Background: It was unclear whether subunit NuoK housing the essential Glu-36 residue participated in proton translocation in NDH-1.
Results: An indispensable role of Glu-36 and the importance of charged residues in loop-1 of NuoK were shown.
Conclusion: NuoK is involved in the proton translocation.
Significance: A bundle of NuoAJKH may act as a proton pump machinery of NDH-1.
Keywords: Bioenergetics/Electron Transfer Complex, Dehydrogenase, Membrane Enzymes, Proton Pumps, Site-directed Mutagenesis, NADH Dehydrogenase, Complex I, Energy Coupling
Abstract
The bacterial H+-translocating NADH:quinone oxidoreductase (NDH-1) catalyzes electron transfer from NADH to quinone coupled with proton pumping across the cytoplasmic membrane. The NuoK subunit (counterpart of the mitochondrial ND4L subunit) is one of the seven hydrophobic subunits in the membrane domain and bears three transmembrane segments (TM1–3). Two glutamic residues located in the adjacent transmembrane helices of NuoK are important for the energy coupled activity of NDH-1. In particular, mutation of the highly conserved carboxyl residue (KGlu-36 in TM2) to Ala led to a complete loss of the NDH-1 activities. Mutation of the second conserved carboxyl residue (KGlu-72 in TM3) moderately reduced the activities. To clarify the contribution of NuoK to the mechanism of proton translocation, we relocated these two conserved residues. When we shifted KGlu-36 along TM2 to positions 32, 38, 39, and 40, the mutants largely retained energy transducing NDH-1 activities. According to the recent structural information, these positions are located in the vicinity of KGlu-36, present in the same helix phase, in an immediately before and after helix turn. In an earlier study, a double mutation of two arginine residues located in a short cytoplasmic loop between TM1 and TM2 (loop-1) showed a drastic effect on energy transducing activities. Therefore, the importance of this cytosolic loop of NuoK (KArg-25, KArg-26, and KAsn-27) for the energy transducing activities was extensively studied. The probable roles of subunit NuoK in the energy transducing mechanism of NDH-1 are discussed.
Introduction
The proton-translocating NADH-quinone oxidoreductase (complex I,6 EC 1.6.5.3) is the first enzyme in the respiratory chain in mitochondria (1). It catalyzes oxidation of NADH in the mitochondrial matrix and reduction of quinone in the membrane, coupled to proton translocation through the inner mitochondrial membrane (2, 3). Complex I is by far the largest enzyme of the respiratory chain with a molecular mass of ∼1000 kDa and 45 different polypeptides (1, 4, 5). It has been implicated in several human neurodegenerative disorders and is believed to be the principal source of reactive oxygen species in mitochondria (6, 7). The bacterial counterpart enzyme (NDH-1) that consists of only 13–14 subunits is ideally suited to study complex I as all these subunits are homologs to the central core subunits of the mitochondrial enzyme (8, 9). Complex I/NDH-1 possess a characteristic L-shaped form with two clearly defined domains (10). The hydrophilic peripheral arm is projected into the mitochondrial matrix/bacterial cytoplasm and houses all the cofactors (one flavin mononucleotide and eight to nine iron-sulfur (Fe/S) clusters) that may participate in the electron transfer (11–13). The transfer of electron from NADH starts at the tip of the peripheral arm with FMN being the primary electron acceptor and then through a chain of seven conserved Fe/S clusters to the likely quinone-binding site expected to be present at the interface with the membrane domain (11–14). The hydrophobic arm is embedded in the inner mitochondrial/cytoplasmic membrane and is believed to participate in the proton translocation and in the binding of quinone and inhibitors (9, 15–18).
For functional study of complex I/NDH-1, it is a prerequisite to clarify sites and mechanism of the proton translocation. The stoichiometry of the proton translocation in NDH-1/complex I is considered to be four H+ per NADH oxidized (19). Recently, it has been reported that under certain conditions H+/2e− of complex I are decreased to three (2). These suggest that complex I/NDH-1 contains maximum of four proton translocation sites. The three-dimensional structural model revealed a common topology for three Na+/H+ antiporter-like subunits (NuoL (ND5), NuoM (ND4), and NuoN (ND2)) that are located side by side (10, 13, 20–22). These three subunits are considered to be good candidates for the proton translocation sites. In fact, biochemical and mutation analyses strongly suggest that NuoL and NuoM are involved in the proton translocation (23–26). Recent research on the efficiency of proton translocation indicates that NDH-1/complex I lacking both NuoL and NuoM can pump protons at H+/2e− = 2 (27, 28). These data suggest that in addition to NuoL, NuoM, and NuoN, one additional proton translocation site is present in NDH-1/complex I. It seems likely that at least one of the four remaining hydrophobic subunits (NuoA, NuoJ, NuoK, and NuoH) participates in the proton translocation. The three-dimensional structural model confirmed our earlier prediction that the hydrophobic subunits NuoA, NuoJ, and NuoK form a multisubunit helix bundle similar to subunits NuoL, NuoM, and NuoN and are in contact with the hydrophilic domain subunits NuoCD and NuoB at the quinone-binding interface (29–32). Of these subunits, only NuoK contains an essential charged residue in the center of the transmembrane (TM) segment (29, 33–36).
Subunit NuoK (counterpart of the mitochondrial ND4L subunit) is the smallest subunit of NDH-1 and is proposed to show sequence similarity to the MrpC subunit of the multisubunit Na+/H+ antiporters (37). The three-dimensional structural model shows that the NuoK subunit spans the membrane with three linearly arranged α-helices, connected by short loops. The subunit was also shown to have extensive interaction with the NuoN subunit with its C terminus extending between NuoN and helix HL (an α-helix of NuoL that spans multiple subunits in the membrane domain anchoring near NuoN), forming many inter-subunit links (29). Previously, Kervinen et al. (38) and Kao et al. (31) have reported that two conserved glutamic residues located in adjacent TMs of NuoK are important for the energy coupled activity of NDH-1. In particular, mutation of highly conserved KGlu-36 in TM2 to Ala/Gln led to a complete loss of the NDH-1 activities, whereas mutation of the other conserved Glu, KGlu-72 in TM3, caused moderate reduction in the activities (31). Kervinen et al. (38) also observed a severely impaired electron transfer activity and cell growth when these two glutamates of the Escherichia coli NDH-1 were mutated. To clarify the mechanism of proton translocation of NDH-1, we have investigated the importance of a specific location of both carboxylic residues, KGlu-36 and KGlu-72, in the NuoK subunit. Earlier, we have also shown that a double mutant of the two arginine residues (R25A/R26A) located in a small loop (loop-1) between TM1 and TM2 had a drastic effect on activities, with greatly reduced electron transfer rates and a diminished electrochemical gradient (31). In this study, we have further probed the role of the short cytoplasmic side loop (25RRN27) of this smallest subunit of NDH-1.
EXPERIMENTAL PROCEDURES
Materials
PCR product, DNA gel extraction, and plasmid purification kits were from Qiagen (Valencia, CA). The endonucleases were from New England Biolabs (Beverly, MA). The site-directed mutagenesis kit (QuikChange®II XL kit), the Herculase® Enhanced DNA polymerase, and the pCRScript vector were purchased from Stratagene (Cedar Creek, TX). The pKO3 vector was generously provided by Dr. George M. Church (Harvard Medical School, Boston). p-Nitro blue tetrazolium was from EMD Bioesciences (La Jolla, CA). Sigma was the major source of all other chemicals, including dNADH, NADH, and the antibiotics. E. coli MC4100 (F−, araD139, Δ(arg F-lac)U169, ptsF25, relA1, flb5301, rpsL 150.λ−) was used to generate nuoK site-specific mutations.
Mutagenesis of the E. coli nuoK Gene and Preparation of Mutant Cells
The strategy used to obtain the NuoK mutants was essentially the same as reported previously (31). In summary, pCRScript(nuoKE36A) and pCRScript(nuoKE72A) previously obtained in our laboratory (31) were used as template to generate the NuoK EAE mutants. All other NuoK mutants were made using pCRScript(nuoK) as the template. The mutated nuoK fragments were then cloned in pKO3/nuoK-T generating pKO3(nuoK mutants) or pKO3(nuoKEAE mutants). The above pKO3 plasmids were then used to replace the spc gene for the mutated nuoK genes in the NuoK KO in E. coli chromosome as described earlier (31). The mutations were finally confirmed by PCR and sequencing using specific oligonucleotides.
Growth and Membrane Preparation of E. coli NuoK Mutants
E. coli NuoK mutants were grown, and inverted membrane vesicles were prepared according to the method described previously in our laboratory (14, 31).
Immunoblots and Blue-native Electrophoresis
The antibodies against various E. coli NDH-1 subunits, namely NuoB (26), NuoCD (39), NuoE, NuoF, NuoG, and NuoI (40), NuoK (31), NuoM (23), and NuoL (41), were obtained previously in our laboratory. The above antibodies were used to analyze the content of the NDH-1 subunits by Western blotting. The procedures for performing blue-native PAGE (BN-PAGE) has been described earlier (42). In BN-PAGE, certain mutants were sensitive to detergent extraction. We performed the extraction using 0.5 or 1.0% dodecyl maltoside as indicated in the figure legend. The assembly of NDH-1 was examined by NADH dehydrogenase activity staining as reported (42).
Activity Analysis
The activity analysis of different NDH-1 mutants was performed using membrane vesicles according to the method described previously (43). Briefly, the dNADH-K3Fe(CN)6 reductase activity was performed at 30 °C with 80 μg of protein/ml in 10 mm potassium phosphate (pH 7.0), 1 mm EDTA containing 10 mm KCN, and 1 mm K3Fe(CN)6. The samples were preincubated for 1 min before starting the reaction with the addition of 150 μm dNADH. The signal was followed at 420 nm. K3Fe(CN)6 was replaced by 50 μm DB as an electron acceptor, and the signal was monitored at 340 nm for the dNADH-DB reductase activity in the same buffer. Similarly, 50 μm UQ1 was used to measure the dNADH-UQ1 reductase activity. Capsaicin-40 was added to inhibit the reaction (44). The dNADH oxidase activity was measured under the same conditions minus KCN and DB. The extinction coefficients used for activity calculations were ϵ340 = 6.22 mm−1 cm−1 for dNADH and ϵ420 = 1.00 mm−1 cm−1 for K3Fe(CN)6. Apart from the usual pH 7.0, the dNADH oxidase, dNADH-DB, and dNADH-UQ1 reductase activities were also monitored at various pH values for some of the NuoK mutants as described under “Results.” The NDH-1 proton pump activity was followed by ACMA fluorescence quenching using 200 μm dNADH as the substrate (45). All activity measurements were done 2–3 times.
Other Analytical Procedures
The BCA protein assay kit (Pierce) with bovine serum albumin as standard was used to determine the protein concentrations according to the manufacturer's instructions. Any variations from the procedures and details are described in the figure legends.
RESULTS
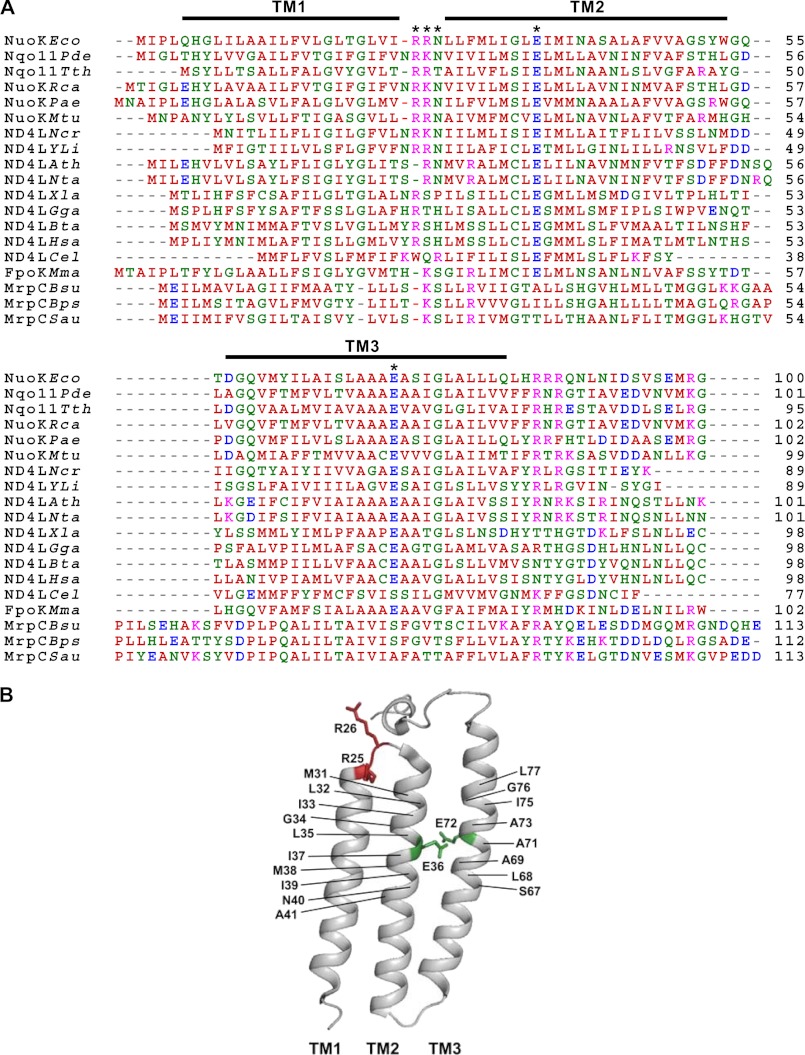
The NuoK subunit is one of the least conserved subunits of NDH-1. This subunit has been speculated to also show sequence similarity to the MrpC subunit of the multisubunit Na+/H+ antiporters (Fig. 1A) (37). Two Glu residues in subunit NuoK, KGlu-36 and KGlu-72, are present in the middle of TM2 and TM3, respectively (Fig. 1, A and B). KGlu-36 is perfectly conserved among all species, and KGlu-72 is almost perfectly conserved. However, neither one is conserved in the MrpC subunit of the multisubunit Na+/H+ antiporters (Fig. 1A) (34). Earlier, we have shown that in the NuoK subunit, mutations E36A and E36Q resulted in almost total abolishment of energy transducing NDH-1 activities, whereas E72A and E72Q mutations caused a partial but significant loss of activities (31). Furthermore, we have proposed that charged residues KGlu-36 and KGlu-72 participate, directly or indirectly, in the coupling mechanism of NDH-1 in conjunction with NuoA and NuoJ subunits (29). In this work, we studied how these two carboxyl residues in the NuoK subunit play a role in H+ translocation. For this purpose, 18 double site-directed mutants were made (Table 1) by systematically relocating KGlu-36 (EAE1 to EAE10) and KGlu-72 (EAE11 to EAE18) from its original position within TM2 and TM3, respectively (Fig. 1B). In addition, two charged residues, KArg-25 and KArg-26, present in a short loop (loop-1) connecting TM1 and TM2 were also shown to have an important role in the energy-transducing electron transfer and the architecture of NDH-1 (31). We have extensively studied the role of the three residues in loop-1 using various site-specific mutations.
FIGURE 1.
A, comparison of the amino acid sequence of the E. coli NuoK subunit with its homologous subunits. The alignment was carried out using the ClustalW program (59), and the residues are color-coded as per the program's default setup. Sequence sources and their UniProtKB/Swiss-Prot accession numbers are as follows: NuoKEco, E. coli K-12 NuoK subunit (P0AFE4); Nqo11Pde, Paracoccus denitrificans Nqo11 subunit (P29923); Nqo11Tth, Thermus thermophilus Nqo11 (Q56226); NuoKRca, Rhodobacter capsulatus NuoK subunit (P50940); NuoKPae, Pseudomonas aeruginosa NuoK subunit (Q9I0J2); NuoKMtu, Mycobacterium tuberculosis NuoK subunit (H6S7F5); ND4LNcr, Neurospora crassa ND4L subunit (P05509); ND4LYli, Yarrowia lipolytica ND4L subunit (Q9B6D4); ND4LAth, Arabidopsis thaliana ND4L subunit (P26289); ND4LNta, Nicotiana tabacum ND4L subunit (P06261); ND4LXla, Xenopus laevis ND4L subunit (P03904); ND4LGga, Gallus gallus ND4L subunit (P18942); ND4LBta, Bos taurus ND4L subunit (P03902); ND4LHsa, Homo sapiens ND4L subunit (P03901); ND4LCel, Caenorhabditis elegans ND4L subunit (P24886); FpoKMma, M. mazei FpoK subunit (Q9P9F6); MrpCBsu, Bacillus subtilis MrpC subunit; MrpCBps, Bacillus pseudofirmus MrpC subunit (Q9RGZ3); and MrpCSau, Staphylococcus aureus MrpC subunit (P60682). The positions of the three transmembrane segments are highlighted with bars above the sequence alignment. KGlu-36 in TM2 and KGlu-72 in TM3 and KArg-25, KArg-26 and KAsn-27 (all in loop-1) are indicated by the asterisks. B, mutated residues displayed in a schematic representation of the NuoK subunit of E. coli NDH-1. The three-dimensional structure of NuoK was taken from Protein Data Bank code 3RKO and a schematic representation was generated using PyMOL version 1.5.0.1 (60). KGlu-36 and KGlu-72 in the two transmembrane helices are shown with their side chains. KArg-25 and KArg-26, present in the short cytoplasmic loop, are also shown. The residues that were mutated in this study were marked with the E. coli numbering.
TABLE 1.
Enzymatic activities of the wild type and various NuoK mutants of E. coli NDH-1
| Mutanta | dNADH-O2b | dNADH-DBb | dNADH-UQ1b | dNADH-K3Fe(CN)6b | Cap-40 IC50c |
|---|---|---|---|---|---|
| WT | 100 ± 6%d | 100 ± 1%e | 100 ± 0%f | 100 ± 1%g | 0.05 |
| NuoK-KO | 2 ± 0% | 3 ± 1% | 5 ± 2% | 14 ± 2% | |
| KO-Revert | 94 ± 2% | 97 ± 4% | 99 ± 2% | ||
| E36A | 3 ± 0% | 5 ± 0% | 7 ± 2% | 86 ± 2% | |
| E36A/M31E (EAE1) | 3 ± 1% | 4 ± 0% | 5 ± 2% | 94 ± 1% | |
| E36A/L32E (EAE2) | 52 ± 1% | 57 ± 1% | 56 ± 5% | 101 ± 5% | 0.05 |
| E36A/I33E (EAE3) | 2 ± 0% | 2 ± 0% | 4 ± 1% | 85 ± 5% | |
| E36A/G34E (EAE4) | 3 ± 0% | 2 ± 0% | 94 ± 1% | ||
| E36A/L35E (EAE5) | 2 ± 0% | 3 ± 0% | 7 ± 0% | 80 ± 2% | |
| E36A/I37E (EAE6) | 5 ± 0% | 3 ± 1% | 8 ± 1% | 98 ± 0% | |
| E36A/M38E (EAE7) | 69 ± 1% | 68 ± 2% | 65 ± 7% | 87 ± 4% | 0.14 |
| E36A/I39E (EAE8) | 75 ± 2% | 49 ± 0% | 60 ± 7% | 101 ± 2% | 0.08 |
| E36A/N40E (EAE9) | 47 ± 6% | 57 ± 3% | 77 ± 3% | 78 ± 1% | 0.03 |
| E36A/A41/E (EAE10) | 3 ± 0% | 4 ± 1% | 4 ± 0% | 79 ± 0% | |
| E72A | 52 ± 1% | 54 ± 1% | 47 ± 9% | 105 ± 14% | 0.04 |
| E72A/S67E (EAE11) | 16 ± 0% | 15 ± 0% | 11 ± 4% | 84 ± 0% | 0.13 |
| E72A/L68E (EAE12) | 57 ± 0% | 66 ± 1% | 73 ± 10% | 94 ± 1% | 0.05 |
| E72A/A69E (EAE13) | 63 ± 2% | 68 ± 1% | 74 ± 10% | 114 ± 2% | 0.05 |
| E72A/A71E (EAE14) | 30 ± 0% | 27 ± 0% | 23 ± 9% | 94 ± 19% | 0.17 |
| E72A/A73E (EAE15) | 33 ± 0% | 28 ± 1% | 39 ± 2% | 77 ± 3% | 0.04 |
| E72A/I75E (EAE16) | 54 ± 0% | 59 ± 1% | 62 ± 5% | 99 ± 3% | 0.04 |
| E72A/G76E (EAE17) | 68 ± 1% | 81 ± 2% | 84 ± 1% | 106 ± 1% | 0.05 |
| A72A/L77E (EAE18) | 45 ± 0% | 46 ± 2% | 55 ± 0% | 93 ± 0% | 0.05 |
| R25A | 55 ± 4% | 54 ± 1% | 73 ± 6% | 85 ± 2% | |
| R25K | 67 ± 4% | 58 ± 2% | 78 ± 7% | 97 ± 1% | |
| R25C | 69 ± 5% | 53 ± 3% | 81 ± 8% | 91 ± 1% | 0.05 |
| R25S | 58 ± 3% | 51 ± 1% | 64 ± 1% | 87 ± 1% | 0.09 |
| R26A | 72 ± 2% | 68 ± 4% | 73 ± 6% | 98 ± 0% | |
| R26K | 80 ± 6% | 81 ± 2% | 95 ± 8% | 106 ± 1% | |
| R26C | 81 ± 7% | 70 ± 2% | 88 ± 7% | 106 ± 5% | 0.05 |
| R26S | 73 ± 3% | 70 ± 4% | 89 ± 10% | 92 ± 1% | 0.05 |
| N27C | 82 ± 8% | 69 ± 2% | 67 ± 4% | 95 ± 2% | 0.05 |
| N27S | 59 ± 8% | 59 ± 3% | 67 ± 4% | 75 ± 7% | 0.09 |
a Short names are given in parentheses.
b The means and standard errors are shown.
c Concentration of capsaicin-40 required to inhibit dNADH-O2 reductase by 50% (μm).
d Percentage of dNADH-O2 reductase activity (100% activity of WT was 0.73 μmol of dNADH/mg protein/min).
e Percentage of dNADH-DB reductase activity (100% activity of WT was 1.31 μmol of dNADH/mg of protein/min).
f Percentage of dNADH-UQ11 reductase activity (100% activity of WT was 0.92 μmol of dNADH/mg of protein/min).
g Percentage of dNADH-K3Fe(CN) 6 reductase activity (100% activity of WT was 1.51 μmol K3Fe(CN)6/mg protein/min).
NDH-1 Subunit Content and Assembly in E. coli NuoK Mutants
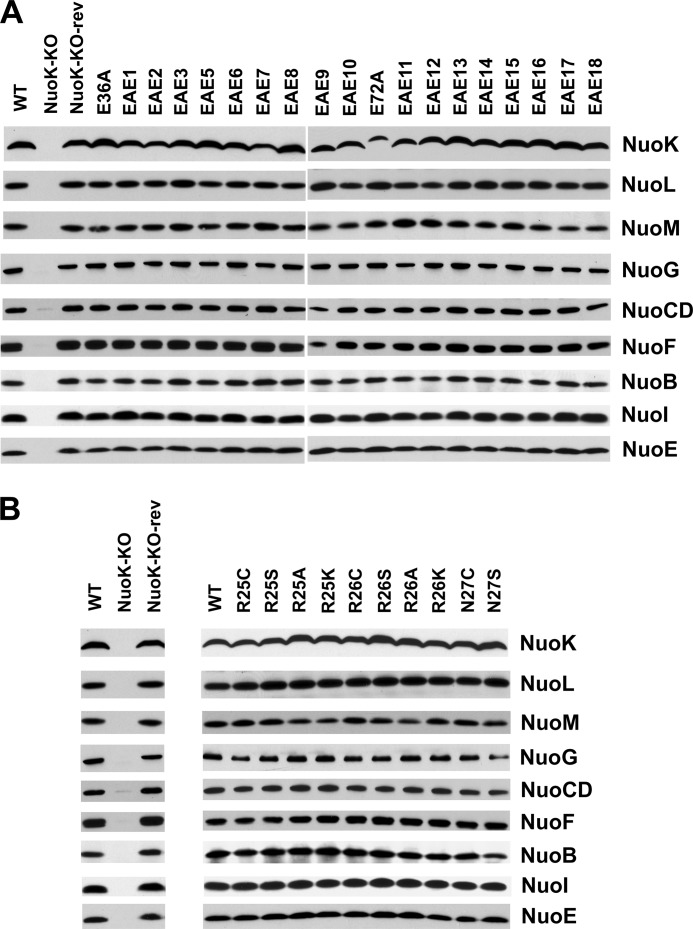
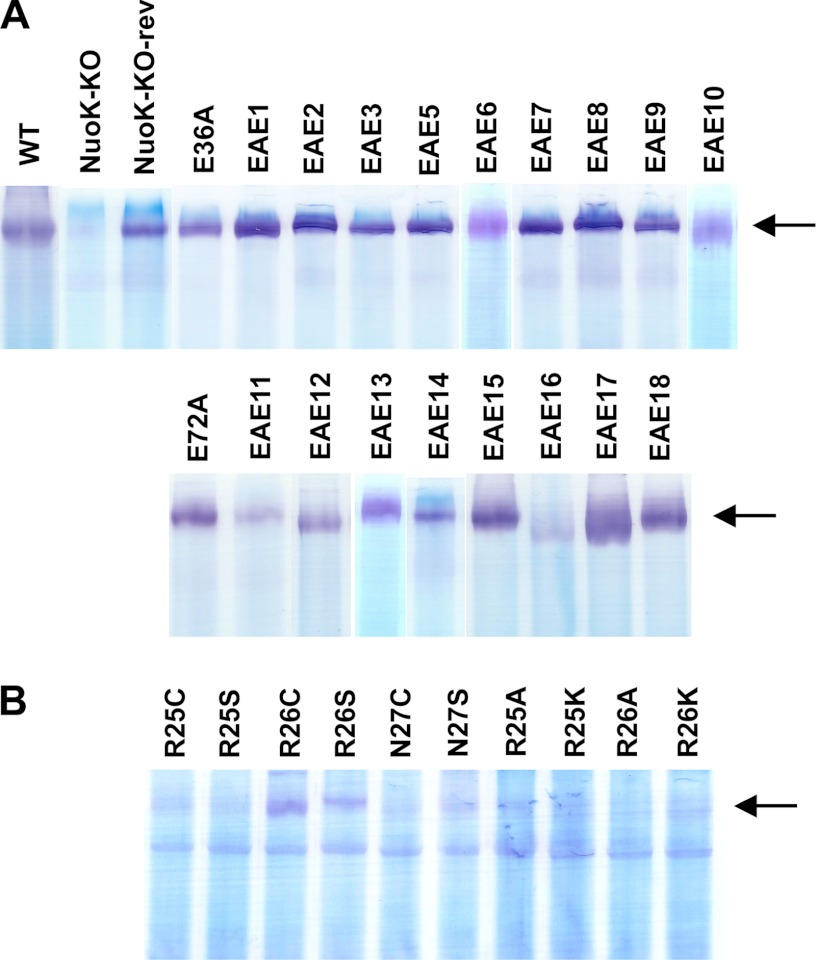
The effects of double site-specific EAE mutations on the NDH-1 subunit content were studied by immunoblotting. Antibodies raised against three membrane subunits (NuoK, NuoL, and NuoM), and six peripheral subunits (NuoB, NuoCD, NuoE, NuoF, NuoG, and NuoI) were used for this purpose (Fig. 2A). The NuoK-KO mutant did not show the presence of any of the analyzed subunits, confirming an essential role of this subunit in stabilization/assembly of NDH-1. The apparent ratios of the subunits of E36A, E72A, or any of the 18 EAE mutants seemed to be similar to those of the wild type (Fig. 2A). The assembly of the NDH-1 from the wild type, NuoK-KO, and NuoK-EAE mutants was verified through BN-PAGE. As expected, no NDH-1 was observed in the NuoK-KO mutant (Fig. 3A). Membranes isolated from the wild type and all site-specific NuoK mutants, including EAE mutants, seemed to contain fully assembled NDH-1 (Fig. 3A).
FIGURE 2.
Immunoblotting of membrane preparations of the NuoK mutants. A, WT and NuoK EAE mutants. Antibodies specific to all of the six hydrophilic subunits NuoB, NuoCD, NuoE, NuoF, NuoG, and NuoI and three membrane domain subunits NuoK, NuoL, and NuoM were used. 15 μg of protein from E. coli membranes per lane were loaded on a 4–20% Tris/glycine gels (Invitrogen). The secondary antibody used was goat anti-rabbit IgG horseradish peroxidase conjugate (Pierce). B, WT and NuoK KArg-25, KArg-26, and KAsn-27 mutants. The primary and secondary antibodies used and the gel conditions are the same as in A.
FIGURE 3.
BN-PAGE of membrane preparation of the NuoK mutants. A, WT and NuoK EAE mutants. B, NuoK KArg-25, KArg-26, and KAsn-27 mutants. Extraction of NDH-1 from the membrane samples was carried out using dodecyl maltoside at 0.5% (w/v) (A) or 1.0% (w/v) (B). For activity staining, the gels were incubated with 2.5 μg/ml p-nitro blue tetrazolium and 0.15 mm NADH in 2 mm Tris buffer (pH 7.5) at 37 °C. The reaction was stopped by 7% acetic acid. NDH-1 bands are shown by arrows.
To further analyze the correct assembly of the peripheral arm of NDH-1, we measured the dNADH-K3Fe(CN)6 reductase activity in inverted membrane vesicles of the mutants (42). Most of the site-directed NuoK EAE mutants constructed in this study showed dNADH-K3Fe(CN)6 reductase activity similar to the wild type (Table 1). A slightly reduced dNADH-K3Fe(CN)6 reductase activity (∼80%) was seen in the following cases: E36A/L35E (EAE5), E36A/N40E (EAE9), E36A/A41E (EAE10), and E72A/A73E (EAE15). Taken together, we conclude that none of the EAE mutations introduced in this study affected the assembly of the hydrophilic domain of E. coli NDH-1.
Measurements of NDH-1 Activities in NuoK EAE Mutants
dNADH oxidase, dNADH-DB, and dNADH-UQ1 reductase activities of NDH-1 were measured in E. coli membrane vesicles, using dNADH as a substrate for measuring NDH-1 specific activity (46). A complete inhibition of the energy-coupled NDH-1 activities was seen in the case of mutation E36A, which was similar to the previous study (31). The original energy coupled activities of the NDH-1 could not be retained when the glutamic acid was moved within the TM2 to positions Met-31 (EAE1), Ile-33 (EAE3), Gly-34 (EAE4), Leu-35 (EAE5), Ile-37 (EAE6), and Ala-41 (EAE10) (Table 1). In fact, the dNADH oxidase and dNADH-DB reductase activities observed were almost nullified for these mutants similar to the E36A mutant. Interestingly, the double mutants E36A/L32E (EAE2), E36A/M38E (EAE7), E36A/I39E (EAE8), and E36A/N40E (EAE9), all showed about 50–80% of the wild type activities (Table 1). Thus, repositioning of KGlu-36 either to a few residues upstream or downstream resulted in significant retention of the energy transducing NDH-1 activities. In the same way, energy transducing NDH-1 activities were significantly affected when KGlu-72 was relocated to positions either immediately preceding or immediately after the helix turn. An almost 80% retention of the energy coupled NDH-1 activities were observed for the EAE17 mutant in which KGlu-72 was shifted to position 76. The double mutants E72A/L68E (EAE12), E72A/A69E (EAE13), E72A/I75E (EAE16), and E72A/L77E (EAE18) all showed about 50–70% of the wild type activities. The double mutants EAE11, EAE14, and EAE15 in which the glutamic acid was moved within TM3 to positions 67, 71, and 73, respectively, showed reduced energy coupled activities compared with those of E72A mutant (15–35% of the wild type activities with EAE11 being the least active). The dNADH-UQ1 reductase activity of most of the EAE mutants was consistent with their dNADH oxidase and dNADH-DB reductase activities. Capsaicin-40 is a competitive inhibitor for quinone in NDH-1/complex I that inhibits energy coupled activities (44, 47, 48). No significant difference between wild type and NuoK EAE mutants was found for the IC50 values of capsaicin-40 (0.05 to 0.15 μm, see Table 1) for the dNADH oxidase activity, suggesting that these mutations barely affected the quinone-binding site(s).
Measurements of NDH-1 Activities in Mutants of Cytoplasmic Loop-1 of Subunit NuoK
The small cytosolic side loop-1 of the NuoK subunit consists of three residues, KArg-25, KArg-26, and KAsn-27. Among them, KArg-25 is well conserved. Site-specific mutations R25A, R25K, R25C, R25S, R26A, R26K, R26C, R26S, N27C, and N27S were made to assess the importance of the individual residues in this loop. First, the subunit contents of NDH-1 were studied by immunoblotting for the aforementioned mutants of KArg-25, KArg-26, and KAsn-27. All of the studied mutants showed subunit contents similar to the wild type (Fig. 2B). The correct assembly of the NuoK loop-1 mutants was also verified through BN-PAGE and NADH dehydrogenase activity staining. Unlike the EAE mutants, the NDH-1 band was barely visible for the loop-1 mutants when the gel was incubated for about 5 min with the substrate. However, upon increasing the incubation time to 1–2 h, all the loop-1 mutants showed the NDH-1 band, albeit with a reduced signal intensity (Fig. 3B). Likewise, the dNADH-K3Fe(CN)6 reductase activity was comparable with the wild type in the membrane vesicles for a majority of these mutants (Table 1). We observed a slightly reduced dNADH-K3Fe(CN)6 reductase activity (∼75%) for the N27S mutant (Table 1). Taken together, we conclude that none of the mutations of the three residues located in the cytoplasmic side loop-1 affected the assembly of E. coli NDH-1.
Mutation of KArg-25 (R25A, R25K, R25C, and R25S) resulted in about 50–70% dNADH oxidase and dNADH-DB reductase activities compared with the wild type (Table 1). Mutation of the other positively charged residue KArg-26 (R26A, R26K, R26C, and R26S) had less appreciable effects with the activities being in the range of 65–85% (Table 2). The dNADH-UQ1 reductase activity for most of the mutants corresponded well with their respective dNADH oxidase and dNADH-DB reductase activities.
TABLE 2.
Mutation of the residues in cytoplasmic loop-1
| Mutant | dNADH-O2a |
dNADH-DBa |
dNADH-UQ1a |
||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 7.0 | pH 7.5 | pH 8.0 | pH 7.0 | pH 7.5 | pH 8.0 | pH 7.0 | pH 7.5 | pH 8.0 | |
| WT | 100 ± 5%b1 | 100 ± 5%b2 | 100 ± 6%b3 | 100 ± 1%c1 | 100 ± 3%c2 | 100 ± 2%c3 | 100 ± 0%d1 | 100 ± 0%d2 | 100 ± 6%d3 |
| R25A | 55 ± 4% | 50 ± 14% | 42 ± 9% | 54 ± 2% | 50 ± 2% | 28 ± 2% | 73 ± 6% | 46 ± 10% | 25 ± 3% |
| R25K | 67 ± 4% | 50 ± 0% | 43 ± 3% | 58 ± 2% | 61 ± 3% | 48 ± 4% | 78 ± 7% | 49 ± 6% | 27 ± 4% |
| R25C | 69 ± 5% | 55 ± 9% | 44 ± 3% | 53 ± 3% | 38 ± 6% | 30 ± 5% | 81 ± 8% | 50 ± 1% | 31 ± 7% |
| R25S | 58 ± 3% | 50 ± 1% | 37 ± 1% | 51 ± 1% | 35 ± 2% | 20 ± 2% | 64 ± 1% | 34 ± 6% | 26 ± 3% |
| R26A | 72 ± 2% | 63 ± 7% | 52 ± 11% | 68 ± 4% | 61 ± 3% | 45 ± 11% | 73 ± 6% | 57 ± 4% | 29 ± 6% |
| R26K | 81 ± 6% | 92 ± 11% | 100 ± 6% | 81 ± 2% | 89 ± 5% | 98 ± 7% | 95 ± 8% | 92 ± 9% | 79 ± 2% |
| R26C | 82 ± 7% | 68 ± 0% | 69 ± 1% | 70 ± 2% | 67 ± 4% | 51 ± 5% | 88 ± 7% | 70 ± 3% | 52 ± 3% |
| R26S | 74 ± 3% | 70 ± 14% | 70 ± 4% | 70 ± 4% | 59 ± 2% | 38 ± 4% | 89 ± 10% | 64 ± 4% | 33 ± 5% |
| N27C | 83 ± 8% | 66 ± 1% | 66 ± 2% | 69 ± 2% | 57 ± 2% | 33 ± 2% | 67 ± 4% | 53 ± 3% | 28 ± 0% |
| N27S | 60 ± 8% | 50 ± 5% | 50 ± 3% | 59 ± 3% | 48 ± 2% | 24 ± 1% | 67 ± 4% | 47 ± 3% | 23 ± 2% |
a The means and standard errors are shown.
b 1–3Percentage of dNADH-O2 reductase activity (100% activity of WT at pH 7.0, pH 7.5, and pH 8.0 were 0.73, 0.78, and 0.76 μmol dNADH/mg of protein/min, respectively).
c 1–3Percentage of dNADH-DB reductase activity (100% activity of WT at pH 7.0, pH 7.5, and pH 8.0 were 1.31, 1.29, and 1.15 μmol dNADH/mg of protein/min, respectively).
d 1–3Percentage of dNADH-UQ1 reductase activity (100% activity of WT at pH 7.0, pH 7.5, and pH 8.0 were 0.92, 0.96, and 0.97 μmol dNADH/mg of protein/min, respectively).
The dNADH oxidase and dNADH-DB reductase activities of all these mutants were checked under different pH conditions. The two activities did not behave in a similar manner among the same set of mutants. There was no significant difference in the level of either dNADH oxidase or dNADH-DB reductase activity for the wild type, at pH 7.0, 7.5, and 8.0 (Table 2). In the KArg-25 mutants, a decline in the activities was observed as the pH was raised. In mutants R25A, R25C, and R25S that had 50–55% of the wild type dNADH-DB reductase activity at neutral pH showed a considerably lower activity (20–30%) at higher pH 8.0. The pH-dependent change was less drastic in case of the R25K mutant in which the positive charge was preserved (58% of the wild type at pH 7 versus 48% at pH 8.0). Mutants R26S, N27S, and N27C that had 60–70% of the wild type dNADH-DB reductase activity at neutral pH, exhibited a lower activity (20–40%) at pH 8.0. Reduced dNADH-DB reductase activity at higher pH value was also seen with mutants R26A and R26C but to a lesser extent (Table 2). The dNADH-DB reductase activity of the R26K mutant in which the positive charge was preserved remained the same at different pH values. When UQ1 was used as the acceptor, a similar trend in the results, i.e. activity reduction of R25A, R25C, R25S, R26A, R26C, R26S, N27C, and N27S mutants at high pH, was also observed (Table 2). However, it should be noted that, in contrast to their dNADH-DB reductase activity at higher pH, the R25K mutant showed a marked activity reduction with an increase in pH with UQ1. The dNADH-UQ1 reductase activities of the R26K mutant remained essentially unchanged like those of the wild type irrespectively of assay pH (from 7.0 to 8.0) (Table 2).
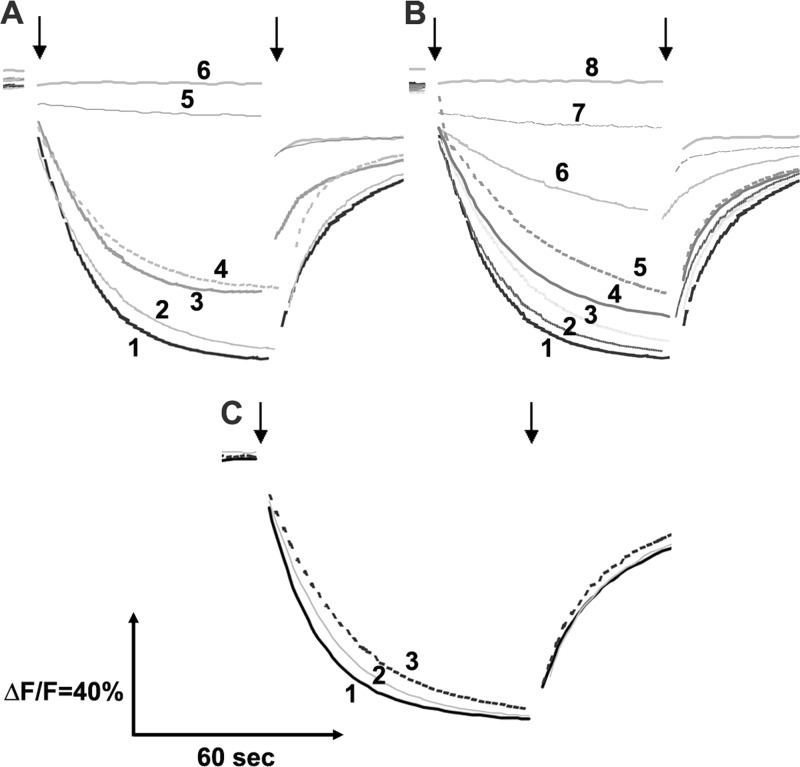
Proton Translocation by NuoK EAE Mutants
One of the primary functions of NDH-1 is the translocation of protons across the cytoplasmic membrane, which generates a proton gradient. Thus, we monitored the generation of a proton gradient in the inverted membrane vesicles of different NuoK EAE mutants using ACMA as the reporter and dNADH as the substrate. As expected, no proton gradient was observed for the membrane vesicle of the E36A mutant. Likewise, the double mutants EAE1, EAE3, EAE4, EAE5, EAE6, and EAE10 of E36A series that earlier showed very little energy coupled activity (Table 1) also lacked proton translocation ability (Fig. 4A). It was of particular interest to observe the proton translocation capacity of the EAE2, EAE7, EAE8, and EAE9 mutants that earlier showed significant retention of energy coupled activities (Table 1). In accord with their dNADH oxidase and dNADH-DB reductase activities, membrane vesicles from the EAE2, EAE7, EAE8, and EAE9 mutants behaved differently in their ability to generate ΔpH, with a maximum response seen in the case of NuoK EAE7, slightly lower for EAE2 and EAE8 and a further lower response observed for the EAE9 mutant (Fig. 4A). Similar trends were observed for the double mutants (EAE11–18) of E72A. Akin to their dNADH oxidase and DB reductase activities, EAE17 and EAE13 mutants exhibited maximum proton translocation, although ΔpH generation was very low for the mutant EAE11. The mutants E72A, EAE12, and EAE16 all showed a moderate level of proton translocation, with NuoK EAE15 and EAE18 mutants being slightly lower and EAE14 showing a further lower level of proton translocation (Fig. 4B). It seems conceivable that in the EAE mutants the proton uptake is correlating well with electron transfer of NDH-1 coupled to energy transduction.
FIGURE 4.
Generation of pH gradient (ΔpH) by dNADH oxidation in E. coli NuoK mutants. ΔpH was monitored by quenching of the fluorescence of ACMA at room temperature. Excitation wavelength was 410 nm, and emission wavelength was 480 nm. The 1st arrow indicates the addition of 0.2 mm dNADH, and the 2nd arrow shows the addition of 10 μm carbonyl cyanide-p-trifluoromethoxy-phenylhydrazone. Assay mixture contains 50 μg of protein/ml membrane vesicles in 50 mm MOPS (pH 7.3), 10 mm MgCl2, 50 mm KCl, and 2 μm ACMA. A, 1, WT; 2, EAE7; 3, EAE2 and EAE8; 4, EAE9; 5, E36A, EAE1, EAE3, EAE4, EAE5, EAE6, and EAE10; 6, NuoK KO. B, 1, WT, EAE17; 2, EAE13; 3, E72A, EAE12, and EAE16; 4, EAE15; 5, EAE18; 6, EAE14; 7, EAE11; 8, NuoK KO. C, 1, WT, R25K, R26K; 2, R25C, R26A R26C, R26S, N27C; 3, N27S, R25A, and R25S.
Proton Translocation by KArg-25, KArg-26, and KAsn-27 Mutants
The proton translocation ability of KArg-25, KArg-26, and KAsn-27 mutants in the absence of external DB is shown in Fig. 4C. The R25K and R26K mutants showed maximum proton translocation ability similar to the wild type. A slightly reduced proton translocation was observed for R25C, R26A, R26C, R26S, and N27C, which was further reduced for R25A, R25S, and N27S (Fig. 4C). We also measured the generation of pH gradient by dNADH oxidation in E. coli NuoK KArg-25, KArg-26, and KAsn-27 mutants in the presence of external DB (data not shown). In each case, external DB could not stimulate the rate of proton translocation driven by dNADH. Again, the mutations of the loop-1 residues hardly affected the energy-coupled NDH-1 electron transfer.
DISCUSSION
The importance of the presence of conserved charged amino acids in various membrane subunits of NDH-1/complex I has been well documented (23, 49). Moreover, a carboxylic amino acid located in the membrane is considered to be important for both proton pump and ion translocation activities in the membrane-bound enzyme complexes (50–52). The three-dimensional structural model of the E. coli membrane segment indicates that KGlu-36 forms a hydrogen bond to Tyr-59 in NuoJ TM3 (JTyr-59) (see Fig. 5B) (29). Our mutagenesis experiments using Y59C and Y59F in NuoJ illustrated that the mutations of JTyr-59 moderately affected the energy transducing NDH-1 activities, suggesting that JTyr-59 or the hydrogen bond between KGlu-36 and JTyr-59 is not essential for the energy transducing NDH-1 activities (30). In previous work, we have also described the presence of a well conserved transmembrane KGlu-36 whose replacement by a noncharged amino acid, but not by Asp in NuoK, led to a complete loss in the energy transducing NDH-1 activities. This indicates that KGlu-36 is an essential residue for H+-translocating NDH-1 with activities similar to MGlu144 and LGlu144 (31).
FIGURE 5.
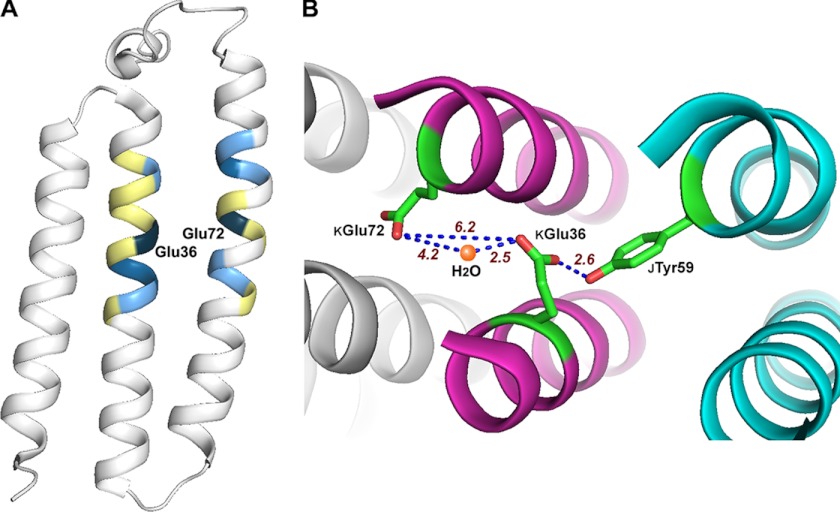
A, relocation of the two Glu residues and the activities of NDH-1. A schematic representation of NuoK was drawn as in Fig. 1B. The position to which the KGlu-36 or KGlu-72 was relocated is colored based on the % activity relative to the wild type as follows: dark blue, 100% (native positions); medium blue, 70–100%; light blue, 40–70%; yellow, < 40%. B, schematic of the interaction among KGlu-36, KGlu-72, and a water molecule. A schematic representation of the E. coli NDH-1 was generated using PyMOL version 1.5.0.1 (60) from the Protein Data Bank file 3RKO. Portions of subunits NuoK (magenta), NuoJ (blue), and NuoN (gray) are displayed. Side chains of KGlu-72, KGlu-36, and JTyr-59 are colored in green. Position of a water molecule in NuoK is shown as an orange ball. Distances between the highlighted residues/water are given in red numbers.
The energy transducing activities were almost completely suppressed for several of the NuoK EAE mutants of KGlu-36 in TM2, showing values similar to the E36A mutant (Table 1) (31). Remarkably, in cases of mutations EAE2, EAE7, EAE8, and EAE9, which resulted in relocation of KGlu-36 to positions 32, 38, 39, and 40, respectively, the NDH-1 activities were largely preserved (Fig. 5A). Positions 32 and 39 are one helix turn downstream and upstream, respectively, of KGlu-36. These results suggest that KGlu-36 has site flexibility. Similar results were obtained when the other conserved residue KGlu-72 was relocated along TM3. Most of the mutants, except for EAE17, displayed the energy coupled activities close to the E72A mutant (Table 1 and Fig. 5A). The EAE17 mutant in which KGlu-72 was shifted upstream by one helix turn sustained the NDH-1 activities like that of the four mutants of KGlu-36. We have previously studied the effect of shifting MGlu144, which is absolutely essential for proton translocating in the NuoM subunit, to all positions within TM5 on the function of NDH-1 (25). Most of the repositioning mutants of MGlu144 lost their energy transducing NDH-1 activities. Only two mutants, E144A/F140E and E144A/L147E, located one helix turn downstream and upstream in the TM5 helix retained the energy transducing activities of NDH-1 (25). These data indicate that the two conserved Glu residues in NuoK are both akin to MGlu144 concerning site flexibility. This characteristic was also reported earlier for essential Asp-61 of the dicyclohexylcarbodiimide-binding subunit of E. coli ATP synthase (53).
In the DCCD-binding subunit (so called c-ring) of the ATP synthase (52, 54), electron density for a solvent molecule (most likely a water molecule) was observed within 2.9 Å of a Glu residue known to be essential for proton translocation. It is speculated that a proton is present between the atom positions of the essential Glu (oxygen) and the water oxygen and may be involved in the proton pump (52). The protonated water cluster has also been reported for bacteriorhodopsin (55). According to the three-dimensional structural model of the E. coli NDH-1 membrane arm currently available, KGlu-36 coordinates at least one water molecule at a distance of 2.5 Å (Fig. 5B) (29). From structural similarity, we may envision that the water molecule coordinated by KGlu-36 functionally resembles that associated with the essential Glu in the DCCD-binding subunit. If the involvement of water molecules is indeed a common feature in the mechanism of proton translocation, it will explain the site flexibility described above. Specifically, the essential Glu residues may be relocatable depending on the (re)distribution of water molecules that participate in proton translocation. This notion may be verified when more water molecules are identified as the structural analysis of NDH-1 is further advanced.
There exists a short loop (loop-1) between TM1 and TM2 composed of three residues (25RRN27). In most mutants of loop-1, dNADH-DB reductase activities were inhibited to a greater extent at pH 8.0, but pH dependence of dNADH oxidase activities (endogenous UQ8-linked) was barely observed (Table 2). Stronger inhibition at high pH was also observed by using UQ1 as the acceptor. There was no significant difference in IC50 of capsaicin-40 (Q analog inhibitor (44, 47)) between the wild type and the loop-1 mutants, suggesting that loop-1 is not involved in the association of UQ with NDH-1. A salt bridge between anionic (Asp, Glu) and cationic amino acids (Lys, Arg) is known to contribute toward the alkaline stability in various proteins, as typically seen in alkalophilic enzymes (56). Because KArg-26 is located close to LArg-592 in the HL-helix of NuoL and also KAsp-93, we can envision that the loss of charge at KArg-26 by site-directed mutagenesis might decrease the stability of NDH-1 at alkaline pH. A speculative mechanistic difference between endogenous UQ8 and exogenous UQ1 and DB reaction is that endogenous UQ is recycled between NDH-1 and quinol oxidase, whereas reduced exogenous UQ that is relatively hydrophilic is released into the aqueous phase from the respiratory chain system. Therefore, it seems possible that the three residues of loop-1 or the salt bridge formed by them are involved in the reduced exogenous UQ release but not endogenous UQ8H2.
There seems to be another possible role of loop-1. NhaA, the main Na+/H+ antiporter of E. coli and many enterobacteria, is essential for cytoplasmic pH regulation and was reported to have a “pH sensor” contributed by charged residues (Glu-241, Lys-249, and Glu-252) (57, 58). This so-called pH sensor, in which a pH signal results in alteration of the protonation state, is located at the entrance of the cytoplasmic passage (similar to KArg-26), and it elicits conformational changes that are transmitted to activate NhaA. The N terminus of the activator structural element (helix IX) contributing to the pH sensor is in direct contact with the essential part of the Na+/H+ exchange machinery present in the TMIV/XI assembly at the center of the membrane, which itself undergoes a pH-induced conformational change. A similar function can be envisioned for the arginine residues KArg-25 and KArg-26 that are present near the cytoplasmic surface. KArg-26 of the NuoK subunit closely interacts with the HL helix of NuoL through LArg-592 in NDH-1 (29). Thus, the mutation of positively charged arginines in the NuoK subunit to uncharged residues may result in disruption of electrostatic interaction between NuoK and the “movable” structural element HL. This in turn would render the whole complex rigid, thus disturbing the conformational change by preventing the dynamic movements that drive proton translocation. Positively charged arginines (KArg-26 in particular) present in the short cytoplasmic loop are additional key residues in the proton translocation channel located at the interface of the membrane and peripheral arm and might be important in regulating the function of NDH-1.
This study strongly suggests that NuoK is involved in proton translocation in addition to the antiporter-like subunits NuoL (16 TMs), NuoM (14 TMs), and NuoN (14 TMs) in NDH-1. Given that NuoK has only three TMs, it most likely would not constitute a pumping machine by itself. The three-dimensional structural model of the membrane arm indicates that NuoK interacts with NuoA (3 TMs), NuoJ (5 TMs), and probably NuoH (8 TMs according to topological studies). A bundle of NuoAJKH might act as a proton pump machine, and KGlu-36 of NuoK plays a critical role in the unit. This speculation should be experimentally examined in the future.
Acknowledgment
We thank Dr. Jennifer Barber-Singh for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant GM33712 from the United States Public Health Service (to T. Y.).
- complex I
- mitochondrial proton-translocating NADH-quinone oxidoreductase
- NDH-1
- bacterial proton-translocating NADH-quinone oxidoreductase
- DB
- 2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinone
- dNADH
- reduced nicotinamide hypoxanthine dinucleotide
- ACMA
- 9-amino-6-chloro-2-methoxyacridine
- BN-PAGE
- blue-native PAGE
- TM
- transmembrane
- UQ
- ubiquinone.
REFERENCES
- 1. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 2. Wikström M., Hummer G. (2012) Stoichiometry of proton translocation by respiratory complex I and its mechanistic implications. Proc. Natl. Acad. Sci. U.S.A. 109, 4431–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galkin A. S., Grivennikova V. G., Vinogradov A. D. (2001) H+/2e− stoichiometry of the NADH:ubiquinone reductase reaction catalyzed by submitochondrial particles. Biochemistry 66, 435–443 [DOI] [PubMed] [Google Scholar]
- 4. Carroll J., Fearnley I. M., Shannon R. J., Hirst J., Walker J. E. (2003) Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2, 117–126 [DOI] [PubMed] [Google Scholar]
- 5. Carroll J., Fearnley I. M., Skehel J. M., Shannon R. J., Hirst J., Walker J. E. (2006) Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 281, 32724–32727 [DOI] [PubMed] [Google Scholar]
- 6. Lu J., Sharma L. K., Bai Y. (2009) Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res. 19, 802–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharma L. K., Lu J., Bai Y. (2009) Mitochondrial respiratory complex I. Structure, function, and implication in human diseases. Curr. Med. Chem. 16, 1266–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yagi T., Yano T., Di Bernardo S., Matsuno-Yagi A. (1998) Procaryotic complex I (NDH-1), an overview. Biochim. Biophys. Acta 1364, 125–133 [DOI] [PubMed] [Google Scholar]
- 9. Yagi T., Matsuno-Yagi A. (2003) The proton-translocating NADH-quinone oxidoreductase in the respiratory chain. The secret unlocked. Biochemistry 42, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 10. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 11. Hinchliffe P., Sazanov L. A. (2005) Organization of iron-sulfur clusters in respiratory complex I. Science 309, 771–774 [DOI] [PubMed] [Google Scholar]
- 12. Sazanov L. A. (2007) Respiratory complex I. Mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46, 2275–2288 [DOI] [PubMed] [Google Scholar]
- 13. Sazanov L. A., Hinchliffe P. (2006) Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311, 1430–1436 [DOI] [PubMed] [Google Scholar]
- 14. Sinha P. K., Nakamaru-Ogiso E., Torres-Bacete J., Sato M., Castro-Guerrero N., Ohnishi T., Matsuno-Yagi A., Yagi T. (2012) Electron transfer in subunit NuoI (TYKY) of Escherichia coli NDH-1 (NADH:quinone oxidoreductase). J. Biol. Chem. 287, 17363–17373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinha P. K., Castro-Guerrero N., Matsuno-Yagi A., Yagi T., Torres-Bacete J. (2009) Bacterial complex I (NDH-1). Functional roles of the hydrophobic domain. Curr. Top. Biochem. Res. 11, 79–90 [Google Scholar]
- 16. Murai M., Sekiguchi K., Nishioka T., Miyoshi H. (2009) Characterization of the inhibitor binding site in mitochondrial NADH-ubiquinone oxidoreductase by photoaffinity labeling using a quinazoline type inhibitor. Biochemistry 48, 688–698 [DOI] [PubMed] [Google Scholar]
- 17. Shiraishi Y., Murai M., Sakiyama N., Ifuku K., Miyoshi H. (2012) Fenpyroximate binds to the interface between PSST and 49-kDa subunits in mitochondrial NADH-ubiquinone oxidoreductase. Biochemistry 51, 1953–1963 [DOI] [PubMed] [Google Scholar]
- 18. Schuler F., Yano T., Di Bernardo S., Yagi T., Yankovskaya V., Singer T. P., Casida J. E. (1999) NADH-quinone oxidoreductase: PSST subunit couples electron transfer from iron-sulfur cluster N2 to quinone. Proc. Natl. Acad. Sci. U.S.A. 96, 4149–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galkin A. S., Grivennikova V. G., Vinogradov A. D. (1999) →H+/2e− stoichiometry in NADH-quinone reductase reactions catalyzed by bovine heart submitochondrial particles. FEBS Lett. 451, 157–161 [DOI] [PubMed] [Google Scholar]
- 20. Berrisford J. M., Sazanov L. A. (2009) Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 284, 29773–29783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baranova E. A., Holt P. J., Sazanov L. A. (2007) Projection structure of the membrane domain of Escherichia coli respiratory complex I at 8 Å resolution. J. Mol. Biol. 366, 140–154 [DOI] [PubMed] [Google Scholar]
- 22. Hunte C., Zickermann V., Brandt U. (2010) Functional modules and structural basis of conformational coupling in mitochondrial complex I. Science 329, 448–451 [DOI] [PubMed] [Google Scholar]
- 23. Torres-Bacete J., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2007) Characterization of the NuoM (ND4) subunit in Escherichia coli NDH-1. Conserved charged residues essential for energy-coupled activities. J. Biol. Chem. 282, 36914–36922 [DOI] [PubMed] [Google Scholar]
- 24. Euro L., Belevich G., Verkhovsky M. I., Wikström M., Verkhovskaya M. (2008) Conserved lysine residues of the membrane subunit NuoM are involved in energy conversion by the proton-pumping NADH:ubiquinone oxidoreductase (complex I). Biochim. Biophys. Acta 1777, 1166–1172 [DOI] [PubMed] [Google Scholar]
- 25. Torres-Bacete J., Sinha P. K., Castro-Guerrero N., Matsuno-Yagi A., Yagi T. (2009) Features of subunit NuoM (ND4) in Escherichia coli NDH-1. Topology and implication of conserved Glu-144 for coupling site 1. J. Biol. Chem. 284, 33062–33069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakamaru-Ogiso E., Kao M. C., Chen H., Sinha S. C., Yagi T., Ohnishi T. (2010) The membrane subunit NuoL(ND5) is involved in the indirect proton pumping mechanism of E. coli complex I. J. Biol. Chem. 285, 39070–39078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steimle S., Bajzath C., Dörner K., Schulte M., Bothe V., Friedrich T. (2011) The role of subunit NuoL for proton translocation by the respiratory complex I. Biochemistry 50, 3386–3393 [DOI] [PubMed] [Google Scholar]
- 28. Dröse S., Krack S., Sokolova L., Zwicker K., Barth H. D., Morgner N., Heide H., Steger M., Nübel E., Zickermann V., Kerscher S., Brutschy B., Radermacher M., Brandt U. (2011) Functional dissection of the proton pumping modules of mitochondrial complex I. PLoS Biol. 9, e1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Efremov R. G., Sazanov L. A. (2011) Structure of the membrane domain of respiratory complex I. Nature 476, 414–420 [DOI] [PubMed] [Google Scholar]
- 30. Kao M. C., Di Bernardo S., Nakamaru-Ogiso E., Miyoshi H., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoJ (ND6) of the NADH-quinone oxidoreductase from Escherichia coli by chromosomal DNA manipulation. Biochemistry 44, 3562–3571 [DOI] [PubMed] [Google Scholar]
- 31. Kao M. C., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2005) Characterization of the membrane domain subunit NuoK (ND4L) of the NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 44, 9545–9554 [DOI] [PubMed] [Google Scholar]
- 32. Kao M. C., Di Bernardo S., Perego M., Nakamaru-Ogiso E., Matsuno-Yagi A., Yagi T. (2004) Functional roles of four conserved charged residues in the membrane domain subunit NuoA of the proton-translocating NADH-quinone oxidoreductase from Escherichia coli. J. Biol. Chem. 279, 32360–32366 [DOI] [PubMed] [Google Scholar]
- 33. Kao M. C., Di Bernardo S., Matsuno-Yagi A., Yagi T. (2003) Characterization and topology of the membrane domain Nqo10 subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry 42, 4534–4543 [DOI] [PubMed] [Google Scholar]
- 34. Kao M. C., Di Bernardo S., Matsuno-Yagi A., Yagi T. (2002) Characterization of the membrane domain Nqo11 subunit of the proton-translocating NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry 41, 4377–4384 [DOI] [PubMed] [Google Scholar]
- 35. Bernardo S. D., Yano T., Yagi T. (2000) Exploring the membrane domain of the reduced nicotinamide adenine dinucleotide-quinone oxidoreductase of Paracoccus denitrificans. Characterization of the NQO7 subunit. Biochemistry 39, 9411–9418 [DOI] [PubMed] [Google Scholar]
- 36. Zickermann V., Barquera B., Wikström M., Finel M. (1998) Analysis of the pathogenic human mitochondrial mutation ND1/3460, and mutations of strictly conserved residues in its vicinity, using the bacterium Paracoccus denitrificans. Biochemistry 37, 11792–11796 [DOI] [PubMed] [Google Scholar]
- 37. Mathiesen C., Hägerhäll C. (2003) The “antiporter module” of respiratory chain complex I includes the MrpC/NuoK subunit–a revision of the modular evolution scheme. FEBS Lett. 549, 7–13 [DOI] [PubMed] [Google Scholar]
- 38. Kervinen M., Pätsi J., Finel M., Hassinen I. E. (2004) A pair of membrane-embedded acidic residues in the NuoK subunit of Escherichia coli NDH-1, a counterpart of the ND4L subunit of the mitochondrial Complex I, are required for high ubiquinone reductase activity. Biochemistry 43, 773–781 [DOI] [PubMed] [Google Scholar]
- 39. Castro-Guerrero N., Sinha P. K., Torres-Bacete J., Matsuno-Yagi A., Yagi T. (2010) Pivotal roles of three conserved carboxyl residues of the NuoC (30k) segment in the structural integrity of proton-translocating NADH-quinone oxidoreductase from Escherichia coli. Biochemistry 49, 10072–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takano S., Yano T., Yagi T. (1996) Structural studies of the proton-translocating NADH-quinone oxidoreductase (NDH-1) of Paracoccus denitrificans. Identity, property, and stoichiometry of the peripheral subunits. Biochemistry 35, 9120–9127 [DOI] [PubMed] [Google Scholar]
- 41. Torres-Bacete J., Sinha P. K., Matsuno-Yagi A., Yagi T. (2011) Structural contribution of C-terminal segments of NuoL (ND5) and NuoM (ND4) subunits of complex I from Escherichia coli. J. Biol. Chem. 286, 34007–34014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sinha P. K., Torres-Bacete J., Nakamaru-Ogiso E., Castro-Guerrero N., Matsuno-Yagi A., Yagi T. (2009) Critical roles of subunit NuoH (ND1) in the assembly of peripheral subunits with the membrane domain of Escherichia coli NDH-1. J. Biol. Chem. 284, 9814–9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yagi T. (1986) Purification and characterization of NADH dehydrogenase complex from Paracoccus denitrificans. Arch. Biochem. Biophys. 250, 302–311 [DOI] [PubMed] [Google Scholar]
- 44. Satoh T., Miyoshi H., Sakamoto K., Iwamura H. (1996) Comparison of the inhibitory action of synthetic capsaicin analogs with various NADH-ubiquinone oxidoreductases. Biochim. Biophys. Acta 1273, 21–30 [DOI] [PubMed] [Google Scholar]
- 45. Amarneh B., Vik S. B. (2003) Mutagenesis of subunit N of the Escherichia coli complex I. Identification of the initiation codon and the sensitivity of mutants to decylubiquinone. Biochemistry 42, 4800–4808 [DOI] [PubMed] [Google Scholar]
- 46. Matsushita K., Ohnishi T., Kaback H. R. (1987) NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26, 7732–7737 [DOI] [PubMed] [Google Scholar]
- 47. Yagi T. (1990) Inhibition by capsaicin of NADH-quinone oxidoreductases is correlated with the presence of energy-coupling site 1 in various organisms. Arch. Biochem. Biophys. 281, 305–311 [DOI] [PubMed] [Google Scholar]
- 48. Shimomura Y., Kawada T., Suzuki M. (1989) Capsaicin and its analogs inhibit the activity of NADH-coenzyme Q oxidoreductase of the mitochondrial respiratory chain. Arch. Biochem. Biophys. 270, 573–577 [DOI] [PubMed] [Google Scholar]
- 49. Michel J., DeLeon-Rangel J., Zhu S., Van Ree K., Vik S. B. (2011) Mutagenesis of the L, M, and N subunits of complex I from Escherichia coli indicates a common role in function. PLoS ONE 6, e17420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller M. J., Oldenburg M., Fillingame R. H. (1990) The essential carboxyl group in subunit c of the F1F0 ATP synthase can be moved and H+-translocating function retained. Proc. Natl. Acad. Sci. U.S.A. 87, 4900–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Padan E. (2008) The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem. Sci. 33, 435–443 [DOI] [PubMed] [Google Scholar]
- 52. Preiss L., Yildiz O., Hicks D. B., Krulwich T. A., Meier T. (2010) A new type of proton coordination in an F1FoATP synthase rotor ring. PLoS Biol. 8, e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Langemeyer L., Engelbrecht S. (2007) Essential arginine in subunit a and aspartate in subunit c of FoF1 ATP synthase. Effect of repositioning within helix 4 of subunit a and helix 2 of subunit c. Biochim. Biophys. Acta 1767, 998–1005 [DOI] [PubMed] [Google Scholar]
- 54. Symersky J., Pagadala V., Osowski D., Krah A., Meier T., Faraldo-Gómez J. D., Mueller D. M. (2012) Structure of the c(10) ring of the yeast mitochondrial ATP synthase in the open conformation. Nat. Struct. Mol. Biol. 19, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Garczarek F., Brown L. S., Lanyi J. K., Gerwert K. (2005) Proton binding within a membrane protein by a protonated water cluster. Proc. Natl. Acad. Sci. U.S.A. 102, 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Umemoto H., Ihsanawati, Inami M., Yatsunami R., Fukui T., Kumasaka T., Tanaka N., Nakamura S. (2009) Improvement of alkaliphily of Bacillus alkaline xylanase by introducing amino acid substitutions both on catalytic cleft and protein surface. Biosci. Biotechnol. Biochem. 73, 965–967 [DOI] [PubMed] [Google Scholar]
- 57. Hunte C., Screpanti E., Venturi M., Rimon A., Padan E., Michel H. (2005) Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 58. Arkin I. T., Xu H., Jensen M. Ø., Arbely E., Bennett E. R., Bowers K. J., Chow E., Dror R. O., Eastwood M. P., Flitman-Tene R., Gregersen B. A., Klepeis J. L., Kolossváry I., Shan Y., Shaw D. E. (2007) Mechanism of Na+/H+ antiporting. Science 317, 799–803 [DOI] [PubMed] [Google Scholar]
- 59. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 60. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]