Background: Pharmaceutical application of human IgA requires a highly specific IgA purification system.
Results: A peptide affinity ligand for IgA was designed and optimized for affinity/specificity using randomized phage libraries and mutational studies.
Conclusion: The designed IgA-binding peptide has high affinity and specificity for human IgA.
Significance: This IgA-binding peptide can be used for specific purification of human IgA.
Keywords: Antibodies, Computer Modeling, Molecular Dynamics, Peptides, Phage Display, Protein Purification, IgA, T7 Phage, Peptide Library
Abstract
Phage display system is a powerful tool to design specific ligands for target molecules. Here, we used disulfide-constrained random peptide libraries constructed with the T7 phage display system to isolate peptides specific to human IgA. The binding clones (A1–A4) isolated by biopanning exhibited clear specificity to human IgA, but the synthetic peptide derived from the A2 clone exhibited a low specificity/affinity (Kd = 1.3 μm). Therefore, we tried to improve the peptide using a partial randomized phage display library and mutational studies on the synthetic peptides. The designed Opt-1 peptide exhibited a 39-fold higher affinity (Kd = 33 nm) than the A2 peptide. An Opt-1 peptide-conjugated column was used to purify IgA from human plasma. However, the recovered IgA fraction was contaminated with other proteins, indicating nonspecific binding. To design a peptide with increased binding specificity, we examined the structural features of Opt-1 and the Opt-1-IgA complex using all-atom molecular dynamics simulations with explicit water. The simulation results revealed that the Opt-1 peptide displayed partial helicity in the N-terminal region and possessed a hydrophobic cluster that played a significant role in tight binding with IgA-Fc. However, these hydrophobic residues of Opt-1 may contribute to nonspecific binding with other proteins. To increase binding specificity, we introduced several mutations in the hydrophobic residues of Opt-1. The resultant Opt-3 peptide exhibited high specificity and high binding affinity for IgA, leading to successful isolation of IgA without contamination.
Introduction
IgA is most abundantly produced in the human body, particularly in the mucous membrane, and plays an important role in mucosal immunity, which is the first line of defense against bacterial and viral invasion (1). This protein is also the second most abundant immunoglobulin in human plasma after IgG (2). IgA is secreted in the mucus as a dimer (secretory form of IgA) connected by a joining chain via intermolecular disulfide bridges, whereas the major form of IgA in plasma is monomeric (3). IgA has two subclasses: IgA1 and IgA2. The latter lacks the 13-amino acid proline-rich sequence present in the hinge region of IgA1 and therefore is more resistant to bacterial proteases (4).
Immunological studies on IgA have focused on vaccine development for mucosal immunity to control bacterial and viral infections at the mucosal site (5, 6). The Fc receptor for IgA (CD89 or FcαRI) has been identified and characterized (7, 8). This receptor is constitutively expressed on the surface of immune cells, including monocytes/macrophages, polymorphonuclear leukocytes, neutrophils, and eosinophils (9). The IgA receptor can mediate the various activities, including phagocytosis, oxidative burst, cytokine release, and antibody-dependent cellular cytotoxicity performed by these immune effector cells (10). Although IgG1 is popularly used as anti-cancer therapeutic antibody at present, IgA may also be a potential candidate for anti-tumor therapeutics given its antibody-dependent cellular cytotoxicity ability. Indeed, a recombinant IgA specific to tumor cell surface markers kills tumor cells in the presence of whole blood or neutrophils (11, 12). Furthermore, human IgA (hIgA)4 induces antibody-dependent cellular cytotoxicity more effectively against solid tumor cells or lymphoma than the relevant IgG (11, 13). These findings encourage the development of production methodologies, including cell culture (14) and purification systems, for the clinical use of IgA.
Several IgA purification methods have been reported. Jacalin, a lectin targeting IgA1-specific sugar (a galactosyl β-1,3-N-acetylgalactosamine) has been used as an affinity ligand for IgA purification (15). A conventional method using ion exchange and size exclusion chromatography has also been reported (16). However, these methods are not suitable for antibody purification on an industrial scale, like the protein A column for IgG purification. On the other hand, specific IgA-binding proteins have been identified in Staphylococcus group A (Staphylococcus pyogenes) and group B bacteria (17, 18). Typically, the M proteins from S. pyogenes, M22, Arp4, or Sir22, exhibit binding activities toward IgA1 and A2 or the monomeric and secretory forms of IgA. These proteins recognize the interdomain region between CH2 and CH3 of IgA-Fc and can inhibit the interaction between IgA and IgA receptor (19–22). A simple and highly efficient affinity purification method using IgA-binding proteins was previously described (16). However, the use of bacterial proteins for pharmaceutical antibody purification requires careful attention to prevent contamination with endotoxin or bacterial proteins, given their highly toxic and antigenic nature, as described for the protein A/G column used in IgG purification for pharmaceutical use (23).
To solve this problem, synthetic low molecular weight ligands, such as TG19318 (24) and protein A mimetic compounds (25), have been investigated. However, these compounds still suffer from insufficient specificity and affinity for use in IgA purification. Sandin et al. (26) reported the purification of hIgA using a synthetic peptide comprising 50 residues extracted from the IgA-binding domain (Sap) of the Sir22 M protein. Using a dimerized form of this peptide (peptide M), Sandin et al. successfully detected and purified hIgA; however, the dimerized peptide of 100 residues is too large for industrial applications.
Here, we report a novel hIgA-binding peptide that was isolated from random peptide T7 phage libraries by biopanning against hIgA. The essential residues in the peptide were identified, and the peptide was optimized for affinity/specificity. Our peptide is only 16 residues long and exhibits high specificity/affinity for hIgA, which is suitable for hIgA purification.
EXPERIMENTAL PROCEDURES
Materials
Polyclonal hIgA1/IgA2, IgE, and IgG were purchased from Acris Antibodies GmbH (Herford, Germany), Athens Research & Technology (Athens, GA), and ICN/Cappel Biomedicals (Aurora, OH), respectively. Anti-HER2 IgG1 humanized antibody, Trastuzumab (Herceptin), and anti-human IL13-specific hIgA2 were from Chugai Pharmaceutical Corp. Ltd. (Tokyo, Japan) and Invivogen (San Diego, CA). The recombinant human FcαR/CD89 was obtained from R&D Systems (Minneapolis, MN). Mouse IgG and IgE were from PharMingen (San Diego, CA).
Construction of the T7 Phage Display Library and Biopanning
The T7 phage libraries displaying typically X3CX7–10CX3 random peptides, where X represents the randomized amino acid positions generated using mixed oligonucleotides on template DNA, were constructed using the T7 Select vector 10–3b from Merck (Tokyo, Japan), according to methods described previously (27).
Microplate wells (Nunc Maxisorp) were coated with polyclonal hIgA (1 μg/300 μl/well) and blocked with 0.5% BSA in PBS. The T7 phage libraries (5 × 1010 pfu) of X3CX7–10CX3 were incubated for 1 h in wells coated with hIgG and HSA to remove nonspecific phages and were then added to hIgA-coated wells. After incubation for 1 h, the plate was washed 5–30 times with PBS containing 0.1% Tween 20 (PBST). Escherichia coli BLT5615 cells (300 μl) (Novagen) in log phase growth were added to the wells, infected with phages for 10 min, and propagated in 2TY medium at 37 °C. After bacteriolysis, the phages were recovered from the culture supernatant by centrifugation (15,000 rpm for 10 min). The recovered phage solution was used for the next round of biopanning.
Preparation of Synthetic Peptides
Synthetic peptides were prepared by solid phase synthesis using Fmoc chemistry. All peptides were C-terminally amidated. After removal of the protecting groups, the peptides were mildly oxidized to form intramolecular disulfide bonds. The generated disulfide-constrained peptides were purified by reversed phase HPLC. After lyophilization, the peptides were dissolved in the appropriate buffers and used for assay after centrifugation. Amino or biotinylated PEG spacer-armed peptides were chemically synthesized by coupling the protected peptides on the resin with N-Fmoc-amido-PEG4-COOH and biotin. Purity and disulfide bond formation of the peptides were confirmed by mass spectrometry on the Acquity SQD ultra performance liquid chromatography system (Waters Corp., Milford, MA).
Detection of Phages or Synthetic Peptide Binding to hIgA by ELISA
The wells of a microplate (Nunc Maxisorp) were coated with hIgA and other proteins (50g/50 μl/well) and blocked with 0.25% BSA in PBS. Phage solution was added to each well and incubated for 2 h. After washing the plate, the bound phages were detected with biotinylated anti-T7 phage antibody (Novagen) and HRP-conjugated streptavidin (Vector Laboratories, Peterborough, UK) or alkaline phosphatase-conjugated streptavidin (Vector Laboratories) using the detection reagents tetramethylbenzidine (Wako Pure Chemicals) and p-nitrophenyl phosphate (Wako Pure Chemicals), respectively.
For peptide binding, biotinylated peptide (40 nm) was preincubated with HRP-conjugated streptavidin (10 nm) to form a tetrameric peptide complex. The mixture was added to hIgA-coated wells in a plastic plate. After 1 h of incubation, the wells were washed five times with PBST, and binding was detected with tetramethylbenzidine reagent.
Purification of hIgA from Human Plasma
Approximately 500 nmol of amino PEG4-Opt1 or -Opt3 peptide were immobilized on a HiTrap NHS-activated HP column (1 ml; GE Healthcare) according to the manufacturer's instructions. Heparin-treated human plasma was injected into the column (1 ml), which was connected to Profinia purification system (Bio-Rad). After washing the column with PBS, hIgA was eluted with 0.1 m glycine-HCl (pH 2.5). The column was re-equilibrated with PBS and stored at 4 °C until use.
For analysis of eluted fractions, the samples were mixed with SDS sample buffer and subjected to SDS-PAGE on a 4–20% gradient gel. After electrophoresis, the gel was stained with GelCode Blue Stain reagent (Thermo Scientific).
Surface Plasmon Resonance (SPR) Analysis
SPR analysis was performed on a BIAcore T100 (GE Healthcare) at 25 °C. All reagents and sensor chips were purchased from GE Healthcare. IgA1 and IgA2 were immobilized on a CM5 sensor chip using the amine coupling protocol, according to the manufacturer's instructions. The amount of the immobilized IgA was adjusted to fall within 5000–6000 response units. The association reaction was monitored by injecting the peptides into the sensor chip at a flow rate of 50 μl/min for 180 s. The dissociation reaction was performed by running the HBS-EP buffer (10 mm HEPES, pH 7.4, containing 400 mm NaCl, 3 mm EDTA, and 0.005% Tween 20). The binding kinetic parameters were calculated using the BIAcore T100 evaluation software (GE Healthcare).
Molecular Dynamics (MD) Simulations and Trajectory Analysis
All MD simulations on the Opt-1 peptide and its complex with hIgA-Fc were performed using the SANDER module of the AMBER 10 simulation package (28) with the force field ff99SB (29). Each system was explicitly solvated with TIP3P water molecules and neutralized by counter ions. The SHAKE algorithm was applied to constrain all bonds linking hydrogen atoms, and the particle mesh Ewald method was used for treating long range electrostatic interactions. For Opt-1 simulation, the system was subjected to 1000 steps of steepest decent minimization followed by 1000 steps of conjugate gradient minimization with 500 kcal·(mol·Å2)−1 harmonic constrains on the peptide to remove unfavorable van der Waals contacts. Then the whole system was minimized using 5000 steps of steepest decent minimization without harmonic restraints. The system was gradually heated from 0 to 310 K over 20 ps for equilibration. Production runs were carried out for 100 ns with 2-fs steps. A total of six independent MD trajectories were generated from various starting structures, i.e., three unfolded conformations and three β-hairpin conformations.
For simulation of the Opt-1 complex with hIgA-Fc, the initial structure for the complex was modeled with the simulated Opt-1 structure and x-ray-determined hIgA-Fc structure (Protein Data Bank code 2QEJ). To remove unfavorable van der Waals contacts, the system was subjected to 6000 steps of steepest decent minimization followed by 6000 steps of conjugate gradient minimization, whereas the complex was constrained by 500 kcal·(mol·Å2)−1 harmonic potential. The whole system was then minimized using 20,000 steps of steepest decent minimization without harmonic restraints. The system was gradually heated from 0 to 310 K over 50 ps for equilibration. Then the production run was carried out for 5 ns with 2-fs steps. A total of three independent simulations were performed for the complex with various relative orientations between Opt-1 and hIgA-Fc. All of the MD simulations were carried out with the NPT ensemble, i.e., a constant number of particles (N), pressure (P), and temperature (T), and the trajectories were recorded every 2 ps.
RESULTS
Isolation of hIgA-specific Phage Clones from Phage-displayed Random Peptide Libraries
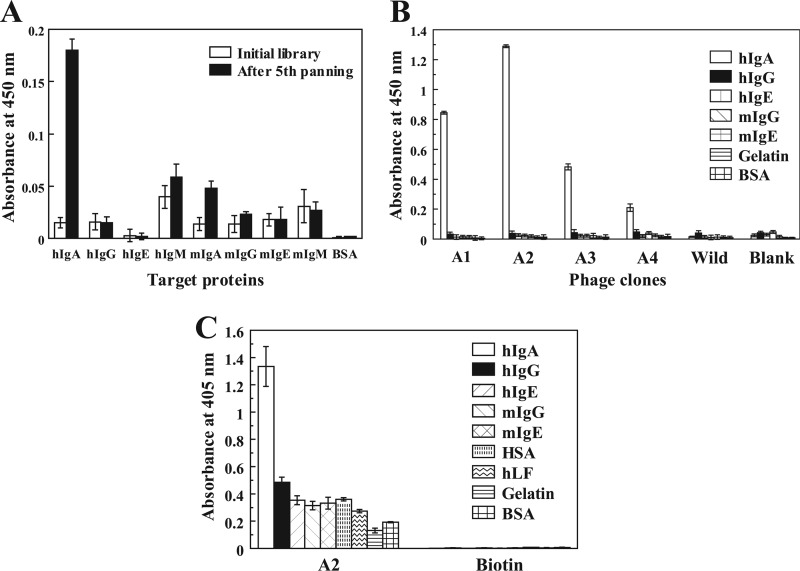
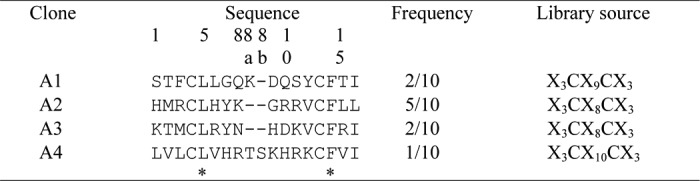
We chose disulfide-constrained cyclic peptide libraries to isolate the specific peptides, because it is generally known that cyclic peptides have higher affinity than its linear form because of the reduction of conformational chain entropy (30). Five rounds of biopanning were performed against hIgA using T7 phage-displayed random peptide libraries of X3CX7–10CX3, where X represents randomized amino acid positions. The binding activities of the phages after the fifth round of biopanning are shown in Fig. 1A. Compared with the initial library, the phages after biopanning clearly exhibited increased binding to hIgA but not other immunoglobulins or BSA. The phages were cloned and screened using ELISA. Among the 20 clones isolated, 10 clones exhibited binding to hIgA. Sequence analysis of their displayed peptides revealed four individual motifs (A1–A4), as shown in Table 1. A low similarity, involving the two conserved residues Leu-5 and Phe-14 but not the two fixed cysteine residues, was found among them. The binding specificities of the selected clones are shown in Fig. 1B. All four clones exhibited binding specificities toward hIgA.
FIGURE 1.
Isolation of hIgA-specific phage clones from T7 phage-displayed random peptide libraries. A, binding specificities of the phages from the initial library and the X3CX7–10CX3 library enriched by five rounds of biopanning against hIgA were examined by ELISA. B, binding specificities of the four phage clones (A1–A4) isolated by biopanning were confirmed by ELISA. C, binding specificities of the A2 synthetic peptide were examined by ELISA. hIg, human immunoglobulin; mIg, mouse immunoglobulin; HSA, human serum albumin; hLF, human lactoferrin.
TABLE 1.
Comparison of the amino acid sequences of hIgA-binding peptides
The amino acid positions are numbered on the basis of the X3CX8CX3 sequence. The inserted positions in the A1 and A4 clones are labeled as 8a and 8b, based on the alignment of the four sequences. Asterisks indicate completely conserved positions.
The synthetic peptide derived from the A2 motif (A2 peptide) was prepared, and its affinity for hIgA was analyzed by SPR. The equilibrium constant for the dissociation (Kd) between the A2 peptide and hIgA2 immobilized on the CM5 sensor chip was estimated to be 1.3 μm, which is not sufficient for an affinity ligand. The specificity of this peptide evaluated by ELISA indicated a distinct binding to hIgA but with a somewhat high background (Fig. 1C).
Identification of A2 Motif Residues Important for hIgA Binding
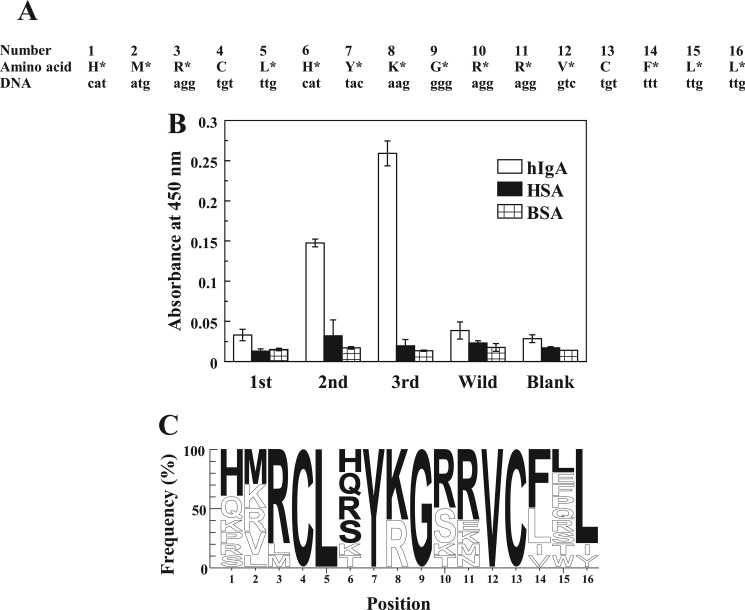
To characterize the peptide motifs for IgA binding, the important residues in A2 motif were identified using a second library constructed on the basis of the A2 motif. The partially randomized gene library of the A2 motif was generated by synthesizing the gene using mixed nucleotides comprising 70% authentic nucleotides of the A2 motif and 10% each of the other three nucleotides (Fig. 2A), which led to the appearance of authentic amino acids in the A2 motif at frequencies of ∼34–49%. This second library was used for biopanning (Fig. 2B), and the phages cloned after three rounds of biopanning were screened for hIgA binding. Ten binding clones were selected and sequenced (Fig. 2C). Sequence comparison clearly revealed the complete conservation of Leu-5, Tyr-7, Gly-9, and Val-12, indicating that they were essential for binding.
FIGURE 2.
The second library used to identify the important residues for hIgA binding. A, the library design to identify the important residues for binding is shown. Partial random mutations were introduced at the sites (other than the two Cys) indicated by the asterisks by generating a synthetic gene for the library using mixed nucleotides containing 70% of the indicated (authentic) nucleotide and 10% each of the other three nucleotides. B, binding specificities of the phages after the first, second, and third biopanning were examined by ELISA. Blank indicates measurement without the phages. Wild indicates wild-type phages with no displaying peptides. C, the peptide sequences of the clones isolated after three rounds of biopanning are represented as a logo plot. The height of each character of amino acid indicates its appearing frequency, and the black characters represent the most frequently appearing amino acid at each position. Sequence logos were done using WebLogo.
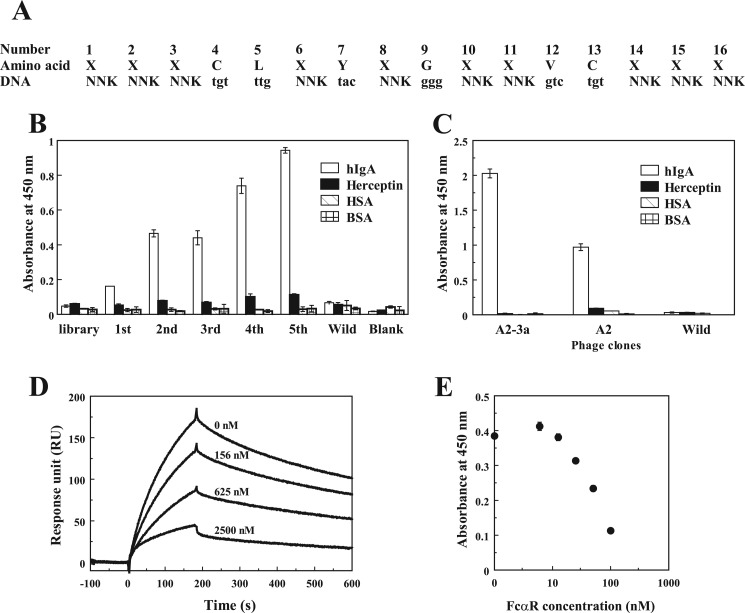
Construction of the Third Library for Affinity Improvement
We then attempted to improve the affinity of the A2 peptide by using a third library, which was designed to fix the four conserved residues Leu-5, Tyr-7, Gly-9, and Val-12 and randomize the other amino acid positions (except the two cysteines) (Fig. 3A). The third library was biopanned against hIgA under strict conditions. After repeated biopanning, hIgA-binding phages were clearly enriched (Fig. 3B). The phages after the fifth round of biopanning were cloned and evaluated for binding. Among the 80 selected clones, 29 strong binders were selected and subjected to sequence analysis. The most abundant clone A2-3a exhibited an apparent increase in hIgA binding activity compared with the original A2 clone (Fig. 3C). Indeed, the synthetic peptide of A2-3a exhibited a 2.6-fold stronger binding (Kd = 0.5 μm) than the A2 peptide (Kd = 1.3 μm).
FIGURE 3.
The design of the third library and specific phage selection for affinity maturation. A, the third library designed for affinity maturation is shown. The codons of the conserved residues in Fig. 2A were fixed, and the others were randomized using NNK-mixed oligonucleotides. B, the third library was biopanned against hIgA. Specific phages were enriched by repeated biopanning, and the binding of the phages in each round was measured by ELISA. C, binding of the isolated phage clones A2-3a and A2 to hIgA was examined by ELISA. D, the interactions between hIgA and the recombinant FcαR immobilized on a sensor chip were examined by SPR analysis in the presence of different concentrations of the A2-3a peptide (0–2,500 nm). E, inhibition of the binding between A2-3a peptide and IgA by FcαR. Biotinylated peptide/streptavidin-HRP complex was reacted with hIgA coated on ELISA plates in the presence of different concentrations of FcαR (0–100 nm).
To obtain information on the binding site of the peptide, the inhibitory effect of the A2-3a peptide on the binding between Fcα receptor (FcαR) and hIgA was examined by SPR analysis. The binding of IgA to FcαR immobilized on a sensor chip was decreased by the A2-3a peptide in a dose-dependent manner (Fig. 3D). This inhibitory activity of the peptide was confirmed by competitive ELISA, as shown in Fig. 3E. These results indicate that the binding site of the A2-3a peptide is located in the marginal region between CH2 and CH3 of IgA-Fc, which is a common binding site for bacterial IgA-binding proteins as well as FcαR (31, 32).
Confirmation of the Roles of the Conserved Residues of A2-3a
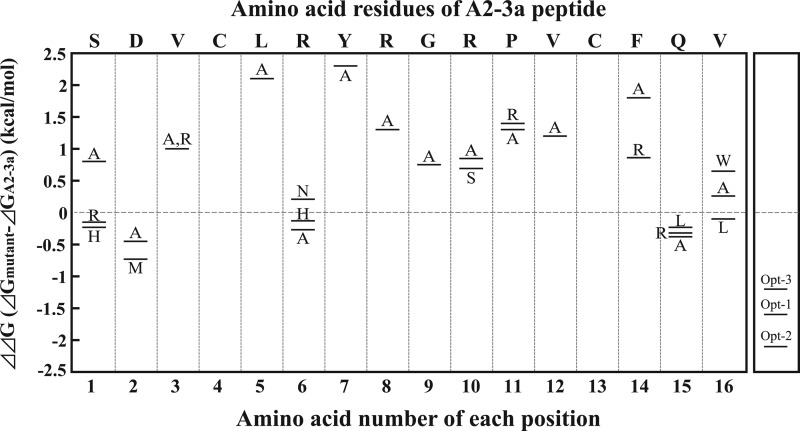
As shown in Fig. 2C, four conserved residues, Leu-5, Tyr-7, Gly-9, and Val-12, were found among the peptide motifs selected from the second library. To confirm their roles in binding, Ala mutations were introduced in the A2-3a synthetic peptide. The Kd values and the changes in the binding free energy (ΔΔG) are summarized in Table 2 and Fig. 4. Ala mutations of the four conserved residues resulted in large decreases in binding energy (0.75–2.3 kcal/mol), suggesting the critical roles of these conserved residues in binding.
TABLE 2.
Changes in the affinity and binding energy of hIgA-binding peptides following Ala scanning or additional mutations for affinity maturation
All the peptides used were C-terminally amidated. The binding affinity between the peptide and hIgA was measured at 25 °C and pH 7.0 on BIAcore T100 (GE Healthcare). ΔΔG indicates the difference in the binding free energy between A2-3a peptide and its derivatives. The underlined mutations were additively introduced into the A2-3a sequence to design the Opt-1 peptide. The bold type in the sequences of Opt-1 and Opt-2 represents the mutations added to the A2-3a sequence for affinity improvement.
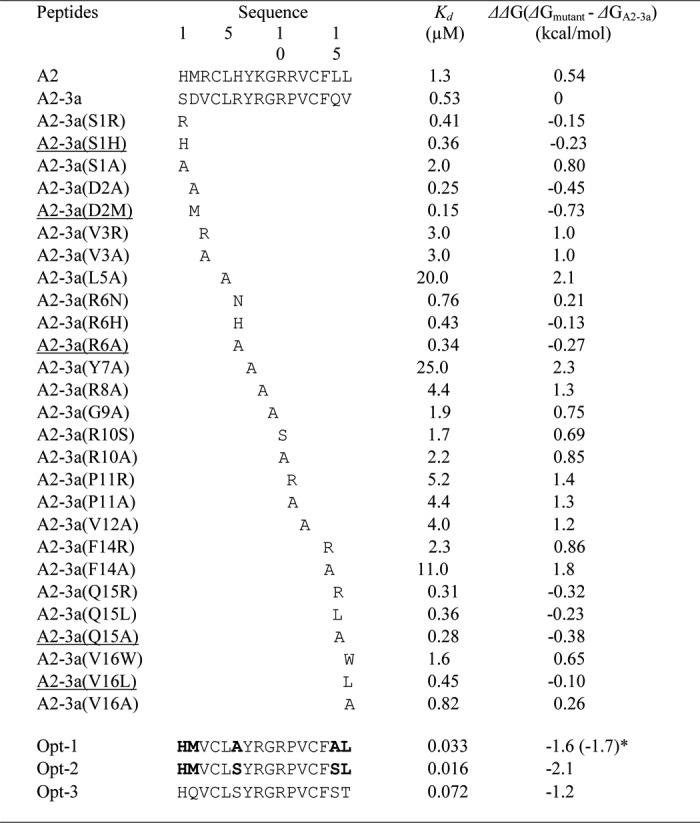
FIGURE 4.
The schematic view of the changes in the binding energy of the mutational peptides. The changes of binding free energies by the mutations on A2-3a peptide summarized in Table 2 were plotted against amino acid positions.
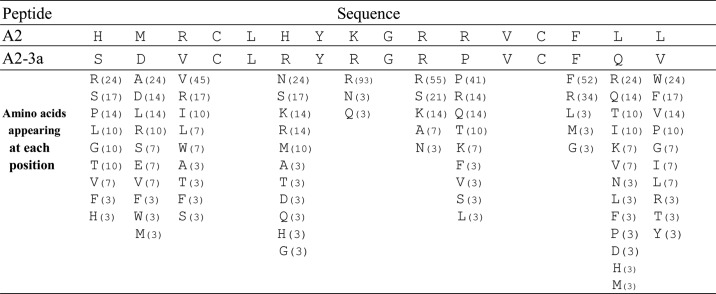
Table 3 shows the frequency (%) with which each amino acid appeared at the randomized positions of the peptide motifs obtained from the third library. We assumed that the amino acids appearing at a high frequency at each position would contribute to the affinity. To confirm this, the highly frequent residues (>41%) Val-3, Arg-8, Arg-10, Pro-11, and Phe-14 were mutated to Ala. As shown in Table 2 and Fig. 4, the Ala mutations elicited large decreases (0.8–1.8 kcal/mol) in binding energy, suggesting their critical roles in IgA binding.
TABLE 3.
The frequencies of amino acids at each randomized site appearing in the clones selected from the third library
The numbers in parentheses indicates the frequencies (%) of amino acids appearing at each position.
Affinity Maturation of A2-3a by Additional Substitutions in Less Conserved Sites
We further examined the roles of the residues at positions 1, 2, 6, 15, and 16. The substitutions of Arg-6 and Val-16 with Asn and Trp (both appearing at a frequency of 24%) decreased the binding energy by 0.21 and 0.65 kcal/mol, respectively. On the other hand, the substitutions of Ser-1, Asp-2, and Gln-15 with the corresponding high frequent residues (Arg, Ala, and Arg) slightly increased the binding energy by 0.15, 0.45, and 0.32 kcal/mol, respectively. We also found that substitutions of Ser-1, Asp-2, and Val-16 with the amino acids on the A2 motif (His, Met, and Leu) slightly increased the binding energy by 0.23, 0.73, and 0.1 kcal/mol, respectively. Furthermore, the most favorable mutations at Arg-6 and Gln-15 positions were Ala mutations, which increased the binding energy by 0.27 and 0.38 kcal/mol, respectively.
Based on the above mutational studies, the Opt-1 peptide was designed by combining the mutations suitable for affinity improvement (S1H, D2M, R6A, Q15A, and V16L). This Opt-1 peptide exhibited a high affinity for hIgA2 with a Kd of 33 nm, which is 16- and 39-fold lower than those of A2-3a and A2 peptides, respectively.
Application of the Opt-1 Peptide in IgA Affinity Purification
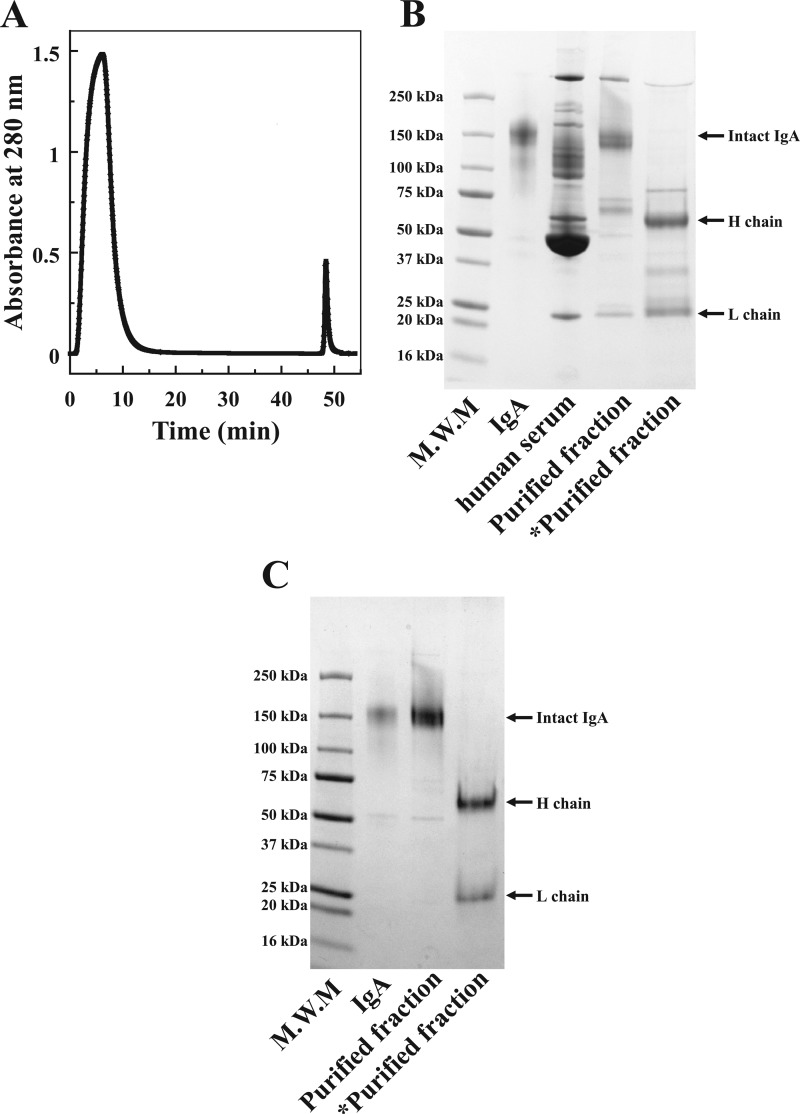
To examine the utility of the Opt-1 peptide in IgA purification, we prepared an Opt-1 peptide-conjugated column by immobilizing the amino PEG4 spacer-armed Opt-1 peptide on a HiTrap NHS-activated HP column (1 ml) using amine-coupling protocol. The column was used to purify IgA from human plasma. The eluted fraction from the column was subjected to SDS-PAGE, followed by protein staining to evaluate the purity of IgA (Fig. 5, A and B). However, the eluted fraction from the Opt-1 column contained other proteins besides IgA, indicating the occurrence of nonspecific interactions.
FIGURE 5.
Purification of hIgA from human plasma using peptide-conjugated column. A, affinity chromatography for hIgA purification from human plasma on Opt-1 peptide-conjugated column. B and C, SDS-PAGE of the fractions eluted from the column of Opt-1 (B) and Opt-3 peptide (C). Asterisks indicate the samples reduced with 2-mercaptoethanol. M.W.M., the molecular weight maker.
MD Simulations of the Opt-1 Peptide
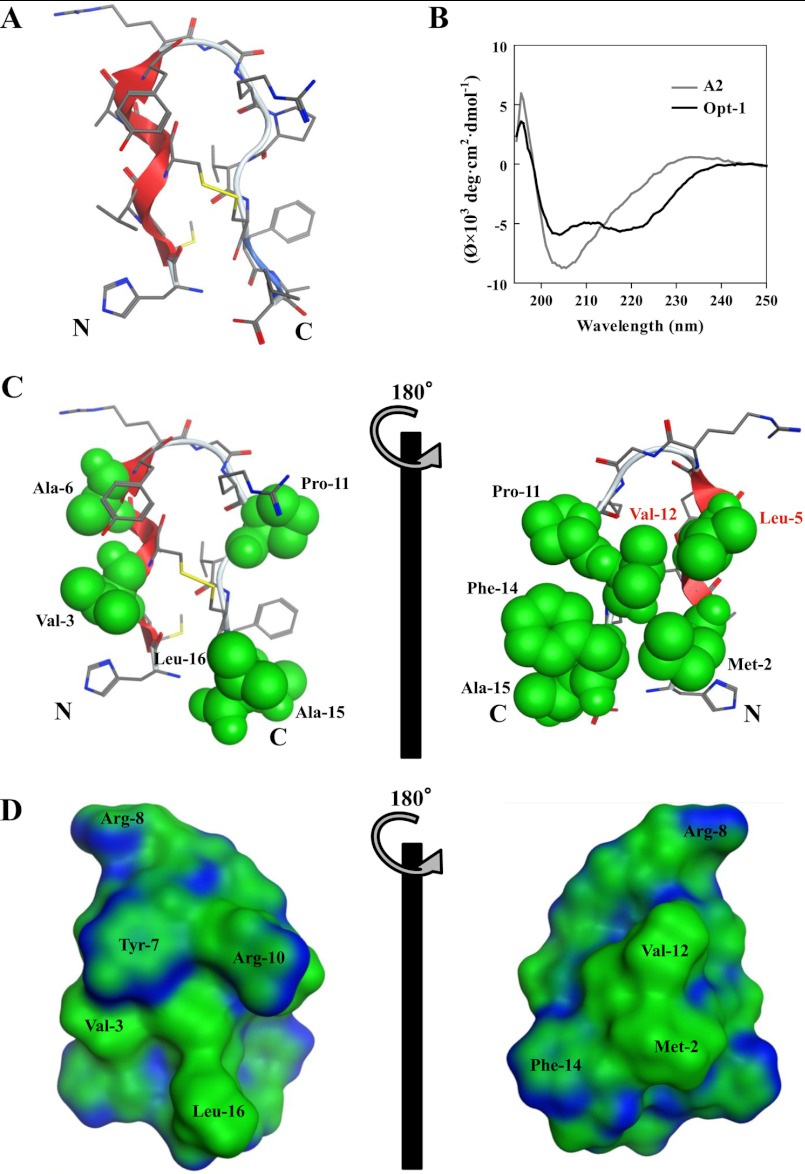
To elucidate the structural features of the Opt-1 peptide for designing a peptide with increased specificity, we performed fully atomistic, explicit water MD simulations of the Opt-1 peptide, starting from various random conformations at 310 K. Each MD simulation was conducted for 100 ns. The equilibrium structure was characterized by the final 20-ns structures of each MD trajectory. One structure from a representative trajectory is shown in Fig. 6A. Interestingly, the Opt-1 peptide displayed a partial helical conformation in the N-terminal region spanning residues 2–7, in good agreement with the experimental CD spectrum (Fig. 6B). Helix-favoring residues in the N terminus, i.e., Met-2, Leu-5, Ala-6, and Tyr-7, contribute to helical structure formation, and the helical content was computed to be 18%. As shown in Fig. 6 (C and D), the hydrophobic residues Met-2, Leu-5, Pro-11, Val-12, and Phe-14 are oriented toward the same side of the peptide, forming a hydrophobic cluster.
FIGURE 6.
Structural characterization of Opt-1peptide. A, the equilibrium structure of Opt-1 derived from MD simulations at 310 K. The side chain and main chain are displayed in line representation. Gray, carbon; red, oxygen; yellow, sulfur; blue, nitrogen. B, CD spectrum of the A2 and Opt-1 peptide. Peptides were diluted in phosphate buffer containing 8.1 mm Na2HPO4, 1.5 mm KH2PO4, 2.7 mm KCl, and 68 mm NaCl (pH 7.4) and measured on a JASCO J-820 spectropolarimeter (Jasco Corp., Tokyo, Japan) at 25 °C. C, the locations of hydrophobic residues on both the surfaces of Opt-1. The hydrophobic residues are shown in green with a space filling model. The red characters indicate the important residues for IgA binding. D, surface map of Opt-1.
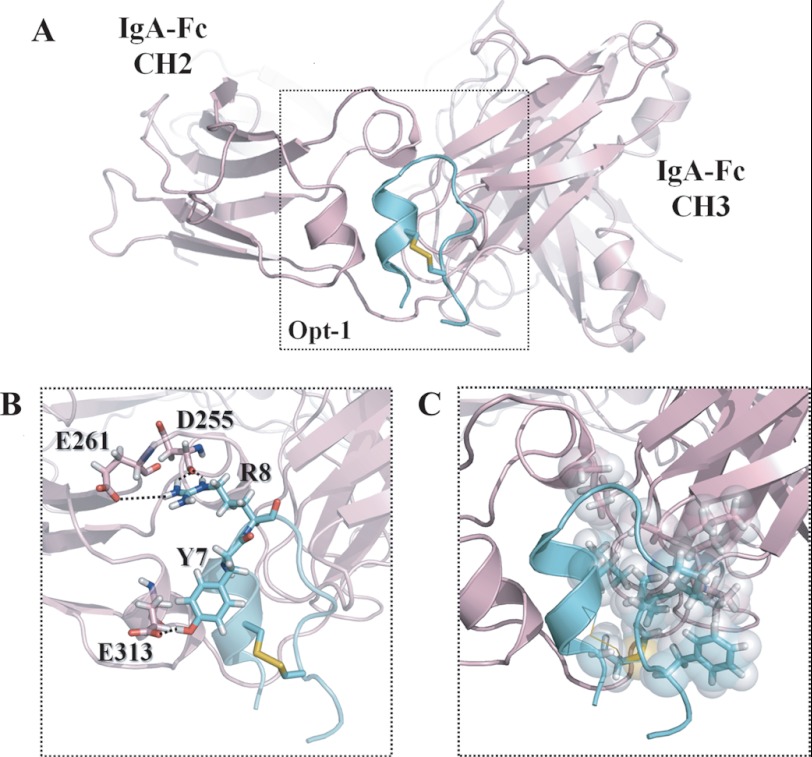
MD Simulations of the Opt-1-IgA-Fc Complex
We performed further MD simulations to examine the structure of the complex, as well as the binding modes of the Opt-1 peptide with IgA-Fc. Based on the above finding that the binding site of the A2-3a peptide is located in the marginal region between CH2 and CH3 of IgA-Fc, we constructed various initial conformations of the complex by placing the Opt-1 peptide near this common binding site of IgA-Fc. Fig. 7 shows the simulated structure of the complex and the binding modes of the Opt-1 peptide with IgA-Fc. As shown in Fig. 7B, Tyr-7 and Arg-8 of Opt-1 significantly contribute to the binding by forming H-bonds with Asp-255, Glu-261, and Glu-313 of IgA-Fc. Additionally, the hydrophobic interactions between Met-2, Leu-5, Pro-11, Val-12, and Phe-14 of Opt-1 and Leu-257, Ala-438, Pro-440, Leu-441, and Ala-442 of IgA-Fc (Fig. 7C) play an important role in tight binding to form the complex. Importantly, the conserved residues of Opt-1, i.e., Leu-5, Tyr-7, and Val-12, play a significant role in IgA-Fc recognition.
FIGURE 7.
The complex structure of Opt-1 and IgA-Fc. A, structure of the complex and binding modes of Opt-1 peptide (cyan) and IgA-Fc (light pink) derived from MD simulations at 310 K. B, electrostatic interactions in the complex. H-bonding is represented by black dotted lines. C, hydrophobic interactions between the Opt-1 peptide and IgA-Fc. Residues (Opt-1: Met-2, Leu-5, Pro-11, Val-12, and Phe-14; IgA-Fc: Leu-257, Ala-438, Pro-440, Leu-441, and Ala-442) contributing to hydrophobic interactions are shown in stick as well as sphere representation.
Improvement of Opt-1 Peptide Specificity
Based on the above MD simulation results, most of the hydrophobic residues in Opt-1 are located on the peptide surface exposed to water (Fig. 6, C and D) and are beneficial for tight binding to IgA-Fc (Fig. 7C). However, these hydrophobic residues would also contribute to the nonspecific binding to other proteins, as shown in Fig. 5B. Therefore, to reduce the hydrophobicity of Opt-1 without perturbing the binding affinity, we designed the Opt-2 peptide by introducing two mutations, A6S and A15S, in the Opt-1 peptide. Interestingly, the Opt-2 peptide exhibited an affinity for hIgA with a Kd of 16 nm (supplemental Fig. S1A), which is higher than the Opt-1 peptide (Kd = 33 nm), demonstrating that the Opt-2 peptide is the strongest binder among our peptides. However, the nonspecific binding to other proteins was observed in ELISA and in the purification of IgA from human plasma on Opt-2 peptide-conjugated column (supplemental Fig. S1, B and C).
To improve the specificity of the Opt-2 peptide, the Opt-3 peptide was designed by introducing the two mutations M2Q and L16T into the Opt-2 peptide. The design strategy was to minimize the perturbation of the binding affinity and maximize the specificity based on the MD-simulated structures shown in Figs. 6 and 7. Notably, the Opt-3 peptide exhibited not only high binding affinity for IgA2 (Kd = 72 nm) but also demonstrated specific binding to IgA in the peptide-conjugated column, indicated by a single band with a molecular mass of 150,000 on SDS-PAGE after purification from human plasma (Fig. 5C).
DISCUSSION
Several bacterial IgA-binding proteins, such as the Staphylococcus aureus superantigen-like protein 7 (33) and the Sap peptide (a peptide from the IgA-binding domain of the streptococcal M protein) (26), have been used as affinity ligands for IgA purification. These molecules, the Fcα receptor, and our peptide share the binding site on IgA (31, 32) (Fig. 3, D and E). The interdomain region contains the hydrophobic surface and may therefore represent a hotspot region commonly targeted or recognized by many IgA-binding molecules.
Our peptides bound both IgA1 and IgA2 (supplemental Fig. S1B), similar to SSL7 and the Sap peptide. The IgA purified from human saliva using our system exhibited a peak of the dimeric secretory form of IgA with a molecular mass of 400,000 (supplemental Fig. S2). These results indicate that our peptide is suitable for the purification of both subclasses/subtypes of IgA from various body fluids, in the same way as bacterial IgA-binding proteins or their derived peptides.
The most distinctive characteristic of our peptide is its low molecular mass. In contrast to SSL7 (molecular mass of ∼23,000) and the Sap peptide (molecular mass of ∼10,000), our peptide is composed of only 16 amino acids (molecular mass of ∼1,800). Despite the low molecular mass, Opt-3 exhibited sufficient functionality in IgA purification. It is known that bacterial IgA-binding proteins, such as SSL7 or M22, possess the ability to bind to other proteins such as complement C5 (33) or its regulator protein C4b-binding protein (34), which activate or perturb the human complement system (19). Therefore, contamination of the purified IgA with these proteins should be avoided, particularly for pharmaceutical use. In this regard, our peptide is highly advantageous for IgA purification for pharmaceutical use, given its small molecular size.
The first isolated peptides (A1–A4) shared only two conserved residues, Leu-5 and Phe-14 (Table 1). Therefore, we sought to identify the important residues for IgA binding using the second library (Fig. 2). Four residues, Leu-5, Tyr-7, Gly-9, and Val-12, were identified as important for binding (Fig. 2C), which was confirmed by Ala mutations of these residues (Table 2 and Fig. 4). The third library was used to improve the affinity of the A2 peptide. The strict selection conditions enabled us to isolate a better binder, A2-3a, and analysis of the other motifs revealed additional critical residues for binding. The residues Val-3, Arg-8, Arg-10, Pro-11, and Phe-14, which appeared at a frequency of >40% (Table 3), largely contributed to IgA binding with the free energy of 0.85–1.8 kcal/mol (Table 2 and Fig. 4). However, substitutions of residues at less conserved sites (positions 1, 2, 6, 15, and 16) yielded somewhat controversial results (Table 2 and Fig. 4). The S1R, D2A, and Q15R substitutions in A2-3a enhanced the binding energy by 0.15, 0.45, and 0.32 kcal/mol, respectively, but R6N and V16W reduced the binding energy by 0.21 and 0.65 kcal/mol, respectively. This suggests that the highly frequent residues have the tendency to contribute to the binding, whereas the residues appearing with low frequency (<24%) do not necessarily enhance affinity, probably because of unfavorable conflicts or contradictory effects of the other substitutions.
The Opt-1 peptide was designed by combining the different substitutions (S1H, D2M, R6A, Q15A, and V16L), which contributed to affinity improvement and increased the binding energy by 1.6 kcal/mol. Interestingly, the sum of the binding energies resulting from all the substitutions in the A2-3a peptide is 1.7 kcal/mol. This indicates that each substitution independently contributes to the binding. To further characterize the structure and binding properties of Opt-1, we performed MD simulations of the solution structure of Opt-1 and its complex with IgA-Fc. Notably, Opt-1 displayed a helical structure (Fig. 6A) that is consistent with its CD spectrum (Fig. 6B). The residual helical conformation in the N terminus of Opt-1 may play an important role in recognizing IgA. Indeed, the A2 peptide with low affinity did not exhibit helical properties in the CD spectrum (Fig. 6B).
On the other hand, to confirm the importance of the intramolecular disulfide bond, the linear form of Opt-1 (Opt-1L), Cys-4 and Cys-13 of which were substituted with Ser, was prepared. The affinity of Opt-1L peptide was reduced ∼750 times as compared with Opt-1, indicating the intramolecular cross-link is essential for high binding. The CD spectrum of Opt-1L peptide indicated the loss of the helical property (data not shown), which probably caused the large decrease of the affinity.
Opt-1 exhibited nonspecific interactions in the purification of IgA from plasma (Fig. 5B). To solve this problem, the hydrophobicity of the peptide was reduced by Ala/Ser substitutions. Surprisingly, the designed Opt-2 peptide exhibited a higher affinity (Kd = 16 nm) than Opt-1 (Kd = 33 nm), although the specificity was not improved (supplemental Fig. S1C). We speculate that the electrostatic interaction between Ser6 of Opt-2 and IgA may contribute to the tighter binding of Opt-2 compared with Opt-1. To resolve nonspecific binding, we designed the Opt-3 peptide by introducing additional hydrophilic substitutions (M2Q and L16T) in Opt-2. Notably, the Opt-3 peptide elicited specific purification of IgA without any contamination with other plasma proteins, and the binding affinity was comparable with that of Opt-1 and Opt-2. These results suggest that the hydrophobic residues in the peptide contribute to tight binding with IgA, whereas the hydrophilic residues may modulate the specificity toward the target protein.
In summary, we successfully developed an IgA-binding peptide from A2 to Opt-3 with high binding affinity, as well as specificity. This was achieved through the identification of the essential/important residues for IgA binding using phage libraries, subsequent confirmation of their roles, and affinity/specificity maturation by combining the substitutions in the synthetic peptides. Our novel IgA-binding peptide is not only compact but also highly functional as an affinity ligand. Furthermore, the strategy of combining a phage library with a synthetic peptide is a conventional and efficient way to engineer functional peptides.
Supplementary Material
This work was supported by funds from the Japan Science and Technology Agency. This work was also supported by the Basic Science Research Program of the National Research Foundation of Korea funded by Ministry of Education, Science and Technology Grant 2011-0012096 and by Japan Society for the Promotion of Science Grant-in-Aid 238036 (to T. H.).

This article contains supplemental Figs. S1 and S2.
- h
- human
- MD
- molecular dynamics
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- SPR
- surface plasmon resonance
- FcαR
- Fcα receptor.
REFERENCES
- 1. Fagarasan S., Honjo T. (2003) Intestinal IgA synthesis. Regulation of front-line body defences. Nat. Rev. Immunol. 3, 63–72 [DOI] [PubMed] [Google Scholar]
- 2. Durrer P., Glück U., Spyr C., Lang A. B., Zurbriggen R., Herzog C., Glück R. (2003) Mucosal antibody response induced with a nasal virosome-based influenza vaccine. Vaccine 21, 4328–4334 [DOI] [PubMed] [Google Scholar]
- 3. Kerr M. A. (1990) The structure and function of human IgA. Biochem. J. 271, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plaut A. G., Wistar R., Jr., Capra J. D. (1974) Differential susceptibility of human IgA immunoglobulins to streptococcal IgA protease. J. Clin. Invest. 54, 1295–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Holmgren J., Czerkinsky C. (2005) Mucosal immunity and vaccines. Nat. Med. 11, S45-S53 [DOI] [PubMed] [Google Scholar]
- 6. Holmgren J. (1991) Mucosal immunity and vaccination. FEMS Microbiol. Immunol. 89, 1–9 [DOI] [PubMed] [Google Scholar]
- 7. Albrechtsen M., Yeaman G. R., Kerr M. A. (1988) Characterization of the IgA receptor from human polymorphonuclear leucocytes. Immunology 64, 201–205 [PMC free article] [PubMed] [Google Scholar]
- 8. Monteiro R. C., Kubagawa H., Cooper M. D. (1990) Cellular distribution, regulation, and biochemical nature of an Fc alpha receptor in humans. J. Exp. Med. 171, 597–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morton H. C., van Egmond M., van de Winkel J. G. (1996) Structure and function of human IgA Fc receptors (FcαR). Crit. Rev. Immunol. 16, 423–440 [PubMed] [Google Scholar]
- 10. Monteiro R. C., Van De Winkel J. G. (2003) IgA Fc receptors. Annu Rev. Immunol. 21, 177–204 [DOI] [PubMed] [Google Scholar]
- 11. Dechant M., Beyer T., Schneider-Merck T., Weisner W., Peipp M., van de Winkel J. G., Valerius T. (2007) Effector mechanisms of recombinant IgA antibodies against epidermal growth factor receptor. J. Immunol. 179, 2936–2943 [DOI] [PubMed] [Google Scholar]
- 12. Zhao J., Kuroki M., Shibaguchi H., Wang L., Huo Q., Takami N., Tanaka T., Kinugasa T., Kuroki M. (2008) Recombinant human monoclonal igA antibody against CEA to recruit neutrophils to CEA-expressing cells. Oncol. Res. 17, 217–222 [DOI] [PubMed] [Google Scholar]
- 13. Huls G., Heijnen I. A., Cuomo E., van der Linden J., Boel E., van de Winkel J. G., Logtenberg T. (1999) Antitumor immune effector mechanisms recruited by phage display-derived fully human IgG1 and IgA1 monoclonal antibodies. Cancer Res. 59, 5778–5784 [PubMed] [Google Scholar]
- 14. Beyer T., Lohse S., Berger S., Peipp M., Valerius T., Dechant M. (2009) Serum-free production and purification of chimeric IgA antibodies. J. Immunol. Methods 346, 26–37 [DOI] [PubMed] [Google Scholar]
- 15. Kondoh H., Kobayashi K., Hagiwara K. (1987) A simple procedure for the isolation of human secretory IgA of IgA1 and IgA2 subclass by a jackfruit lectin, jacalin, affinity chromatography. Mol. Immunol. 24, 1219–1222 [DOI] [PubMed] [Google Scholar]
- 16. Pack T. D. (2001) in Current Protocols in Immunology (Coligan J. E., Bierer B. E., Margulies D. H., Shevach E. M., Strober W., eds) John Wiley and Sons, Inc., New York [Google Scholar]
- 17. Russell-Jones G. J., Gotschlich E. C., Blake M. S. (1984) A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J. Exp. Med. 160, 1467–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lindahl G., Akerström B., Vaerman J. P., Stenberg L. (1990) Characterization of an IgA receptor from group B streptococci. Specificity for serum IgA. Eur. J. Immunol. 20, 2241–2247 [DOI] [PubMed] [Google Scholar]
- 19. Carlsson F., Berggård K., Stålhammar-Carlemalm M., Lindahl G. (2003) Evasion of phagocytosis through cooperation between two ligand-binding regions in Streptococcus pyogenes M protein. J Exp. Med. 198, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pleass R. J., Areschoug T., Lindahl G., Woof J. M. (2001) Streptococcal IgA-binding proteins bind in the Cα2-Cα3 interdomain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 276, 8197–8204 [DOI] [PubMed] [Google Scholar]
- 21. Stenberg L., O'Toole P. W., Mestecky J., Lindahl G. (1994) Molecular characterization of protein Sir, a streptococcal cell surface protein that binds both immunoglobulin A and immunoglobulin G. J. Biol. Chem. 269, 13458–13464 [PubMed] [Google Scholar]
- 22. Frithz E., Hedén L. O., Lindahl G. (1989) Extensive sequence homology between IgA receptor and M proteins in Streptococcus pyogenes. Mol. Microbiol. 3, 1111–1119 [DOI] [PubMed] [Google Scholar]
- 23. Food and Drug Administration (2003) Control of Microbiological Contamination, Code of Federal Regulations 21, 211.113, U.S. Government Printing Office, Washington, D.C [Google Scholar]
- 24. Palombo G., De Falco S., Tortora M., Cassani G., Fassina G. (1998) A synthetic ligand for IgA affinity purification. J. Mol. Recognit. 11, 243–246 [DOI] [PubMed] [Google Scholar]
- 25. Li R., Dowd V., Stewart D. J., Burton S. J., Lowe C. R. (1998) Design, synthesis, and application of a protein A mimetic. Nat. Biotechnol. 16, 190–195 [DOI] [PubMed] [Google Scholar]
- 26. Sandin C., Linse S., Areschoug T., Woof J. M., Reinholdt J., Lindahl G. (2002) Isolation and detection of human IgA using a streptococcal IgA-binding peptide. J. Immunol. 169, 1357–1364 [DOI] [PubMed] [Google Scholar]
- 27. Krumpe L. R., Atkinson A. J., Smythers G. W., Kandel A., Schumacher K. M., McMahon J. B., Makowski L., Mori T. (2006) T7 lytic phage-displayed peptide libraries exhibit less sequence bias than M13 filamentous phage-displayed peptide libraries. Proteomics 6, 4210–4222 [DOI] [PubMed] [Google Scholar]
- 28. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott J. K. (1992) Discovering peptide ligands using epitope libraries. Trends Biochem. Sci. 17, 241–245 [DOI] [PubMed] [Google Scholar]
- 31. Ramsland P. A., Willoughby N., Trist H. M., Farrugia W., Hogarth P. M., Fraser J. D., Wines B. D. (2007) Structural basis for evasion of IgA immunity by Staphylococcus aureus revealed in the complex of SSL7 with Fc of human IgA1. Proc. Natl. Acad. Sci. U.S.A. 104, 15051–15056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herr A. B., Ballister E. R., Bjorkman P. J. (2003) Insights into IgA-mediated immune responses from the crystal structures of human FcαRI and its complex with IgA1-Fc. Nature 423, 614–620 [DOI] [PubMed] [Google Scholar]
- 33. Langley R., Wines B., Willoughby N., Basu I., Proft T., Fraser J. D. (2005) The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-FcαRI binding and serum killing of bacteria. J. Immunol. 174, 2926–2933 [DOI] [PubMed] [Google Scholar]
- 34. Persson J., Lindahl G. (2005) Single-step purification of human C4b-binding protein (C4BP) by affinity chromatography on a peptide derived from a streptococcal surface protein. J. Immunol. Methods 297, 83–95 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.