Abstract
Recent genetic association studies have made progress in uncovering components of the genetic architecture of the body mass index (BMI). We used the ITMAT-Broad-Candidate Gene Association Resource (CARe) (IBC) array comprising up to 49 320 single nucleotide polymorphisms (SNPs) across ∼2100 metabolic and cardiovascular-related loci to genotype up to 108 912 individuals of European ancestry (EA), African-Americans, Hispanics and East Asians, from 46 studies, to provide additional insight into SNPs underpinning BMI. We used a five-phase study design: Phase I focused on meta-analysis of EA studies providing individual level genotype data; Phase II performed a replication of cohorts providing summary level EA data; Phase III meta-analyzed results from the first two phases; associated SNPs from Phase III were used for replication in Phase IV; finally in Phase V, a multi-ethnic meta-analysis of all samples from four ethnicities was performed. At an array-wide significance (P < 2.40E-06), we identify novel BMI associations in loci translocase of outer mitochondrial membrane 40 homolog (yeast) - apolipoprotein E - apolipoprotein C-I (TOMM40-APOE-APOC1) (rs2075650, P = 2.95E-10), sterol regulatory element binding transcription factor 2 (SREBF2, rs5996074, P = 9.43E-07) and neurotrophic tyrosine kinase, receptor, type 2 [NTRK2, a brain-derived neurotrophic factor (BDNF) receptor gene, rs1211166, P = 1.04E-06] in the Phase IV meta-analysis. Of 10 loci with previous evidence for BMI association represented on the IBC array, eight were replicated, with the remaining two showing nominal significance. Conditional analyses revealed two independent BMI-associated signals in BDNF and melanocortin 4 receptor (MC4R) regions. Of the 11 array-wide significant SNPs, three are associated with gene expression levels in both primary B-cells and monocytes; with rs4788099 in SH2B adaptor protein 1 (SH2B1) notably being associated with the expression of multiple genes in cis. These multi-ethnic meta-analyses expand our knowledge of BMI genetics.
INTRODUCTION
Obesity is a complex disorder affecting more than one-third of the US adult population (1,2) and approximately half a billion people worldwide (3). Obesity increases the risk of metabolic conditions such as cardiovascular diseases (CVDs) (4,5), type 2 diabetes (T2D) (6), hyperlipidemia (7) as well as certain cancers (8). Although sedentary behaviors and poor nutrition certainly contribute to the pathogenesis of obesity, genetic variation also plays a role, with estimated heritability ranging from 40% to as high as 90% (9–11). As of March 2012, common genetic variants in 32 human loci from genome-wide association studies (GWAS) have been reported to be associated with the body mass index (BMI) in individuals of European ancestry (EA) (12–16). These studies have specifically searched for and identified common genetic variants that associate with BMI in individuals of EA and follow an additive genetic model. Variants that are rare in individuals of EA but may be more common in other ancestries, or those loci that require environmental exposures that differ by the population subgroup, exist and require approaches beyond standard GWAS in only European-based samples to be identified (17,18).
In this study we aimed to identify BMI-associated variants that may have been missed by previous studies and sought to confirm previously reported associations. We used the ITMAT-Broad-CARe (IBC) array (19), also referred to as the CardioChip or the human CVD BeadChip (Illumina, San Diego, CA, USA), which comprises up to 49 320 single nucleotide polymorphisms (SNPs) selected across ∼2100 metabolic and cardiovascular-related loci with variation in most targeted genes captured at a density greater or equal to the standard genome-wide genotyping arrays. Content was selected for the IBC array using data from first waves of CVD-related GWAS results, additional high-priority candidate genes of interest and analysis of cardiovascular, inflammatory and metabolic pathways. Robust associations have been shown on the IBC array for a range of phenotypes including coronary artery disease (20,21), heart failure (22,23), lipids (24), height (25) and T2D (26).
Ten of the 32 GWAS-identified BMI loci reported to date are included on the IBC array; however, many genes with plausible roles in metabolism and in the etiology of obesity, but which have not previously been implicated, are specifically tagged on the array. SNP selection included the use of resequencing data from the SeattleSNPs and National Institute of Environmental Health Sciences (NIEHS) SNP consortia, as well as the International HapMap Consortium, to ensure deeper capture of variation within genes of interest. The IBC array was enriched for uncommon SNPs, as >17 000 included SNPs have minor allele frequency (MAF) <0.05 in test populations of EA, with the inclusion of putative functional SNPs from the literature and non-synonymous SNPs (7.7% of the total number of IBC array SNPs) (19).
Conditional analysis in additional complex phenotypes, such as height (25) and plasma lipids (27), has revealed loci containing multiple independent signals which are strong plausible sources to explain portions of the ‘missing’ genetic variance. Suitable sample sizes and dense loci coverage are required to perform conditional analysis. Furthermore the consistency of an association signal observed across multiple ethnicities increases confidence in the validity of the signals (28). Finally, variability in signal strength between populations, e.g. due to different allele frequencies or patterns of linkage disequilibrium (LD), may allow for novel signals to be identified in different ethnic groups.
In the current study, we perform large-scale BMI meta-analysis on 92 903 individuals of EA, 12 297 African-Americans, 2625 Hispanics and 1087 East Asians across 46 separate studies. We directly queried common and uncommon genetic variants within this targeted array with the aim of validating known BMI loci in these different ethnicities as well as discovering novel BMI-associated loci. We performed primary analyses (Fig. 1; left shaded panel) of discovery and replication for BMI-associated signals by using population-specific and multi-ethnic approaches, and we also conducted secondary calculations (Fig. 1; right shaded panel) including conditional analysis to search for multiple independent signals within associated loci, sex-specific associations testing for sex-related differences in BMI association and two other additional meta-analyses in an attempt to further validate our findings.
Figure 1.
Schematic design of study for genetic association between BMI and genetic markers in the ITMAT-Broad-CARe (IBC) array. The work flow includes three parts, which are within study procedures, primary analyses and secondary analyses. Details can be found in the text.
RESULTS
Meta-analysis of individuals of European ancestry replicates known BMI signals and reveals novel BMI associations in the TOMM40-APOE-APOC1, SREBF2 and NTRK2 loci
BMI meta-analysis using the IBC array was initially performed in cohorts of EA which provided individual-level data (Phase I in Fig. 1; n = 50 933). We observed six array-wide significant lead signals (Table 1): rs1421085 in the fat mass and obesity associated (FTO) locus, rs2272903 in transcription factor activating enhancer binding protein 2 beta (TFAP2B), rs10767664 in brain-derived neurotrophic factor (BDNF), rs12617233 in Fanconi anemia, complementation group L (FANCL)-FLJ30838, rs2075650 in translocase of outer mitochondrial membrane 40 homolog (yeast)-apolipoprotein E-apolipoprotein C-I (TOMM40-APOE-APOC1) and rs2229616 (also known as V103I) in melanocortin 4 receptor(MC4R). One of the findings, the TOMM40-APOE-APOC1 locus is previously unreported. The uncommon SNP rs2229616 (MAF = 0.02) in MC4R was reported to be associated with obesity in two independent studies (29,30), but never reached genome-wide significance (P < 5.00E-08). Cohorts providing unpublished summary-level association results (Phase II in Fig. 1; n = 27 503) provided nominal evidence for replication (P ≤ 2.45E-03) in four of the six array-wide significant associated SNPs from the individual-level data: the FTO, BDNF, MC4R and TOMM40-APOE-APOC1 SNPs (Table 1).
Table 1.
Primary results for BMI association analysis using the ITMAT-Broad-CARe (IBC) array
| Locus rank | Candidate gene(s) | Chr. | SNP | Genomic position | Effect allele | Eff. All. Freq.a | Phase I meta-analysisb |
Phase II meta-analysisc |
Phase III meta-analysisd |
Phase IV meta-analysise |
Phase V meta-analysisf P-valve | Reference (PMID; year) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | % var | Effect | P-value | Effect | P-value | I2 | Effect | P-value | I2 | |||||||||
| 1 | FTO | 16q12.2 | rs1421085 | 52 358 455 | C | 0.42 | 0.086 | 7.75E-40 | 0.37 | 0.097 | 1.38E-27 | 0.090 | 1.57E-65 | 16 | 0.089 | 1.26E-73 | 18 | 1.35E-69 | 17434869; 2007 |
| 2 | BDNF | 11p13 | rs10767664 | 27 682 562 | A | 0.79 | 0.041 | 3.48E-07 | 0.03 | 0.044 | 2.85E-05 | 0.042 | 4.57E-11 | 0 | 0.043 | 3.40E-13 | 0 | 7.87E-14 | 19079260; 2008 |
| 3 | MC4R | 18q22 | rs2229616 | 56 190 256 | C | 0.98 | 0.111 | 1.96E-06 | 0.04 | 0.130 | 6.11E-05 | 0.118 | 5.28E-10 | 10 | 0.112 | 2.82E-10 | 0 | 6.39E-12 | 18454148; 2008g |
| 4 | TOMM40-APOE-APOC1 | 19q13 | rs2075650 | 50 087 459 | A | 0.86 | 0.045 | 6.70E-07 | 0.09 | 0.037 | 2.47E-03 | 0.042 | 6.87E-09 | 11 | 0.042 | 2.95E-10 | 1 | 5.99E-11 | — |
| 5 | SH2B1 | 16p11.2 | rs4788099 | 28 763 228 | G | 0.39 | 0.031 | 2.20E-05 | 0.06 | 0.022 | 2.39E-02 | 0.028 | 1.92E-06 | 13 | 0.031 | 7.30E-09 | 12 | 1.87E-07 | 20935630; 2010 |
| 6 | FANCL-FLJ30838 | 2p16.1 | rs12617233 | 58 893 502 | C | 0.60 | 0.035 | 1.23E-07 | 0.06 | 0.015 | 8.67E-02 | 0.028 | 1.41E-07 | 9 | 0.028 | 8.25E-09 | 3 | 5.54E-09 | 20935630; 2010 |
| 7 | TFAP2B | 6p12 | rs2272903 | 50 894 530 | G | 0.90 | 0.056 | 1.07E-07 | 0.09 | 0.023 | 1.43E-01 | 0.045 | 2.84E-07 | 30 | 0.043 | 7.61E-08 | 23 | 4.53E-09 | 20935630; 2010 |
| 8 | MAP2K5 | 15q23 | rs997295 | 65 803 397 | T | 0.59 | 0.024 | 3.08E-04 | 0.04 | 0.039 | 1.44E-05 | 0.029 | 4.09E-08 | 10 | 0.026 | 2.48E-07 | 10 | 1.43E-08 | 20935630; 2010 |
| 9 | COL4A3BP-HMGCR | 5q13.3 | rs4704220 | 74 793 312 | G | 0.61 | 0.029 | 9.99E-06 | 0.04 | 0.023 | 9.80E-03 | 0.027 | 3.68E-07 | 1 | 0.025 | 4.19E-07 | 0 | 1.04E-07 | 20935630; 2010 |
| 10 | SREBF2 | 22q13 | rs5996074 | 40 566 283 | A | 0.70 | 0.022 | 1.68E-03 | 0.05 | 0.027 | 5.03E-03 | 0.024 | 2.72E-05 | 15 | 0.026 | 9.43E-07 | 8 | 1.93E-07 | — |
| 11 | NTRK2 | 9q22.1 | rs1211166 | 86 475 812 | A | 0.80 | 0.029 | 3.75E-04 | 0.01 | 0.025 | 2.66E-02 | 0.027 | 2.97E-05 | 0 | 0.029 | 1.04E-06 | 0 | 2.87E-08 | — |
Chr., chromosome; % var, average variance explained (adjusted R2) in Phase I cohorts weighted by cohort sample size; I2, heterogeneity measurement.
Eleven signals reached the array-wide significance level after Phase IV meta-analysis of European ancestry samples from individual-level, summary-level and look-up studies.
The table is sorted by P-values in Phase IV meta-analysis. All SNPs mapped back to NCBI build 36 (UCSC hg18) human reference genome. P-values passing the array-wide significance cut-off of 2.40E-06 are highlighted in bold, and P-values less than the conventional genome-wide significance of 5.00E-08 are further annotated in Italics.
aEff. All. Freq., effect allele frequency estimated from Phase IV meta-analysis.
bSample size up to 50 933.
cSample size up to 27 503.
dSample size up to 78 436.
eSample size up to 92 903.
fSample size up to 108 912.
gThe reference has reported a SNP rs7227255, which is in perfect linkage disequilibrium with our lead SNP rs2229616, as an independent signal within the MC4R region.
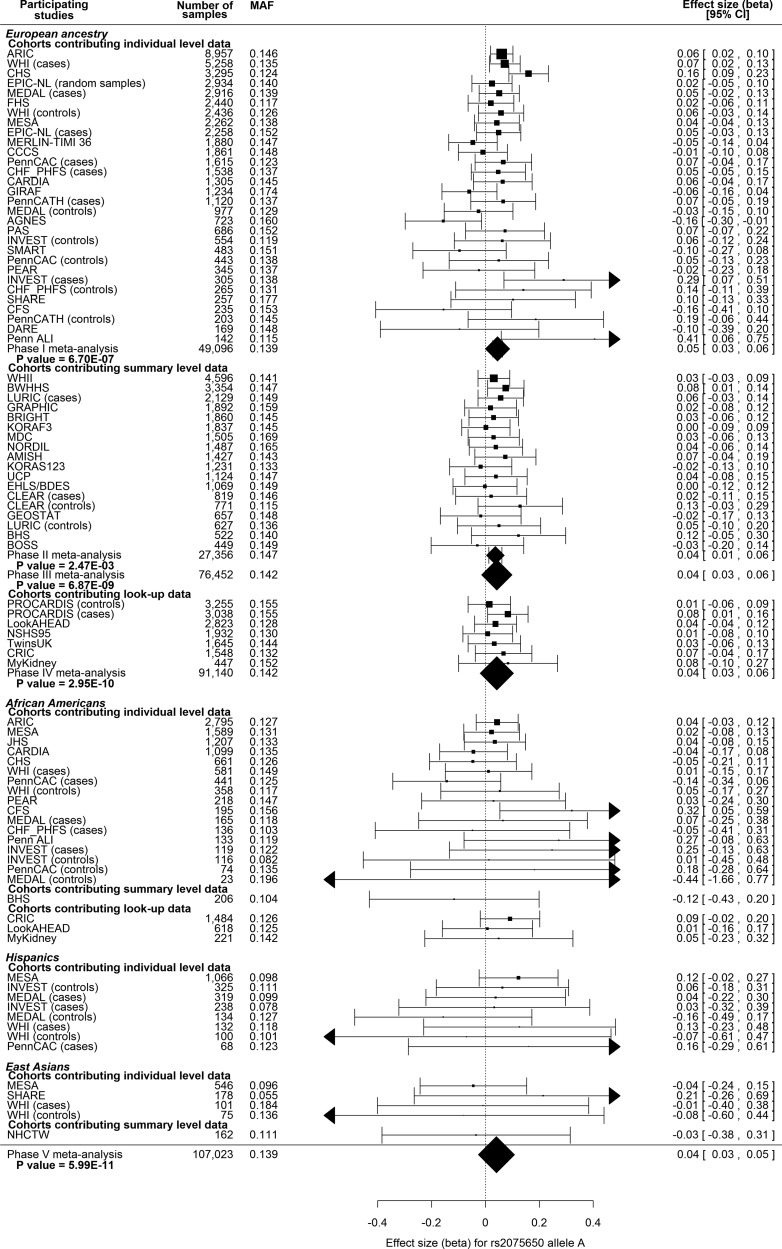
Meta-analysis was then performed including both individual- and summary-level cohorts of EA (Phase III in Fig. 1; n = 78 436). All signals of association identified in individual-level cohorts remained in this phase, with three additional signals revealed: rs997295 in mitogen-activated protein kinase kinase 5 (MAP2K5), rs4704220 in collagen, type IV, alpha 3 (Goodpasture antigen) binding protein (COL4A3BP) - 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and rs4788099 in SH2B adaptor protein 1 (SH2B1). In total, nine loci were associated with BMI at array-wide significance (P < 2.40E-06, shown in bold in Table 1) with five loci surpassing the traditional genome-wide significance threshold (P < 5.00E-08, shown in bold italic in Table 1). These results provide replication of the well-established BMI association signals in FTO (12–16) and BDNF (14,16), which serves as robust positive controls of our study design. Further examination of the novel SNP in TOMM40-APOE-APOC1 is shown for each cohort using a forest plot in Figure 2. Forest plots for all the other eight top signals in Phase III can be found in Supplementary Material, Figures S1–S8.
Figure 2.
Forest plots for the novel finding rs2075650 at the translocase of outer mitochondrial membrane 40 homolog (yeast)-apolipoprotein E-apolipoprotein C-I (TOMM40-APOE-APOC1) locus of genetic association between BMI and ITMAT-Broad-CARe (IBC) SNPs. Name of participating study, number of samples entering the meta-analysis, minor allele frequency (MAF) and effect size together with 95% confidence interval (CI) are shown as indicated by the header line of each plot. Effect sizes of meta-analysis results are shown at the bottom of each plot. Studies are sorted by sample size and are grouped according to ethnicities and data-contributing type (individual level or summary level). Sub-meta-analysis results are also shown where applicable.
In Phase IV we selected a list of 29 look-up SNPs including positive controls, borderline signals and those in LD with SNPs in previously reported BMI loci (see Methods Section), and interrogated them in six additional look-up studies serving as another replication data set. After performing meta-analysis incorporating the new data, two novel signals, rs5996074 in sterol regulatory element binding transcription factor 2 (SREBF2; P = 9.43E-07) and rs1211166 in neurotrophic tyrosine kinase, receptor, type 2 [NTRK2, a brain-derived neurotrophic factor (BDNF) receptor gene; P = 1.04E-06], became associated with BMI at array-wide significance (Table 1; for forest plots, see Supplementary Material, Figs S9 and S10). All the other Phase III top SNPs showed stronger evidence of association, except SNPs rs997295 in MAP2K5 and rs4704220 in the COL4A3BP-HMGCR locus, for which P-values weakened slightly (Table 1). Moreover, all heterogeneity measures (I2 values) are less for the Phase IV meta-analysis than for those in Phase III (Table 1), except for the case of FTO.
As all cohort-specific analysis was conducted in a sex-stratified manner, meta-analysis of each sex was performed and the results were compared for concordance between males and females. All signals identified had concordant direction of effect in males and females. Marginal heterogeneity was observed at rs997295 (MAP2K5), where the genetic signal was stronger in males (males P = 5.50E-06; females P = 8.03E-02; heterogeneity P = 0.04; Supplementary Material, Table S1).
The Phase IV meta-analysis results provide replication and validate five loci SH2B1, MAP2K5, TFAP2B, FANCL-FLJ30838 and COL4A3BP-HMGCR, which were recently identified by Speliotes et al. (16) in the Genetic Investigation of Anthropometric Traits (GIANT) consortium (Table 1), in addition to the three previously reported BMI loci: FTO, MC4R and BDNF. Further examination provided replication in 8 of the 10 GIANT loci that are represented on the IBC array with adequate coverage. Two loci, TNNI3 interacting kinase (TNNI3K) and gastric inhibitory polypeptide receptor (GIPR) (with previously reported lead SNPs rs1514175 and rs2287019, respectively) did not reach array-wide significance in the Phase III meta-analysis, but marginal significance was observed with consistent direction of effect (Table 2 and Supplementary Material, Table S2). To calculate the extent of variation explained by the associated and validated SNPs from Phase IV, we returned to Phase I cohorts with individual-level phenotype and genotype data available. The variation explained by each SNP individually was limited ranging from 0.01 to 0.37% per SNP (Table 1).
Table 2.
Established loci that are represented on the ITMAT-Broad-CARe (IBC) array at equal or greater coverage than GWAS arrays but that did not show array-wide significant P-value in Phase IV meta-analysis (n = 92 903)
| Chr. | Candidate gene | Previous lead SNP | IBC SNP with strongest P-value |
IBC SNP of highest r2 with previous lead SNP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Effect | r2 with previous SNPa | P-value | Same direction of effect | SNP | Effect | r2 with previous SNPa | P-value | Same direction of effect | |||
| 1p31.1 | TNNI3K | rs1514175 | rs7553158 | 0.015 | 0.97 | 1.49E-03 | Yes | rs1514176 | 0.014 | 1.00 | 3.45E-03 | Yes |
| 19q13.3 | GIPR | rs2287019 | rs11672660 | 0.028 | 0.67 | 2.74E-06 | Yes | rs11672660 | 0.028 | 0.67 | 2.74E-06 | Yes |
Chr., chromosome.
aLinkage disequilibrium measure r2 with the previous lead SNP in column three.
Conditional analysis reveals two independent BMI signals in MC4R and BDNF
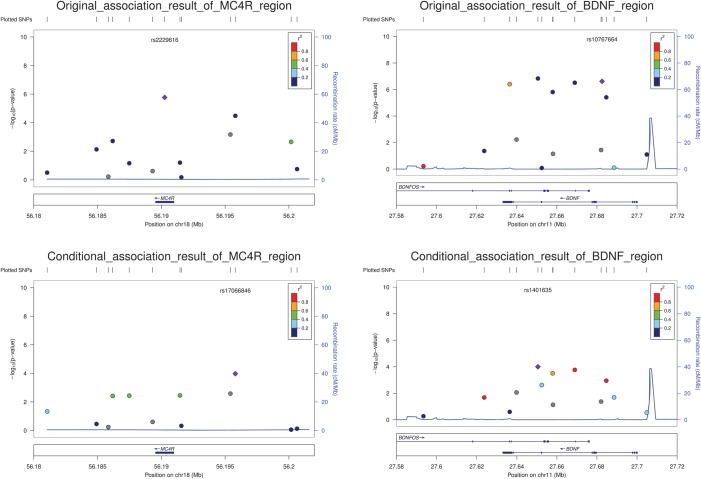
We performed conditional analysis to identify loci containing multiple variants independently influencing BMI. In cohorts where individual-level data were available, regression analysis was repeated including an additional term to adjust for the effect of the lead SNP identified in the overall EA meta-analysis. Two loci, BDNF and MC4R, were shown to contain an additional independent signal (Table 3; Fig. 3). In BDNF, after conditioning on SNP rs10767664 a second SNP rs1401635 showed locus-wide significance (P = 1.00E-04; between-SNP LD measures: r2 = 0.115, D′ = 0.995). Similarly in MC4R, after conditioning on rs2229616, an additional SNP, rs17066846, showed locus-wide significance (P = 1.05E-04; between-SNP LD measures: r2 = 0.005, D′ = 1.000). For each locus, when both independent signals were incorporated in the regression model, no further signals became stronger than the locus-wide significance threshold. Interestingly, in both loci the lower MAF SNP was the lead SNP and the minor allele in the lead SNP was never observed on the same haplotype as the minor allele at the secondary SNP signal (resulting in a high level of D′). However, in both cases the genotype at one SNP was not a good predictor of the genotype at the other SNP (resulting in the low r2).
Table 3.
Conditional analysis showing two loci harbor independent signals in BMI association with ITMAT-Broad-CARe (IBC) SNPs after conditioning on the lead SNP
| Locus | # SNPs in the locus | SNP | Position | Effect allele | Eff. All. Freq. | Phase I meta-analysis |
Meta-analysis of conditional associations |
r2 with lead SNP | D′ with lead SNP | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | Effect | P-value | ||||||||
| MC4R | 16 | rs2229616 | 56190256 | C | 0.98 | 0.111 | 1.71E-06 | — | — | — | — |
| rs17066846 | 56195798 | G | 0.19 | 0.040 | 3.29E-05 | 0.038 | 1.05E-04 | 0.005 | 1 | ||
| BDNF | 13 | rs10767664 | 27682562 | A | 0.21 | 0.041 | 2.41E-07 | — | — | — | — |
| rs1401635 | 27650567 | C | 0.30 | 0.037 | 1.46E-07 | 0.029 | 1.00E-04 | 0.115 | 0.995 | ||
Eff. All. Freq., effect allele frequency.
Data based on all Phase I European ancestry samples (n = 50 933). All SNPs mapped back to NCBI.
Figure 3.
Regional plots for the melanocortin 4 receptor (MC4R) and brain-derived neurotrophic factor (BDNF) region before and after doing conditional analysis. Two MC4R sub-figures are to the left panel and two BDNF sub-figures are to the right. For each locus, the top part shows unconditional meta-analysis results for individual-level samples of European ancestry (EA) and the bottom part shows meta-analysis results conditioning on lead SNP, rs2229616 in MC4R and rs10767664 in BDNF, respectively. Figures were generated using LocusZoom (31).
Meta-analysis of additional ethnicities reveals the consistency and strengthened the significance of BMI-associated variants identified in European ancestry
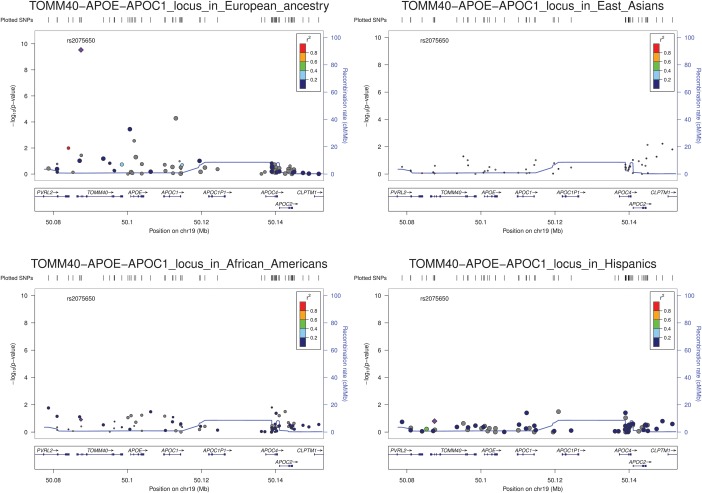
BMI meta-analysis limited to the cohorts containing African-Americans, Hispanics and East Asian individuals did not reveal any array-significant association, the likely result of lower power afforded by the smaller sample sizes. Comparison of meta-analysis results in those populations for variants associated with BMI in EA samples are shown in Supplementary Material, Table S3. Regional plots representing genomic context around loci with top BMI-association SNPs were generated using LocusZoom (31) for TOMM40-APOE-APOC1 (Fig. 4) as well as FTO, BDNF, MC4R, COL4A3BP−HMGCR and SH2B1 loci (Supplementary Material, Figs S11–S15). Histograms of the distribution of observed effects are outlined in Supplementary Material, Figure S16. The width of the distribution of observed effects was larger in the non-European ethnicities due to fewer cohorts and the smaller sample sizes. Of 11 SNPs identified in EA cohorts in Phase IV, 6 (FTO, BDNF, MC4R, FANCL-FLJ30838, TFAP2B and NTRK2) were observed to be nominally associated, with the same direction of effect, in the African-American cohorts (P < 0.05; Supplementary Material, Table S3). Only one of the signals (rs4788099 in SH2B1) is of opposite effect direction in samples of African ancestry, but the effect is very close to zero. No variants were associated in Hispanics or East Asians, which is again likely due to their limited power due to small sample sizes. The estimated direction of effect was concordant for 7 of 11, and 6 of 11, lead SNPs in Hispanics and East Asians, respectively (Supplementary Material, Table S3).
Figure 4.
Regional plots of the TOMM40-APOE-APOC1 region for multi-ethnic association between BMI and ITMAT-Broad-CARe (IBC) array SNPs. Meta-analysis regional results of overall European ancestry individuals (top left), African-American (bottom left), East Asians (top right) and Hispanics (bottom right) are shown using LocusZoom (31). In each sub-figure, each spot represents one IBC SNP and its y-axis coordinate indicates significance level in association. For each SNP, linkage disequilibrium (LD) measure r2 with the lead SNP can be determined from the inset color scheme. Genome recombination rates (in cM/Mb) are shown by blue lines and gene annotation information can be found at the bottom.
In the global multi-ethnic meta-analysis, we interrogated all 92 903 individuals of EA, 12 297 African-Americans, 2625 Hispanics and 1087 East Asians (total n = 108 912). Of the 11 SNPs found in the Phase IV meta-analysis including every cohort of EA, nine achieved greater evidence for association in the global multi-ethnic meta-analysis. As for the remaining two signals, the top BMI-associated SNP rs1421085 (in the FTO gene) showed weaker evidence for association, but still of P-value ∼ 2.00 E-68, and rs2904880 (in the SH2B1 locus) became weaker but still met array-wide significance (Table 1; Fig. 1). We observed one additional variant surpassing the array-wide significance, rs11672660 in GIPR (effect allele C, effect = 0.027 and P-value = 1.58E-06) which is the same index SNP of borderline significance in the Phase IV meta-analysis (Table 2). This result also ensured validation of the previously reported BMI association of GIPR.
Exclusion of CVD enriched sample confirms the robust nature of the observed BMI associations identified from our cohorts of European ancestry
As some participating studies in this meta-analyses recruited individuals using case–control or clinical trial ascertainment criteria, we sought to reduce the potential bias effects of enrichment for CVD status. We repeated the BMI meta-analysis but included only samples from the population- or community-based and control-only cohorts using individuals of EA (n = 48 241). Compared with Phase III results, five of the nine signals remain at array-wide significance, whereas four others were only marginally associated with BMI (P ≤ 1.18E-04; Supplementary Material, Table S4). The relative effect size and effect direction remained concordant in all nine SNPs, suggesting the reduction in statistical evidence is most likely a result of the reduction in sample size.
As some cohorts in this study also participated in the recent GIANT BMI GWAS meta-analysis (16), we performed an additional analysis to assess BMI associations within cohorts not represented in the GIANT meta-analysis. Five of the nine loci were significant at the array-wide threshold in the IBC ‘non-GIANT’ meta-analysis while the remaining four SNPs showed marginal evidence for association (P ≤ 1.51E-04; Supplementary Material, Table S2). Interestingly a previously undescribed signal in CADM1 (rs3802858) was still near the array-wide significance threshold, with P = 9.26E-06 (versus 4.45E-06 when we include IBC data from GIANT cohorts; see look-up SNP list in Supplementary Material, Table S5) and intriguingly a gene in the same functional group, CADM2, was documented to be associated with BMI in the GIANT study (Supplementary Material, Table S2). However after performing association meta-analysis including the look-up studies, this particular SNP did not replicate.
eQTL analyses of the eleven BMI-associated loci reveals multi-loci cis-acting eQTLs for the associated SH2B1 SNP in both purified primary B-cells and monocytes
We attempted to assess functionality of the 11 BMI SNPs found to be associated at array-wide significance using expression QTL (eQTL) analysis in two distinct primary cell types: B-cells and monocytes. The cis eQTL findings for the array-wide significant SNPs with the Spearman test P ≤ 7.00E-05 are shown in Supplementary Material, Table S6. We observed that three of the BMI-associated SNPs either form, or are in LD to SNPs that are eQTL. Most notable of these eQTL include the rs4788099 variant in SH2B1 which forms multi-loci cis eQTLs in both primary B-cells and primary monocytes. Supplementary Material, Figure S17 illustrates that rs4788099 forms a multi-loci cis eQTL to probes in spinster homolog 1 (Drosophila) (SPNS1) with PB-Cell = 4.25E-10 and also to probes in Tu translation elongation factor, mitochondrial (TUFM) with PB-Cell = 4.69E-37. A multi-loci cis eQTL is also evident for rs4788099 in primary monocytes to probes in five loci (Supplementary Material, Fig. S17): coiled-coil domain containing 101 (CCDC101; Pmonocyte = 1.14E-06 and 4.90E-06); SPNS1 (Pmonocyte = 3.46E-10); sul-fotransferase family, cytosolic, 1A, phenol-preferring, member 1 (SULT1A1; Pmonocyte = 5.34E-06); sul-fotransferase family, cytosolic, 1A, phenol-preferring, member 4 (SULT1A4; Pmonocyte = 1.43E-10) and TUFM (Pmonocyte = 1.83E-29).
DISCUSSION
In a meta-analysis of gene-centric association studies of BMI encompassing 92 903 individuals of EA (Phase IV as illustrated in Fig. 1), we identified three novel BMI associations: the TOMM40-APOE-APOC1 locus at genome-wide significance (P ≤ 1.49E-08), and SREBF2 and NTRK2 genes at array-wide significance (P ≤ 1.04E-06). We also observed association in three previously established BMI genes (FTO, BDNF and MC4R) and replicated five loci (FANCL-FLJ30838, SH2B1, TFAP2B, MAP2K5 and COL4A3BP-HMGCR) first identified in the recent GIANT BMI analysis (16). Two additional loci described in GIANT and captured on the array showed nominal evidence for association with BMI (TNNI3K, GIPR). Conditional analysis identified two loci each containing two independent signals within BDNF and MC4R. Utilizing SNP data in up to 108 912 individuals and considering ancestry as a covariate, we also conducted a multi-ethnic meta-analysis (Phase V in Fig. 1), with the result showing stronger signals for all but two top findings in the Phase IV result.
Among the novel findings in the current investigation, the lead BMI SNP (rs2075650) lies within an intron of TOMM40. The LD structure spanning the TOMM40-APOE-APOC1 locus precludes more discrete localization of the causal signal(s) with the available data. The G allele of rs2075650, associated with lower BMI in the current report, has been reported to be associated with elevated Alzheimer's disease risk (32) increased total cholesterol concentrations (33) and longevity (34,35). In data from the 1000 Genomes Project, no LD is observed between rs2075650 and either of the APOE isoform-determining SNPs in Europeans. Associations between the functional APOE ɛ2/ɛ3/ɛ4 isoforms and circulating lipid concentrations have been known for over 25 years (36). Subsequently, association between the APOE isoforms and elevated risk for CVD and Alzheimer's disease was identified. The three isoforms are captured by SNPs rs7412 and rs429358, but unfortunately the SNPs either failed quality control metrics in this study (and generally do not perform well in microarray-based genotyping due to nearby nucleotide sequences, and as such are often not included genome-wide data sets) or was not included in the SNP panel used to perform meta-analysis and were thus not directly available in the current investigation. No BMI association was observed for a SNP in LD with the APOE isoform-determining SNPs. It remains unclear at this point if APOE or one of the other gene products in the locus is responsible for the observed effects.
While the TOMM40-APOE-APOC1 locus has not been previously associated with BMI, the forest plot (Fig. 2) of rs2075650 shows the association is strongly consistent across the cohorts investigated, including across ethnicities, increasing our confidence in the authenticity of the association. Inadequate coverage of the area in previous BMI association studies is not likely responsible, as rs2075650 is included in the main Illumina genome-wide genotyping products and the International HapMap Project, and a P-value of 0.0237 with effective sample size of 111 450 was observed in GIANT consortium data (16) (obtained via online GIANT consortium data files at http://www.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files; date last accessed September 2012). Heterogeneity of the signal in different populations is a possible reason for no previous report, but the wide range of study types included in this meta-analysis argues against this. Analysis by Speliotes et al. (16) indicates that over 250 common variant loci are predicted to contribute to the distribution of BMI across the population. Even with extremely large sample sizes the power to identify variants with small effects is not complete (37). Thus, random chance may be the most likely explanation why association between BMI and the TOMM40-APOE-APOC1 locus has not been previously discovered.
The rs5996074 intronic variant lies within the ubiquitously expressed SREBF2 gene . The protein product, SREBF2, has many known roles for modulating transcription of genes involved in fatty acid and cholesterol metabolism including 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase and ATP-binding cassette transporter A1(ABCA1). As well, the locus contains the microRNA-33a/b, further controlling lipid metabolism (38). However, no genetic variant within SREBF2 has previously been associated with either plasma lipid concentrations or BMI in high-throughput analyses.
The rs1211166 intronic variant lies in NTRK2, which is a receptor of BDNF. Like BDNF, NTRK2 has been implicated in the regulation of mammalian eating behavior and energy balance (39). Mouse models with an Ntrk2 mutation resulting in only 25% of typical expression levels have been shown to develop obesity (40). BDNF and NTRK2 have several additional roles in processes such as learning, sensation, memory and locomotor behavior (41). Furthermore, Yeo et al. (42) have described a de novo loss-of-function mutation in NTRK2 associated with severe early-onset obesity, hyperphagia, developmental delay and other defects in higher neurological functions in a child patient.
Rare mutations within the MC4R were first identified to be segregating in families with extreme obesity in a monogenic fashion. Subsequently, many functional mutations have been identified in children selected for sequencing due to extreme obesity, accounting for up to 6% of childhood obesity cases (43). Common SNPs have been repeatedly reported to be associated with multiple measures of obesity with small effect throughout the population (15). Interestingly, the SNP showing the strongest MC4R association in the current study was the functional (44) and uncommon rs2229616, with an MAF of 2% in individuals of EA. The observed effect size of rs2229616 is modest in size (β = −0.12 per minor allele T), but was the largest effect of all associated variants in the current study. The allele frequency and effect was similar across the other three examined ethnicities. SNP rs2229616 represents an uncommon polymorphism with modest effect that could only be successfully replicated at the genome-wide significance level through the large sample sizes and dense genotyping provided by the study design of the current investigation.
Conventional GWAS studies have lacked direct coverage or effective imputation of many low frequency variants. Imputation is often performed to increase the number of queried SNPs in GWAS efforts by using phased haplotype data from panels of densely genotyped individuals, such as those provided by the HapMap and 1000 Genomes Project (45). Because the IBC array contains many SNPs with MAFs <5% and a large number of individuals have been genotyped on the array, it provides a unique opportunity and sufficient power to directly test for the association of lower frequency variants with relatively small effect sizes. In the current meta-analysis, one low frequency variant in MC4R with MAF ∼0.02 was shown to be associated with BMI.
The results of large-scale genetic association meta-analyses are important for identifying robust and valid associations that shed new light onto important genes and biological pathways, and that the results can be combined to have a larger effect on predicted heritability. The results of large-scale genetic association meta-analyses are important for identifying robust and valid associations that shed new light onto important genes and biological pathways, and that the results can be combined to have a larger effect on predicted heritability. It is a statistical inevitability that if extremely large sample sizes are required to confidently identify and confirm a signal, the effect of the identified signals need be either small, relatively heterogeneous in effect or that are from variants with lower minor allele frequency. It is clear that a wide spectrum of variant types and frequencies, and interactions between them and the environment, as well as the possibility of new discoveries, will be required to fully explain the heritability of BMI.
Genes identified from monogenic disorders influencing height and lipid levels have been subsequently shown to contain common SNPs associated with the respective phenotypes across its distribution in the population. As the power of genetic association studies improves with increasing sample size, more examples of this pattern have become apparent. Common variants that are not identified within genes associated with monogenic disorders and genes with common variants that do not appear to be mutated in monogenic forms of the disease, may provide additional clues to the role of the particular gene in the etiology of the disease. Of 21 genes mutated in monogenic forms of obesity (46), 16 were adequately represented on the IBC array (Supplementary Material, Table S7), but only MC4R and BDNF were found to be associated with BMI, providing insight into the role of common and uncommon variants in the genetic architecture of obesity. Why there appears to be fewer obesity-associated common SNPs in ‘monogenic obesity’ genes, when compared with other traits, is yet to be understood. Several current initiatives will further assess whether obese individuals have an over-representation of disruptive rare variants in genes with common obesity-associated SNPs.
Association testing conditioned on the genotype of the identified lead SNPs revealed two loci, MC4R and BDNF, that each appear to harbor at least two independent signals of association. The observed independently associated SNPs are physically close to the lead SNPs (5 and 32 kb, respectively), and no recombination hotspot lies between them. In both loci, the two SNPs represent mutations on an ancestral haplotype that have become more common in the population, thus the rare alleles at the two SNPs are never observed on the same haplotype yielding D′ = 1. As the common genotype at either SNP gives little information regarding the genotype at the other SNP, the r2 remains low (with r2 of 0.005 and 0.115, respectively). Association of two independent, though different, SNPs with BMI has been previously reported within MC4R (16). With >30 functional mutations reported in MC4R (47), allelic heterogeneity at the locus is well established. Similarly, independent BMI-associated SNPs have also been reported within BDNF (14); thus the current study replicated findings of allelic heterogeneity in the genetic architecture of BMI.
Using eQTL analyses in two distinct primary cell-types with divergent functions, B-cells and monocytes, we found strong multi-loci cis eQTLs for the rs4788099 variant in SH2B1 with both SULT1A1 and TUFM. Interestingly Gutierrez-Aguilar et al. (48) recently demonstrated that in high-fat diet versus chow-fed rats SH2B1 is down-regulated in the hypothalamus and that the mRNA levels of SULT1A1 were down-regulated in the adipose tissue of rats fed on high-fat diet, whereas TUFM was up-regulated. Furthermore, SULT1A1 and TUFM were shown to be up-regulated in the livers of high-fat diet rats versus chow-fed rats.
In conclusion, in the current study of 108 912 individuals from 46 cohorts including four ethnicities, we identify three novel BMI signals in TOMM40-APOE-APOC1, NTRK2 and SREBF2, and confirmed genetic associations in an additional eight loci recently reported by the GIANT consortium. Two independent signals of association were identified in both MC4R and BDNF. Although individual examinations of the additional ethnic groups were not powered to adequately replicate specific findings from European cohorts, combined analyses did increase the strength of associations.
MATERIALS AND METHODS
Study design
Figure 1 illustrates the overall study design. Five primary analyses were undergone, including discovery (Phase I) and replication (Phase II, III and IV) of BMI-associated signals in European descendants, and meta-analysis including additional ethnicities (Phase V). Quality control of phenotype (BMI), genotype (SNPs) and covariates (age, ethnicity, disease status etc.) was performed in each cohort independently, regardless of whether providing individual-level data (Phase 1), cohort association results (Phase II) or lookup SNP results (Phase IV). Four secondary analyses were performed including conditional association, sex-specific tests, analyses limited to healthy controls and meta-analysis of cohorts not present in the recent GIANT BMI GWAS (16).
Participating studies
In total, 24 studies contributed individual-level genotype, BMI and covariate data (self-reported ethnicity, sex, age at measurement, study site information) for a total of 64 440 participants (Phase I, Supplementary Material, Table S8). Sixteen studies contributed summary-level association results from a standardized analysis guideline, representing an additional 27 868 subjects (Phase II, Supplementary Material, Table S9). Finally six studies of 16 604 samples served as a further layer of replication (Phase IV, Supplementary Material, Table S10), where top association SNPs from Phase III meta-analysis were interrogated as look-ups, creating an overall total sample size of 108 912. Participating data sets included population-based cohorts, collections of cases and controls for a variety of metabolic and cardiovascular phenotypes, and individuals collected for clinical trials (Supplementary Material, Tables S8–10). All participating studies obtained informed consent for DNA analysis and received approval from local institutional review boards. Detailed summary statistics for participants' BMI and age for each study is shown in Supplementary Material, Tables S11–13.
Genotyping and quality control
Genotyping was performed using the gene-centric IBC array, whose design and use has been described in detail elsewhere (19). Up to 49 320 SNPs were clustered into genotype calls using the BeadStudio software (Illumina) and subjected to quality control filters at the sample and SNP level, separately within each cohort. Samples were excluded where individual call rates were <90% and where sex mismatches between the genetic data and reported value were observed. SNPs were removed for call rates of <95% or for the Hardy–Weinberg equilibrium (HWE) cut-off P = 1.00E-07. Because of the large number of low frequency SNPs included in the design of the IBC array, and the desire to capture low MAF variants of large effect across the large data set, no filtering was performed based on MAF.
Statistical analyses
Evaluation of relatedness
To ensure the removal of cryptic relatedness and duplicate samples from cohorts composed of unrelated individuals, pairwise identity-by-descent proportions were estimated between all subjects within each participating study based on identity-by-state (IBS) sharing and sample allele frequencies using PLINK (49). For each set of duplicates or monozygotic twins, and for those samples with an estimated pairwise IBS threshold of >0.3, was retained the sample with the highest genotyping call rate for analysis. In cohorts with extensive family structure except the Amish study and PROCARDIS, only founders were kept to the following steps of analysis with a pairwise IBS check also implemented to ensure the removal of related individuals. In the Amish study, family structure was taken into account and the analysis was carried out using the Mixed Model Analysis for Pedigree (MMAP) software (J. O'Connell, 2008, Annual Meeting of The American Society of Human Genetics, Philadelphia, PA, USA, abstract). When performing association computations in the PROCARDIS study, family structures was also considered as described elsewhere (20).
Evaluation of population stratification
Within each participating study, self-reported ethnicity was verified by multidimensional scaling analysis of IBS distances as implemented in PLINK, with HapMap CEU, YRI and CHB/JPT samples included as reference standards. After performing a prune of SNPs in LD (r2 > 0.3), we used Eigenstrat to compute 10 principal components (PCs) on the subset of non-excluded individuals for use as covariates in the regression analyses (50,51). Self-reported ethnicity in the African, Hispanic and East Asians individuals was also verified using Eigenstrat.
BMI residuals
For each cohort, baseline BMI values were first regressed on age, age-squared, and study site within ethnicity-sex stratum and case–control status for CVD or T2D case–control studies. Since test statistics for low frequency SNPs are better calibrated if the phenotype is normally distributed, we normal quantile transformed the residuals by obtaining the rank order of the residual values within stratum, dividing the rank value by (n + 1) and then taking the probit (inverse normal) of this value. For sex-combined analysis, we combined the male- and female-specific normal quantile transformed residuals.
Association testing and meta-analysis (in Phase I through V of the primary analyses)
The primary association analyses were performed using linear regression with normal quantile transformed BMI residuals as a continuous trait, as implemented in PLINK (49). We assumed an additive genetic model and included covariate adjustment for the top 3 PCs of ancestry for individuals of EA and 10 PCs for all other population subgroups. All analyses were stratified by sex, ethnicity and CVD and/or T2D case–control status if applicable. All meta-analyses were performed using sample–size based as well as inverse-variance (standard error)-based models in METAL (52), and between-study heterogeneity was assessed using the I2 metric (53). Additional ethnicities were evaluated using the same approach as described for individuals of EA. We then performed global multi-ethnic meta-analysis on all available cohorts.
Different significance cut-off thresholds have been used in previous studies utilizing the IBC array-based studies (20,24). The standard Bonferroni approach for calculating a significance threshold to declare a SNP associated is inappropriate considering the variants on the array were selected to densely cover hypothesis-driven loci. In the CARe IBC array studies, we utilized the ethnicity-specific local LD structures to estimate the effective numbers of independent tests. We observed ∼26 500 and ∼20 500 independent observations yielding statistical thresholds of P = 1.89E-06 and P = 2.44E-06 in African-Americans and European Americans, respectively, to maintain a false positive rate of 5% (54).
Selection of look-up SNPs for additional replication studies
After Phase III meta-analysis of EA samples from individual- and summary-level data sets, we generated a list of 29 SNPs (Supplementary Material, Table S5) for look up replications in six additional cohorts (Supplementary Material, Table S10). We selected (i) two top BMI-associated SNPs in Phase III, rs2229616 in MC4R (also an uncommon variant with an MAF of 0.02 in EA individuals) and rs2075650 in TOMM40-APOE-APOC1 (the novel finding up to Phase III); (ii) four array-wide, yet not genome-wide significant SNPs (those in FANCL-FLJ30838, TFAP2B, COL4A3BP-HMGCR and SH2B1); (iii) 16 marginal signals in Phase III; and (iv) seven SNPs that are of highest LD value (r2) with any signal in each established BMI GWAS associated locus for which IBC array has equal or better coverage. We then performed association testing for the selected SNP set within the six look-up studies.
Conditional analysis
We further examined all loci harboring evidence for association for additional independent signals. Conditional analyses was performed in PLINK (49) using a regression model that included the lead SNP as a covariate, and surrounding SNPs were evaluated for significance. Conditional analyses were performed in 50 933 individuals of EA for which individual-level data were available. We used a locus-wide Bonferroni-corrected significance threshold, i.e. 0.05/(number of SNPs tested within a given locus), when evaluating the conditional analyses results. To directly assess the LD at each associated locus, we used all the individual-level genotype data and resolved haplotype structure using PHASE version 2.1.1 (55,56).
Sex-specific analysis
To test for sex-related differences in BMI-associated signals, we performed sex-specific analysis after Phase III of our primary analysis. We meta-analyzed individual and summary level data for males only and females only, conducted a second round meta-analysis including those two sets of outputs, and checked heterogeneity measurement of each SNP deemed with evidence for association in the Phase III results. We used heterogeneity P-value <0.05 between males and females as SNP selection criteria.
Meta-analyses restricted to subsets of participating efforts
We then aggregated data sets with CVD cases only from the case–control study designs, and we also grouped together population- or community-based, and control-only studies contributing individual- and summary-level data (specified in Supplementary Material, Tables S8 and S9) and performed a separate meta-analysis. Similarly, excluding participating studies appearing in the recent GIANT BMI effort (16) (specified in Supplementary Material, Tables S8 and S9), a meta-analysis on non-GIANT cohorts in the Phase I and II stages was performed using the IBC array data sets.
Variance explained
The variance explained (adjusted R2) by associated SNPs was calculated within EA cohorts with individual-level genotype and phenotype data available (phase I) using a linear regression model incorporating the age- and age-squared-adjusted inverse normal transformed BMI residuals as outcome, as well as 10 PCs as covariates. The average variance explained, weighted by the sample size of each contributing study, is reported.
Expression QTL analysis
eQTL analyses were conducted on array-wide significant loci using monocyte and primary B-cells from 288 healthy blood donors of European origin as recently described (57). Positive selection was used to enrich CD19 + B cells and CD14+ monocytes from peripheral blood mononuclear cells prepared from the whole blood of 288 healthy Europeans. Sample purity was confirmed with flow cytometry and was found to 90–95% for the primary B-cells and ∼99% for monocytes. We performed array based genome-wide gene expression profiling using the HumanHT-12 v4 Expression BeadChips (Illumina) and whole genome genotyping was performed using Human OmniExpress-12v1.0 BeadChips (Illumina). Following standard processing and quality control filtering, we performed eQTL mapping at 651 210 SNPs for 283 samples that passed QC criteria. Probes were tested against SNPs residing within a 2.5 Mb window on either side of the probe as designated by Illumina co-ordinates for association using a Spearman rank model. All values presented represent uncorrected significance levels. Statistics were analyzed using R and appropriate packages.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health of the United States of America (contract number HHSN268200960009C). Y.G. is supported by the Hilda and Preston Davis Foundation through the Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research. M.B.L. is supported by a Canadian Institutes of Health Research (CIHR) MD-, PhD Studentship Award.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the scientists, technicians and participants of all of the contributing cohorts. Specific study acknowledgements are cited in the Supplementary material, Acknowledgements.
Conflict of interest statement. None declared.
APPENDIX
Yiran Guo*1, Matthew B. Lanktree*2, Kira C. Taylor*3, Benjamin P. Fairfax4, Clara C. Elbers5, John Barnard6, Martin Farrall7, Sandosh Padmanabhan8, Jens Baumert9, Berta A. Castillo10, Tom R. Gaunt11, Yan Gong12, Ramakrishnan Rajagopalan13, Simon PR Romaine14, Meena Kumari15, Suzanne Rafelt16, Erin N. Smith17, Yun R. Li18, Suthesh Sivapalaratnam19, Erik PA van Iperen20, Elizabeth K. Speliotes21, Elina Toskala22, Li Zhang23, Heather M. Ochs-Balcom24, Tushar R. Bhangale25, Hareesh R. Chandrupatla26, Fotios Drenos27, Christian Gieger28, Jayanta Gupta29, Toby Johnson30, Marcus E. Kleber31, Seiko Makino32, Massimo Mangino33, Yan Meng34, Christopher P. Nelson35, James S. Pankow36, Nathan Pankratz37, Tom S. Price38, Jonathan Shaffer39, Haiqing Shen40, Sam Tischfield41, Maciej Tomaszewski42, Larry D. Atwood43, Kristian M. Bailey44, Ashok Balasubramanyam45, Clinton T. Baldwin46, Hanneke Basart47, Florianne Bauer48, Elijah R. Behr49, Amber L. Beitelshees50, Gerald S. Berenson51, Shirley AA Beresford52, Connie R. Bezzina53, Deepak L. Bhatt54, Jolanda MA Boer55, Peter S. Braund56, Gregory L. Burke57, Ben Burkley58, Cara Carty59, Wei Chen60, Robert Clarke61, Rhonda M. Cooper-DeHoff62, Sean P. Curtis63, Paul IW de Bakker64, Jonas S. de Jong65, Christian Delles66, Anna F. Dominiczak67, David Duggan68, Harold I. Feldman69, Clement E. Furlong70, Mathias M. Gorski71, John G. Gums72, Robert Hardwick73, Claire Hastie74, Iris M. Heid75, Guan-Hua Huang76, Gordon S. Huggins77, Steve E. Humphries78, Susan A. Kirkland79, Mika Kivimaki80, Ronald Klein81, Barbara E. Klein82, William C. Knowler83, Kandice Kottke-Marchant84, Andrea Z. LaCroix85, Taimour Y. Langaee86, Mingyao Li87, Helen N. Lyon88, Steffi Maiwald89, Julieann K. Marshall90, Amar Mehta91, Matthijs FL Meijs92, Olle Melander93, Nuala Meyer94, Nandita Mitra95, Cliona M. Molony96, David A. Morrow97, Gurunathan Murugesan98, Stephen J. Newhouse99, Javier F. Nieto100, N. Charlotte Onland-Moret101, Willem H. Ouwehand102, Jutta Palmen103, Carl J. Pepine104, Jane Ranchalis105, Sylvia E. Rosas106, Elisabeth A. Rosenthal107, Hubert Scharnagl108, Nicholas J. Schork109, Pamela J. Schreiner110, Tina Shah111, Michael Shashaty112, Daichi Shimbo113, Sathanur R. Srinivasan114, Fridtjof Thomas115, Martin D. Tobin116, Michael Y. Tsai117, W.M.Monique Verschuren118, Lynne E. Wagenknecht119, Bernhard R. Winkelmann120, Taylor Young121, Salim Yusuf122, Mohammad H. Zafarmand123, Joseph M. Zmuda124, Aeilko H. Zwinderman125, Sonia S. Anand126, Anthony J. Balmforth127, Bernhard O. Boehm128, Eric Boerwinkle129, Paul R. Burton130, Thomas P. Cappola131, Juan P. Casas132, Mark J. Caulfield133, David C. Christiani134, Jason Christie135, Karen J. Cruickshanks136, George Davey-Smith137, Karina W. Davidson138, Ian N. Day139, Pieter A. Doevendans140, Gerald W. Dorn II141, Garret A. FitzGerald142, Alistair S. Hall143, Aroon D. Hingorani144, Joel N. Hirschhorn145, Marten H. Hofker146, Kees G. Hovingh147, Thomas Illig148, Yalda Jamshidi149, Gail P. Jarvik150, Julie A. Johnson151, Peter A. Kanetsky152, John JP Kastelein153, Wolfgang Koenig154, Debbie A. Lawlor155, Winfried März156, Jeanne McCaffery157, Jessica L. Mega158, Braxton D. Mitchell159, Sarah S. Murray160, Jeffery R. O'Connell161, Sanjay R. Patel162, Annette Peters163, Mary Pettinger164, Daniel J. Rader165, Susan Redline166, Muredach P. Reilly167, Marc S. Sabatine168, Eric E. Schadt169, Alan R. Shuldiner170, Roy L. Silverstein171, Tim D. Spector172, Herman A. Taylor173, Barbara Thorand174, Mieke D. Trip175, Hugh Watkins176, H.-Erich Wichmann177, Caroline S. Fox178, Struan FA Grant179, Inga Peter180, Philippa J. Talmud181, Patricia B. Munroe182, James G. Wilson183, Julian C. Knight184, Nilesh J. Samani185, Robert A. Hegele186, Folkert W. Asselbergs187, Keri L. Monda188, Yvonne T. van der Schouw189, Ellen W. Demerath190, Cisca Wijmenga191, Nicholas J. Timpson192, Alex P. Reiner193, Kari E. North194, George J. Papanicolaou195, Hakon Hakonarson196, Leslie A. Lange†197, Brendan J. Keating†198
1Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA; BGI-Shenzhen, Beishan Industrial Zone, Yantian District, Shenzhen, 518083, China, 2Departments of Medicine and Biochemistry, Schulich School of Medicine and Dentistry, University of Western Ontario, London, Ontario, Canada, 3Department of Epidemiology, School of Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC 27514, USA; Department of Epidemiology and Population Health, School of Public Health and Information Sciences, University of Louisville, Louisville, KY 40292, USA, 4Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK., 5Complex Genetics Section, Department of Medical Genetics (DBG), University Medical Center Utrecht, Utrecht, the Netherlands; Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, The Netherlands, 6Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA, 7Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, OX3 7BN, UK; Department of Cardiovascular Medicine, University of Oxford, Level 6 West Wing, John Radcliffe Hospital, Headley Way, Headington, Oxford, OX3 9DU, UK, 8BHF Glasgow Cardiovascular Research Centre; Institute of Cardiovascular and Medical Sciences; College of Medical, Veterinary and Life Sciences; University of Glasgow; Glasgow G12 8TA; UK, 9Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany, 10Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 11MRC Centre for Causal Analyses in Translational Epidemiology, School of Social and Community Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol, UK, 12Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, Gainesville, FL, USA, 13Department of Medicine, Division of Medical Genetics, University of Washington, Seattle, WA, USA, 14Leeds Institute of Genetics, Health & Therapeutics, University of Leeds, Leeds, LS2 9JT, UK, 15Research Department of Epidemiology & Public Health, UCL Institute of Epidemiology & Health Care, University College London, 1-19 Torrington Place, London WC1E 6BT, UK, 16Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK, 17Department of Pediatrics and Rady's Children's Hospital, University of California at San Diego, School of Medicine, La Jolla, CA 92093, USA, 18Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 19Department of Vascular Medicine, Academic Medical Center, 1105 AZ Amsterdam, The Netherlands., 20Durrer Center for Cardiogenetic Research, Amsterdam, The Netherlands; Department of Clinical Epidemiology, Biostatistics and Bioinformatics, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands., 21Metabolism Initiative and Program in Medical and Population Genetics, Broad Institute, Cambridge, MA 02142, USA, 22Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 23Department of Quantitative Health Sciences, Lerner Research Institute, Cleveland Clinic, Cleveland, OH, USA, 24Department of Social and Preventive Medicine, University at Buffalo, Buffalo, NY, USA, 25Department of Bioinformatics and Computational Biology, Genentech, South San Francisco, CA, USA, 26Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 27Centre for Cardiovascular Genetics, Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London, 5 University Street, London WC1E 6JF, UK, 28Institute of Genetic Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany, 29Center for Clinical Epidemiology and Biostatistics, Univeresity of Pennsylvania School of Medicine, Philadelphia, PA 19104-6021, USA, 30Clinical Pharmacology and Barts and the London Genome,Centre, William Harvey Research Institute, Barts and the London School of Medicine, Queen Mary University of London, London EC1M 6BQ, UK, 31LURIC Study nonprofit LLC, Freiburg, Germany and Mannheim Institute of Public Health, Social and Preventive Medicine, Mannheim Medical Faculty, University of Heidelberg, Mannheim, Germany, 32Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK., 33Department of Twin Research and Genetic Epidemiology, King's College London, London, UK, 34Program for Medical and Population Genetics, Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA, USA, 35Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Cardiovascular Biomedical Research Unit, Glenfield Hospital, Groby Road, Leicester, LE3 9QP, UK, 36Division of Epidemiology & Community Health. University of Minnesota. Minneapolis, MN, USA, 37Institute of Human Genetics, Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN, USA, 38The Institute for Translational Medicine and Therapeutics, School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvannia, USA, 39Department of Medicine, Columbia University, New York, NY, USA, 40Division of Endocrinology, Diabetes and Nutrition, University of Maryland School of Medicine, Baltimore, MD, USA, 41Tri-Institutional Training Program in Computational Biology and Medicine, Weill Cornell Medical College, New York, NY, USA, 42Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK, 43Framingham Heart Study, Boston University School of Medicine, Boston, MA 02118-2526, 44Leeds Institute of Genetics, Health & Therapeutics, University of Leeds, Leeds, LS2 9JT, UK, 45Translational Metabolism Unit, Division of Diabetes, Endocrinology and Metabolism, Baylor College of Medicine, Houston, TX, 46School of Medicine. Boston University School of Medicine. Boston, MA, 47Department of Vascular Medicine, University of Amsterdam, Amsterdam, NL, 48Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, the Netherlands; Complex Genetics Section, Department of Medical Genetics (DBG), University Medical Center Utrecht, Utrecht, the Netherlands, 49Cardivascular Sciences Research Centre, St George's University of London, London, UK, 50Department of Medicine, University of Maryland School of Medicine, Baltimore, MD, USA, 51Department of Epidemiology, 1440 Canal Street, Suite 1829, Tulane University, New Orleans, LA, USA, 52Department of Epidemiology, Box 357236, University of Washington, Seattle, WA 98195-7236, USA, 53Heart Failure Research Center, Department of Clinical and Experimental Cardiology, Academic Medical Center, Amsterdam, the Netherlands; Molecular and Experimental Cardiology Group, Academic Medical Centre, Amsterdam, the Netherlands , 54VA Boston Healthcare System, Brigham and Women's Hospital, and Harvard Medical School, Boston, MA, USA, 55National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands, 56Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Cardiovascular Biomedical Research Unit, Glenfield Hospital, Groby Road, Leicester, LE3 9QP, UK, 57Division of Public Health Sciences, Wake Forest University School of Medicine, Winston-Salem, NC, USA, 58Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, Gainesville, FL, USA, 59Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA, 60Department of Epidemiology, 1440 Canal Street, Suite 1829, Tulane University, New Orleans, LA, USA, 61Clinical Trial Service Unit (CTSU), University of Oxford, Oxford, UK, 62Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, Gainesville, FL, USA, 63Merck Research Laboratories, P.O. Box 2000, Rahway, NJ 07065, USA, 64Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, The Netherlands; Department of Medical Genetics, Biomedical Genetics, University Medical Center, Utrecht, The Netherlands, 65Heart Failure Research Center, Department of Clinical and Experimental Cardiology, Academic Medical Center, Amsterdam, the Netherlands, 66BHF Glasgow Cardiovascular Research Centre; Institute of Cardiovascular and Medical Sciences; College of Medical, Veterinary and Life Sciences; University of Glasgow; Glasgow G12 8TA; UK, 67BHF Glasgow Cardiovascular Research Centre; Institute of Cardiovascular and Medical Sciences; College of Medical, Veterinary and Life Sciences; University of Glasgow; Glasgow G12 8TA; UK, 68Translational Genomics Research Institute, Phoenix, AZ, USA, 69Renal Division, Department of Medicine, Center for Clinical Epidemiology and Biostatistics, and Leonard Davis Institute, Univeresity of Pennsylvania, Philadelphia, PA, USA, 70Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington, Seattle, WA, USA, 71Institute of Epidemiology and Preventive Medicine, University Hospital Regensburg, Germany; Institute of Epidemiology I, Helmholtz Zentrum München - German Research Center for Environmental Health, Neuherberg, Germany, 72Departments of Pharmacotherapy and Translational Research and Community Health and Family Medicine, University of Florida, FL, USA, 73Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK, 74BHF Glasgow Cardiovascular Research Centre; Institute of Cardiovascular and Medical Sciences; College of Medical, Veterinary and Life Sciences; University of Glasgow; Glasgow G12 8TA; UK, 75Institute of Epidemiology and Preventive Medicine, University Hospital Regensburg, Germany; Institute of Epidemiology I, Helmholtz Zentrum München - German Research Center for Environmental Health, Neuherberg, Germany, 76Institute of Statistics. National Chiao Tung University. Hsinchu 300, Taiwan, 77Molecular Cardiology Research Institute, Center for Translational Genomics, Tufts Medical Center and Tufts University, Boston, MA, USA, 78Centre for Cardiovascular Genetics, Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London, 5 University Street, London WC1E 6JF, UK, 79Department of Community Health and Epidemiology, Dalhousie University, Canada., 80Research Department of Epidemiology & Public Health, UCL Institute of Epidemiology & Health Care, University College London, 1-19 Torrington Place, London WC1E 6BT, UK, 81Department of Ophthalmology & Visual Sciences. University of Wisconsin-Madison. Madison, WI, USA, 82Department of Ophthalmology & Visual Sciences. University of Wisconsin-Madison. Madison, WI, USA, 83National Institute of Diabetes and Digestive and Kidney Diseases, Phoenix, AZ, 84Pathology and Laboratory Medicine Institute, Cleveland Clinic, Cleveland, OH, USA, 85Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA, 86Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, Gainesville, FL, USA, 87Cardiovascular Institute, Univeresity of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA., 88Program in Medical and Population Genetics, Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA, USA; Program in Genomics and Divisions of Genetics and Endocrinology, Children's Hospital, Boston, MA, USA, 89Department of Vascular Medicine, University of Amsterdam, Amsterdam, The Netherlands, 90Departments of Medicine (Medical Genetics), University of Washington, Seattle, WA, USA, 91Environmental and Occupational Medicine and Epidemiology Program, Harvard School of Public Health, Boston, MA, USA, 92Department of Cardiology, Division Heart and Lungs, University Medical Center Utrecht, Utrecht, The Netherlands, 93Clinical Research Center (CRC), Malmö University Hospital, Malmö SE-205 02, Sweden, 94Univeresity of Pennsylvania Medical Center, Pulmonary, Allergy & Critical Care Division, Philadelphia, PA 19104-6160, USA, 95Center for Clinical Epidemiology and Biostatistics, Univeresity of Pennsylvania School of Medicine, Philadelphia, PA 19104-6021, USA, 96Department of Genetics, Rosetta Inpharmatics, Seattle, WA, USA, 97TIMI Study Group, Cardiovascular Division, Brigham and Women's Hospital, Boston, MA, USA, 98Pathology and Laboratory Medicine Institute, Cleveland Clinic, Cleveland, OH, USA, 99Clinical Pharmacology and Barts and the London Genome,Centre, William Harvey Research Institute, Barts and the London School of Medicine, Queen Mary University of London, London EC1M 6BQ, UK, 100Department of Population Health Sciences. University of Wisconsin-Madison. Madison, WI, USA, 101Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, the Netherlands; Complex Genetics Section, Department of Medical Genetics (DBG), University Medical Center Utrecht, Utrecht, the Netherlands, 102Department of Haematology, University of Cambridge & Wellcome Trust Sanger Institute, Cambridge, UK, 103Centre for Cardiovascular Genetics, Dept Medicine University College London, 5 University St London, WC1E 6JF, UK, 104Division of Cardiovascular Medicine, University of Florida College of Medicine, Gainesville, FL, USA, 105Departments of Medicine (Medical Genetics), University of Washington, Seattle, WA, USA, 106Univeresity of Pennsylvania School of Medicine, Renal-Electrolyte and Hypertension Division, 1 Founders, 3400 Spruce Street, Philadelphia, PA 19104, USA, 107Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington, Seattle, WA, USA, 108Clinical Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Graz, Austria, 109The Scripps Translational Science Institute and The Scripps Research Institute, 3344 N. Torrey Pines Ct. Ste 300, La Jolla, CA, USA, 110Division of Epidemiology & Community Health, University of Minnesota, Minneapolis, MN 55454, USA, 111Research Department of Epidemiology & Public Health, UCL Institute of Epidemiology & Health Care, University College London, 1-19 Torrington Place, London WC1E 6BT, UK, 112Univeresity of Pennsylvania Medical Center, Pulmonary, Allergy & Critical Care Division, Philadelphia, PA 19104-6160, USA, 113Department of Medicine, Columbia University, New York, NY, USA, 114Department of Epidemiology, 1440 Canal Street, Suite 2000, Tulane University, New Orleans, LA, USA, 115Department of Preventive Medicine College of Medicine, The University of Tennessee Health Science Center, 66 N. Pauline, Suite 633 Memphis, TN 38105, USA, 116Dept of Health Sciences, University of Leicester, Leicester, UK, 117Department of Laboratory Medicine and Pathology, University of Minnesota, MN, USA, 118National Institute for Public Health and the Environment (RIVM), Bilthoven, the Netherlands, 119Division of Public Health Sciences, Wake Forest School of Medicine, Winston-Salem, NC, 120Cardiology Group, Frankfurt-Sachsenhausen, Germany, 121Program for Medical and Population Genetics, Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA, USA, 122Population Health Research Institute, Hamilton Health Sciences, McMaster University, Hamilton, Ontario L8L 2X2, Canada, 123Department of Cardiology, Division Heart and Lungs, University Medical Center Utrecht, Utrecht, The Netherlands, 124Department of Epidemiology, Graduate School of Public Health, University of Pittsburgh, 130 DeSoto St, Pittsburgh, PA 15261, USA, 125Department of Clinical Epidemiology, Biostatistics and Bioinformatics, AMC, Amsterdam, The Netherlands, 126Population Health Research Institute, Hamilton Health Sciences, McMaster University, Hamilton, Ontario L8L 2X2, Canada, 127Leeds Institute of Genetics, Health & Therapeutics, University of Leeds, Leeds, LS2 9JT, UK, 128Division of Endocrinology, Ulm University Medical Centre, Ulm, Germany, 129Human Genetics Center, University of Texas Health Science Center and Human Genome Sequencing Center at Baylor College of Medicine, Houston, TX, USA, 130Dept of Health Sciences, University of Leicester, Leicester, UK, 131Penn Cardiovascular Institute, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, 132Faculty of Epidemiology and Population Health, London School of Hygiene and Tropical Medicine, UK; Genetic Epidemiology Group, Department of Epidemiology and Public Health, University College London, UK, 133Clinical Pharmacology and Barts and the London Genome,Centre, William Harvey Research Institute, Barts and the London School of Medicine, Queen Mary University of London, London EC1M 6BQ, UK, 134Environmental and Occupational Medicine and Epidemiology Program, Harvard School of Public Health, Boston, MA, USA, 135Univeresity of Pennsylvania Medical Center, Pulmonary, Allergy & Critical Care Division, Philadelphia, PA 19104-6160, USA, 136Department of Population Health Sciences. Department of Ophthalmology & Visual Sciences. University of Wisconsin-Madison. Madison, WI, USA, 137MRC Centre for Causal Analyses in Translational Epidemiology, Department of Social Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol BS8 2BN, UK, 138Departments of Medicine & Psychiatry, Columbia University, New York, NY, USA, 139MRC Centre for Causal Analyses in Translational Epidemiology, School of Social and Community Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol, UK, 140Department of Cardiology, Division Heart and Lungs, University Medical Center Utrecht, Utrecht, The Netherlands, 141Center for Pharmacogenomics, Department of Medicine, Washington University School of Medicine, St. Louis, MO, USA, 142The Institute for Translational Medicine and Therapeutics, School of Medicine, Univeresity of Pennsylvania, Philadelphia, Pennsylvannia, USA, 143Leeds Institute of Genetics, Health & Therapeutics, University of Leeds, Leeds, LS2 9JT, UK, 144Research Department of Epidemiology & Public Health, UCL Institute of Epidemiology & Health Care, University College London, 1-19 Torrington Place, London WC1E 6BT, UK, 145Metabolism Initiative and Program in Medical and Population Genetics, Broad Institute, Cambridge, MA 02142, USA; Divisions of Genetics and Endocrinology and Program in Genomics, Children's Hospital, Boston, MA 02115, USA; Department of Genetics, Harvard Medical School, Boston, MA 02115, USA, 146Dept. Pathology and Medical Biology, Medical Biology division, Molecular Genetics, University Medical Center Groningen and Groningen University, Groningen, The Netherlands, 147Department of Vascular Medicine, AMC, Amsterdam, The Netherlands, 148Research Unit of Molecular Epidemiology, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany; Hannover Unified Biobank, Hannover Medical School, Hannover, Germany, 149Division of Clinical Developmental Sciences, St George's University of London, London, UK, 150Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington, Seattle, WA, USA, 151Department of Pharmacotherapy and Translational Research and Center for Pharmacogenomics, University of Florida, Gainesville, FL, USA, 152Center for Clinical Epidemiology and Biostatistics, Univeresity of Pennsylvania School of Medicine, Philadelphia, PA 19104-6021, USA, 153Department of Vascular Medicine, AMC, Amsterdam, The Netherlands, 154Department of Internal Medicine II – Cardiology, University of Ulm Medical Centre, Ulm, Germany, 155MRC Centre for Causal Analyses in Translational Epidemiology, School of Social and Community Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol, UK, 156Synlab Academy, Mannheim, Germany and Mannheim Institute of Public Health, Social and Preventive Medicine, Mannheim Medical Faculty, University of Heidelberg, Mannheim, Germany, 157Weight Control and Diabetes Research Center, The Miriam Hospital and Warren Alpert School of Medicine at Brown University, Providence, RI, USA, 158TIMI Study Group, Cardiovascular Division, Brigham and Women's Hospital, Boston, MA, USA, 159Division of Endocrinology, Diabetes and Nutrition, University of Maryland School of Medicine, Baltimore, MD, USA, 160Scripps Translational Science Institute and Scripps Health, 3344 N. Torrey Pines Ct. Ste 300, La Jolla, CA, USA, 161Division of Endocrinology, Diabetes and Nutrition, University of Maryland School of Medicine, Baltimore, MD, USA, 162Department of Medicine, Brigham and Women's Hospital, Boston MA, USA, 163Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany, 164Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA, 165Cardiovascular Institute, the Perelman School of Medicine at the Univeresity of Pennsylvania, Philadelphia, PA, USA, 166Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA, USA, 167Cardiovascular Institute, the Perelman School of Medicine at the Univeresity of Pennsylvania, Philadelphia, PA, USA, 168TIMI Study Group, Cardiovascular Division, Brigham and Women's Hospital, Boston, MA, USA, 169Sage Bionetworks, Seattle, WA 98109, USA., 170Division of Endocrinology, Diabetes and Nutrition, University of Maryland School of Medicine, Baltimore, MD, USA; Geriatric Research and Education Clinical Center, Veterans Administration Medical Center, Baltimore, MD, USA, 171Department of Medicine, Medical College of Wisconsin, Milwaukee, WI, USA, 172Department of Twin Research and Genetic Epidemiology, King's College London, London, UK, 173Department of Medicine, University of Mississippi Medical Center, Jackson, MS, USA, 174Institute of Epidemiology II, Helmholtz Zentrum München, German Research Center for Environmental Health, Neuherberg, Germany, 175Department of Cardiology, AMC, Amsterdam, The Netherlands, 176Department of Cardiovascular Medicine, The Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK, 177Institute of Epidemiology I, Helmholtz Zentrum München - German Research Center for Environmental Health, Neuherberg, Germany, 178Framingham Heart Study, Boston University School of Medicine, Boston, MA 02118-2526, USA, 179Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 180Department of Genetics and Genomic Sciences; Mount Sinai School of Medicine, New York, NY, USA, 181Centre for Cardiovascular Genetics, Institute of Cardiovascular Science, Faculty of Population Health Sciences, University College London, 5 University Street, London WC1E 6JF, UK, 182Clinical Pharmacology and Barts and the London Genome,Centre, William Harvey Research Institute, Barts and the London School of Medicine, Queen Mary University of London, London EC1M 6BQ, UK, 183Department of Physiology and Biophysics, University of Mississippi Medical Center, Jackson, MS, USA, 184Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK., 185Dept of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Cardiovascular Biomedical Research Unit, Glenfield Hospital, Groby Road, Leicester, LE3 9QP, UK, 186Robarts Research Institute, University of Western Ontario, London, Ontario, Canada N6A5K8, 187Department of Cardiology, Division Heart and Lungs, University Medical Center Utrecht, Utrecht, The Netherlands; Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, The Netherlands; Department of Medical Genetics, Biomedical Genetics, University Medical Center, Utrecht, The Netherlands, 188Department of Genetics, University of North Carolina School of Medicine at Chapel Hill, Chapel Hill, NC 27514, USA, 189Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht, the Netherlands, 190Department of Epidemiology and Community Health, University of Minnesota, Minneapolis, MN, USA, 191University Medical Center Groningen, and University of Groningen, Groningen, the Netherlands, 192MRC Centre for Causal Analyses in Translational Epidemiology, School of Social and Community Medicine, University of Bristol, Oakfield House, Oakfield Grove, Bristol BS8 2BN, UK, 193Division of Public Health Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA, USA, 194Department of Epidemiology, School of Public Health, University of North Carolina at Chapel Hill, Chapel Hill, NC 27514, USA, 195National Heart, Lung, and Blood Institute (NHLBI), Division of Cardiovascular Sciences, Bethesda, MD 20892, USA, 196Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA, 197Department of Genetics, University of North Carolina School of Medicine at Chapel Hill, Chapel Hill, NC 27514, USA, 198Center for Applied Genomics, Abramson Research Center, The Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA
*: Co-first authors (Y.G., M.B.L. and K.C.T.). †: Joint corresponding authors (L.A.L. and B.J.K.).
REFERENCES
- 1.Ogden C.L., Carroll M.D., Curtin L.R., McDowell M.A., Tabak C.J., Flegal K.M. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. doi:10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 2.International Association for Study of Obesity (IASO) Global prevalence of adult obesity. IASO; 2011. http://www.iaso.org/site_media/uploads/Prevalence_of_Adult_Obesity_April_2011_New.pdf. (date last accessed September 2012) [Google Scholar]
- 3.World Health Organization (WHO) Obesity and overweight fact sheet. WHO; 2011. http://www.who.int/mediacentre/factsheets/fs311/en/index.html. (date last accessed September 2012) [Google Scholar]
- 4.Center for Disease Control and Prevention (CDC) Heart Disease Facts. CDC; 2010. http://www.cdc.gov/heartdisease/facts.htm. (date last accessed September 2012) [Google Scholar]
- 5.Wormser D., Kaptoge S., Di Angelantonio E., Wood A.M., Pennells L., Thompson A., Sarwar N., Kizer J.R., Lawlor D.A., Nordestgaard B.G., et al. Separate and combined associations of body-mass index and abdominal adiposity with cardiovascular disease: collaborative analysis of 58 prospective studies. Lancet. 2011;377:1085–1095. doi: 10.1016/S0140-6736(11)60105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy M.I. Genomics, type 2 diabetes, and obesity. N. Engl. J. Med. 2010;363:2339–2350. doi: 10.1056/NEJMra0906948. doi:10.1056/NEJMra0906948. [DOI] [PubMed] [Google Scholar]
- 7.Miller W.M., Nori-Janosz K.E., Lillystone M., Yanez J., McCullough P.A. Obesity and lipids. Curr. Cardiol. Rep. 2005;7:465–470. doi: 10.1007/s11886-005-0065-8. doi:10.1007/s11886-005-0065-8. [DOI] [PubMed] [Google Scholar]
- 8.Wolin K.Y., Carson K., Colditz G.A. Obesity and cancer. Oncologist. 2010;15:556–565. doi: 10.1634/theoncologist.2009-0285. doi:10.1634/theoncologist.2009-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maes H.H., Neale M.C., Eaves L.J. Genetic and environmental factors in relative body weight and human adiposity. Behav. Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. doi:10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 10.Hjelmborg J.B., Fagnani C., Silventoinen K., McGue M., Korkeila M., Christensen K., Rissanen A., Kaprio J. Genetic influences on growth traits of BMI: a longitudinal study of adult twins. Obesity (Silver Spring) 2008;16:847–852. doi: 10.1038/oby.2007.135. doi:10.1038/oby.2007.135. [DOI] [PubMed] [Google Scholar]
- 11.Wardle J., Carnell S., Haworth C.M., Plomin R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am. J. Clin. Nutr. 2008;87:398–404. doi: 10.1093/ajcn/87.2.398. [DOI] [PubMed] [Google Scholar]
- 12.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. doi:10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. doi:10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. doi:10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 15.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. doi:10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speliotes E.K., Willer C.J., Berndt S.I., Monda K.L., Thorleifsson G., Jackson A.U., Allen H.L., Lindgren C.M., Luan J., Magi R., et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42:937–948. doi: 10.1038/ng.686. doi:10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleiman P.M., Flory J., Imielinski M., Bradfield J.P., Annaiah K., Willis-Owen S.A., Wang K., Rafaels N.M., Michel S., Bonnelykke K., et al. Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. doi:10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 18.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. doi:10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 19.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. doi:10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarke R., Peden J.F., Hopewell J.C., Kyriakou T., Goel A., Heath S.C., Parish S., Barlera S., Franzosi M.G., Rust S., et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N. Engl. J. Med. 2009;361:2518–2528. doi: 10.1056/NEJMoa0902604. doi:10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 21.IBC 50K CAD Consortium. Large-scale gene-centric analysis identifies novel variants for coronary artery disease. PLoS Genet. 2011;7:e1002260. doi: 10.1371/journal.pgen.1002260. doi:10.1371/journal.pgen.1002260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stark K., Esslinger U.B., Reinhard W., Petrov G., Winkler T., Komajda M., Isnard R., Charron P., Villard E., Cambien F., et al. Genetic association study identifies HSPB7 as a risk gene for idiopathic dilated cardiomyopathy. PLoS Genet. 2010;6:e1001167. doi: 10.1371/journal.pgen.1001167. doi:10.1371/journal.pgen.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cappola T.P., Li M., He J., Ky B., Gilmore J., Qu L., Keating B., Reilly M., Kim C.E., Glessner J., et al. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ. Cardiovasc. Genet. 2010;3:147–154. doi: 10.1161/CIRCGENETICS.109.898395. doi:10.1161/CIRCGENETICS.109.898395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talmud P.J., Drenos F., Shah S., Shah T., Palmen J., Verzilli C., Gaunt T.R., Pallas J., Lovering R., Li K., et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am. J. Hum. Genet. 2009;85:628–642. doi: 10.1016/j.ajhg.2009.10.014. doi:10.1016/j.ajhg.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanktree M.B., Guo Y., Murtaza M., Glessner J.T., Bailey S.D., Onland-Moret N.C., Lettre G., Ongen H., Rajagopalan R., Johnson T., et al. Meta-analysis of dense genecentric association studies reveals common and uncommon variants associated with height. Am. J. Hum. Genet. 2011;88:6–18. doi: 10.1016/j.ajhg.2010.11.007. doi:10.1016/j.ajhg.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxena R., Elbers C.C., Guo Y., Peter I., Gaunt T.R., Mega J.L., Lanktree M.B., Tare A., Castillo B.A., Li Y.R., et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am. J. Hum. Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. doi:10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi:10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulit S.L., Voight B.F., de Bakker P.I. Multiethnic genetic association studies improve power for locus discovery. PLoS ONE. 2010;5:e12600. doi: 10.1371/journal.pone.0012600. doi:10.1371/journal.pone.0012600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stutzmann F., Vatin V., Cauchi S., Morandi A., Jouret B., Landt O., Tounian P., Levy-Marchal C., Buzzetti R., Pinelli L., et al. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum. Mol. Genet. 2007;16:1837–1844. doi: 10.1093/hmg/ddm132. doi:10.1093/hmg/ddm132. [DOI] [PubMed] [Google Scholar]
- 30.Young E.H., Wareham N.J., Farooqi S., Hinney A., Hebebrand J., Scherag A., O'Rahilly S., Barroso I., Sandhu M.S. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int. J. Obes. (Lond.) 2007;31:1437–1441. doi: 10.1038/sj.ijo.0803609. doi:10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R., Willer C.J. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. doi:10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert J.C., Heath S., Even G., Campion D., Sleegers K., Hiltunen M., Combarros O., Zelenika D., Bullido M.J., Tavernier B., et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat. Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. doi:10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 33.Aulchenko Y.S., Ripatti S., Lindqvist I., Boomsma D., Heid I.M., Pramstaller P.P., Penninx B.W., Janssens A.C., Wilson J.F., Spector T., et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat. Genet. 2009;41:47–55. doi: 10.1038/ng.269. doi:10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sebastiani P., Solovieff N., Dewan A.T., Walsh K.M., Puca A., Hartley S.W., Melista E., Andersen S., Dworkis D.A., Wilk J.B., et al. Genetic signatures of exceptional longevity in humans. PLoS ONE. 2012;7:e29848. doi: 10.1371/journal.pone.0029848. doi:10.1371/journal.pone.0029848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deelen J., Beekman M., Uh H.W., Helmer Q., Kuningas M., Christiansen L., Kremer D., van der Breggen R., Suchiman H.E., Lakenberg N., et al. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. doi:10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehnholm C., Lukka M., Kuusi T., Nikkila E., Utermann G. Apolipoprotein E polymorphism in the Finnish population: gene frequencies and relation to lipoprotein concentrations. J. Lipid. Res. 1986;27:227–235. [PubMed] [Google Scholar]
- 37.Park J.H., Wacholder S., Gail M.H., Peters U., Jacobs K.B., Chanock S.J., Chatterjee N. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 2010;42:570–575. doi: 10.1038/ng.610. doi:10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rayner K.J., Suarez Y., Davalos A., Parathath S., Fitzgerald M.L., Tamehiro N., Fisher E.A., Moore K.J., Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. doi:10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kernie S.G., Liebl D.J., Parada L.F. BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000;19:1290–1300. doi: 10.1093/emboj/19.6.1290. doi:10.1093/emboj/19.6.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu B., Goulding E.H., Zang K., Cepoi D., Cone R.D., Jones K.R., Tecott L.H., Reichardt L.F. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat. Neurosci. 2003;6:736–742. doi: 10.1038/nn1073. doi:10.1038/nn1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang E.J., Reichardt L.F. Neurotrophins: roles in neuronal development and function. Annu. Rev. Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. doi:10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeo G.S., Connie Hung C.C., Rochford J., Keogh J., Gray J., Sivaramakrishnan S., O'Rahilly S., Farooqi I.S. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat. Neurosci. 2004;7:1187–1189. doi: 10.1038/nn1336. doi:10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 43.Stutzmann F., Cauchi S., Durand E., Calvacanti-Proenca C., Pigeyre M., Hartikainen A.L., Sovio U., Tichet J., Marre M., Weill J., et al. Common genetic variation near MC4R is associated with eating behaviour patterns in European populations. Int. J. Obes. (Lond.) 2009;33:373–378. doi: 10.1038/ijo.2008.279. doi:10.1038/ijo.2008.279. [DOI] [PubMed] [Google Scholar]
- 44.Xiang Z., Litherland S.A., Sorensen N.B., Proneth B., Wood M.S., Shaw A.M., Millard W.J., Haskell-Luevano C. Pharmacological characterization of 40 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists and the agouti-related protein (AGRP) antagonist. Biochemistry. 2006;45:7277–7288. doi: 10.1021/bi0600300. doi:10.1021/bi0600300. [DOI] [PubMed] [Google Scholar]
- 45.Kolz M., Johnson T., Sanna S., Teumer A., Vitart V., Perola M., Mangino M., Albrecht E., Wallace C., Farrall M., et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5:e1000504. doi: 10.1371/journal.pgen.1000504. doi:10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahituv N., Kavaslar N., Schackwitz W., Ustaszewska A., Martin J., Hebert S., Doelle H., Ersoy B., Kryukov G., Schmidt S., et al. Medical sequencing at the extremes of human body mass. Am. J. Hum. Genet. 2007;80:779–791. doi: 10.1086/513471. doi:10.1086/513471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stutzmann F., Tan K., Vatin V., Dina C., Jouret B., Tichet J., Balkau B., Potoczna N., Horber F., O'Rahilly S., et al. Prevalence of melanocortin-4 receptor deficiency in Europeans and their age-dependent penetrance in multigenerational pedigrees. Diabetes. 2008;57:2511–2518. doi: 10.2337/db08-0153. doi:10.2337/db08-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez-Aguilar R., Kim D.H., Woods S.C., Seeley R.J. Expression of new loci associated with obesity in diet-induced obese rats: from genetics to physiology. Obesity (Silver Spring) 2012;20:306–312. doi: 10.1038/oby.2011.236. doi:10.1038/oby.2011.236. [DOI] [PubMed] [Google Scholar]
- 49.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 51.Price A.L., Butler J., Patterson N., Capelli C., Pascali V.L., Scarnicci F., Ruiz-Linares A., Groop L., Saetta A.A., Korkolopoulou P., et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 2008;4:e236. doi: 10.1371/journal.pgen.0030236. doi:10.1371/journal.pgen.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. doi:10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. doi:10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 54.Lo K.S., Wilson J.G., Lange L.A., Folsom A.R., Galarneau G., Ganesh S.K., Grant S.F., Keating B.J., McCarroll S.A., Mohler E.R., 3rd, et al. Genetic association analysis highlights new loci that modulate hematological trait variation in Caucasians and African Americans. Hum. Genet. 2010;129:307–317. doi: 10.1007/s00439-010-0925-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephens M., Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am. J. Hum. Genet. 2005;76:449–462. doi: 10.1086/428594. doi:10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. doi:10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fairfax B.P., Makino S., Radhakrishnan J., Plant K., Leslie S., Dilthey A., Ellis P., Langford C., Vannberg F.O., Knight J.C. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat. Genet. 2012;44:502–510. doi: 10.1038/ng.2205. doi:10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.