Background: RPS3 is an essential component that confers transcriptional specificity to NF-κB.
Results: A 21–186 truncation of p65 prevents NF-κB activation-induced nuclear translocation of RPS3 by competing it off full-length p65.
Conclusion: The 21–186 truncation of p65 selectively blocks RPS3-dependent NF-κB gene transcription.
Significance: These findings uncover a new function for the N-terminal domain of p65 and provide a novel strategy for selective inhibition of NF-κB.
Keywords: Gene Regulation, NF-κ B (NF-KB), Protein Domains, Signal Transduction, T Cell Receptor, NF-κB, Ribosomal Protein S3 (RPS3), Regulatory Specificity
Abstract
NF-κB is a pleiotrophic transcription factor that plays a prominent regulatory role in various cellular processes. Although previous efforts have focused on its activation, how NF-κB selects specific target genes in response to discrete signals remains puzzling. In addition to the well defined Rel protein components of NF-κB, the ribosomal protein S3 (RPS3) was identified to be an essential component of specific NF-κB complexes. RPS3 synergistically interacts with the NF-κB p65 subunit to achieve optimal binding and transactivation of a subset of NF-κB target genes, thus providing regulatory specificity. Emerging evidence suggests an important role for the RPS3-p65 interaction in context-specific NF-κB gene transcription. The food-borne pathogen Escherichia coli O157:H7 impacts the transcription of a subset of NF-κB target genes encoding proinflammatory cytokines and chemokines in host cells by preventing the nuclear translocation of RPS3, but not p65. The N terminus of p65 is crucial for RPS3 binding. Although several p65 N-terminal fragments are generated by either protease cleavage or alternative mRNA splicing under certain pathophysiological conditions, the role of these fragments in modulating NF-κB signaling, in particular RPS3-dependent selective gene transcription, has not been fully characterized. Here we report that an N-terminal fragment of p65 (amino acids 21–186) can selectively modulate NF-κB gene transcription by competing for RPS3 binding to p65. This 21–186 fragment preferentially localizes in the cytoplasm where it delays stimuli-induced RPS3 nuclear translocation, without affecting the nuclear translocation of p65. Our findings thus uncover a new cytoplasmic function for the N-terminal domain of p65 and provide a novel strategy for selective inhibition of NF-κB gene transcription.
Introduction
The Nuclear Factor-κ B (NF-κB) signal pathway plays a critical role in a variety of vital cellular procedures (1–5). Despite an extensive study on the signaling that leads to the activation of NF-κB over past years (6–11), it remains puzzling how NF-κB specifically transcribes its target genes (12–15). In addition to the best characterized Rel family proteins (RelA (p65), RelB, c-Rel, p50, and p52), ribosomal protein S3 (RPS3)2 was identified as an essential component in certain native NF-κB complexes (16). RPS3 can complex with p65 in the cytoplasm and in the nucleus; its subcellular localization, however, is actively regulated by the NF-κB activation signaling cascade (16, 17). More specifically, we recently showed that the IκB kinase β (IKKβ)-mediated phosphorylation of RPS3 at serine 209 is required for its nuclear translocation and function during NF-κB activation (17). In the nucleus, RPS3 has been proposed to synergistically interact with p65 achieving optimal DNA binding capacity and conferring the transcriptional selectivity of the NF-κB complex (13, 16). Collectively, RPS3-conferred NF-κB signaling provides a novel clue to NF-κB specific gene transcription.
The significance of RPS3-dependent specific NF-κB gene transcription has been illustrated by an increasing number of key pathophysiological processes. Previous studies demonstrated that preventing the RPS3-p65 interaction via knockdown of RPS3 dramatically attenuated cell receptor engagement-induced cytokine production and proliferation in T cells and immunoglobulin κ light chain gene expression in B cells (16). Moreover, the RPS3-p65 interaction was reported to play an important role in cell receptor editing during B cell development (18) and human islet cell death (19). Furthermore, Snyder and co-workers (20) showed that TNF-induced sulfhydration of p65 at cysteine 38 promoted its interaction with RPS3 and binding to the κB promoters of cIAP-2 (baculoviral IAP repeat-containing protein 3), Bcl-XL (B-cell lymphoma-extra large), and XIAP (X-linked inhibitor of apoptosis protein) genes, which suggests the RPS3-p65 interaction serves as a physiologic determinant of the anti-apoptotic transcriptional activity in macrophages (21). Our recent studies also underscored the key role of RPS3-dependent NF-κB signaling in host defense against intestinal attaching/effecting pathogens including enterohemorrhagic Escherichia coli, enteropathogenic E. coli, and Citrobacter rodentium (17, 22, 23). In particular, the type III secretion system effector protein NleH1 from E. coli O157:H7 specifically blocks the transcription of NF-κB-mediated proinflammatory cytokine genes (e.g., TNF and IL8), by attenuating the IKKβ-mediated phosphorylation of RPS3 at serine 209 and RPS3 nuclear translocation, without affecting NF-κB stimuli-triggered p65 nuclear translocation (17, 22). Therefore diminishing the RPS3-p65 interaction only inhibited the transcription of RPS3-dependent, but not all p65 required, NF-κB target genes. This represents a novel strategy to selectively, rather than globally, inhibit NF-κB activity.
Our previous structure-function study demonstrated that the N-terminal portion, in particular amino acids 21–186, of p65 was necessary for binding to RPS3, whereas the dimerization domain and C-terminal transactivation domain (TAD) were not involved (16). Of note, a host of p65 N-terminal truncated fragments has been reported to be produced through protease cleavage or alternative mRNA splicing in pathogen-infected cells, apoptotic cells, and cancer cells (24–30). For instance, NleC, an anti-inflammatory bacterial zinc metalloprotease from enteropathogenic and enterohemorrhagic E. coli, cleaves host p65 within the N-terminal portion during infection (24–27). Moreover, an N-terminal fragment of p65 was also reported in cells undergoing cytochrome c/caspase-mediated apoptosis (28, 29). These p65 cleavages were proposed to dampen NF-κB-mediated inflammatory response and anti-apoptosis; however, the mechanisms are not clear. In particular, the generated N-terminal truncations of p65 have not been carefully characterized. It remains largely unknown whether they could play an important role in regulating NF-κB signaling, in particular the RPS3-dependent selective transcription of NF-κB target genes.
In this work, we discovered that an N-terminal fragment (amino acids 21–186) of p65, which has high binding affinity to RPS3, specifically impairs RPS3-dependent NF-κB gene transcription through competing RPS3 off endogenous p65. The 21–186 truncated mutant of p65 predominantly resides in the cytoplasm where it selectively retards the stimuli-induced nuclear translocation of RPS3, but not p65. Our results reveal a novel mechanism for the N-terminal 21–186 fragment of p65, which has putative nucleic acid binding function in the nucleus, to selectively modulate the cytoplasmic NF-κB signaling and certain branches of NF-κB target gene transcription.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
Jurkat A3, HEK293T, and mouse embryonic fibroblasts (MEF) cells (ATCC) were cultured in RPMI 1640 and DMEM, respectively, supplemented with 10% fetal calf serum, 2 mm glutamine, and 100 units/ml each of penicillin and streptomycin. IκBα (C-21, sc-371), C-terminal p65 (C-20, sc-372), N-terminal p65 (F-6, sc-8008x), and Sam68 (Src-associated in mitosis 68 kDa; C-20, sc-333) antibodies were from Santa Cruz Biotechnology; GFP (7.1 and 13.1, 11814460001) antibody was from Roche Applied Science; RNA polymerase II (CTD4H8, 05-623) antibody was from Millipore; β-actin (AC-15, A5441) and importin-α (IM-75, I1784) antibodies were from Sigma; poly(ADP-ribose) polymerase (C2–10, 556362), Hsp90 (68, 610418), caspase 3 (19, 610322), CD3 (HIT3a, 550367), and CD28 (CD28.2, 555726) antibodies were from BD; phospho-IκBα (5A5, 9246) and cleaved caspase 3 (Asp-175, 9661) antibodies were from Cell Signaling Technology; RPS3 and Ser-209 phosphorylated RPS3 antibodies were generated and affinity-purified by Primm Biotech, as previously described (16, 17).
Plasmid Constructs
The GFP-Sam68 and GFP-Sam68 (ΔNLS) constructs (31) were kindly provided by S. Richard (McGill University, Montreal, Canada). The GFP, GFP-p65, GFP-p65 (21–186), GFP-p65 (311–551), Ig κB-driven luciferase, AP-1-driven luciferase, and Renilla luciferase plasmids were described previously (16). The GFP-p65 (21–186) (M1A) mutant, with the first methionine encoded by the start codon ATG of EGFP substituted with an alanine to avoid the alternative splicing variant for EGFP, was used in the fluorescence imaging studies. The point mutant of EGFP was generated by site-directed mutagenesis using the QuikChange kit (Stratagene) with primers forward 5′-CCACCGGTCGCCACCGCGGTGAGCAAGGGCGAG-3′ and reverse 5′-CTCGCCCTTGCTCACCGCGGTGGCGACCGGTGG-3′ for M1A and verified by DNA sequencing.
Transient Transfection
DNA constructs were transfected into Jurkat A3 cells with electroporation by the Gene Pulser Xcell electroporation systems (Bio-Rad) using a 0.2-cm cuvette under the following condition: 140 V, 1000 microfarad, and ∞Ω. The plasmids were transiently transfected into HEK293T cells using TurboFect in vitro transfection reagent (Thermo Scientific) according to the manufacturer's instructions.
Luciferase Reporter Gene Assays
Luciferase reporter gene assays were performed as previously described (16). Briefly, the cells were co-transfected with either five copies of Ig κB or AP-1 promoter-driven firefly luciferase constructs and the Renilla luciferase pTKRL plasmid (ratio 10:1), together with appropriate plasmids. The cells were cultured for 1–2 days and then stimulated in triplicate before harvest. The lysates were analyzed using a dual-luciferase kit (Promega).
Immunofluorescence Microscopy
Immunofluorescence microscopy was performed as previously described (16, 17). Briefly, HEK293T cells were seeded on the poly-l-lysine-coated coverslips and transfected with appropriate plasmids. Following stimulation, the cells were fixed with 4% paraformaldehyde in PBS, permeabilized with 0.05% Triton X-100 in PBS, and then stained with Alexa Fluor 594-conjugated rabbit anti-RPS3 antibody (Primm Biotech) or anti-p65 antibody (Santa Cruz) for 40 min together with 1 μg/ml of Hoechst 33342 (Sigma) for 5 min at 25 °C. The coverslips were then rinsed with PBS three times, and covers were mounted for fluorescence microscopy. Immunofluorescence images were obtained with a LSM510 fluorescence confocal microscope (Carl Zeiss) under a ×40 oil immersion lens.
Subcellular Fractionation
Subcellular fractionation was performed as previously described (16, 17). Briefly, the cells were resuspended in chilled Buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 1.5 mm MgCl2, 0.1 mm EDTA, 0.5 mm DTT, 0.4% Nonidet P-40, 0.5 mm PMSF, complete protease inhibitor mixture) at 4 °C for 5 min. The lysates were centrifuged at 4 °C, 500 × g for 3 min, and supernatants were collected as cytosolic fractions. The pellets were incubated in Buffer C (20 mm HEPES, pH 7.9, 420 mm NaCl, 1.5 mm MgCl2, 25% glycerol, 0.5 mm PMSF, 0.2 mm EDTA, 0.5 mm DTT, complete protease inhibitor mixture) at 4 °C for 10 min, followed by a centrifuge at 4 °C, 13,000 × g for 10 min. Supernatants were collected as nuclear fractions.
Immunoprecipitation and Immunoblot
The cells were lysed on ice by 0.4 ml of the TNTG buffer (30 mm Tris, pH 8.0, 150 mm NaCl, 1% Triton X-100, 10% glycerol, complete protease inhibitor mixture) for 30 min. The lysates were centrifuged at 13,000 × g at 4 °C for 10 min to remove insoluble material. After normalizing protein concentrations, the lysates were subjected to immunoprecipitation by adding 10 mg/ml appropriate antibody plus 30 ml of protein G-agarose (Roche Applied Science) and rotated for 2 h at 4 °C. The precipitates were washed five times with cold lysis buffer followed by SDS-PAGE separation under reduced and denaturing conditions. Nitrocellulose membranes were blocked in 5% nonfat milk in 0.1% PBS-Tween 20 (PBS-T) and probed with appropriate antibodies. Immunoblotting of phosphorylated proteins was carried out as described previously (17, 32). Briefly, the gels were transferred to methanol-treated polyvinylidene fluoride membranes, retreated with methanol, and dried for 30 min. The blots were blocked in 5% bovine serum albumin in 0.1% Tris-buffered saline with Tween 20 and probed with specific antibodies. Bands were imaged by the Super Signaling system (Pierce) according to the manufacturer's instructions.
Real Time PCR
Total RNA was isolated, and cDNA was prepared as previously described (16). Real time PCRs were performed in triplicate using the SYBR Green PCR Master Mix (Qiagen) in a 7900HT sequence detection system (Applied Biosystems) according to the manufacturer's protocol, with the following primers: IL2-F, 5′-CCAAGAAGGCCACAGAACTGAAACATC-3′; IL2-R, 5′-GGTTGCTGTCTCATCAGCATATTCACAC-3′; CD25-F, 5′-CCAGAGATCCCACACGCCACATTCAAAG-3′; and CD25-R, 5′-GCATTGACATTGGTTGTCCCAGGACGAG-3′; GAPDH-F, 5′-CACATCAAGAAGGTGGTG-3′; and GAPDH-R, 5′-TGTCATACCAGGAAATGA-3′. The relative transcription level was calculated using the ΔΔCt method (16).
RESULTS
Expression of 21–186 Truncated Mutant of p65 Inhibits Stimuli-induced NF-κB Activation
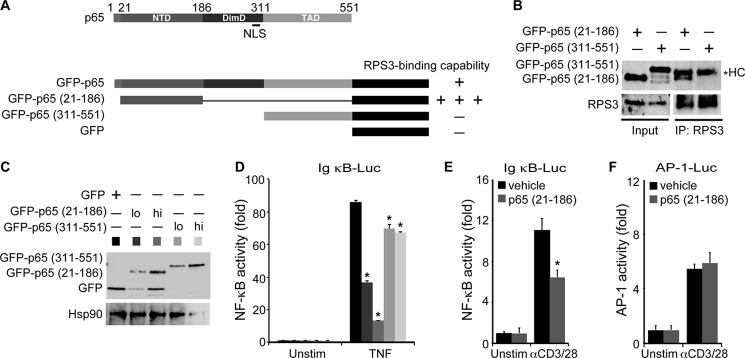
We recently discovered that the E. coli O157:H7 type III secretion system effector NleH1 selectively attenuates host NF-κB gene transcription through its specific interaction with RPS3 (17, 22). A recent structure-function study of the p65-RPS3 interaction showed that the N-terminal portion of p65 (amino acids 21–186) is necessary for binding to RPS3, whereas the dimerization domain and TAD domain are not involved (16). The N-terminal portion of p65 seemingly possesses an even stronger binding affinity to RPS3, relative to the full-length p65 (16) (Fig. 1A), leading us to hypothesize that this truncated mutant of p65 could potentially interfere with NF-κB signaling through this strong interaction with RPS3. We verified that the p65 (21–186) fragment bound to endogenous RPS3 when transfected into HEK293T cells (Fig. 1B). In contrast, the C-terminal TAD (amino acids 311–551) of p65 did not associate with RPS3 (Fig. 1, A and B) (16). We then determined the effect of the p65 (21–186) truncated mutant on NF-κB activation by co-transfecting either the 21–186 or 311–551 truncated mutant of p65 together with an NF-κB luciferase reporter construct into HEK293T cells. Compared with the vehicle EGFP-transfected cells, overexpressing the p65 (21–186) fragment dramatically decreased 60–85% of TNF-induced NF-κB activation (Fig. 1, C and D). Conversely, overexpression of the p65 (311–551) fragment only reduced the TNF-induced NF-κB activation by roughly 20%, although they were statistically significant (Fig. 1D). To explore the potential effect of the 21–186 truncated p65 on the other stimuli-induced NF-κB signaling, we co-transfected either vehicle GFP control or GFP-tagged p65 (21–186) fragment into Jurkat A3 cells. The expression of the p65 (21–186) fragment significantly inhibited anti-CD3/CD28-triggered NF-κB reporter gene expression (Fig. 1E), which suggests that the 21–186 truncated mutant of p65 negatively impacts NF-κB function for multiple stimuli. Moreover, T cell receptor engagement is well known to activate multiple signal pathways including NF-κB, AP-1, and others (33), which provided the stage to assess the impact of p65 (21–186) on the other transcription factor-mediated signal pathways. Indeed, we observed that the anti-CD3/CD28-induced AP-1 reporter expression in the p65 (21–186) fragment-expressing cells was identical to that in the GFP-expressing cells (Fig. 1F), suggesting that the inhibitory function of the 21–186 truncated mutant of p65 appears to be more specific for NF-κB rather than the other signal pathways.
FIGURE 1.
p65 (21–186) fragment inhibits NF-κB gene expression. A, diagram of human p65 and the EGFP-fused full-length and indicated truncation mutants of p65. NTD, N-terminal domain; DimD, dimerization domain. The association capacities of the full-length and indicated truncated mutants of p65 to RPS3 are indicated as follows: −, no detectable interaction; +, detectable interaction; and +++, detectable strong interaction. B, whole cell lysates (Input) from HEK293T cells, transfected with either of the indicated EGFP-fused p65 truncated mutants, were immunoblotted directly or after immunoprecipitation (IP) with RPS3 antibody for the indicated proteins. The asterisk indicates nonspecific heavy chain (HC). C, HEK293T cells were transiently transfected with low dose (lo) or high dose (hi) of the 21–186 or 311–551 truncated mutants of p65 or EGFP vehicle control. After 24 h, the whole cell lysates were derived and immunoblotted directly with GFP antibody for the indicated EGFP fusion proteins. Hsp90 was used as a loading control. D, HEK293T cells were transfected as in C together with 5× κB-Luc reporter and pTKRL plasmids. After 24 h, the cells were left untreated (Unstim) or stimulated with TNF (50 ng/ml) and analyzed for luciferase. Shown are the means ± S.D. of relative NF-κB-driven luciferase activity (fold increase) (n = 3). *, p < 0.001 (Student's t test). E and F, Jurkat A3 cells were transiently transfected with EGFP vehicle control or p65 (21–186) truncated mutant, together with 5× κB-Luc reporter (E) or AP-1-Luc reporter (F) and pTKRL plasmids. After 48 h, the cells were left untreated (Unstim) or stimulated with CD3 and CD28 antibodies (1 μg/ml each, αCD3/28) and analyzed for luciferase. Shown are the means ± S.D. of relative NF-κB-driven (E) or AP-1-driven (F) luciferase activity (fold increase) (n = 3). *, p < 0.001 (E) and not significant (F) by Student's t test.
The 21–186 Truncated Mutant of p65 Localizes to the Cytoplasm
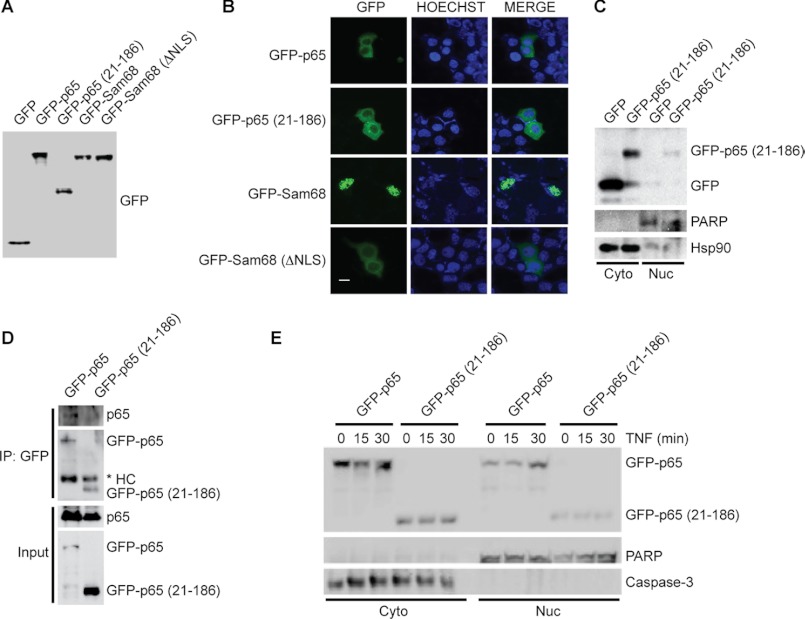
It was previously shown that RPS3 associates with p65 in the inhibitory p65-p50-IκBα complex in the cytoplasm of resting cells, and upon NF-κB activation, RPS3 translocates to the nucleus, where it facilitates NF-κB binding to relevant DNA targets via a synergistic interaction with p65 (12, 13, 16). To identify the step at which the 21–186 truncated mutant of p65 disturbs NF-κB signaling, we examined the subcellular localization of EGFP-tagged p65 (21–186) fusion protein in both resting and stimulated cells. We used Sam68, a known strictly nuclear protein, and its mutant ΔNLS that localizes predominantly in the cytoplasm because of the loss of its NLS (31), to differentiate cytoplasmic and nuclear subcellular localization. We first verified that the fluorescence detected in transfected cells originated entirely from the EGFP-fused proteins rather than any trace of EGFP potentially generated via alternative splicing, because only a single EGFP band with expected molecular weight was detected for each EGFP-fused protein (Fig. 2A). In line with the previous report, EGFP-Sam68 preferentially resided in the nucleus, whereas the Sam68 (ΔNLS) mutant localized exclusively in the cytoplasm (Fig. 2B). We found that the majority of the transfected p65 (21–186) truncated mutant was present in the cytoplasm, similarly to EGFP-p65 and the Sam68 (ΔNLS) mutant but distinct from EGFP-Sam68 (Fig. 2B). The preferential localization of the p65 (21–186) truncated mutant to the cytoplasm in resting HEK293T cells is consistent with the notion that the 21–186 truncation does not possess a functional NLS (Fig. 1A). We further confirmed this result biochemically through subcellular fractionation and detected significantly more p65 (21–186) truncated protein in the cytoplasmic rather than the nuclear fraction derived from transfected HEK293T cells (Fig. 2C). These results demonstrate that the 21–186 truncated mutant of p65 preferentially localizes in the cytoplasm in resting cells.
FIGURE 2.
p65 (21–186) truncated mutant localizes in the cytoplasm. A, whole cell lysates from HEK293T cells, transiently transfected with either EGFP or indicated EGFP fusion proteins, were immunoblotted directly with GFP antibody. B, immunofluorescence micrographs of HEK293T cells transfected as in A. The nuclei were counterstained with Hoechst 33342. The scale bar equals 10 μm. C, immunoblot analysis of cytosolic (Cyto) and nuclear (Nuc) subcellular fractions of HEK293T cells overexpressing EGFP or EGFP-tagged p65 (21–186). Hsp90 and poly(ADP-ribose) polymerase (PARP) serve as cytosolic and nuclear markers, respectively, and/or loading controls throughout. D, whole cell lysates (Input) from HEK293T cells, transfected with either full-length or 21–186 truncated mutant p65 fused to EGFP, were immunoblotted directly or after IP with GFP antibody for GFP fusion proteins and endogenous p65. The asterisk indicates nonspecific heavy chain (HC). E, immunoblot analysis of cytosolic (Cyto) and nuclear (Nuc) subcellular fractions of HEK293T cells overexpressing either EGFP-fused full-length or 21–186 truncated mutant p65 and stimulated with TNF (50 ng/ml) for the indicated periods. Caspase-3 and poly(ADP-ribose) polymerase serve as cytosolic and nuclear markers, respectively, and/or loading controls throughout. The results are representative of two to four experiments.
We next tested whether endogenous p65 could complex with ectopically expressed full-length or truncated p65 to rule out the possibility that such an interaction would account for our findings. As expected, the EGFP-tagged full-length p65 associated with endogenous p65 (Fig. 2D). In contrast, and as could be predicted because of the lack a dimerization domain in the p65 (21–186) fragment (Fig. 1A), there was virtually no detectable interaction between the 21–186 truncated mutant of p65 and endogenous p65 (Fig. 2D). Therefore, it is unlikely that the 21–186 truncated mutant of p65 complexes with endogenous p65 and thereby dominantly interferes with its function.
We then examined whether the p65 (21–186) fragment translocates from the cytoplasm to the nucleus upon NF-κB activation. As a positive control, a significant portion of transfected EGFP-tagged full-length p65 accumulated in the nucleus 30 min after TNF stimulation (Fig. 2E). However, TNF stimulation failed to trigger the nuclear translocation of the overexpressed 21–186 truncated mutant of p65 (Fig. 2E). Collectively, these results suggest that the p65 (21–186) fragment remains in the cytoplasm in both resting and activated cells rather than impairing the functions of endogenous p65 and RPS3 in the nucleus.
The 21–186 Truncated Mutant of p65 Does Not Impair Stimuli-induced p65 Nuclear Translocation
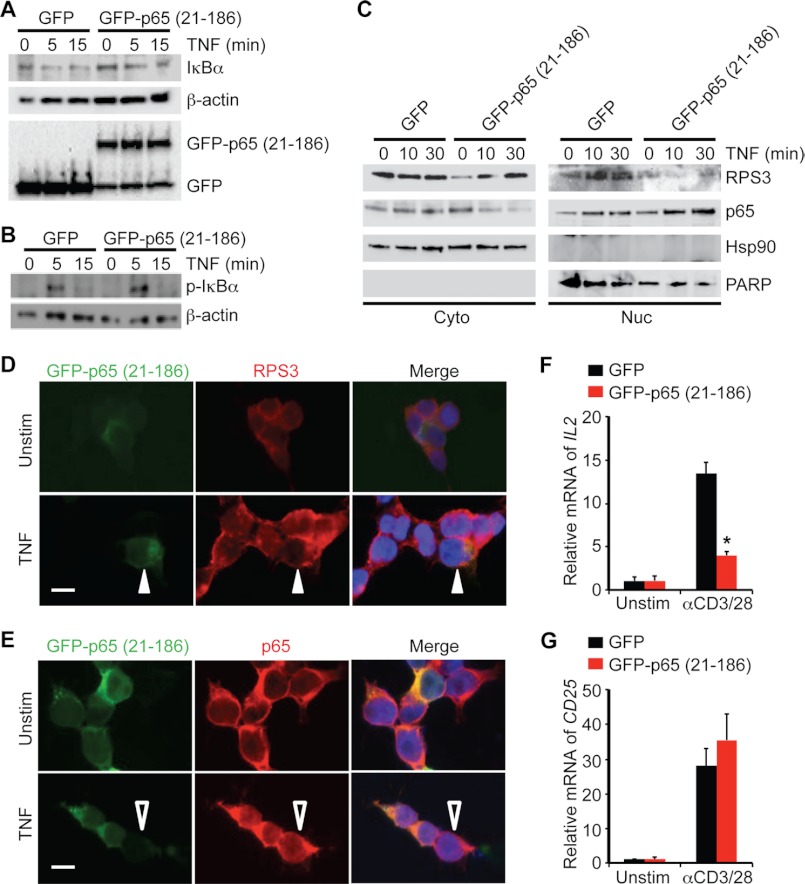
To determine how the 21–186 truncated mutant of p65 inhibits NF-κB gene expression, we examined the TNF-induced cytoplasmic NF-κB signaling in HEK293T cells transfected with either the p65 (21–186) truncated mutant or the vehicle EGFP. In EGFP-expressing cells, as expected, TNF stimulation induced serine phosphorylation of IκBα that peaked at 5 min and degradation of IκBα (Fig. 3, A and B). Overexpression of the 21–186 truncated mutant of p65 did not significantly impair TNF-induced IκBα phosphorylation or degradation (Fig. 3, A and B). These results suggest that the 21–186 truncated mutant of p65 does not globally attenuate signaling events upstream from NF-κB activation.
FIGURE 3.
p65 (21–186) fragment selectively attenuates RPS3 nuclear translocation and gene transcription, without affecting p65 nuclear accumulation. A and B, HEK293T cells were transiently transfected with either EGFP vehicle control or EGFP-tagged p65 (21–186) truncated mutant, followed by stimulation with TNF (50 ng/ml) for the indicated period. Whole cell lysates were derived, separated, and immunoblotted directly for IκBα (A) or serine 32 and serine 36 phosphorylated IκBα (p-IκBα) (B) with β-actin serving as a loading control. C, immunoblot analysis of cytosolic (Cyto) and nuclear (Nuc) subcellular fractions of HEK293T cells that were transfected with either EGFP or EGFP-tagged p65 (21–186) mutant and stimulated with TNF (50 ng/ml) for indicated period. Hsp90 and poly(ADP-ribose) polymerase (PARP) served as cytosolic and nuclear markers, respectively, and/or loading controls. D, HEK293T cells, transfected with EGFP-tagged p65 (21–186) truncated mutant, were left untreated (Unstim) or stimulated with TNF for 30 min. Shown are micrographs of transfected EGFP-fused p65 (21–186), endogenous RPS3, and Hoechst 33342-counterstained nuclei. TNF-triggered RPS3 nuclear translocation was attenuated in the p65 (21–186) fragment transfected cells (indicated by filled triangles), compared with adjacent nontransfected cells. E, HEK293T cells were treated as in D. Shown are confocal micrographs of transfected EGFP-tagged p65 (21–186), endogenous p65, and Hoechst 33342-counterstained nuclei. TNF-triggered p65 nuclear translocation was identical in the p65 (21–186) truncated mutant transfected cells, compared with adjacent nontransfected cells (indicated by open triangles). F and G, Jurkat cells transiently transfected with either EGFP vehicle or EGFP-tagged p65 (21–186) truncated mutant were left untreated (Unstim) or stimulated with CD3 and CD28 antibodies (1 μg/ml each, αCD3/28) for 6 h (F) and 3 h (G), respectively. RT-PCR analysis of mRNA for IL2 (F) and CD25 (G) was normalized to expression of GAPDH and presented relative to the expression in untreated cells. *, p < 0.001 (F) and not significant (G) by Student's t test.
Because the translocation of p65 from the cytoplasm to the nucleus is essential for activation of NF-κB, we next used subcellular fractionation to assess the impact of the p65 (21–186) fragment expression on NF-κB activation-induced nuclear accumulation of endogenous p65. In the EGFP-expressing control cells, TNF triggered substantial nuclear translocation of endogenous p65 as expected (Fig. 3C). Similarly, the TNF-induced p65 nuclear translocation in the 21–186 truncated mutant of p65-transfected cells appeared identical, if not increased, to that in control cells (Fig. 3C). Consistent with our observation that ectopic expression of the p65 (21–186) fragment did not attenuate the phosphorylation and degradation of IκBα (Fig. 3, A and B), these results demonstrate that the 21–186 truncated mutant of p65 does not impair stimuli-induced p65 nuclear translocation.
To verify our biochemical result, we then applied immunofluorescence microscopy to visualize the subcellular localization of endogenous p65 protein in HEK293T cells expressing the 21–186 truncated mutant of p65. Consistent with previous studies (16, 17), the endogenous p65 resided mainly in the cytoplasm in the resting HEK293T cells, independently of the presence of the p65 (21–186) fragment (Fig. 3E). After TNF stimulation, the endogenous p65 translocated to the nucleus to an identical level in both p65 (21–186)-transfected and nontransfected cells (Fig. 3E). This confirms that the 21–186 truncated mutant of p65 does not affect the p65 nuclear accumulation during NF-κB activation.
The 21–186 Truncated Mutant of p65 Selectively Attenuates RPS3 Nuclear Translocation
The stimuli-induced nuclear translocation of RPS3 has been shown to be critical for the optimal transcription of specific RPS3-dependent NF-κB target genes important for inflammatory response and cell survival (17, 20, 22). We therefore assessed the impact of the p65 (21–186) fragment expression on the NF-κB-activation-induced nuclear accumulation of endogenous RPS3 by both subcellular fractionation and immunofluorescence microscopy. In EGFP-expressing control cells, TNF stimulation drove a significant portion of RPS3 to translocate from the cytoplasm to the nucleus, whereas the TNF-induced RPS3 nuclear accumulation was markedly diminished in cells expressing the 21–186 truncated mutant of p65 (Fig. 3C). As reported previously (16, 22), in unstimulated HEK293T cells endogenous RPS3 largely resided in the cytoplasm. This localization was not affected in cells transfected with or without the p65 (21–186) fragment (Fig. 3D). After TNF stimulation, a large fraction of RPS3 migrated into the nucleus in those cells that were not transfected by the 21–186 truncated mutant of p65. Conversely, RPS3 remained in the cytoplasm in those p65 (21–186) fragment-transfected cells (Fig. 3D). These results demonstrate that in contrast to not affecting the nuclear translocation of p65, the expression of the 21–186 truncated mutant of p65 selectively attenuates the nuclear translocation of RPS3 during NF-κB activation.
The 21–186 Truncated Mutant of p65 Specifically Blocks RPS3-dependent NF-κB Gene Transcription in T Cell Activation
We previously showed that during receptor engagement-induced T cell activation, the optimal induction of a subset of NF-κB target genes (such as IL2, encoding interleukin 2) required RPS3, whereas the transcription of other NF-κB target genes (such as CD25, encoding Interleukin 2 receptor α chain) did not (16). We therefore transfected either vehicle EGFP or the EGFP-tagged p65 (21–186) fragment into Jurkat cells and analyzed the effect of the 21–186 truncated mutant of p65 on receptor ligation-induced transcription of IL2 and CD25 genes. As expected, stimulation with CD3 and CD28 antibodies robustly up-regulated both IL2 and CD25 genes in EGFP-transfected cells (Fig. 3, F and G). Ectopic expression of the p65 (21–186) fragment, however, significantly attenuated T cell receptor engagement-induced IL2 gene transcription, compared with the vehicle control (Fig. 3F). In contrast, the receptor engagement-induced CD25 gene transcription was identical, if not higher, in the cells expressing the 21–186 truncated mutant of p65 (Fig. 3F). Thus, consistent with the findings that the 21–186 truncated mutant of p65 specifically attenuated RPS3 nuclear translocation without affecting p65 nuclear accumulation (Fig. 3, C–E), these findings establish that expression of the 21–186 truncated mutant of p65 selectively inhibits RPS3-dependent gene transcription during T cell receptor ligation-mediated NF-κB activation.
The 21–186 Truncated Mutant of p65 Specifically Blocks the NF-κB Signaling Required for RPS3 Nuclear Translocation
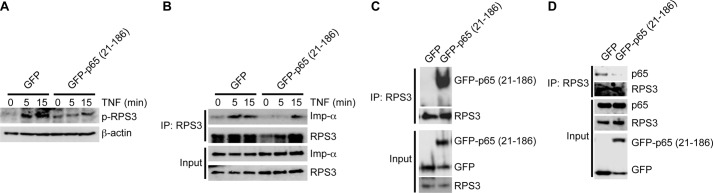
In a recent study, we showed that the IKKβ-mediated phosphorylation of RPS3 at Ser-209 facilitated its association with importin-α in the karyopherin pathway, a key event for the nuclear translocation of RPS3 during NF-κB activation (17). Given that expression of the 21–186 truncated mutant of p65 attenuated TNF-triggered RPS3 nuclear translocation (Fig. 3, C and D), we examined Ser-209 phosphorylation in cells expressing the 21–186 truncated mutant of p65 or an EGFP control. Relative to EGFP-transfected cells, phosphorylation of RPS3 was dramatically diminished in cells expressing the 21–186 truncated mutant of p65 (Fig. 4A). In support of this, we observed an enhanced association of RPS3 to importin-α upon NF-κB activation in control cells, whereas their interaction was significantly impaired in the presence of the 21–186 truncated mutant of p65 (Fig. 4B). Together our results show that the 21–186 truncated mutant of p65 specifically and selectively attenuates RPS3-related cytoplasmic NF-κB signaling during NF-κB activation.
FIGURE 4.
p65 (21–186) fragment inhibits the NF-κB signaling required for RPS3 nuclear translocation via competing RPS3 off p65. A, HEK293T cells were transiently transfected with either EGFP vehicle control or EGFP-tagged p65 (21–186) truncated mutant, followed by stimulation with TNF (50 ng/ml) for indicated period. Whole cell lysates were derived, separated, and immunoblotted directly for serine 209 phosphorylated RPS3 (p-RPS3), with β-actin as a loading control. B, whole cell lysates (Input) from HEK293T cells, transiently transfected with either EGFP or EGFP-tagged p65 (21–186) truncated mutant and stimulated as indicated, were immunoblotted directly or after IP with RPS3 antibody for RPS3 and importin-α (Imp-α). C and D, whole cell lysates (Input) from HEK293T cells, transiently transfected with either EGFP vehicle control or EGFP-tagged p65 (21–186) truncated mutant, were immunoblotted directly or after IP with RPS3 antibody for GFP fusion proteins with GFP antibody (C) or for endogenous p65 with p65 (C-20) antibody (D).
The 21–186 Truncated Mutant of p65 Competes RPS3 off Endogenous p65 in the Cytoplasm
Phosphorylation of RPS3 at serine 209 by IKKβ during NF-κB activation has been proposed to occur shortly after IKKβ phosphorylates IκBα at serines 32 and 36 in the cytoplasmic inhibitory p65-p50-IκBα complex (17). Given that the 21–186 truncated mutant of p65 remained in the cytoplasm in both resting and stimulated cells (Fig. 2, B, C, and E) and also specifically blocked Ser-209 phosphorylation and importin-α association (Fig. 4, A and B), we hypothesized that the 21–186 truncated mutant of p65 could compete RPS3 off the endogenous p65-p50-IκBα inhibitory complex in the cytoplasm. To test this hypothesis, we determined the association between RPS3 and endogenous p65/EGFP-tagged fusion proteins in the resting HEK293T cells transfected with either EGFP vehicle or the EGFP-fused 21–186 truncated mutant of p65. As expected, we observed a robust interaction between RPS3 and the p65 (21–186) fragment, whereas there was no detectable association of EGFP to RPS3 (Fig. 4C). We also detected a significant interaction between endogenous p65 and RPS3 in the control EGFP-expressing cells (Fig. 4D), in line with our previous finding that RPS3 occurs in the cytoplasmic p65-p50-IκBα inhibitory complex (16, 17). In contrast, the interaction between RPS3 and endogenous p65 was nearly abolished in cells expressing the 21–186 truncated mutant of p65 (Fig. 4D). These results suggest that the p65 (21–186) fragment competes RPS3 off endogenous full-length p65.
The Caspase 3-cleaved 1–97 Fragment of p65 Shares a Similar Function to the p65 (21–186) Truncated Mutant in Attenuating NF-κB Activation-induced RPS3 Nuclear Translocation
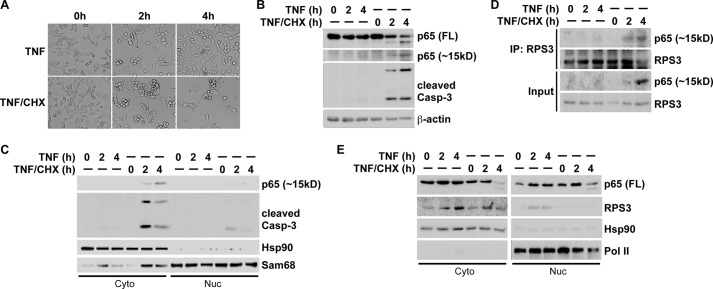
Previous studies demonstrate that p65 is cleaved by caspase 3 at Asp-97 in cells undergoing apoptosis (28, 29); despite this, the function of the generated 1–97 fragment of p65 has not been characterized. Of note, the 1–97 fragment largely overlaps with the 21–186 truncation, in particular they both contain Cys-38, a key residue that was recently identified to be critical for RPS3-p65 interaction (20, 21). We therefore examined whether they share similar functions in modulating RPS3-dependent NF-κB signaling. To this end, we first verified that a combination of TNF and cycloheximide treatment, but not TNF treatment alone, induced apoptosis in MEF cells, indicated by cell morphology (Fig. 5A) and activated caspase 3 (Fig. 5B). We observed that a cleaved N-terminal fragment of p65, migrating around 15 kDa in SDS-PAGE gel, coincided with activated caspase 3 (Fig. 5B), similar to the previously reported caspase 3-cleaved p65 (1–97) fragment (28, 29). A further subcellular fractionation experiment revealed that this endogenous p65 (1–97) fragment predominately localized in the cytoplasm of MEF cells undergoing apoptosis (Fig. 5C), which led us to examine the potential interaction between endogenous RPS3 and the cleaved p65 (1–97) fragment in the cytosolic subcellular fraction. Indeed, we detected a strong interaction between the cleaved p65 (1–97) fragment and RPS3 in the cytoplasm (Fig. 5D). To examine whether the cleaved p65 (1–97) fragment attenuates NF-κB activation-induced RPS3 nuclear translocation, we immunoblotted subcellular fractions derived from MEF cells treated with TNF alone or TNF plus cycloheximide. As expected, after 2–4 h of treatment, TNF alone triggered significant nuclear accumulation of p65 and RPS3 in MEF cells (Fig. 5E). In contrast, in the MEF cells undergoing apoptosis by TNF plus cycloheximide treatment, the TNF-induced RPS3 nuclear translocation was nearly abolished, despite an almost identical p65 nuclear translocation (Fig. 5E), suggesting that the caspase 3-cleaved p65 (1–97) fragment associates to RPS3 in the cytoplasm and selectively attenuates TNF-induced RPS3 nuclear translocation in apoptotic MEF cells. Hence these results support the notion that the N-terminal 21–186 truncation of p65 competes RPS3 off endogenous p65, thus protecting RPS3 from nuclear translocation and its subsequent function in a subset of NF-κB target gene transcription (Fig. 6). Altogether, our results reveal a novel mechanism for the 21–186 truncated mutant of p65 to impair the p65-RPS3 interaction in the cytoplasm, thus selectively attenuating RPS3-dependent NF-κB gene transcription.
FIGURE 5.
Caspase 3-cleaved N-terminal 1–97 fragment of p65 interacts with RPS3 and attenuates NF-κB activation-induced RPS3 nuclear translocation in cells undergoing apoptosis. A, representative images of MEF cells, stimulated with TNF (50 ng/ml) alone or combined with cycloheximide (CHX, 10 μg/ml) for indicated period. B, whole cell lysates from MEF cells, stimulated as in A, were immunoblotted for full-length (FL), the C-terminal fragment, and the smaller N-terminal fragment (∼15 kDa) of p65 with the antibody specifically recognizing the N terminus of p65, and cleaved caspase 3, with β-actin as a loading control. C, immunoblot analysis of cytosolic (Cyto) and nuclear (Nuc) subcellular fractions of MEF cells, stimulated as in A, for cleaved N-terminal p65 and cleaved caspase-3. Hsp90 and Sam68 serve as cytosolic and nuclear markers, respectively. D, cytosolic subcellular fractions (Input) from MEF cells, stimulated as indicated, were immunoblotted directly or after IP with RPS3 antibody for RPS3 and the p65 N-terminal fragment (∼15 kDa). E, immunoblot analysis of cytosolic (Cyto) and nuclear (Nuc) subcellular fractions of MEF cells, stimulated as indicated, for full-length (FL) p65 and RPS3. Hsp90 and RNA polymerase II (pol II) serve as cytosolic and nuclear markers and loading controls, respectively.
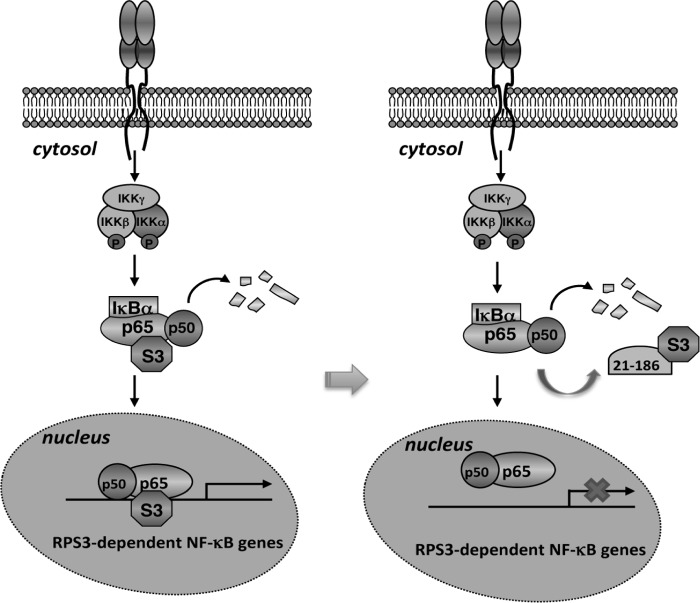
FIGURE 6.
Schematic model of selective attenuation of NF-κB gene transcription by the 21–186 truncated mutant of p65. Left panel, RPS3 exists in the cytoplasmic p65-p50-IκBα inhibitory complex in resting cells. NF-κB activation stimuli trigger the phosphorylation and degradation of IκBα, allowing p65, p50, and RPS3 to translocate to the nucleus for the transcription of a subset of NF-κB target genes. Right panel, because of its high affinity to RPS3, the 21–186 truncated mutant of p65 competes RPS3 off the p65-p50-IκBα inhibitory complex in the cytoplasm. Upon NF-κB activation, the 21–186 truncated mutant of p65 sequesters RPS3 in the cytoplasm but leaves p65 nuclear translocation to occur normally. Lacking RPS3, the binding of the NF-κB complex to the RPS3-dependent NF-κB target genes is diminished. Thus, the 21–186 truncated mutant of p65 selectively inhibits NF-κB gene transcription by targeting RPS3.
DISCUSSION
We previously found that RPS3 can interact with p65 in both the cytoplasmic p65-p50-IκBα inhibitory complex and the nuclear NF-κB DNA binding complexes (16, 17). Moreover, structure-function analyses demonstrated that the 21–186 domain of p65 was essential for its interaction with RPS3, and conversely the hnRNP K homolog domain in RPS3 had the strongest binding affinity to p65 (16). Given that both the 21–186 domain in p65 and the hnRNP K homolog domain in RPS3 possess nucleic acid binding capacity, their interactions were proposed to form a stable platform with protein-protein and protein-DNA interactions for the synergistic binding of NF-κB complex to target κB DNA in the nucleus (12, 13, 16). Two possibilities emerge from the observation that ectopic expression of the 21–186 truncated mutant of p65 specifically blocked stimuli-induced NF-κB activation (Fig. 1). It is possible for the p65 (21–186) fragment to dominantly interfere with NF-κB gene transcription in the nucleus by competing the endogenous p65 off the target κB DNA, provided its strong interactions with both RPS3 and double-stranded DNA. The 21–186 truncated mutant of p65 lacks a functional NLS; however, it still could accumulate in the nucleus through the association with RPS3, which was previously shown to translocate to the nucleus with its own NLS (16). Unexpectedly, we found that the 21–186 truncated mutant of p65 preferentially resided in the cytoplasm, whether cells were stimulated or not. These findings favor the second possibility that the 21–186 truncated mutant of p65 executes its modulatory function for specific NF-κB gene transcription in the cytoplasm. Indeed, the 21–186 truncated mutant of p65 associated with RPS3 in the cytoplasm and attenuated the stimuli-induced RPS3 nuclear translocation. In support of this, the expression of the p65 (21–186) fragment diminished the Ser-209 phosphorylation of RPS3 and the association of RPS3 to importin-α, which are known to be required for NF-κB activation-induced RPS3 nuclear translocation (17). Furthermore, we observed that the 21–186 truncated mutant of p65 competed RPS3 off endogenous p65, so that RPS3 could not be phosphorylated by IKKβ during NF-κB activation. This result is consistent with our recent finding that Ser-209 phosphorylation is a prerequisite for RPS3 nuclear translocation (17), which also accounts for why the 21–186 truncated mutant of p65 could not utilize the NLS of RPS3 to enter the nucleus during NF-κB activation. It is notable that the 21–186 truncated mutant of p65 selectively retarded RPS3 nuclear translocation, without affecting p65 nuclear translocation. Consistently, only the RPS3-dependent but not all p65-required NF-κB target genes were inhibited by introducing the 21–186 truncated mutant of p65 into Jurkat cells that were stimulated through T cell receptor engagement. Such selective and dominant effects on blocking the RPS3-dependent, but not all, NF-κB signaling by the 21–186 truncated mutant of p65, are similar to those caused by knockdown of RPS3 with siRNA, mutation on the Ser-209 phosphorylation site, or retarding RPS3 in the cytoplasm by pathogen-injected effector proteins (16, 17, 22). Together, our results assign a previously unknown cytoplasmic function of the 21–186 domain of p65, which is critical for κB DNA binding and putatively functions in the nucleus, to selectively modulate the specific NF-κB gene transcription through targeting RPS3.
NF-κB p65 has emerged as a new target for a variety of proteases to minimalize NF-κB activation, especially under the pathophysiological settings such as pathogen infection and apoptosis (24–29, 34–40). Despite the majority of the reported protease cleavage sites on p65 locating in the TAD and dimerization domains, at least two proteases cleave p65 within its N terminus. The type III secretion system effector NleC, an anti-inflammatory bacterial zinc metalloprotease, was shown to cleave host p65 during infections with enteropathogenic E. coli and enterohemorrhagic E. coli (24–27). An N-terminal fragment of p65 was also reported to be cleaved by caspase 3 in cytochrome c/caspase 9-mediated apoptosis (28, 29). Almost all previous research focused on characterizing the larger C-terminal fragments of p65 generated by protease cleavage (24–29), whereas the smaller N-terminal fragments of p65 have not been carefully characterized. The N-terminal domain of p65 is largely believed to function in DNA binding in the nucleus; however, most of the proteases were reported to cleave p65 in the cytoplasm (24–29, 34–40). Moreover, the NF-κB complex, in particular the p65 homodimer-containing one, has been shown to be critical for the transcription of the target genes encoding inflammatory cytokines (e.g., IL-8, macrophage chemotactic protein-1, and cyclooxygenase), stress response molecules (e.g., NAD(P)H quinone oxidoreductase), and anti-apoptotic molecules (e.g., cIAP-2, Bcl-XL, and XIAP) (17, 41–47), making them potential targets to be impaired by the p65-RPS3 interaction. Indeed, the proteases efficiently suppress the transcription of the NF-κB target genes, despite only a small fraction of p65 being cleaved (24–29, 38). It will be interesting for further studies to determine under which pathophysiological setting(s) NF-κB p65 can be cleaved to generate an N-terminal 21–186 fragment by certain protease(s). Of note, we also report here that the endogenous caspase 3-cleaved p65 (1–97) fragment interacts with RPS3 and attenuates NF-κB activation-induced RPS3 nuclear translocation, without affecting p65 nuclear accumulation, in MEF cells undergoing apoptosis. These results indicate that the caspase 3-cleaved N-terminal fragment of p65 shares a similar function to the 21–186 truncated mutant of p65 to selectively modify NF-κB gene transcription by targeting RPS3. Given the critical role of RPS3 in anti-apoptosis (20) and host inflammatory response to pathogen infection (17, 22, 23), further studies are needed to investigate whether and how the NleC-cleaved and the caspase 3-cleaved N-terminal fragment of p65 selectively modify NF-κB gene transcription by targeting RPS3.
Accumulating evidence suggests that alternative splicing of NF-κB signaling components has emerged as another means of regulating NF-κB signaling (30). So far, three alternative splice variants of p65, i.e. p65Δ, p65Δ2, and p65Δ3, have been identified, of which p65Δ encodes a protein lacking amino acids 222–231, p65Δ2 lacks N-terminal amino acids 13–22, and p65Δ3 lacks amino acids 187–293 (30). Previous study on the genomic organization of the RELA gene encoding p65 suggests that multiple variants of the p65 protein could be generated by alternative splicing (48). In particular, a 10-nucleotide sequence (TCTCCTGCAG), which contains a pyrimidine stretch (TCTCCT) and the AG dinucleotide, forms the alternative acceptor splice site within intron 5 (48). This suggests that intron 5 could be a novel splicing “hot spot” to generate the alternative splicing variant with exons 1–5, which encodes the 1–186 truncated mutant of p65. It is certainly worth identifying such p65 mRNA splicing variants containing only exons 1–5 in future studies. Given that the expression profile and activity of the identified p65 splice variants have not been fully characterized, it will also be interesting to examine their subcellular localization and interaction with RPS3, especially the p65Δ3, which contains the 1–186 fragment and TAD.
Given that RPS3 is crucial for specific NF-κB gene transcription, an emerging role of RPS3 in tumorigenesis has been supported by an increasing number of studies (49–52). In particular, RPS3 expression is increased in adenocarcinomas and in the majority of adenomatous polyps (50). We speculate that the interaction of RPS3 and the 21–186 truncated mutant of p65 may be amenable to peptide-based inhibition strategies, and the 21–186 fragment as a prototypic backbone could be further narrowed down to more efficient peptides. Interestingly, such approaches are predicted to selectively block only the RPS3-dependent subset of NF-κB target genes encoding proinflammatory and anti-apoptotic molecules but not all NF-κB-induced gene transcription. Because NF-κB has many vital roles in normal cellular function, developing cell permeable peptides that selectively target the RPS3-dependent transcription of a crucial panel of NF-κB target genes without blocking NF-κB activity itself could be a very exciting prospect.
Acknowledgments
We are grateful to S. Richard (McGill University) and R. Barnitz (Harvard Medical School) for providing reagents; other members in the Wan laboratory for discussions; P. Coulombe for encouragement, suggestions, use of equipment, and critical reading of the manuscript; and M. Matunis for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R00CA137171 (to F. W.). This work was also supported by National Cancer Institute Training Grant T32CA009110 (to E. M. W.).
- RPS3
- ribosomal protein S3
- EGFP
- enhanced green fluorescent protein
- Hsp90
- heat shock protein 90
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblasts
- NLS
- nuclear localization sequence
- IKKβ
- inhibitor of κB kinase β
- TAD
- transactivation domain.
REFERENCES
- 1. Ben-Neriah Y., Karin M. (2011) Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12, 715–723 [DOI] [PubMed] [Google Scholar]
- 2. Oeckinghaus A., Hayden M. S., Ghosh S. (2011) Crosstalk in NF-κB signaling pathways. Nat. Immunol. 12, 695–708 [DOI] [PubMed] [Google Scholar]
- 3. Hoffmann A., Baltimore D. (2006) Circuitry of nuclear factor κB signaling. Immunol. Rev. 210, 171–186 [DOI] [PubMed] [Google Scholar]
- 4. Lenardo M. J., Baltimore D. (1989) NF-κB. A pleiotropic mediator of inducible and tissue-specific gene control. Cell 58, 227–229 [DOI] [PubMed] [Google Scholar]
- 5. Li Q., Verma I. M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 [DOI] [PubMed] [Google Scholar]
- 6. Israël A. (2010) The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb. Perspect. Biol. 2, a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu T., Stark G. R. (2010) Use of forward genetics to discover novel regulators of NF-κB. Cold Spring Harb. Perspect. Biol. 2, a001966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harhaj E. W., Dixit V. M. (2011) Deubiquitinases in the regulation of NF-κB signaling. Cell Res. 21, 22–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Natoli G., Chiocca S. (2008) Nuclear ubiquitin ligases, NF-κB degradation, and the control of inflammation. Sci Signal. 1, pe1. [DOI] [PubMed] [Google Scholar]
- 10. Perkins N. D. (2007) Integrating cell-signalling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 [DOI] [PubMed] [Google Scholar]
- 11. Skaug B., Jiang X., Chen Z. J. (2009) The role of ubiquitin in NF-κB regulatory pathways. Annu. Rev. Biochem. 78, 769–796 [DOI] [PubMed] [Google Scholar]
- 12. Wan F., Lenardo M. J. (2009) Specification of DNA binding activity of NF-κB proteins. Cold Spring Harb. Perspect. Biol. 1, a000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan F., Lenardo M. J. (2010) The nuclear signaling of NF-κB. Current knowledge, new insights, and future perspectives. Cell Res. 20, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sen R., Smale S. T. (2010) Selectivity of the NF-κB response. Cold Spring Harb. Perspect. Biol. 2, a000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smale S. T. (2011) Hierarchies of NF-κB target-gene regulation. Nat. Immunol. 12, 689–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wan F., Anderson D. E., Barnitz R. A., Snow A., Bidere N., Zheng L., Hegde V., Lam L. T., Staudt L. M., Levens D., Deutsch W. A., Lenardo M. J. (2007) Ribosomal protein S3. A KH domain subunit in NF-κB complexes that mediates selective gene regulation. Cell 131, 927–939 [DOI] [PubMed] [Google Scholar]
- 17. Wan F., Weaver A., Gao X., Bern M., Hardwidge P. R., Lenardo M. J. (2011) IKKβ phosphorylation regulates RPS3 nuclear translocation and NF-κB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 12, 335–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cadera E. J., Wan F., Amin R. H., Nolla H., Lenardo M. J., Schlissel M. S. (2009) NF-κB activity marks cells engaged in receptor editing. J. Exp. Med. 206, 1803–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mokhtari D., Barbu A., Mehmeti I., Vercamer C., Welsh N. (2009) Overexpression of the nuclear factor-κB subunit c-Rel protects against human islet cell death in vitro. Am. J. Physiol. Endocrinol. Metab. 297, E1067–E1077 [DOI] [PubMed] [Google Scholar]
- 20. Sen N., Paul B. D., Gadalla M. M., Mustafa A. K., Sen T., Xu R., Kim S., Snyder S. H. (2012) Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. Mol. Cell 45, 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Perkins N. D. (2012) Cysteine 38 holds the key to NF-κB activation. Mol. Cell 45, 1–3 [DOI] [PubMed] [Google Scholar]
- 22. Gao X., Wan F., Mateo K., Callegari E., Wang D., Deng W., Puente J., Li F., Chaussee M. S., Finlay B. B., Lenardo M. J., Hardwidge P. R. (2009) Bacterial effector binding to ribosomal protein S3 subverts NF-κB function. PLoS Pathog. 5, e1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pham T. H., Gao X., Tsai K., Olsen R., Wan F., Hardwidge P. R. (2012) Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infect. Immun. 80, 2133–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shames S. R., Bhavsar A. P., Croxen M. A., Law R. J., Mak S. H., Deng W., Li Y., Bidshari R., de Hoog C. L., Foster L. J., Finlay B. B. (2011) The pathogenic Escherichia coli type III secreted protease NleC degrades the host acetyltransferase p300. Cell Microbiol. 13, 1542–1557 [DOI] [PubMed] [Google Scholar]
- 25. Pearson J. S., Riedmaier P., Marchès O., Frankel G., Hartland E. L. (2011) A type III effector protease NleC from enteropathogenic Escherichia coli targets NF-κB for degradation. Mol. Microbiol. 80, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yen H., Ooka T., Iguchi A., Hayashi T., Sugimoto N., Tobe T. (2010) NleC, a type III secretion protease, compromises NF-κB activation by targeting p65/RelA. PLoS Pathog. 6, e1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baruch K., Gur-Arie L., Nadler C., Koby S., Yerushalmi G., Ben-Neriah Y., Yogev O., Shaulian E., Guttman C., Zarivach R., Rosenshine I. (2011) Metalloprotease type III effectors that specifically cleave JNK and NF-κB. EMBO J. 30, 221–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coiras M., López-Huertas M. R., Mateos E., Alcamí J. (2008) Caspase-3-mediated cleavage of p65/RelA results in a carboxy-terminal fragment that inhibits IκBα and enhances HIV-1 replication in human T lymphocytes. Retrovirology 5, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang K. H., Lee K. H., Kim M. Y., Choi K. H. (2001) Caspase-3-mediated cleavage of the NF-κB subunit p65 at the NH2 terminus potentiates naphthoquinone analog-induced apoptosis. J. Biol. Chem. 276, 24638–24644 [DOI] [PubMed] [Google Scholar]
- 30. Leeman J. R., Gilmore T. D. (2008) Alternative splicing in the NF-κB signaling pathway. Gene 423, 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lukong K. E., Larocque D., Tyner A. L., Richard S. (2005) Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J. Biol. Chem. 280, 38639–38647 [DOI] [PubMed] [Google Scholar]
- 32. Barnitz R. A., Wan F., Tripuraneni V., Bolton D. L., Lenardo M. J. (2010) Protein kinase A phosphorylation activates Vpr-induced cell cycle arrest during human immunodeficiency virus type 1 infection. J. Virol. 84, 6410–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Isakov N., Altman A. (2002) Protein kinase Cθ in T cell activation. Annu. Rev. Immunol. 20, 761–794 [DOI] [PubMed] [Google Scholar]
- 34. Neznanov N., Chumakov K. M., Neznanova L., Almasan A., Banerjee A. K., Gudkov A. V. (2005) Proteolytic cleavage of the p65-RelA subunit of NF-κB during poliovirus infection. J. Biol. Chem. 280, 24153–24158 [DOI] [PubMed] [Google Scholar]
- 35. Neuzil J., Schröder A., von Hundelshausen P., Zernecke A., Weber T., Gellert N., Weber C. (2001) Inhibition of inflammatory endothelial responses by a pathway involving caspase activation and p65 cleavage. Biochemistry 40, 4686–4692 [DOI] [PubMed] [Google Scholar]
- 36. Christian J., Vier J., Paschen S. A., Häcker G. (2010) Cleavage of the NF-κB family protein p65/RelA by the chlamydial protease-like activity factor (CPAF) impairs proinflammatory signaling in cells infected with Chlamydiae. J. Biol. Chem. 285, 41320–41327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mühlen S., Ruchaud-Sparagano M. H., Kenny B. (2011) Proteasome-independent degradation of canonical NFκB complex components by the NleC protein of pathogenic Escherichia coli. J. Biol. Chem. 286, 5100–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lad S. P., Li J., da Silva Correia J., Pan Q., Gadwal S., Ulevitch R. J., Li E. (2007) Cleavage of p65/RelA of the NF-κB pathway by Chlamydia. Proc. Natl. Acad. Sci. U.S.A. 104, 2933–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gregory D. J., Godbout M., Contreras I., Forget G., Olivier M. (2008) A novel form of NF-κB is induced by Leishmania infection. Involvement in macrophage gene expression. Eur. J. Immunol. 38, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 40. Neves B. M., Silvestre R., Resende M., Ouaissi A., Cunha J., Tavares J., Loureiro I., Santarém N., Silva A. M., Lopes M. C., Cruz M. T., Cordeiro da Silva A. (2010) Activation of phosphatidylinositol 3-kinase/Akt and impairment of nuclear factor-κB. Molecular mechanisms behind the arrested maturation/activation state of Leishmania infantum-infected dendritic cells. Am. J. Pathol. 177, 2898–2911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ueda A., Okuda K., Ohno S., Shirai A., Igarashi T., Matsunaga K., Fukushima J., Kawamoto S., Ishigatsubo Y., Okubo T. (1994) NF-κ B and Sp1 regulate transcription of the human monocyte chemoattractant protein-1 gene. J. Immunol. 153, 2052–2063 [PubMed] [Google Scholar]
- 42. Yamamoto K., Arakawa T., Ueda N., Yamamoto S. (1995) Transcriptional roles of nuclear factor κ B and nuclear factor-interleukin-6 in the tumor necrosis factor α-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J. Biol. Chem. 270, 31315–31320 [DOI] [PubMed] [Google Scholar]
- 43. Kunsch C., Rosen C. A. (1993) NF-κ B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell. Biol. 13, 6137–6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao K. S., O'Dwyer P. J. (1995) Involvement of NF-κB in the induction of NAD(P)H:quinone oxidoreductase (DT-diaphorase) by hypoxia, oltipraz and mitomycin C. Biochem. Pharmacol. 49, 275–282 [DOI] [PubMed] [Google Scholar]
- 45. Stehlik C., de Martin R., Kumabashiri I., Schmid J. A., Binder B. R., Lipp J. (1998) Nuclear factor (NF)-κB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumor necrosis factor α-induced apoptosis. J. Exp. Med. 188, 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee R. M., Gillet G., Burnside J., Thomas S. J., Neiman P. (1999) Role of Nr13 in regulation of programmed cell death in the bursa of Fabricius. Genes Dev. 13, 718–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turner D. J., Alaish S. M., Zou T., Rao J. N., Wang J. Y., Strauch E. D. (2007) Bile salts induce resistance to apoptosis through NF-κB-mediated XIAP expression. Ann. Surg. 245, 415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deloukas P., van Loon A. P. (1993) Genomic organization of the gene encoding the p65 subunit of NF-κB. Multiple variants of the p65 protein may be generated by alternative splicing. Hum. Mol. Genet. 2, 1895–1900 [DOI] [PubMed] [Google Scholar]
- 49. Gao X., Hardwidge P. R. (2011) Ribosomal protein S3. A multifunctional target of attaching/effacing bacterial pathogens. Front. Microbiol. 2, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pogue-Geile K., Geiser J. R., Shu M., Miller C., Wool I. G., Meisler A. I., Pipas J. M. (1991) Ribosomal protein genes are overexpressed in colorectal cancer. Isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol. Cell. Biol. 11, 3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu N. X., Zheng S., Xu R. Z., Gao R. L., Shen J. P., Yu R. X. (2003) [Identification of multidrug resistance related genes in leukemia by suppression subtractive hybridization]. Zhonghua Xue Ye Xue Za Zhi 24, 14–17 [PubMed] [Google Scholar]
- 52. McDoniels-Silvers A. L., Nimri C. F., Stoner G. D., Lubet R. A., You M. (2002) Differential gene expression in human lung adenocarcinomas and squamous cell carcinomas. Clin. Cancer Res. 8, 1127–1138 [PubMed] [Google Scholar]