Background: Cervical carcinogenesis mechanism remains to be further addressed along with papillomavirus infection.
Results: ZNF268b2 is overexpressed in human cervical cancer. It promotes HeLa cell proliferation in vitro and tumor growth in vivo by up-regulating NF-κB activity.
Conclusion: ZNF268b2 overexpression contributes to cervical cancer via enhancing NF-κB signaling.
Significance: ZNF268 may be a novel therapeutic target or a diagnostic marker for cervical cancer.
Keywords: Cancer Biology, Molecular Cell Biology, NF-κB Transcription Factor, Signal Transduction, Zinc Finger, Cervical Cancer, ZNF268
Abstract
Cervical cancer is one of the most common tumors affecting women's health worldwide. Although human papillomavirus can be detected in nearly all cases, the mechanism of cervical carcinogenesis remains to be further addressed. Here, we demonstrated that ZNF268, a Krüppel-associated box-containing zinc finger protein, might contribute to the development of cervical cancer. We found that ZNF268b2, an isoform of ZNF268, was overexpressed in human squamous cervical cancer specimens. Knockdown of ZNF268 in cervical cancer cells caused cell cycle arrest at the G0/G1 phase, reduced colony formation, and increased sensitivity to TNFα-induced apoptosis. In addition, HeLa cell growth in xenograft nude mice was suppressed by ZNF268 knockdown, with increased apoptosis. Furthermore, ZNF268b2 was shown to increase NF-κB signaling in vitro and in vivo. Reconstitution of NF-κB activity restored proliferation in ZNF268 knockdown HeLa cells. Of note, we observed a high frequency of NF-κB activation in ZNF268-overexpressing cervical cancer tissues, suggesting a pathological coincidence of ZNF268b2 overexpression and NF-κB activation. Taken together, our results reveal a novel role of ZNF268b2 that contributes to cervical carcinogenesis in part through enhancing NF-κB signaling.
Introduction
The Krüppel-associated box (KRAB)4-containing zinc finger proteins are the largest single family of transcriptional regulators in mammals and represent approximately one-third of all C2H2 zinc finger proteins (1). ZNF268, a typical KRAB-containing zinc finger protein (2), encodes two isoforms, ZNF268a and ZNF268b2 (3). ZNF268a contains the KRAB and zinc finger domains, which may function as a transcriptional repressor. ZNF268b2 consists of only the zinc finger domain and has been reported to be an IκB kinase (IKK)-associated candidate of the NF-κB signaling pathway (4). The ZNF268 promoter is located in the first exon of the gene, and cAMP-response element-binding protein 2 (CREB2) binds the promoter region to regulate ZNF268 expression (5). Previous studies suggest that ZNF268 may play a role in human fetal liver development (6), blood cell development, and hematological diseases (7–9). However, the function of ZNF268 remains largely unknown.
Recently, ZNF268 has been suggested to be involved with cervical cancer development. Cervical cancer is the third most commonly diagnosed cancer and the fourth leading type of cancer among women worldwide (10). Environmental risk factor such as human papillomavirus infection is necessary, but not sufficient, for cervical cancer development (11). Genetic risk factors contribute about 27% of the underlying factors for carcinogenesis and the progress of cervical cancer (12). A genome-wide linkage scan identified three chromosomal regions, 9q32, 12q24, and 16q24, as cervical cancer susceptible loci (13). ZNF268, by virtue of its localization on chromosome 12q24.33, is proposed to be one of the cervical cancer-related genes. This was supported by a microarray study showing that differential expression of ZNF268 mRNA was observed between normal cervical tissues and cervical carcinomas (14). Taken together, these findings suggest that ZNF268 may function in the development of cervical cancer.
NF-κB is a multifunctional transcription factor that regulates expression of numerous genes involved in both proliferation and apoptosis (15). Two major signaling pathways that activate NF-κB, termed the classic pathway and alternative pathway, have been identified based on the stimuli and signaling molecules involved (16). The classical NF-κB pathway is triggered by proinflammatory cytokines, such as tumor necrosis factor α (TNFα), which subsequently triggers IKK, degrades IκB, and activates the p65 and p50 subunits (17). The NF-κB pathway has been implicated in the development and progression of various cancer types (18), including cervical cancer (19).
In this study, we investigate the role of ZNF268 in cervical cancer development. We show that ZNF268 is overexpressed in cervical cancer, and knockdown of ZNF268 suppresses HeLa cell growth in vitro as well as in vivo. In the HeLa cell model, knockdown of ZNF268 enhances the sensitivity of HeLa cells to TNFα-induced apoptosis. Further study show that ZNF268 activates NF-κB by participating IKK complex assembly and promoting IKK phosphorylation. In addition, restoration of NF-κB by overexpression of constitutively active p65 rescues the growth impairment induced by ZNF268 deficiency. Finally, we show that NF-κB activation frequently coincides with ZNF268 overexpression in cervical cancer tissues. Taken together, our data suggest that aberrant expression of ZNF268 in cervical cancer may enhance NF-κB signaling, subsequently alter cell growth, and contribute to carcinogenesis of cervical cancer.
EXPERIMENTAL PROCEDURES
Tissue Specimens, Histology, and Immunohistochemistry
Paraffin sections (MC5003) containing 22 normal human and carcinoma specimens were purchased from US Biomax and immunohistochemically stained with the ZNF268 antibodies. The SD antibody detected total ZNF268 proteins (ZNF268a and ZNF268b2, two predominant products of ZNF268 gene), and antibody E3 specifically recognized ZNF268a but not ZNF268b2 as described previously (3). In addition, four paraffin-embedded normal cervical squamous epithelium specimens and 30 paraffin-embedded cervical squamous carcinoma specimens were obtained from the Department of Pathology, Zhongnan Hospital of Wuhan University in China, to detect ZNF268 and IκBα expression or p65 nuclear translocation. In total, we collected seven normal squamous epithelium and 47 cervical carcinomas. All cervical carcinomas were squamous cell carcinomas in which six specimens were classified as well differentiated squamous cell carcinoma, 32 specimens of moderately differentiated SCC, and eight specimens of poorly differentiated SCC. The classification of one specimen remained unknown. Our study was approved by the Regional Committee of Medical Research Ethics in China, and informed consent was obtained from each patient. The expression level of examined proteins was scored by two independent pathologists without knowledge of the clinical information of each patient. The expression levels were scored as 0 point (−) for no detectable staining, 1 point (+) for weak staining, 2 points (++) for clear but not strong staining, and 3 points (+++) for strong staining. For p65 translocation, nucleus staining positivity scored 1 and negativity was 0.
Specimens of HeLa xenografts were fixed in 4% paraformaldehyde and embedded with paraffin. All immunohistochemistry staining assays were performed by Jiayuan Quantum Dots Co. (Wuhan, China) following a standard procedure. In addition to the above DAB staining, ZNF268a and total ZNF268 protein were also labeled with 605- and 545-nm QDs, respectively, and the expression signal in cervical tissue was observed and analyzed under microscopy with UV excitation light as described previously (20).
Establishment of Stable ZNF268 Knockdown HeLa Cells
HeLa cells (CCTCC, Wuhan, China) were grown in DMEM supplemented with 10% FBS (Invitrogen), penicillin (100 units/ml), and streptomycin (100 μg/ml) at 37 °C in a 5% CO2 incubator. Lentivirus expressing two shRNA hairpins specifically for ZNF268 (shZNF268) or control lentivirus was purchased from Cyagen Biosciences, China. HeLa cells were infected by lentivirus and selected for GFP positivity by sorting. Down-regulation of ZNF268 was confirmed at the mRNA and protein level. The stem sequences for two shZNF268 hairpins are 5′-CGGGAAAGACTTCAGTAGTAAA-3′ and 5′-GCACGCATGGAAAGAGTTTGAT-3′, respectively. The stem sequence of control shRNA is 5′-GCGCGCTTTGTAGGATTCG-3′, which is widely used in other researches and does not match any known human-coding cDNAs or human EST sequences by blasting the GenBankTM.
Cell Cycle, Apoptosis, and Cell Growth Assay
Cell cycle and apoptosis were assessed through propidium iodide staining and analyzed by flow cytometry. Briefly, HeLa cells were fixed in ice-cold 70% ethanol, treated with RNase A (1 mg/ml), and stained with propidium iodide (PI, 5 μg/ml). Cell growth was measured through MTT assay. In short, HeLa cells (2 × 103/well) were cultured in a 96-well plate, incubated with 20 μl of MTT dye (5 mg/ml), followed by solubilization in DMSO (100 μl/well). The absorbance was determined at 570 nm using a Microplate reader (Biotechnology) and normalized to day 1 (1 day after plating). The data were presented as proliferation rate.
Immunofluorescent Staining
HeLa cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and incubated with the cleaved caspase 3 antibody overnight at 4 °C. The secondary TRITC-conjugated antibody (Pierce) was then applied for 1 h at room temperature. Finally, nuclei were stained with DAPI (Roche Applied Science). Fluorescent images were obtained using FV1000 configuration with a BX61 microscope (Olympus).
Colony Forming Assay in Soft Agar
Cells were trypsinized, suspended in top agar containing 10% FBS and 0.3% agarose, and seeded into 60-mm dishes containing bottom agar with 10% FBS and 0.6% agar. After 3 weeks, 1 ml of 2-(p-iodophenyl)-3-(p-nitrophenyl)-5-phenyl tetrazolium chloride (1 mg/ml) was added. After 4 h, the number of colonies was counted, and the plates were photographed.
Nude Mouse Tumor Formation Assay
Male nude mice (BALB/c nu/nu, 5 weeks) were purchased from SJA Lab Animal Ltd. Co. (China) and maintained in a barrier mouse facility at College of Life Sciences, Wuhan University. HeLa cells (5 × 106) in exponential growth phase were injected subcutaneously into nude mice. All mice were maintained for 30 days before they were sacrificed to collect tumors. Tumor volume was calculated using the following formula: tumor volume = ½ × (longer diameter) × (shorter diameter)2. All animal experiments were approved by the Animal Research Ethics Board of Wuhan University in China and were in compliance with institutional guidelines on the care of experimental animals.
Plasmid Construction
The mammalian two-hybrid system, including plasmids pM, pVP16, and the GAL4-dependent luciferase reporter (Gal4-luci), were kind gifts of Dr. Ying Zhu (Wuhan University, China). The NF-κB-dependent luciferase reporter (NF-κB-luc) and TNFR1, TRAF2, RIP, IKKα, IKKβ, and p65 expression plasmids were kind gifts of Dr. Hong-Bing Shu (Wuhan University, China). Other plasmids, including pcDNA-GFP, pcDNA-ZNF268b2, pM-ZNF268b2, pM-IKKα, pM-IKKβ, pM-IKKγ, pVP16/IKKα, pVP16-IKKβ, and pVP16-IKKγ, were constructed using a PCR strategy. These plasmids were confirmed by DNA sequencing.
Transfection and Luciferase Assay
All luciferase assays were performed in HeLa cells. Briefly, 3.0 × 104 HeLa cells were seeded in 48-well plates the day before transfection. When cells reached about 70% confluence, NF-κB firefly luciferase reporter (NF-κB-luc) and other plasmids were transfected together with pRL-TK, which expresses Renilla luciferase as internal control. Luciferase activity was assayed according to the manufacturer's instructions (Dual-Luciferase Reporter Assay System, Promega). All of the transfection assays were performed with Lipofectamine 2000 reagent (Invitrogen).
Cytoplasmic and Nuclear Protein Extraction
Briefly, cells were collected and incubated in 5 volumes of cytoplasmic extraction reagent (Boster, China) for 30 min on ice, followed by centrifugation at 12,000 rpm at 4 °C. The collected supernatants were the cytoplasmic proteins. The cell pellets were incubated with 2 volumes of nuclear extraction reagent (Boster, China) for 30 min, with rough rotation every 5 min, and then centrifuged at 12,000 rpm at 4 °C. Nuclear proteins existed in the supernatant.
Electrophoresis Mobility Shift Assays (EMSAs)
Equal amounts of nuclear extracts (3 μg) were used for EMSA analysis. Biotin 5′ end-labeled NK-κB consensus oligonucleotides (NF-κB sense, biotin-AGTTGAGGGGACTTTCCCAGGC; NF-κB antisense, biotin-GCCTGGGAAAGTCCCCTCAACT) were synthesized (Sangon Biotech, China). EMSA was performed according to the manufacturer's instructions (Pierce). Competition assays were performed with a 200-fold excess of unlabeled NF-κB consensus oligonucleotides.
Immunoprecipitation
Briefly, 5.0 × 106 cells were collected and lysed in 800 μl of RIPA buffer. Precleared cell extracts were mixed with 2 μg of the appropriate antibodies plus 20 μl of protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) and rotated overnight at 4 °C. The immunoprecipitated complexes were obtained by centrifugation, washed three times with RIPA lysis buffer, and boiled for 10 min for Western blotting analysis.
Western Blotting
Cells were collected and lysed in RIPA buffer on ice for 15 min, followed by centrifugation at 12,000 rpm at 4 °C for 10 min. Supernatants were collected, and the concentration of whole cell lysates (WCL) was determined by BCA method (Pierce). The WCL were boiled in an equal volume of SDS-PAGE 2× sample buffer, and Western blotting was performed according to standard procedure. Antibodies used for Western blotting analysis were as follows: actin, IKKα, IKKβ, IKKγ, and mouse control IgG antibodies were purchased from Santa Cruz Biotechnology. Cyclin D1, cyclin E2, PCNA, procaspase 3, p-IKKα/β, p65, IκBα, p53, c-Myc, CD31 and Bcl-xL antibodies were purchased from Cell Signaling Technology. Other antibodies include SD and E3 antibody for ZNF268, a GFP antibody purchased from EarthOx, and a p50 antibody purchased from Millipore.
Statistics
For cell proliferation, luciferase assay, and gene expression, data were represented as mean ± S.D. of samples. All experiments were repeated three times. Two-tailed Student's t test was used to compare differences between two experimental groups. A p value less than 0.05 was considered statistically significant. To compare ZNF268 expression in human normal and cancer tissues, a two-tailed Student's t test was also performed using immunohistochemistry scores of samples.
RESULTS
ZNF268 Is Overexpressed in Human Cervical Cancer
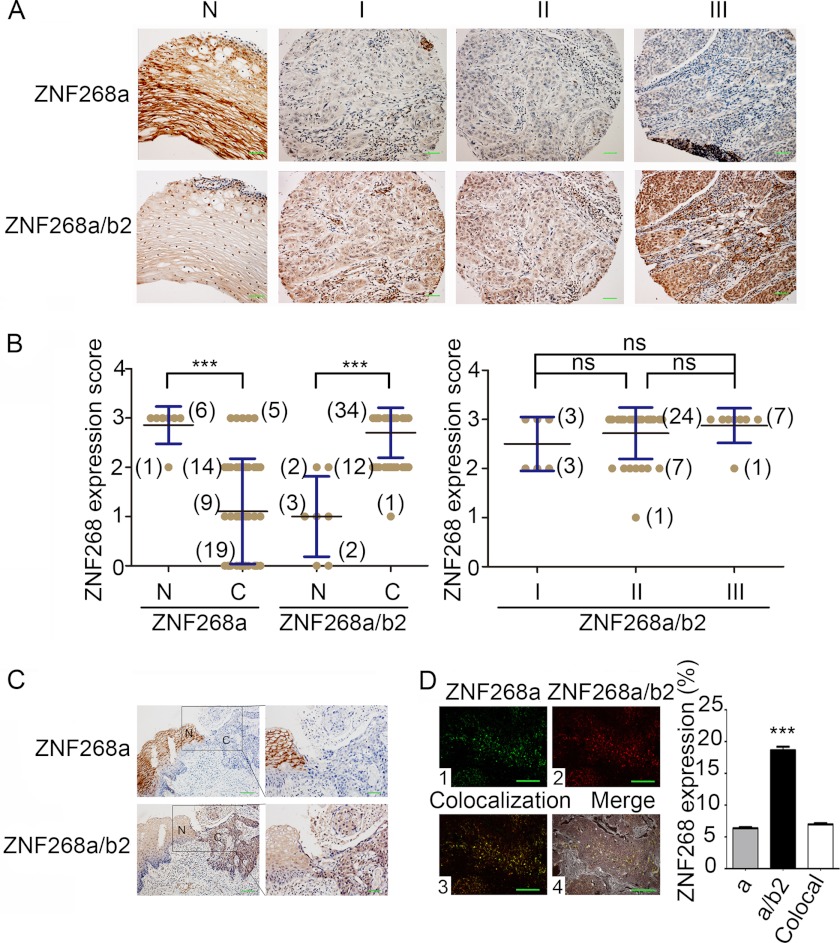
ZNF268 has been implied to play a role in cervical cancer. To further investigate this, we first examined ZNF268 protein expression in cervical cancer species by immunohistochemistry. We used SD antibody to detect total ZNF268 proteins (ZNF268a and ZNF268b2, two predominant products of the ZNF268 gene) and antibody E3 that specifically recognized ZNF268a but not ZNF268b2 as described previously (3). We observed higher expression of total ZNF268 in SCC than that in normal squamous epithelium tissues (p < 0.001) (Fig. 1, A and B). However, we also detected high expression of ZNF268a in all of the normal samples, although about 40% (19 out of 47) of the carcinomas were completely negative (p < 0.001) (Fig. 1, A and B). This expression pattern of ZNF268 was further verified by immunohistochemistry analysis of three samples containing both normal squamous epithelium and squamous carcinoma tissue adjacent to each other (Fig. 1C). These findings suggest that ZNF268 overexpression is possibly involved in cervical cancer development. To support this, we also performed a ZNF268 expression pattern profiling assay through screening a large scale tissue microarray by immunohistochemistry. We observed that the up-regulation of ZNF268 in cervical cancer was the most significant one in all cancer tissues examined (supplemental Fig. S1). Some tissues showed reverse expression patterns (supplemental Fig. S1). To test whether ZNF268 overexpression is associated with cervical cancer grade, we further classified squamous cervical carcinoma samples into three groups based on cell differentiation status: well differentiated SCC, moderately differentiated SCC, and poorly differentiated SCC. Then we compared the expression level of ZNF268. We did not observe a statistically significant difference in all three groups (NS, p > 0.05) (Fig. 1, A and B), suggesting that ZNF268 overexpression may not be associated with squamous cervical cancer cell differentiation status or malignancy.
FIGURE 1.
ZNF268 is overexpressed in cervical cancer tissues. A, representative image of ZNF268 immunohistochemistry staining in cervical tissues. E3 and SD antibodies were used for immunohistochemistry to detect ZNF268a and ZNF268a/ZNF268b2 proteins, respectively. N indicates normal cervical squamous epithelium; C indicates squamous cell carcinoma; panels I–III indicate well differentiated squamous cell carcinoma, moderately differentiated squamous cell carcinoma, and poorly differentiated squamous cell carcinoma, respectively. Scale bars, 50 μm. B, ZNF268 expression score from each group of immunohistochemistry was presented as mean ± S.D. Two-tailed Student's t test was performed to compare ZNF268 expression in normal and cervical cancer tissues (left panel). The ZNF268 expression in different differentiation grades of squamous cervical carcinomas (right panel) was also compared. Numbers in parentheses indicate the numbers of the specimens studied. *** indicates p < 0.001; ns indicates p > 0.05. C, representative immunohistochemistry image of ZNF268 expression in two cervical tissues containing both normal squamous epithelium (N) and squamous carcinomas (C). Scale bars, 100 μm (left panel) and 50 μm (right panel). Right panel is the magnified picture of the boxed field in left panel. D, relative expression of ZNF268a and ZNF268b2 in cervical carcinoma was measured by QD technique. Panel 1 (green) is for ZNF268a. Panel 2 (red) is for ZNF268a/ZNF268b2. Panels 3 and 4 indicated the colocalization (colocal) signal (green and red signals) and the merged result, respectively. The statistic of expression signal is shown in the right panel. Scale bars, 50 μm.
Considering the high expression of total ZNF268 and low expression of ZNF268a in SCC as described above, we inferred that ZNF268b2 accounted for the high expression of total ZNF268 protein in SCC. To quantitatively test this, we used the QD technique to label E3 (ZNF268a) or SD (total ZNF268) and analyzed their relative expression in the squamous cervical carcinoma samples. ZNF268a and total ZNF268 protein showed 6.36 and 18.63% expression (Fig. 1D, panels 1 and 2), respectively, with the 6.96% colocalization signal (Fig. 1D, panel 3) almost equal to that of ZNF268a. Hence, by subtracting the colocalization signals, we could estimate the expression signal of ZNF268b2 to be 12.27%. This was nearly twice the expression level of ZNF268a and confirmed that ZNF268b2 was the major isoform that was overexpressed in SCC. Such an expression pattern suggests that alteration of the relative expression level of ZNF268a and ZNF268b2 may contribute to human cervical cancer development.
ZNF268 Knockdown Impairs HeLa Cell Proliferation
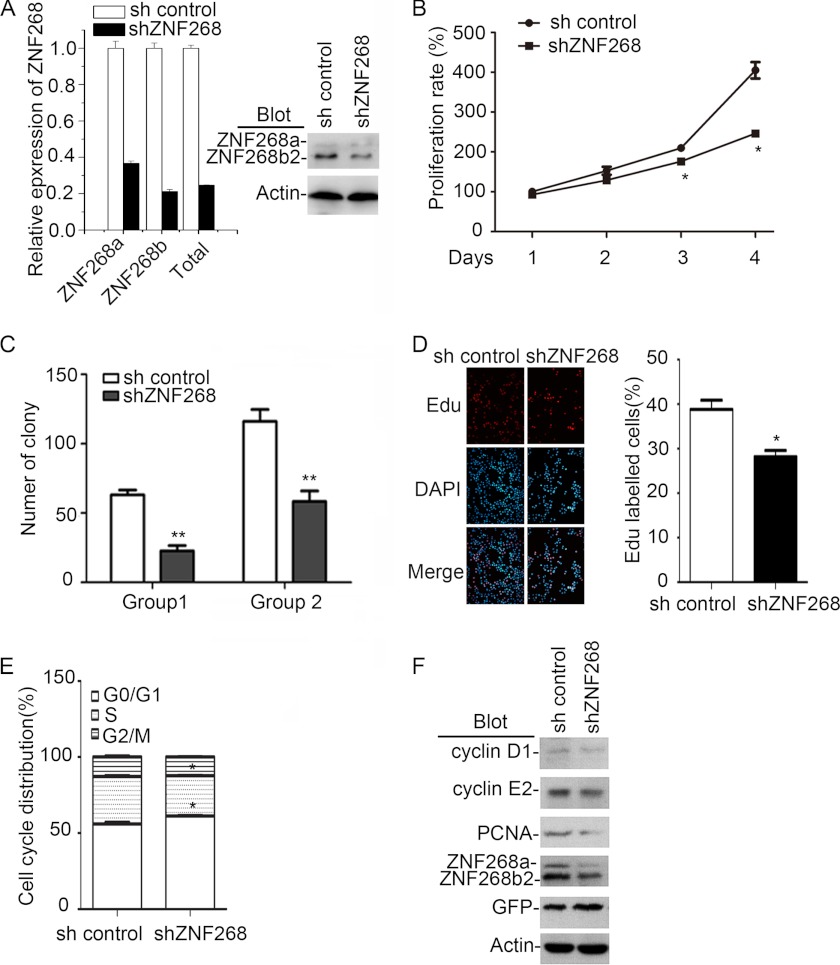
To investigate the biological significance of ZNF268 overexpression in cervical cancer, we utilized the relevant cervical cancer cell line HeLa that showed relatively high expression of ZNF268 compared with other cancer cells (supplemental Fig. S2). We established a new line in which both ZNF268a and ZNF268b2 were knocked down by shRNA (shZNF268) (Fig. 2A). As expected, we observed that ZNF268 knockdown reduced cell proliferation in MTT assays in 2 days and more significantly in 3 days after plating (p < 0.05, compared with sh control cells) (Fig. 2B). To exclude the possibility that the observed phenomenon was due to the off-target effect of the shRNA, a second shZNF268 hairpin was also used to knock down ZNF268 expression in HeLa cells. Again, ZNF268 knockdown in HeLa cells impaired cell proliferation (supplemental Fig. S3), confirming the specific effects of ZNF268 shRNA. These results suggest that proliferation inhibition in shZNF268 HeLa cells is due to specific down-regulation of ZNF268 expression by shZNF268, but not an off-target effect. Thus, we used the first shRNA hairpin for the rest of the studies to further explore the effects of ZNF268 knockdown on HeLa cells. Consistent with the results of the MTT assays, shZNF268 HeLa cells also exhibited a significant reduction of colony-forming ability in soft agar as evidenced by an approximate 2-fold decrease of colony number in shZNF268 HeLa cells compared with that of sh control cells (p < 0.01) (Fig. 2C). Finally, we examined cell proliferation using an EdU incorporation assay, a sensitive and specific assay that labels replicating cells. The number of EdU-labeled cells in the shZNF268 HeLa cells were significantly (p < 0.05) decreased compared with that in the sh control HeLa cells (Fig. 2D), again confirming that the knockdown of ZNF268 impaired HeLa cell proliferation.
FIGURE 2.
ZNF268 knockdown impairs HeLa cell proliferation in vitro. A, HeLa cells were infected with lentivirus expressing an shRNA specific for ZNF268 (shZNF268) or control vector (sh control) and selected for GFP positivity by sorting. ZNF268 knockdown were measured by quantitative RT-PCR (left panel) and Western blot (right panel). B, sh control and shZNF268 HeLa cells were seeded in a 96-well plate. An MTT assay was performed at the indicated time points. Data were normalized to day 1 (1 day after plating) and presented as proliferation rate. C, sh control and shZNF268 HeLa cells (4.0 × 103 cells/dish in Group 1 and 8.0 × 103 cells/dish in Group 2) were also seeded in 35-mm soft agar dishes. The colony number was counted 2 weeks post-inoculation to determine colony-forming ability. D, sh control and shZNF268 HeLa cells were labeled with EdU (50 μm). The EdU incorporation was measured by immunofluorescent staining and counted under fluorescence microscopy. Left panel was the representative results, and right panel was the statistics of EdU labeling experiment. E, sh control and shZNF268 HeLa cells were collected during the exponential phase of growth, stained with PI, and analyzed by flow cytometry. The cell cycle profiles of sh control and shZNF268 HeLa cells were analyzed by ModFit software. F, The expression of cyclin D1, cyclin E2, and PCNA in sh control and shZNF268 HeLa cells were detected by Western blot. Actin was used as a protein loading control. * indicates p < 0.05; ** indicates p < 0.01.
To further determine the potential mechanism by which ZNF268 knockdown caused cell proliferation inhibition, we measured cell apoptosis in sh control or ZNF268 knockdown HeLa cells by propidium iodide (PI) staining and analyzed by flow cytometry. We failed to observe a pronounced sub-G1/G0 cell population in either cell group (data not shown). However, ZNF268 knockdown did alter cell cycle distribution. In shZNF268 HeLa cells, the G0/G1 cell population reproducibly increased while the cells in S phase decreased accordingly (p < 0.05, compared with that of sh control HeLa cells) (Fig. 2E). Consistent with the cell cycle profile, the expression of cyclin D1 and cyclin E2 that promotes G1/S transition (21) was decreased in shZNF268 HeLa cells (Fig. 2F). In addition, PCNA, a proliferation marker that assists in DNA replication, was decreased in shZNF268 HeLa cells (Fig. 2F). Taken together, these results demonstrated that ZNF268 knockdown suppressed HeLa cell growth in vitro in part by affecting cell cycle progress.
ZNF268 Knockdown Increases TNFα/CHX-induced Apoptosis in HeLa Cells
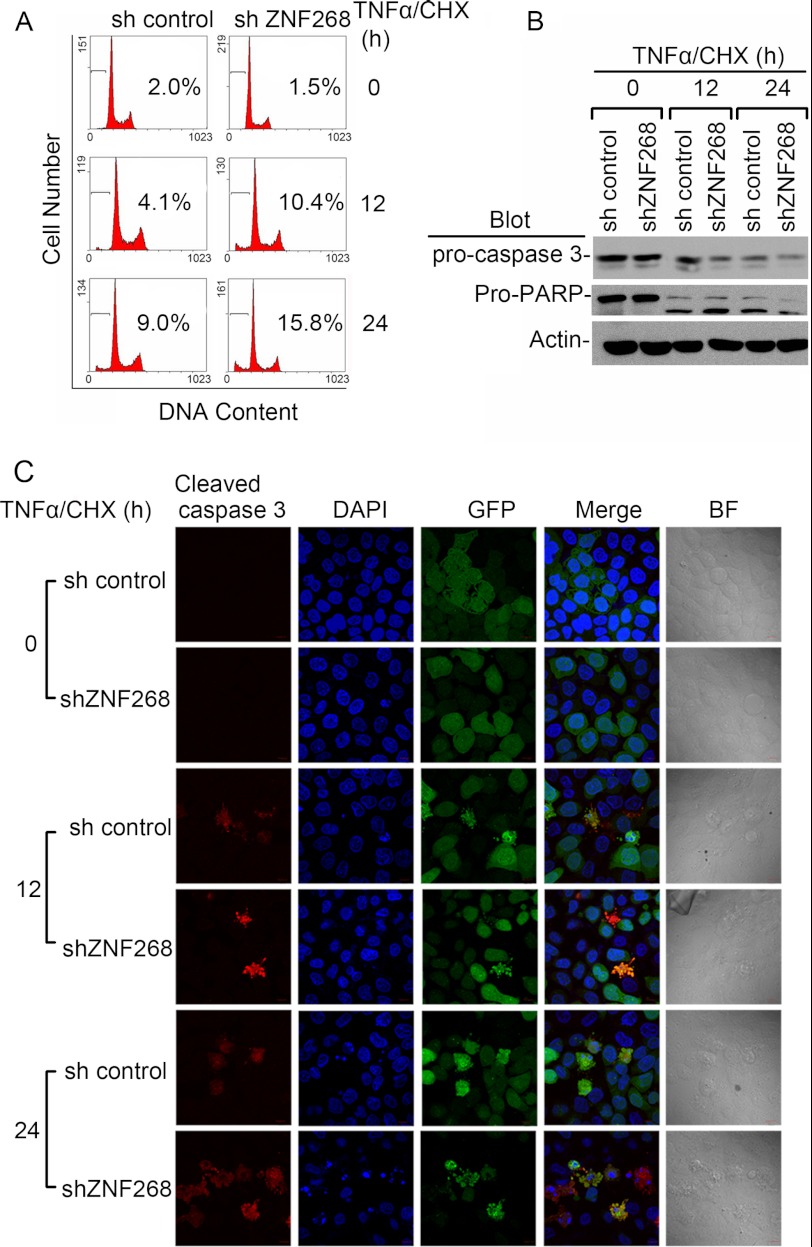
Minor alterations in cell cycle distribution may not fully explain the cell proliferation inhibition. We further tested whether ZNF268 functioned under apoptosis-inducing conditions. TNFα is a pleiotropic, proinflammatory cytokine that has also been implicated in cervical pathogenesis (22, 23). As expected, TNFα plus cycloheximide (TNFα/CHX) treatment potently induced apoptosis at both 12 and 24 h post-treatment as evidenced by the appearance of a sub-G1/G0 peak (Fig. 3A). Notably, ZNF268 knockdown significantly increased the proportion of the sub-G1/G0 cell population (10.4 and 15.8% in shZNF268 HeLa cells compared with 4.1 and 9.0% in sh control HeLa cells at 12 and 24 h post-treatment, respectively). Consistent with flow data, shZNF268 HeLa cells also showed increased caspase 3 and PARP cleavage measured by Western blot and immunofluorescent staining (Fig. 3, B and C). Morphology markers of apoptosis, including cell shrinkage and nuclear and cellular fragmentation, were also readily observed under light microscopy in TNFα/CHX treatment conditions, and shZNF268 HeLa cells displayed a worse phenotype than that in sh control (Fig. 3C). These findings suggest that ZNF268 knockdown may promote apoptosis through a caspase 3, and PARP-dependent manner and ZNF268 may influence apoptosis under inflammatory conditions that frequently occur in cancer tissues.
FIGURE 3.
ZNF268 knockdown promotes apoptosis induced by TNFα/CHX stimulation in HeLa cells. A, sh control and shZNF268 HeLa cells were treated with TNFα plus CHX for the time indicated (0, 12, and 24 h). Apoptosis was measured by PI staining to detect the sub-G1 cell population and analyzed by flow cytometry. Numbers indicate the percentage of cells undergoing apoptosis. B, apoptosis in sh control and shZNF268 HeLa cells treated with TNFα plus CHX was also evidenced by pro-caspase 3 and pro-PARP cleavage measured by Western blot. C, pro-caspase 3 cleavage was further measured by immunofluorescent with an antibody specific for carved caspase 3 and labeled with TRITC. DNA was stained with DAPI to detect apoptotic nuclei. GFP was expressed in both sh control and shZNF268 cells. The merged pictures of three colors were also shown. BF indicates bright field. Scale bars, 10 μm.
ZNF268b2 Enhances NF-κB Activation Induced by TNFα Treatment
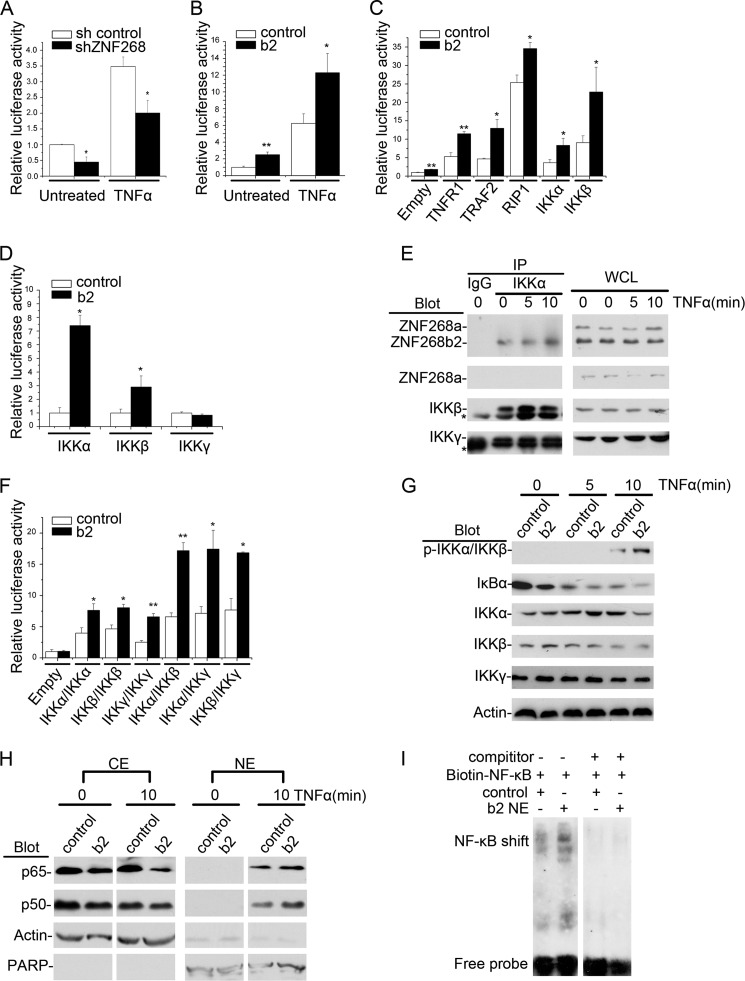
Previously, ZNF268b2 was reported to associate with the IKK complex. Thus, we investigated whether ZNF268 regulated NF-κB activity in HeLa cells. We transfected HeLa cells with a typical NF-κB-responsive luciferase reporter construct NF-κB-luc to indicate NF-κB activation. In ZNF268 knockdown cells (shZNF268), the luciferase activity was lower than that in sh control HeLa cells under TNFα-treated or -untreated conditions, suggesting that ZNF268 knockdown suppressed both background NF-κB activity and TNFα-induced NF-κB activation (Fig. 4A). Consistent with ZNF268 knockdown, ZNF268b2 overexpression (b2) under TNFα-treated or -untreated conditions increased luciferase activities, suggesting ZNF268b2 overexpression increased basal NF-κB activity as well as potentiated TNFα-induced activation (Fig. 4B). In addition, the effect of ZNF268b2 on NF-κB activation was dose-dependent (supplemental Fig. S4).
FIGURE 4.
ZNF268 enhances NF-κB activation by TNFα induction. A, sh control or shZNF268 HeLa cells were transfected with NF-κB-luc and treated with or without TNFα (20 ng/ml) for 6 h. The NF-κB activation in the untreated and treated (TNFα) cells was monitored by measuring luciferase activities. B, HeLa cells were transfected with NF-κB-luc plus control vector or ZNF268b2 (b2) and treated (TNFα) or untreated with TNFα (20 ng/ml) for 6 h. The NF-κB activation was monitored by measuring luciferase activities. C, HeLa cells were transfected with NF-κB-luc plus control vector or ZNF268b2 (b2) in combination with other vectors as indicated. Cells were further treated with TNFα for 6 h, and NF-κB activities were monitored. D, HeLa cells were transfected with Gal4-luc and the indicated bait (control or ZNF268b2) and prey constructs (IKKα, IKKβ, or IKKγ). ZNF268b2 interaction with IKKα, IKKβ, or IKKγ was determined by monitoring the luciferase activities. E, HeLa cells were treated with TNFα (20 ng/ml) for the times indicated. Whole cell lysates (WCL) were then immunoprecipitated (IP) with the IKKα antibody or control IgG and then immunoblotted with the indicated antibodies. Whole cell lysates were also immunoblotted to verify equal amounts of protein for immunoprecipitation. Asterisk indicates nonspecific binds. F, HeLa cells were cotransfected with Gal4-luc with combination of ZNF268b2 (b2) or control and IKKα, IKKβ, or IKKγ (bait) and IKKα, IKKβ, or IKKγ (prey) as indicated. The formation of homo- or hetero-complex was detected by monitoring luciferase activities. G, HeLa cells were transfected with ZNF268b2 (b2) or control, stimulated with TNFα (20 ng/ml) for the indicated time (0, 5, and 10 min) and blotted with antibodies as indicated. H, HeLa cells were transfected with ZNF268b2 (b2) or control and stimulated with TNFα (20 ng/ml) for the times indicated. Cytoplasmic extracts (CE) and nuclear extracts (NE) were separated, followed by Western blotting analysis. PARP and actin were used as internal controls for the quality of nuclear extracts and cytoplasmic extract separation. I, nuclear extracts from control vector or ZNF268b2 (b2 NE) transfected HeLa cells treated with TNFα were used for EMSA to detect NF-κB DNA binding activity. ZNF268b2 overexpression increased DNA binding of NF-κB. * indicates p < 0.05; ** indicates p < 0.01.
Various signaling molecules, including TNFR1, TRAF2, RIP1, IKKα, and IKKβ are involved in TNFα-induced NF-κB activation (24). Cotransfection of ZNF268b2 and these molecules increased luciferase activity, suggesting that ZNF268b2 can synergize with TNFR1, TRAF2, RIP1, IKKα, and IKKβ to activate NF-κB (Fig. 4C). These findings suggest that ZNF268b2 is involved in the classic TNFα-induced NF-κB signaling pathway.
We further investigated which subunit of IKK complex interacts with ZNF268b2 using a mammalian two-hybrid system. We used ZNF268b2 as a bait and IKKα, IKKβ, or IKKγ as prey. Interaction of bait and prey will lead to luciferase gene expression. As shown in Fig. 4D, cells transfected with ZNF268b2 plus IKKα or IKKβ but not IKKγ caused strong luciferase activities, suggesting ZNF268b2 interacts with IKKα and IKKβ but not IKKγ. The interaction of IKKα and ZNF268b2 was confirmed by coimmunoprecipitation assay. The presence of IKKβ and ZNF268b2 could be readily detected by Western blot in protein complex immunoprecipitated by an IKKα antibody (Fig. 4E). Moreover, TNFα induction further increased the amount of ZNF268b2 protein that was recruited to the IKK complex. However, ZNF268a protein was neither associated with nor recruited to the IKK complex after TNFα induction (Fig. 4E).
To determine the effect of ZNF268b2 on IKK complex formation, IKKα, IKKβ, or IKKγ cDNAs was inserted into the pM vectors as bait as well as the pVP16 vectors as prey. The combination of these constructs allowed us to test homo- and hetero-complexes in IKKα, IKKβ, or IKKγ. We found that all possible combinations efficiently increase luciferase activities as expected (25), and the presence of ZNF268b2 further up-regulated luciferase activities. These results suggest that ZNF268b2 overexpression may facilitate all homo- and hetero-complex formation (Fig. 4F).
Given the effect of ZNF268b2 on IKK complex formation, ZNF268b2 may affect IKK activation. In response to TNFα treatment, the amount of phosphorylated IKKα/β increased substantially in HeLa cells, followed by the degradation of one of its substrates, IκBα (Fig. 4G). Notably, ZNF268b2 overexpression (b2) enhanced IKK phosphorylation and led to more IκBα degradation measured by Western blot (Fig. 4G). Increased phosphorylation of IKK by ZNF268b2 subsequently caused increased NF-κB subunits (p65/p50) in the nuclear extract, suggesting NF-κB subunits translocation into the nucleus (Fig. 4H). More NF-κB subunits in the nuclear extract from ZNF268b2 overexpression cells (b2) ultimately increased target DNA binding ability evidenced by a stronger NF-κB shift in EMSA compared with that of control (Fig. 4I). In summary, our data suggest that TNFα stimulation allows ZNF268b2 protein to be recruited to the IKK complex and facilitates the formation of IKK complex. This subsequently leads to increased IKK kinase activity and NF-κB activation as evidenced by more nuclear translocation and target DNA binding ability.
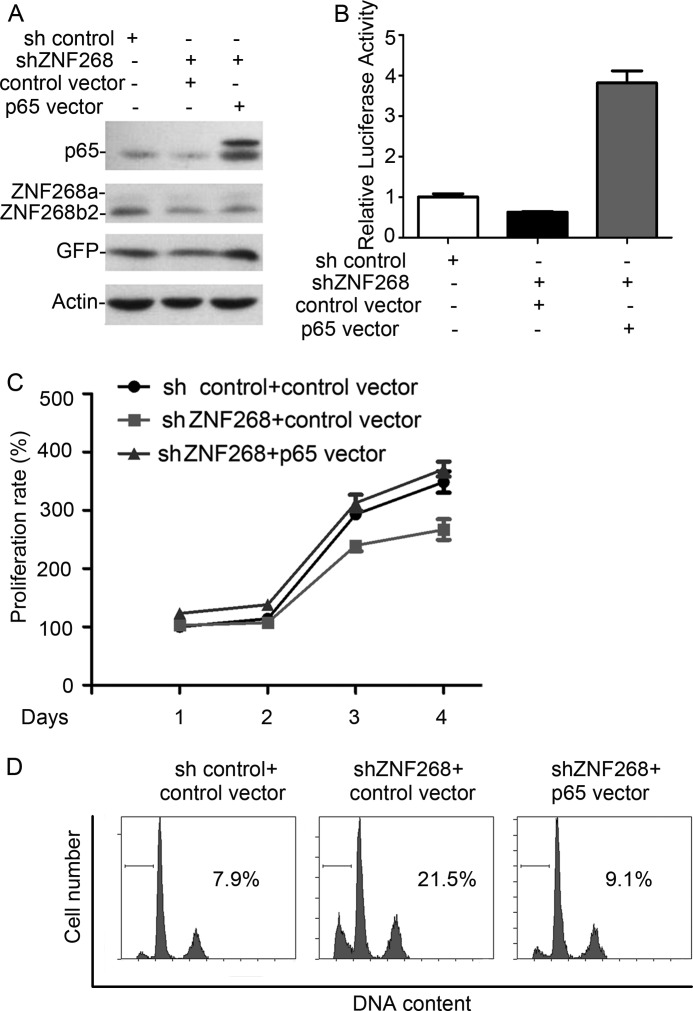
To further verify the role of NF-κB in mediating the function of ZNF268 on cell proliferation and apoptosis, we reconstituted NF-κB activity by overexpressing the constitutively active form of p65 in ZNF268 knockdown cells (Fig. 5A). As expected, overexpression of p65 significantly increased luciferase activity (Fig. 5B). Furthermore, restoration of NF-κB activity in ZNF268 knockdown cells normalized cell proliferation as well as apoptosis upon TNFα/CHX induction comparable with that of control cells (Fig. 5, C and D). These results confirmed that NF-κB might be the major downstream molecule that mediates the function of ZNF268b2.
FIGURE 5.
Reconstitution of NF-κB activity rescues proliferation inhibition and apoptosis induced by ZNF268 knockdown. A, sh control or shZNF268 HeLa cells were transfected with constitutively active p65 and selected for puromycin resistance. The p65 and ZNF268 expression in HeLa cells was measured by Western blot. B, p65 overexpression restored NF-κB activity that was impaired by ZNF268 knockdown. HeLa cells were transfected with plasmids as indicated and subjected to luciferase assay 48 h post-transfection. C, p65 overexpression in ZNF268 knockdown HeLa cells rescued cell proliferation inhibition. sh control or shZNF268 HeLa cells were transfected with control vector or constitutively active p65. The MTT assay was performed at the indicated time points. Data were normalized to day 1 (1 day after plating) and presented as proliferation rate. D, p65 overexpression in ZNF268 knockdown HeLa cells significantly reduced cell death. Cells were transfected with plasmids as indicated. After 24 h, the cells were treated with TNFα plus CHX for another 24 h. Cell apoptosis was measured by PI staining and analyzed by flow cytometry.
ZNF268 Knockdown Impairs Tumorigenesis of HeLa Cells in Xenograft Nude Mouse Model
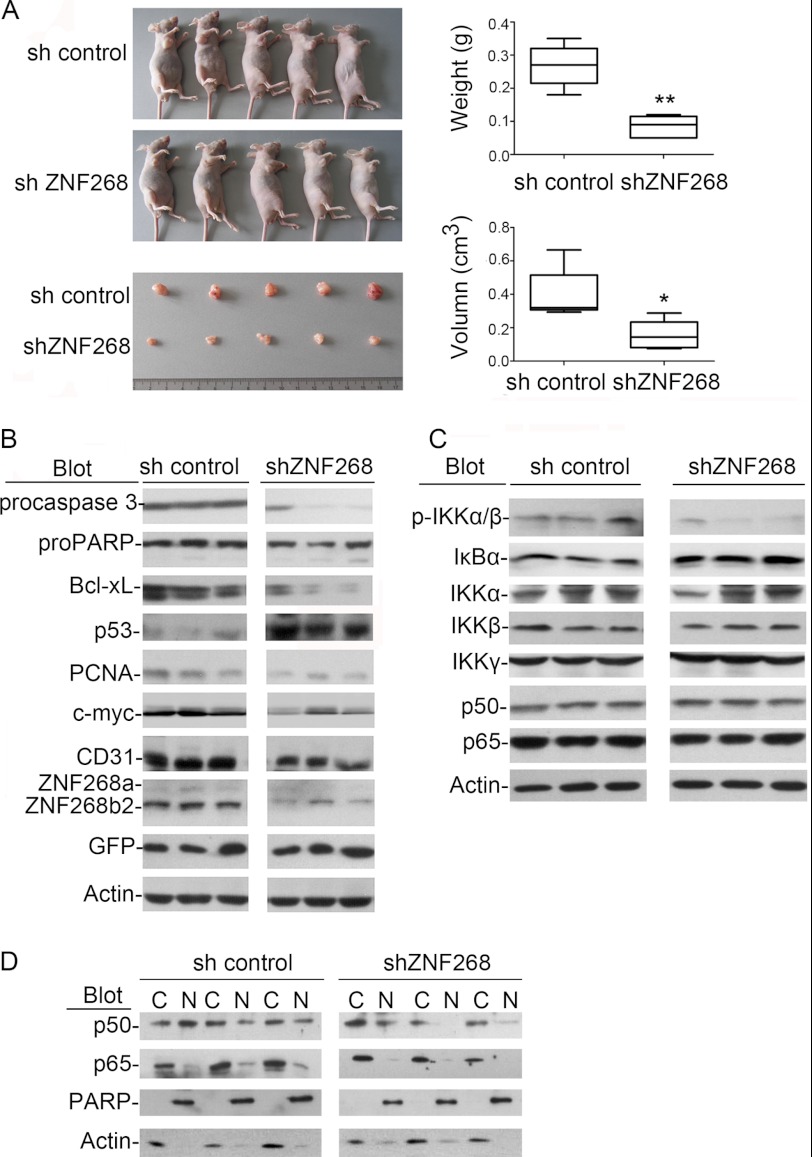
To study the in vivo function of ZNF268, we used the xenograft mouse model. Subcutaneous injection of HeLa cells in nude mice led to tumor growth. Consistent with in vitro findings, tumors collected from recipient mice engrafted with shZNF268 HeLa cells exhibited significantly reduced tumor weight (p < 0.01) and size (p < 0.05) compared with that of the sh control group (Fig. 6A). Tumors from recipient mice engrafted with shZNF268 HeLa cells also showed increased procaspase 3 cleavages, less PCNA and CD31 expression, three markers for apoptosis, proliferation, and microvessels, respectively (Fig. 6B). These results were confirmed by immunohistochemistry experiments (supplemental Fig. S5A) and were supported by other markers, including decreased expression of the oncogene c-myc, anti-apoptotic gene Bcl-xL, and increased expression of the tumor suppressor p53 (Fig. 6B). Increased cleavage of caspase 3 subsequently resulted in more PARP cleavage (Fig. 6B). H&E staining also showed that tumors from the shZNF268 group had more cells undergoing apoptosis (supplemental Fig. S5B). These data suggest that ZNF268 knockdown inhibits tumor growth by increasing apoptosis in vivo.
FIGURE 6.
ZNF268 knockdown impairs tumor growth and NF-κB activation in HeLa cell xenograft nude mouse model. A, sh control or shZNF268 HeLa cells (5 × 106) were subcutaneously injected into nude mice. Tumors formed in recipient mice were collected 30 days post-injection. Tumors from recipient mice engrafted with ZNF268 knockdown cells showed reduced size and weight (n = 5). Right panel was the statistics of xenograft experiment in the left panel. * indicates p < 0.05; ** indicates p < 0.01. B and C, tumors from three randomly selected sh control or shZNF268 recipient mice were subject to Western blot analysis with the antibodies indicated. D, nuclear (N) and cytoplasm (C) extracts were prepared from tumors collected from recipient mice engrafted with sh control or shZNF268 HeLa cells. Subcellular localization of p65 and p50 in tumors were also analyzed by Western blot. PARP and actin were used as internal controls for the quality of nuclear and cytoplasmic proteins fraction. Three tumors were randomly selected from the sh control group and shZNF268 group for the Western blot analysis. Results are representative blots from three independent experiments.
Down-regulation of NF-κB Activity in Tumors from shZNF268 Xenograft Group and Its Implication in Human Cervical Cancer
To further confirm the effect of ZNF268 on NF-κB activation in vivo, we examined NF-κB activity in tumors from the shZNF268 xenograft group. As shown in Fig. 6C, tumors from the shZNF268 xenograft group exhibited less IKK protein phosphorylation and less IκBα degradation than that in the sh control xenograft group. In addition, we observed less p65 and p50 translocation into the nucleus in tumors from the shZNF268 xenograft group than that in the sh control group as measured by Western blot (Fig. 6D). This was confirmed by immunohistochemistry experiments as evidenced by a decrease of IKKα/β phosphorylation staining and reduction of p50 and p65 nuclear translocation (supplemental Fig. S5, C and D). These data suggest that ZNF268 overexpression may promote tumor growth in vivo by activating NF-κB under pathological conditions.
Constitutively active NF-κB has been reported in cervical cancer (19). Considering our findings that ZNF268b2 activates NF-κB signaling in HeLa cells, these two events could pathologically coincide in human cervical cancer. Thus, we measured IκBα expression level and p65 nucleus translocation in 34 paraffin-embedded cervical tissue specimens by immunohistochemistry. As expected, we observed 86.7% cervical carcinoma specimens with a high expression (indicated as +++) level of ZNF268, and the rest showed a medium level (indicated as ++), whereas only half of the normal tissues showed medium expression of ZNF268a and were negative for the rest (indicated as − in Table 1 and supplemental Fig. S6). A high IκBα expression was observed in normal squamous cervical epithelium specimens compared with that in SCC. Twenty five percent of normal squamous cervical epithelium specimens had nuclear p65 staining, whereas 73% of the SCC specimens had nuclear p65 (Table 1 and supplemental Fig. S6). These findings were consistent with our results obtained from the xenograft nude mouse model and strongly supported the idea that ZNF268b2 overexpression and NF-κB activation might pathologically concur in human cervical cancer and contribute to carcinogenesis.
TABLE 1.
Immunohistochemistry analysis of ZNF268, IκBα, and p65 expression in cervical cancer tissues
n indicates the number of samples used for immunohistochemistry staining. −, +, ++, and +++ indicate negative, weak, medium, or strong staining intensity, respectively. Numbers in parentheses of staining intensity represent sample numbers in that category. Percentages in parentheses represent the percentage of samples with positive staining in cytoplasm (C) or nucleus (N). Normal indicates normal squamous cervical epithelium; SCC indicates squamous cervical carcinoma.
| Histology | ZNF268 | IκBα | p65 |
|---|---|---|---|
| Normal (n = 4) | ++ (2) | ++ (3) | C (75%) |
| − (2) | + (1) | N (25%) | |
| SCC (n = 30) | +++ (26) | ++ (6) | C (27%) |
| ++ (4) | + (11) | N (73%) | |
| − (13) |
DISCUSSION
Although ZNF268 was isolated more than a decade ago, little is known about its physiological function. This is due, in part, to the fact that there is no mouse ortholog. ZNF268 contains a KRAB domain and 24 zinc fingers. There is evidence showing that poly-zinc finger proteins tend to increase the number of zinc finger repeats during the evolutionary process (26). Such unique characteristics suggest that ZNF268 evolved to provide a novel function in human development. In fact, our previous studies have implied that ZNF268 may function in human fetal liver development, blood cell development, and hematological diseases. In this study, we present evidence that suggests a role of ZNF268 in cervical cancer development.
One novel point we have revealed in this report is that the differential expression pattern of the ZNF268 isoforms may contribute to cervical cancer development. Alternative splicing of many oncogenes and suppressors has been known to be different in normal and abnormal physiological processes and has been associated with many kinds of human disease (27). Alternative splicing of many cancer-related genes may also be useful biomarkers for diagnoses (28). For example, CD44 (29) and human papillomavirus type-16 E6/E6 (30) are aberrantly spliced during development of cervical cancer. In this study, we have presented clear evidence that ZNF268 may play different functions in cancer or normal cervical cells due to the production of two isoforms of ZNF268 through alternative splicing. We showed lower expression of ZNF268a and higher ZNF268b2 expression in cervical carcinomas compared with normal tissue control. Moreover, down-regulation of ZNF268 in HeLa cells impaired its tumor formation in the xenograft. This suggests that ZNF268a may function in the normal cellular processes of cervical tissues, whereas ZNF268b2 may function as an aberrantly expressed variant that contributes to cervical cancer. These results also suggest that ZNF268b2 may be a new risk factor of cervical caner and may serve as a marker for target therapy or diagnosis.
How ZNF268a and ZNF268b2 may function differently remains unknown. Our preliminary study suggests that different subcellular distribution may attribute to a functional difference, which has been shown to be one possible mechanism in carcinogenesis (31). We recently found that ZNF268a exclusively localized in the nucleus of HeLa cells (data not shown), consistent with our finding that ZNF268a did not interact with IKK and functioned as a transcription repressor in the nucleus (32). However, ZNF268b2 localized both in the cytoplasm and nucleus of HeLa cells (data not shown). In cytoplasm, ZNF268b2 may facilitate activation of the NF-κB pathway by interacting with the IKK complex as described in this study and subsequently promotes cell proliferation or survival. In the nucleus, one can imagine that ZNF268b2 may retain the DNA-binding ability with the entire zinc finger domain. Without the KRAB domain, it may only partially maintain the function of ZNF268a as a weak repressor. Thus, the role of ZNF268b2 is the net effect of both functions. Moderate overexpression of ZNF268b2 in cervical carcinoma may disrupt the balance and bias to enhance the NF-κB signaling pathway in cytoplasm over transcription repression through DNA binding in the nucleus as we observed in cervical carcinoma in this study.
Our study also suggests that NF-κB signaling mediates the effect of ZNF268b2 in human cervical cancer development. Considerable evidence indicates NF-κB is constitutively activated in cervical cancer (19, 33). However, the mechanisms of NF-κB activation in cancer development are not fully understood (34). In our study, aberrant expression ZNF268b2 contributed to NF-κB activation by interacting with the IKK complex and increasing the IKK phosphorylation. TNFα induction further increased the interaction of ZNF268b2 and IKK (Fig. 4). By further supporting NF-κB as the mediator of ZNF268b2, reconstitution of NF-κB activities in shZNF268 HeLa cells normalized cell proliferation (Fig. 5). Moreover, NF-κB target genes were also altered in shZNF268 HeLa cells. For instance, the decreased expression of cyclin D1 in shZNF268 HeLa cells is an important factor preventing G1 to S cell cycle transition. Recent studies have shown that NF-κB binds to the promoter of cyclin D1 gene and is directly involved in cell cycle control (35–37). The anti-apoptosis gene BCL-XL, a target gene of NF-κB (38), was down-regulated in tumors from shZNF268 xenograft group. Together, our results suggest that aberrant expression of ZNF268b2 may deteriorate NF-κB activation that contributes to cervical cancer development.
Finally, our results also present a novel mechanism by which human cancer cells may modulate the seemingly adverse inflammation microenvironment. TNFα is a well known pro-inflammatory cytokine and induces both survival and apoptosis signals by activating NF-κB and caspase cascade, respectively (39). In two elegant mouse experiments, the TNF-signaling pathway was required for induction of skin tumors (40, 41). Moreover, studies also show that NF-κB had a protective role for TNFα-induced apoptosis (15). In our study, TNFα plus CHX induced apoptosis in HeLa cells and down-regulation of ZNF268b2 further deteriorated apoptosis. However, ectopic expression of ZNF268b2 potently promoted IKK complex assembly and increased phosphorylation. Thus ZNF268b2 overexpression may shift the balance of TNFα signaling from apoptosis to survival. Considering the high expression of ZNF268b2 and the implication of TNFα in cervical cancer pathogenesis, we have identified a novel pathway that cervical cancer cells may use to promote survival and proliferation in response to inflammation microenvironment.
To our knowledge, this is the first study to investigate the function of ZNF268 in human solid tumors. Our results demonstrate that ZNF268 was overexpressed in SCC, and knockdown of ZNF268 suppressed HeLa cell growth both in vitro and in vivo xenografts via NF-κB signaling. Thus aberrantly expressed ZNF268 may contribute to human cervical cancer development, and ZNF268 may serve as a novel therapeutic target or diagnostic marker.
Supplementary Material
Acknowledgment
We thank Dr. John Crispino from Northwestern University for critical reading of the manuscript.
This work was supported by National High Technology Research and Development Program of China (863 Program) Grant 2006AA02A306 (to W. L.), National Natural Science Foundation of China Grants 30871245 and 31271511 (to M. G.) and 30700412 and 81070406 (to Z. H.), and Specialized Research Fund for the Doctoral Program of Higher Education of China Grant 200804861004 (to M. G.).

This article contains supplemental Figs. S1–S6.
- KRAB
- Krüppel-associated box
- SCC
- squamous cervical cancer
- WCL
- whole cell lysate
- TRITC
- tetramethylrhodamine isothiocyanate
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PI
- propidium iodide
- QD
- quantum dot
- CHX
- cycloheximide
- IKK
- IκB kinase
- PARP
- poly(ADP-ribose) polymerase
- PCNA
- proliferating cell nuclear antigen
- EdU
- 5-ethynyl-2′-deoxyuridine.
REFERENCES
- 1. Urrutia R. (2003) KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gou D. M., Sun Y., Gao L., Chow L. M., Huang J., Feng Y. D., Jiang D. H., Li W. X. (2001) Cloning and characterization of a novel Krüppel-like zinc finger gene, ZNF268, expressed in early human embryo. Biochim. Biophys. Acta 1518, 306–310 [DOI] [PubMed] [Google Scholar]
- 3. Shao H., Zhu C., Zhao Z., Guo M., Qiu H., Liu H., Wang D., Xue L., Gao L., Sun C., Li W. (2006) KRAB-containing zinc finger gene ZNF268 encodes multiple alternatively spliced isoforms that contain transcription regulatory domains. Int. J. Mol. Med. 18, 457–463 [PubMed] [Google Scholar]
- 4. Chun J. N., Song I. S., Kang D. H., Song H. J., Kim H. I., Seo J., Suh J. W., Lee K. J., Lee K. J., Kim J., Kang S. W. (2008) A splice variant of the C2H2-type zinc finger protein, ZNF268s, regulates NF-κB activation by TNF-α. Mol. Cells 26, 175–180 [PubMed] [Google Scholar]
- 5. Guo M. X., Wang D., Shao H. J., Qiu H. L., Xue L., Zhao Z. Z., Zhu C. G., Shi Y. B., Li W. X. (2006) Transcription of human zinc finger ZNF268 gene requires an intragenic promoter element. J. Biol. Chem. 281, 24623–24636 [DOI] [PubMed] [Google Scholar]
- 6. Sun Y., Shao H., Li Z., Liu J., Gao L., Peng X., Meng Y., Li W. (2004) ZNF268, a novel Kruppel-like zinc finger protein, is implicated in early human liver development. Int. J. Mol. Med. 14, 971–975 [PubMed] [Google Scholar]
- 7. Krackhardt A. M., Witzens M., Harig S., Hodi F. S., Zauls A. J., Chessia M., Barrett P., Gribben J. G. (2002) Identification of tumor-associated antigens in chronic lymphocytic leukemia by SEREX. Blood 100, 2123–2131 [DOI] [PubMed] [Google Scholar]
- 8. Wang D., Guo M. X., Hu H. M., Zhao Z. Z., Qiu H. L., Shao H. J., Zhu C. G., Xue L., Shi Y. B., Li W. X. (2008) Human T-cell leukemia virus type 1 oncoprotein tax represses ZNF268 expression through the cAMP-responsive element-binding protein/activating transcription factor pathway. J. Biol. Chem. 283, 16299–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng Y., Wang W., Ma J., Wang X., Guo M., Li W. (2012) Knockdown of ZNF268, which is transcriptionally down-regulated by GATA-1, promotes proliferation of K562 cells. PLoS ONE 7, e29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 11. Walboomers J. M., Jacobs M. V., Manos M. M., Bosch F. X., Kummer J. A., Shah K. V., Snijders P. J., Peto J., Meijer C. J., Muñoz N. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 [DOI] [PubMed] [Google Scholar]
- 12. Magnusson P. K., Lichtenstein P., Gyllensten U. B. (2000) Heritability of cervical tumours. Int. J. Cancer 88, 698–701 [DOI] [PubMed] [Google Scholar]
- 13. Engelmark M. T., Ivansson E. L., Magnusson J. J., Gustavsson I. M., Beskow A. H., Magnusson P. K., Gyllensten U. B. (2006) Identification of susceptibility loci for cervical carcinoma by genome scan of affected sib-pairs. Hum. Mol. Genet. 15, 3351–3360 [DOI] [PubMed] [Google Scholar]
- 14. Wilting S. M., de Wilde J., Meijer C. J., Berkhof J., Yi Y., van Wieringen W. N., Braakhuis B. J., Meijer G. A., Ylstra B., Snijders P. J., Steenbergen R. D. (2008) Integrated genomic and transcriptional profiling identifies chromosomal loci with altered gene expression in cervical cancer. Genes Chromosomes Cancer 47, 890–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barkett M., Gilmore T. D. (1999) Control of apoptosis by Rel/NF-κB transcription factors. Oncogene 18, 6910–6924 [DOI] [PubMed] [Google Scholar]
- 16. Hayden M. S., Ghosh S. (2004) Signaling to NF-κB. Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 17. Chen G., Goeddel D. V. (2002) TNF-R1 signaling: a beautiful pathway. Science 296, 1634–1635 [DOI] [PubMed] [Google Scholar]
- 18. Karin M. (2006) Nuclear factor-κB in cancer development and progression. Nature 441, 431–436 [DOI] [PubMed] [Google Scholar]
- 19. Nair A., Venkatraman M., Maliekal T. T., Nair B., Karunagaran D. (2003) NF-κB is constitutively activated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the human uterine cervix. Oncogene 22, 50–58 [DOI] [PubMed] [Google Scholar]
- 20. Chen C., Peng J., Xia H., Wu Q., Zeng L., Xu H., Tang H., Zhang Z., Zhu X., Pang D., Li Y. (2010) Quantum-dot-based immunofluorescent imaging of HER2 and ER provides new insights into breast cancer heterogeneity. Nanotechnology 21, 095101 [DOI] [PubMed] [Google Scholar]
- 21. Sherr C. J. (1996) Cancer cell cycles. Science 274, 1672–1677 [DOI] [PubMed] [Google Scholar]
- 22. Tjiong M. Y., van der Vange N., ter Schegget J. S., Burger M. P., ten Kate F. W., Out T. A. (2001) Cytokines in cervicovaginal washing fluid from patients with cervical neoplasia. Cytokine 14, 357–360 [DOI] [PubMed] [Google Scholar]
- 23. Vieira K. B., Goldstein D. J., Villa L. L. (1996) Tumor necrosis factor α interferes with the cell cycle of normal and papillomavirus-immortalized human keratinocytes. Cancer Res. 56, 2452–2457 [PubMed] [Google Scholar]
- 24. Häcker H., Karin M. (2006) Regulation and function of IKK and IKK-related kinases. Sci. STKE 2006, re13. [DOI] [PubMed] [Google Scholar]
- 25. Ran R., Lu A., Zhang L., Tang Y., Zhu H., Xu H., Feng Y., Han C., Zhou G., Rigby A. C., Sharp F. R. (2004) Hsp70 promotes TNF-mediated apoptosis by binding IKKγ and impairing NF-κB survival signaling. Genes Dev. 18, 1466–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Looman C., Abrink M., Mark C., Hellman L. (2002) KRAB zinc finger proteins. An analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol. Biol. Evol. 19, 2118–2130 [DOI] [PubMed] [Google Scholar]
- 27. Faustino N. A., Cooper T. A. (2003) Pre-mRNA splicing and human disease. Genes Dev. 17, 419–437 [DOI] [PubMed] [Google Scholar]
- 28. Brinkman B. M. (2004) Splice variants as cancer biomarkers. Clin. Biochem. 37, 584–594 [DOI] [PubMed] [Google Scholar]
- 29. Woerner S. M., Givehchian M., Dürst M., Schneider A., Costa S., Melsheimer P., Lacroix J., Zöller M., Doeberitz M. K. (1995) Expression of CD44 splice variants in normal, dysplastic, and neoplastic cervical epithelium. Clin. Cancer Res. 1, 1125–1132 [PubMed] [Google Scholar]
- 30. Rosenberger S., De-Castro Arce J., Langbein L., Steenbergen R. D., Rösl F. (2010) Alternative splicing of human papillomavirus type-16 E6/E6* early mRNA is coupled to EGF signaling via Erk1/2 activation. Proc. Natl. Acad. Sci. U.S.A. 107, 7006–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S. C., Hung M. C. (2005) Cytoplasmic/nuclear shuttling and tumor progression. Ann. N.Y. Acad. Sci. 1059, 11–15 [DOI] [PubMed] [Google Scholar]
- 32. Sun Y., Gou D. M., Liu H., Peng X., Li W. X. (2003) The KRAB domain of zinc finger gene ZNF268: a potential transcriptional repressor. IUBMB Life 55, 127–131 [DOI] [PubMed] [Google Scholar]
- 33. Branca M., Giorgi C., Ciotti M., Santini D., Di Bonito L., Costa S., Benedetto A., Bonifacio D., Di Bonito P., Paba P., Accardi L., Mariani L., Ruutu M., Syrjänen S., Favalli C., Syrjänen K. (2006) Up-regulation of nuclear factor-κB (NF-κB) is related to the grade of cervical intraepithelial neoplasia, but is not an independent predictor of high-risk human papillomavirus or disease outcome in cervical cancer. Diagn. Cytopathol. 34, 555–563 [DOI] [PubMed] [Google Scholar]
- 34. Van Waes C. (2007) Nuclear factor-κB in development, prevention, and therapy of cancer. Clin. Cancer Res. 13, 1076–1082 [DOI] [PubMed] [Google Scholar]
- 35. Guttridge D. C., Albanese C., Reuther J. Y., Pestell R. G., Baldwin A. S., Jr. (1999) NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 19, 5785–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baldin V., Lukas J., Marcote M. J., Pagano M., Draetta G. (1993) Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 7, 812–821 [DOI] [PubMed] [Google Scholar]
- 37. Weinberg R. A. (1995) The retinoblastoma protein and cell cycle control. Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 38. Lee H. H., Dadgostar H., Cheng Q., Shu J., Cheng G. (1999) NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 96, 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Philip M., Rowley D. A., Schreiber H. (2004) Inflammation as a tumor promoter in cancer induction. Semin. Cancer Biol. 14, 433–439 [DOI] [PubMed] [Google Scholar]
- 40. Suganuma M., Okabe S., Marino M. W., Sakai A., Sueoka E., Fujiki H. (1999) Essential role of tumor necrosis factor α (TNF-α) in tumor promotion as revealed by TNF-α-deficient mice. Cancer Res. 59, 4516–4518 [PubMed] [Google Scholar]
- 41. Moore R. J., Owens D. M., Stamp G., Arnott C., Burke F., East N., Holdsworth H., Turner L., Rollins B., Pasparakis M., Kollias G., Balkwill F. (1999) Mice deficient in tumor necrosis factor-α are resistant to skin carcinogenesis. Nat. Med. 5, 828–831 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.