage product resulting from alkaline hydrolysis of an rNMP-containing extension product is indicated (black arrowhead). Products resulting from 3′ excision of the substrate by Pol ϵ are indicated (white arrowhead). All measurements were performed in triplicate with mean values reported.
Exonuclease-proficient human Pol ϵ readily extended from an rGMP-terminal primer (Fig. 5, left). The appearance of the n − 1 (white arrowhead) and to a lesser extent n − 2 and n − 3 bands indicated that excision also occurred in the presence of dNTPs. When the reaction products were treated with KOH, 52% of the extended reaction products were cleaved (Fig. 5, compare Cl with OH lanes). This indicated that half of the observed extension products were due to Pol ϵ extending from an rGMP at the primer terminus. We observed a similar pattern with a rUMP-terminal primer (Fig. 5, right panel). In this case, 37% of the extended products were alkaline-sensitive.
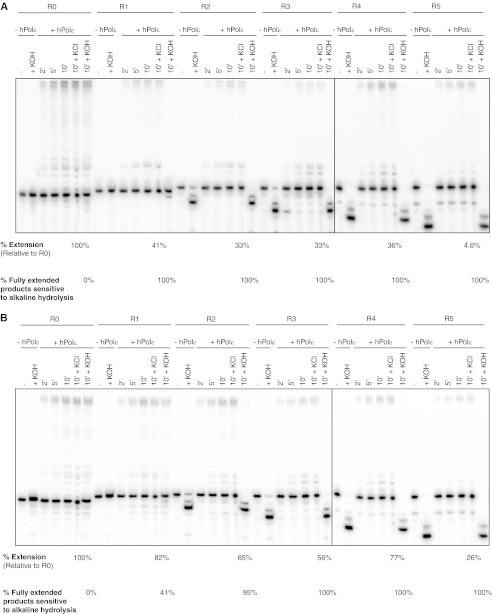
The ability to efficiently extend from a single ribonucleotide raised the possibility that Pol ϵ might extend from two and possibly more ribonucleotides. To test this possibility, chimeric primers were made that were identical in sequence to those described above but with increasing numbers of ribonucleotides substituted at the 3′-end (Table 2). We first measured the ability of proofreading-deficient, Exo− Pol ϵ to extend from these primers. Enzyme and substrate concentrations were held constant for all substrates to compare their relative extension efficiencies. Exo− human Pol ϵ was able to extend from each of the ribonucleotide-terminating primers under these identical reaction conditions (Fig. 6A, R1–R5). For the R1–R4 substrates, the efficiencies were between 33 and 41% that of extension from the fully DNA-containing primer (Fig. 6A, R0). The extension products from the each of the rNMP-terminal primers were all completely sensitive to alkaline hydrolysis (Fig. 6A, 10′ + KOH for each substrate), indicating that the observed products were all extended from ribonucleotide-containing substrates. As expected, the R0 extension products were insensitive to alkaline hydrolysis.
FIGURE 6.
Pol ϵ extension from increasing numbers of 3′-terminal ribonucleotides. Human Pol ϵ was incubated with substrates containing primer oligonucleotides with increasing numbers of 3′-terminal ribonucleotides (Table 2). Primers have sequences identical to the fully DNA primers described above (R0) but with one, two, three, four, or five consecutive ribonucleotides at the 3′-end (R1–R5). Reactions were carried out at 37 °C for the indicated times using 25 μm dNTPs, 100 nm substrate, and either 1 nm Exo− (A) or Exo+ Pol ϵ (B). Unreacted substrates (− hPol ϵ) were either untreated or treated with KOH to demonstrate alkaline hydrolysis of the ribonucleotide-containing substrates. Aliquots of Pol ϵ extension products were taken at the indicated times (+ hPol ϵ) and treated with either salt (+ KCl) or alkali (+ KOH). All measurements were performed in triplicate with mean values reported. ′, minutes.
Because terminal ribonucleotides could be excised prior to extension by the proofreading-proficient enzyme, the ability of Exo+ Pol ϵ to extend from multiple terminal ribonucleotides was measured. Products resulting from excision alone will migrate faster than the unreacted substrate. Products resulting from extension directly from ribonucleotides will migrate more slowly than the substrate but will be sensitive to alkali and generate faster migrating products after KOH treatment. Products resulting from excision of any ribonucleotide(s) followed by extension without enzyme dissociation will also migrate more slowly than the unreacted substrates but will instead be resistant to alkaline hydrolysis and will remain after KOH treatment. The difference in product intensities before and after alkaline hydrolysis can thus help resolve these different reactions. As with the Exo− enzyme, Exo+ Pol ϵ was able to extend from each of the ribonucleotide-terminating primers under identical reaction conditions (Fig. 6B, R1–R5), and none of the R0 extension products were sensitive to alkali (Fig. 6B, R0). As seen above, approximately half of the R1 extension products were sensitive to alkali treatment. The fraction of alkaline-sensitive extension products increased to 95% when the primer terminated in two ribonucleotides. When the primer terminated in three to five ribonucleotides, 100% of the observed extension products were sensitive to alkali. For the R1–R4 substrates, the extension efficiency remained within 1.8-fold of the fully deoxyribonucleotide primer. A 4-fold decrease in extension product formation was observed for the R5 substrate.
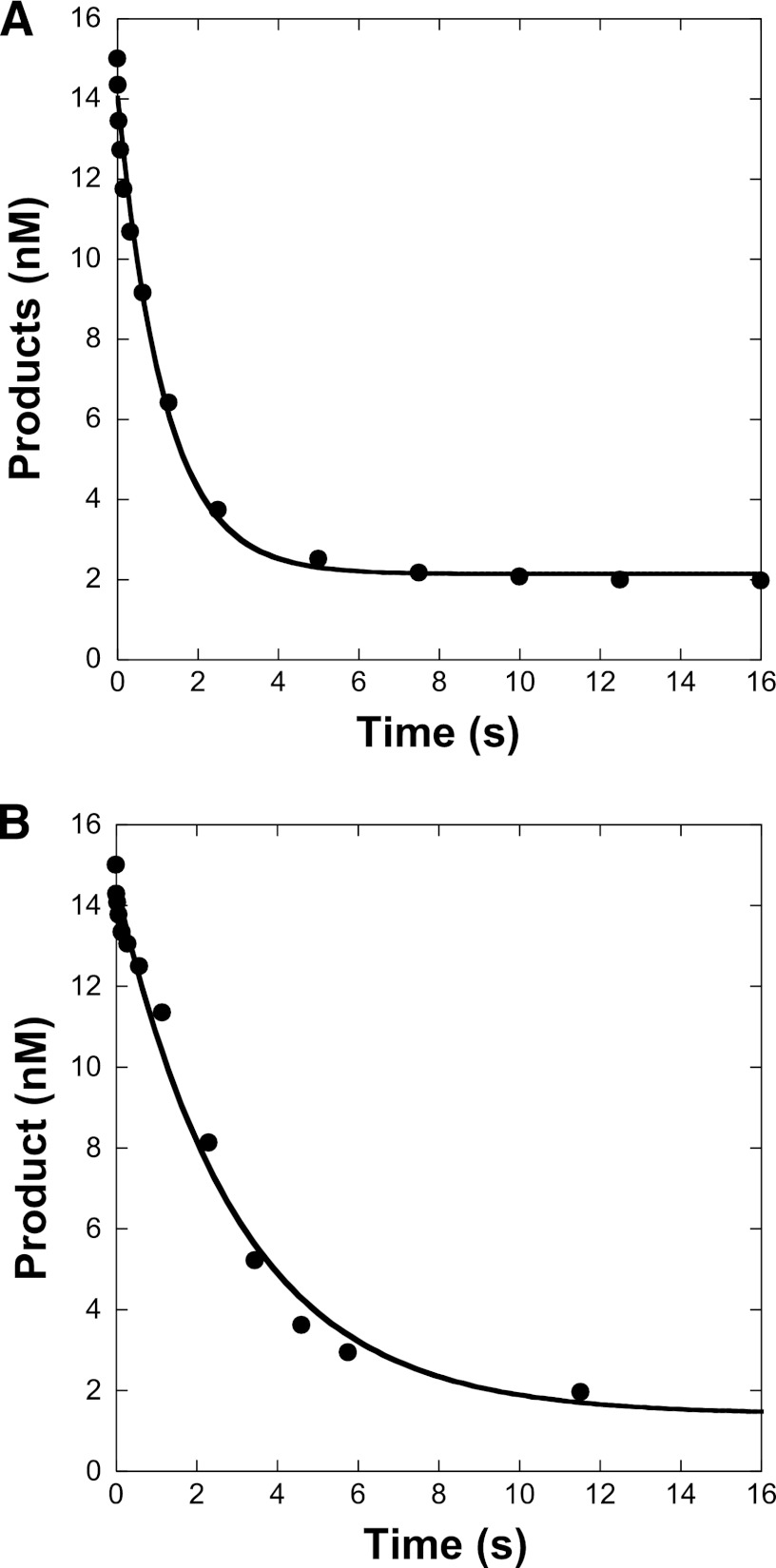
To assess the effect of increasing primer terminal ribonucleotides on the 3′ → 5′ exonuclease activity of wild-type Pol ϵ, we determined the primer excision rate with each of the DNA substrates in Table 2 in the absence of dNTPs. Under single turnover kinetic conditions, a rate of excision of 0.86 s−1 was determined for the correctly paired, dNMP-terminal primer in R0 (Fig. 7A). This rate increased to 2.8 s−1 when the same primer terminated with a single ribonucleotide (Table 3). Further substitutions of two to five ribonucleotides in the primer 3′ terminus of R2–R5 significantly reduced kexo, which was determined to be 0.15–0.63 s−1 (Fig. 7B and Table 3).
FIGURE 7.
Excision rate of the 3′ → 5′ exonuclease activity of wild-type hPol ϵ. A preincubated solution of wild-type hPol ϵ (150 nm) and DNA (15 nm) was rapidly mixed with Mg2+ (8 mm) for various reaction times. Remaining substrate was plotted as a function of time. The solid lines represent the best fit to Equation 2 (see “Experimental Procedures”). hPol ϵ catalyzed the excision of the primers of DNA substrates R0 (A) and R4 (B) at a rate of 0.86 ± 0.08 and 0.33 ± 0.02 s−1, respectively.
TABLE 3.
Excision rates of the 3′ → 5′ exonuclease activity of hPol ϵ
| Substrate | kexo | -Fold change |
|---|---|---|
| s−1 | ||
| R0 | 0.86 ± 0.08 | 1× |
| R1 | 2.8 ± 0.7 | 3.3× |
| R2 | 0.63 ± 0.03 | 0.71× |
| R3 | 0.15 ± 0.02 | 0.18× |
| R4 | 0.33 ± 0.02 | 0.38× |
| R5 | 0.29 ± 0.03 | 0.33× |
DISCUSSION
We sought to examine the effects of ribonucleotide on the polymerase and exonuclease activities of human Pol ϵ, one of the major human replicative DNA polymerases. We found that human Pol ϵ is able to readily incorporate each of the four ribonucleotides even when present along with physiologically observed concentrations of dNTPs. This incorporation varied considerably depending on the sequence context. Additionally, we found that human Pol ϵ is able to efficiently bypass a ribonucleotide in the template. Exonuclease-proficient human Pol ϵ extended from almost half of incorporated ribonucleotides, highlighting the potential for ribonucleotide incorporation by Pol ϵ in human cells. Human Pol ϵ was also able to efficiently extend from multiple consecutive ribonucleotides at the 3′ primer terminus. Substituting ribonucleotides at three or more of the 3′-terminal sugars slowed the rate of 3′ → 5′ excision by Pol ϵ.
Three factors govern the abundance of ribonucleotides incorporated by an exonuclease-proficient DNA polymerase in vivo. The first factor is the intrinsic selectivity of the polymerase or how well the Pol is able to discriminate between the presence and absence of a 2′-OH group on the sugar. The second factor is how efficiently the ribonucleotide now located at the primer terminus is shifted to the exonuclease active site and excised. The third factor is the concentration of each ribonucleotide present in the cell. Higher absolute NTP concentrations as well as high concentrations of NTPs relative to dNTPs will cooperate to increase the probability of NMP incorporation. Studies from different eukaryotic organisms have shown ribonucleotides to be present in great excess over deoxyribonucleotides in cycling cells (5, 8, 10). In general, dNTPs are present at 5–30 μm, whereas NTPs range from 280 to 5400 μm. This 10–350-fold relative abundance of NTPs greatly increases the probability of NTP incorporation in vivo. The situation is even more asymmetric in non-cycling cells with NTPs ranging from 160- to 2000-fold more than dNTPs (8, 10).
We find that human Pol ϵ has the lowest discrimination factor for ribonucleotide insertion of any eukaryotic replicative polymerase yet measured. The most extreme difference is observed for insertion of the dT/rU pair. Yeast Pol δ, the most highly selective of the yeast Pols, discriminates between dT and rU insertion by over 106-fold, whereas yeast Pols α and ϵ discriminate close to 104-fold each (5). Human Pol ϵ discriminates between dT and rU by only 210-fold, a 50-fold decrease over the yeast enzyme. This low discrimination is not confined to the dT/rU pair as similar low factors are observed for the dC/rC and dG/rG pairs. The exception is the dA/rA pair where rA insertion is more strongly discriminated against.
Once a ribonucleotide is inserted, its persistence in genomic DNA will be dictated in large part by how efficiently it is recognized and removed by the exonuclease activity. Previous studies have shown that φ29, T4, and T7 Pols each excise primer 3′-terminal rNMPs with an efficiency equal to that of removing a terminal dNMP (37, 38). In comparison, human Pol ϵ excised 3′-terminal rGMP in R1 (Table 2) with a 3-fold higher rate (Table 3) than it cleaved a corresponding 3′-terminal dGMP in R0 (Table 2). When the ribonucleotide was instead placed in the second or third position from the primer terminus, however, the exonuclease activities of T4 and T7 Pols were strongly reduced (38). This trend was also found with human Pol ϵ based on significantly lower primer excision rates (Table 3) with DNA substrates R2–R5 (Table 2) than with R1. Structural and kinetic studies with T4 DNA Pol suggested that this decrease is due to steric clashes between the 2′-OH in the second and third sugars from the primer terminus and amino acid residues. Based on structural data, these residues are unlikely to be repositioned, and this would destabilize single-stranded DNA binding to the Exo active site and reduce excision. The primer terminal 2′-OH also clashes sterically with amino acids, but repositioning can be accommodated and allow excision to proceed. Thus, it would be interesting to see how human Pol ϵ binds to R0–R5 in their binary or ternary structures.
We found that in the presence of dNTPs human Pol ϵ is able to recognize and excise up to 50% of 3′-terminal ribonucleotides. This means that almost half of all ribonucleotides inserted by human Pol ϵ in vivo could escape proofreading and require subsequent editing. Yeast Pol ϵ, which has been shown to incorporate ribonucleotides in vivo, shows similar extension properties from single rNMP-containing primers (39). Excess ribonucleotides incorporated by Pol ϵ would make genomic DNA more vulnerable to strand breaks and thus might account for the different mutation rates and tumor spectra observed in exonuclease-deficient Pol ϵ and exonuclease-deficient Pol δ mice (40).
Under normal physiological conditions, any ribonucleotides incorporated by the polymerase into the genome are essentially all removed. However, conditions that disrupt the normal equilibrium between insertion and removal cause increased genomic ribonucleotide incorporation and are mutagenic. Yeast mutants that cause elevated levels of genomic rNMPs lead to increases in two- to five-nucleotide deletions in repeat sequences (4, 19). Persistent ribonucleotides can also disrupt synthesis during subsequent rounds of replication (5). To help guard against this, RNase H2 is able to recognize and remove rNMPs in duplex DNA. Consequently, conditions that either inactivate RNase H2 or otherwise cause an increase in ribonucleotide incorporation are mutagenic. Because a subset of patients with Aicardi-Goutières syndrome are deficient in RNase H2 (27, 41), ribonucleotide incorporation associated with Pol ϵ may play a significant role in disease development. Although RNase H2 physically interacts with proliferating cell nuclear antigen and colocalizes with active replication forks (42), the mismatch repair system, which corrects replication errors (43) and colocalizes with sites of replication (44), does not appear to correct rNMPs incorporated by Pol ϵ in yeast (45).
In addition to ribonucleotide incorporation, the ability of Pol ϵ to efficiently extend from multiple consecutive ribonucleotides raises questions about the conditions and substrates for this activity in vivo. Pol ϵ would require access to an RNA hybridized to DNA to carry out this activity. Several replication systems, including the M13 bacteriophage, ColE1 plasmids, and mitochondrial DNA, use RNA primers made by RNA polymerases to initiate DNA replication (46–48). Purified Escherichia coli replisome, which includes the replicative DNA polymerase, is able to use the RNA transcript from a co-directional paused RNA polymerase to continue leading strand synthesis (49). It could be speculated that that this novel property of Pol ϵ might uniquely position it to enable direct leading strand restart at a replication fork that collides with RNA polymerase transcribing in the same direction, although it should be emphasized that no direct evidence in support of this yet exists. Understanding how human Pol ϵ incorporates and extends from ribonucleotides in vitro will now enable experiments designed to test the extent to which this occurs in vivo and to examine subsequent physiological consequences.
Acknowledgments
We thank Dr. Jim Karam and Dr. Art Lustig for critical discussions and thoughtful comments on the manuscript and Dr. Jason Fowler for purification of wild-type hPol ϵ.
This work was supported, in whole or in part, by National Institutes of Health Grants ES016780 and RR020152 (to Z. F. P.) and GM079403 (to Z. S.). This work was also supported by Tulane University start-up funds (to Z. F. P.).
- Pol
- polymerase
- Exo
- exonuclease
- hPol
- human Pol.
REFERENCES
- 1. Burgers P. M. (2009) Polymerase dynamics at the eukaryotic DNA replication fork. J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kunkel T. A., Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kunkel T. A. (2011) Balancing eukaryotic replication asymmetry with replication fidelity. Curr. Opin. Chem. Biol. 15, 620–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nick McElhinny S. A., Kumar D., Clark A. B., Watt D. L., Watts B. E., Lundström E. B., Johansson E., Chabes A., Kunkel T. A. (2010) Genome instability due to ribonucleotide incorporation into DNA. Nat. Chem. Biol. 6, 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kasiviswanathan R., Copeland W. C. (2011) Ribonucleotide discrimination and reverse transcription by the human mitochondrial DNA polymerase. J. Biol. Chem. 286, 31490–31500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cavanaugh N. A., Beard W. A., Wilson S. H. (2010) DNA polymerase β ribonucleotide discrimination: insertion, misinsertion, extension, and coding. J. Biol. Chem. 285, 24457–24465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ferraro P., Franzolin E., Pontarin G., Reichard P., Bianchi V. (2010) Quantitation of cellular deoxynucleoside triphosphates. Nucleic Acids Res. 38, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar D., Abdulovic A. L., Viberg J., Nilsson A. K., Kunkel T. A., Chabes A. (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 39, 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Traut T. W. (1994) Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 11. Brown J. A., Suo Z. (2011) Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 50, 1135–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Joyce C. M. (1997) Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. U.S.A. 94, 1619–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moon A. F., Garcia-Diaz M., Batra V. K., Beard W. A., Bebenek K., Kunkel T. A., Wilson S. H., Pedersen L. C. (2007) The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair 6, 1709–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruiz J. F., Juárez R., García-Díaz M., Terrados G., Picher A. J., González-Barrera S., Fernández de Henestrosa A. R., Blanco L. (2003) Lack of sugar discrimination by human Pol μ requires a single glycine residue. Nucleic Acids Res. 31, 4441–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown J. A., Fiala K. A., Fowler J. D., Sherrer S. M., Newmister S. A., Duym W. W., Suo Z. (2010) A novel mechanism of sugar selection utilized by a human X-family DNA polymerase. J. Mol. Biol. 395, 282–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pavlov Y. I., Shcherbakova P. V., Kunkel T. A. (2001) In vivo consequences of putative active site mutations in yeast DNA polymerases α, ϵ, δ, and ζ. Genetics 159, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lippert M. J., Kim N., Cho J. E., Larson R. P., Schoenly N. E., O'Shea S. H., Jinks-Robertson S. (2011) Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 108, 698–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takahashi T., Burguiere-Slezak G., Van der Kemp P. A., Boiteux S. (2011) Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 108, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim N., Huang S. N., Williams J. S., Li Y. C., Clark A. B., Cho J. E., Kunkel T. A., Pommier Y., Jinks-Robertson S. (2011) Mutagenic processing of ribonucleotides in DNA by yeast topoisomerase I. Science 332, 1561–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frank P., Braunshofer-Reiter C., Pöltl A., Holzmann K. (1998) Cloning, subcellular localization and functional expression of human RNase HII. Biol. Chem. 379, 1407–1412 [DOI] [PubMed] [Google Scholar]
- 21. Eder P. S., Walder J. A. (1991) Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization, and substrate specificity. J. Biol. Chem. 266, 6472–6479 [PubMed] [Google Scholar]
- 22. Jeong H. S., Backlund P. S., Chen H. C., Karavanov A. A., Crouch R. J. (2004) RNase H2 of Saccharomyces cerevisiae is a complex of three proteins. Nucleic Acids Res. 32, 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen J. Z., Qiu J., Shen B., Holmquist G. P. (2000) Mutational spectrum analysis of RNase H(35) deficient Saccharomyces cerevisiae using fluorescence-based directed termination PCR. Nucleic Acids Res. 28, 3649–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu J., Qian Y., Frank P., Wintersberger U., Shen B. (1999) Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol. Cell. Biol. 19, 8361–8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hiller B., Achleitner M., Glage S., Naumann R., Behrendt R., Roers A. (2012) Mammalian RNase H2 removes ribonucleotides from DNA to maintain genome integrity. J. Exp. Med. 209, 1419–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reijns M. A., Rabe B., Rigby R. E., Mill P., Astell K. R., Lettice L. A., Boyle S., Leitch A., Keighren M., Kilanowski F., Devenney P. S., Sexton D., Grimes G., Holt I. J., Hill R. E., Taylor M. S., Lawson K. A., Dorin J. R., Jackson A. P. (2012) Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell 149, 1008–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crow Y. J., Leitch A., Hayward B. E., Garner A., Parmar R., Griffith E., Ali M., Semple C., Aicardi J., Babul-Hirji R., Baumann C., Baxter P., Bertini E., Chandler K. E., Chitayat D., Cau D., Déry C., Fazzi E., Goizet C., King M. D., Klepper J., Lacombe D., Lanzi G., Lyall H., Martínez-Frías M. L., Mathieu M., McKeown C., Monier A., Oade Y., Quarrell O. W., Rittey C. D., Rogers R. C., Sanchis A., Stephenson J. B., Tacke U., Till M., Tolmie J. L., Tomlin P., Voit T., Weschke B., Woods C. G., Lebon P., Bonthron D. T., Ponting C. P., Jackson A. P. (2006) Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nat. Genet. 38, 910–916 [DOI] [PubMed] [Google Scholar]
- 28. Coffin S. R., Hollis T., Perrino F. W. (2011) Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutières syndrome. J. Biol. Chem. 286, 16984–16991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Perrino F. W., Harvey S., Shaban N. M., Hollis T. (2009) RNaseH2 mutants that cause Aicardi-Goutieres syndrome are active nucleases. J. Mol. Med. 87, 25–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., Thelander L. (2003) Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112, 391–401 [DOI] [PubMed] [Google Scholar]
- 31. Korona D. A., Lecompte K. G., Pursell Z. F. (2011) The high fidelity and unique error signature of human DNA polymerase ϵ. Nucleic Acids Res. 39, 1763–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kokoska R. J., McCulloch S. D., Kunkel T. A. (2003) The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 278, 50537–50545 [DOI] [PubMed] [Google Scholar]
- 33. Goodman M. F., Creighton S., Bloom L. B., Petruska J. (1993) Biochemical basis of DNA replication fidelity. Crit. Rev. Biochem. Mol. Biol. 28, 83–126 [DOI] [PubMed] [Google Scholar]
- 34. Nick McElhinny S. A., Kissling G. E., Kunkel T. A. (2010) Differential correction of lagging-strand replication errors made by DNA polymerases α and δ. Proc. Natl. Acad. Sci. U.S.A. 107, 21070–21075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Watt D. L., Johansson E., Burgers P. M., Kunkel T. A. (2011) Replication of ribonucleotide-containing DNA templates by yeast replicative polymerases. DNA Repair 10, 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Syväoja J., Suomensaari S., Nishida C., Goldsmith J. S., Chui G. S., Jain S., Linn S. (1990) DNA polymerases α, δ, and ϵ: three distinct enzymes from HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 87, 6664–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bonnin A., Lázaro J. M., Blanco L., Salas M. (1999) A single tyrosine prevents insertion of ribonucleotides in the eukaryotic-type φ29 DNA polymerase. J. Mol. Biol. 290, 241–251 [DOI] [PubMed] [Google Scholar]
- 38. Lin T. C., Wang C. X., Joyce C. M., Konigsberg W. H. (2001) 3′–5′ exonucleolytic activity of DNA polymerases: structural features that allow kinetic discrimination between ribo- and deoxyribonucleotide residues. Biochemistry 40, 8749–8755 [DOI] [PubMed] [Google Scholar]
- 39. Williams J. S., Clausen A. R., Nick McElhinny S. A., Watts B. E., Johansson E., Kunkel T. A. (2012) Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ϵ. DNA Repair 11, 649–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Albertson T. M., Ogawa M., Bugni J. M., Hays L. E., Chen Y., Wang Y., Treuting P. M., Heddle J. A., Goldsby R. E., Preston B. D. (2009) DNA polymerase ϵ and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc. Natl. Acad. Sci. U.S.A. 106, 17101–17104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rice G., Patrick T., Parmar R., Taylor C. F., Aeby A., Aicardi J., Artuch R., Montalto S. A., Bacino C. A., Barroso B., Baxter P., Benko W. S., Bergmann C., Bertini E., Biancheri R., Blair E. M., Blau N., Bonthron D. T., Briggs T., Brueton L. A., Brunner H. G., Burke C. J., Carr I. M., Carvalho D. R., Chandler K. E., Christen H. J., Corry P. C., Cowan F. M., Cox H., D'Arrigo S., Dean J., De Laet C., De Praeter C., Dery C., Ferrie C. D., Flintoff K., Frints S. G., Garcia-Cazorla A., Gener B., Goizet C., Goutières F., Green A. J., Guet A., Hamel B. C., Hayward B. E., Heiberg A., Hennekam R. C., Husson M., Jackson A. P., Jayatunga R., Jiang Y. H., Kant S. G., Kao A., King M. D., Kingston H. M., Klepper J., van der Knaap M. S., Kornberg A. J., Kotzot D., Kratzer W., Lacombe D., Lagae L., Landrieu P. G., Lanzi G., Leitch A., Lim M. J., Livingston J. H., Lourenco C. M., Lyall E. G., Lynch S. A., Lyons M. J., Marom D., McClure J. P., McWilliam R., Melancon S. B., Mewasingh L. D., Moutard M. L., Nischal K. K., Ostergaard J. R., Prendiville J., Rasmussen M., Rogers R. C., Roland D., Rosser E. M., Rostasy K., Roubertie A., Sanchis A., Schiffmann R., Scholl-Burgi S., Seal S., Shalev S. A., Corcoles C. S., Sinha G. P., Soler D., Spiegel R., Stephenson J. B., Tacke U., Tan T. Y., Till M., Tolmie J. L., Tomlin P., Vagnarelli F., Valente E. M., Van Coster R. N., Van der Aa N., Vanderver A., Vles J. S., Voit T., Wassmer E., Weschke B., Whiteford M. L., Willemsen M. A., Zankl A., Zuberi S. M., Orcesi S., Fazzi E., Lebon P., Crow Y. J. (2007) Clinical and molecular phenotype of Aicardi-Goutières syndrome. Am. J. Hum. Genet. 81, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bubeck D., Reijns M. A., Graham S. C., Astell K. R., Jones E. Y., Jackson A. P. (2011) PCNA directs type 2 RNase H activity on DNA replication and repair substrates. Nucleic Acids Res. 39, 3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kunkel T. A., Erie D. A. (2005) DNA mismatch repair. Annu. Rev. Biochem. 74, 681–710 [DOI] [PubMed] [Google Scholar]
- 44. Kleczkowska H. E., Marra G., Lettieri T., Jiricny J. (2001) hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 15, 724–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clark A. B., Lujan S. A., Kissling G. E., Kunkel T. A. (2011) Mismatch repair-independent tandem repeat sequence instability resulting from ribonucleotide incorporation by DNA polymerase ϵ. DNA Repair 10, 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clayton D. A. (1982) Replication of animal mitochondrial DNA. Cell 28, 693–705 [DOI] [PubMed] [Google Scholar]
- 47. Fusté J. M., Wanrooij S., Jemt E., Granycome C. E., Cluett T. J., Shi Y., Atanassova N., Holt I. J., Gustafsson C. M., Falkenberg M. (2010) Mitochondrial RNA polymerase is needed for activation of the origin of light-strand DNA replication. Mol. Cell 37, 67–78 [DOI] [PubMed] [Google Scholar]
- 48. Zenkin N., Severinov K. (2008) RNA polymerase—the third class of primases. Cell. Mol. Life Sci. 65, 2280–2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pomerantz R. T., O'Donnell M. (2008) The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 456, 762–766 [DOI] [PMC free article] [PubMed] [Google Scholar]