Background: Structures of functional K+ channels in lipid membranes are lacking.
Results: We dissected the pore module from the intact subunit, demonstrated its functional competence, and determined its crystal structure in membranes.
Conclusion: The structure is unprecedented, because it embodies a transient conformation trapped between closed and open states.
Significance: The findings highlight the deterministic role of surface match between sensor and pore underpinning their functional coupling.
Keywords: Ion Channels, Membrane Proteins, Membrane Reconstitution, Structural Biology, X-ray Crystallography, Lipidic Cubic Phase, Channel Gating, Droplet Interface Bilayers, Ion Selectivity, Lipid Bilayers
Abstract
Voltage-gated K+ channels underlie the electrical excitability of cells. Each subunit of the functional tetramer consists of the tandem fusion of two modules, an N-terminal voltage-sensor and a C-terminal pore. To investigate how sensor coupling to the pore generates voltage-dependent channel opening, we solved the crystal structure and characterized the function of a voltage-gated K+ channel pore in a lipid membrane. The structure of a functional channel in a membrane environment at 3.1 Å resolution establishes an unprecedented connection between channel structure and function. The structure is unique in delineating an ion-occupied ready to conduct selectivity filter, a confined aqueous cavity, and a closed activation gate, embodying a dynamic entity trapped in an unstable closed state.
Introduction
Ion channels allow the selective and regulated diffusion of ions across membranes. Voltage-gated K+ channels (Kv)3 are pivotal underpins of electrically excitable cells (1). Kvs are tetrameric proteins organized around a central pore. Each subunit consists of two tandemly arranged structural modules: an N-terminal voltage sensor module encompassing four transmembrane segments (TMs) S1–S4 and a C-terminal pore (PM) consisting of TMs S5 and S6 (2–6). The two modules are connected through a cytoplasmic linker (S4-S5 linker) between TMs S4 and S5.
Molecular dissection of KvLm, a Kv from Listeria monocytogenes, led to the isolation of a sensorless PM that exhibits the ionic selectivity, permeation, and blocker sensitivity properties characteristic of the intact KvLm, namely a functional PM (7). Here, we present the structure of such PM obtained from diffraction quality crystals using the lipidic cubic phase approach (8, 9).
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
A single amino acid mutation (A98C) was introduced in the KvLm PM (7) using the QuikChange I site-directed mutagenesis kit (Agilent), according to the manufacturer's instructions. Mutant channel protein was expressed as previously described (7). Following centrifugation, the cells were resuspended in 50 mm Hepes, pH 7.5, 50 mm KCl, 5 mm MnCl2, 0.1 mm CaCl2, 10% (v/v) glycerol, 0.02% (w/v) NaN3 (denoted as low salt buffer) supplemented with bovine serum albumin, lysozyme (Sigma-Aldrich), DNase, and protease inhibitor tablets (Roche Applied Science) and lysed with a microfluidizer. The membranes were pelleted by centrifugation at 125,000 × g, resuspended in the low salt buffer, and incubated for 1 h at 4 °C. The procedure was repeated with a high salt membrane wash buffer (same as the low salt buffer but with 0.5 m KCl). Following ultracentrifugation, the cells were resuspended in solubilization buffer (50 mm Hepes, pH 7.5, 50 mm KCl, 5 mm imidazole) prior to the addition of n-decyl-β-d-maltopyranoside (DM) (Anatrace) to 20 mm. The membranes were solubilized for 3–3.5 h at 4 °C under stirring, followed by the addition of KCl to a final concentration of 400 mm and ultracentrifugation at 125,000 × g for 1 h. The supernatant was passed through a cobalt (Co2+) IMAC resin (Clontech) three times before rigorous washing in three steps: first, with 20 column volumes of 50 mm Hepes, pH 7.5, 400 mm KCl, 20 mm imidazole, 10% glycerol, 15 mm DM followed by 20 column volumes with 50 mm Hepes, pH 7.5, 300 mm KCl, 20 mm imidazole, 5% (v/v) glycerol, 10 mm DM, and a final wash with 20 column volumes of 50 mm Hepes, pH 7.5, 200 mm KCl, 20 mm imidazole, 1% (v/v) glycerol, 5 mm DM. Purified KvLm was eluted in 50 mm Hepes, pH 7.5, 200 mm KCl, 500 mm imidazole, 1% glycerol, 5 mm DM. Pooled fractions were desalted in PD-10 desalting column (GE Healthcare) and eluted in crystallization buffer (50 mm Hepes, pH 7.5, 200 mm KCl, 3 mm DM, 0.1 mm TCEP). Protein digestion with l-(tosylamido-2-phenyl)ethyl chloromethyl ketone modified trypsin (New England Biolabs) was carried out overnight at ∼20 °C by incubation with trypsin at a trypsin to PM mass ratio of 50:1. Analysis by nano-LC/MS/MS (The Scripps Research Institute Center for Mass Spectrometry) showed that the enzymatic digestion of KvLm-PM removed the C-terminal 22 residues, including the His6 tag, to yield a truncated PM construct at residue Arg137 with a molecular mass of 15.6 kDa. The digested PM was isolated by passing the enzymatic reaction first through 1 ml of nickel-nitrilotriacetic acid IMAC resin (Qiagen) and second through 100 μl of benzamidine-Sepharose resin (GE Healthcare) and collecting the flow through. The sample was then concentrated to 40–45 mg ml−1 using a 100-kDa cutoff concentrator (Sartorius). Analytical Size exclusion chromatography and SDS-PAGE were used to monitor sample homogeneity and purity, respectively.
Crystallization and Data Collection
The protocol used for lipidic cubic phase crystallization has been described in detail previously (8, 9). Diffraction quality crystals were obtained in 26–30% (v/v) PEG-MME-550, 0.5–0.6 m ammonium sulfate, and 50 mm ADA, pH 7.0 (ammonium condition), and in 25% (v/v) PEG-MME-550, 0.35 m sodium nitrate, 50 mm ADA, pH 6.25, 50 mm sodium malonate, pH 7.0 (sodium condition). The approximate final cation concentrations in each crystallization condition are 27 mm K+ and 0.4 m NH4+ in the ammonium condition and 27 mm K+ and 0.3 m Na+ in the sodium condition.
Crystals appeared after 1–2 weeks; the final average crystal size was 80 μm × 20 μm × 5 μm. Data collection was as previously described (10). Diffraction data were collected to an overall resolution of 3.1 Å from a total of 32 microcrystals for the ammonium condition and to a resolution of 3.4 Å from a total of 10 microcrystals for the sodium condition (Table 1). The data were integrated per condition, scaled, and merged using HKL2000 (11). Because of the strong anisotropy along the three principal axes: a* (3.1 Å resolution), b* (3.1 Å resolution), and c* (4.2 Å resolution), the scaled P4212 data sets containing 2,994 reflections from the ammonium condition were processed by the UCLA anisotropy server (12), and the anisotropically scaled 2,674 structure factors were used for subsequent molecular replacement and refinement. The P21212 data sets were also very anisotropic along the three principal axes: a* (4.1 Å resolution), b* (3.3 Å resolution), and c* (4.0 Å resolution). However, ∼30% of total reflections were discarded by the anisotropy server because they fell outside the specified ellipsoid with dimensions 4.1, 3.3, and 4.0 Å along a*, b*, and c*, respectively. To avoid the loss of these reflections, we skipped the anisotropic scaling procedure for the P21212 data sets for the sodium condition.
TABLE 1.
Data collection and refinement statistics
The highest resolution shell is shown in parentheses.
| Structure | KvLm |
|
|---|---|---|
| Ammonium condition | Sodium condition | |
| Data collection | ||
| Space group | P4212 | P21212 |
| Cell dimensions | a = b = 53.37 Å, c = 108.16 Å | a = 56.10 Å, b = 80.67 Å, c = 55.52 Å |
| Number of reflections measured | 21,139 | 22,028 |
| Number of unique reflections | 2,994 | 3,799 |
| Resolution (Å) | 50–3.10 (3.21–3.10) | 50–3.35 (3.47–3.35) |
| Rmerge | 0.132 (0.592) | 0.116 (0.660) |
| Mean I/σ(I) | 18.6 (2.45) | 15.8 (1.10) |
| Completeness (%) | 94.4 (74.5) | 95.2 (86.0) |
| Redundancy | 7.1 (5.5) | 5.8 (4.8) |
| Refinement | ||
| Resolution (Å) | 16.4–3.10 | 32.6–3.35 |
| Number of reflections (test set) | 2,674 (369) | 3,736 (534) |
| Rwork/Rfree (%) | 28.1/30.5 | 31.4/34.1 |
| Number of atoms | ||
| Protein | 673 | 650 (A chain), 653 (B chain) |
| K+ ion | 3 | 4 |
| Lipids | 57 | |
| Average B value (Å2) | ||
| Protein | 115 | 191 (A chain), 194 (B chain) |
| K+ ion | 154 | 167 |
| Lipids | 103 | |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.01 | 0.94 |
| Ramachandran plot statistics (%)a | ||
| Ramachandran favored regions | 98 | 99 |
| Additionally allowed regions | 2 | 0 |
| Ramachandran outliers | 0 | 1 |
a The Ramachandran plot was calculated by using Molprobity (version 3.17).
Structure Determination and Refinement
The initial phase information was obtained by molecular replacement with the program PHASER (13), using ensemble structures designated with Protein Data Bank codes 3EFF (14), 2ATK (15), 2A79 (5), and 1ORQ (4) as search models. Refinements were performed with REFMAC5 (16), CNS (17), and autoBUSTER (18), followed by manual examination and rebuilding of the refined coordinates in the program COOT (19) using both sigma-A weighted 2|Fo| − |Fc| and |Fo| − |Fc| maps, as well as omit maps. The data collection and refinement statistics are summarized in Table 1.
To determine the best fit to electron density at the filter (Thr57–Tyr60), we positioned the carbonyl group of residue Val58 in the highest resolution structure facing away from the permeation path, because it was observed in the low K+ KcsA structure (Val76 in Protein Data Bank code 1K4D (20)). After refinement, the Val58 side chain reverted back to the conductive conformation, with the carbonyl oxygen facing the permeation path (R/Rfree = 0.302/0.335). We also attempted to refine the structure with the flipped carbonyl of Val58 without an ion fitted at S3; again after refinement, Val58 reverted to the conductive conformation with the carbonyl oxygen facing the permeation pathway (R/Rfree = 0.275/0.313). This suggests that the Val58 carbonyl is indeed best fit in the conductive conformation and that this conformation is independent of whether there is an ion modeled at the S3 position. We justify the modeling of an ion in S3, despite the small difference in R/Rfree when refining with or without an ion, with the presence of a clear peak at this position in the |Fo| − |Fc| electron density map contoured at 3 σ. The ions found in the selectivity filter of both structures could be NH4+ (Fig. 1, B and C) or Na+ (Fig. 1, D and E), the majority cations in each crystallization solution. However, this is unlikely, given 1) that 27 mm K+ is the only constant in the two conditions and 2) the observed significant increase in the R/Rfree upon NH4+ ion substitution. For simplicity, we therefore surmise that the ions in the filter are K+.
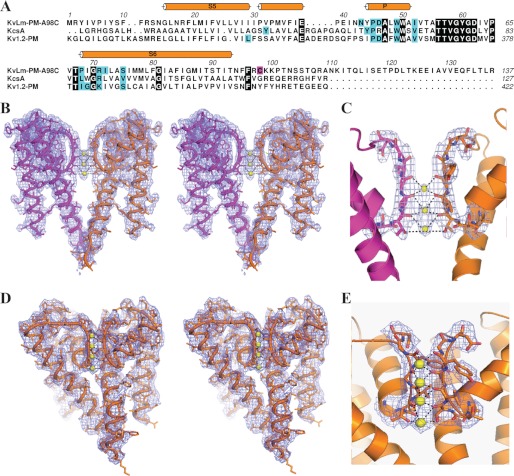
FIGURE 1.
Structure of the KvLm PM. A, sequence alignment of KvLm-PM with KcsA (Protein Data Bank code 1K4C) (20) and Kv1.2 PM (Protein Data Bank code 2R9R) (6) highlighting conserved residues (black). The alignment is annotated to display secondary structure and lipid-binding sites (cyan). The mutation (A98C) is highlighted in magenta. Residue numbering is according to Protein Data Bank; for KvLm it is indicated above the sequence. B and D, stereo profiles of two diametrically facing subunits displaying the electron density map (2|Fo| − |Fc|) contoured at 1 σ used to fit activation gate closed for crystals obtained in 27 mm K+ and 0.4 m ammonium sulfate (B) or 0.3 m sodium nitrate (D). Yellow spheres depict K+ coordinated by the residues 57TVG. C and E, view of the selectivity filter showing the experimental electron density map (2|Fo| − |Fc|) contoured at 1 σ used to fit the position of the bound ions and the filter in a conductive conformation with the carbonyl oxygens for residues Thr57, Val58, Gly59, and Tyr60 and the hydroxyl of residue Thr57 facing the ions in two different crystal conditions (“Experimental Procedures”).
Structure Analysis
All structural analysis was performed using PyMOL (Schrodinger) and YASARA (YASARA Biosciences) on the highest resolution structure (3.1 Å) obtained in ammonium sulfate, unless otherwise indicated. A Cα alignment of the two KvLm PM structures shows that they deviate by ∼0.61 Å. A Cα alignment of the PMs of KvLm and of the Kv1.2-Kv2.1 chimera (Protein Data Bank code 2R9R (6)) has an root mean square deviation of 1.6 Å. To calculate the electrostatic surface potential in a lipid membrane, the preoriented protein coordinates for KvLm PM were obtained from the PPM web server (21) and provided as input to the pbeq solver web server (22). The cavity profiles were obtained using the MOLE server (23).
Reconstitution in Droplet Interface Lipid Bilayers, Single-channel Recordings, and Analysis
Droplet interface lipid bilayers (24) were formed between two lipid monolayer-encased aqueous nanoliter droplets submerged in hexadecane containing 0.5 mg ml−1 monoolein (1-oleoyl-rac-glycerol) (Avanti Polar Lipids). Liposomes were composed of 0.5 mm 1,2-diphytanoyl-sn-glycero-3-phosphocholine (Avanti Polar Lipids). Purified KvLm PM was diluted into the liposome suspension to a final concentration of 2–5 ng ml−1. The electrode carrying the droplet with KvLm PM and desired buffer-lipid mix (0.5 m or 27 mm KCl, 10 mm Hepes, pH 7.4) was connected to the grounded end of the amplifier head stage (Axopatch 200B). The second electrode, in a droplet containing exclusively the buffer-lipid mix, was connected to the working end of the head stage. All of the reconstitution experiments were done at room temperature. The data were acquired at 10 kHz and filtered at 2 kHz before digitization. Transitions were detected by the segmental k-means method implemented in QuB (25). For analysis, records acquired in 27 mm KCl were filtered off-line to 0.2 kHz with a low pass Gaussian filter. Transitions shorter than 0.3 ms (0.5 m KCl) or 1.0 ms (27 mm KCl) were excluded from the analysis. The data were analyzed using Clampfit v.9.2 (Axon Instruments), QuB, Excel 2007 (Microsoft), and IGOR Pro (Wavemetrics). Single-channel conductance (γ) was calculated from Gaussian fits to current histograms. The statistical values represent the means ± S.E., unless otherwise indicated. n and N denote the number of experiments and the number of events, respectively. For presentation, records were filtered to 1 kHz (0.5 m KCl) or 0.2 kHz (27 mm KCl) using a low pass Gaussian filter.
RESULTS
Building on the discovery that a sensorless PM dissected from the bacterial KvLm (Fig. 1A) recapitulates the permeation properties of the full-length KvLm (7), we pursued its structural characterization. A crystal structure solved at 3.1 Å (Fig. 1, B and C) in a monoolein (1-oleoyl-rac-glycerol) membrane shows that the KvLm PM shares the funnel architecture of other K+ channels (5, 6, 20, 26–28). Each subunit contributes two transmembrane helices (outer helix or TM S5 (Gly15–Ile7) inner helix or TM S6 (Pro-68 to Thr-93) tilted with respect to the lipid bilayer normal, a pore helix (Tyr44–Ala55) with the C-end directed to the pore lumen, and the selectivity filter (Thr56–Asp62) formed at the center of the tetrameric channel (Figs. 1, A and B, and 2A). The inner helix is α-helical, whereas the outer helix is α-helical up to the C-terminal two turns, in which it adopts an irregular α-helical conformation unique to KvLm, coincident with the presence of a PVP motif (31PVP) (Fig. 1, A and B). The selectivity filter is occupied by three equidistant (3.1 Å) K+ at positions (S2–S4) (29) (Fig. 1C) (“Experimental Procedures”). Despite low ion occupancy, the ion binding sites in the filter are mostly intact: the backbone carbonyls of residues Thr57, Val58, Gly59, and Tyr60 and the hydroxyl of Thr57 are facing the permeation path, positioning the filter in a ready to conduct conformation (Fig. 1, B and C). In agreement, a PM structure determined at 3.4 Å in a different condition, with two subunits in the asymmetric unit, shows a fully occupied selectivity filter (Fig. 1, D and E). A narrow central cavity appears at the center of the ion channel delimited on the extracellular side by the filter and on the intracellular boundary by the crossing of the inner helices, the activation gate (30, 31).
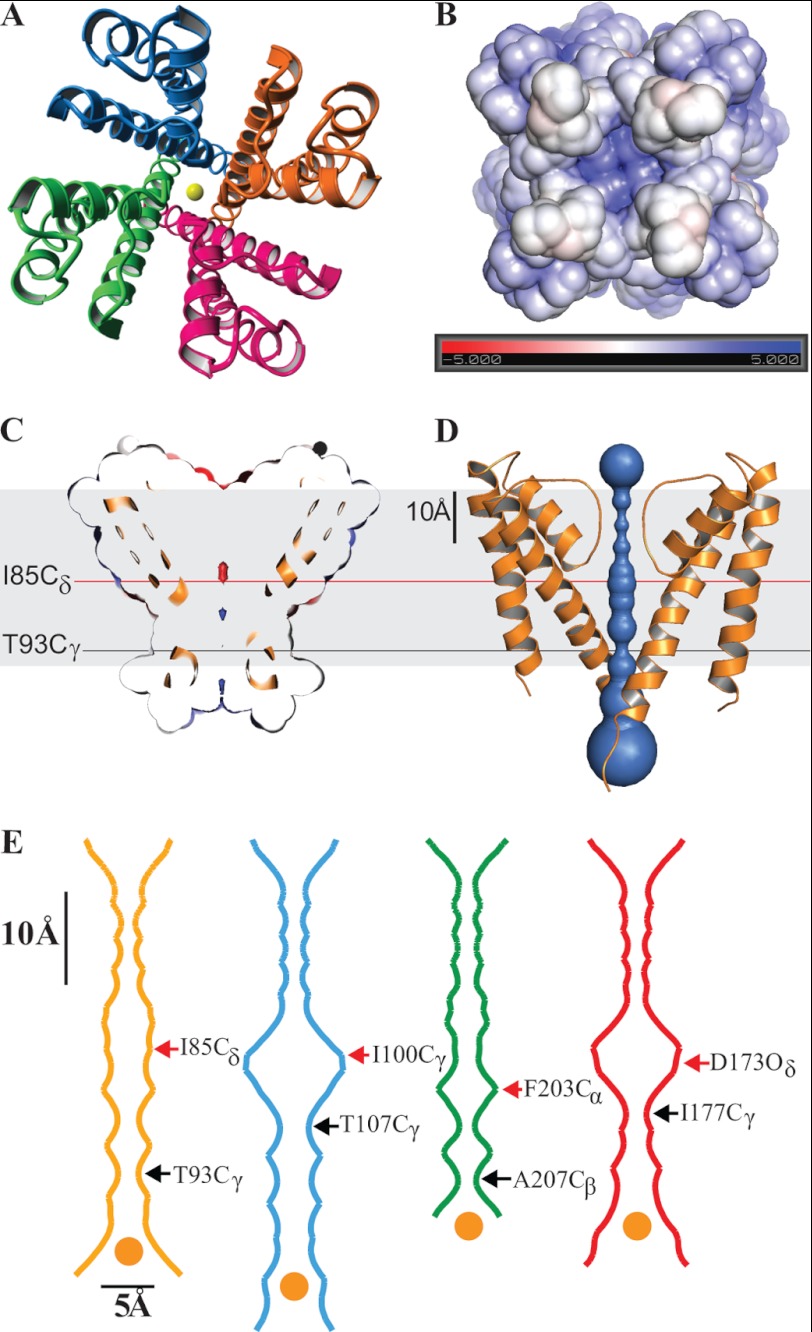
FIGURE 2.
KvLm PM has its filter gate in a conductive conformation and its cytoplasmic activation gate closed. A, crystal structure of the KvLm PM tetramer seen from the extracellular surface with subunits in red, green, blue, and orange. K+ ions at the center of the pore are in yellow. B, end view of the tetramer from the cytoplasm. The electrostatic surface potential of the KvLm tetramer computed in a bilayer environment (22) that mimics the crystallization condition (0.5 m KCl) rendered on a solvent-accessible surface representation. The electrostatic scaling is −5 kcal/mole (red) to +5 kcal/mole (blue). C and D, side view of a cross-section through the middle of the tetramer exposing the channel lumen. The indicated residues form the tighter and wider constrictions. The contour depicted in gray represents the lipid bilayer. E, cavity radius profile of KvLm (gold) and three other K+ channels crystallized in a closed conformation: KcsA (20) (Protein Data Bank code 1K4C, blue), MlotiK1 (26) (Protein Data Bank code 3BEH, green), and an inward rectifier (27) (Protein Data Bank code 3JYC, red). The indicated residues form the tighter and wider constrictions. The dehydrated K+ ion depicted in yellow at the entrance to the activation gate is drawn to scale (1.33 Å radius) (39). The contour depicted in gray represents the lipid bilayer.
Mapping the electrostatic potential on the solvent-accessible surface of the activation gate at the channel bundle crossing reveals that the gate is closed and positively charged (Fig. 2B). The large positive potential at the intracellular entrance is generated by residues 97RCKK that cap the inner helix and presumably act as an electrostatic barrier to ion access into the channel vestibule. Residues Ile85, Thr89, and Thr93 face the pore lumen resulting in a neutral electrostatic surface. Near the entryway to the filter is a negatively charged surface (Fig. 2C), most likely arising from the focusing of the pore helices dipoles.
The solvent-accessible ion-conducting pathway displays a narrow infundibulum with several constrictions (Fig. 2D). At the selectivity filter, four constrictions are created by rings of the carbonyl oxygens of the residues forming the filter (Thr57, Val58, Gly59, and Tyr60). The central cavity reveals three prominent constrictions generated by rings of Ile, Thr, and Thr side chains at positions 85, 89, and 93, respectively. The constrictions range from 1.9 Å diameter at the narrowest (Thr93, Cγ–Cγ) to 3.3 Å at its widest (Ile85, Cγ–Cγ). The funnel-like assembly (Fig. 2, D and E) measures 38 Å in length, with a wide entry way into the pore at the cytoplasmic side (8.8 Å at Arg97), a narrow channel vestibule (4.2 Å at Ile85 Cδ), and a wide exit (7.6 Å at Gly61 O). A comparative analysis of the cavity profile highlights that the KvLm PM diverges from the stand-alone 2-TM channels KcsA and the inward rectifier Kir2.2, in that the central water accessible cavity is narrower (4.2 Å versus 9.1 Å in KcsA, and 8.9 Å in the inward rectifier) (Fig. 2E). Surprisingly, a similarly tight central cavity occurs in the structure of the cyclic nucleotide gated channel (MlotiK1), which, akin to KvLm, is assembled from four 6-TM subunits. The constricted central cavity in KvLm and MlotiK1 may account for the absence of K+ within the cavity of the closed structure: the cavity is too narrow to harbor a K+ ion octa-coordinated to water as observed in KcsA (Protein Data Bank code 1K4C (20)). In addition, the presence of K+ in the central cavity in both of these structures is predicted to be less favorable than in KcsA because of the positive charge clustering at the cytoplasmic pore entry (97RCKK in KvLm and Arg219 in MlotiK1) (32). Thus, the closed structure of KvLm is unique in that it excludes ions from the cavity and couples a wide electrostatic gate at the permeation entrance with a sterically occluded access to the central cavity that prevents ion conduction.
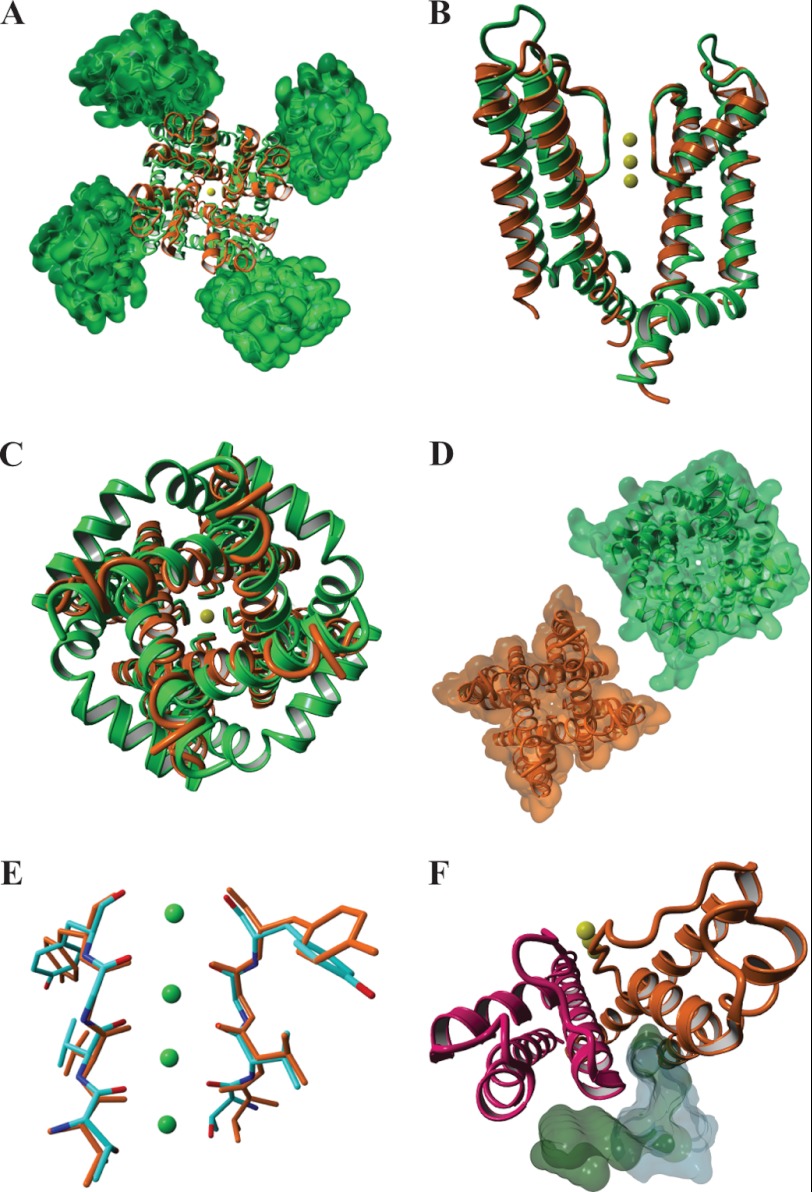
A comparison of the KvLm PM structure with that of the chimera Kv1.2-Kv2.1 (6) (Fig. 3A), a tandem fusion of two eukaryotic Kv modules (a sensor and a pore), is informative because it underscores significant differences and similarities attributable in part to the removal of the sensor from the KvLm PM. First, the crystallized KvLm-PM contained the S4-S5 linker, yet no density was observed for any of its residues suggesting that the linker was not immobilized at the position found in the eukaryotic chimera (6) (Fig. 3B). For the chimera, the inner helices are more distant and disclose an open activation gate that leads to an unobstructed pore lumen (Fig. 3, B–D); by contrast, in the KvLm PM a narrower permeation path appears occluded by a closed activation gate. Therefore, we surmise that the structure of the activation gate is sensor-dependent. At variance, the near identical conformation of the selectivity filters in a conductive conformation (Fig. 3E), and the conservation of the lipid immobilization surface at the intersubunit crevice (Fig. 3F) suggests that the presence of a voltage sensor module is not required for conservation of these PM features. Importantly, the conservation of the lipid-binding sites (Fig. 3F) implies that both full length and PM are permissive to modulation by membrane lipid composition. Hence, the KvLm PM structures underscore the essential role of the voltage sensor in stabilizing the conductive state of the channel.
FIGURE 3.
A structural alignment of KvLm PM (orange) and the eukaryotic Kv1.2-Kv2.1 channel chimera (green) in an open conformation discloses a conserved conductive selectivity filter and a lipid-binding site at the subunit interface. A, end view from the extracellular milieu displaying the structural similarity between the pores of KvLm (orange) and the eukaryotic chimera (green) and the position of the sensors in the chimera (rendered as a molecular surface). B, side view of two opposing subunits. C and D, end view of the pores from cytoplasm (backbone) (C) and their molecular surface representations omitting all ions in the structures (D). E, a structural alignment of the Cα atoms in the residues forming the K+ binding sites of the two selectivity filters: KvLm (orange) and eukaryotic chimera (colored by element) (root mean square deviation = 0.2 Å). F, in the chimera, co-crystallized lipids (green molecular surface) localize at the subunit interface as they do in KvLm (gray molecular surface) notwithstanding a significant increase in the crevice width.
There are conspicuous differences between the PM of KvLm and that of the chimera that are not attributable to the missing sensors. Salient among them is that, for KvLm, the extracellular facing surface is flat, whereas in Kv1.2-Kv2.1, the extra-long linker that connects the outer helix and the pore helix protrudes ∼15 Å out toward the extracellular milieu and contributes both sterically and electrostatically to the docking of other interacting proteins, including toxins (33–35). In contrast, access to the selectivity filter in KvLm is unobstructed, implying that the structural element connecting outer helix and pore helix may be of modular nature, a site adaptable to respond to different gating modifying signals.
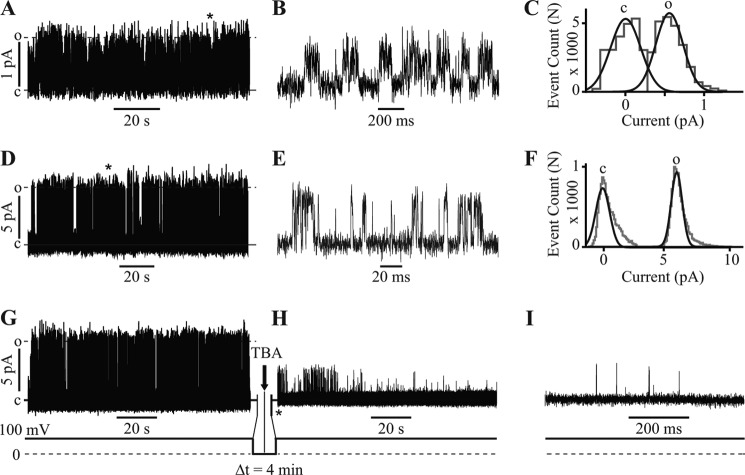
To establish a rigorous connection between structure and function, the KvLm PM was reconstituted in lipid bilayers composed of the lipid used for crystallization (monoolein) and bathed in symmetric 27 mm KCl, thereby mimicking the crystallization condition (“Experimental Procedures”). The single-channel conductance (γ) is 4 ± 2 pS (Fig. 4, A–C, and Table 2), with the channels opening primarily at positive membrane potentials. Higher signal to noise ratio single-channel currents recorded in 0.5 m KCl exhibit the characteristic discrete channel transitions between the closed and open state (Fig. 4D), which are clearly discerned by displaying the segment indicated by the asterisk in Fig. 4I at higher time resolution in Fig. 4E: γ is 68 ± 9 pS (Fig. 4F and Table 2). To explore the gating dynamics, we assessed whether at 0 mV the pore lumen was accessible to the open channel blocker tetrabutylammonium. Fig. 4 (G–I) demonstrates that tetrabutylammonium enters the permeation pathway and blocks the current at 0 mV. This observation establishes that the structure of the KvLm PM in monoolein membranes is that of a pore in transit, quickly sampling multiple conformations before being captured in one of them.
FIGURE 4.
KvLm PM is functional as crystallized and conserves structural features of the eukaryotic PM. A, reconstitution of KvLm PM in monoolein lipid bilayers. B, single-channel currents recorded at 150 mV in 27 mm KCl. The section marked by the asterisk in A is displayed in B at a 100-fold higher time resolution; γ = 4 ± 2 pS. C, all event current amplitude histogram of a 6-min-long recording in symmetric 27 mm KCl, 10 mm Hepes, pH 7.4, at 150 mV. The total number of opening events (N) analyzed was 34,225; γ = 4 ± 2 pS. γ was calculated from Gaussian fits (black lines) to current amplitude histograms (gray lines); c and o denote closed and open states. D, currents at 100 mV in 0.5 m KCl. E, the section marked by the asterisk in D is displayed in E at a 1000-fold higher time resolution; γ = 68 ± 9 pS. F, all event current amplitude histogram of a 5-min-long recording in symmetric 0.5 m KCl, 10 mm Hepes, pH 7.4, at 100 mV. n = 20,200 and γ = 68 ± 9 pS. G, currents at 100 mV in 0.5 m KCl before addition of blocker. After 2 min at 0 mV, 100 μm tetrabutylammonium (TBA) was injected onto one side of the bilayer (arrow) followed by a 2-min waiting time at 0 mV. H, the voltage was stepped to 100 mV evoking the single-channel currents. I, the segment immediately succeeding the application of the second 100-mV step indicated by the asterisk in H, displayed at 100-fold higher time resolution, demonstrates that the channel is already blocked at the onset of the voltage step. Δt signifies the time interval at 0 mV.
TABLE 2.
Single-channel characteristics of KvLm PM reconstituted in monoolein bilayers
Single-channel current recordings in which only one channel was open at any given time were analyzed to determine the channel dwell times in the open and closed states. The experimental conditions were 0.5 m KCl, 10 mm Hepes, pH 7.4, recorded at 100 mV, and 27 mm KCl, 10 mm Hepes, pH 7.4, recorded at 150 mV. For τ1 closed and τ2 closed, the relative area under the fitted curve is indicated in parentheses and represents the frequency of occurrence of each state. The results are cumulative from three different experiments; error indicates S.E.
| Experimental condition | 0.5 m KCl | 27 mm KCl |
|---|---|---|
| Conductance (pS) | 68 ± 9 | 4 ± 2 |
| Open occupancy | 0.1 ± 0.1 | 0.3 ± 0.2 |
| Closed occupancy | 0.9 ± 0.1 | 0.7 ± 0.2 |
| τopen (ms) | 1.3 ± 0.3 | 12 ± 5 |
| τ1 closed (ms) | 1.8 ± 0.5 (0.62) | 13 ±5 (0.66) |
| τ2 closed (ms) | 350 ± 40 (0.38) | 113 ± 50 (0.34) |
| Burst length (ms) | 14 ± 3 | 70 ± 10 |
| Total openings analyzed | 20,200 | 35,202 |
DISCUSSION
Functional assays showing PM channel block at 0 mV by the open channel blockers tetrabutylammonium (36) (Fig. 4, D–F) and 4-aminopyridine (37) (data not shown) demonstrate that despite the crystal structure displaying an unequivocally closed activation gate, the reconstituted PM in lipid bilayers gates open at 0 mV long enough for the blocker to enter the cavity and abort permeation. This implies that the PM structure (a composite of 32 individual crystals) depicts a pore with a mobile activation gate able to open and close yet captured, on average, in a closed conformation. As such, the structure of KvLm PM represents the first structure of a Kv channel pore trapped with the activation gate closed. Comparison with other Kv channel structures that contain both the voltage sensor module and PM and exhibit an open PM implies that the voltage sensor module determines the stability of the open conformation of the PM (Fig. 3).
What is the significance of these data? Three main points emerge from our analysis: 1) The two new structures are the first structures of a channel obtained by crystallization of the protein in its native environment: a lipid bilayer. Specifically for Kvs, it represents the first module dissected out from a whole channel protein subunit that recapitulates function and structure in its native and identical environment: the epitome of a folded functional unit in a membrane. 2) The new structures of KvLm show for the first time a Kv channel pore in a closed conformation. The structural similarity of the KvLm pore to other reported K+ channel pores is indeed striking: they share the same fold and display a conformation of the two gates that has been depicted in another K+ channel pore that of KcsA. However, KcsA is not a Kv channel, nor is it a structural pore module, because both gating sensor and pore are present in the structure. Moreover, the closed structure of KcsA, determined at pH 7.5, has been demonstrated to be closed and stable in such conformation (38). In contrast, the pore of KvLm is indeed a module given that, in the absence of its gating sensor, it resides in an unstable closed conformation. This may appear at first sight as structurally similar to KcsA, yet it is, in essence, energetically distinct. A comparison with the two available structures of the eukaryotic Kv1.2 pore (Protein Data Bank codes 2R9R and 2A79) (5, 6) reveals another crucial difference: in KvLm, the pore gates are closed, whereas in the other two existing structures the pores are open. 3) The fundamental tenet behind the modular design of Kvs prompts the question of how do the sensor and pore modules interact? One way to answer this question about how the machine operates is to determine whether the structure of the pore alone, in the absence of an activating signal, is closed or open. The demonstration that, in the absence of sensor, the KvLm pore in a lipid bilayer resides in an unstable and functional closed state indicates that the closed and open structures are in close energetic equilibrium. The implication is that the work that the sensor module must do on its attached pore module to elicit the transition is limited. By the same token, it also demonstrates that to keep the channel stable open or closed, the sensor must play an important role. Therefore the new information explains how the two independent modules in a Kv channel interact to generate the observable signal.
Our findings provide compelling evidence that the structure of a K+ channel in lipidic cubic phase can be determined close to atomic resolution. The new findings give further support to nature's conservation of the pillar principles of protein science in the context of the unique environment conferred by the lipid bilayer for protein folding.
Acknowledgments
We thank Prof. Luis Cuello for insightful comments on the manuscript and Janet Smith, Robert Fischetti, and Nukri Sanishvili at the GM/CA-CAT beamline at the Advanced Photon Source for assistance in development and use of the minibeam and beamtime.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-073197 and GM-49711. This work was also supported by National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Sciences Grant Y1-GM-1104.
- Kv
- voltage-gated K+ channels
- KvLm
- a Kv from L. monocytogenes
- PM
- pore module
- TM
- transmembrane segment
- DM
- n-decyl-β-d-maltopyranoside
- ADA
- [(carbamoylmethyl)imino]diacetic acid.
REFERENCES
- 1. Bezanilla F. (2000) The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80, 555–592 [DOI] [PubMed] [Google Scholar]
- 2. Montal M. (1990) Molecular anatomy and molecular design of channel proteins. FASEB J. 4, 2623–2635 [DOI] [PubMed] [Google Scholar]
- 3. Montal M. (1995) Design of molecular function. Channels of communication. Annu. Rev. Biophys. Biomol. Struct. 24, 31–57 [DOI] [PubMed] [Google Scholar]
- 4. Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. (2003) X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41 [DOI] [PubMed] [Google Scholar]
- 5. Long S. B., Campbell E. B., MacKinnon R. (2005) Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903 [DOI] [PubMed] [Google Scholar]
- 6. Long S. B., Tao X., Campbell E. B., MacKinnon R. (2007) Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 [DOI] [PubMed] [Google Scholar]
- 7. Santos J. S., Grigoriev S. M., Montal M. (2008) Molecular template for a voltage sensor in a novel K+ channel. III. Functional reconstitution of a sensorless pore module from a prokaryotic Kv channel. J. Gen. Physiol. 132, 651–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caffrey M., Cherezov V. (2009) Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cherezov V., Peddi A., Muthusubramaniam L., Zheng Y. F., Caffrey M. (2004) A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr. D Biol. Crystallogr. 60, 1795–1807 [DOI] [PubMed] [Google Scholar]
- 10. Cherezov V., Hanson M. A., Griffith M. T., Hilgart M. C., Sanishvili R., Nagarajan V., Stepanov S., Fischetti R. F., Kuhn P., Stevens R. C. (2009) Rastering strategy for screening and centring of microcrystal samples of human membrane proteins with a sub-10 microm size x-ray synchrotron beam. J. R. Soc. Interface 6, (Suppl. 5) S587–S597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 12. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes. Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uysal S., Vásquez V., Tereshko V., Esaki K., Fellouse F. A., Sidhu S. S., Koide S., Perozo E., Kossiakoff A. (2009) Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. U.S.A. 106, 6644–6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cordero-Morales J. F., Cuello L. G., Zhao Y., Jogini V., Cortes D. M., Roux B., Perozo E. (2006) Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13, 311–318 [DOI] [PubMed] [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 18. Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Y., Morais-Cabral J. H., Kaufman A., MacKinnon R. (2001) Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 [DOI] [PubMed] [Google Scholar]
- 21. Lomize M. A., Pogozheva I. D., Joo H., Mosberg H. I., Lomize A. L. (2012) OPM database and PPM web server. Resources for positioning of proteins in membranes. Nucleic Acids Res. 40, D370–D376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jo S., Vargyas M., Vasko-Szedlar J., Roux B., Im W. (2008) PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 36, W270–W275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petrek M., Kosinová P., Koca J., Otyepka M. (2007) MOLE. A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure 15, 1357–1363 [DOI] [PubMed] [Google Scholar]
- 24. Bayley H., Cronin B., Heron A., Holden M. A., Hwang W. L., Syeda R., Thompson J., Wallace M. (2008) Droplet interface bilayers. Mol. Biosyst. 4, 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qin F., Auerbach A., Sachs F. (1996) Estimating single-channel kinetic parameters from idealized patch-clamp data containing missed events. Biophys. J. 70, 264–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clayton G. M., Altieri S., Heginbotham L., Unger V. M., Morais-Cabral J. H. (2008) Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel. Proc. Natl. Acad. Sci. U.S.A. 105, 1511–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tao X., Avalos J. L., Chen J., MacKinnon R. (2009) Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jiang Y., Lee A., Chen J., Cadene M., Chait B. T., MacKinnon R. (2002) Crystal structure and mechanism of a calcium-gated potassium channel. Nature 417, 515–522 [DOI] [PubMed] [Google Scholar]
- 29. Zhou Y., MacKinnon R. (2003) The occupancy of ions in the K+ selectivity filter. Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates. J. Mol. Biol. 333, 965–975 [DOI] [PubMed] [Google Scholar]
- 30. Armstrong C. M., Hille B. (1972) The inner quaternary ammonium ion receptor in potassium channels of the node of Ranvier. J. Gen. Physiol. 59, 388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu Y., Holmgren M., Jurman M. E., Yellen G. (1997) Gated access to the pore of a voltage-dependent K+ channel. Neuron 19, 175–184 [DOI] [PubMed] [Google Scholar]
- 32. Jogini V., Roux B. (2005) Electrostatics of the intracellular vestibule of K+ channels. J. Mol. Biol. 354, 272–288 [DOI] [PubMed] [Google Scholar]
- 33. Goldstein S. A., Pheasant D. J., Miller C. (1994) The charybdotoxin receptor of a Shaker K+ channel. Peptide and channel residues mediating molecular recognition. Neuron 12, 1377–1388 [DOI] [PubMed] [Google Scholar]
- 34. MacKinnon R., Cohen S. L., Kuo A., Lee A., Chait B. T. (1998) Structural conservation in prokaryotic and eukaryotic potassium channels. Science 280, 106–109 [DOI] [PubMed] [Google Scholar]
- 35. Lange A., Giller K., Hornig S., Martin-Eauclaire M.-F., Pongs O., Becker S., Baldus M. (2006) Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature 440, 959–962 [DOI] [PubMed] [Google Scholar]
- 36. Zhou M., Morais-Cabral J. H., Mann S., MacKinnon R. (2001) Potassium channel receptor site for the inactivation gate and quaternary amine inhibitors. Nature 411, 657–661 [DOI] [PubMed] [Google Scholar]
- 37. Armstrong C. M., Loboda A. (2001) A model for 4-aminopyridine action on K channels. Similarities to tetraethylammonium ion action. Biophys. J. 81, 895–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heginbotham L., LeMasurier M., Kolmakova-Partensky L., Miller C. (1999) Single streptomyces lividans K+ channels. Functional asymmetries and sidedness of proton activation. J. Gen. Physiol. 114, 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pauling L. (1960) The Nature of the Chemical Bond and the Structure of Molecules and Crystals: An Introduction to Modern Structural Chemistry, 3rd Ed., p. 513, Cornell University Press, Ithaca, NY [Google Scholar]