Abstract
The insertion of a stable soluble protein into loops of transmembrane proteins has proved to be a successful approach for enhancing their stabilities and crystallization, and may also be useful in contexts where the inserted proteins can modulate or report on the activities of membrane proteins. While the use of T4 lysozyme to replace portions of the third intracellular loops of G protein-coupled receptors (GPCRs) has allowed determination of the structures of members of this important class of receptors, the creation of such fusion proteins generally leads to loss of signaling function of the resulting fusion protein, since the third intracellular loops of GPCRs play critical roles in their interactions with G proteins. We describe here a random screening approach allowing insertion of T4 lysozyme into diverse positions in the third loop of the yeast α-pheromone receptor, a GPCR encoded by the yeast STE2 gene. Insertions were accompanied by varying extents of deletion or duplication of the loop. A set of phenotypic screens allow detection of potentially rare variant receptors that are expressed, bind to agonist and are capable of signal transduction via activation of the cognate G protein. A large fraction of screened full-length receptor variants containing at least partial duplications of the loop on either side of the inserted T4 lysozyme retain the ability to activate the downstream signaling pathway in response to binding of ligand. However, we were unable to identify any receptors with truncated C-termini that retain significant signaling function in the presence of inserted T4 lysozyme. Our results establish the feasibility of creating functional receptors containing insertions of T4 lysozyme in their third intracellular loops.
Keywords: G protein-coupled receptors, pheromone receptor, receptor stabilization, T4 Lysozyme, yeast
Despite the importance of transmembrane proteins in physiology and medicine, and the fact that they make up 20–30% of the proteins encoded in most genomes, the number of such proteins for which high-resolution structures has been determined remains relatively small (White). In particular, at this writing, the number of known high-resolution structures of different transmembrane proteins derived from eukaryotic sources (not including mitochondria and chloroplasts) is currently only about two dozen, of which about half are members of the G protein-coupled receptor (GPCR) superfamily.
A major technical innovation that has contributed to most of the available structures of GPCRs has been the engineering of T4 lysozyme (T4L) as an insertion into, or replacement for, the third intracellular (IC3) loops of the receptors. The rationale for this replacement, first suggested by Prive and co-workers (Prive et al., 1994; Engel et al., 2002a,b), is that the insertion of an intact soluble protein as a replacement for a flexible loop of a structurally dynamic transmembrane protein could stabilize the protein in a single conformation that would be more likely to crystallize than the unmodified protein. In addition, insertion of a small, readily crystallizeable, soluble protein into membrane proteins that lack aqueous-facing protein-interacting surfaces may provide new protein–protein contacts for crystallization. To date, the creation of T4 lysozyme-containing fusions has facilitated the structure determination of the β2-adrenergic (Cherezov et al., 2007; Rosenbaum et al., 2007), A2a adenosine (Jaakola et al., 2008), dopamine D3 (Chien et al., 2010); CXCR4 chemokine (Wu et al., 2010), histamine H1 (Shimamura et al., 2011), lyso-phospholipid S1P (Hanson et al., 2012), M2 and M3 muscarinic acetylcholine (Haga et al., 2012; Kruse et al., 2012), and δ-, κ- and μ-opioid (Granier et al., 2012; Manglik et al., 2012; Wu et al., 2012) receptors. Rhodopsin is the only GPCR for which a structure was initially determined without introduction of any stabilizing substitutions into the protein. The structure of the β1-adrenergic and A2a adenosine receptors has been determined using mutated receptors containing multiple stabilizing point mutations and deletions, but no insertion of a fusion protein (Warne et al., 2008; Tate and Schertler, 2009; Lebon et al., 2011). A structure of the β2-adrenergic receptor has also been solved without fusing to T4 lysozyme by stabilizing the protein in complexes with an antibody (Rasmussen et al., 2007). Recently, a structure of the β2-adrenergic receptor has been solved in complex with G protein. The receptor construct used in this study contained no fusion protein in the intracellular loops but did contain T4 lysozyme as a replacement for the extracellular N-terminal of the receptor (Rasmussen et al., 2011b).
In addition to being useful for X-ray crystallography, the insertion of soluble proteins into loops of transmembrane proteins may prove to have other applications, such as the incorporation of fluorescent sensor proteins into functionally important parts of membrane proteins, or promoting novel protein–protein interactions by insertion of soluble interaction domains into loops of receptors and transporters. Insertion of such soluble domains into loop regions may have advantages over the fusion of the domains into flexible or extended N- or C-termini of membrane proteins.
The GPCR/T4L fusion constructs that have been used for structure determination retain the ability to bind ligand with high affinity and exhibit other properties indicating that they adopt a native-like structure. However, GPCR IC3 loops have, in many cases, been implicated in receptor–G protein coupling, thus, modification of the loop could be expected to interfere with G protein activation. Insertion of T4L into the β2-adrenergic receptor rendered the altered receptor incapable of catalyzing nucleotide exchange by the cognate Gs protein (Rosenbaum et al., 2007). Assays of G protein activation by other GPCR/T4L fusion constructs have not generally been reported. This raises uncertainties about the details of models of receptor activation based on structures of receptor-T4L fusions.
The particular constructs used in previous structure determinations of receptor-T4L fusions have, to date, been identified based on trial-and-error approaches involving cycles of design and testing of multiple individual constructs (Rosenbaum et al., 2007). As an alternative approach, we have developed and tested a procedure for screening receptor constructs in which T4L has been inserted at random positions in IC3 as a replacement for variable lengths of the endogenous loop sequence. We applied this procedure in the creation of T4L fusions with the yeast α-factor receptor, Ste2p, a member of the GPCR family that is, in some cases, functionally interchangeable with mammalian GPCRs, despite sharing little sequence similarity to the mammalian receptors (King et al., 1990; Dowell and Brown, 2009). Screening libraries of random insertions allowed rapid identification of Ste2p-T4L fusions that maintain optimal ligand-binding properties, as well as, surprisingly, fusions of T4L in the receptor third intracellular loop that retain full competence to activate downstream signaling responses.
Materials and methods
Yeast plasmids and strains
Starting plasmids for insertional mutagenesis were created based on the previously described multicopy plasmids pMD1154 and pMD1167 encoding, respectively, truncated (Δ305–431) and full-length versions of Ste2p fused to GFP2 (Gehret et al., 2006) (Note that all discussion of GFP in this manuscript refers to the GFP2 variant used for bioluminescence resonance energy transfer BRET2 (Bertrand et al., 2002)). Deletion of the third intracellular loop of Ste2p and insertion of BsaBI and SmaI sites was accomplished by site-directed mutagenesis using oligonucleotide ON1311 to create plasmids pMD1858 and pMD1859 encoding truncated and full-length receptors, respectively. (The SmaI site can be used to improve the efficiency of generating insert-containing plasmid by removing any plasmid that was not cleaved at both BsaBI sites prior to transformation into E.coli.) (Sequences of referenced oligonucleotides are available from the authors upon request.)
Plasmid pMD1887, containing full duplication of the 16-residues IC3 loop, but no inserted T4L, was created by oligonucleotide-directed mutagenesis (oligonucleotide ON 1424) of plasmid pMD1886, recovered from the library screen, that contained 16 residues of IC3, followed by T4L, followed by 15 residues of IC3. Yeast strain A2953 (Taslimi et al., 2012) was used for screening of insertional libararies. Strain A575 (Sommers et al., 2000) was used for retransformation with identified STE2 alleles for further characterization. Plasmids pMD4277, pMD4279, pMD4283 and pMD4284 expressing C-terminally truncated receptors with IC3 duplications and T4L insertions were created by cloning regions of STE2 genes containing the insertion from full-length receptor alleles isolated from screening of libraries as a replacement for the corresponding region of a truncated STE2 gene. In particular, the region encompassing IC3 was excised from the library plasmid by digestion with NheI and KpnI, then ligated into similarly cut plasmid pMD1230 encoding a truncated STE2 gene with a C-terminal triple myc epitope. Plasmid pMD1230, in turn, was created by excising the NheI-KpnI fragment from pMD1003, encoding a full-length STE2 allele and ligating it into NheI-KpnI-cut pMD1055, encoding the C-terminally truncated form of the receptor (Gehret et al., 2006).
For expression testing and immunoblotting, full-length alleles containing IC3 insertions were inserted into plasmid pSGP46 by ligation-independent cloning, as described previously (Clark et al., 2010). This plasmid provides expression under control of the inducible ADH2 promoter and fusion of cleavable ZZ (protein A) and RGSHis10 tags (Clark et al., 2010). Expression testing was conducted following transformation of the relevant plasmids into the protease-deficient yeast strain BJ5460 (ATCC 208285; A3702) (Jones, 2002; Clark et al., 2010).
A table listing the yeast strains and plasmids created for this study is presented in the Supplementary material for this manuscript as Supplementary Table SI.
Creation of the insertional library
A schematic representation of the creation of the mutational library is presented in Fig. 2. A version of the T4 lysozyme gene containing the mutations Cys54Thr and Cys97Ala (provided by the laboratory of Joseph Wedekind) encoded on pMD1854 (pHS1403)) was subjected to amplification using primers (ON1309 and ON1310) that added flanking BbsI sites, as well as a sequence encoding the amino acid sequence Ser–Gly and a HindIII site at the 5′ end and the sequence Gly–Ser and a SacI site at the 3′ end. Pairs of complementary oligonucleotides corresponding to different lengths of N- and C-terminal linkers were mixed (1 μM each), heated to ∼95°C and allowed to slowly cool to 5°C. They were then ligated to the amplified T4 lysozyme gene that had been digested with BbsI. The ligation products ranging in length from ∼500 to 700 bp) were then subjected to gel purification using Purelink gel extraction kits (Invitrogen). Plasmids pMD1858 and pMD1859 encoding truncated or full-length forms of Ste2p with BsaBI sites inserted in place of the third intracellular loop were digested with BsaBI and subjected to gel purification. The digested plasmids (final concentration 2.5 ng/μl) were mixed with the linker-ligated T4 lysozyme gene (final concentration 0.5 ng/μl) and incubated for 1 h at 22°C. The mixtures were then used to transform NOVABlue cells (Novagen). Totals of 7500 and 6000 transformants were obtained for the full-length and truncated libraries, respectively. Single colonies were picked for sequencing of random clones, then the libraries were pooled and cultured for preparation of plasmid DNA. The pooled plasmid DNA was used to transform the indicated yeast strain (A2953), yielding 2400 and 2000 colonies for the full-length and truncated libraries, respectively.
Fig. 2.
Schematic diagram of procedure for random insertion of T4L into the third intracellular loop of the α-factor receptor. (a) PCR product following amplification of the gene for T4 lysozyme. Amplification was conducted using primers that added HindIII and BbsI sites at the 5′ end and BbsI and SacI sites at the 3′ end. The BbsI sites were used for insertion into the STE2 gene. The additional sites were added for ligation into cloning vectors. Underlined sequences correspond to the indicated restriction sites. (b) Amplified T4 lysozyme-containing PCR product following cleavage at the flanking BbsI sites. (c) Product of ligation of cleaved T4 lysozyme-containing PCR product with the library of variable-length linkers. The bases indicated in bold are derived from the linkers. ‘NXoligo’ refers to the N-terminal oligonucleotides listed in Supplementary Table SII. ‘CXoligo’ refers to the C-terminal oligonucleotides listed in Supplementary Table SIII. (d) Nucleotide and amino acid sequences of the region of STE2 including the third intracellular loop. The boxes indicate the positions of predicted transmembrane segments. (e) Sequence of the modified form of STE2 from which the third intracellular loop has been deleted. The converging BsaBI sites are indicated. Recognition sequences for BsaBI (consisting of the sequence GATNNNNATC, where N can be any base) are underlined. Note that the cleavage site for this enzyme is outside the actual recognition sequence. ‘Ter’ refers to a termination sequence that would be encountered if this form of the plasmid lacking a T4 lysozyme insertion were to be transcribed. The indicated SmaI site can be used to linearized uncleaved plasmids in case that the BsaBI cleavage is not efficient. (f) Product of BsaBI cleavage of the modified STE2 gene. (g) Formation of single-stranded overhangs by T4 DNA polymerase in the presence of dCTP. (h) Products of ligation independent cloning of variable length linkers ligated to T4 lysozyme into cleaved T4-DNA polymerase-treated plasmids encoding modified STE2 genes.
Synthesis and fluorescent properties of [K7(bodipy-TR),Nle12]α-factor
The fluorescent α-factor analog [K7(bodipy-TR),Nle12] α-factor was used to detect expression of binding-competent receptors at the yeast cell surface. This reagent was used instead of the previously described NBD-labeled α-factor ([K7(NBD),Nle12] α-factor) (Ding et al., 2001, 2002; Bajaj et al., 2004) because of the similarity between the fluorescence emission of NBD-labeled ligand and the GFP2 fused to the STE2 alleles used for screening. The synthesis of [K7(bodipy-TR),Nle12]α-factor followed procedures previously reported to derivatize the ɛ-amine of Lys7 of α-factor starting with Fmoc[Nle12]α-factor (Ding et al., 2001, 2002). To a solution of Fmoc-WHWLQLKPGQPNleY (17.4 mg, 8.22 μmol) in N,N-dimethylformamide (DMF) (1 ml) and sodium tetraborate (50 mM, 1 ml) at 4°C, was added a solution of 6-(((4-(4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-yl)phenoxy)acetyl)amino)hexanoic acid, succinimidyl ester (BODIPY TR-X, SE; Life Technologies Corp.) (5 mg) in 0.5 ml DMF. The mixture was stirred at 4°C for 30 min. Reverse phase high pressure liquid chromatography (HPLC) analysis indicated that no starting material remained. Piperidine (0.1 ml) in 0.4 ml DMF was added to the reaction and the resulting mixture was stirred at 4°C for 1 h, at which point HPLC indicated complete cleavage of the Fmoc protecting group. The solution was then neutralized with 2% HCl (ca. 1.6 ml), and after filtration the filtrate was purified twice (2.3 ml each) by preparative HPLC (Delta Pak, 15 μ, 19 mm × 300 mm) using 20–70% acetonitrile (0.1%TFA) over 70 min. Pure fractions were combined and lyophilized. The final product [K7(bodipy-TR),Nle12]α-factor (15 mg) was highly homogeneous when analyzed by reverse-phase HPLC (Supplementary Fig. S1) and had the expected MW (calculated: 2185; found: 2185).
The [K7(bodipy-TR),Nle12]α-factor binds to Ste2p with essentially the same affinity (see below) as the previously characterized fluorescent ligand [K7(NBD),Nle12]α-factor (Bajaj et al., 2004; Bajaj et al., 2007). However, in comparison with the NBD derivative, [K7(bodipy-TR),Nle12]α-factor does not display as great a difference between the emission intensities of the bound and free ligand. Thus, flow cytometry-based saturation binding experiments with the bodipy-TR derivative exhibit a greater contribution to fluorescence emission from non-specifically bound ligand than was case for [K7(NBD),Nle12]α-factor.
Flow cytometry
Screening for clones expressing receptors competent to bind ligands was conducted using the fluorescent α-factor agonist, [K7(bodipy-TR),Nle12]α-factor, in which Lys7 is derivatized with the fluorophore bodipy-TR (6-(((4-(4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-yl)phenoxy)acetyl)amino)hexanoic acid). Cells expressing the library of T4-lysozyme-containing receptors were cultured in SD-ura to an OD600 of ∼1, incubated on ice with 100 or 500 nM [K7(bodipy-TR),Nle12]α-factor, then subjected to FACS using a Becton Dickenson FACSAria instrument with excitation at 532 nm and detection at 610 nm (20 nm bandwidth). Individual clones obtained from sorting were plated on SD-ura medium, then subcloned to single colonies. Plasmid DNA from each clone was used for DNA sequencing, and used to re-transform fresh cells of the strain A575 for further testing of receptor properties. The ligand-binding properties of individual clones were assayed by conducting saturation binding experiments using a Becton Dickinson LSRII flow cytometer with excitation at 532 nm and detection at 660 nm using [K7(bodipy-TR),Nle12]α-factor essentially as described previously (Mathew et al., 2011). The data were fit by non-linear least squares procedure to single site binding with a non-specific component using the ligand-binding module of Sigmaplot (SPSS). GFP expressed in the cells was excited at 488 nm and was detected at 515 nm.
Screens and assays of signaling function
Screening for signaling-competent receptors was performed by replica plating libraries of yeast transformants on SD-ura-his medium containing 300 nM α-factor. The presence of the FUS1-HIS3 reporter in strain A2953 allows colonies expressing signaling-competent receptors to grow when the pheromone response pathway is activated. Individual clones exhibiting growth in the presence of α-factor were used for the isolation of plasmid DNA, which was subjected to DNA sequencing and used to re-transform a fresh yeast host (A575) for quantitation of α-factor responses. α-Factor responses were assayed based on induction of a FUS1-lacZ reporter using the fluorogenic substrate fluorescein di-β-d-galactopyranoside as described previously (Celic et al., 2004). A small number of transformants from the library of truncated receptors initially exhibited growth on medium lacking histidine. However, upon re-transformation into a fresh host, plasmid derived from these cells failed to confer α-factor-dependent signaling as judged by the lack of induction of the FUS1-lacZ reporter. Levels of FUS1-lacZ induction in response to α-factor were fit to sigmoidal functions by a non-linear least squares procedure using the ligand-binding module of Sigmaplot (SPSS). Because of day-to-day variability in assays of FUS1-lacZ responses, all assays included a strain expressing receptors without IC3 inserts, to which the responses of experimental strains were compared.
Immunoblotting
Cells were cultured in SD-ura medium to an OD600 of ∼3. The cells were then diluted 4-fold in YPD medium and were regrown to an OD600 of ∼10. The cells (5 OD × ml) were then harvested, washed with cold, sterile water and resuspended in 300 μl of 20 mM sodium acetate (pH 4.6), 0.1 mM EDTA and 1 mM PMSF. Cells were disrupted by vortexing with glass beads in 10 cycles of 20 s of vortexing and 2 min of cooling on ice, followed by centrifugation to pellet the beads. The cell extract (8 μl) was mixed with 22 μl of a solution containing 100 mM Tris pH 6.8, 0.1 mM EDTA, 9 M urea, 5% sodium dodecyl sulfate (SDS), 0.02 mg/ml bromophenol blue, warmed at 37°C for 5 min and subjected to SDS-polyacrylamide gel electrophoresis on 10% gels, transferred to nitrocellulose filters and probed as recommended by the manufacturer using anti-RGS-HRP antibodies (Qiagen) diluted 1 : 100 000 or anti-enolase antibodies diluted 1 : 30 000 (a generous gift of Michael Holland of the University of California, Davis). An horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Sigma), diluted 1 : 10 000, was used to detect the enolase. The proteins bands were visualized using SuperSignal reagent (Pierce).
Results
Previous strategies for fusing T4L to receptors have relied on construction and testing of individual T4L fusion constructs containing various extents of deletion of the third intracellular loop (Rosenbaum et al., 2007). We describe here the development of an approach in which T4L is randomly inserted at different sites in the third intracellular loop of the yeast α-factor receptor Ste2p, accompanied by variable extents of deletion of loop sequences (see Fig. 1). The combination of the receptor construct used and the choice of yeast host strain for expression allows direct and exhaustive screening for cells expressing libraries of receptor variants to identify those with the following three characteristics: (i) highest levels of total receptor protein. Green fluorescent protein (GFP) was appended to the C-terminus of all Ste2p receptor constructs containing the T4L insertions, allowing quantitation of levels of fusion protein by flow cytometry and determination of receptor subcellular location by fluorescence microscopy. Ste2p-GFP fusions have previously been shown to retain full signaling function (Li et al., 1999; Gehret et al., 2006). (ii) Highest cell surface expression of receptor that is competent to bind ligand. Screening for this property was conducted by fluorescence-activated cell sorting (FACS) using a fluorescently labeled analog of α-factor (Bajaj et al., 2004; Mathew et al., 2011), following approaches that have previously been applied to GPCRs and other membrane- and surface-expressed proteins in yeast and other types of cells (Sklar et al., 1984; Nolan and Sklar, 1998; Boder and Wittrup, 2000; Sklar et al., 2002; Sarkar et al., 2008). (iii) Maximal signaling function of T4L-containing receptors. G protein activation by mutant receptors was assayed based on α-factor-dependent induction of expression of a HIS3 reporter gene fused to the pheromone-responsive FUS1 reporter. This provided the cells expressing functional receptors with the ability to grow on medium lacking histidine in the presence of agonist (Manfredi et al., 1996).
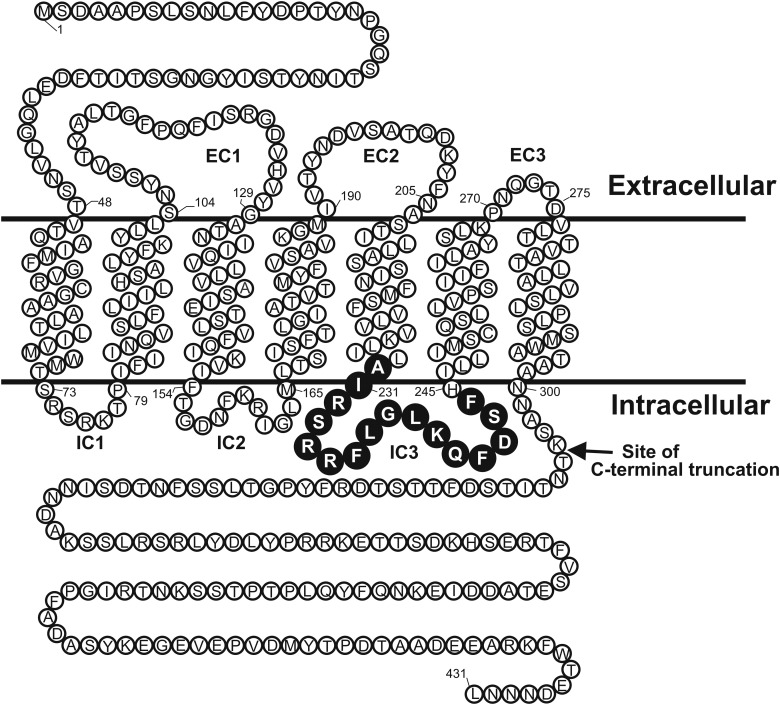
Fig. 1.
Schematic diagram of the predicted topology of Ste2p. Residues in the predicted IC3 region targeted for deletion and duplication associated with insertion of T4 lysozyme are shown as black circles with white letters. The site of C-terminal truncation is indicated.
The approach for random insertion of T4L into the IC3 loop of Ste2p is based on ligation of variable length linkers into restriction sites introduced at the 5′ and 3′ ends of the T4L gene, followed by insertion of the ligated products into Ste2p via ligation independent cloning (Aslanidis and de Jong, 1990; Dieckman et al., 2002; Alexandrov et al., 2004) (Fig. 2). Two sets of 17 different variable-length double-stranded linkers containing single stranded 5′ overhanging regions (Supplementary Tables SII and SIII) were prepared corresponding to various positions of insertion at the N- and C-terminal ends of the IC3 loop. These linkers were pooled and ligated to the ends of the amplified T4L gene. The linkers encoded Ser-Gly and Gly-Ser sequences in frame with the T4L reading frame at its N- and C-terminal ends, respectively. A plasmid to serve as a recipient for the T4L insertions was prepared by modifying the STE2 gene so as to contain pairs of converging BsaBI sites replacing the region encoding the third IC3 loop. Cleavage of the plasmid at these sites creates a linear piece of DNA that can be treated with T4 DNA polymerase in the presence of dCTP (but no other deoxynucleoside triphosphates) to generate single-stranded 5′ overhangs complementary to those in the linker sequences ligated to T4L. The T4 polymerase-treated linearized vector was incubated with the library of T4L-containing inserts, then transformed into Escherichia coli, allowing cellular enzymes to ligate the pieces together. The described procedures yield alleles containing T4L insertions at different sites in the IC3 loop of Ste2p. Insertion can be accompanied by duplication or deletion of sequences from the third intracellular loop.
Following their transformation into E.coli, libraries of transformants were pooled and subjected to plasmid isolation. The resulting libraries of mutated plasmids were used to transform a yeast strain containing a deletion of the normal chromosomal copy of STE2. Following testing for α-factor binding and signaling function, receptor-encoding plasmids were isolated from strains exhibiting relevant phenotypes. These plasmids were subjected to DNA sequencing and were re-transformed into a fresh ste2-Δ host in order to determine whether the phenotype was plasmid dependent.
Two separate insertional libraries were created, starting with either full-length STE2 or with a C-terminally truncated form STE2-Δ305-431. In the absence of T4L, such truncated receptors exhibit full signaling function and, compared with full-length receptor, enhanced sensitivity to α-factor and an increased number of cell surface binding sites (Konopka et al., 1988; Reneke et al., 1988), presumably due to removal of sites of phosphorylation and ubiquitination (Hicke, 1997; Hicke et al., 1998). Insertion of T4L into truncated receptors was, thus, pursued as a way of enhancing receptor function and abundance in case detrimental effects of insertion of T4L were encountered. Sequencing of 20 random clones from each of the plasmid libraries confirmed that they all contained T4L inserted within the expected range of positions in Ste2p (Supplementary Table SIV). Based on the distribution of insertions recovered in the random clones, the shortest ligation products (corresponding to the largest deletions of loop sequences) appear to have been cloned less frequently than longer products, as the shortest loop sequences recovered contained four residues at the N-terminal side of the T4L and two residues at the C-terminal side.
Full-length receptors
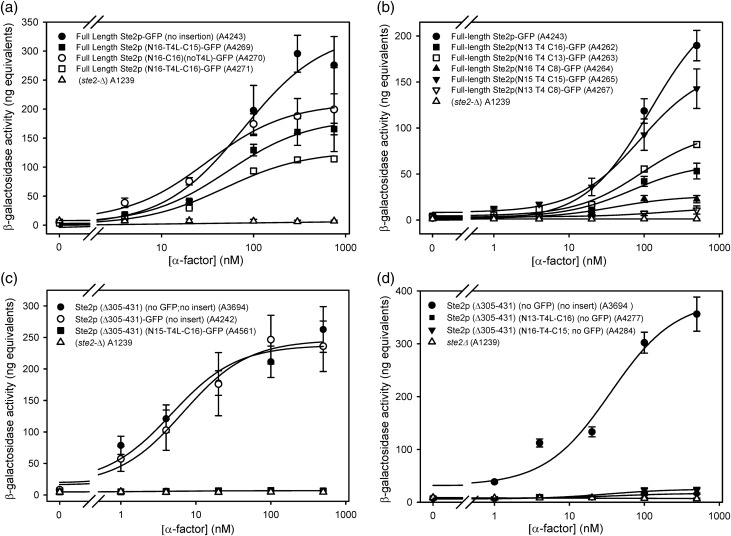
Approximately 40% of yeast transformed with the library of full-length Ste2p-T4L receptors exhibited growth in the presence of α-factor on medium lacking histidine, indicative of retention of signaling function. This α-factor-responsive phenotype was maintained following isolation of receptor-encoding plasmids and re-transformation into fresh yeast host cells. Sequencing and functional characterization of plasmids from the α factor-responsive clones revealed the following properties of the functional T4L-containing receptors (Table I). (Representative α-factor responses of some of the T4L-containing receptors are shown in Fig. 3a and b.):
They were capable of maximal signaling responses up to 66% of the maximal levels exhibited by normal receptors with no insertions in IC3. The EC50s for activation of T4L-containing receptors differed by less than 2-fold from those of native receptors lacking IC3 insertions.
They all contained at least partial duplications of the third intracellular loop incorporating IC3 sequences on both the N- and C-terminal sides of the T4L insertion.
The functional Ste2p-T4L fusion with the smallest number of amino acids in its IC3 loop contained 21 total amino acid residues in IC3 in addition to the T4L insertion. (The parent Ste2p receptor contains 16 residues in the corresponding region of IC3.) The shortest IC3 region recovered at the N-terminal side of the T4L insertion in a receptor with detectable signaling function was 11 amino acids in length (in strain A4266). The shortest sequence at the C-terminal of the T4L insertion in a functional receptor was eight amino acids (in strain A4267). Thus, the presence of a full-length contiguous IC3 loop sequence is not required for signaling. However, receptors containing the short sequences flanking the T4L insertion exhibit very low maximal responses to the α-factor.
Receptors with longer inserted regions flanking T4L, including examples containing the full complement of 16 residues on each side of T4L, exhibited the highest maximal α-factor responses.
Larger deletions of IC3 at the N-terminal side of T4L were more deleterious than comparable deletions at the C-terminal side. The largest deletion at the N-terminal side recovered in a functional receptor variant removed five IC3 residues. The largest deletion at the C-terminal side of T4L in a functional variant removed eight residues. Cells expressing receptors with either no deletion or deletion of one residue in the N-terminal region accompanied by deletion of three residues in the C-terminal regions exhibited maximal responses to α-factor of 39 and 49%, respectively, compared with cells expressing native receptors. In contrast cells expressing receptors with the opposite configuration, three residues deleted from the N-terminal region and none from the C-terminal, exhibited maximal responses to α-factor that were only 25% of those seen with native receptors.
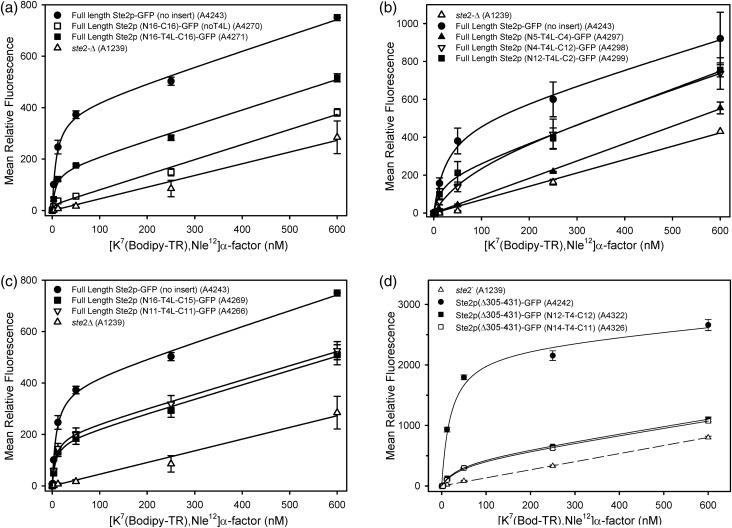
Based on saturation binding of the fluorescent ligand [K7(NBD),Nle12] α-factor (Table I, Fig. 4a, b and c), receptors exhibiting detectable signaling responses to α-factor were expressed at the cell surface at levels ranging from 41–82% of levels of similarly expressed unfused native full-length receptors. Among these receptors, no correlation was observed between levels of signaling and levels of cell surface expression, consistent with previous reports that wide variations in expression levels of Ste2p do not affect signaling responses (Shah and Marsh, 1996; Leavitt et al., 1999).
Table I.
Full-length Ste2p receptors containing inserted T4 lysozyme
| Strain | IC3 residues: N-terminal | IC3 residues: C-terminal | Relative GFP fluorescencea | [K7(Bdpy-Tr,Nle12]α factor Binding |
β-Galactosidase assay (FUS1-lacZ) |
||
|---|---|---|---|---|---|---|---|
| Relative Kdb | Relative Bmaxb | Relative EC50c | Relative maximum responsec | ||||
| A4243 (WT) | WT IC3 | 1.0 | 1.0 | 100 | 1.0 | 100 | |
| A1239 (ste2Δ) | – | – | 0 | – | – | – | – |
| A4270 (no T4) | 16 | 16 | 0.8 ± 0.1 | 0.2 ± 0.6 | 6 ± 3 | 0.4 ± 0.02 | 61 ± 9 |
| A4271 | 16 | 16 | 1.6 ± 0.1 | 0.8 ± 0.4 | 41 ± 5 | 0.6 ± 0.02 | 38 ± 6 |
| A4269 | 16 | 15 | 1.7 ± 0.1 | 0.8 ± 0.4 | 45 ± 5 | 0.7 ± 0.02 | 54 ± 8 |
| A4263 | 16 | 13 | 1.9 ± 0.3 | 0.7 ± 0.2 | 77 ± 3 | 0.8 ± 0.01 | 39 ± 5 |
| A4264 | 16 | 8 | 1.7 ± 0.1 | 0.7 ± 0.2 | 82 ± 4 | 0.3 ± 0.02 | 10 ± 2 |
| A4330 | 16 | 5 | 1.6 ± 0.1 | 0.9 ± 0.5 | 35 ± 6 | NRd | NR |
| A4265 | 15 | 15 | 2.2 ± 0.1 | 0.8 ± 0.2 | 73 ± 3 | 0.7 ± 0.01 | 66 ± 8 |
| A4327 | 15 | 13 | 1.6 ± 0.1 | 0.9 ± 0.4 | 49 ± 6 | 1.2 ± 0.03* | 49 ± 3 |
| A4262 | 13 | 16 | 2.0 ± 0.2 | 0.8 ± 0.3 | 52 ± 4 | 0.6 ± 0.01 | 25 ± 4 |
| A4267 | 13 | 8 | 1.5 ± 0.1 | 0.8 ± 0.2 | 69 ± 4 | 1.6 ± 0.02 | 4 ± 1 |
| A4268 | 12 | 16 | 1.8 ± 0.2 | 0.7 ± 0.3 | 45 ± 4 | 0.3 ± 0.02 | 16 ± 2 |
| A4299 | 12 | 2 | 1.5 ± 0.1 | 0.6 ± 0.5 | 41 ± 9 | NR | NR |
| A4266 | 11 | 11 | 1.7 ± 0.1 | 0.8 ± 0.4 | 51 ± 5 | 0.5 ± 0.01 | 7 ± 1 |
| A4295 | 11 | 3 | 1.8 ± 0.1 | 1.1 ± 0.3 | 45 ± 2 | ND | NR |
| A4335 | 9 | 12 | 2.0 ± 0.3 | 0.5 ± 0.5 | 39 ± 6 | NR | NR |
| A4296 | 9 | 5 | 1.9 ± 0.1 | 0.8 ± 0.3 | 43 ± 2 | ND | NR |
| A4338 | 9 | 4 | 2.0 ± 0.3 | 0.8 ± 0.6 | 46 ± 9 | NR | NR |
| A4334 | 9 | 3 | 2.3 ± 0.4 | 0.6 ± 0.5 | 21 ± 4 | NR | NR |
| A4331 | 8 | 15 | 1.8 ± 0.1 | 1.0 ± 0.5 | 37 ± 5 | NR | NR |
| A4332 | 8 | 8 | 1.8 ± 0.1 | 1.2 ± 0.6 | 39 ± 6 | NR | NR |
| A4328 | 8 | 8 | 1.7 ± 0.1 | 0.9 ± 0.4 | 38 ± 4 | NR | NR |
| A4329 | 8 | 7 | 1.7 ± 0.1 | 0.9 ± 0.4 | 45 ± 6 | NR | NR |
| A4336 | 8 | 5 | 2.0 ± 0.4 | 0.6 ± 0.5 | 43 ± 8 | NR | NR |
| A4337 | 7 | 4 | 2.0 ± 0.3 | 0.7 ± 0.5 | 43 ± 8 | NR | NR |
| A4333 | 6 | 14 | 2.3 ± 0.4 | 0.6 ± 0.5 | 34 ± 5 | NR | NR |
| A4518 | 6 | 9 | 1.9 ± 0.2 | 0.5 ± 0.2 | 43 ± 3 | NR | NR |
| A4297 | 5 | 4 | 1.5 ± 0.1 | -e | <3e | NR | NR |
| A4298 | 4 | 12 | 1.9 ± 0.1 | 1.9 ± 1.3 | 39 ± 20 | NR | NR |
aGFP Fluorescence was calculated from the mean fluorescence in a flow cytometry channel excited at 488 nm and emitting at 525 nm. Values are presented relative to the fluorescence of strain A4243, expressing full-length Ste2p. The background corresponding to the autofluorescence of a strain lacking Ste2p and GFP was subtracted from each measurement. The listed error is the propagated standard error of the mean derived from triplicate measurements of three independent yeast transformants. Note that all the Ste2p receptors listed in this table are fused to a C-terminal GFP moiety.
bThe Kd for [K7(bodipy-TR),Nle12]α-factor of native full-length receptors lacking any IC3 insertion, but containing C-terminally fused GFP (yeast strain A4243) is 13 nM (± 2 nM SEM, n = 7). The indicated uncertainties in the Kd's and Bmax's of the insertion-containing receptors represent propagated errors derived from non-linear least squares fitting to single-site binding with a non-specific component. The fitting was conducted on triplicate experiments using three independent yeast transformants. Relative Bmax refers to the observed mean fluorescence of the analyzed population of cells relative to the mean fluorescence of cells expressing receptors without insertions.
cThe indicated uncertainties represent propagated errors derived from non-linear least squares fitting to a sigmoidal dose–response curve. The fitting was conducted on triplicate experiments using three independent yeast transformants.
dNR, no response, defined as <3% of the maximal response of cells expressing native full-length Ste2p-GFP with no insertion.
eThe very low fluorescence levels for strain A4297 precluded determination of Kd or Bmax values.
Fig. 3.
FUS1-lacZ responses of full-length and C-terminally truncated Ste2p alleles. (a) Comparison of FUS1-lacZ responses to α-factor by strains expressing full-length Ste2p with no insertion in IC3 (A4243), full-length Ste2p with insertions of T4L with large duplications of IC3 sequences (A4269 and A 4271), and full-length Ste2 with an insertion of the fully duplicated IC3 without T4L (A4270). The response of a strain that expresses no Ste2p (A1239) is shown for reference. (b) Comparison of FUS1-lacZ responses to α-factor by strains expressing full-length Ste2p with the indicated extents of duplications of IC3 regions, in comparison to strains expressing full-length Ste2p with no insertion (A4243) and or no Ste2p (A1239). (c) Comparison of FUS1-lacZ responses to α-factor by strains expressing C-terminally truncated Ste2p without any IC3 insertion in the presence (A4242) or absence (A3694) of C-terminally fused GFP with strains expressing the Ste2p with a N14-T4L-C16 insertion (A4561) and a strain expressing no Ste2p (A1239). (d) FUS1-lacZ responses to α-factor by strains expressing C-terminally truncated Ste2p without C-terminally fused GFP. Strains expressing Ste2p with IC3 insertions (A4277 and A4284) are shown in comparison with a strain expressing Ste2p with no insertion (A3694) and a strain expressing no Ste2p (A1239).
Fig. 4.
Saturation binding of [K7(bodipy-TR),Nle12]α-factor to full-length and C-terminally truncated Ste2p receptors. (a) Comparison of the binding of [K7(bodipy-TR),Nle12]α-factor to cells expressing native full-length Ste2p (A4243), cells expressing full-length receptors containing full duplications of the IC3 sequence with (A4271) or without (A4270) T4L, and cells expressing no receptors (A1239). (b) Comparison of binding to cells expressing native full length Ste2p (A4243), no Ste2p (A1239) or full-length forms of Ste2p with the indicated lengths of duplications of IC3 and T4L insertions (A4297, A4298 and A4299). (c) Comparison of binding to cells expressing native full length Ste2p (A4243), no Ste2p (A1239) or full-length forms of Ste2p with the indicated lengths of duplications of IC3 and T4L insertions (A4266 and A4269). (d) Binding of [K7(bodipy-TR), Nle12]α-factor to cells expressing C-terminally truncated, but otherwise native Ste2p (A4242), cell expressing two truncated Ste2p receptors with insertions of IC3 duplications and T4L (A4322 and A4326), and cells expressing no Ste2p (A1239). All Ste2p alleles presented in this figure were expressed as fusions to GFP.
In addition to screening for functional T4L-containing full-length receptors, we also characterized a number of receptors picked at random from the library (listed in Table I) that did not confer α-factor-dependent induction of the FUS1-HIS3 reporter sufficient to allow growth on medium lacking histidine. Saturation binding curves for cells expressing representative examples of these alleles are shown in Fig. 4. Analysis of these receptors led to the following conclusions:
Cells expressing the characterized receptors that could not elicit expression of the FUS1-HIS3 reporter exhibited cell surface-binding sites for the fluorescent ligand [K7(NBD),Nle12] α-factor (based on fitting to saturation binding curves) up to 46% of the levels of binding to cells expressing un-fused full-length native Ste2p. Since numbers of binding sites were determined based on the mean fluorescence of ligand bound to cells, analyzed by flow cytometry, it is theoretically possible that the relatively low fluorescence exhibited by cells expressing the insertion-containing alleles could be the result of an alteration in the environment of the bound fluorophore leading to lower emission, rather than different numbers of cell surface receptors. However, based on previous extensive characterization of mutations affecting the fluorescence emission of the bound fluorescent agonist (Mathew et al., 2011), modifications of the IC3 region are unlikely to affect the emission of bound fluorescent ligand.
Affinities for [K7(bodipy-TR),Nle12]α-factor of the mutant receptors that exhibited detectable binding differed by less than 2-fold from the binding affinities of native receptors lacking IC3 insertions (Table I, Fig. 4a, b and c).
Duplicated IC3 sequences were not required for retention of ligand binding. The shortest total number of IC3 residues (N- plus C-terminal to T4L) in a T4L-containing receptor exhibiting detectable ligand binding was 15, representing a one amino acid deletion compared to the native sequence of the loop (A4518; Supplementary Fig. S2a). The shortest N-terminal sequence of a T4L-containing receptor exhibiting binding activity was four residues (A4298; Fig. 4b). The shortest C-terminal region was three residues (A4334; Supplementary Fig. S2b).
Most of the sequence of IC3 must be present in order for a T4L-receptor fusion to confer detectable ligand binding when expressed in cells. For example, an allele encoding five residues at the N-terminal and four residues at the C-terminal (A4297) exhibits no detectable binding, either because of very low affinity for ligand or because of low receptor abundance at the cell surface (Table I, Fig. 4b).
Based on GFP fluorescence detected by flow cytometry, all T4L-containing full-length receptors are expressed in cells at slightly higher levels (up to 2-fold greater) than their non-T4L-containing counterparts (Table I). This is true of all three classes of receptors, those that are competent for signaling and binding, those that are competent for binding but not signaling, and those that do not bind or signal. Comparison of GFP levels to the numbers of cell surface ligand binding sites for the T4L fusions indicates that the fusion proteins either have reduced efficiency of receptor folding to a binding-competent conformation or reduced efficiency of targeting to the cell surface compared with native receptors. However, precise measurements of levels based on GFP fluorescence are confounded by the significant, and variable autofluorescence of yeast cells that overlaps significantly with the spectrum of GFP.
Receptor fusions that fail to activate the pheromone response pathway tend to be expressed at the cell surface in lower numbers than signaling-competent fusions; Bmax values for non-functional Ste2p-T4L fusions were all <46% of those for unfused full-length receptors, whereas Bmaz values for fusions exhibiting detectable signaling function were all >41% of those for unfused receptors. Low levels of cell surface expression can not be a general explanation for the loss of signaling function by non-functional Ste2p-T4L fusions, since reduction of expression of native receptors to levels at least 40-fold below the levels of native receptors expressed from multicopy plasmids (well below the levels of most of the tested non-functional fusions) has little effect on responses to α-factor (Shah and Marsh, 1996).
The observation that all signaling-competent alleles of the T4L-containing receptors encode duplicated regions of IC3, raised the question of whether a Ste2p receptor with a duplicated IC3 region could function in the absence of inserted T4 lysozyme. This was tested by site-directed creation of an allele encoding a full duplication of the IC3 loop but with no insertion of T4L. Yeast strain A4270, expressing this construct, exhibited a maximal response to α-factor that was approximately 60% of the maximal response of native receptors, similar to the highest activity that was observed for any of the receptors containing T4L. The EC50 for signaling by strain A4270 was slightly lower than for native receptors (Table I, Fig. 3a). However, based on the Bmax extracted from the binding data, the strain expressing the receptor with a duplicate loop and no T4L exhibited an ∼7-fold lower number of cell surface binding sites than strains expressing any of the T4L-containing receptors (Table I, Fig. 4a).
A sampling of receptors with insertions in the IC3 loop that allowed retention of signaling function was transferred into an expression vector suitable for large-scale purifications (Clark et al., 2010). The C-terminal GFP moiety was removed from the receptors during this step. Small-scale growths of cells expressing these alleles were subjected to immunoblotting (Supplementary Fig. S3). The results confirmed that the overall expression levels of some of the T4L-containing receptors in cells were comparable with levels of receptors without inserts expressed from the same vector. Furthermore, the T4L-containing receptors appeared to be expressed as intact proteins that transit at least part way through the secretory pathway, based on the appearance of differentially migrating species known to reflect differential glycosylation of Ste2p (Mentesana and Konopka, 2001). Differences in the apparent abundances of particular Ste2p-T4L fusions depending on whether abundance is assayed via GFP fluorescence, immunoblotting, or ligand binding at the cell surface, could arise from: (i) effects of removing the GFP moiety from the genes tested by immunoblotting; (ii) different inefficiencies of different fusions in transiting the secretory pathway; (iii) preferential endocytosis of certain T4L-containing receptors from the cell surface; (iv) C-terminal proteolysis of some fusions, leading to loss of the C-terminal epitope tag; or (v) persistence of the GFP moiety in cases where the rest of the receptor is degraded.
C-terminally truncated receptors
Screening of a library of C-terminally truncated receptors containing IC3 insertions failed to yield any receptor variants capable of α-factor-dependent responses sufficient to allow growth on medium lacking histidine via expression of the FUS1-HIS3 reporter. Thus, this library of truncated receptors was subjected to a screen using FACS to identify truncated T4L-containing receptor alleles that give rise to the highest ligand-binding capacity. A population of cells in the library exhibited enhanced ligand binding, compared with cells that do not express receptors (see Supplementary Fig. S4). Plasmids isolated from cells exhibiting the highest levels of fluorescent ligand binding (using the gating limits shown in Supplementary Fig. S4) were re-transformed into a fresh yeast host and the re-transformed strains were used for saturation binding assays using [K7(bodipy-TR),Nle12]α-factor (Table II, Fig. 4d). The selected alleles encoded receptors that bound fluorescent ligand with affinities that were similar (within ∼2-fold) to the affinities for this ligand exhibited by native unfused receptors. The numbers of binding sites at the surface of cells expressing these T4L-containing truncated receptors ranged from 3 to 18% of the numbers for truncated receptors with native IC3 loops. In contrast, native Ste2p receptors truncated after Lys304 are expressed at the cell surface at abundances 3–7 times greater than native full-length receptors encoded on similar vectors (Konopka et al., 1988; Reneke et al., 1988; Bajaj et al., 2004). Among the C-terminally truncated Ste2p-T4L fusions, the highest levels of cell surface binding were seen for fusions with modest deletions of IC3 sequences—the three variant receptors exhibiting the highest Bmax's for [K7(bodipy-TR),Nle12]α-factor contained 12, 14 and 16 residues from IC3 at the N-terminal side of the T4L and 12, 11 and 10 residues from IC3, respectively, at the C-terminal of T4L. Saturation binding curves for two of the truncated variant receptors with the highest relative Bmax values are shown in Fig. 4d. Some C-terminally truncated receptors with nearly complete duplications of IC3 flanking T4L exhibited lower levels of ligand binding than those with less complete duplications. Overall expression levels of the T4L-containing receptors in cells, based on detection of cellular GFP fluorescence, were generally similar to the levels of native receptors lacking T4L (<2-fold increased or decrease (Table II)).
Table II.
Receptor variants recovered from screening of C-terminally truncated Ste2p receptors containing inserted T4 lysozyme
| Strain | IC3 residues: N-terminal | IC3 residues: C-terminal | GFP fluorescence | Relative Kda | Relative Bmaxa | Relative EC50b | Relative maximum responseb |
|---|---|---|---|---|---|---|---|
| Ste2p (Δ305-431)-GFP | |||||||
| A4242 (Trunc WT) | WT IC3 | WT IC3 | 1.0 | 1.0 | 100 | 1.0 | 100.0 |
| A1239 (ste2Δ) | – | – | 0 | – | <3 | NR | |
| A4325 | 16 | 10 | 1.1 ± 0.1 | 1.1 ± 0.7 | 14 ± 3 | NDc | ND |
| A4562 | 16 | 9 | 0.5 ± 0.0 | 0.4 ± 0.5 | 5 ± 1 | NRd | NR |
| A4323 | 16 | 6 | 1.1 ± 0.1 | 0.8 ± 0.6 | 11 ± 2 | NR | NR |
| A4561 | 15 | 16 | 0.6 ± 0.0 | 0.5 ± 0.5 | 6 ± 1 | NR | NR |
| A4560 | 15 | 14 | 0.5 ± 0.0 | 0.4 ± 0.6 | 5 ± 1 | NR | NR |
| A4326 | 14 | 11 | 1.1 ± 0.1 | 2.0 ± 0.9 | 17 ± 3 | ND | ND |
| A4322 | 12 | 12 | 1.2 ± 0.1 | 1.9 ± 0.8 | 18 ± 3 | ND | ND |
| A4324 | 10 | 5 | 1.3 ± 0.2 | 1.0 ± 0.7 | 11 ± 2 | ND | ND |
| A4559 | 4 | 10 | 0.5 ± 0.0 | 0.4 ± 0.6 | 3 ± 1 | NR | NR |
| Ste2p (Δ305-431) without GFP | |||||||
| A3694 | WT IC3 | WT IC3 | ND | ND | ND | 1.0 | 100.0 |
| A4284 | 16 | 15 | ND | ND | ND | 0.9 ± 0.1 | 5 ± 1 |
| A4279 | 16 | 8 | ND | ND | ND | 0.2 ± 0.1 | NR |
| A4277 | 13 | 16 | ND | ND | ND | 1.2 ± 0.1 | 3 ± 1 |
| A4283 | 12 | 16 | ND | ND | ND | 2.4 ± 0.1 | 6 ± 1 |
aThe Kd for [K7(bodipy-TR),Nle12]α-factor of native C-terminally truncated receptors lacking any IC3 insertion, but containing C-terminally fused GFP (yeast strain A4242) is 23 nM (± 4 nM SEM, n = 3). The indicated uncertainties represent propagated errors derived from non-linear least squares fitting to single-site binding with a non-specific component. The fitting was conducted on triplicate experiments using three independent yeast transformants. Relative Bmax refers to the observed mean fluorescence of the analyzed population of cells relative to the mean fluorescence of cells expressing receptors without insertions.
bThe indicated uncertainties represent propagated errors derived from non-linear least squares fitting to a sigmoidal dose–response curve. The fitting was conducted on triplicate experiments using three independent yeast transformants.
cND, not determined.
dNR, no response, defined as less than 3% of the maximal response of cells expressing truncated (Δ305-431) but otherwise native Ste2p-GFP with no insertion.
The unexpected lack of signaling function by any C-terminally truncated Ste2p-T4L fusions could be the result of inherent instability, misfolding or defective intracellular trafficking of the truncated receptor fusions, such that any insertion of T4L renders them incapable of undergoing activation. Alternatively, since the Ste2p-T4L proteins subjected to the original screen actually contained GFP fused at their C-termini, the presence of GFP fused immediately adjacent to the transmembrane region of the tail-less receptors could have been interfering with folding or function of the fusions, possibly by interacting directly with the inserted T4L. To distinguish between these possibilities, we removed both the C-terminal tail sequences and the fused GFP from four full-length alleles that encoded receptors with significant signaling function. These C-terminally truncated fusions lacking GFP exhibited very weak α-factor-responses (Table II, Fig. 3d). Thus, their failure to signal appears to result from deletion of the C-terminal tail and not from proximity of the GFP to the transmembrane sequences of the receptor.
Discussion
Fusion of the third intracellular loop of GPCRs to T4L has proven to be a successful strategy for obtaining well-diffracting crystals of these receptors. Structures of such fusions have been determined both in antagonist- and agonist-bound states (Cherezov et al., 2007; Rosenbaum et al., 2007; Jaakola et al., 2008; Chien et al., 2010; Wu et al., 2010; Rasmussen et al., 2011a,b; Shimamura et al., 2011; Xu et al., 2011; Haga et al., 2012; Hanson et al., 2012; Kruse et al., 2012). However, in no case has such a receptor-T4L fusion been reported to be capable of activating a G protein in a ligand-dependent manner. Furthermore, rigidification of receptors induced by the presence of T4 lysozyme has been invoked to explain apparent anomalies in the behavior of the ‘ionic lock’ between the conserved DRY motif in TM3 and a conserved glutamic acid in TM6 of GPCRs (Dror et al., 2009; Salon et al., 2011). Although the ionic interactions comprising the ‘lock’ are believed to play a role in maintaining receptors in inactive states, the lock is observed to be broken in a number of structures of GPCRs bound to inverse agonists and antagonists. This is consistent with the fact that some receptor-T4L fusions exhibit elevated agonist affinities (compared to the native receptors), a characteristic that is associated with enhanced agonist-independent receptor activity of some receptor variants (that do not contain T4L) (Rosenbaum et al., 2007).
Understanding of mechanisms of receptor activation would be enhanced by the availability of structures of receptors that are capable of activation of the cognate G-protein. Furthermore, introduction of full-length fluorescent proteins or protein interaction domains could provide tools useful for understanding GPCR action outside of the area of structure determination. To date, the primary approaches for identifying optimal sites of T4L insertion have involved cycles of design and testing of fusion constructs. As an alternative approach for identifying optimal receptor-T4L fusions, we used created a library of receptor variants containing insertions of T4L into all possible positions within a selected region of the third loop of the yeast α-factor receptor Ste2p, accompanied by varying extents of deletion and duplication of loop sequences. Screening of the complete set of receptor-T4L fusions in this library, using screens to identify alleles providing maximal receptor expression, ligand binding and signaling function, led to the identification of receptors with insertions of T4L in the third intracellular loop that exhibit robust ligand-dependent activation of the downstream G protein pathway. The functional fusions we identified all contain duplication of IC3 loop sequences flanking the T4L insertion created via the particular cloning strategy used for introduction of T4L sequences.
Previous studies have implicated the third intracellular loops of various GPCRs in the activation of G proteins (Strader et al., 1994; Taylor et al., 1994; Erlenbach and Wess, 1998; Thompson et al., 1998; Wess, 1998). The IC3 loop has also been specifically implicated in the signaling function of the α-factor receptor (Clark et al., 1994; Stefan and Blumer, 1994; Celic et al., 2003). This raises the question of how a receptor can maintain competence to activate G protein with T4L inserted into the critical IC3 region. One rationale for using T4L as a fusion partner is the proximity of its N- and C-termini, which reside <11 Å apart in crystal structures of the isolated enzyme (Rosenbaum et al., 2007). Since closely packed helices are ∼10–15 Å (center to center), properly placed T4L may fit into loops between helices of transmembrane proteins with minimal perturbation of the helical core conformations. (Insertion of T4L into a hydrophilic loop of the lac permease was compatible with retention of lactose transport (Engel et al., 2002a)). However, even if insertion of T4L into the third intracellular loop of a GPCR does not interfere with the loop's conformation or dynamics, it is surprising that the presence of a ∼20 kD protein inserted at this critical position does not sterically interfere with critical receptor-G protein contacts. The simplest explanation of retention of signaling activity by Ste2p variants with the T4L insertions is that the flexible attachment and the extra chain length of the duplicated IC3 sequences in these fusions allow the lysozyme to move out of the way of the critical interactions between IC3 loop sequences and Gα. The role of IC3 flexibility in allowing productive interactions between T4L-containing receptors and G proteins is supported by the observation that all recovered functional T4L-inserted receptors contained extra lengths of IC3 insertion compared with the normal IC3 loop of Ste2p.
Since we find that different regions of IC3 sequences are duplicated in different functional Ste2p-T4L fusion proteins, the retention of native-like interactions with G protein appears to depend more on the presence of extra amino acids than on a role for a specific duplicated sequence. The third intracellular loop of Ste2p has been previously shown to be tolerant of a wide variety of amino acid substitutions (Celic et al., 2003) and even of a discontinuity of the peptide chain in this region, since fragments of Ste2p comprising the first five predicted transmembrane segments and the last two segments are capable of associating to form an active α-factor receptor (Martin et al., 1999). Consistent with the current results, successful assembly occurred when the fragments retained intact sequences adjacent to the cytoplasmic ends of TM5 and TM6, but not when the discontinuity was close to the cytoplasmic end of TM5.
Although C-terminal truncation of the cytoplasmic tail of native α-factor receptors results in increased receptor sensitivity to ligand and an increased number of ligand-binding sites at the cell surface (Konopka et al., 1988; Reneke et al., 1988), we were unable to recover any tail-less Ste2p-T4L fusions that retained the ability to activate G proteins. Since removing both the receptor's C-terminal tail and C-terminal GFP tag from active full-length Ste2p-T4L fusions resulted in loss of signaling function, the inability of the truncated receptor-T4L fusions to signal must be a direct consequence of the lack of the tail rather than any effect of the fusion to GFP. This suggests that, although it is not directly required for signal transduction, an interaction between the third intracellular loops and the tail may serve to stabilize or promote correct folding or trafficking of altered forms of the receptor. Alternatively, the tail may undergo interactions with the G protein that are not essential for activation of the G protein but may strengthen the interactions with G protein by receptors with altered IC3 loops (Dosil et al., 2000). The role of the C-terminal tail in formation of the receptor-G protein complex is of particular interest in view of the fact that most of this region has been removed from the receptors for which structures are available from X-ray crystallography or has been found (with the exception of the eighth helix) to be disordered (Choe et al., 2011).
Two rationales have been provided for the benefits of using T4L fusions for promoting crystallization of transmembrane proteins. One of these is the reduction in structural heterogeneity and the protection against denaturation that are expected to result from insertion of a stable, soluble protein into a region that is likely to connect two mobile regions of a membrane protein. The other rationale is that the presence of the T4L simply provides additional hydrophilic surfaces that, based on the tendency of T4L to crystallize as an isolated soluble protein, are likely to promote crystallization of the membrane proteins into which it is inserted. The procedures we describe have been used to recover insertions of T4L into third intracellular loops of receptors that maintain strong signaling function. The observation that duplication of IC3 sequences flanking an inserted T4L results in expression of many more cell surface-binding sites than are observed for Ste2p variants containing the duplicated IC3 sequences without T4L raises the possibility that the presence of the lysozyme may actually enhance the receptor's stability in cells. However, the increase in binding sites in the presence of lysozyme could also reflect enhanced folding or intracellular trafficking of T4L-containing receptors.
We do not yet know whether insertion of T4L into signaling-competent receptors will provide stabilization useful for enhancing crystallization. While flexibility of the extended IC3 loop sequences may reduce the rigidity of the T4L-containing receptors, the presence of T4L is expected to provide interacting surfaces that could enhance crystallization. The recent determinations of the structure of the β2-adrenergic receptor-G protein complex using a receptor construct containing T4L fused at its extreme N-terminal (Rasmussen et al., 2011b) and the structure of the nociceptin/orphanin opioid receptor using a construct containing a stabilized apocytochrome b562 fused at the receptor's N-terminal (Thompson et al., 2012) suggest that the addition of the interacting surfaces in the absence of receptor stabilization may be sufficient to significantly promote receptor crystallization. We anticipate that the protocols we have developed and that the knowledge that GPCR function can be maintained even when a soluble protein is inserted into the third intracellular loop will be useful in further structural and functional studies of GPCRs.
Supplementary data
Funding
This work was supported by National Institutes of Health grants GM084083 to M.E.D and GM22086 to F.N.
Supplementary Material
Acknowledgements
We thank the University of Rochester Flow Cytometry Core facility for assistance with cell sorting and flow cytometry.
References
- Alexandrov A., Vignali M., LaCount D.J., et al. Mol Cell Proteomics. 2004;3:934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- Aslanidis C., de Jong P.J. Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj A., Celic A., Ding F.X., Naider F., Becker J.M., Dumont M.E. Biochemistry. 2004;43:13564–13578. doi: 10.1021/bi0494018. [DOI] [PubMed] [Google Scholar]
- Bajaj A., Connelly S.M., Gehret A.U., Naider F., Dumont M.E. Biochim. Biophys. Acta. 2007;1773:707–717. doi: 10.1016/j.bbamcr.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L., Parent S., Caron M., et al. J. Recept. Signal Transduct. Res. 2002;22:533–541. doi: 10.1081/rrs-120014619. [DOI] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Methods Enzymol. 2000;328:430–444. doi: 10.1016/s0076-6879(00)28410-3. [DOI] [PubMed] [Google Scholar]
- Celic A., Connelly S.M., Martin N.P., Dumont M.E. Methods Mol. Bio.l. 2004;237:105–120. doi: 10.1385/1-59259-430-1:105. [DOI] [PubMed] [Google Scholar]
- Celic A., Martin N.P., Son C.D., Becker J.M., Naider F., Dumont M.E. Biochemistry. 2003;42:3004–3017. doi: 10.1021/bi0269308. [DOI] [PubMed] [Google Scholar]
- Cherezov V., Rosenbaum D.M., Hanson M.A., et al. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien E.Y., Liu W., Zhao Q., et al. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H.W., Kim Y.J., Park J.H., Morizumi T., Pai E.F., Krauss N., Hofmann K.P., Scheerer P., Ernst O.P. Nature. 2011;471:651–655. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Clark C.D., Palzkill T., Botstein D. J. Biol. Chem. 1994;269:8831–8841. [PubMed] [Google Scholar]
- Clark K.M., Fedoriw N., Robinson K., Connelly S.M., Randles J., Malkowski M.G., DeTitta G.T., Dumont M.E. Protein Expr. Purif. 2010;71:207–223. doi: 10.1016/j.pep.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckman L., Gu M., Stols L., Donnelly M.I., Collart F.R. Protein Expr. Purif. 2002;25:1–7. doi: 10.1006/prep.2001.1602. [DOI] [PubMed] [Google Scholar]
- Ding F.X., Lee B.K., Hauser M., Davenport L., Becker J.M., Naider F. Biochemistry. 2001;40:1102–1108. doi: 10.1021/bi0021535. [DOI] [PubMed] [Google Scholar]
- Ding F.X., Lee B.K., Hauser M., Patri R., Arshava B., Becker J.M., Naider F. J. Pept. Res. 2002;60:65–74. doi: 10.1034/j.1399-3011.2002.21004.x. [DOI] [PubMed] [Google Scholar]
- Dosil M., Schandel K.A., Gupta E., Jenness D.D., Konopka J.B. Mol Cell Biol. 2000;20:5321–5329. doi: 10.1128/mcb.20.14.5321-5329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S.J., Brown A.J. Methods Mol. Biol. 2009;552:213–229. doi: 10.1007/978-1-60327-317-6_15. [DOI] [PubMed] [Google Scholar]
- Dror R.O., Arlow D.H., Borhani D.W., Jensen M.O., Piana S., Shaw D.E. Proc. Natl Acad. Sci. USA. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C.K., Chen L., Prive G.G. Biochim. Biophys. Acta. 2002a;1564:38–46. doi: 10.1016/s0005-2736(02)00398-x. [DOI] [PubMed] [Google Scholar]
- Engel C.K., Chen L., Prive G.G. Biochim. Biophys. Acta. 2002b;1564:47–56. doi: 10.1016/s0005-2736(02)00397-8. [DOI] [PubMed] [Google Scholar]
- Erlenbach I., Wess J. J. Biol. Chem. 1998;273:26549–26558. doi: 10.1074/jbc.273.41.26549. [DOI] [PubMed] [Google Scholar]
- Gehret A.U., Bajaj A., Naider F., Dumont M.E. J. Biol. Chem. 2006;281:20698–20714. doi: 10.1074/jbc.M513642200. [DOI] [PubMed] [Google Scholar]
- Granier S., Manglik A., Kruse A.C., Kobilka T.S., Thian F.S., Weis W.I., Kobilka B.K. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga K., Kruse A.C., Asada H., et al. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson M.A., Roth C.B., Jo E., et al. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L. FASEB J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- Hicke L., Zanolari B., Riezman H. J. Cell Biol. 1998;141:349–358. doi: 10.1083/jcb.141.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola V.P., Griffith M.T., Hanson M.A., Cherezov V., Chien E.Y., Lane J.R., Ijzerman A.P., Stevens R.C. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E.W. Methods Enzymol. 2002;351:127–150. doi: 10.1016/s0076-6879(02)51844-9. [DOI] [PubMed] [Google Scholar]
- King K., Dohlman H.G., Thorner J., Caron M.G., Lefkowitz R.J. Science. 1990;250:121–123. doi: 10.1126/science.2171146. [DOI] [PubMed] [Google Scholar]
- Konopka J.B., Jenness D.D., Hartwell L.H. Cell. 1988;54:609–620. doi: 10.1016/s0092-8674(88)80005-9. [DOI] [PubMed] [Google Scholar]
- Kruse A.C., Hu J., Pan A.C., et al. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt L.M., Macaluso C.R., Kim K.S., Martin N.P., Dumont M.E. Mol. Gen. Genet. 1999;261:917–932. doi: 10.1007/s004380051039. [DOI] [PubMed] [Google Scholar]
- Lebon G., Warne T., Edwards P.C., Bennett K., Langmead C.J., Leslie A.G., Tate C.G. Nature. 2011;474:521–525. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kane T., Tipper C., Spatrick P., Jenness D.D. Mol. Cell Biol. 1999;19:3588–3599. doi: 10.1128/mcb.19.5.3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfredi J.P., Klein C., Herrero J.J., Byrd D.R., Trueheart J., Wiesler W.T., Fowlkes D.M., Broach J.R. Mol. Cell. Biol. 1996;16:4700–4709. doi: 10.1128/mcb.16.9.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A., Kruse A.C., Kobilka T.S., et al. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N.P., Leavitt L.M., Sommers C.M., Dumont M.E. Biochemistry. 1999;38:682–695. doi: 10.1021/bi982062w. [DOI] [PubMed] [Google Scholar]
- Mathew E., Bajaj A., Connelly S.M., Sargsyan H., Ding F.X., Hajduczok A.G., Naider F., Dumont M.E. J Mol. Biol. 2011;409:513–528. doi: 10.1016/j.jmb.2011.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentesana P.E., Konopka J.B. Biochemistry. 2001;40:9685–9694. doi: 10.1021/bi0108507. [DOI] [PubMed] [Google Scholar]
- Nolan J.P., Sklar L.A. Nat. Biotechnol. 1998;16:633–638. doi: 10.1038/nbt0798-633. [DOI] [PubMed] [Google Scholar]
- Prive G.G., Verner G.E., Weitzman C., Zen K.H., Eisenberg D., Kaback H.R. Acta Crystallogr. D Biol. Crystallogr. 1994;50:375–379. doi: 10.1107/S0907444993014301. [DOI] [PubMed] [Google Scholar]
- Rasmussen S.G., Choi H.J., Fung J.J., et al. Nature. 2011a;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.G., Choi H.J., Rosenbaum D.M., et al. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Rasmussen S.G., DeVree B.T., Zou Y., et al. Nature. 2011b;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneke J.E., Blumer K.J., Courchesne W.E., Thorner J. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- Rosenbaum D.M., Cherezov V., Hanson M.A., et al. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- Salon J.A., Lodowski D.T., Palczewski K. Pharmacol. Rev. 2011;63:901–937. doi: 10.1124/pr.110.003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar C.A., Dodevski I., Kenig M., Dudli S., Mohr A., Hermans E., Pluckthun A. Proc. Natl Acad. Sci. USA. 2008;105:14808–14813. doi: 10.1073/pnas.0803103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A., Marsh L. Biochem. Biophys. Res. Commun. 1996;226:242–246. doi: 10.1006/bbrc.1996.1340. [DOI] [PubMed] [Google Scholar]
- Shimamura T., Shiroishi M., Weyand S., et al. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar L.A., Edwards B.S., Graves S.W., Nolan J.P., Prossnitz E.R. Annu. Rev. Biophys. Biomol. Struct. 2002;31:97–119. doi: 10.1146/annurev.biophys.31.082901.134406. [DOI] [PubMed] [Google Scholar]
- Sklar L.A., Finney D.A., Oades Z.G., Jesaitis A.J., Painter R.G., Cochrane C.G. J Biol Chem. 1984;259:5661–5669. [PubMed] [Google Scholar]
- Sommers C.M., Martin N.P., Akal-Strader A., Becker J.M., Naider F., Dumont M.E. Biochemistry. 2000;39:6898–6909. doi: 10.1021/bi992616a. [DOI] [PubMed] [Google Scholar]
- Stefan C.J., Blumer K.J. Mol. Cell Biol. 1994;14:3339–3349. doi: 10.1128/mcb.14.5.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader C.D., Fong T.M., Tota M.R., Underwood D., Dixon R.A. Annu. Rev. Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- Taslimi A., Mathew E., Celic A., Wessel S., Dumont M.E. J. Mol. Biol. 2012;418:367–378. doi: 10.1016/j.jmb.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C.G., Schertler G.F. Curr. Opin. Struct. Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Taylor J.M., Jacob-Mosier G.G., Lawton R.G., Remmers A.E., Neubig R.R. J. Biol. Chem. 1994;269:27618–27624. [PubMed] [Google Scholar]
- Thompson A.A., Liu W., Chun E., et al. Nature. 2012;485:395–399. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.B., Wade S.M., Harrison J.K., Salafranca M.N., Neubig R.R. J. Pharmacol. Exp. Ther. 1998;285:216–222. [PubMed] [Google Scholar]
- Warne T., Serrano-Vega M.J., Baker J.G., Moukhametzianov R., Edwards P.C., Henderson R., Leslie A.G., Tate C.G., Schertler G.F. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J. Pharmacol. Ther. 1998;80:231–264. doi: 10.1016/s0163-7258(98)00030-8. [DOI] [PubMed] [Google Scholar]
- White S.H. http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html .
- Wu B., Chien E.Y., Mol C.D., et al. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Wacker D., Mileni M., et al. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Wu H., Katritch V., Han G.W., Jacobson K.A., Gao Z.G., Cherezov V., Stevens R.C. Science. 2011;332:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.