Background: UDP-galactose:ceramide galactosyltransferase (CGalT) is a glycoprotein that synthesizes galactosylceramides at the endoplasmic reticulum (ER).
Results: Molecular chaperone sigma-1 receptors promote degradation of CGalT by forming a complex with Insig.
Conclusion: CGalT and its activity are post-translationally regulated by ER-associated degradation (ERAD) involving sigma-1 receptor chaperones.
Significance: The sterol-sensing ERAD system controls the enzyme involved in glycosphingolipid biosynthesis.
Keywords: Ceramide, Endoplasmic Reticulum (ER), ER-associated Degradation, Lipid Metabolism, Neurodegeneration, Sigma Receptor, Sphingolipid, Sterol, Galactosylseramide, Sigma-1 Receptor
Abstract
The glycosphingolipid biosynthesis is initiated by monoglycosylation of ceramides, the action of which is catalyzed either by UDP-glucose:ceramide glucosyltransferase or by UDP-galactose:ceramide galactosyltransferase (CGalT). CGalT is expressed predominantly at the endoplasmic reticulum (ER) of oligodendrocytes and is responsible for synthesizing galactosylceramides (GalCer) that play an important role in regulation of axon conductance. However, despite the importance of ceramide monoglycosylation enzymes in a spectrum of cellular functions, the mechanism that fine tunes activities of those enzymes is largely unknown. In the present study, we demonstrated that the sigma-1 receptor (Sig-1R) chaperone, the mammalian homologue of a yeast C8-C7 sterol isomerase, controls the protein level and activity of the CGalT enzyme via a distinct ER-associated degradation system involving Insig. The Sig-1R forms a complex with Insig via its transmembrane domain partly in a sterol-dependent manner and associates with CGalT at the ER. The knockdown of Sig-1Rs dramatically prolonged the lifetime of CGalT without affecting the trimming of N-linked oligosaccharides at CGalT. The increased lifetime leads to the up-regulation of CGalT protein as well as elevated enzymatic activity in CHO cells stably expressing CGalT. Knockdown of Sig-1Rs also decreased CGalT degradation endogenously expressed in D6P2T-schwannoma cells. Our data suggest that Sig-1Rs negatively regulate the activity of GalCer synthesis under physiological conditions by enhancing the degradation of CGalT through regulation of the dynamics of Insig in the lipid-activated ER-associated degradation system. The GalCer synthesis may thus be influenced by sterols at the ER.
Introduction
Glycosphingolipids play a variety of roles in cell biology, not only as structural components of membranes, but also as regulators of signal transductions, cell death, cell adhesion, protein/lipid trafficking, cellular differentiation, receptor clustering, and conduction of electrical impulses at axons (1–6). Ceramide, the backbone of glycosphingolipids, is synthesized at the endoplasmic reticulum (ER)3 and acquires glucosylation or galactosylation catalyzed by specific transferases at the ER or the Golgi (7–9). The monoglycosylation step of ceramides influences the synthesis activity of gangliosides and sulfatides, intramembrane flip-flop movements of sphingolipids, membrane curvature, and cytotoxic activity of ceramides (9–11). Glucosylation is catalyzed at the outer surface of the Golgi apparatus by the UDG-glucose:ceramide glucosyltransferase for the synthesis of glucosylceramides (GlcCer) (12). Conversely, galactosylation of ceramides is catalyzed by the UDG-galactose:ceramide galactosyltransferase (CGalT) at the lumen of the ER (7, 13).
The CGalT amino sequence contains a KKVK motif, the potential ER retrieval signal, at the C terminus (14, 15). A study demonstrated that CGalT remains sensitive to endoglycosidase H, supporting its ER localization (7). Immunocytochemistry using specific antibodies also showed that CGalT immunoreactivity is found exclusively at the ER and nuclear envelope and not at the Golgi and plasma membrane (7, 13). These series of studies confirmed that CGalT is a class I integral ER protein possessing a long ER lumenal catalytic domain and a single transmembrane domain at the C terminus (7). A study using knock-out mice of CGalT confirms that there is only one galactosylceramide (GalCer)-synthesizing enzyme in the brain. CGalT is highly and specifically expressed in oligodendrocytes and in human epithelia (14). GalCer synthesized by CGalT comprise a major lipid in the myelin sheath of oligodendrocytes or Schwann cells that insulates axons to regulate electric impulses conducted by neuronal depolarization (4). Although highly enriched in myelins, GalCer is shown to serve as a negative regulator of oligodendrocyte differentiation and myelin formation (16).
The sigma-1 receptor (Sig-1R) is an integral membrane protein ubiquitously expressed in multiple organs, including the brain (17, 18). The Sig-1R binds a variety of structurally different drugs ((+)-isoforms of benzomorphans, haloperidol, and fluvoxamine) as well as endogenous molecules, such as sterols (e.g. progesterone) and simple sphingolipids (e.g. d-erythro-sphingosine, ceramide, and GalCer) (18–22). The structure of the Sig-1R shares no similarity with those of any mammalian proteins but shares 70% similarity with that of a yeast C8-C7 sterol isomerase (23). In animals, Sig-1Rs have been shown to promote neuronal survival under ischemia or β-amyloid depositions, potentiate morphine-induced analgesia, improve learning and memory, and contribute to the development of addictive behaviors induced by psychostimulants (24–29). A recent study identified the Sig-1R as being a novel ligand-operated molecular chaperone that can regulate ER stress and signal transductions (30). When ER Ca2+ is depleted, Sig-1Rs chaperone inositol 1,4,5-trisphosphate receptors localized at the interface between ER and mitochondria to ensure proper Ca2+ transmission between the two organelles (30, 31). Whether Sig-1Rs stabilize any other ER proteins or regulate ER-associated degradation (ERAD) has not been investigated in detail.
Sig-1Rs are highly expressed in oligodendrocytes of rat brains (32, 33). We previously demonstrated that Sig-1Rs colocalize with GalCer at the ER and up-regulate in oligodendrocyte-type 2 astrocyte progenitors during their oligodendrocyte differentiation (33). Knockdown of Sig-1Rs almost completely blocks the differentiation and formation of myelin sheets in oligodendrocytes (33). Because both Sig-1Rs and CGalT are localized at the ER, and GalCer synthesized by CGalT is a major regulator of oligodendrocyte differentiation, we hypothesized that Sig-1Rs may regulate CGalT activity at the ER and may thus regulate cellular differentiation. Here we employed a simple cellular model (i.e. CHO cell line stably expressing CGalT) to examine whether Sig-1Rs regulate the activity of CGalT. We found that Sig-1R chaperones, in lieu of stabilizing CGalT proteins, promote instead the acceleration of CGalT degradation by utilizing a distinct class of ERAD systems. Sig-1Rs do so by forming a protein complex with the sterol-sensing protein Insig to regulate degradation of CGalT.
EXPERIMENTAL PROCEDURES
Materials
Antibodies against Sig-1R and CGalT were raised as described previously (7, 30). Sources of other antibodies are as follows: anti-actin, anti-SREBP cleavage-activating protein (SCAP), and anti-extracellular signal-regulated kinase 1 (ERK) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); polyclonal anti-FLAG, monoclonal anti-FLAG, anti-HA, and polyclonal anti-Myc antibodies from Sigma; monoclonal anti-Myc from Cell Signaling (Boston, MA); anti-GM130, anti-Mcl-1, and anti-BiP from BD Biosciences; anti-α1-antitrypsin from DakoCytomation (Glastrup, Denmark); Alexa480- or Alexa590-labeled goat secondary antibodies and anti-V5 antibody from Invitrogen; and anti-GalCer monoclonal antibody from Millipore (Billerica, MA). [14C]Serine and [14C]UDP-galactose were purchased from GE Healthcare, and [35S]methionine was from MP Biomedical (Solon, OH). Vectors for expression of rat Sig-1R, mouse Sig-1R C-terminally tagged with enhanced yellow fluorescent protein (Sig-1R-EYFP), rat Sig-1R C-terminally tagged with FLAG (Sig-1R-FLAG), control siRNA (siCon), Sig-1R siRNA (siSig-1R), rat CGalT, and rat CGalT-Myc were constructed as described previously (7, 33, 34). Vectors for expression of CD3-δ-HA, CD3-δΔ-HA, and misfolded α1-antitrypsin variant, null (Hong Kong) (NHK) were kindly donated by Dr. Molinari (Institute for Research in Biomedicine, Bellinzona, Switzerland). Vectors for expression of SCAP and Insig1-Myc were purchased from ATCC (Manassas, VA). Castanospermine, kifnensine, and swainsomine were purchased from Toronto Research Chemicals (York, Canada). (+)-Pentazocine was synthesized at the Division of Basic Research at NIDA, National Institutes of Health. Other lipids and chemicals were purchased from Sigma.
Cell Culture
Wild-type Chinese hamster ovary (CHO) cells and CHO cell lines stably expressing EYFP, Sig-1R-EYFP, or CGalT were cultured as described previously (7, 30). D6P2T myeloma cells were purchased from ATCC and maintained in Dulbecco's modified Eagle's medium with 5% fetal calf serum with 5% CO2.
Plasmid Construction and Transfection
cDNAs encoding full-length or C-terminally truncated rat Sig-1R were amplified by PCR using pCR3.0-rat Sig-1R as a template (35) with the following primer sets: 5′-GAATTCATGCCGTGGGGCTGTGGGCT-3′ (forward) and 5′-GGGGTCTTGGCCAAAGAGGT-3′ (reverse for Sig-1R(1–223)), 5′-ACCCCGGCCATACTCCAC-3′ (reverse for ΔSig-1R(1–176)), 5′-GACAACCGTCTCTCCTGG-3′ (reverse for ΔSig-1R(1–153)), 5′-ATGGCCATGGGAGCCCAG-3′ (reverse for ΔSig-1R(1–116)), 5′-CATCCAGCCGCCCGCGTT-3′ (reverse for ΔSig-1R(1–90)), or 5′-CGCGTACTGTCGAGCAAG-3′ (reverse for ΔSig-1R(1–50)); 5′-ATGTCGGGACGATACTGGGCT-3′ (forward) and 5′-GGGGTCTTGGCCAAAGAGGT-3′ (reverse for Sig-1R(117–223)) or 5′-TCACAGCTCGTCCTTGGTACGCGTAGAATCGAGACC-3′ (reverse for ΔSig-1R(117–223)-KDEL). Mouse cDNA of 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase was amplified by PCR using pCMV6-Kan/Neo-MC206001 as a template (OriGene Technology, Rockville, MD; accession number NM_008255) and the following primer set: 5′-GCATGCGGCCGCCCACCATGTTGTCAAGACTTTTCCGG-3′ (forward) and 5′-ACTAGAATTCCCAGCTGCCTTCTTGGTGCA-3′ (reverse). Amplified PCR products were ligated into pcDNA3.1/V5-His TOPO vectors (Invitrogen). For transfection, 4 μl of Lifopectamine-2000 (Invitrogen) was mixed with expression vectors (rat Sig-1R, rat Sig-1R-V5, and mouse Sig-1R-EYFP at 0.5 μg/dish; Insig1-Myc at 0.02 μg/dish; SCAP at 0.03 μg/dish; others at 2 μg/dish) and incubated with cells in 6-cm dishes for 6 h. Cells were maintained for 2 days before experiments. siRNAs against Insig1 was purchased from Thermo Dharmacon (Lafayette, CO) and transfected by PepMute siRNA Transfection reagent according to the company's instructions (SignaGen Laboratories, Ijamsville, MD).
Immunocytochemistry and Confocal Microscopy
CHO cells grown on poly-d-lysine-coated 1-cm round coverslips (Fisher) were fixed with 4% paraformaldehyde in phosphate-buffered saline (pH 7.4) for 20 min and permeabilized with 0.2% Triton X-100 for 5 min at room temperature (for CGalT immunostaining alone) or 0.01% Triton X-100 for 1 min at 4 °C (for GalCer immunostaining). Permeabilization at the lower temperature with the low concentration of Triton X-100 was used to preserve immunoreactive GalCer in CHO cells. After blocking with 10% nonfat dry milk (1 h), samples were incubated with anti-CGalT (1:200 in PBS containing 4% BSA and 0.5% Nonidet P-40) or with a mixture of anti-CGalT (1:300) and anti-GalCer (1:300) in PBS containing 4% BSA at 4 °C overnight. After labeling with secondary antibodies (Alexa480 or Alexa590-conjugated goat IgG), immunofluorescence was observed by UltraView confocal microscopy (PerkinElmer Life Sciences) as previously reported (30).
Immunoblotting
Cells were quickly harvested in ice-cold PBS and centrifuged at 1500 × g for 10 min at 4 °C. Cell pellets were dissolved in 2× SDS sample buffer (100 mm Tris-HCl, pH 6.8, 20% glycerol, 4% SDS) with sonication (two times for 10 s each). After measuring protein concentrations by a micro-BCA reagent (Thermo), cell lysates were boiled for 5 min in the presence of β-mercaptoethanol and bromphenol blue. For HMG-CoA reductase-V5 immunoblotting, cells were lysed in the SDS buffer (10 mm Tris-HCl (pH 6.8), 1% (w/v) SDS, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, and protease inhibitor (Sigma)). After protein assays, an equal volume of urea buffer (62.5 mm Tris-HCl (pH 6.8), 15% SDS, 8 m urea, 10% (v/v) glycerol, and 100 mm dithiothreitol) was added, and lysates were incubated at 37 °C for 20 min. Total lysates (20–50 μg/lane) were resolved by 13% SDS-PAGE and analyzed by immunoblotting, as described previously (22, 30, 33). Protein bands were densitometrically analyzed with a Kodak Image Station 440 CF (Eastman Kodak Co.).
Thin Layer Chromatography (TLC)
Total lipids were extracted by Bligh and Dyer partitioning as described previously (22, 36). Lipid extracts dried under an N2 flow were dissolved in a chloroform/methanol mixture (2:1) and spotted on a high performance thin layer chromatography plate (Merck). After resolution on TLC plates with a chloroform/methanol/water mixture (13:5:0.5), lipids were visualized by a diphenylamine-aniline or 0.2% 8-anilinonaphthalene-1-sulfonate spray. Respective lipids were identified by resolving purified lipids on the same TLC plate. Lipids were quantified by a Kodak Image Station 440 CF.
[14C]Serine Labeling for Analysis of Lipid Synthesis
After washing with serum-free medium, cells in 6-cm dishes were incubated at 37 °C in 1 ml of minimum essential medium containing 0.5 μCi/ml [14C]serine/ml. Cells harvested on ice were suspended in 200 μl of ice-cold H2O. Following vortex (15 times for 1 s each), 5 μl of cell suspensions were transferred into a 96-well plate (duplicate) for the Bio-Rad protein assay. Cells were lysed by adding 20 μl of 1% SDS (on ice for 5 min). Lipids in the lysates were extracted by successively adding the following reagents: ∼563 μl of CHCl3/methanol (1:2) (vortex for 10 min), ∼188 μl of CHCl3 (vortex for 1 min), and 187.5 μl of H2O (vortex for 1 min). Following a centrifugation for 3 min at 5000 × g, the lower phase was transferred to a new tube. After adding ∼280 μl of CHCl3 to the upper water phase in the original tubes, samples were vortexed for 1 min and centrifuged for 3 min at 5000 × g. The yielded lower phase was combined with the previously obtained lower phase in a new tube. Lipids in the lower phase were dried under a N2 flow and resolved by TLC as described above. The TLC plate was air-dried and exposed to a β-Max film (Amersham Biosciences) for 5 days at room temperature.
CGalT Activity Assay
CHO cell pellets containing 100 μg of total proteins were suspended in 150 μl of reaction buffer (20 mm HEPES (pH 7.4), 120 mm potassium glutamate, 15 mm KCl, 5 mm NaCl, 0.8 mm CaCl2, 2 mm MgCl2, 2 mm MnCl2, 1.6 mm EGTA, 1 mm UDP-galactose, 2 μCi/ml [14C]UDP-galactose, 100 nm BSA-conjugated C6-ceramides) containing 1 mg/ml saponin and incubated at 37 °C with horizontal shaking (150 rpm) for 0–120 min. Lipids were extracted and resolved by TLC as described above. The level of synthesized GalCer was quantified with a Kodak Image Station 440 CF.
RT-PCR
Total RNA was extracted with a NucleoSpin II kit (Clontech) from CHO cells. The level of CGalT or Insig1 mRNA was measured by using RT-PCR (Titanium One-Step RT-PCR kit, Clontech). The primer set and PCR conditions for amplification of the CGalT mRNA were as follows: 5′-GCTAAAATCATCATTGTGCCG-3′ (forward) and 5′-CCAACTTCAGCAGGATACCAAAGG-3′ (reverse), Tm = 55 °C, 25 cycles. Primers for the Insig1 mRNA were as follows: 5′-GGCTTGTGGTGGACATTCG-3′ (forward) and 5′-GGCGATGGTGATCCCAAGT-3′ (reverse), Tm = 58 °C, 23 cycles. Primers for β-actin mRNA were purchased from Clontech. The mRNA levels were normalized to β-actin mRNA.
Sucrose Fractionation
CHO cells grown on a 15-cm dish were homogenized with a Dounce glass homogenizer (30 strokes) in 400 μl of homogenization buffer (10 mm HEPES (pH 7.4), 15 mm KCl, 1.5 mm MgCl2, protease inhibitor mixture (Sigma)). Cell homogenate was centrifuged at 380 × g for 10 min at 4 °C. The supernatant was pooled. The pellet was homogenized (20 strokes) again in 200 μl of homogenization buffer. After centrifugation (380 × g, 10 min), the supernatant was combined with one obtained after the first centrifugation. 300 μl of the supernatant was placed on top of a discontinuous sucrose gradient (0.5, 0.7, 0.9, 1.1, 1.3, and 1.5 m sucrose; 600 μl for each fraction). The sample was centrifuged in a SW 55i rotor at 180,000 × g for 3 h at 4 °C. 12 fractions were obtained from the top (325 μl/fraction) and stored at −80 °C until the assays.
Immunoprecipitation
Cell pellets from 6-cm dishes were suspended in the lysis buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.2% sodium deoxycholate, 0.1% SDS, 1% Triton X-100, and protease inhibitor mixture). After rotation at 4 °C for 30 min, lysates were centrifuged at 12,000 × g for 15 min. 300 μg of supernatants were precleared by incubating with 30 μl of either Sepharose-protein A beads (GE Healthcare) or Protein A/G PLUS agarose (Santa Cruz Biotechnology, Inc.) for 60 min. Precleared cell lysates were incubated with specific antibodies for immunoprecipitation at 4 °C overnight (1:300 for anti-CGalT; 1:200 for polyclonal anti-Myc, anti-α1-antitrypsin, and polyclonal anti-FLAG; 1:500 for anti-HA and polyclonal anti-GFP; 1:1000 for anti-V5). Immunocomplexes were precipitated with 30 μl of Sepharose-protein A beads or Protein A/G PLUS agarose and analyzed by SDS-PAGE followed by immunoblotting as described previously (30). In the CGalT-Sig-1R-V5 co-immunoprecipitation assay, harvested cells were cross-linked by 0.1 mg/ml DSP (30 min at 4 °C) in 50 mm HEPES (pH 7.4).
Pulse-Chase Experiment
A day before experiments, cells were seeded onto 6-cm (CHO-CGalT cells) or 10-cm (D6P2T cells) dishes at 90–100% confluence. After a washing with PBS, cells were incubated for 30–60 min in methionine-free DMEM containing 3% dialyzed calf serum. Cells were pulse-labeled with [35S]methionine (CHO-CGalT at 50 μCi/ml for 10 min; D6P2T at 200 μCi/ml for 30 min). After pulsing, cells were chased in culture medium containing 10% serum and 0.75 mg/ml methionine. 35S-Labeled CGalT was immunoprecipitated and analyzed by SDS-PAGE followed by autoradiography.
Cholesterol Deprivation and Drug Treatment
The intracellular cholesterol level was reduced according to the previously established procedure (37, 38). Briefly, 6 h after transfection of plasmids, CHO cells were seeded onto 6-cm dishes with normal culture medium. On the next day, medium was replaced with 3 ml of minimum essential medium containing 2% lipoprotein-deficient serum (LPDS), 10 μm compactin, and 50 μm mevalonate with or without sterols. After 16 h, cells were incubated in the MEM-α containing 2% LPDS, 25 μm compactin, and 5 mm mevalonate with/without sterols or (+)-pentazocine for 4 h. Sterols and compactin were dissolved in ethanol. Mevalonate and (+)-pentazocine were dissolved in H2O. Controls received the same vehicles without drugs.
RESULTS
CHO Cell Line Stably Expressing CGalT
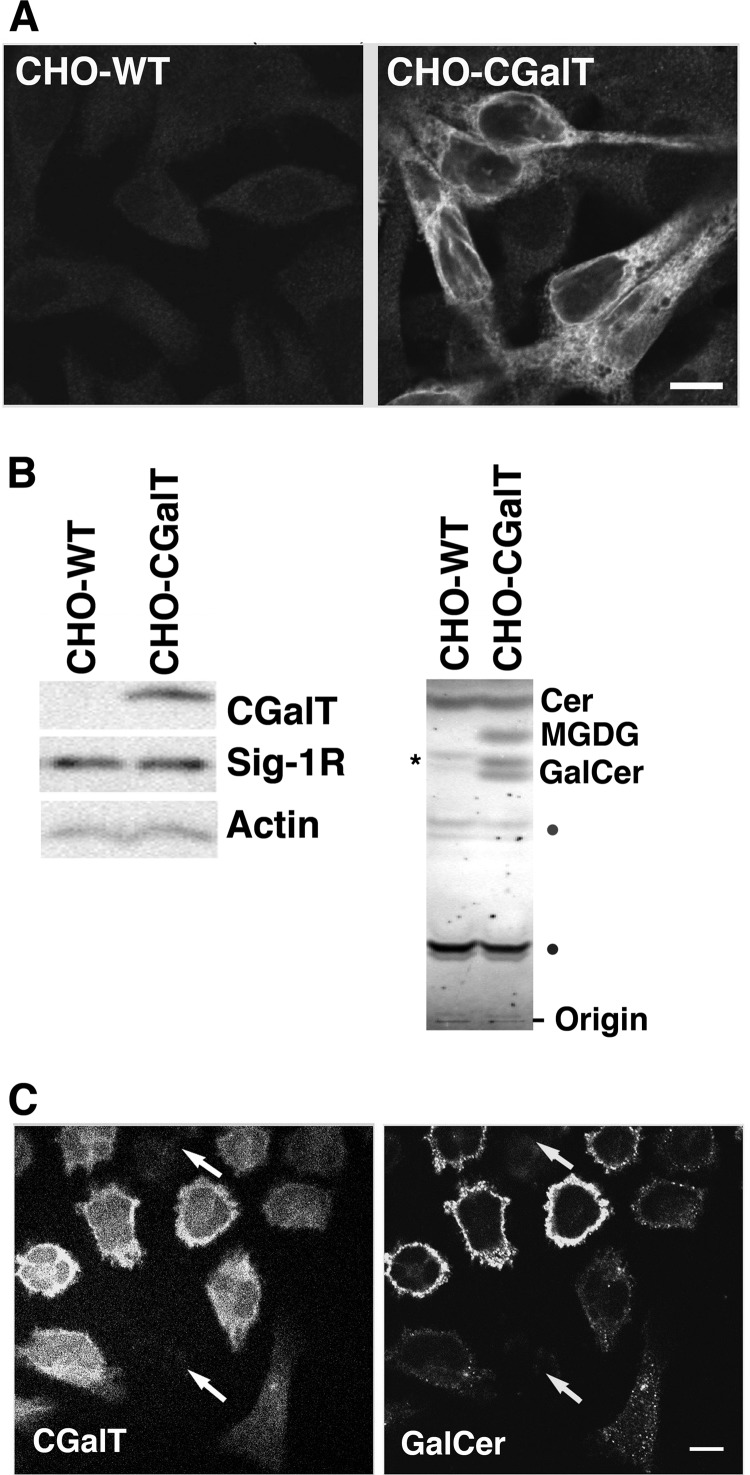
To examine the possibility that Sig-1Rs regulate the CGalT activity, the CHO cell line stably expressing rat CGalT (CHO-CGalT) was employed (7). This cell line constitutively expresses a considerably high level of endogenous Sig-1Rs (22, 30), and stably transfected rat CGalT is shown to behave normally regarding its subcellular distribution and enzymatic activity (7, 13). The CGalT antibody (7) revealed strong CGalT-like immunoreactivity in this line but not in wild-type CHO cells (CHO-WT) (Fig. 1A). CGalT distributed in the cytoplasmic region, showing a typical ER pattern (Fig. 1A).
FIGURE 1.
Expression and distribution of CGalT in CHO-CGalT cells. A, CGalT immunocytochemistry of wild-type CHO cells (CHO-WT) or CHO cells stably expressing rat CGalT (CHO-CGalT). Both cell lines fixed with paraformaldehyde were permeabilized with Triton X-100 (0.2%, 5 min) and immunostained with anti-CGalT antiserum. Images were captured by a confocal microscope. Bar, 10 μm. B, profile of protein and lipid expressions in CHO-CGalT. In the left panels, 20 μg of total cell lysates prepared from CHO-WT or CHO-CGalT were resolved by 13% SDS-PAGE and immunoblotted with respective specific primary antibodies. In the right panels, total lipid extracts were resolved by TLC followed by a 0.2% 8-anilinonaphthalene-1-sulfonate spray. Cer, ceramide. The asterisk in the CHO-WT represents endogenous GlcCer. Lipids marked with dots were not identified. The images represent the result of 4–5 repeated experiments. C, immunostaining of CGalT and GalCer expressed in CHO-CGalT. CHO-CGalT cells were fixed with paraformaldehyde. Following a mild permeabilization with Triton X-100 (0.01%, 1 min), samples were immunostained with anti-CGalT antiserum and anti-GalCer antibodies. Images were captured by a confocal microscope. The arrows indicate CHO-CGalT expressing very low levels of CGalT. Note that the level of GalCer in individual cells is fairly proportional to that of CGalT. Bars, 10 μm.
In immunoblotting, anti-CGalT antibodies detected a single band with a 55-kDa molecular mass (7). Expression of Sig-1Rs was similar between CHO-WT and CHO-CGalT cells (Fig. 1B). Because CGalT accepts both ceramides and diacylglycerols as substrates (7), both GalCer (doublets in Fig. 1B) and monogalactosylated diacylglycerols were present in lipid extracts from CHO-CGalT cells but not in those from CHO-WT (Fig. 1B). The upper and lower bands of GalCer represent those containing non-hydroxylated and hydroxylated acyl-chains, respectively (40).
Monoclonal antibodies against GalCer expressed strong GalCer-like immunoreactivities only in CHO-CGalT (Fig. 1C). The level of GalCer immunoreactivities is fairly correlated with the CGalT level in most CHO-CGalT cells (Fig. 1C). However, there are some cells that express a high level of CGalT but express less GalCer immunoreactivity. The lipid environment that may vary between individual cells and/or cell permeabilization procedures during immunofluorescence may affect the immunoreactivity of anti-GalCer antibodies.
Sig-1Rs Negatively Regulate Production of GalCer
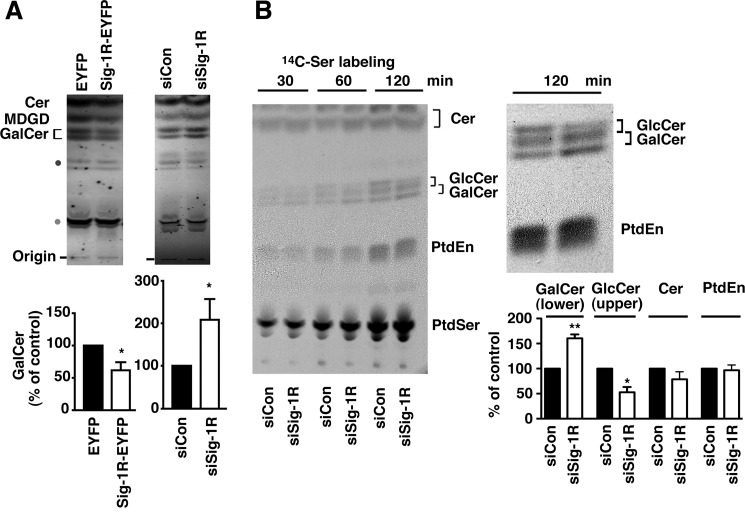
As shown by TLC (Fig. 2A), we found that CHO-CGalT cells overexpressing Sig-1R-EYFP, when compared with those expressing EYFP, contained lower GalCer. In contrast, knockdown of Sig-1Rs caused an increase in GalCer (Fig. 2A). The same tendency was also observed in the level of monogalactosylated diacylglycerols (Fig. 2A), suggesting that Sig-1Rs negatively regulate the enzymatic activity of CGalT.
FIGURE 2.
Sig-1Rs regulate CGalT activity. A, TLC analysis of total GalCer and MDGD levels in CHO-CGalT cells. CHO-CGalT cells were transfected for 2 days with either EYFP or Sig-1R-EYFP or with either siCon or siSig-1R. Cer, ceramide; SM, sphingomyelin. Intensities of GalCer bands were densitometrically measured and shown in bar graphs as mean ± S.E. (error bars) (n = 6). *, p < 0.05, Student's t test. B, effect of the Sig-1R knockdown on the synthesis of [14C]Ser-labeled lipids in CHO-CGalT cells. Two days after transfection with siRNAs, CHO-CGalT cells were incubated in the MEM-α containing 0.5 μCi/ml of [14C]serine/ml for the indicated periods of time. Lipids were extracted and resolved by TLC. 14C-Labeled lipids were visualized by direct autoradiography. PtdEn, phosphatidylethanolamine; PtdSer, phosphatidylserine. Respective lipids were identified based on Rf values of purified lipids resolved under the same TLC system. Identities of lipids marked with dots were not confirmed. The right panel enlarges the autoradiographic image at the 120-min time point. The graph represents mean ± S.E. (n = 4) of the level of the lower and upper bands of [14C]GalCer, [14C]ceramide, and [14C]phosphatidylethanolamine at 120 min. *, p < 0.05; **, p < 0.01, paired t test.
Next, sphingolipid synthesis between control and Sig-1R knockdown cells was monitored in [14C]serine-labeled CHO-CGalT cells. Because serine is also utilized for phospholipid synthesis, autoradiography of TLC plates revealed both 14C-labeled phospholipids and 14C-labeled sphingolipids. Because CHO-CGalT cells possess endogenous UDP-glucose:ceramide glucosyltransferase activity, both [14C]GalCer and [14C]GlcCer were detected in [14C]serine-labeled cells (both showing doublets in TLC). Under our TLC conditions, the lower band of GlcCer and upper band of GalCer were overlapped, thus hampering quantification of the upper bands of GalCer. Nonetheless, data clearly demonstrated that CHO-CGalT cells lacking Sig-1Rs increase the synthesis of the lower band of [14C]GalCer. There was no apparent difference in the synthesis of ceramides and phospholipids between control and Sig-1R knockdown cells (Fig. 2B). In this study, we also serendipitously found that knockdown of Sig-1Rs decreased the GlcCer synthesis (see the upper band of [14C]GlcCer in Fig. 2B). Because the GlcCer synthase, which shares no homology with CGalT, localizes mainly at the Golgi complex, this result may suggest that Sig-1Rs regulate stability and/or the transport of the enzyme to the Golgi complex. Although this is a striking finding, we focused solely on exploring the regulation of CGalT by Sig-1Rs in this paper.
Sig-1Rs Down-regulate CGalT Proteins
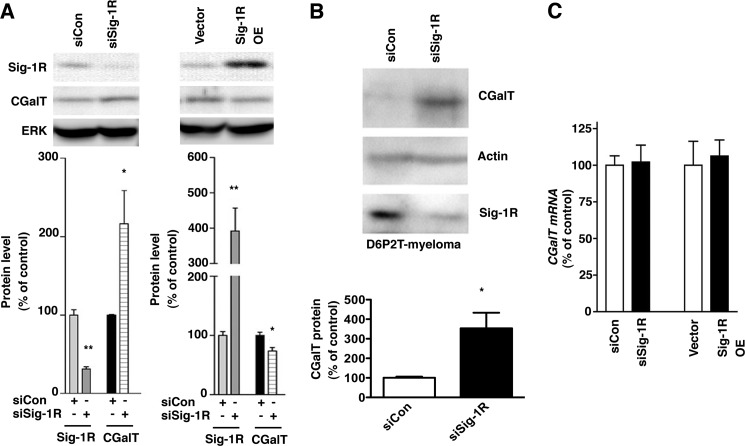
ER chaperones are known to promote protein folding but also serve as subcomponents in ERAD systems (41, 42). We therefore examined whether overexpression or knockdown of Sig-1Rs affects CGalT protein levels. Immunoblotting demonstrated that the level of CGalT was significantly increased by Sig-1R knockdown, whereas it was decreased by overexpression of Sig-1Rs (Fig. 3A). Knockdown of Sig-1Rs also caused an increase of CGalT proteins that are endogenously expressed in D6P2T-schwannoma cells (Fig. 3B), indicating the physiological relevance of our finding. Neither knockdown nor overexpression of Sig-1Rs affected the mRNA level of CGalT in CHO-CGalT (Fig. 3C). Therefore, Sig-1Rs may post-translationally regulate the protein level of CGalT.
FIGURE 3.
Sig-1Rs negatively regulate the protein level of CGalT. A, Western blotting analysis of CGalT in CHO-CGalT cells knocking down or overexpressing Sig-1Rs. Control plasmids or plasmids for expression of Sig-1R siRNA or wild-type rat Sig-1Rs were transfected 2 days before the assay. Protein levels of Sig-1Rs or CGalT were measured by immunoblotting using specific antibodies. ERK immunoblotting serves as a loading control. Intensities of respective proteins were measured and normalized to ERK. Graphs represent means ± S.E. (error bars) from five independent experiments. *, p < 0.05; **, p < 0.01 compared with the corresponding controls. B, CGalT protein level endogenously expressed in D6P2T myeloma. Control or Sig-1R siRNA was transfected 2 days before Western blotting. Protein levels were measured by immunoblotting. The graph represents mean ± S.E. (n = 4). C, RT-PCR analysis of CGalT mRNA. siRNAs were transfected 2 days before the extraction of total RNA. Levels of CGalT mRNA were measured by RT-PCR. The graph represents mean ± S.E. of six independent samples.
Sig-1Rs Associate with CGalT at the ER of CHO Cells
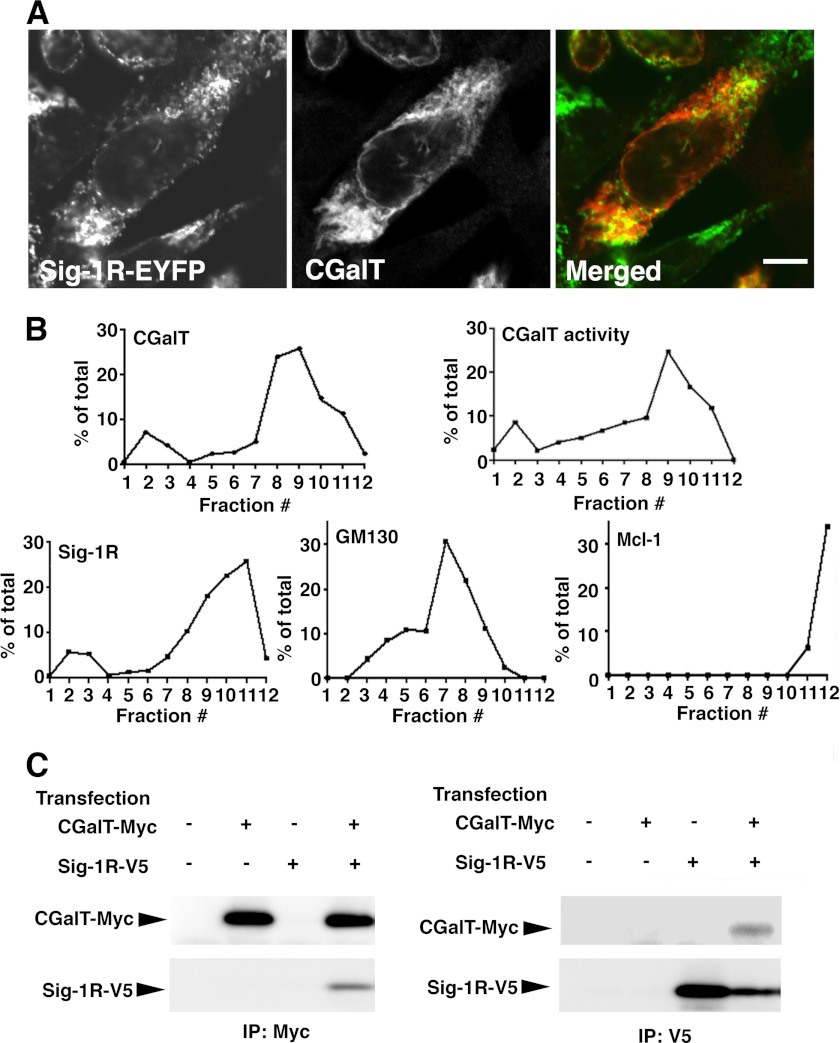
Because Sig-1Rs physically associate with structurally different proteins to regulate degradation of associated proteins (20, 30), we next examined whether Sig-1Rs also associate with CGalT. Confocal microscopy demonstrated that both Sig-1R-EYFP (expressed at a low level with 0.5 μg of plasmid/6-cm dish) and CGalT distribute over the punctate/reticular ER structures and nuclear envelopes (Fig. 4A).
FIGURE 4.
Subcellular distribution and association of CGalT with Sig-1Rs. A, confocal microscopic images of CGalT and Sig-1R-EYFP. The small amount of Sig-1R-EYFP vectors (0.5 μg/6-cm dish) were transfected into CHO-CGalT 2 days before the observation. CGalT was immunocytochemically labeled with anti-rat CGalT antiserum, followed by labeling with Alexa 590 goat anti-rabbit IgG secondary antibodies. Bar, 10 μm. The image represents the results from five independent immunolabelings. B, subcellular fractionation of CHO-CGalT cells. Fractions 1–12 were obtained by a sucrose gradient centrifugation of cell homogenates. The levels of respective proteins were measured by immunoblotting in each fraction. The percentage of the total in fraction x was calculated by the equation, 100 × (level of the protein in fraction x)/(the sum of the protein levels in 12 fractions). CGalT activity was measured by incubating each fraction with 2 μCi/ml [14C]UDP-galactose and 100 nm BSA-conjugated C6-ceramides at 37 °C for 120 min, as described in the legend to Fig. 2C. The data represent three independent experiments. C, immunoprecipitation assay for the detection of the CGalT-Sig-1R association. Two days before immunoprecipitation assays (IP), CHO cells were transfected with empty vectors (−), CGalT-Myc (2 μg/6-cm dish), and/or rat Sig-1R-V5 (0.5 μg/6-cm dish). After cross-linking with DSP, cell lysates were prepared as described under “Experimental Procedures.” Each lysate was then divided in half to use one half for Myc immunoprecipitation (top two panels) and the other for V5 immunoprecipitation (bottom two panels). The images represent results from three independent experiments.
Similarity of subcellular distribution of these proteins was also examined by using subcellular fractionation. The result showed that distribution of membranes containing CGalT possesses two peaks: the small peak at low density fractions (F2 and F3) and the large peak at high density fractions (F8–F11). Membranes containing Sig-1Rs also showed a similar pattern (Fig. 4B). Sig-1Rs were also contained in fractions F10 and F11 (Fig. 4B). Fractionation patterns of GM130 (cis-Golgi marker) and Mcl-1 (mitochondria marker) are completely different from that of CGalT.
Although confocal microscopy and membrane fractionation have certain limitations in determining precise subcellular localization of molecules, the above data suggest that Sig-1Rs and CGalT may co-localize at the ER. Accordingly, we performed an immunoprecipitation assay using CHO cells expressing CGalT-Myc and Sig-1R-V5. Expressed CGalT-Myc showed subcellular distribution identical to that of CGalT in CHO-CGalT cells (data not shown). As shown in Fig. 4C, anti-Myc antibodies immunoprecipitated CGalT-Myc together with Sig-1R-V5 (Fig. 4C, lane 4 of the upper panels). Anti-Myc antibodies failed to pull down Sig-1R-V5 without co-expression of CGalT-Myc (lane 3 of the upper panels), verifying the specific interaction between CGalT-Myc and Sig-1R-V5. Similarly, specific anti-V5 antibodies immunoprecipitated Sig-1R-V5 together with CGalT-Myc (Fig. 4C, lower panels). Sig-1Rs failed to co-immunoprecipitate several other ER-resident proteins (e.g. GRP94 and calnexin (30)).
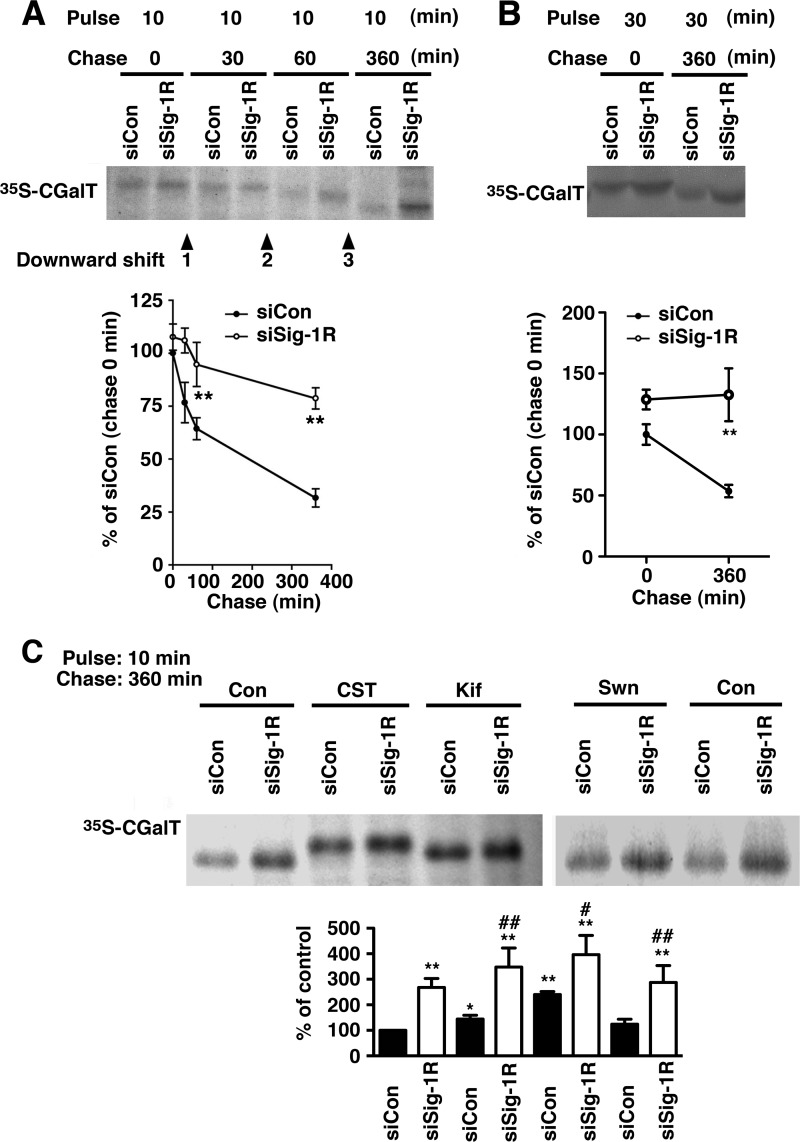
Knockdown of Sig-1Rs Prolonged the Lifetime of CGalT
To delineate the molecular mechanism by which Sig-1Rs regulate the CGalT protein level, the pulse-chase experiment was performed. As shown in Fig. 5A, CGalT was shown to have a t½ of approximately 200 min. The knockdown of Sig-1R did not affect the synthesis of [35S]CGalT during the 10-min pulse-labeling (chase 0 min in Fig. 5A) but significantly delayed the decline of the [35S]CGalT during chasing (Fig. 5A). The knockdown of Sig-1Rs thus prolongs the lifetime of CGalT. The similar effect of Sig-1R knockdown was seen with endogenously expressed CGalT in D6P2T cells (Fig. 5B).
FIGURE 5.
The knockdown of Sig-1Rs prolongs the lifetime of CGalT. A, pulse-chase experiment of CGalT. CHO-CGalT cells were pulse-labeled with [35S]methionine (50 μCi/dish) for 10 min at 37 °C and then chased for the indicated periods of time with 10% FCS and 0.75 mg/ml methionine. 35S-Labeled CGalT was immunoprecipitated and visualized with autoradiography. The graph represents the mean ± S.E. (error bars) of six independent studies. **, p < 0.01, paired t test. B, the inhibition of CGalT degradation by Sig-1R knockdown in D6PT2 cells. D6P2T schwannoma cells in 10-cm dishes were pulse-labeled with [35S]methionine for 30 min and chased for 360 min. Bands were visualized and quantified as in A. **, p < 0.01, paired t test (n = 5). C, inhibition of GalCer downward shift by inhibitors of ER glucosidase or mannosidase. CGalT pulse-labeled with [35S]methionine for 10 min was chased for 360 min. [35S]CGalT was immunoprecipitated and detected by autoradiography. Inhibitors were applied 60 min prior to the pulse labeling. CST, castanospermine (1 mm); Kif, kifnensine (2 μg/ml); Swm, swainsomine (100 μm). *, p < 0.05; **, p < 0.01 compared with siCon without drug treatment (first column). #, p < 0.05; ##, p < 0.01 compared with siCon with the same drug treatment; paired t test, n = 4–5.
We previously demonstrated that N-linked oligosaccharides on CGalT remain sensitive to endoglycosidase H digestion even 16 h after chasing, suggesting that CGalT is an ER-resident glycoprotein and is not transported through the Golgi complex (7). Here, we found that in the pulse-chase experiments, CGalT bands are downward shifted slightly at the 30-min chasing point and then shifted further at 60- and 360-min chasing points (arrowheads in Fig. 5A), indicating the active trimming of the N-linked oligosaccharides being processed on CGalT. In fact, the downward shifts were partially blocked by castanospermine (ER glucosidase I and II inhibitor) and kifnensine (ER mannosidase I inhibitor) but not by swainsomine, an ER mannosidase II inhibitor (Fig. 5C). Notably, the effect of Sig-1R knockdown on delaying degradation of CGalT was still observed in the presence of those inhibitors. Thus, the action of Sig-1Rs regulating the degradation of CGalT is probably independent of the trimming status of CGalT (i.e. in apparent contrast to that seen in well defined ERAD of glycoproteins that is operated by lectin chaperones, such as calnexin) (41, 42).
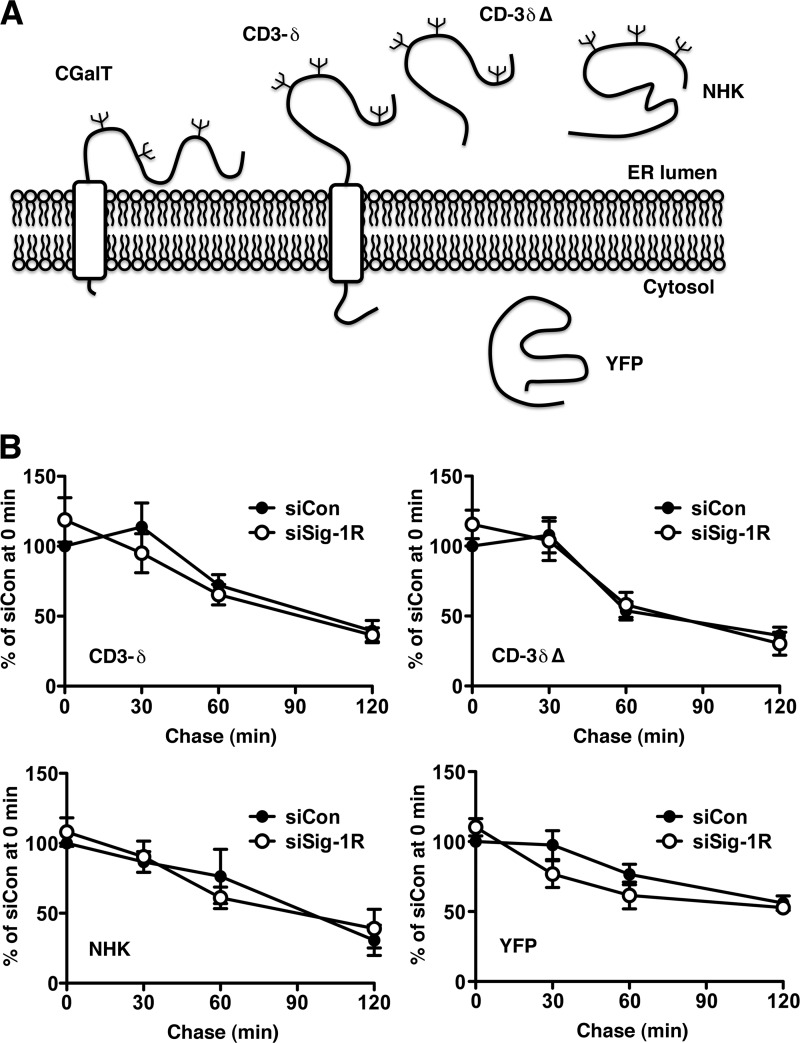
A specific ERAD pathway is selected based on topological and structural configurations of individual proteins to be degraded (41). For example, ER proteins with a defect on the cytosolic domain utilize ERAD-C, whereas ER proteins having defects at the ER lumenal domain utilize ERAD-L for degradation. The latter is further classified into ERAD-LM and ERAD-LS, based on whether the substrate proteins possess a transmembrane domain (i.e. membrane proteins) or not (i.e. soluble protein) (41, 43). Because CGalT is a glycoprotein having nearly its entire sequence inside the ER (Fig. 6A), Sig-1Rs might promote degradation of CGalT via ERAD-L. To test this possibility, we employed well characterized substrates of ERAD-LM (CD3-δ) and ERAD-LS (NHK; CD3-δΔ lacking its transmembrane domain) (43) and examined whether knockdown of Sig-1Rs compromises degradation of those proteins. As shown in Fig. 6B, we found that knockdown of Sig-1Rs does not affect degradation of any ERAD-L substrates tested. Knockdown of Sig-1Rs also did not affect the lifetime of the cytosolic protein YFP (Fig. 6B).
FIGURE 6.
Sig-1Rs are not involved in degradation of ERAD-L substrates. A, a scheme depicting membrane topology of ERAD-L substrates, YFP, and CGalT. B, summary of pulse-chase experiments showing the effect of the Sig-1R knockdown on degradation of ERAD-LM substrate CD3-δ and ERAD-LS substrates CD-3δΔ and NHK. YFP was chosen to represent cytosolic proteins. The graphs indicate means ± S.E. (error bars) of 3–5 independent experiments.
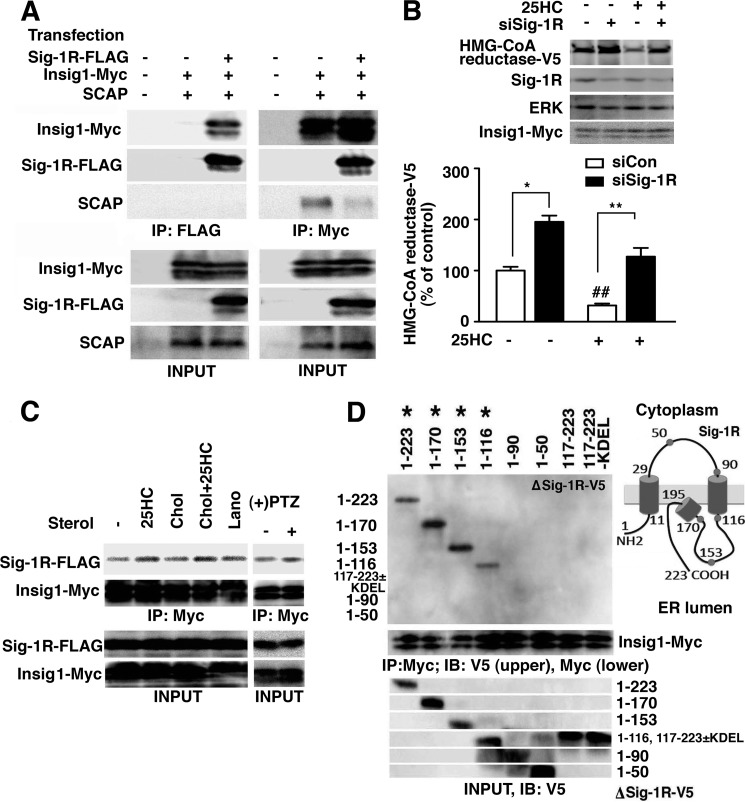
Sig-1Rs Form a Protein Complex with Insig1
There is a distinct subclass of ERAD complexes that processes protein degradation by sensing lipid levels at the ER membrane. When the cholesterol level is high at the ER, Insig1 or -2 delivers a specific E3 ligase to HMG-CoA reductase to degrade the enzyme (37). Insig also forms a different protein complex with SCAP to regulate the translocation of the transcription factor SREBPs from the ER to Golgi (44). In both systems, Insig interacts with counterpart proteins when they bind sterols (44). Because the Sig-1R possesses a putative sterol-binding site, we speculated that the Sig-1R might form a complex with Insig, thus regulating ERAD of CGalT.
We designed an immunoprecipitation study (i) to examine the potential interaction of Sig-1Rs with Insig1 and (ii) to clarify which Insig complex (i.e. Insig associating with SCAP or Insig coupling to ERAD) associates with Sig-1Rs. As shown in Fig. 7A, anti-FLAG antibodies immunoprecipitated Sig-1R-FLAG together with Insig1-Myc but not with SCAP. Insig1-Myc was pulled down with anti-FLAG antibodies only when Sig-1R-FLAG was co-expressed (Fig. 7A), verifying the specific co-immunoprecipitation of Insig1-Myc by Sig-1R-FLAG proteins. The result also indicates that Sig-1R-FLAG does not associate with SCAP. Conversely, anti-Myc antibodies immunoprecipitated Insig1-Myc together with SCAP and Sig-1R-FLAG, suggesting the existence of two Insig1 complexes: one associating with SCAP and the other with Sig-1Rs. Notably, when Sig-1R-FLAG was expressed, thus leading to the increase of Sig-1R·Insig1 complexes, SCAP associating with Insig1 was significantly reduced (Fig. 7A, lane 5 versus lane 6 in the third panel from the top). Therefore, Sig-1Rs associating with Insig1 may prevent the interaction of Insig1 with SCAP.
FIGURE 7.
Sig-1Rs form complexes with the Insig-mediated ERAD machinery. A, immunoprecipitation (IP) of the Sig-1R·Insig1 complexes. Plasmids expressing Sig-1R-FLAG (0.5 μg/6-cm dish), Insig1-Myc (0.02 μg/6-cm dish), and/or SCAP (0.03 μg/6-cm dish) were transfected 2 days before experiments. −, empty vectors were transfected. Note the decrease of SCAP co-immunoprecipitated with Insig1-Myc (lane 6). B, effects of the Sig-1R knockdown on sterol-induced degradation of HMG-CoA reductase. At day 0, CHO cells were transfected with HMG-CoA reductase-V5 and Insig1-Myc together with either control or Sig-1R siRNA. At day 1, growth medium was replaced with the sterol-reducing medium (see “Experimental Procedures”). At day 2, cells were incubated in the MEM-α containing 2% LPDS, 25 μm compactin, and 5 mm mevalonate with/without 25-hydroxycholesterol (25HC; 1 μg/ml) for 4 h. The level of HMG-CoA reductase-V5 was measured by immunoblotting. The graph represents the mean ± S.E. (error bars) of six independent experiments. *, p < 0.05; **, p < 0.01, paired t test. ##, p < 0.01 compared with siCon without 25HC. C, effects of sterols and (+)-pentazocine on Sig-1R·Insig1 association. At day 0, CHO cells were transfected with Sig-1R-FLAG (0.5 μg/6-cm dish) and Insig1-Myc (0.02 μg/6-cm dish). At day 1, the growth culture medium was replaced with the sterol-reducing medium (see “Experimental Procedures”), and 25-HC (1 μg/ml for 16 h), cholesterol (Chol; 10 μg/ml for 16 h), or lanosterol (Lano; 10 μg/ml for 16 h) was added. At day 2, (+)-pentazocine ((+)PTZ; 1 μm) was applied for 2 h. Myc immunoprecipitation were performed as shown in A. Images represent the results of 3–5 independent experiments. D, immunoprecipitation for detection of the association between truncated Sig-1R-V5 and Insig1-Myc. V5-tagged rat Sig-1Rs was immunoprecipitated with anti-V5 antibodies. Co-immunoprecipitated Insig1-Myc was detected by anti-Myc antibodies. *, truncated Sig-1R-V5 that co-immunoprecipitates Insig1-Myc. The same result was consistently obtained from four independent experiments. The scheme depicts the membrane topology of the Sig-1R at the ER. IB, immunoblot.
Sig-1Rs Are Involved in Insig-regulated ERAD
To test whether Sig-1Rs are involved in Insig-mediated ERAD, we employed a well characterized model of Insig-induced protein degradation. HMG-CoA reductase overexpressed in CHO cells is stable under sterol-reducing conditions, whereas 25-hydroxycholesterol promotes a rapid down-regulation of the enzyme via Insig (37). In this assay, Insig was routinely co-overexpressed to enhance Insig-induced protein degradation (37). We confirmed that the application of 25-hydroxycholesterol to sterol-reduced CHO cells indeed causes a reduction of V5-tagged HMG-CoA reductase (67.6% reduction, n = 4). Interestingly, knockdown of Sig-1Rs per se increased the level of HMG-CoA reductase-V5 (Fig. 7B). Further, in CHO cells knocking down Sig-1Rs, the 25-hydroxycholesterol-induced decrease of HMG-CoA reductase-V5 was significantly smaller (Fig. 7B; 30.6% reduction; *, p < 0.05 compared with the control siRNA samples by paired t test, n = 6), indicating that Sig-1Rs play a role in Insig-mediated ERAD.
Sig-1Rs Associate with Insig1 via the Second Transmembrane Domain in a 25-Hydroxycholesterol/Ligand-sensitive Manner
To gain insight into the functional relevance of the interaction between Sig-1Rs and Insig, the effect of sterols on their association was examined. We found that Sig-1Rs constitutively associate with Insig1 to a certain degree even under sterol-reducing conditions (Fig. 7C, lane 1), where the association of SCAP with Insig was completely abolished (45). 25-Hydroxycholesterol (1 μg/ml for 16 h), the potent inducer of Insig·SCAP association, potentiated the association between Sig-1R-FLAG and Insig1-Myc (Fig. 7C). The association of Sig-1R-FLAG with Insig1-Myc was unaffected by cholesterol (10 μg/ml for 16 h) or lanosterol (10 μg/ml for 16 h), the relatively less potent inducer of the Insig·SCAP association (Fig. 7C). Sig-1Rs bind sterols but also diverse classes of hydrophobic or amphipathic drugs (18). Notably, the prototypic Sig-1R ligand (+)-pentazocine potentiates the association of Sig-1R-FLAG with Insig1-Myc (Fig. 7C).
The domain of the Sig-1R responsible for the association with Insig was also explored by expressing various truncated Sig-1R-V5 (ΔSig-1R-V5). ΔSig-1R-V5 lacking a part of or the entire ER-lumenal domain (amino acids 116–223) associated with Insig1-Myc to a similar degree as shown with the full-length Sig-1R-V5 (Fig. 7D). The Sig-1R-V5 mutants that lack the second transmembrane domain (amino acids 90–116) no longer co-immunoprecipitated Insig1-Myc. Thus, the second transmembrane domain, which possesses a putative sterol-binding pocket (23), seems to be essential for the formation of the Sig-1R·Insig1 complex. In contrast, the ER lumenal domain (Sig-1R-V5(117–223) with/without the KDEL ER retrieval sequence) that exhibits innate chaperone activity of the Sig-1R (30) failed to associate with Insig1. We also found that knockdown of Sig-1Rs did not affect the protein level of Insig1-Myc (data not shown), suggesting that Sig-1Rs do not associate with Insig1 merely to stabilize/degrade Insig1.
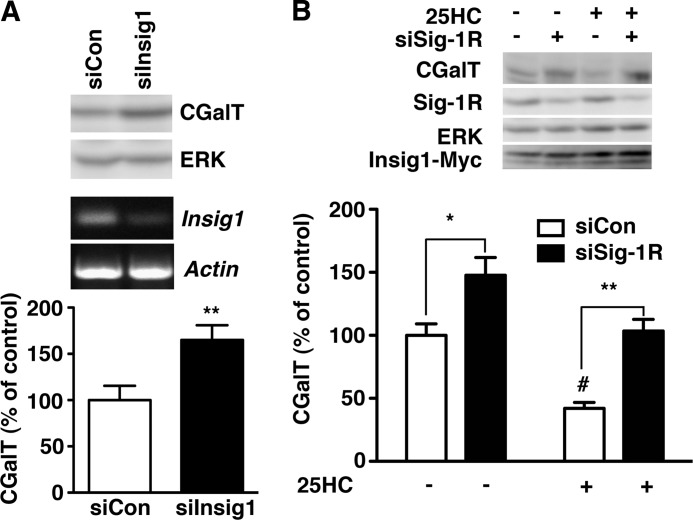
Sig-1Rs Promote the Down-regulation of CGalT Proteins via Insig-operated ERAD
Finally, we tested whether the Sig-1R·Insig1 complex plays a role in controlling the CGalT level. Similar to knockdown of Sig-1Rs, knockdown of Insig1 in CHO-CGalT significantly increased the protein level of CGalT (Fig. 8A) without affecting the mRNA level of CGalT (control siRNA-transfected cell, 100 ± 4.8%; Insig1 siRNA-transfected cell, 93.9 ± 11.4%; n = 5). We knocked down only Insig1 and not Insig2 in this study because Insig1 is known to play a primary role in sterol-induced ERAD in CHO cells (38).
FIGURE 8.
CGalT is degraded by Insig-mediated ERAD. A, effect of Insig1 knockdown on the protein level of CGalT. Control or active siRNA against rat Insig1 was transfected 2 days before the assay. The level of CGalT was measured by immunoblotting and normalized to ERK. The effect of Insig1 siRNA (siInsig1) was verified by RT-PCR (bottom two panels). The graph represents mean ± S.E. (error bars) (n = 5). **, p < 0.01, paired t test. B, sterol-sensitive down-regulation of CGalT. CHO-CGalT was transfected with Insig1-Myc in combination with control or Sig-1R siRNA. At day 2, the culture medium was replaced with the sterol-reducing medium as described in B. At day 1, 25HC (1 μg/ml) was applied to cells in the MEM-α containing 2% LPDS, 25 μm compactin, and 5 mm mevalonate for 4 h. The level of CGalT was measured by immunoblotting and normalized to ERK. The graph represents mean ± S.E. (n = 7). *, p < 0.05; **, p < 0.01 compared with siCon in the same treatment, paired t test. #, p < 0.05 compared with siCon without 25HC.
Adapting the experimental paradigm used in Fig. 7B, we examined whether 25-hydroxycholesterol induces the down-regulation of CGalT under sterol-reducing conditions. The application of 25-hydroxycholesterol causes a significant reduction of CGalT in control cells (Fig. 8B, 54.5% reduction, n = 7). In Sig-1R knockdown cells, the 25-hydeoxycholesterol-induced reduction of CGalT was significantly smaller when compared with CHO-CGalT transfected with control siRNA (Fig. 8B; 29.3% reduction in siSig-1R cells; *, p < 0.05 compared with the control by paired t test, n = 7). Taken together, these results suggest that similar to HMG-CoA reductase, the CGalT protein level is regulated by sterol-sensing ERAD that involves Sig-1Rs and Insig.
DISCUSSION
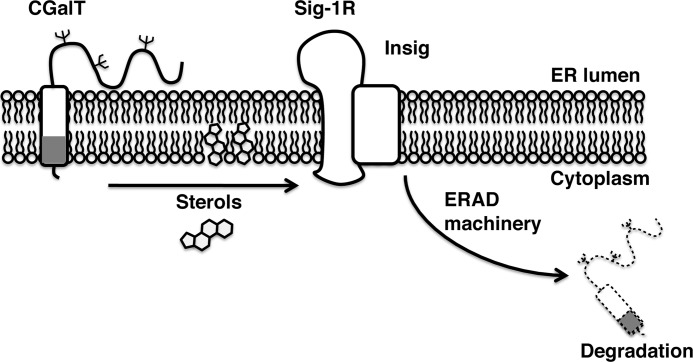
This study demonstrated for the first time that Insig-mediated ERAD involves the novel ER chaperone Sig-1R and that this ERAD system regulates the degradation of the sphingolipid enzyme CGalT. Our findings therefore suggest that expression of the CGalT enzyme and the production of GalCer can be regulated by sterols inside of the cell. Exactly how the Sig-1R, particularly its chaperone activity, is involved in the Insig-mediated ERAD machinery needs to be examined in the future at a detailed molecular level. Also, how Insig-mediated ERAD recognizes CGalT is unclear at present. Nevertheless, according to our amino acid sequence analysis, the transmembrane domain of rat CGalT contains a cholesterol-binding domain motif of the sequence (L/V)X1–5YX1–5(K/R) (amino acids V495TKFIY500RK502), as described in the benzodiazepine receptor (25). Further, we previously found that CGalT is photoaffinity-labeled with [3H]azicholesterol.4 Thus, it is plausible to speculate that CGalT interacting with sterols may be recognized by Insig (Fig. 9). It is also noteworthy that progesterone receptor membrane component 1 (PGRMC1) was recently found to constitute the binding activity of the sigma-2 receptor (46), another subtype of sigma receptors (47). Interestingly, PGRMC1 was previously found to associate with Insig (48). Thus, the two pharmacologically defined subtypes of sigma receptors, namely Sig-1R and sigma-2 receptor, may form heteropolymeric complexes with Insig to regulate the lipid-sensing ERAD.
FIGURE 9.
A scheme depicting a putative molecular action of Sig-1Rs regulating the CGalT level. The second transmembrane domain of the Sig-1R associates with Insig to form an ERAD complex at the ER membrane. The association is strengthened by 25-hydroxycholesterol or Sig-1R ligands, such as (+)-pentazocine. In the presence of high sterols (e.g. 25-hydroxysterol), CGalT is recruited to the Sig-1R-Insig machinery for degradation. Other components involved in the Sig-1R-regulated ERAD machinery (e.g. ubiquitin ligases) are not defined. The transmembrane domain of CGalT (a gray box) contains a putative sterol-binding motif that, upon binding to sterols, might be recognized by the Insig-mediated ERAD complex.
Because the level of endogenous CGalT in primary cells and schwannoma cell lines is considerably low, experiments for exploration of detailed molecular mechanisms of CGalT regulation are substantially limited with those cell types. Therefore, we used the CHO cell line stably expressing CGalT as a model to examine the post-translational modification of the enzyme. Therefore, it remains possible that the processing of ectopically expressed CGalT in CHO cells might be different from that of endogenously expressed CGalT. Nonetheless, our data from D6P2T schwannoma (Figs. 3B and 5B) support the notion that the mechanism we found in this study is utilized in cells endogenously expressing CGalT. Furthermore, because amino acid sequences of both Sig-1R and CGalT are highly conserved between mammals (>90.1% for the Sig-1R and >93.7% for CGalT between human, mouse, and rat; NCBI COBALT alignment analysis) and the cholesterol and sphingolipid metabolism is omnipresent in vertebrates, the mechanism found in our CHO cell model could be extrapolated to other mammalian cells, such as the human oligodendrocyte.
It is known that CGalT expression begins at the late progenitor stage of the oligodendrocyte-type 2 astrocyte lineage. The enzymatic product GalCer, which together with sulfatide comprises 27% of total myelin lipids (49), is transported from the ER to the outer leaflet of the oligodendrocyte plasma membrane to serve as a negative regulator of myelination (16). It is interesting that myelin is also highly enriched in cholesterol, the synthesis of which is increased at the same stage of oligodendrocyte differentiation (49). In contrast to GalCer, however, sterols are known to stimulate oligodendrocyte differentiation (50). Sterols and GalCer may therefore serve as an accelerator and a brake, respectively, on the oligodendrocyte differentiation, and our data suggest that the two systems might cross-talk via the Sig-1R-Insig complex. A future study using primary oligodendrocyte progenitors is certainly necessary to support this possibility and to explore a direct relationship between sterol levels and GalCer levels.
Previous studies have demonstrated that the second transmembrane domain of the Sig-1R is involved in the formation of the sterol/ligand-binding domain (23, 51). Notably, amino acids that are identical between Sig-1R and yeast C8-C7 sterol isomerase are in fact clustered within the second transmembrane domain of the Sig-1R (23). The segment of those amino acids corresponds to the sterol-binding pocket of a yeast C8-C7 sterol isomerase (23). In the presence of sterols, Insig associates specifically with the so-called sterol-sensing domain of SCAP or HMG-CoA reductase that binds 25-hydroxycholesterol (44). The association between Sig-1Rs and Insig1 is similarly potentiated by 25-hydroxicholesterol (Fig. 7C). Thus, it is conceivable that the second transmembrane domain of the Sig-1R may serve as a sterol-sensing domain that promotes the Insig association (Fig. 7A) and that 25-hydroxicholesterol may serve as a high affinity endogenous Sig-1R ligand. Importantly, however, there are some biochemical differences between Sig-1R-Insig and SCAP-Insig associations. First, under sterol-deprived conditions that can promote nearly complete dissociation of Insig from SCAP (45), the Sig-1R is still able to hold a certain degree of association with Insig. Second, cholesterol and lanosterol have no effect on the Sig-1R-Insig association, at least under our assay condition. Third, the association between Sig-1R and Insig can be altered by Sig-1R ligands as shown by the prototypic Sig-1R ligand (+)-pentazocine. The Sig-1R binds a variety of hydrophobic/amphipathic drugs (e.g. antidepressants) as well as certain endogenous compounds, such as progesterone, sphingolipids (e.g. d-erythro-sphingosine, and monoglycosylceramides), and N,N-dimethyltryptamine (20–22, 51). Although more studies are needed to confirm whether these synthetic and endogenous compounds similar to (+)-pentazocine also contribute to tightening the association between Sig-1R and Insig, the unique drug/lipid-binding profile of the Sig-1R may explain why Sig-1Rs still maintain a certain degree of the association with Insig under sterol-depriving conditions.
Our results indicate that the Insig1·Sig-1R complex is devoid of SCAP. The increased association of Sig-1R with Insig1 leads to the decrease of SCAP associating with Insig1 (Fig. 7A). This may suggest that the association of Sig-1Rs with Insig keeps Insig away from SCAP, thus influencing the equilibrium in the Insig·SCAP association. Because the dissociation of Insig from SCAP is a critical step for the departure of the SCAP·SREBP complex from ER to Golgi for SREBP activation (44), Sig-1R-mediated sequestration of Insig from SCAP may potentially result in activation of SREBP. Particularly under ER stress, where Sig-1Rs are highly up-regulated (30), the inhibitory action of Sig-1Rs on the Insig·SCAP association might become relevant. Indeed, we have found that knockdown of Sig-1Rs decreases the active form of SREBP1 (i.e. the form cleaved at Golgi), whereas the overexpression increases activated SREBP1 in CHO cells.3 This mechanism may also partly explain why ER stress activates SREBPs (52).
In summary, we found that a novel ERAD complex involving Sig-1R chaperones and Insig regulates the lifetime of CGalT under physiological conditions. The identified ERAD components may provide a mechanistic insight into understanding of the cross-talk between sterol and sphingolipid metabolisms. Last, we would like to mention that CGalT, based on its structure, belongs to the glucuronyltransferase family of ER enzymes (7, 39). In collaboration with cytochrome p450 that comprises the phase I drug metabolism pathway, glucuronyltransferases comprise the phase II drug metabolism pathway that plays a crucial role in biotransformation of drugs, sterols, and xenobiotics (39). At present, it is unknown why Sig-1R possesses the unusual binding profile that allows the protein to bind numerous types of drugs and sterols. It will be interesting to see whether Sig-1Rs, which bind xeno-/endobiotics, regulate the lifetime of drug metabolism enzymes that include the glucuronyltransferase family at the ER.
Acknowledgments
We thank Dr. Gerrit van Meer (University of Utrecht) for comments on this work and for providing valuable materials. We thank Mary Pfeiffer for critical editing of the manuscript.
This work was supported, in whole or in part, by the National Institutes of Health, NIDA, Intramural Research Program.
H. Sprong, unpublished data.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- CGalT
- UDP-galactose:ceramide galactosyltransferase
- Sig-1R
- sigma-1 receptor
- GlcCer
- glucosylceramide(s)
- GalCer
- galactosylceramide(s)
- SREBP
- sterol regulatory element-binding protein
- SCAP
- SREBP cleavage-activating protein
- NHK
- α1-antitrypsin variant, null (Hong Kong)
- EYFP
- enhanced yellow fluorescent protein
- MEM
- minimum essential medium
- HMG-CoA reductase
- 3-hydroxy-3-methylglutaryl-coenzyme A reductase
- TLC
- thin layer chromatography
- 25HC
- 25-hydroxycholesterol
- LPDS
- lipoprotein-deficient serum
- PGRMC1
- progesterone receptor membrane component 1
- siCon
- control siRNA
- siSig-1R
- sigma-1 receptor siRNA.
REFERENCES
- 1. van Meer G., Holthuis J. C. (2000) Sphingolipid transport in eukaryotic cells. Biochim. Biophys. Acta 1486, 145–170 [DOI] [PubMed] [Google Scholar]
- 2. Sprong H., Degroote S., Claessens T., van Drunen J., Oorschot V., Westerink B. H., Hirabayashi Y., Klumperman J., van der Sluijs P., van Meer G. (2001) Glycosphingolipids are required for sorting melanosomal proteins in the Golgi complex. J. Cell Biol. 155, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simons K., Vaz W. L. (2004) Model systems, lipid rafts, and cell membranes. Annu. Rev. Biophys. Biomol. Struct. 33, 269–295 [DOI] [PubMed] [Google Scholar]
- 4. Boggs J. M., Gao W., Hirahara Y. (2008) Myelin glycosphingolipids, galactosylceramide and sulfatide, participate in carbohydrate-carbohydrate interactions between apposed membranes and may form glycosynapses between oligodendrocyte and/or myelin membranes. Biochim. Biophys. Acta 1780, 445–455 [DOI] [PubMed] [Google Scholar]
- 5. Lopez P. H., Schnaar R. L. (2009) Gangliosides in cell recognition and membrane protein regulation. Curr. Opin. Struct. Biol. 19, 549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hakomori S. I. (2008) Structure and function of glycosphingolipids and sphingolipids. Recollections and future trends. Biochim. Biophys. Acta 1780, 325–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sprong H., Kruithof B., Leijendekker R., Slot J. W., van Meer G., van der Sluijs P. (1998) UDP-galactose:ceramide galactosyltransferase is a class I integral membrane protein of the endoplasmic reticulum. J. Biol. Chem. 273, 25880–25888 [DOI] [PubMed] [Google Scholar]
- 8. Halter D., Neumann S., van Dijk S. M., Wolthoorn J., de Mazière A. M., Vieira O. V., Mattjus P., Klumperman J., van Meer G., Sprong H. (2007) Pre- and post-Golgi translocation of glucosylceramide in glycosphingolipid synthesis. J. Cell Biol. 179, 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kohyama-Koganeya A., Sasamura T., Oshima E., Suzuki E., Nishihara S., Ueda R., Hirabayashi Y. (2004) Drosophila glucosylceramide synthase. A negative regulator of cell death mediated by proapoptotic factors. J. Biol. Chem. 279, 35995–36002 [DOI] [PubMed] [Google Scholar]
- 10. Sprong H., van der Sluijs P., van Meer G. (2001) How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2, 504–513 [DOI] [PubMed] [Google Scholar]
- 11. Sot J., Aranda F. J., Collado M. I., Goñi F. M., Alonso A. (2005) Different effects of long- and short-chain ceramides on the gel-fluid and lamellar-hexagonal transitions of phospholipids. A calorimetric, NMR, and x-ray diffraction study. Biophys. J. 88, 3368–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids. Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sprong H., Degroote S., Nilsson T., Kawakita M., Ishida N., van der Sluijs P., van Meer G. (2003) Association of the Golgi UDP-galactose transporter with UDP-galactose:ceramide galactosyltransferase allows UDP-galactose import in the endoplasmic reticulum. Mol. Biol. Cell 14, 3482–3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coetzee T., Fujita N., Dupree J., Shi R., Blight A., Suzuki K., Suzuki K., Popko B. (1996) Myelination in the absence of galactocerebroside and sulfatide. Normal structure with abnormal function and regional instability. Cell 86, 209–219 [DOI] [PubMed] [Google Scholar]
- 15. Schulte S., Stoffel W. (1993) Ceramide UDPgalactosyltransferase from myelinating rat brain. Purification, cloning, and expression. Proc. Natl. Acad. Sci. U.S.A. 90, 10265–10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bansal R., Winkler S., Bheddah S. (1999) Negative regulation of oligodendrocyte differentiation by galactosphingolipids. J. Neurosci. 19, 7913–7924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hashimoto K. (2009) Sigma-1 receptors and selective serotonin reuptake inhibitors. Clinical implications of their relationship. Cent. Nerv. Syst. Agents Med. Chem. 9, 197–204 [DOI] [PubMed] [Google Scholar]
- 18. Hayashi T., Tsai S. Y., Mori T., Fujimoto M., Su T. P. (2011) Targeting ligand-operated chaperone sigma-1 receptors in the treatment of neuropsychiatric disorders. Expert Opin. Ther. Targets 15, 557–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su T. P., London E. D., Jaffe J. H. (1988) Steroid binding at sigma receptors suggests a link between endocrine, nervous, and immune systems. Science 240, 219–221 [DOI] [PubMed] [Google Scholar]
- 20. Fontanilla D., Johannessen M., Hajipour A. R., Cozzi N. V., Jackson M. B., Ruoho A. E. (2009) The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 323, 934–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramachandran S., Chu U. B., Mavlyutov T. A., Pal A., Pyne S., Ruoho A. E. (2009) The sigma1 receptor interacts with N-alkyl amines and endogenous sphingolipids. Eur. J. Pharmacol. 609, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hayashi T., Fujimoto M. (2010) Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol. Pharmacol. 77, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanner M., Moebius F. F., Flandorfer A., Knaus H. G., Striessnig J., Kempner E., Glossmann H. (1996) Purification, molecular cloning, and expression of the mammalian sigma1-binding site. Proc. Natl. Acad. Sci. U.S.A. 93, 8072–8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maurice T., Su T. P., Privat A. (1998) Sigma1 (sigma 1) receptor agonists and neurosteroids attenuate B25–35-amyloid peptide-induced amnesia in mice through a common mechanism. Neuroscience 83, 413–428 [DOI] [PubMed] [Google Scholar]
- 25. Meunier J., Ieni J., Maurice T. (2006) The anti-amnesic and neuroprotective effects of donepezil against amyloid β25–35 peptide-induced toxicity in mice involve an interaction with the sigma1 receptor. Br. J. Pharmacol. 149, 998–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsumoto R. R., McCracken K. A., Pouw B., Zhang Y., Bowen W. D. (2002) Involvement of sigma receptors in the behavioral effects of cocaine. Evidence from novel ligands and antisense oligodeoxynucleotides. Neuropharmacology 42, 1043–1055 [DOI] [PubMed] [Google Scholar]
- 27. Sabino V., Cottone P., Zhao Y., Iyer M. R., Steardo L., Jr., Steardo L., Rice K. C., Conti B., Koob G. F., Zorrilla E. P. (2009) The sigma-receptor antagonist BD-1063 decreases ethanol intake and reinforcement in animal models of excessive drinking. Neuropsychopharmacology 34, 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim F. J., Kovalyshyn I., Burgman M., Neilan C., Chien C. C., Pasternak G. W. (2010) Sigma 1 receptor modulation of G-protein-coupled receptor signaling. Potentiation of opioid transduction independent from receptor binding. Mol. Pharmacol. 77, 695–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nuwayhid S. J., Werling L. L. (2003) Sigma1 receptor agonist-mediated regulation of N-methyl-d-aspartate-stimulated [3H]dopamine release is dependent upon protein kinase C. J. Pharmacol. Exp. Ther. 304, 364–369 [DOI] [PubMed] [Google Scholar]
- 30. Hayashi T., Su T. P. (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca2+ signaling and cell survival. Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 31. Hayashi T., Rizzuto R., Hajnoczky G., Su T. P. (2009) MAM. More than just a housekeeper. Trends Cell Biol. 19, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palacios G., Muro A., Vela J. M., Molina-Holgado E., Guitart X., Ovalle S., Zamanillo D. (2003) Immunohistochemical localization of the sigma1-receptor in oligodendrocytes in the rat central nervous system. Brain Res. 961, 92–99 [DOI] [PubMed] [Google Scholar]
- 33. Hayashi T., Su T. P. (2004) Sigma-1 receptors at galactosylceramide-enriched lipid microdomains regulate oligodendrocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 101, 14949–14954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meunier J., Hayashi T. (2010) Sigma-1 receptors regulate Bcl-2 expression by reactive oxygen species-dependent transcriptional regulation of nuclear factor κB. J. Pharmacol. Exp. Ther. 332, 388–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hayashi T., Su T. P. (2001) Regulating ankyrin dynamics. Roles of sigma-1 receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 37. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 38. Sever N., Lee P. C., Song B. L., Rawson R. B., Debose-Boyd R. A. (2004) Isolation of mutant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 279, 43136–43147 [DOI] [PubMed] [Google Scholar]
- 39. Iyanagi T. (2007) Molecular mechanism of phase I and phase II drug-metabolizing enzymes. Implications for detoxification. Int. Rev. Cytol. 260, 35–112 [DOI] [PubMed] [Google Scholar]
- 40. Fewou S. N., Büssow H., Schaeren-Wiemers N., Vanier M. T., Macklin W. B., Gieselmann V., Eckhardt M. (2005) Reversal of non-hydroxy:α-hydroxy galactosylceramide ratio and unstable myelin in transgenic mice overexpressing UDP-galactose:ceramide galactosyltransferase. J. Neurochem. 94, 469–481 [DOI] [PubMed] [Google Scholar]
- 41. Bernasconi R., Molinari M. (2011) ERAD and ERAD tuning. Disposal of cargo and of ERAD regulators from the mammalian ER. Curr. Opin. Cell Biol. 23, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith M. H., Ploegh H. L., Weissman J. S. (2011) Road to ruin. Targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bernasconi R., Galli C., Calanca V., Nakajima T., Molinari M. (2010) Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J. Cell Biol. 188, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goldstein J. L., DeBose-Boyd R. A., Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 [DOI] [PubMed] [Google Scholar]
- 45. Hua X., Nohturfft A., Goldstein J. L., Brown M. S. (1996) Sterol resistance in CHO cells traced to point mutation in SREBP cleavage-activating protein. Cell 87, 415–426 [DOI] [PubMed] [Google Scholar]
- 46. Xu J., Zeng C., Chu W., Pan F., Rothfuss J. M., Zhang F., Tu Z., Zhou D., Zeng D., Vangveravong S., Johnston F., Spitzer D., Chang K. C., Hotchkiss R. S., Hawkins W. G., Wheeler K. T., Mach R. H. (2011) Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat. Commun. 2, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bowen W. D. (2000) Sigma receptors. Recent advances and new clinical potentials. Pharm. Acta Helv. 74, 211–218 [DOI] [PubMed] [Google Scholar]
- 48. Suchanek M., Radzikowska A., Thiele C. (2005) Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods 2, 261–267 [DOI] [PubMed] [Google Scholar]
- 49. Dietschy J. M., Turley S. D. (2001) Cholesterol metabolism in the brain. Curr. Opin. Lipidol. 12, 105–112 [DOI] [PubMed] [Google Scholar]
- 50. Baulieu E. E., Robel P., Schumacher M. (2001) Neurosteroids. Beginning of the story. Int. Rev. Neurobiol. 46, 1–32 [DOI] [PubMed] [Google Scholar]
- 51. Pal A., Hajipour A. R., Fontanilla D., Ramachandran S., Chu U. B., Mavlyutov T., Ruoho A. E. (2007) Identification of regions of the sigma-1 receptor ligand binding site using a novel photoprobe. Mol. Pharmacol. 72, 921–933 [DOI] [PubMed] [Google Scholar]
- 52. Kammoun H. L., Chabanon H., Hainault I., Luquet S., Magnan C., Koike T., Ferré P., Foufelle F. (2009) GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest. 119, 1201–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]