Background: Oxidized forms of benzylisoquinoline alkaloids occur in plants.
Results: In vitro and in vivo characterization of flavoprotein oxidases led to the isolation of a novel alkaloid biosynthetic enzyme in opium poppy.
Conclusion: The final conversions in sanguinarine and papaverine biosynthesis are catalyzed by a flavoprotein oxidase.
Significance: We have extended the importance of flavoprotein oxidases in benzylisoquinoline alkaloid metabolism.
Keywords: Enzyme Kinetics, Functional Genomics, Gene Silencing, Metabolism, Plant Biochemistry, Plant Molecular Biology, Plant Physiology, Transcriptomics
Abstract
Benzylisoquinoline alkaloids are a diverse class of plant specialized metabolites that includes the analgesic morphine, the antimicrobials sanguinarine and berberine, and the vasodilator papaverine. The two-electron oxidation of dihydrosanguinarine catalyzed by dihydrobenzophenanthridine oxidase (DBOX) is the final step in sanguinarine biosynthesis. The formation of the fully conjugated ring system in sanguinarine is similar to the four-electron oxidations of (S)-canadine to berberine and (S)-tetrahydropapaverine to papaverine. We report the isolation and functional characterization of an opium poppy (Papaver somniferum) cDNA encoding DBOX, a flavoprotein oxidase with homology to (S)-tetrahydroprotoberberine oxidase and the berberine bridge enzyme. A query of translated opium poppy stem transcriptome databases using berberine bridge enzyme yielded several candidate genes, including an (S)-tetrahydroprotoberberine oxidase-like sequence selected for heterologous expression in Pichia pastoris. The recombinant enzyme preferentially catalyzed the oxidation of dihydrosanguinarine to sanguinarine but also converted (RS)-tetrahydropapaverine to papaverine and several protoberberine alkaloids to oxidized forms, including (RS)-canadine to berberine. The Km values of 201 and 146 μm for dihydrosanguinarine and the protoberberine alkaloid (S)-scoulerine, respectively, suggested high concentrations of these substrates in the plant. Virus-induced gene silencing to reduce DBOX transcript levels resulted in a corresponding reduction in sanguinarine, dihydrosanguinarine, and papaverine accumulation in opium poppy roots in support of DBOX as a multifunctional oxidative enzyme in BIA metabolism.
Introduction
Benzylisoquinoline alkaloids (BIAs)4 are a large and diverse group of ∼2500 plant specialized metabolites, many of which possess potent pharmacological properties (1), including the narcotic analgesics morphine and codeine, the cough suppressant and potential anticancer drug noscapine (2), the antimicrobial agents sanguinarine and berberine, and the vasodilator papaverine (Fig. 1). BIA biosynthesis begins with the condensation of two tyrosine derivatives, dopamine and 4-hydroxyphenylacetaldehyde, yielding 1-benzylisoquinoline (S)-norcoclaurine as the central precursor to BIA biosynthesis (Fig. 1). Internal carbon-carbon coupling of the (S)-norcoclaurine derivative and branch point intermediate (S)-reticuline results in the formation of various BIA structural subgroups, including morphinan (e.g. morphine), protoberberine (e.g. berberine), and benzophenanthridine (e.g. sanguinarine) alkaloids. Further carbon-oxygen coupling and functional group modifications within each branch pathway yield a multitude of structurally related compounds. Several of the enzymes responsible for the formation of novel backbone structures, hydroxylation of aromatic rings, and the modification of functional groups are oxidoreductases, including cytochromes P450, 2-oxoglutarate/Fe2+-dependent dioxygenases, and flavoproteins (1, 3).
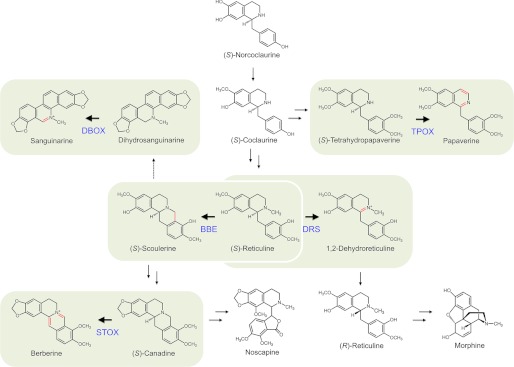
FIGURE 1.
Established and putative reactions catalyzed by flavoprotein oxidases in BIA metabolism. BIA biosynthesis begins with the formation of (S)-norcoclaurine, a precursor to the central intermediate (S)-reticuline. (S)-Reticuline can be oxidized to (S)-scoulerine by BBE, a pivotal step toward the formation of either benzophenanthridine (e.g. sanguinarine) or protoberberine (e.g. berberine) alkaloids. The final steps in these pathways are catalyzed by DBOX and STOX, respectively. Alternatively, (S)-reticuline can be oxidized by DRS, leading to formation of (R)-reticuline and the morphinan alkaloid pathway. (S)-Coclaurine is a key intermediate for papaverine biosynthesis, with tetrahydropapaverine oxidase (TPOX) catalyzing the final step. Corresponding genes have been reported for BBE and STOX.
Berberine bridge enzyme (BBE) is a well characterized flavoprotein oxidase that catalyzes the stereo-specific conversion of the central intermediate (S)-reticuline to (S)-scoulerine (4) (Fig. 1). The formation of (S)-scoulerine is the first step in the biosynthesis of the benzophenanthridine and protoberberine alkaloids sanguinarine and berberine, respectively. (S)-Tetrahydroprotoberberine oxidase (STOX) is another flavoprotein that catalyzes the four-electron oxidation of (S)-canadine to berberine. Isolated STOX from Berberis wilsonae cell cultures accepted a variety of tetrahydroprotoberberine alkaloid substrates, including (S)-scoulerine, (S)-tetrahydrocolumbamine, and (S)-tetrahydropalmatine, along with certain 1-benzylisoquinoline alkaloids, including (S)-norreticuline (5). Recently, BBE from California poppy (Eschscholzia californica) was also shown to oxidize (S)-scoulerine in a manner similar to STOX, highlighting the catalytic similarities between the two enzymes (6).
The conversion of (S)-scoulerine to sanguinarine involves the closure of two methylenedioxy bridges by cytochromes P450 (7) followed by N-methylation (8), yielding the intermediate (S)-cis-N-methylstylopine. Two subsequent cytochrome P450-dependent oxidations coupled with spontaneous intramolecular rearrangement convert the protoberberine (S)-cis-N-methylstylopine through a protopine backbone to the benzophenanthridine dihydrosanguinarine (9). The two-electron oxidation of dihydrosanguinarine to sanguinarine is catalyzed by dihydrobenzophenanthridine oxidase (DBOX) (Fig. 1). Partially purified and characterized DBOX from E. californica cell cultures shares certain features with BBE, such as molecular weight (56000 for DBOX versus 57000 for BBE) and the nature of the catalyzed reaction. However, whether or not E. californica DBOX is a flavoprotein oxidase has not been determined. A distinct dihydrobenzophenanthridine oxidase, designated DHBO to distinguish it from DBOX, was also isolated from bloodroot (Sanguinaria canadensis) cell cultures (10). In contrast to DBOX, DHBO was a copper-dependent protein with a molecular weight of 77000 (11). Corresponding genes for neither DBOX nor DHBO have been isolated.
Both sanguinarine and berberine are quaternary ammonium cations, which also occur at other points in BIA metabolism, including the oxidation of (S)-reticuline to 1,2-dehydroreticuline (Fig. 1). The introduction of a double bond into (S)-reticuline, catalyzed by 1,2-dehydroreticuline synthase (DRS), is proposed to initiate the formation of (R)-reticuline required for morphine biosynthesis due to the stereospecificity of the subsequent cytochrome P450, yielding the promorphinan intermediate salutaridine (12). DRS was purified 5-fold from opium poppy (Papaver somniferum) seedlings and was suggested to require a covalently linked cofactor, such as FAD (13). However, unlike the final oxidations in the formation of sanguinarine, berberine, and papaverine, the conversion of (S)-reticuline to 1,2-dehydroreticuline does introduce an additional aromatic ring system (Fig. 1). The major metabolic route to papaverine does not proceed through (S)-reticuline, but rather involves a series of N-desmethylated intermediates (14–16). In a final step, (S)-tetrahydropapaverine is dehydrogenated to papaverine (Fig. 1). Although a dedicated (S)-tetrahydropapaverine oxidase has not been isolated from a papaverine-producing species, such as opium poppy, STOX from Berberis wilsonae, a plant not known to produce papaverine, was reported to accept (S)-tetrahydropapaverine as a substrate (5).
Flavoprotein oxidases catalyze the enantio- and regio-specific oxidation of a substrate with a concomitant reduction of molecular oxygen, yielding hydrogen peroxide, and are involved in a diverse array of biological processes (17). Many flavoprotein oxidases act on amine substrates and can have either a broad or a highly restricted substrate range (18). Members of the vanillyl-alcohol oxidase flavoprotein family favor the covalent binding of FAD, a feature that increases the redox potential of the cofactor (19). BBE, STOX, and related vanillyl-alcohol oxidase flavoproteins, such as the marijuana (Cannabis sativa) enzymes tetrahydrocannabinolic acid synthase (20) and cannabidiolic acid synthase (21), and the sunflower (Helianthus annua) carbohydrate oxidase (22) are of interest due to their unique bivalent 8α-histidyl-6-S-cysteinyl mode of FAD attachment. Our work describes the isolation and functional characterization of an opium poppy cDNA encoding DBOX, a flavoprotein oxidase catalyzing the terminal oxidations in sanguinarine and papaverine biosynthesis.
EXPERIMENTAL PROCEDURES
Plants and Chemicals
Opium poppy (P. somniferum) plants were cultivated as described previously (16). Plant materials for gene expression and alkaloid analyses were harvested 1 day before anthesis (60–80 days after germination) and stored at −80 °C. (RS)-Norlaudanosoline was purchased from Sigma-Aldrich. (S)-Norreticuline was purchased from Toronto Research Chemicals Inc. Dihydrosanguinarine and dihydrochelerythrine were prepared by NaBH4 reduction of sanguinarine and chelerythrine, respectively (23). (S)-Scoulerine was prepared by the enzymatic oxidation of (S)-reticuline (100 mg) using recombinant opium poppy BBE secreted from Pichia pastoris cells and purified by two rounds of thin layer chromatography. All other chemicals were synthesized or purchased as described previously (3, 8, 16, 24).
Selection of FADOX Candidates and Phylogenic Analysis
Using the translated opium poppy BBE sequence (GenBankTM accession number AAC61839) as a query, local sequence databases for opium poppy stem, root, and cell cultures were searched to identify homologous sequences encoding putative FAD-dependent oxidoreductase (FADOX) candidates. Available resources include individual Roche 454-based pyrosequencing databases for sanguinarine-producing elicitor-treated cell cultures and eight plant chemotypes (25, 26), in addition to Sanger sequencing databases for stem from a high morphine variety (3) and elicitor-treated cell cultures (27). RNA-seq databases for root and stem of the opium poppy variety Bea's Choice were generated using Illumina HiSeq and Genome Analyzer Ilx technologies, respectively. Following RNA extraction (26), poly(A)+ RNA purification, cDNA library preparation, emulsion-based PCR and short-read sequencing were performed by the Genome Québec at the McGill University Innovation Center. Quality control, de novo transcript assembly, annotation, and gene expression analysis were performed at the University of Calgary Visual Genomics Center. Initial quality assessment of Illumina data were conducted based on FastQC statistics (Brabraham Bioinformatics). Cutadapt (28) was employed for adapter/primer trimming, and in-house scripts were written to perform quality score conversion, trimming of reads based on a minimum quality score cut-off of 25, and removal of read pairs where at least one member is shorter than 35 base pairs. Short-read data were assembled using Trinity (29), and contigs were annotated in MAGPIE. Relative expression levels in RNA-seq databases were calculated as fragments per kilobase of exon model per million mapped reads (FPKM). Database queries yielded six unique, full-length FADOX sequences and eight partial sequences. Phylogeny (Fig. 2) and amino acid alignments (supplemental Fig. S1) were performed using ClustalX (30), and phylogenetic data were displayed using TREEVIEW (31).
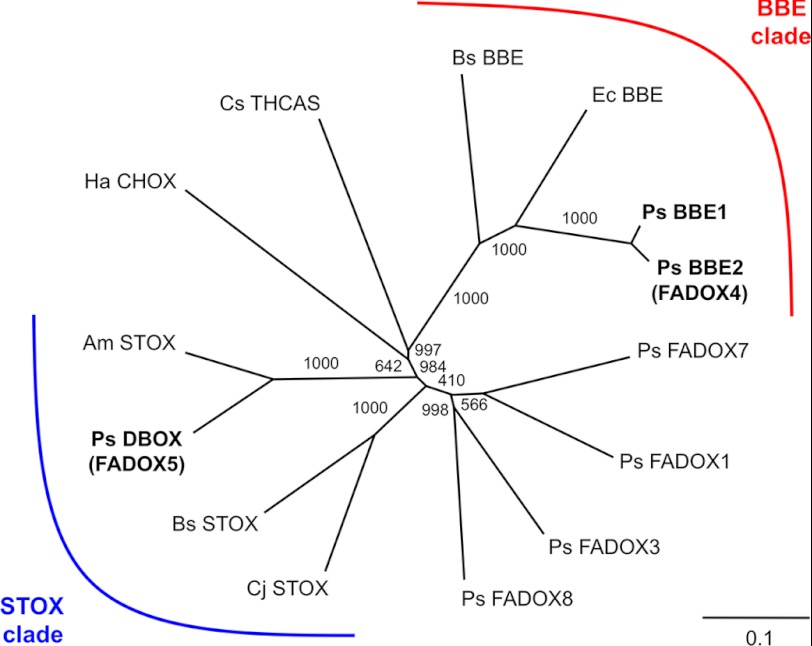
FIGURE 2.
Unrooted neighbor-joining phylogenetic tree for opium poppy FADOXs and related flavoproteins. Bootstrap frequencies for each clade are percentages of 1000 iterations. Species and associated GenBankTM accession numbers for phylogenetic tree construction are as follows: P. somniferum (Ps) BBE1, AAC61839; B. stolonifera (Bs) BBE, AAD17487; E. californica (Ec) BBE, AAC39358; C. sativa (Cs) Δ1-tetrahydrocannabinolic acid synthase (THCAS), AB057805; H. annua (Ha) carbohydrate oxidase (CHOX), AAL77103; A. mexicana (Am) (S)-tetrahydroprotoberberine oxidase (STOX), ADY15027; B. wilsoniae (Bw) STOX, ADY15026; C. japonica (Cj) STOX, BAJ40864.
Expression Vector Construction
The PichiaPink Expression System (Invitrogen) was used for recombinant protein production in P. pastoris. Open reading frames (ORFs) encoding FADOX and BBE1 proteins were amplified using Phusion High-Fidelity DNA polymerase (Thermo Scientific) and sense and antisense primers with flanking StuI and KpnI restriction sites, respectively (supplemental Table S1), from cDNA prepared from opium poppy stem tissues. Sense primers were designed to exclude known (32) or predicted signal peptides targeting the endoplasmic reticulum and to create fusion products with the plasmid-encoded Saccharomyces cerevisiae α-mating factor presequence. Fusion with the α-mating factor presequence permitted the secretion of targeted gene products from P. pastoris cells into the culture medium, circumventing cell lysis prior to enzyme analysis and yielding a fraction highly enriched in recombinant protein. PCR amplicons were inserted between the StuI and KpnI sites of pPinkα-HC. The same approach was used to assemble constructs for the expression of His6-tagged BBE1, BBE2, and FADOX5, except that different primers were used for ORF amplification (supplemental Table S1). For these constructs, a flanking region encoding six histidines followed by a stop codon was added to the reverse primer. Plasmid propagation was performed in Escherichia coli strain XL1-Blue, and expression constructs were transformed into PichiaPink Strain 4, a double knock-out strain for endogenously secreted proteases prb1 and pep4. For recombinant protein expression in S. cerevisiae, primers used to assemble expression construct components are listed in supplemental Table S1. The partial Kozak site AAAACA was introduced upstream of all ORFs, which were cloned into pYES2 (Invitrogen). pYES2 was amplified using primers pYES2 HA Tag F and pYES2 HA Tag R to eliminate a putative partial Kozak site and introduce an HA tag. The DBOX ORF was amplified using primers DBOX F and DBOX HA Tag R, introducing regions of homology to the modified pYES2 vector. pYES2 and DBOX PCR amplicons were transformed into the yeast strain CEN.PK113–13D (MATα ura3-52, MAL2-8C, SUC2) resulting in the assembly of the vector pGC487 (pYES-2μ- ura3PGAL1-FADOX5-HAtag-TCYC1). A further modified pYES2 vector, whereby the URA3 auxotrophic marker was replaced with LEU2, was used for the expression of 6OMT and 4′OMT2. This pYES2 vector was amplified using primers pYES F and pYES R, whereas 6OMT and 4′OMT2 codon-optimized synthetic genes (DNA2.0) were amplified using primers 6OMT F-6OMT R and 4OMT F-4OMT R, respectively. Yeast promoters and terminators were isolated from CEN.PK genomic DNA using primers PMA1 F-PMA1 R for PPMA1, TDH3 F-TDH3 R for PTDH3, CYC1 F-CYC1 R for Tcyc1, and ADH1 F-ADH1 R for TADH1, which added 25-bp linker sequences at their 5′ and 3′ termini. Promoters and terminators with linker sequences were used as templates to amplify components for homologous recombination using primers pYES:C1 F and PMA1 R2 for PPMA1, CYC1 F2 and C6:H1 R for TCYC1, C1:H1 F and TDH3 R2 for PTDH3, and ADH1 F2 and pYES:C6 R for TADH1. The seven PCR products were co-transformed into yeast strain CEN.PK113-16B (MATα leu2-3_112, MAL2–8C, SUC2), resulting in assembly of vector pGC634 (pYES2–2μ-leu2 PPMA1-6OMT-TCYC1-PTDH3-4′OMT2- TADH1). Constructs pGC487 and pGC634 were co-transformed into the haploid CEN.PK113 17A (MATα ura3-52, leu2-3_112, MAL2–8C, SUC2). CEN.PK113 13D (MATα ura3-52, MAL2-8C, SUC2) was transformed separately with pGC487 for DBOX expression or the empty plasmid pYES2 for use as a negative control.
Heterologous Expression in Yeast
For P. pastoris, recombinant proteins were generated according to the manufacturer's instructions (PichiaPink Expression System, Invitrogen). Briefly, freshly transformed cells were cultured in 500 ml of growth medium to an OD between 2 and 6 and then transferred to 100 ml of induction medium containing 0.5% (v/v) methanol. Following a 100-h induction period at 28 °C, the medium fraction containing secreted, recombinant protein was centrifuged to remove cells and applied in batches to Amicon Ultra-15 centrifugal filter units (Millipore) to concentrate the protein (final volume of 1 ml/100 ml of induction medium) and exchange to appropriate buffering conditions (100 mm Tris-HCl, pH 8.8). Desalted, concentrated fractions were stored at 4 °C for no longer than 48 h prior to enzyme analysis. For Saccharomyces cerevisiae, strains were grown overnight in YNB supplemented with synthetic dropout medium lacking uracil, histidine, tryptophan, and leucine but containing 0.2% (w/v) glucose and 1.8% (w/v) galactose. After 24 h, yeast cultures were diluted 20-fold in 10 ml of fresh medium containing 2% (w/v) galactose. After 6 h at 30 °C and 200 rpm, cells were collected by centrifugation at 2000 × g for 5 min and resuspended in 200 μl of medium containing 2 mm norlaudanosoline. Cells were incubated at 30 °C and 200 rpm for 44 h and harvested by centrifugation. Alkaloid content of cell extracts and culture medium was determined using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Immunoblot analysis was performed on SDS-PAGE-fractionated cell lysate protein (50 μg) using anti-HA DyLight antibodies (Rockland). Immunoblots were visualized using a Typhoon imager (GE Healthcare).
Enzyme Assays
To examine substrate range, triplicate enzyme assays were performed in 100 mm Tris-HCl, pH 8.8, using 500 μm alkaloid substrate and desalted, concentrated recombinant enzyme in a total volume of 80 μl. Assays were incubated for 2 h at 37 °C, stopped by adding 1 ml of quenching solution (20% (v/v) 10 mm ammonium acetate, pH 5.5, in ethanol), and stored at −20 °C until analyzed. Immediately prior to LC-MS/MS analysis, assays were centrifuged at room temperature for 5 min to remove insoluble debris. For Km and relative Vmax determinations, identical assay conditions were used with the exception that alkaloid substrate concentrations were varied (supplemental Fig. S2). Saturation curves and kinetic constants were calculated based on Michaelis-Menten kinetics using GraphPad Prism 5 (GraphPad Software). To account for the spontaneous oxidation of the substrates dihydrosanguinarine and dihydrochelerythrine, results from control enzyme assays using culture medium proteins from P. pastoris cells harboring the pPinkα-HC vector were subtracted from corresponding assays performed with recombinant enzyme extracts. Empty vector control data were also subtracted from recombinant enzyme assays using (S)-norreticuline due to the conversion of this substrate by endogenous P. pastoris proteins.
Virus-induced Gene Silencing
Gene-specific silencing constructs were designed for in planta functional analysis of FADOX1, FADOX3, FADOX5, and FADOX8 using unique 3′-UTR and/or 3′-ORF sequences. Due to a high sequence identity (94%) between BBE1 and BBE2 (FADOX4), a single VIGS construct was designed based on the BBE1 sequence, which contained numerous conserved stretches of >24 nucleotides capable of silencing both BBE1 and BBE2 (supplemental Fig. S3). Full-length cDNAs were used as templates to amplify gene-specific fragments (supplemental Fig. S3). Forward and reverse PCR primers were designed with flanking BamHI and XhoI restriction sites, respectively (supplemental Table S1). Amplicons generated using Phusion High-Fidelity DNA polymerase (Thermo Scientific) were ligated into the BamHI and XhoI sites of pTRV2. Constructs in pTRV2 along with pTRV1 were independently mobilized in Agrobacterium tumefaciens strain GV3101 (33). Infiltration of 2-week-old seedlings was performed as described previously (3). Plants were harvested just prior to anthesis and screened by PCR to identify individuals harboring viral coat protein transcripts (supplemental Table S1; supplemental Fig. S4). Infected plants were analyzed by real-time quantitative PCR (RT-qPCR) to determine relative target gene transcript levels. For BBE- and DBOX-silenced plants, roots were ground to a fine powder under liquid N2 and extracted three times with 4 ml of methanol, and pooled extracts were reduced to dryness under reduced pressure. Residue was reconstituted in 1500 μl of methanol and stored at −80 °C until LC-MS/MS analysis. Latex alkaloids of FADOX1, FADOX3, and FADOX8-silenced plants were analyzed by HPLC. Briefly, latex samples were suspended in 30 μl of methanol, and 15 μl was further diluted with 235 μl of methanol, vortexed, and centrifuged to remove insoluble debris. 100 μl of the supernatant was analyzed as described previously (34). Major alkaloids were identified based on retention times and UV spectra compared with those of authentic standards. Statistical analysis of transcript and metabolite data were performed using GraphPad InStat 3.1a (GraphPad Software). For three-way comparisons, Tukey-Kramer multiple comparison tests were used to determine statistical significance at p values of <0.05, <0.01, and <0.001. For two-way comparisons, paired t tests were performed using the same confidence intervals.
LC-MS/MS
Routine enzyme assay analyses were performed using a 6410 Triple Quadropole LC-MS/MS system (Agilent Technologies, Santa Clara, CA). Two microliters of quenched reactions were subjected to liquid chromatography using a Zorbax Eclipse Plus C18 column (2.1 mm × 50 mm, 1.8-μm particle size; Agilent Technologies) at a flow rate of 0.2 ml/min. For protoberberine alkaloids, the column was equilibrated with solvent A (95:5 ratio of 10 mm ammonium acetate, pH 5.5, to acetonitrile), and elution and washing were performed using the following gradient: 0–5 min, 0–30% solvent B (100% acetonitrile); 5–10 min, 30–100% solvent B; 10–15 min, 100% solvent B; 15–15.1 min, 100–0% solvent B; 15.1–16 min, 0% solvent B. For benzophenanthridine alkaloids, gradient conditions were as follows: 0–8 min, 0–100% solvent B; 8–12 min, 100% solvent B; 12–12.1 min, 100–0% solvent B; 12.1–16 min, 0% solvent B. For 1-benzylisoquinoline alkaloids, the flow was adjusted to 0.4 ml/min, and the gradient conditions were as follows: 0–1 min, 0% solvent B; 1–10 min, 0–35% solvent B; 10–11 min, 35–100% solvent B; 11–13 min, 100% solvent B; 13–13.1 min, 100–0% solvent B; 13.1–16 min, 0% solvent B. Injection into the mass analyzer was performed using an electrospray ionization probe inlet. Ions were generated and focused using an electrospray ionization voltage of 4000 V, 10 liter/min gas flow, 50 p.s.i. nebulizing pressure, and gas temperature of 350 °C. MS data acquisition was performed in positive ion mode over 100–700 m/z. Product identification was based on comparisons of retention times and collision-induced dissociation (CID) mass spectra of authentic standards or published data (supplemental Table S2). Collision energies used for CID spectra were either −30 or −35 eV, as noted in supplemental Fig. S5, with the argon collision gas at a pressure of 1.8 × 10−3 torr. For the analysis of VIGS experiments, 10 μl of root extract were subjected to liquid chromatography at a flow rate of 0.5 ml/min with the following gradient: 0–10 min, 0–50% solvent B; 10–12 min, 50–99% solvent B; 12–13 min, 99% solvent B; 13–13.1 min, 99–0% solvent B; 13.1–17.1 min, 0% solvent B. Ion source and MS parameters were identical to those used for enzyme assays except that CID spectra were generated with collision energies listed in supplemental Table S2.
Real-time Quantitative PCR Analysis
For gene expression analysis in different opium poppy organs, tissues (∼0.1 g) from three plants were collected just prior to anthesis, and total RNA was isolated with TRIzol (Invitrogen). Reverse transcription was performed at 42 °C for 60 min using 2.5 mm anchored oligo(dT) primer (dT20VN), 0.5 mm dNTP, 10–40 ng/μl RNA, and 5 microunits/μl reverse transcriptase (Fermentas, Burlington, Canada) after denaturing of the RNA/primer mix at 70 °C for 5 min. Real-time quantitative PCR using SYBR Green detection was performed using a 7300 real-time PCR system (Applied Biosystems). Each 10-μl PCR included 1 μl of cDNA (taken directly from the RT reaction in the case of stem or diluted 50% (v/v) with water for bud, leaf, and root), 300 nm forward and reverse primers (supplemental Table S1), and 1× Power SYBR Green PCR Master Mix (Applied Biosystems). Reactions were subjected to 40 cycles of template denaturation, primer annealing, and primer extension. To evaluate RT-qPCR specificity, the amplicons of all primer pairs were subjected to melt-curve analysis using the dissociation method as suggested by the manufacturer. The 2−ΔΔCt method was used to determine relative gene expression levels (35). The gene encoding ubiquitin was used as the internal control (supplemental Table S1), and the plant line showing the highest expression level served as the calibrator for each target gene. The same method was used to analyze VIGS experiments, except that nine plants per plant line were used to calculate mean transcript abundances, and alkaloid profiles were determined for the same plants.
RESULTS
Isolation of FADOX Gene Candidates
Opium poppy genes encoding FADOXs were identified from stem and elicitor-treated cell culture transcriptome databases queried using opium poppy BBE. 16 non-redundant contigs were identified, including eight full-length or nearly full-length sequences (supplemental Fig. S6). FADOX9 and FADOX10 were partial clones found only in a cell culture database generated by Sanger sequencing (27). FADOX clones 11–16 all consisted of partial sequences represented by few or single reads from individual 454 pyrosequencing databases (25, 26). Except for FADOX2 and FADOX6, which contain stop codons and a deletion in the ORF, respectively, amino acid sequences of other FADOX candidates were aligned to opium poppy BBE1 (supplemental Fig. S1). Opium poppy BBE1 contains an N-terminal ER-targeting peptide (32), and online software predicted signal peptides in all FADOX candidates. Both BBE1 and FADOX4 (BBE2) contain Glu-421 as an equivalent of Glu-417, a key catalytic residue in berberine bridge formation, found in E. californica BBE (4). All other FADOXs were substituted with other residues at this position (supplemental Fig. S1). A phylogenetic tree based on an alignment of protein sequences corresponding to functionally characterized plant flavoproteins shows high bootstrap support for a monophyletic clade containing BBE homologues from E. californica (36), barberry (B. wilsoniae) (37), and opium poppy (38) (Fig. 2). The nearest neighbor to FADOX5 (DBOX) is an enzyme from Mexican prickly poppy (Argemone mexicana), a member of the Papaveraceae along with opium poppy, that was recently shown to exhibit STOX activity (39). An additional clade was formed by partially characterized STOX enzymes from B. wilsoniae and Japanese goldthread (Coptis japonica), which are members of the Berberidaceae and Ranunculaceae, respectively. Monophylogeny between the remaining FADOXs, carbohydrate oxidase, tetrahydrocannabinolic acid synthase and either the BBE or STOX clade was not supported. The relative transcript levels of full-length FADOX candidates were compared in nine 454-pyrosequencing databases for opium poppy cell culture and stems from different chemotypes (supplemental Fig. S7). All poppy chemotypes produce morphine except for T, which accumulates the morphinan pathway intermediate thebaine (3), and Przemko, which is a nearly alkaloid-free variety (34). In addition to morphine, varieties Roxanne and Veronica accumulate papaverine and the phthalideisoquinoline alkaloid noscapine, whereas Marianne, Natasha, and Deborah accumulate only noscapine or the related compound narcotoline. Elicited cell cultures of opium poppy are devoid of morphinan alkaloids but accumulate sanguinarine (27). Only BBE1 and FADOX4 (BBE2) transcripts were relatively abundant in all databases, including cell cultures. In contrast, FADOX5 and FADOX7 transcripts were absent in stem, occurring exclusively in cell cultures. Transcript levels for the remaining FADOXs were sporadic, with FADOX1, for example, represented in all databases except for T and FADOX8 in five of eight chemotypes. Read counts for FADOX3 were also variable and notably absent in cell cultures.
In Vitro Characterization
Flavoproteins containing a covalently linked FAD cofactor require eukaryotic expression systems to generate active proteins. Of the several expression systems previously employed for the production of E. californica BBE, the methylotrophic yeast P. pastoris has proven most successful (4, 6, 40). The secretory pathway of P. pastoris was used to produce opium poppy BBE and FADOX proteins. Replacing the native endoplasmic reticulum secretion signal sequence with S. cerevisiae α-mating factor has been shown to yield higher expression levels of E. californica BBE (6). For this reason, existing or predicted signal peptides were removed from opium poppy homologues prior to expression. Proteins with predicted molecular weight (57200 for BBE1 and BBE2 (FADOX4); 57500 for FADOX5) were routinely observed in concentrated and desalted medium fractions harvested 100 h postinduction (supplemental Fig. S8). These proteins were never observed in empty vector controls, and enzyme activity was only associated with samples containing these proteins. Recombinant FADOX1, FADOX3, and FADOX8 proteins were not detected by SDS-PAGE, and enzyme assays performed on concentrated and desalted medium fractions showed no activity using available substrates. For the purposes of detecting BBE1, BBE2, and FADOX5 by immunoblot analysis and pursuing affinity chromatography-based protein purification, we attempted to generate His6-tagged proteins in P. pastoris. However, recombinant proteins were not detected by either SDS-PAGE or immunoblot analysis, and no enzyme activities were observed.
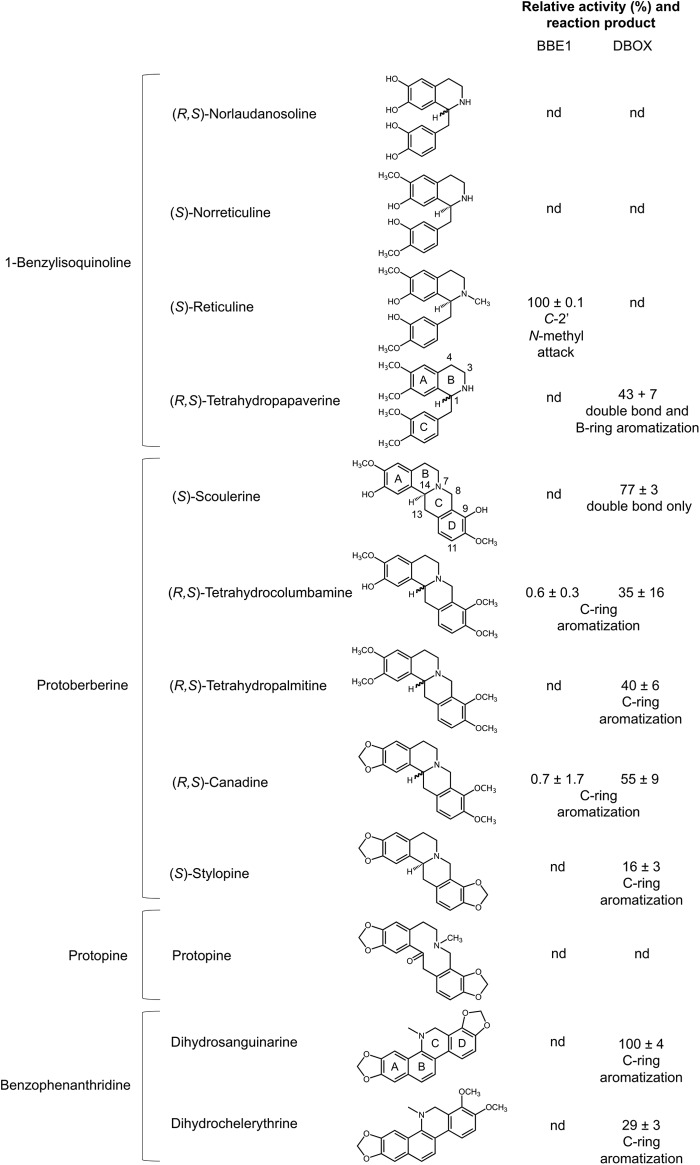
As a positive control for assay conditions, recombinant BBE1 and BBE2 (FADOX4) were tested for enzyme activity using 23 BIAs as potential substrates (Fig. 3 and supplemental Fig. S5). As expected, both BBE1 and BBE2 catalyzed the efficient transformation of the N-methyl group of (S)-reticuline into the C-8 berberine bridge carbon of (S)-scoulerine (supplemental Fig. S9). In addition, columbamine and berberine were produced at low levels from (RS)-tetrahydrocolumbamine and (RS)-canadine, respectively, which represents a four-electron oxidation characteristic of STOX activity. No other alkaloids were accepted as substrates (Fig. 3 and supplemental Fig. S5). Although reported for E. californica BBE (40), the four-electron oxidation of (S)-scoulerine to dehydroscoulerine was not detected with opium poppy BBE1 or BBE2.
FIGURE 3.
Relative conversion of selected substrates by BBE and DBOX (FADOX5) under standard assay conditions. Values represent mean ± S.D. of three independent experiments. The nature of each reaction is indicated below the activity values. The activity value for (RS)-tetrahydropapaverine includes both dihydropapaverine and papaverine products. nd, not detected.
DBOX (FADOX5) was capable of both two- and four-electron oxidations. For example, STOX activity was apparent with most protoberberine substrates, with (RS)-canadine showing the highest turnover rate under standard assay conditions (Figs. 3 and 4). Conversely, DBOX did not fully aromatize the C-ring of (S)-scoulerine but instead catalyzed a two-electron oxidation to yield m/z 326 (Fig. 4). Extracted ion chromatographs revealed two products at m/z 326 with essentially identical CID spectra, which were characterized as dihydroscoulerine isomers (supplemental Fig. S10). The two-electron oxidation of (S)-scoulerine is consistent with a previous characterization of STOX from A. mexicana (39). The formation of both dihydropapaverine and papaverine was observed upon incubation of DBOX with (RS)-tetrahydropapaverine, although other 1-benzylisoquinoline alkaloids, notably (S)-norreticuline, were not accepted as substrates (Fig. 3). CID analysis of dihydropapaverine revealed low abundance, approximately equal intensity ions of m/z 190 and 204, which represent signature fragment masses of 1,2-dihydropapaverine and 3,4-dihydropapaverine, respectively (41). DBOX exhibited the highest relative activity with dihydrosanguinarine, producing the fully conjugated product sanguinarine (Figs. 3 and 4). Dihydrochelerythrine was similarly oxidized with lower efficiency. CID spectra for all products are provided (supplemental Fig. S5). DBOX did not accept any of the tested aporphine, morphinan, phthalideisoquinoline, pavine, or bisbenzylisoquinoline alkaloids (supplemental Fig. S11).
FIGURE 4.

Extracted ion chromatograms showing the substrates and products of DBOX (FADOX5) enzyme assays. In A–H, the tested substrate is indicated in the top left or right corner, the bottom extracted ion chromatogram corresponds to an assay conducted with the empty pPINKα-HC vector control, and the top extracted ion chromatogram shows an assay performed with recombinant DBOX. Parent ion mass-to-charge (m/z) values are indicated beside the substrate and product peaks. The latter were subjected to collision-induced dissociation analysis for identification (supplemental Fig. S5).
Initial characterization of purified, native STOX from B. wilsoniae suggested that (S)-norreticuline was a preferred 1-benzylisoquinoline substrate (5). Assays conducted using opium poppy BBE1, DBOX, and empty expression vector controls all showed the conversion of (S)-norreticuline to two unidentified compounds of m/z 328 that eluted separately during LC-MS/MS analysis. These products were occasionally associated with two unidentified compounds of m/z 326. No turnover of (S)-norreticuline occurred if P. pastoris protein extracts were boiled for 10 min prior to performing assays, suggesting an apparent consumption of (S)-norreticuline by native yeast enzymes. For this reason, DBOX activity with (S)-norreticuline as a substrate was re-evaluated using three different strains of S. cerevisiae (supplemental Fig. S12). In two of these strains, an endogenous supply of (S)-norreticuline was generated by the co-expression of genes encoding (RS)-norcoclaurine 6-O-methyltransferase and (RS)-3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase coupled with the feeding of (RS)-norlaudanosoline (42). One strain also co-expressed a gene encoding DBOX, which presumably could oxidize the endogenous (S)-norreticuline (m/z 316) to expected products of m/z 314 or 312. However, no masses corresponding to (S)-norreticuline reduction products or to byproducts produced by endogenous P. pastoris enzymes (i.e. m/z 328 or 326) were detected (supplemental Fig. S12), supporting the conclusion that (S)-norreticuline is not DBOX substrate.
Km values of 146 ± 9 and 201 ± 67 μm were determined for DBOX using the protoberberine and benzophenanthridine substrates (S)-scoulerine and dihydrosanguinarine, respectively (supplemental Fig. S2). All substrates were assayed using the same enzyme preparations, which enabled the calculation of relative Vmax values of 6.754 and 0.5308 pmol min−1, and relative Vmax Km−1 ratios of 4.63 × 10−8 and 2.64 × 10−9 min−1, for (S)-scoulerine and dihydrosanguinarine, respectively. The insolubility in aqueous solutions at saturating concentrations precluded Km analysis for (RS)-canadine (43). Moreover, the formation of two products, dihydropapaverine and papaverine, precluded a reliable Km determination for (RS)-tetrahydropapaverine due to the possible occurrence of competitive inhibition.
Gene Expression Analysis
Gene expression patterns of DBOX and BBE in different organs of opium poppy were examined using RT-qPCR. Due to the extensive sequence identity between BBE1 and BBE2, RT-qPCR analysis was unable to distinguish individual expression profiles (Fig. 5). However, relative transcript levels were for BBE1 and BBE2 were determined using the FPKM values from Illumina-based stem and root transcriptome databases (supplemental Fig. S13). RT-qPCR results showed the highest DBOX transcript levels in roots compared with other plant organs (Fig. 5). Although direct comparisons of FPKM values are not possible using these databases, DBOX transcripts were not detected in the stem database but were relatively abundant in roots (supplemental Fig. S13). RT-qPCR indicated abundant BBE transcript levels in roots compared with aerial organs (Fig. 5), and FPKM data showed that BBE1 transcripts were restricted to stems, whereas BBE2 mRNAs were only in roots (supplemental Fig. S13).
FIGURE 5.
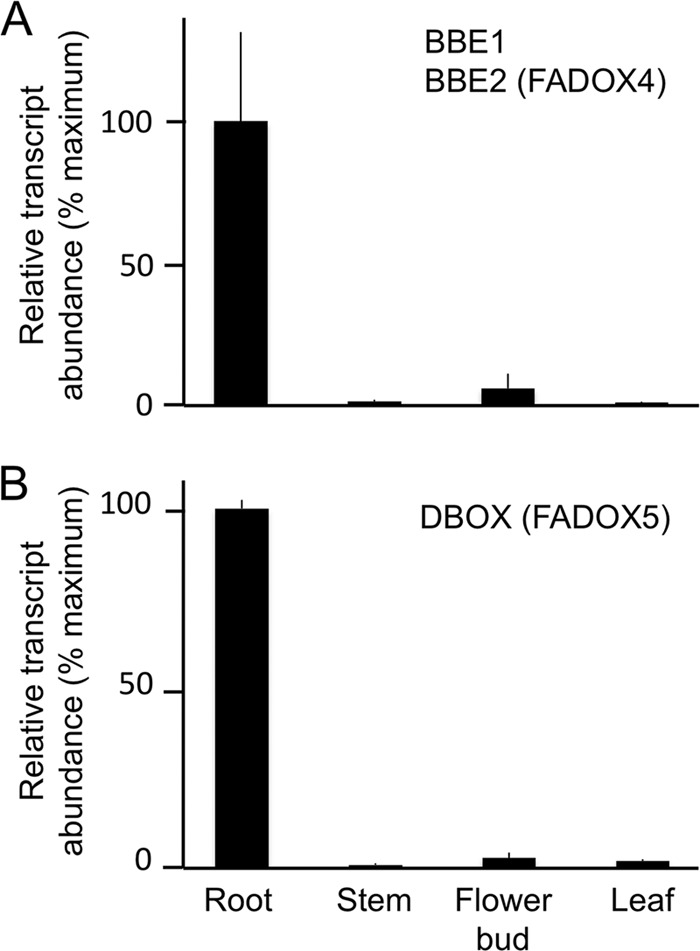
Relative abundance of transcripts encoding BBE (A) and DBOX (B) in opium poppy plant organs. RT-qPCR analysis was performed using cDNA synthesized with RNA isolated from three individual plants. PCR signal normalization was achieved using ubiquitin as the internal control, and the plant organ exhibiting the highest gene expression served as the calibrator for BBE or DBOX. Due to their extensive sequence identity, BBE1 and BBE2 could not be distinguished, and results reflect the cumulative abundance of both transcripts. Error bars, S.D.
VIGS Analysis
The organ-specific occurrence of DBOX transcripts prompted a focus on the metabolic consequences of DBOX gene silencing in roots. Our initial approach attempted to quantify perturbations in the levels of alkaloids representing in vitro substrates or products for DBOX and BBE in silenced versus control plants. However, the low abundance of many compounds, especially protoberberine alkaloids, in opium poppy roots precluded such analysis. Both full ion scanning and multiple reaction monitoring were used, together with CID analysis to confirm compound identities. However, sanguinarine, dihydrosanguinarine, papavarine, and reticuline were readily detected by full ion scanning analysis. Also included in the full ion scanning analyses were thebaine and noscapine, representing end products of the morphinan and pthalideisoquinoline branch pathways, respectively, N-methylcoclaurine, an upstream precursor to (S)-reticuline, and protopine, which serves as a sanguinarine pathway intermediate (supplemental Fig. S14). Sanguinarine and dihydrosanguinarine were significantly reduced in both DBOX- and BBE-silenced plants (Fig. 6). However, spontaneous oxidation of dihydrosanguinarine during the analysis might have contributed to a low dihydrosanguinarine to sanguinarine ratio compared with levels in the plant. In contrast, a reduction in papaverine and noscapine levels was observed only in association with DBOX silencing. Tetrahydropapaverine and dihydropapaverine were not detected in either control or silenced roots. BBE-silenced plants showed an accumulation of N-methylcoclaurine and reticuline compared with empty vector control and DBOX-silenced plants. Protopine and thebaine levels were unaffected by the silencing of either gene. Multiple reaction monitoring analysis yielded data for the relative abundance of the substrate/product pair stylopine/coptisine, which remained unchanged in DBOX-silenced plants (supplemental Fig. S15). The application of VIGS to silence genes encoding FADOX1, FADOX3, and FADOX8 produced no detectable alterations in major latex alkaloid levels compared with empty vector controls (supplemental Fig. S16).
FIGURE 6.
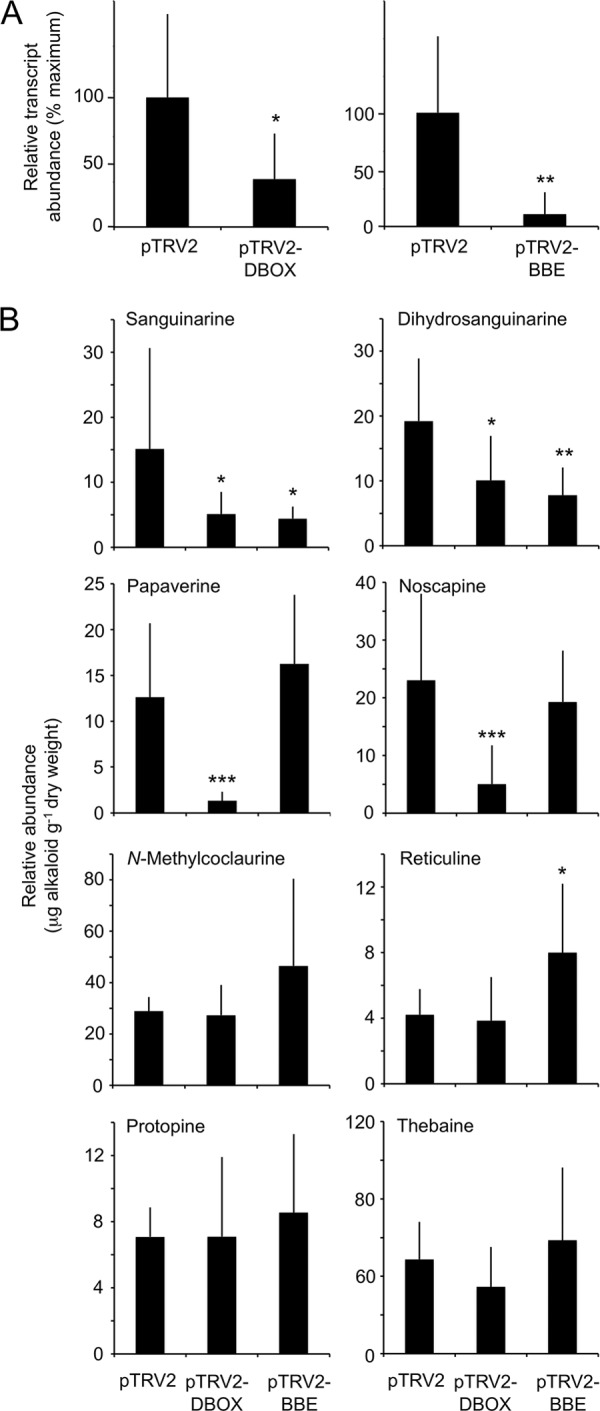
Effect of VIGS on root alkaloid levels. A, relative DBOX and BBE transcript levels in roots as determined by RT-qPCR following VIGS treatment. B, relative abundance of alkaloids in DBOX- or BBE-silenced roots, as determined by LC-MS/MS. Bars, mean ± S.D. (error bars) of nine individual plants, which were used for both gene expression and alkaloid profile analyses. Asterisks denote a statistically significant difference relative to empty vector (pTRV2) at p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***) determined by the Tukey-Kramer multiple comparisons test. pTRV2-BBE targeted both BBE1 and BBE2 transcripts.
DISCUSSION
The isolation and characterization of DBOX as a novel flavoprotein oxidase is a key step toward an understanding of several key reactions within BIA metabolism. Our work demonstrates a functional role for opium poppy FADOX5 as DBOX, an enzyme previously uncharacterized at the genetic level. Results also support a role for FADOX5 in papaverine biosynthesis, although in vitro data (Figs. 3 and 4) combined with the exclusive presence of the transcript in root (Fig. 5), which is the primary site of benzophenanthridine alkaloid accumulation, favored naming the enzyme DBOX. Most genes encoding enzymes in sanguinarine biosynthesis have been isolated (1), with DBOX catalyzing the terminal oxidation (Fig. 1). Opium poppy accumulates relatively large quantities of sanguinarine in roots and elicited cell cultures, a feature shared with other members of the Papaveraceae. Certain opium poppy cultivars also accumulate papaverine in roots and in the latex of stems, leaves, and seed capsules (16). In contrast, (S)-tetrahydroprotoberberine alkaloids are not major alkaloids in opium poppy. In vitro characterization of recombinant DBOX revealed a versatile enzyme capable of accepting a variety of benzophenanthridine, 1-benzylisoquinoline, and (S)-tetrahydroprotoberberine alkaloids (Figs. 3 and 4). Opium poppy DBOX was able to catalyze both two- or four-electron oxidation reactions, depending on the substrate, resulting in the introduction of either one or two double bonds, respectively. The functional versatility of DBOX was corroborated by VIGS analysis, which highlighted changes in the levels of sanguinarine and papaverine in response to DBOX silencing. Surprisingly, the levels of noscapine, which does not appear to be biosynthetically linked to DBOX, were also suppressed (Fig. 6).
Sequence analysis of opium poppy DBOX supported monophylogeny with STOX from A. mexicana (Fig. 2). Opium poppy and A. mexicana are members of the Papaveraceae, which generally accumulate sanguinarine and other benzophenanthridine alkaloids in roots, latex, or elicited cell cultures (44). Previous studies of STOX from A. mexicana and Berberis species did not test benzophenanthridine alkaloids as substrates (5, 39). Opium poppy BBE1 and BBE2 (FADOX4) formed a monophyletic clade with BBE from E. californica (4) and Berberis stolonifera (Fig. 2). No relationship with either the STOX or BBE clades was apparent for other FADOXs, which prompted us to focus on the functional characterization of FADOX5, with BBE1 and BBE2 serving as controls.
Compounds representing nine BIA structural categories were tested, and DBOX accepted certain benzophenanthridine, protoberberine, and 1-benzylisoquinoline alkaloids as substrates (Figs. 3 and 4 and supplemental Fig. S11). Dihydrosanguinarine showed the highest turnover rate compared with other substrates and underwent a two-electron oxidation to complete the C-ring aromatization found in the sanguinarine backbone structure (Fig. 3). DBOX also accepted dihydrochelerythrine at 29% of the turnover rate compared with dihydrosanguinarine, which was in contrast with partially purified DBOX from E. californica that did not convert dihydrochelerythrine (23). Interestingly, purified DHBO from S. canadensis accepted dihydrochelerythrine as a substrate (10). The substrate preference of opium poppy DBOX, combined with the metabolic effects of silencing DBOX in planta, strongly supports the physiological role of the enzyme in sanguinarine biosynthesis. The molecular mass of opium poppy DBOX (58 kDa) (supplemental Fig. S8) was similar to that of E. california DBOX (56 kDa) compared with that of DHBO (77 kDa) (11). E. californica DBOX was suggested as a flavin-dependent enzyme with a covalently linked cofactor based on shared features with STOX from B. wilsoniae, such as inhibition by morin and dicumarol, and the lack of a dissociable cofactor (23, 45). In contrast with opium poppy DBOX, DHBO is not flavinylated and depends instead on copper as a cofactor.
Although the opium poppy and E. californica enzymes are probably sequence orthologues, the Km of opium poppy DBOX for dihydrosanguinarine was substantially higher (201 μm) than that reported for E. californica DBOX (16 μm) and DHBO (6 μm) (supplemental Fig. S2). A high Km for dihydrosanguinarine is not surprising, considering the benzophenanthridine alkaloid levels in the plant. Depending on the opium poppy chemotype, the relative proportion of dihydrosanguinarine and sanguinarine in roots can vary from approximately equal to largely in favor of dihydrosanguinarine. For example, in the Marianne chemotype, 80% of the benzophenanthridine alkaloid content of roots consists of dihydrosanguinarine and 10-hydroxydihydrosanguinarine, whereas sanguinarine and 10-hydroxysanguinarine account for only 20% (46). A high Km value for dihydrosanguinarine would contribute to the accumulation of dihydrosanguinarine.
Opium poppy DBOX yielded fully aromatized alkaloids from most tetrahydroprotoberberine substrates (Fig. 3). Purified STOX from B. wilsoniae showed a strict preference for (S)- over (R)-enantiomers. The available racemic substrates (i.e. (RS)-canadine, (RS)-tetrahydrocolumbamine, and (RS)-tetrahydropalmatine) might have reduced the apparent turnover rate of (S)-enantiomers. (S)-Scoulerine was the preferred protoberberine substrate for opium poppy DBOX, which is in agreement with the reported activity of B. wilsoniae STOX (5). Mechanistic investigation of B. wilsoniae STOX oxidation using (S)-scoulerine showed the formation of a single dihydroprotoberberine with a double bond between N7 and C14. The lack of further oxidation to a fully aromatized dehydroprotoberberine was interpreted as an indication that the initial two-electron oxidation to an iminium ion is enzymatic, whereas a second two-electron oxidation yielding dehydroprotoberberine products, including berberine, columbamine, palmatine, and coptisine (Figs. 3 and 4), occurs spontaneously as the result of iminium ion instability. In contrast, mechanistic studies on E. californica BBE, which exhibits STOX-like activity with (S)-scoulerine, suggested an initial oxidation between N7 and C8 and a four-electron oxidation of (S)-scoulerine to dehydroscoulerine (40). In this case, the second oxidation step did not appear to occur spontaneously but was either enzymatic or the result of iminium ion disproportionation. Opium poppy BBE1 and BBE2 did not accept (S)-scoulerine but did exhibit STOX-like activity with (RS)-tetrahydrocolumbamine and (RS)-canadine, yielding fully aromatized products (Fig. 3). Opium poppy DBOX catalyzed only a two-electron oxidation of (S)-scoulerine, yielding two products with nearly identical CID spectra (supplemental Fig. S10, A–C). We propose that DBOX initially forms a single iminium ion product, which in turn experiences a rearrangement of electrons around the nitrogen to form an isomer (supplemental Fig. S10D). Whether DBOX-catalyzed oxidation occurs between N7 and C14 or between N7 and C8 is not known. Recently, it was shown that feeding (S)-scoulerine or its structural isomer (S)-coreximine to insect cell cultures expressing A. mexicana or B. wilsoniae STOX yielded two-electron oxidation products with a possible double bond at N7 C8 (39). However, evidence supporting the introduction of an N7
C8 (39). However, evidence supporting the introduction of an N7 C8 (versus an N7
C8 (versus an N7 C14) double bond was not provided.
C14) double bond was not provided.
In addition to benzophenanthridine and protoberberine substrates, opium poppy DBOX also accepted the 1-benzylisoquinoline alkaloid (RS)-tetrahydropapaverine (Figs. 3 and 4), yielding two-electron (dihydropapaverine) and four-electron (papaverine) oxidation products. The proposed stepwise mechanism for papaverine formation involves the initial loss of a C3-hydrogen to yield an N2 C3 double bond, followed by deprotonation at C4 to create the enamine 1,2-dihydropapaverine (47). The imine-to-enamine isomerization is followed by the introduction of a double bond at C1
C3 double bond, followed by deprotonation at C4 to create the enamine 1,2-dihydropapaverine (47). The imine-to-enamine isomerization is followed by the introduction of a double bond at C1 N2 to form papaverine. Radioactive tracer experiments with opium poppy seedlings revealed the presence of 1,2-dihydropapaverine upon feeding (RS)-tetrahydropapaverine or (RS)-laudanosine in support of this mechanism (41). Whether DBOX-generated dihydropapaverine represents the 1,2-dihydro or 3,4-dihydro isomer is not known because the purported diagnostic fragment ions following CID analysis (m/z 190 and 204, respectively) (41) were detected at equally low abundance (supplemental Fig. S5A). Tetrahydropapaverine and dihydropapaverine occur naturally in opium poppy at much lower levels than papaverine (16), suggesting that physiological conditions strongly favor papaverine formation. The inability of DBOX to convert (S)-norreticuline (Fig. 3 and supplemental Fig. S12) was unexpected because native STOX was reported to introduce a double bond at C1
N2 to form papaverine. Radioactive tracer experiments with opium poppy seedlings revealed the presence of 1,2-dihydropapaverine upon feeding (RS)-tetrahydropapaverine or (RS)-laudanosine in support of this mechanism (41). Whether DBOX-generated dihydropapaverine represents the 1,2-dihydro or 3,4-dihydro isomer is not known because the purported diagnostic fragment ions following CID analysis (m/z 190 and 204, respectively) (41) were detected at equally low abundance (supplemental Fig. S5A). Tetrahydropapaverine and dihydropapaverine occur naturally in opium poppy at much lower levels than papaverine (16), suggesting that physiological conditions strongly favor papaverine formation. The inability of DBOX to convert (S)-norreticuline (Fig. 3 and supplemental Fig. S12) was unexpected because native STOX was reported to introduce a double bond at C1 N2, yielding 1,2-dehydronorreticuline (5). The acceptance of (RS)-tetrahydropapaverine, but not (S)-norreticuline or (RS)-norlaudanosoline (Fig. 3) suggests a requirement for fully O-methylated 1-benzylisoquinoline substrates.
N2, yielding 1,2-dehydronorreticuline (5). The acceptance of (RS)-tetrahydropapaverine, but not (S)-norreticuline or (RS)-norlaudanosoline (Fig. 3) suggests a requirement for fully O-methylated 1-benzylisoquinoline substrates.
The in vitro capacity of DBOX to convert different BIA structural subgroups suggests that the enzyme participates in more than one biosynthetic pathway. The use of VIGS as a functional genomics tool to investigate the physiological role of BIA biosynthetic genes is well established in opium poppy (3, 16, 48, 49). The restriction of DBOX transcripts to roots (Fig. 5 and supplemental Fig. S13) was in contrast to the occurrence of BBE transcripts, which were most abundant in roots but also present in stems (supplemental Fig. S13). The significant reduction of both sanguinarine and dihydrosanguinarine levels in BBE-silenced opium poppy plants (Fig. 6) was consistent with the action of BBE at an entry point to the benzophenanthridine alkaloid branch pathway (supplemental Fig. S14). The lower sanguinarine content of DBOX-silenced plants was also consistent with the in vitro function of the enzyme (Fig. 6). However, the concomitant reduction of dihydrosanguinarine levels in DBOX-silenced plants suggests additional mechanisms of metabolic control. For example, lower sanguinarine levels or initially higher dihydrosanguinarine levels could trigger regulatory alterations in the activity of other relevant BIA biosynthetic enzymes. Interestingly, levels of upstream pathway intermediates, such as protopine (Fig. 6) and stylopine (supplemental Fig. S15) were unaffected by DBOX silencing.
Papaverine levels were also dramatically reduced in DBOX-silenced plants but were unchanged in BBE-silenced plants (Fig. 6), in support of a role for DBOX in papaverine biosynthesis. Despite a relatively straightforward pathway, many aspects of papaverine biosynthesis remain unresolved (16, 41). The restriction of DBOX transcripts to roots suggests that papaverine biosynthesis does not necessarily occur in the aerial organs in which the compound accumulates. The unaltered levels of stylopine and its aromatized equivalent coptisine suggested that factors besides DBOX are important determinants in regulating metabolic flux toward the formation of protoberberine alkaloids. The spatial separation of DBOX from one or more potential substrates at the cellular or intracellular levels would preclude VIGS-mediated effects on product accumulation. The possibility that DBOX oxidizes substrates beyond the scope of this study cannot be discounted. The suppression of DBOX transcripts correlated with diminished noscapine accumulation, suggesting the possible direct or indirect involvement of DBOX in phthalideisoquinoline alkaloid biosynthesis. However, an off-target shift in BIA metabolism triggered by the knockdown of DBOX transcript is equally plausible.
An initial expectation was that an enzyme exhibiting STOX activity would be responsible for the oxidation of (S)-reticuline to 1,2-dehydroreticuline as the first step toward the generation of (R)-reticuline required for morphinan alkaloid biosynthesis (Fig. 1). Although the nature of the DRS reaction is similar to others catalyzed by DBOX, (S)-reticuline was not accepted in vitro, and silencing DBOX had no apparent effect on reticuline or thebaine levels (supplemental Fig. S14). The lack of alterations in the major alkaloid content levels of FADOX1-, FADOX3-, and FADOX8-silenced poppy plants excluded the possibility that another FADOX enzyme functioned as DRS (supplemental Fig. S16). Although the possibility cannot be ruled out that an additional FADOX homologue could possess DRS activity, it is equally possible that an entirely different enzyme catalyzes this key reaction.
Supplementary Material
Acknowledgments
We thank Ye Zhang and Christoph Sensen (Visual Genomics Centre, University of Calgary) for bioinformatics support.
This work was supported by Genome Canada, Genome Alberta, Genome Québec, the Government of Alberta, and the Canada Foundation for Innovation-Leaders Opportunity Fund.

This article contains supplemental Table 1 and Figs. S1–S16.
- BIA
- benzylisoquinoline alkaloid
- BBE
- berberine bridge enzyme
- CID
- collision-induced dissociation
- DBOX and DHBO
- dihydrobenzophenanthridine oxidase
- DRS
- 1,2-dehydroreticuline synthase
- FADOX
- FAD-dependent oxidoreductases
- STOX
- (S)-tetrahydroprotoberberine oxidase
- VIGS
- virus-induced gene silencing
- FPKM
- fragments per kilobase of exon model per million mapped reads
- contig
- group of overlapping clones
- RT-qPCR
- real-time quantitative PCR.
REFERENCES
- 1. Ziegler J., Facchini P. J. (2008) Alkaloid biosynthesis. Metabolism and trafficking. Annu. Rev. Plant Biol. 59, 735–769 [DOI] [PubMed] [Google Scholar]
- 2. Barken I., Geller J., Rogosnitzky M. (2008) Noscapine inhibits human prostate cancer progression and metastasis in a mouse model. Anticancer Res. 28, 3701–3704 [PubMed] [Google Scholar]
- 3. Hagel J. M., Facchini P. J. (2010) Dioxygenases catalyze the O-demethylation steps of morphine biosynthesis in opium poppy. Nat. Chem. Biol. 6, 273–275 [DOI] [PubMed] [Google Scholar]
- 4. Winkler A., Lyskowski A., Riedl S., Puhl M., Kutchan T. M., Macheroux P., Gruber K. (2008) A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol. 4, 739–741 [DOI] [PubMed] [Google Scholar]
- 5. Amann M., Nagakura N., Zenk M. H. (1988) Purification and properties of (S)-tetrahydroprotoberberine oxidase from suspension-cultured cells of Berberis wilsoniae. Eur. J. Biochem. 175, 17–25 [DOI] [PubMed] [Google Scholar]
- 6. Winkler A., Hartner F., Kutchan T. M., Glieder A., Macheroux P. (2006) Biochemical evidence that berberine bridge enzyme belongs to a novel family of flavoproteins containing a bi-covalently attached FAD cofactor. J. Biol. Chem. 281, 21276–21285 [DOI] [PubMed] [Google Scholar]
- 7. Díaz-Chávez M. L., Rolf M., Gesell A., Kutchan T. M. (2011) Characterization of two methylenedioxy bridge-forming cytochrome P450-dependent enzymes of alkaloid formation in the Mexican prickly poppy Argemone mexicana. Arch. Biochem. Biophys. 507, 186–193 [DOI] [PubMed] [Google Scholar]
- 8. Liscombe D. K., Facchini P. J. (2007) Molecular cloning and characterization of tetrahydroprotoberberine cis-N-methyltransferase, an enzyme involved in alkaloid biosynthesis in opium poppy. J. Biol. Chem. 282, 14741–14751 [DOI] [PubMed] [Google Scholar]
- 9. Takemura T., Ikezawa N., Isawa K., Sato F. (2012) Molecular cloning and characterization of a cytochrome P450 in sanguinarine biosynthesis from Eschscholzia californica cells. Phytochemistry doi: 10.1016/j.phytochem.2012.02.013 [DOI] [PubMed] [Google Scholar]
- 10. Arakawa H., Clark W. G., Psenak M., Coscia C. J. (1992) Purification and characterization of dihydrobenzophenanthridine oxidase from elicited Sanguinaria canadensis cell cultures. Arch. Biochem. Biophys. 299, 1–7 [DOI] [PubMed] [Google Scholar]
- 11. Ignatov A., Neuman M. C., Barg R., Krueger R. J., Coscia C. (1997) Immunoblot analyses of the elicited Sanguinaria canadensis enzyme, dihydrobenzophenanthridine oxidase. Evidence for resolution from a polyphenol oxidase isozyme. Arch. Biochem. Biophys. 347, 208–212 [DOI] [PubMed] [Google Scholar]
- 12. Gesell A., Rolf M., Ziegler J., Díaz Chávez M. L., Huang F. C., Kutchan T. M. (2009) CYP719B1 is salutaridine synthase, the C-C phenol-coupling enzyme of morphine biosynthesis in opium poppy. J. Biol. Chem. 284, 24432–24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hirata K., Poeaknapo C., Schmidt J., Zenk M. H. (2004) 1,2-Dehydroreticuline synthase, the branch point enzyme opening the morphinan biosynthetic pathway. Phytochemistry 65, 1039–1046 [DOI] [PubMed] [Google Scholar]
- 14. Uprety H., Bhakuni D. S., Kapil R. S. (1975) Biosynthesis of papaverine. Phytochemistry 14, 1535–1537 [Google Scholar]
- 15. Brochmann-Hanssen E., Chen C.-H., Chen C. R., Chiang H.-C., Leung A. Y., McMurtrey K. (1975) The biosynthesis of 1-benzylisoquinolines in Papaver somniferum. Preferred and secondary pathways; stereochemical aspects. J. Chem. Soc. Perkin Trans. 1, 1531–1537 [DOI] [PubMed] [Google Scholar]
- 16. Desgagné-Penix I., Facchini P. J. (2012) Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72, 331–344 [DOI] [PubMed] [Google Scholar]
- 17. Joosten V., van Berkel W. J. (2007) Flavoenzymes. Curr. Opin. Chem. Biol. 11, 195–202 [DOI] [PubMed] [Google Scholar]
- 18. Heuts D. P., Scrutton N. S., McIntire W. S., Fraaije M. W. (2009) What's in a covalent bond? On the role and formation of covalently bound flavin cofactors. FEBS J. 276, 3405–3427 [DOI] [PubMed] [Google Scholar]
- 19. Leferink N. G., Heuts D. P., Fraaije M. W., van Berkel W. J. (2008) The growing VAO flavoprotein family. Arch. Biochem. Biophys. 474, 292–301 [DOI] [PubMed] [Google Scholar]
- 20. Sirikantaramas S., Morimoto S., Shoyama Y., Ishikawa Y., Wada Y., Shoyama Y., Taura F. (2004) The gene controlling marijuana psychoactivity. Molecular cloning and heterologous expression of Δ1-tetrahydrocannabinolic acid synthase from Cannabis sativa L. J. Biol. Chem. 279, 39767–39774 [DOI] [PubMed] [Google Scholar]
- 21. Taura F., Sirikantaramas S., Shoyama Y., Yoshikai K., Shoyama Y., Morimoto S. (2007) Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa. FEBS Lett. 581, 2929–2934 [DOI] [PubMed] [Google Scholar]
- 22. Custers J. H.., Harrison S. J., Sela-Buurlage M. B., van Deventer E., Lageweg W., Howe P. W., van der Meijs P. J., Ponstein A. S., Simons B. H., Melchers L. S., Stuiver M. H. (2004) Isolation and characterisation of a class of carbohydrate oxidases from higher plants, with a role in active defence. Plant J. 39, 147–160 [DOI] [PubMed] [Google Scholar]
- 23. Schumacher H.-M., Zenk M. H. (1988) Partial purification and characterization of dihydrobenzophenanthridine oxidase from Eschscholtzia tenuifolia cell suspension cultures. Plant Cell Rep. 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 24. Dang T. T., Facchini P. J. (2012) Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Desgagné-Penix I., Khan M. F., Schriemer D. C., Cram D., Nowak J., Facchini P. J. (2010) Integration of deep transcriptome and proteome analyses reveals the components of alkaloid metabolism in opium poppy cell cultures. BMC Plant Biol. 10, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Desgagné-Penix I., Farrow S. C., Cram D., Nowak J., Facchini P. J. (2012) Integration of deep transcript and targeted metabolite profiles for eight cultivars of opium poppy. Plant Mol. Biol. 79, 295–313 [DOI] [PubMed] [Google Scholar]
- 27. Zulak K. G., Cornish A., Daskalchuk T. E., Deyholos M. K., Goodenowe D. B., Gordon P. M., Klassen D., Pelcher L. E., Sensen C. W., Facchini P. J. (2007) Gene transcript and metabolite profiling of elicitor-induced opium poppy cell cultures reveals the coordinate regulation of primary and secondary metabolism. Planta 225, 1085–1106 [DOI] [PubMed] [Google Scholar]
- 28. Martin J. A., Wang Z. (2011) Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682 [DOI] [PubMed] [Google Scholar]
- 29. Grabherr M. G., Haas B. J., Yassour M., Levin J. Z., Thompson D. A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., di Palma F., Birren B. W., Nusbaum C., Lindblad-Toh K., Friedman N., Regev A. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Page R. D. (1996) TreeView. An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 32. Bird D. A., Facchini P. J. (2001) Berberine bridge enzyme, a key branch-point enzyme in benzylisoquinoline alkaloid biosynthesis, contains a vacuolar sorting determinant. Planta 213, 888–897 [DOI] [PubMed] [Google Scholar]
- 33. Liu Y., Schiff M., Dinesh-Kumar S. P. (2002) Virus-induced gene silencing in tomato. Plant J. 31, 777–786 [DOI] [PubMed] [Google Scholar]
- 34. Hagel J. M., Yeung E. C., Facchini P. J. (2008) Got milk? The secret life of laticifers. Trends Plant Sci. 13, 631–639 [DOI] [PubMed] [Google Scholar]
- 35. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 36. Dittrich H., Kutchan T. M. (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proc. Natl. Acad. Sci. U.S.A. 88, 9969–9973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chou W. M., Kutchan T. M. (1998) Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J. 15, 289–300 [DOI] [PubMed] [Google Scholar]
- 38. Facchini P. J., Penzes C., Johnson A. G., Bull D. (1996) Molecular characterization of berberine bridge enzyme genes from opium poppy. Plant Physiol. 112, 1669–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gesell A., Chávez M. L., Kramell R., Piotrowski M., Macheroux P., Kutchan T. M. (2011) Heterologous expression of two FAD-dependent oxidases with (S)-tetrahydroprotoberberine oxidase activity from Arge mone mexicana and Berberis wilsoniae in insect cells. Planta 233, 1185–1197 [DOI] [PubMed] [Google Scholar]
- 40. Winkler A., Puhl M., Weber H., Kutchan T. M., Gruber K., Macheroux P. (2009) Berberine bridge enzyme catalyzes the six electron oxidation of (S)-reticuline to dehydroscoulerine. Phytochemistry 70, 1092–1097 [DOI] [PubMed] [Google Scholar]
- 41. Han X., Lamshöft M., Grobe N., Ren X., Fist A. J., Kutchan T. M., Spiteller M., Zenk M. H. (2010) The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71, 1305–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hawkins K. M., Smolke C. D. (2008) Production of benzylisoquinoline alkaloids in Saccharomyces cerevisiae. Nat. Chem. Biol. 4, 564–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. O'Neil M. J. (Ed.) (2006) The Merck Index: An Encyclopedia of Chemicals, Drugs, and Biologicals, 14th Ed., Merck, Rahway, NJ [Google Scholar]
- 44. Farrow S. C., Hagel J. M., Facchini P. J. (2012) Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry 77, 79–88 [DOI] [PubMed] [Google Scholar]
- 45. Amann M., Nagakura N., Zenk M. H. (1984) (S)-Tetrahydroprotoberberine oxidase. The final enzyme in protoberberine biosynthesis. Tetrahedron Lett. 25, 953–954 [Google Scholar]
- 46. Frick S., Kramell R., Schmidt J., Fist A. J., Kutchan T. M. (2005) Comparative qualitative and quantitative determination of alkaloids in narcotic and condiment Papaver somniferum cultivars. J. Nat. Prod. 68, 666–673 [DOI] [PubMed] [Google Scholar]
- 47. Battersby A. R., Cheldrake P. W., Staunton J., Summers M. C. (1977) Stereospecificity in the biosynthesis of papaverine. Bioinorg. Chem. 6, 43–47 [Google Scholar]
- 48. Wijekoon C. P., Facchini P. J. (2012) Systematic knockdown of morphine pathway enzymes in opium poppy using virus-induced gene silencing. Plant J. 69, 1052–1063 [DOI] [PubMed] [Google Scholar]
- 49. Winzer T., Gazda V., He Z., Kaminski F., Kern M., Larson T. R., Li Y., Meade F., Teodor R., Vaistij F. E., Walker C., Bowser T. A., Graham I. A. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336, 1704–1708 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.