Background: Tssc3 is a maternally expressed imprinted gene.
Results: TSSC3 regulates Mash2 transcription in TS cells through phosphorylation of AKT and Sp1 translocation from cytoplasm to nucleus.
Conclusion: TSSC3 determines the fate of TS cells in terms of development into trophoblast progenitors and/or labyrinth trophoblasts.
Significance: TSSC3 regulates TS cell differentiation through the AKT/Sp1/MASH2 signaling pathway.
Keywords: Molecular Cell Biology, Placenta, Stem Cells, Transcription Factors, Trophoblast
Abstract
Tssc3 is a maternally expressed/paternally silenced imprinted gene. Recent evidence suggests that the loss of TSSC3 results in placental overgrowth in mice. These findings showed that the TSSC3 gene functions as a negative regulator of placental growth. In this study, we describe the function of TSSC3 and its signaling pathway in mouse trophoblast stem (TS) cell differentiation. First of all, we tested Tssc3 expression levels in TS cells. TS cells expressed Tssc3, and its expression level was the highest from day 1 to 4 but was down-regulated at day 5 after the induction of differentiation. Overexpression of TSSC3 in TS cells up-regulated Gcm1 and Mash2, which are marker genes of mouse trophoblast differentiation. Down-regulation of TSSC3 by siRNA enhanced Pl1 and Tpbpa expression in TS cells cultured under stem cell conditions, suggesting the contribution of TSSC3 to the differentiation from TS to trophoblast progenitors and/or labyrinth trophoblasts. TSSC3 activated the PI3K/AKT pathway through binding with phosphatidylinositol phosphate lipids and enhanced the activity of a promoter containing an E-box structure, which is the binding sequence of the Mash2 downstream target gene promoter. PI3K inhibitor suppressed the promoter activity induced by TSSC3. TSSC3 induced Sp1 translocation from cytoplasm to nucleus through the PI3K/AKT pathway. Nuclear Sp1 activated the Mash2 transcription by Sp1 binding with a consensus Sp1-binding motif. This is the first report describing that TSSC3 plays an important role in the differentiation from TS to trophoblast progenitors and/or labyrinth trophoblasts through the TSSC3/PI3K/AKT/MASH2 signaling pathway.
Introduction
The trophoblast cell lineage is specified at the blastocyst stage of development and contributes exclusively to the epithelial compartment of the placenta. There are four major differentiated cell types that can be derived from trophoblast stem (TS)2 cells in mice. These include trophoblast giant cells, spongiotrophoblasts, syncytiotrophoblasts, and glycogen trophoblast cells (1, 2). Several pathways controlling differentiation of the trophoblast cell lineage are known in mice. The maintenance of TS cells in the early embryo is dependent on FGF signaling involving the ligand FGF4 (3, 4), FGF receptor 2 (5), and downstream transcription factors CDX2 and EOMES (2). The orphan nuclear receptor Err2 is also required for stem cell maintenance (6, 7). MASH2, a member of the basic helix-loop-helix (bHLH) transcription factor family (8–10), is essential for differentiation into spongiotrophoblast cells, because the absence of MASH2 results in the loss of spongiotrophoblast layer, followed by abundant formation of trophoblast giant cells (11). Consistent with this finding, overexpression of MASH2 in Rcho-1 cells, a rat trophoblast tumor (choriocarcinoma) cell line, impairs trophoblast giant cell differentiation (12). Another bHLH factor, Hand1, is required for the formation of trophoblast giant cells (10). The formation of syncytiotrophoblast cells is dependent on Gcm1, which is a distinct type of transcription factor (13). In addition, GCM1 plays a critical role in labyrinth development (14). It is clear from these studies that several key transcription factors are required for the formation or maintenance of a differentiated trophoblast subtype.
The Tssc3 gene is a maternally expressed imprinted gene located at the distal part of chromosome 7 of mice and human chromosome 11p15.5 (15, 16). This 1-Mb chromosomal region contains multiple imprinted gene clusters, including p57Kip2 and Igf2, which are known to regulate fetal and placental growth in mice (17) and humans (18). Similar to the expression profiles of p57 and IGF2, TSSC3 is highly expressed in the extraembryonic ectoderm (19), and its expression persists in the labyrinth trophoblast of the definitive placenta and in the visceral yolk sac endoderm until mid-gestation in mice (19). TSSC3 encodes a cytoplasmic protein with a pleckstrin homology (PH) domain (15); thus, by analogy with other PH domain proteins, TSSC3 modulates cell signaling, intracellular trafficking, or other processes that depend on phosphatidylinositol lipid second messengers. Recent evidence suggests that the loss of TSSC3 expression results in the overgrowth of spongiotrophoblasts in mice (20). TSSC3 is expressed only in cytotrophoblasts in normal human placenta (21). These findings implicate TSSC3 in trophoblast differentiation and further resultant placental formation.
In the present study, we describe the TSSC3 function in the differentiation process from trophoblast stem cells in mice. TSSC3 regulates initial phases of trophoblast differentiation from TS upstream of MASH2 through AKT/Sp1 signaling.
EXPERIMENTAL PROCEDURES
TS Cell Culture
A TS cell line (kindly provided by Satoshi Tanaka, Tokyo University, Japan), derived from a 6.5-day postcoitum conceptus of ICR mice, was cultured in heparin/FGF-4-supplemented culture medium composed of 30% TS medium (RPMI1640 supplemented with 20% FBS, 1 mm sodium pyruvate, 50 μm 2-mercaptoethanol) and 70% mouse embryonic fibroblast-conditioned medium according to methods reported previously (4). Heparin and FGF-4 were added to final concentrations of 1 μg/ml and 25 ng/ml, respectively. Differentiation of the cells was induced by removal of FGF-4, heparin, and the mouse embryonic fibroblast-conditioned medium (3).
Adenoviral Vector and Infection
For the cloning of adenoviral vector, the respective cDNA for mouse Tssc3 or Mash2 inserted into pENTRTM-gus was digested with SalI-XhoI and newly cloned into the adenovirus expression vector of pAd/CMV/V5-DESTTM, which contains the CMV promoter. Sequences of the probes are shown in supplemental Table 1. The correct clone was identified for PacI digestion. 293A cells were transfected using Lipofectamine 2000 reagent (Invitrogen). 293A transfectants as a high titer adenovirus solution were obtained (Takara, Japan). TS cells were infected at a multiplicity of infection of 20 for 2 h at 37 °C and 5% CO2.
RNA Interference of Tssc3, Mash2, and Sp1
Double-stranded small interfering RNA (siRNA) of Tssc3 (siTssc3), Mash2 (siMash2), and SCR, which was used as a negative control, was purchased from Japan Bio Services Co., Ltd. (Saitama, Japan). Sequences of the probes are shown in supplemental Table 2. siRNA of Sp1 (siSp1) was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Transfection of TS cells with siTssc3 and siMash2 was carried out using Oligofectamine reagent according to a detailed protocol from Invitrogen.
Transient Transfection and Luciferase Reporter Assay
A Mash2 promoter (upstream −3000 to −11) construct was generated from the previously characterized mouse Mash2 promoter using PCR. Briefly, genomic DNA from mouse trophoblast stem cells served as a template for PCR using an upstream primer with a KpnI restriction site and a downstream primer with an XhoI restriction site. The amplified product was digested with KpnI and XhoI and ligated into the pGAL2-Luc reporter plasmid. The accuracy of the PCR-generated Mash2 promoter-luciferase reporter construct was verified by DNA sequencing. Primer sequences are shown in supplemental Table 1. TS cells (1 × 105 cells) were transfected with 1 μg of the pGAL2-Luc construct and 200 ng of pcDNA3.1-His-LacZ. The open reading frame (ORF) of Tssc3 or Sp1 was amplified by PCR and was ligated into the p3×FLAG-CMV7.1 expression vector (Sigma-Aldrich). Constructs were transiently transfected into TS cells using Lipofectamine 2000 reagent according to the manufacturer's instructions. pAd/CMV/DEST-TSSC3 (1.0 μg) or siRNA of Tssc3 or Mash2 was cotransfected and used to evaluate infection/transfection efficiency. 48 h after transfection, cells were collected, and lysates were prepared. Luciferase assays were performed using a luciferase assay kit (Promega, Madison, WI) and a β-galactosidase assay kit (CLONTECH).
RNA Extraction, Semiquantitative Reverse Transcription-Polymerase Chain Reaction (RT-PCR), and Real-time RT-PCR
We studied the gene expression of Tssc3, Mash2, Cdx2, Pl1, Tpbpa, or Gcm1 at the mRNA level, using real-time RT-PCR. We extracted total RNA from cultured infected or transfected cells, using ISOGEN (Nippon Gene, Tokyo, Japan). 1 μg of the extracted RNA was used for cDNA synthesis by reverse transcription using an oligo(dT) primer and SuperScriptTM II reverse transcriptase (Invitrogen) and subjected to semiquantitative RT-PCR and real-time RT-PCR. All real-time PCR was performed in triplicate for each sample with the Stratagene MX3000p system. Real-time PCR was next carried out in a total volume of 20 μl using Brilliant 2 Fast SYBR Green QPCR master mix (Stratagene, La Jolla, CA). Primer sequences are shown in supplemental Table 1. Relative expression levels were calculated using the ddCT method (23) after normalization to those of a housekeeping gene, mouse β-actin.
Western Blot Analysis
To examine the expression of various proteins, subconfluent cells were lysed with ice-cold lysis buffer (20 mm Tris-HCl (pH 8.0), 1% Triton X-100, 10% glycerol, 137 mm NaCl, 1.5 m MgCl2, and 1 mm EGTA containing freshly added protease inhibitor mixture; Nacalai Tesque, Kyoto, Japan). After centrifugation at 13,000 × g for 5 min at 4 °C to remove debris, the lysate was subjected to 7.5–15% SDS-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. The extract lysates for immunoprecipitation were incubated overnight with 1 μg of anti-FLAG or AKT antibodies and precipitated with protein A/G PLUS agarose beads (Santa Cruz Biotechnology, Inc.). The membranes were blocked in TBST (10 mm Tris-HCl (pH 7.4), 150 mm NaCl, and 0.05% Tween 20) containing 5% nonfat dry milk and washed in TBST. Anti-TSSC3 or MASH2 antibodies were raised in a rabbit using the synthetic peptide CSEMPSEPGEQSALGP or MDSRALPRPAPPAPGC-NH2, corresponding to the C-terminal region of TSSC3 or MASH2 protein. The blots were then incubated with diluted primary antibodies (GAPDG (FL-335), TSSC3 (E-14), and Sp1 (PEP2) from Santa Cruz Biotechnology, Inc.; AKT (catalog number 9272) and phospho-AKT (Ser-474) (catalog number 9271) from Cell Signaling Technology Inc. (Danvers, MA); FLAG (M2) and HA (HA-7) from Sigma). After incubation with each primary antibody, blots were incubated with horseradish peroxidase-linked anti-mouse, anti-rabbit antibodies (Amersham Biosciences) or anti-goat antibodies (DAKO, Tokyo, Japan) and analyzed with the ECL Plus system (Amersham Biosciences).
Chromatin Immunoprecipitation (ChIP) Assay
TS cells were cultured to 80% confluence in 100-mm dishes and cross-linked with 0.4% (v/v) formaldehyde for 10 min at room temperature. Cells were washed three times with ice-cold PBS with protease inhibitor mixture (Nacalai Tesque, Kyoto, Japan), scraped, and pelleted by centrifugation for 5 min at 2500 rpm at 4 °C. Pellets were treated with immunoprecipitation dilution buffer (20 mm Tris-HCl, pH 8.0, 2 mm EDTA, 150 mm NaCl, 1.0% Triton X-100, 0.01% SDS) with protease inhibitor mixture for 10 min on ice and sonicated to shear DNA. Insoluble fragments were removed by centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant was diluted in immunoprecipitation dilution buffers and then precleared with 1 μg of control IgG, 40 μl of Protein A/G-agarose beads (Santa Cruz Biotechnology, Inc.) for 1 h at 4 °C. The DNA·protein complex was immunoprecipitated using 2 μg of anti-Sp1 or histone H3 antibodies. 40 μl of Protein A/G-agarose beads (Santa Cruz Biotechnology, Inc.) were added to purify the immune complexes, and the DNA was purified by phenol/chloroform extraction. Primers designed for the Mash2 promoter site were made for sequences containing the Sp1 response or non-response regions as shown in supplemental Table 1. The expected products were subsequently resolved on 2% agarose gels.
Data Analysis
Data are represented as the mean ± S.D. and were analyzed by Student's t test. A p value of less than 0.05 was considered statistically significant.
In Situ Hybridization
E10.5 placentas were fixed to ensure integrity of cryostat sections during the in situ hybridization procedure. We prepared 10-μm cryosections of placenta. A plasmid containing a cDNA for mouse Tssc3 was used as template for synthesizing antisense and sense digoxigenin-labeled riboprobes according to the manufacturer's instructions (Roche Applied Science). Tissue sections were air-dried and fixed in ice-cold 4% paraformaldehyde in phosphate-buffered saline. Prehybridization, hybridization, and detection of alkaline phosphatase-conjugated anti-digoxigenin antibody were performed as described previously (24).
RESULTS
Tssc3 Expression Profiles in Trophoblast Stem Cell Differentiation
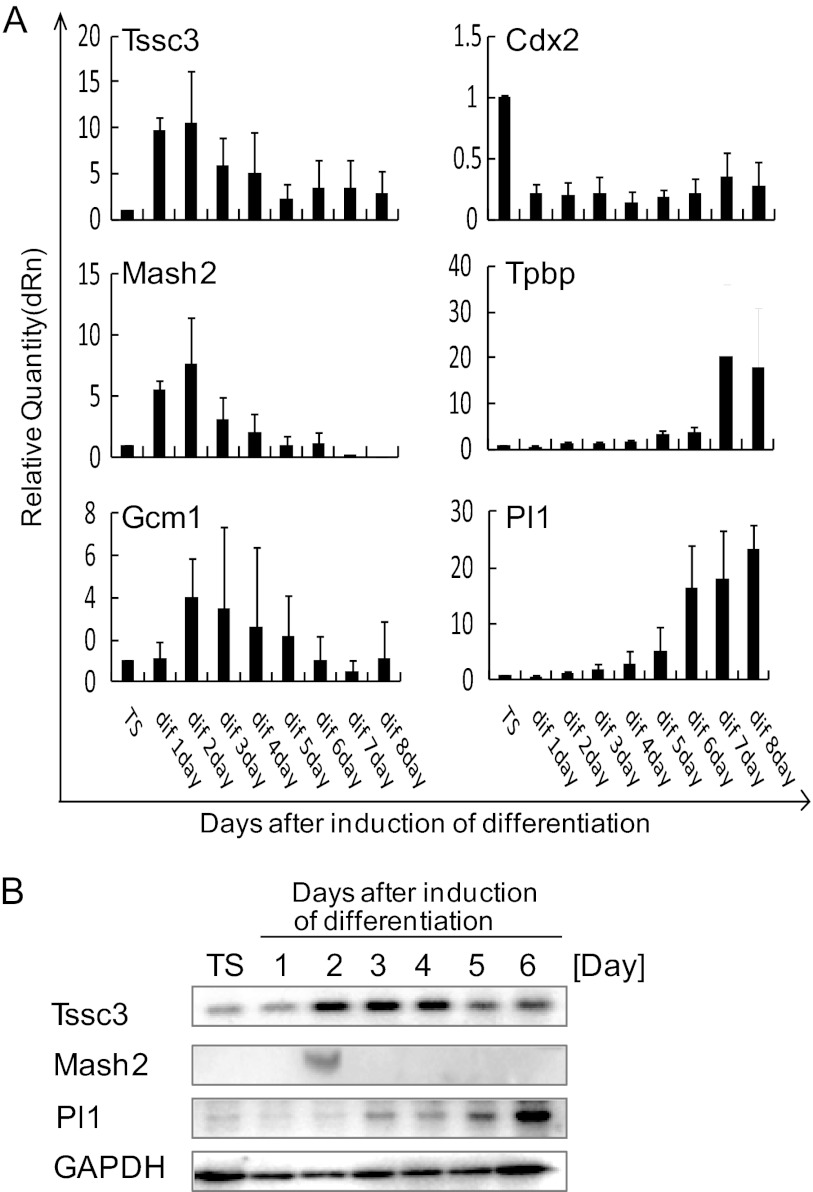
To determine whether Tssc3 is expressed in the process of trophoblast differentiation from TS cells, we performed real-time RT-PCR and Western blot analysis. Tssc3 mRNA and protein were expressed in cultured TS cells in the presence of FGF4 and conditioned medium (CM) (FGF/CM) and feeder cells. Cultured cells retained characteristics of TS cells but lost stem cell features upon withdrawal of FGF/CM (Fig. 1, A and B) (25). From day 1 to day 4 after withdrawal of FGF/CM, the Tssc3 mRNA level was the highest, followed by a decrease after day 5 (Fig. 1A). Mash2, which is known as a marker of trophoblast progenitors (26), was expressed for only a few days after the withdrawal of FGF/CM (Fig. 1, A and B). This alteration of expression profile was similar to that of Tssc3, as determined by real-time RT-PCR (Fig. 1A). Pl1 is known as a marker of trophoblast giant cells (1). Cultured TS cells after withdrawal of FGF/CM started to express Pl1 from day 4 (Fig. 1, A and B). Cdx2 is known as a marker of TS cells (27). TS cells expressed Cdx2 in the stem cell culture conditions, followed by its disappearance upon the withdrawal of FGF/CM (Fig. 1A). Tpbpa is known as a spongiotrophoblast marker (28). Cultured TS cells started to express Tpbpa from day 5 after withdrawal of FGF/CM (Fig. 1A). Gcm1, which is known as a marker of labyrinth (14), was up-regulated 2 days after withdrawal of FGF/CM, followed by a decrease after day 4. These results indicated that TSSC3 was expressed in TS cells, and the prominent up-regulation of Tssc3 at days 1–4 after withdrawal of FGF/CM was followed by a decrease of Tssc3, and withdrawal of FGF/CM from TS cell cultures resulted in the appearance of trophoblast giant cells, spongiotrophoblasts, and labyrinth trophoblasts concomitant with the disappearance of TS cells.
FIGURE 1.
TSSC3 is expressed in mouse TS cells and differentiated cells. A, TSSC3 and trophoblast markers were analyzed by real-time PCR using cDNA, which was reverse-transcribed from total RNA samples. The mRNA levels were normalized using β-actin. Results are the mean ± S.D. (error bars) from three independent experiments. B, alterations in TSSC3, MASH2, and PL1 protein levels after the withdrawal of FGF/CM. GAPDH is provided as a loading control. Results from three independent experiments are shown.
Functional Role of TSSC3 in TS Cell Differentiation
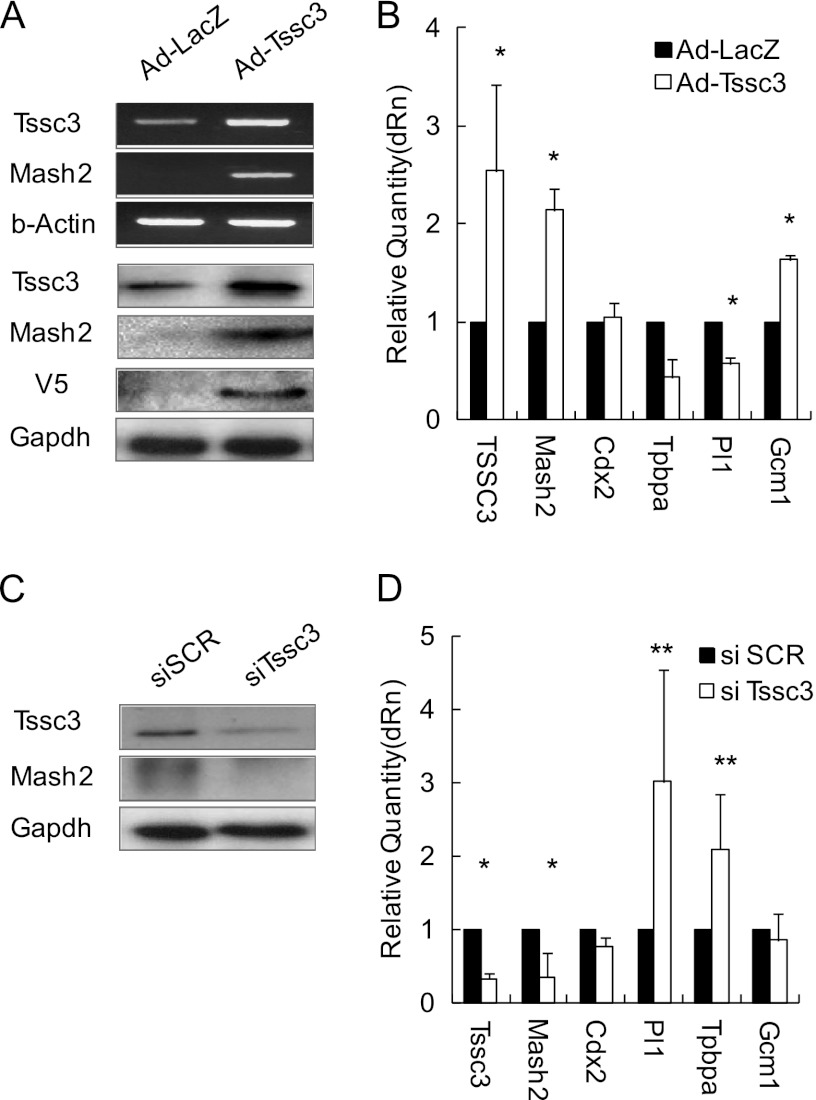
Because Tssc3 was highly expressed from day 1 to day 4 after withdrawal of FGF4/CM, we investigated the TSSC3 function in the trophoblast differentiation process. An adenovirus TSSC3 (Ad-TSSC3) expression vector system was constructed. Three days after infection, total RNA and protein were extracted. TSSC3 expression levels were quantified by RT-quantitative PCR, real-time RT-PCR, and Western blot analysis. Tssc3 expression was up-regulated 2.5-fold by Ad-TSSC3 infection (Fig. 2, A and B). Mash2 expression was examined in control adenovirus-LacZ (Ad-LacZ) and Ad-TSSC3 by real-time RT-PCR and Western blotting (Fig. 2B). Overexpression of TSSC3 led to the 2-fold up-regulation of Mash2 mRNA and protein levels. TSSC3 overexpression resulted in a 2-fold increase of Gcm1 expression in the stem cell culture conditions. In turn, the expression of Pl1 and Tpbpa was down-regulated by TSSC3 overexpression. Ad-LacZ infection failed to alter the expression levels of these markers in the stem cell culture conditions (Fig. 2B). These findings suggested that TSSC3 overexpression elicited progenitor and/or labyrinth trophoblast outgrowth but suppressed the differentiation into trophoblast giant cells and spongiotrophoblasts. Next, we designed siRNA of Tssc3 mRNA (siTssc3) to abrogate TSSC3 expression. TS cells were transfected with control siRNA (siSCR) and siTssc3 at a concentration of 150 nm for 2-day culture under the stem cell culture conditions; mRNA and protein were extracted for real-time RT-PCR and Western blot. Tssc3 expression levels were inhibited about 70% by siTssc3 (Fig. 2, C and D). TSSC3 down-regulation by siRNA resulted in suppression of MASH2 expression and up-regulation of Pl1 and Tpbpa expression levels in the stem cell culture conditions, supporting the possibility that TSSC3 contributed to the propagation of progenitor and/or labyrinth trophoblasts via MASH2 up-regulation. Suppression of Gcm1 by siTssc3 was statistically insignificant.
FIGURE 2.
TSSC3 affects MASH2 expression. A, top panels, mRNA level of Tssc3 and Mash2; bottom panels, protein levels in TSSC3-overexpressing TS cells. B, the mRNA expression levels were assessed by real-time PCR using cDNA from TSSC3-overexpressing TS cells. The mRNA levels were normalized using Gapdh. Results are the mean ± S.D. (error bars) from three independent experiments. The asterisks indicate statistical significance (*, p < 0.01). C, these panels show TSSC3 and Mash2 protein levels in TSSC3 knockdown TS cells. D, the mRNA expression was assessed by real-time PCR using cDNA from TSSC3 knockdown TS cells. The mRNA levels were normalized using Gapdh. Results are the mean ± S.D. from three independent experiments (*, p < 0.01; **, p < 0.05).
Regulation of MASH2 Expression by TSSC3
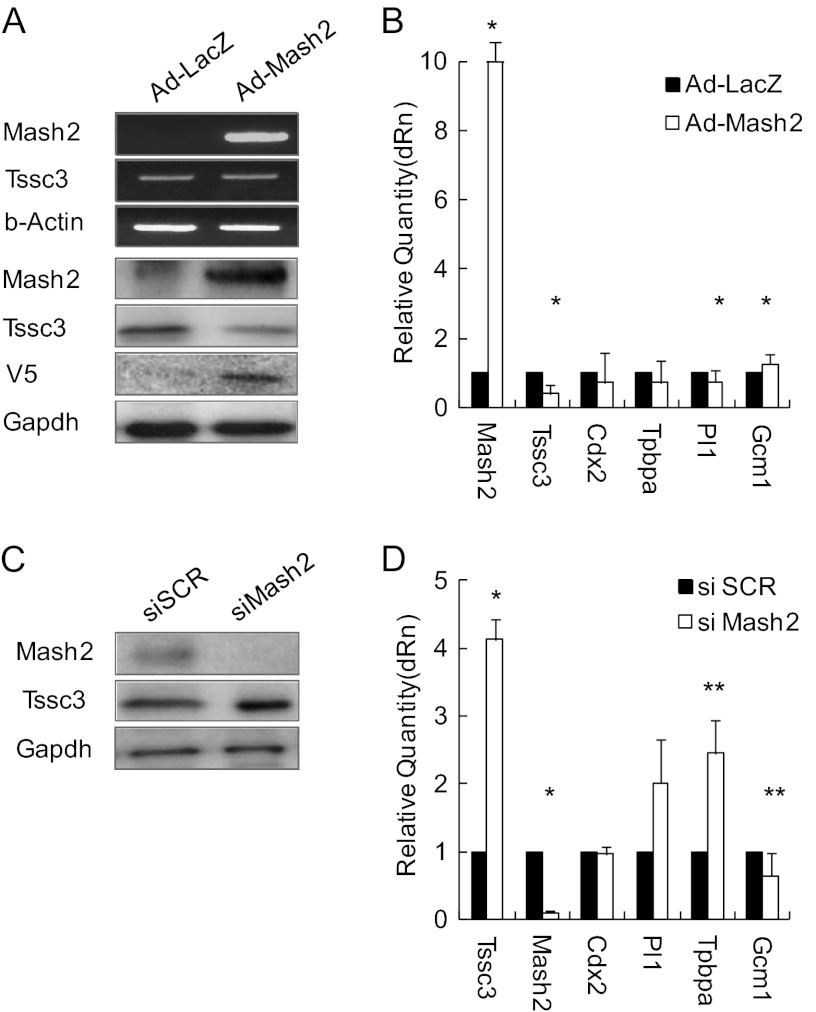
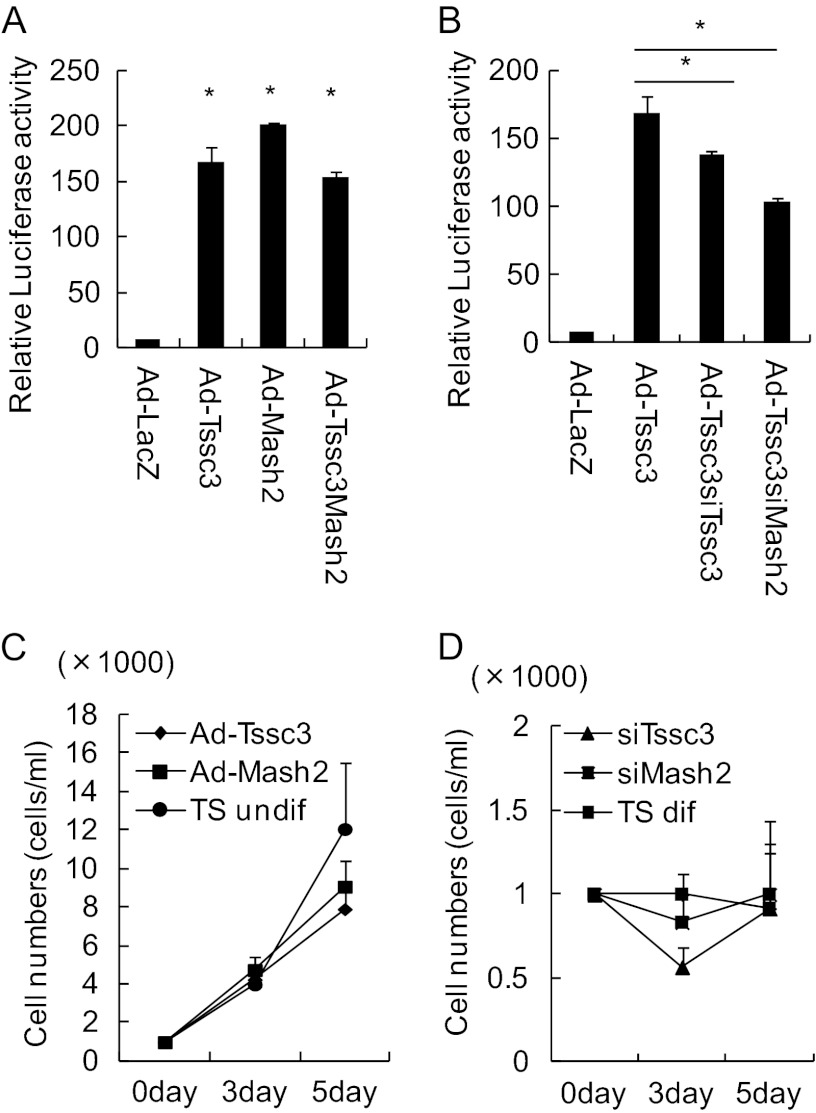
The Mash2 gene, which encodes a transcription factor of the bHLH class, is expressed at high levels in trophoblast progenitors and differentiated progeny trophoblasts (26) and is essential for the morphogenesis and the function of spongiotrophoblasts (11). To examine the interaction of TSSC3 and MASH2, we constructed adenovirus-MASH2 vector in addition to TSSC3 vector to dissect the TSSC3/MASH2 signaling pathway (Fig. 3A). Overexpression of MASH2 promoted Gcm1 expression levels by 1.3-fold but down-regulated Tpbpa and Pl1 expression levels in TS cells cultured under stem cell culture conditions (Fig. 3B). We designed Mash2 siRNA (siMash2) and transfected TS cells with control siSCR and siMash2 (100 nm) under stem cell culture conditions. mRNA and protein were extracted for Western blotting and real-time RT-PCR 2 days after siRNA treatment. Mash2 expression levels were suppressed about 80% by siMash2 but not by control siSCR (Fig. 3, C and D). Down-regulation of MASH2 promoted Tssc3, Pl1 and Tpbpa expression and suppressed Gcm1 expression (Fig. 3D). The TSSC3 up-regulation was compatible with the data that TSSC3 expression was increased in Mash2-null (Mash2−/−) placenta on E9.5 compared with that in wild-type placenta (29). Up-regulation of TSSC3 promoted MASH2 expression, and down-regulation of TSSC3 suppressed MASH2 expression. These findings suggest that TSSC3 is involved upstream of MASH2. In order to confirm the regulation of MASH2 expression by TSSC3, we examined the activity of MASH2 as a transcription factor using the pGL2 construct, including a MASH2 binding site, which is known as the E-box. The bHLH family, including MASH2, binds to this E-box region with E-factor and regulates transcriptional activity (30). The MASH2·E-factor dimer binds to the promoter and activates its activity via the E-box consensus sequence (CANNTG) (30). The promoter activity was promoted by TSSC3 as well as a positive control, MASH2 (Fig. 4A). siTssc3 or siMash2 was transfected to TS cells cultured under stem cell culture conditions, and then the activity of the promoter containing the E-box structure was evaluated. Both siTssc3 and siMash2 suppressed the promoter activity in adeno-infected TS cells (Fig. 4B).
FIGURE 3.
Change of Mash2 expression is similar to TSSC3. A, top panels, Tssc3 and Mash2 mRNA levels in MASH2-overexpressing TS cells. Bottom panels, TSSC3 and Mash2 protein levels in MASH2-overexpressing TS cells. B, mRNA expression levels were assessed by real-time PCR using cDNA from MASH2-overexpressing TS cells. The mRNA levels were normalized using Gapdh. Results are the mean ± S.D. (error bars) from three independent experiments (*, p < 0.01). C, these panels show TSSC3 and MASH2 protein levels in MASH2 knockdown TS cells. D, Mash2 and Tssc3 mRNA expression levels were assessed by real-time PCR using cDNA from MASH2 knockdown TS cells. mRNA levels were normalized using Gapdh. Results are the mean ± S.D. from three independent experiments (*, p < 0.01; **, p < 0.05).
FIGURE 4.
TSSC3 up-regulation of Mash2 as a transcription factor. A, luciferase activity with Ad-TSSC3, Ad-MASH2, Ad-TSSC3, and MASH2. A reporter gene possessing the E-box was constructed. Luciferase activity values were normalized using β-galactosidase activity values. Results are the mean ± S.D. (error bars) from three independent experiments (*, p < 0.01). B, alteration in luciferase activity of E-box reporter by knockdown of TSSC3 or MASH2 by siRNA in TSSC3-overexpressing TS cells. Results are the mean ± S.D. from three independent experiments (*, p < 0.01). C, alteration of cell growth by overexpression of TSSC3 or MASH2 in TS cells is shown. D, alteration of cell growth by knockdown of TSSC3 or MASH2 in TS cells is shown.
Previously, a large placenta was shown in TSSC3-null pregnant mice by expansion of spongiotrophoblasts. However, fetal growth was found to be normal in TSSC3-null mice (20). To demonstrate whether TSSC3 affects trophoblast cell proliferation, we examined the alteration of cell proliferation in Ad-TSSC3-infected and siTssc3-transfected TS cells under stem cell culture conditions. Alteration in TSSC3 expression by Ad-TSSC3 or siTssc3 did not change the cell growth patterns of TS cells in the presence of FGF/CM (Fig. 4C) or by the withdrawal of FGF/CM (Fig. 4D).
Activation of the PI3K-AKT Pathway Signal by TSSC3
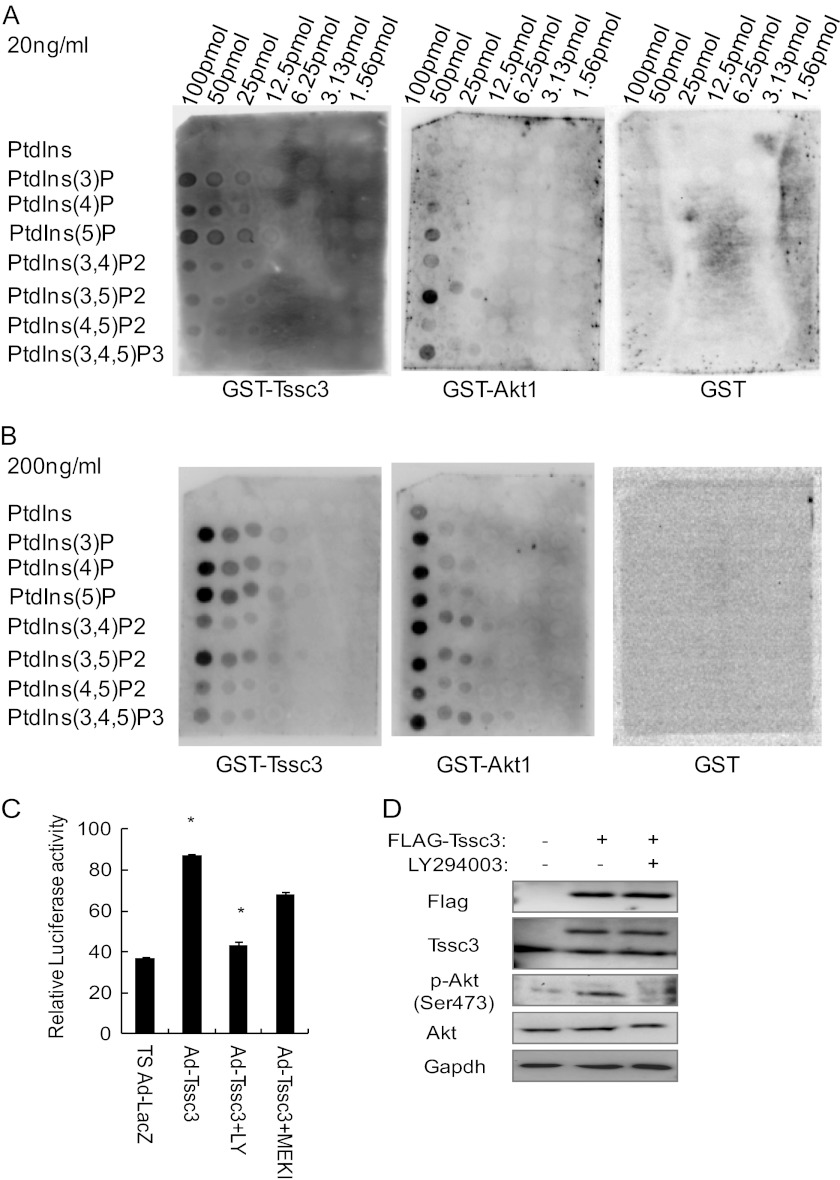
Most PH domain-containing proteins can bind to certain phosphatidylinositol phosphate lipids (PIPs), but their binding affinities for the specific products of phosphoinositide metabolism were found to be variable (31). We made GST fusion TSSC3 recombinant protein to examine whether TSSC3, including the PH domain, binds to PIPs. As shown in Fig. 5A, TSSC3 bound to PIPs weakly at a concentration of 20 ng/ml. However, TSSC3 bound to most PIPs strongly when the concentration of GST-TSSC3 increased to 200 ng/ml (Fig. 5B). GST-AKT1 was used as a positive control. These findings suggested that TSSC3 was involved in the phosphoinositide 3-kinase (PI3K)-dependent pathway by binding with PIPs. E-box promoter activity was examined to demonstrate the pathway between TSSC3 and PI3K by using LY294002, which is a PI3K inhibitor, and PD184161, which is an MEK inhibitor. E-box promoter activity induced by TSSC3 was sharply suppressed by LY294002 but not PD184161 (Fig. 5C). AKT is a serine/threonine kinase downstream of PI3K. TSSC3 overexpression resulted in increases of phosphorylated AKT. PI3K inhibitor suppressed AKT phosphorylation in the presence of TSSC3 overexpression (Fig. 5D).
FIGURE 5.
TSSC3 is involved in the PI3K-AKT pathway. Analysis of PIP arrays using GST-TSSC3. The indicated phospholipids were spotted onto a nitrocellulose membrane at concentrations from 1.6 to 100 pmol/spot. Membranes were incubated with the purified GST fusion protein at 20 (A) or 200 ng/ml (B). GST-AKT is a positive control, and GST is a negative control. C, luciferase activity of E-box reporter was suppressed significantly by PI3K inhibitor but not by MEK inhibitor in TSSC3-overexpressing TS cells. Luciferase activity values were normalized by β-galactosidase activity values. Results are the mean ± S.D. (error bars) from three independent experiments (*, p < 0.05). LY, LY294002, PI3K inhibitor; MEK, MEK inhibitor. D, phosphorylated AKT induced by p3xFLAG-TSSC3 is suppressed by the PI3K inhibitor. GAPDH is provided as a loading control.
Up-regulation of Mash2 Transcription Activated by Sp1 Transcription Factor through TSSC3/AKT Signaling
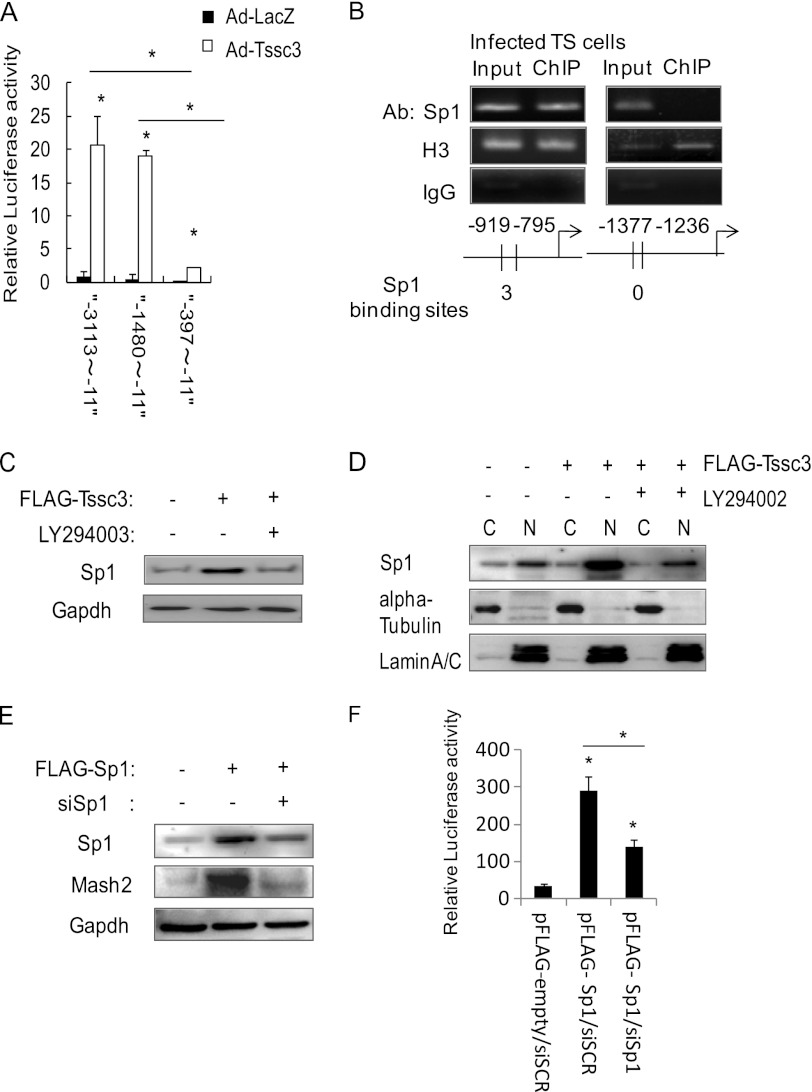
We made luciferase reporter constructs containing the Mash2 promoter in the upstream region −11 to −3113 bp from the transcription start site and their truncated mutants. The constructs were ligated with the luciferase reporter. The reporter possessing a region spanning −11 to −1480 bp showed high transcriptional activity; the level was compatible with that for the −11 to −3113 bp region. The luciferase activity was low in the reporter construct possessing the region of −11 to −397 bp (Fig. 6A). These results showed that the Mash2 promoter was at least in part involved in the region of −397 to −1480 bp from the transcription start site. To demonstrate what kinds of transcription factors bind to Mash2 promoter, we searched for binding sites of transcription factors on the putative Mash2 promoter region using the ALIBABA 2.1 database. We identified consensus Sp1 binding motifs. To examine whether Sp1 binds to the Mash2 promoter and activates its transcription, we carried out ChIP analysis using Sp1 antibody. Consensus Sp1 binding motifs were present at three regions: −373 to −509 bp, −591 to −715 bp, and −795 to −919 bp from the transcription start site. Sp1 was immunoprecipitated abundantly by anti-Sp1 antibody at −795 to −919 bp but weakly at the −373 to −509 bp and −591 to −715 bp regions (Fig. 6B) (data not shown). These findings indicated that Sp1 binds to the Mash2 promoter region at −795 to −919 bp and contributes to its transcription. Our inability to demonstrate Sp1 binding motif at the −373 to −509 bp and −591 to −715 bp regions could indicate that this is not a true Sp1 binding site, or this may just be a DNA region that cannot be amplified by PCR. To investigate whether Sp1 expression is induced by TSSC3, we examined the alteration of Sp1 expression levels using Western blot analysis in FLAG tag TSSC3-expressing TS cells. Sp1 expression was modestly up-regulated by TSSC3 expression and was sharply down-regulated by LY294002 (Fig. 6C). We fractionated the extract of TS cells transfected with FLAG tag TSSC3 in the absence or presence of LY294002 into nuclear and cytoplasmic compartments, followed by Western blot analysis. As shown in Fig. 6D, TSSC3 increased the nuclear Sp1, whereas LY294002 blocked Sp1 translocation from cytoplasm to nucleus. These result suggested that TSSC3 contributed to the Sp1 translocation from cytoplasm to nucleus, resulting in the up-regulation of Mash2 transcription. Next, the Sp1 overexpression system (FLAG-Sp1) was constructed, and we designed siRNA of Sp1 mRNA (siSp1) to abrogate Sp1 expression. Sp1 overexpression resulted in increases of MASH2. Sp1 inhibition by siRNA suppressed MASH2 expression in Sp1-overexpressed TS cells (Fig. 6E). The promoter activity via E-box consensus sequence was examined to demonstrate the pathway between Sp1 and MASH2 by using FLAG-Sp1 and siSp1. The activity was sharply up-regulated by FLAG-Sp1 but suppressed by siSp1 in Sp1-overexpressed TS cells (Fig. 6F). These findings suggested that TSSC3-Sp1 signaling was at least in part one of the MASH2 regulatory expression systems.
FIGURE 6.
PI3K/AKT/Sp1 signaling pathway activated by TSSC3. A, 5′-upstream region of Mash2 spanning from −1480 to −11 from the TSS shows luciferase activity similar to that of −3113 to −11 from TSS. A region spanning −397 to −11 fails to show enhanced activity. Luciferase activity values were normalized by β-galactosidase activity values. Results are the mean ± S.D. (error bars) from three independent experiments (**, p < 0.01). B, ChIP assay shows the association between Sp1 and Sp1 binding sites. Anti-histone H3 antibody (Ab) was used as a loading control. Rabbit control IgG was used as a negative control. C, alterations of Sp1 protein levels by LY294002 in FLAG-TSSC3-overexpressing TS cells. GAPDH is provided as a loading control. D, nuclear translocation of Sp1 induced by FLAG-TSSC3 is suppressed by LY294002. α-Tubulin and lamin A/C are provided as technical controls for cell fractionation (C, cytoplasm; N, nucleus). E, alterations of MASH2 protein levels by knockdown of Sp1 siRNA in Sp1-overexpressing TS cells. GAPDH is provided as a loading control. F, alteration in luciferase activity of E-box reporter by knockdown of Sp1 by siRNA in Sp1-overexpressing TS cells. Results are the mean ± S.D. from three independent experiments (*, p < 0.05).
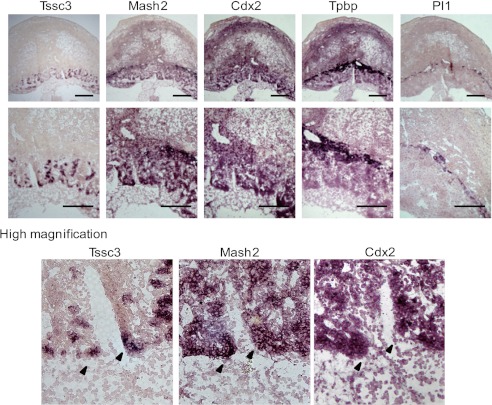
TSSC3 is expressed in labyrinthine layer of mouse placenta (20, 32). MASH2 is expressed in precursor cells located in the Spongiotrophoblast and labyrinthine layers, respectively (26). To examine the expression profile of Tssc3 and Mash2 during wild-type mouse placental development, we performed in situ hybridization using E10.5 placenta. The results demonstrated that Tssc3 was detected in the labyrinthine layer, and Mash2 was detected in the labyrinthine and spongiotrophoblast layers, respectively (Fig. 7, top and middle). Tssc3 was not detected in Tpbp-positive (spongiotrophoblast layer) and Pl1-positive (giant cell) areas (Fig. 7, top and middle). TS cells (Cdx2-positive) were included in the labyrinthine layer (Fig. 7, top and middle). The comparative presentation in Fig. 7 (bottom panels) clearly showed that Tssc3 was detected partly in Mash2-positive, Cdx2-positive, or Mash2/Cdx2-double-positive areas.
FIGURE 7.
Expression profiles of Tssc3 and Mash2 in the wild-type placenta. The expression patterns of trophoblast-specific markers were analyzed by in situ hybridization on a frozen section of wild-type placenta collected at E10.5. Sense probes are not shown. Top panels, all trophoblast-specific markers were detected in E10.5. Middle panels, Tssc3 and Cdx2 located in labyrinthine layer. Mash2 was expressed in labyrinthine and spongiotrophoblast layers. Tpbp was located in the spongiotrophoblast layer. Pl1 was expressed in giant cells. A, scale bar, 500 μm. Bottom panels, Tssc3 was expressed partially in Mash2-positive or Cdx2-positive areas.
DISCUSSION
The present study demonstrated that TS cells express Tssc3, and its expression level is the highest from day 1 to 4 after the withdrawal of FGF/CM, followed by a decline after day 5. These findings implicate TSSC3 in the differentiation processes from TS to various cell lineages. TSSC3 overexpression did not affect Cdx2 expression levels but significantly decreased Pl1 and Tpbpa expression levels in TS cells cultured under stem cell culture conditions. TSSC3 overexpression up-regulated MASH2 and Gcm1 expression levels. In contrast, knockdown of TSSC3 by siRNA resulted in the up-regulation of Pl1 and Tpbpa levels but decreased Mash2 mRNA and protein levels. Gcm1 expression was not affected by TSSC3 knockdown, although the reason for this remains unclear. The findings suggest that TSSC3 elicits the expression of precursor and/or labyrinth trophoblast markers. TSSC3 null mice show placental overgrowth concomitant with an increased spongiotrophoblast layer, although fetal growth and size are not affected (20), which is compatible with the present finding suggesting the negative regulation of differentiation by TSSC3 from TS to spongiotrophoblast and/or trophoblast giant cell lineages. TSSC3 was not associated with TS cell proliferation upon culture under stem cell culture conditions. Thus, placental overgrowth in TSSC3 null mice reflects preferential differentiation from TS into spongiotrophoblast and/or trophoblast giant cell lineages. These findings indicate that TSSC3 plays important roles in the differentiation from TS cells into trophoblast progenitors and/or labyrinth trophoblasts. TSSC3 is expressed in the ectoplacental cone and chorion at E8.0, but its expression is restricted in syncytiotrophoblast layer 1 and is at a lower level in layer 2 of the labyrinth layer at E10.5. In normal human placenta, cytotrophoblasts express TSSC3, but syncytiotrophoblasts fail to express it (21, 33). These localizations of TSSC3 possibly reflect the fact that TSSC3 contributes to the initial phase of differentiation from TS cells to trophoblast progenitors and/or labyrinth trophoblasts.
TSSC3 modulates many functions, such as cell signaling, that depend on phosphatidylinositol lipid second messengers. Phosphoinositides are negatively charged constituents of lipid membranes formed by phosphorylation of hydroxyl groups at position 3′, 4′, or 5′ of the inositol ring of phosphatidylinositol by specific kinases. Phosphatidylinositol 1,4,5-trisphosphate acts as a second messenger by binding to and activating PH domain-containing proteins, including the Ser/Thr kinase AKT/protein kinase B (34–36). The PIP/AKT pathway is a major survival pathway. TSSC3 bound PIPs as well as Phlda3, which is a TSSC3 homolog (37) (Fig. 5). A previous study reported that PHLDA3 competes with the PH domain of AKT for the binding of membrane lipids, thereby inhibiting AKT translocation to the cellular membrane and activation. However, activation of AKT (Ser 473) was induced by trophoblast stem cell differentiation (38). The present study demonstrated that TSSC3 overexpression activated AKT phosphorylation. In addition, PI3K inhibitor inhibited activation of AKT induced by TSSC3 (Fig. 5). Because TSSC3 is expected to induce differentiation from TS cells, our finding that TSSC3 activates the PI3K/AKT pathway through binding with PIPs coincides with previous reports (37). Because the behavior of TSSC3 may be cell type-specific, additional studies are required to resolve further functions of TSSC3 in other cell lineages.
In order to address the interaction between TSSC3 and MASH2, we determined the mouse Mash2 promoter region (Fig. 6A). Among several mutant constructs, the transcriptional activity of the construct containing the region upstream to −1480 bp from the Mash2 transcription start site (TSS) was similar to that of −3113 bp from the TSS. The construct of −397 bp from the TSS failed to show promoter activity. Three Sp1 binding sites were involved in the Mash2 −1480 to −397 bp promoter region. Consensus Sp1 binding motifs were present at −373 to −509 bp, −591 to −715 bp, and −795 to −919 bp from the transcription start site. Sp1 was immunoprecipitated abundantly by anti-Sp1 antibody at −795 to −919 bp but weakly at two other regions. Sp1 overexpression up-regulated MASH2 expression and transcriptional activity by Mash2 (Fig. 6, E and F). These suggest that the Sp1 binding region at −795 to −919 bp from TSS is very important for the Mash2 transcriptional regulation by Sp1 and that Sp1 is a critical transcription factor of Mash2. It has been previously reported that Sp1 regulates various important genes associated with trophoblast cell proliferation and differentiation (39, 40). The present study established for the first time that Sp1 is a transcription factor of Mash2 and regulates Mash2 transcription downstream of TSSC3. TSSC3 overexpression did not affect Sp1 expression levels but promoted Sp1 translocation from cytoplasm to nucleus. In addition, the translocation induced by TSSC3 overexpression was inhibited by LY294002, a PI3K inhibitor. The association of Sp1 activation or translocation has been shown through the MAPK/ERK pathway (42). However, Sp1 translocation was suppressed by LY294002 in MCF7 cells as shown in a previous study (41) and by the present results (Fig. 6). TSSC3 induced the activation of Sp1 by the PI3K/AKT pathway.
An intriguing aspect of Tssc3 expression profiles is co-localization of Mash2- and/or Cdx2-expressing areas in mouse placenta. Cdx2, which is a TS cell marker, is expressed in labyrinthine layer (27). Mash2 is a trophoblast precursor marker and is expressed in labyrinthine and spongiotrophoblast layers (26). Our data showed that Tssc3 is partially expressed in Mash2-positive areas (Fig. 7). High levels of Mash2 expression were also detected in the spongiotrophoblast layer, which did not express Tssc3. The present detection of Tssc3 expression in Cdx2- and/or Mash2-positive areas implicates TSSC3 function in TS to progenitor cell differentiation.
We are particularly interested in the finding that TSSC3 overexpression up-regulated MASH2 expression, but TSSC3 knockdown by siRNA down-regulated Mash2 mRNA and protein levels. Additionally, TSSC3 promoted the transcriptional activity by the MASH2·E-factor dimer (Fig. 5). These findings suggest that TSSC3 functions upstream of MASH2 and regulates MASH2 expression. MASH2, which is known as a marker of trophoblast progenitors (26), is expressed from day 1 to day 4 after withdrawal of FGF/CM, its expression profile being similar to that of TSSC3. These findings suggest that TSSC3 upstream of MASH2 contributes to the differentiation from TS to trophoblast progenitors and/or labyrinth trophoblasts by up-regulating the transcriptional levels of Mash2. In the present study, we demonstrated that TSSC3 activates PI3K/AKT signaling through binding with PIPs, activates the PI3K/AKT pathway, and translocates Sp1 from cytoplasm to nucleus and that nuclear Sp1 binds with the Sp1 binding consensus motif on the Mash2 promoter and up-regulates Mash2 transcription. This TSSC3/AKT/Sp1/MASH2 pathway is important for the initial differentiation processes from TS cells.
In summary, our study demonstrated that TSSC3 enhanced Mash2 transcription in mouse TS cells through the binding to PIP, phosphorylation of AKT, and Sp1 translocation from cytoplasm to nucleus, contributing to the trophoblast cell fate determination. These insights help us to understand the mechanism of trophoblast differentiation processes from mouse TS cells.
Supplementary Material
Acknowledgments
We are very grateful to Dr. Michiko Shirane for help with the technique for the PIP array. We also appreciate the technical support from the Research Support Center, Graduate School of Medical Sciences, Kyushu University.
This work was supported by Grants-in-aid 23249075 and 23592405 from the Ministry of Education, Culture, Sports, Science, and Technology, Japan.

This article contains supplemental Tables 1 and 2.
- TS
- trophoblast stem
- bHLH
- basic helix-loop-helix
- PH
- pleckstrin homology
- Ad
- adenovirus
- PIP
- phosphatidylinositol phosphate lipid
- En
- embryonic day n.
REFERENCES
- 1. Cross J. C., Anson-Cartwright L., Scott I. C. (2002) Transcription factors underlying the development and endocrine functions of the placenta. Recent Prog. Horm. Res. 57, 221–234 [DOI] [PubMed] [Google Scholar]
- 2. Rossant J., Cross J. C. (2001) Placental development. Lessons from mouse mutants. Nat. Rev. Genet. 2, 538–548 [DOI] [PubMed] [Google Scholar]
- 3. Feldman B., Poueymirou W., Papaioannou V. E., DeChiara T. M., Goldfarb M. (1995) Requirement of FGF-4 for postimplantation mouse development. Science 267, 246–249 [DOI] [PubMed] [Google Scholar]
- 4. Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. (1998) Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- 5. Arman E., Haffner-Krausz R., Chen Y., Heath J. K., Lonai P. (1998) Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc. Natl. Acad. Sci. U.S.A. 95, 5082–5087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo J., Sladek R., Bader J. A., Matthyssen A., Rossant J., Giguère V. (1997) Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-β. Nature 388, 778–782 [DOI] [PubMed] [Google Scholar]
- 7. Tremblay G. B., Kunath T., Bergeron D., Lapointe L., Champigny C., Bader J. A., Rossant J., Giguère V. (2001) Diethylstilbestrol regulates trophoblast stem cell differentiation as a ligand of orphan nuclear receptor ERR β. Genes Dev. 15, 833–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guillemot F., Nagy A., Auerbach A., Rossant J., Joyner A. L. (1994) Essential role of Mash-2 in extraembryonic development. Nature 371, 333–336 [DOI] [PubMed] [Google Scholar]
- 9. Riley P., Anson-Cartwright L., Cross J. C. (1998) The Hand1 bHLH transcription factor is essential for placentation and cardiac morphogenesis. Nat. Genet. 18, 271–275 [DOI] [PubMed] [Google Scholar]
- 10. Scott I. C., Anson-Cartwright L., Riley P., Reda D., Cross J. C. (2000) The HAND1 basic helix-loop-helix transcription factor regulates trophoblast differentiation via multiple mechanisms. Mol. Cell Biol. 20, 530–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanaka M., Gertsenstein M., Rossant J., Nagy A. (1997) Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev. Biol. 190, 55–65 [DOI] [PubMed] [Google Scholar]
- 12. Cross J. C., Flannery M. L., Blanar M. A., Steingrimsson E., Jenkins N. A., Copeland N. G., Rutter W. J., Werb Z. (1995) Hxt encodes a basic helix-loop-helix transcription factor that regulates trophoblast cell development. Development 121, 2513–2523 [DOI] [PubMed] [Google Scholar]
- 13. Anson-Cartwright L., Dawson K., Holmyard D., Fisher S. J., Lazzarini R. A., Cross J. C. (2000) The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25, 311–314 [DOI] [PubMed] [Google Scholar]
- 14. Stecca B., Nait-Oumesmar B., Kelley K. A., Voss A. K., Thomas T., Lazzarini R. A. (2002) Gcm1 expression defines three stages of chorio-allantoic interaction during placental development. Mech. Dev. 115, 27–34 [DOI] [PubMed] [Google Scholar]
- 15. Qian N., Frank D., O'Keefe D., Dao D., Zhao L., Yuan L., Wang Q., Keating M., Walsh C., Tycko B. (1997) The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum. Mol. Genet. 6, 2021–2029 [DOI] [PubMed] [Google Scholar]
- 16. Lee M. P., Feinberg A. P. (1998) Genomic imprinting of a human apoptosis gene homologue, TSSC3. Cancer Res. 58, 1052–1056 [PubMed] [Google Scholar]
- 17. Caspary T., Cleary M. A., Perlman E. J., Zhang P., Elledge S. J., Tilghman S. M. (1999) Oppositely imprinted genes p57Kip2 and Igf2 interact in a mouse model for Beckwith-Wiedemann syndrome. Genes Dev. 13, 3115–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maher E. R., Reik W. (2000) Beckwith-Wiedemann syndrome. Imprinting in clusters revisited. J. Clin. Invest. 105, 247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frank D., Mendelsohn C. L., Ciccone E., Svensson K., Ohlsson R., Tycko B. (1999) A novel pleckstrin homology-related gene family defined by Ipl/TSSC3, TDAG51, and Tih1. Tissue-specific expression, chromosomal location, and parental imprinting. Mamm. Genome 10, 1150–1159 [DOI] [PubMed] [Google Scholar]
- 20. Frank D., Fortino W., Clark L., Musalo R., Wang W., Saxena A., Li C. M., Reik W., Ludwig T., Tycko B. (2002) Placental overgrowth in mice lacking the imprinted gene Ipl. Proc. Natl. Acad. Sci. U.S.A. 99, 7490–7495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kato H., Matsuda T., Hirakawa T., Ueda K., Inoue T., Miyanari Y., Asanoma K., Nakano H., Wake N. (2005) Differential diagnosis between complete and partial mole by TSSC3 antibody completely correlates to DNA diagnosis. Diagn. Mol. Pathol. 14, 164–169 [DOI] [PubMed] [Google Scholar]
- 22. Faria T. N., Soares M. J. (1991) Trophoblast cell differentiation. Establishment, characterization, and modulation of a rat trophoblast cell line expressing members of the placental prolactin family. Endocrinology 129, 2895–2906 [DOI] [PubMed] [Google Scholar]
- 23. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Ain R., Canham L. N., Soares M. J. (2003) Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse. Novel endocrine phenotype and regulation. Dev. Biol. 260, 176–190 [DOI] [PubMed] [Google Scholar]
- 25. Natale D. R., Hemberger M., Hughes M., Cross J. C. (2009) Activin promotes differentiation of cultured mouse trophoblast stem cells towards a labyrinth cell fate. Dev. Biol. 335, 120–131 [DOI] [PubMed] [Google Scholar]
- 26. Guillemot F., Caspary T., Tilghman S. M., Copeland N. G., Gilbert D. J., Jenkins N. A., Anderson D. J., Joyner A. L., Rossant J., Nagy A. (1995) Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9, 235–242 [DOI] [PubMed] [Google Scholar]
- 27. Kuckenberg P., Buhl S., Woynecki T., van Fürden B., Tolkunova E., Seiffe F., Moser M., Tomilin A., Winterhager E., Schorle H. (2010) The transcription factor TCFAP2C/AP-2gamma cooperates with CDX2 to maintain trophectoderm formation. Mol. Cell Biol. 30, 3310–3320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iwatsuki K., Shinozaki M., Sun W., Yagi S., Tanaka S., Shiota K. (2000) A novel secretory protein produced by rat spongiotrophoblast. Biol. Reprod. 62, 1352–1359 [DOI] [PubMed] [Google Scholar]
- 29. Oh-McGinnis R., Bogutz A. B., Lefebvre L. (2011) Partial loss of Ascl2 function affects all three layers of the mature placenta and causes intrauterine growth restriction. Dev. Biol. 351, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson J. E., Birren S. J., Saito T., Anderson D. J. (1992) DNA binding and transcriptional regulatory activity of mammalian achaete-scute homologous (MASH) proteins revealed by interaction with a muscle-specific enhancer. Proc. Natl. Acad. Sci. U.S.A. 89, 3596–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemmon M. A., Ferguson K. M. (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350, 1–18 [PMC free article] [PubMed] [Google Scholar]
- 32. Salas M., John R., Saxena A., Barton S., Frank D., Fitzpatrick G., Higgins M. J., Tycko B. (2004) Placental growth retardation due to loss of imprinting of Phlda2. Mech. Dev. 121, 1199–1210 [DOI] [PubMed] [Google Scholar]
- 33. Takao T., Asanoma K., Kato K., Fukushima K., Tsunematsu R., Hirakawa T., Matsumura S., Seki H., Takeda S., Wake N. (2011) Isolation and characterization of human trophoblast side-population (SP) cells in primary villous cytotrophoblasts and HTR-8/SVneo cell line. PLoS One 6, e21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yuan T. L., Cantley L. C. (2008) PI3K pathway alterations in cancer. Variations on a theme. Oncogene 27, 5497–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Engelman J. A., Luo J., Cantley L. C. (2006) The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7, 606–619 [DOI] [PubMed] [Google Scholar]
- 36. Osaki M., Oshimura M., Ito H. (2004) PI3K-Akt pathway. Its functions and alterations in human cancer. Apoptosis 9, 667–676 [DOI] [PubMed] [Google Scholar]
- 37. Saxena A., Morozov P., Frank D., Musalo R., Lemmon M. A., Skolnik E. Y., Tycko B. (2002) Phosphoinositide binding by the pleckstrin homology domains of Ipl and Tih1. J. Biol. Chem. 277, 49935–49944 [DOI] [PubMed] [Google Scholar]
- 38. Kamei T., Jones S. R., Chapman B. M., MCGonigle K. L., Dai G., Soares M. J. (2002) The phosphatidylinositol 3-kinase/Akt signaling pathway modulates the endocrine differentiation of trophoblast cells. Mol. Endocrinol. 16, 1469–1481 [DOI] [PubMed] [Google Scholar]
- 39. Okamoto Y., Sakata M., Yamamoto T., Nishio Y., Adachi K., Ogura K., Yamaguchi M., Takeda T., Tasaka K., Murata Y. (2001) Involvement of nuclear transcription factor Sp1 in regulating glucose transporter-1 gene expression during rat trophoblast differentiation. Biochem. Biophys. Res. Commun. 288, 940–948 [DOI] [PubMed] [Google Scholar]
- 40. Johnson W., Jameson J. L. (1999) AP-2 (activating protein 2) and Sp1 (selective promoter factor 1) regulatory elements play distinct roles in the control of basal activity and cyclic adenosine 3′,5′-monophosphate responsiveness of the human chorionic gonadotropin-β promoter. Mol. Endocrinol. 13, 1963–1975 [DOI] [PubMed] [Google Scholar]
- 41. Mireuta M., Darnel A., Pollak M. (2010) IGFBP-2 expression in MCF-7 cells is regulated by the PI3K/AKT/mTOR pathway through Sp1-induced increase in transcription. Growth Factors 28, 243–255 [DOI] [PubMed] [Google Scholar]
- 42. Udelhoven M., Leeser U., Freude S., Hettich M. M., Laudes M., Schnitker J., Krone W., Schubert M. (2009) Identification of a region in the human IRS2 promoter essential for stress induced transcription depending on SP1, NFI binding and ERK activation in HepG2 cells. J. Mol. Endocrinol. 44, 99–113 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.