Abstract
Purpose.
Evidence supporting the immune system involvement in glaucoma includes increased titers of serum antibodies to retina and optic nerve proteins, although their pathogenic importance remains unclear. This study using an antibody-based proteomics approach aimed to identify disease-related antigens as candidate biomarkers of glaucoma.
Methods.
Serum samples were collected from 111 patients with primary open-angle glaucoma and an age-matched control group of 49 healthy subjects without glaucoma. For high-throughput characterization of antigens, serum IgG was eluted from five randomly selected glaucomatous samples and analyzed by linear ion trap mass spectrometry (LC-MS/MS). Serum titers of selected biomarker candidates were then measured by specific ELISAs in the whole sample pool (including an additional control group of diabetic retinopathy).
Results.
LC-MS/MS analysis of IgG elutes revealed a complex panel of proteins, including those detectable only in glaucomatous samples. Interestingly, many of these antigens corresponded to upregulated retinal proteins previously identified in glaucomatous donors (or that exhibited increased methionine oxidation). Moreover, additional analysis detected a greater immunoreactivity of the patient sera to glaucomatous retinal proteins (or to oxidatively stressed cell culture proteins), thereby suggesting the importance of disease-related protein modifications in autoantibody production/reactivity. As a narrowing-down strategy for selection of initial biomarker candidates, we determined the serum proteins overlapping with the retinal proteins known to be up-regulated in glaucoma. Four of the selected 10 candidates (AIF, cyclic AMP-responsive element binding protein, ephrin type-A receptor, and huntingtin) exhibited higher ELISA titers in the glaucomatous sera.
Conclusions.
A number of serum proteins identified by this immunoproteomic study of human glaucoma may represent diseased tissue-related antigens and serve as candidate biomarkers of glaucoma.
This immunoproteomic study identified antigenic targets of serum antibodies in glaucoma and initiated validation studies to assess their value as disease biomarkers. A number of serum proteins presented may represent diseased tissue-specific antigens and serve as candidate biomarkers of glaucoma.
Introduction
There is increasing awareness that glaucomatous neurodegeneration has an immune-mediated component. Besides multiple evidences supporting local immune/inflammatory responses and complement activation in human glaucoma and animal models (as reviewed by Tezel1), patients with glaucoma exhibit a complex repertoire of serum antibodies reacting with ocular antigens.2–5 Although multiple laboratories worldwide have commonly detected increased serum antibodies in glaucoma, the pathogenic importance of these antibodies is under intensive investigation and debate.6 Similarities in autoantibody production in different subtypes of human glaucoma and experimental animal models with induced ocular hypertension7 suggest that serum antibodies (also evident in many other diseases) may reflect a native response to tissue injury to facilitate phagocytic removal of the opsonized cell debris as a necessary step for tissue cleaning and healing. However, besides histopathologic evidence of immunoglobulin deposition in the glaucomatous human retina,8 there is ex vivo evidence in human donor retinas that supports the possibility of antibody-mediated collateral damage to retinal ganglion cells (RGCs).9 In addition, recent in vivo studies evaluating the possibility of immunogenic injury have included animal models induced by immunization with ocular antigens, and resulted in findings that suggest antibody-mediated RGC loss.10,11 However, a more recent study of Rag1 knockout mice lacking mature T and B lymphocytes has not detected a significant difference in the rate of glaucomatous RGC loss or axon damage relative to wild-type controls.12 Another view pertinent to serum antibodies, which are also present in healthy people, suggests their potential role in maintaining the immune homeostasis.13,14 While the studies evaluating the pathogenic importance of serum antibodies are ongoing,10,11,15,16 an independent research aim related to serum antibodies is the assessment of these antibodies and their target antigens as disease biomarkers in glaucoma.6 Regardless of the causative role of serum antibody response in glaucoma, the potential usefulness of serum antibodies as correlative biomarkers is supported by the unique antibody pattern among glaucoma patients (which exhibits specificity and sensitivity of approximately 93%),17 and the similarities in complex antibody profiles among different ethnic populations.18,19
Identification of glaucoma-specific molecular biomarkers presents great importance to facilitate early disease detection, prognosis prediction, and the follow-up of treatment responses, but exhibits many challenges as recently discussed in the ARVO/Pfizer Ophthalmic Research Institute Conference, 2011 (proceedings upcoming). One of the important challenges of serum biomarker detection is related to much lower abundance of most proteomic biomarkers than some disease-irrelevant serum proteins. However, antibody response holds the relative advantage of signal amplification for biomarker discovery in glaucoma. By using this advantage, we sought to identify antigenic targets of serum antibodies using an antibody-based immunoproteomics approach for high-throughput characterization and initiated validation studies for selected biomarker candidates. In addition, complementary experiments aimed to determine whether glaucoma-related protein modifications affect serum immunoreactivity. Here, we present the proteins that may represent diseased tissue-related antigens and serve as candidate biomarkers for glaucoma.
Materials and Methods
Patients
We studied 111 patients with high- or normal-pressure primary open-angle glaucoma (POAG) (mean age 66.9 ± 12.7 years) and an age-matched control group of 49 healthy volunteers without glaucoma or any other ocular disease (mean age 69.3 ± 10.6 years; P > 0.05). The diagnosis of glaucoma was based on characteristic alterations in the appearance of the optic nerve head and visual field in the absence of alternative causes of optic neuropathy. Maximum recorded intraocular pressure was less than 21 mm Hg in patients with normal-pressure glaucoma; however, since distinction of glaucoma subgroups on the basis of intraocular pressure is an arbitrary process, we studied all patients with a diagnosis of POAG as a single group. In addition to nonglaucomatous controls, to test the specificity of identified biomarker candidates for glaucoma, we also included an additional group of 20 patients with diabetic retinopathy (mean age 56.2 ± 10.5 years) followed by the Retina Clinic at the Department of Ophthalmology & Visual Sciences, University of Louisville, Louisville, KY.
Blood samples were taken after a written informed consent was obtained. The sera separated by centrifugation were stored at −80°C. All human samples were collected after Institutional Review Board approval. All subjects were treated in accordance with the Declaration of Helsinki.
Isolation of Serum IgG
Serum IgG were isolated using two different purification methods: a specific gel-based spin column purification (Melon gel IgG purification resin; Pierce/Thermo Fisher Scientific, Rockford, IL) and protein G-Dynabeads-based purification (Invitrogen, Grand Island, NY).
In the first method, a specific purification kit based on a resin containing a proprietary ligand that only allows IgG to pass through the column support was used to remove nonantibody serum proteins and isolate serum IgG. Briefly, 100 μL sera was added to the mini-spin columns prepared according to the manufacturer's instructions, and IgG was eluted by centrifuge at 2000g.
Dynabeads coupled with protein G (Invitrogen) were alternatively used to capture IgG from serum samples by affinity purification. Briefly, 10 μL sera was added to 100 μL proteinG-Dynabeads prepared according to the manufacturer's instructions and incubated at room temperature for 30 min. The beads were then pulled down using a magnet, and after washing to remove loosely bound components, specifically bound IgG was eluted in reducing conditions.
Eluted fractions were tested by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting with antihuman IgG (1:2000; Sigma-Aldrich, St. Louis, MO).
Mass Spectrometry
IgG elutes obtained from five randomly selected serum samples from each group were subjected to protein analysis by mass spectrometry. Trypsin-digested samples of serum IgG elutes were individually analyzed (not pooled) by two-dimensional capillary liquid chromatography and linear ion trap mass spectrometry (LC-MS/MS) as previously described.20–23 Protein identification from the MS/MS spectra was based on a specific analysis system (Sequest Sorcerer; Sage-N Research, San Jose, CA), which was set up to search a FASTA formatted human protein database with a fragment ion mass tolerance of 1.00 Da and a parent ion tolerance of 1.2 Da. A meta-analysis software (Scaffold; Proteome Software Inc., Portland, OR) was used to validate peptide and protein identifications based on the criteria of >95.0% and 99.0% probability and at least two peptides as specified by the Prophet peptide24 and protein algorithms.25 The abundance of each identified protein was determined by normalizing the number of unique spectral counts matching to the protein by its predicted molecular weight.26
Oxidative protein modification was determined by setting a Sequest search parameter to identify a mass shift corresponding to addition of the oxidation moiety to methionine (+16 Da) from the acquired MS/MS spectra. We normalized the number of methionine oxidation to the number of MS/MS spectra corresponding to the modified protein and expressed the abundance of oxidation as fold difference between glaucomatous and control samples.
Enzyme-Linked Immunosorbent Assay (ELISA)
Serum titers of selected 10 proteins were measured in all samples by sandwich ELISA as previously described.22,27,28 The specific kits used included annexin A1 (sensitivity < 1.25 ng/mL), apoptosis-inducing factor (AIF) (sensitivity < 0.078 ng/mL), baculoviral IAP repeat-containing protein 6 (IAP6) (sensitivity < 0.05 ng/mL), cyclic AMP-responsive element binding protein (CREB)-binding protein (sensitivity < 0.122 ng/mL), dynein heavy chain 1 (sensitivity < 0.112 ng/mL), ephrin type-A1 (sensitivity < 0.064 ng/mL), huntingtin protein (sensitivity < 0.055 ng/mL), neural cell adhesion molecule (sensitivity < 0.67 ng/mL), nucleotide-binding domain, leucine-rich repeat (NLR) containing family-pyrin domain containing 2 (NLRP2) (sensitivity < 0.05 ng/mL), and nuclear factor-kappa B (NF-κB)-binding protein (sensitivity < 0.127 ng/mL) (Uscn Life Science Inc., Wuhan, China). Serum samples were incubated in wells precoated with specific antibodies, and then a horseradish peroxidase-conjugated polyclonal antibody was added to the wells. After addition of a substrate solution, the enzyme reaction was terminated by sulphuric acid solution, and absorbance was measured at 450 nm using an ELISA microplate reader (VersaMax; Molecular Devices, Sunnyvale, CA) supplied with data acquisition and analysis software (SoftMax Pro; Molecular Devices). In addition to duplicate samples, each plate included blank control wells and seven serial dilutions of standards in duplicates. Concentrations were calculated from the standard curve obtained using pre-optimized concentration ranges.
Retinal Protein Sampling
Human retinal protein samples were obtained from donor eyes with or without POAG (kindly provided by Markus H. Kuehn, University of Iowa, as previously documented20–23). Protein lysation utilized a lysis buffer containing 50 mM Hepes-KOH pH 8.0, 100 mM KCl, 2 mM EDTA, 0.10% NP-40, 2 mM dithiothreitol, 10% glycerol, and protease and phosphatase inhibitors as previously described.20,23
Mixed co-cultures of rat retinal cells were prepared by seeding RGCs on the monolayer of macroglial cells as previously described.21,27,28 During the experimental period, cultured cells were incubated in a defined serum-free medium in the absence or presence of oxidative stress. Oxidative stress was generated by treating the co-cultures with H2O2 (50 μM) for 24 hours, while control cultures prepared using an identical passage of cells were simultaneously incubated in the absence of H2O2. This treatment resulted in an approximately 70% decrease in cell survival relative to control cultures.21 Proteins were isolated from the harvested cells and subjected to immunoblotting with serum samples. All animals utilized in in vitro experiments were handled according to the regulations of the Institutional Animal Care and Use Committee. All animals were treated in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research.
Immunoblotting
In addition to retinal proteins, immunoblotting also tested the reactivity of serum samples and isolated IgG elutes to recombinant proteins. The recombinant proteins included AIF (Novoprotein, Shanghai, China), CREB-binding protein (Cayman Chemical, Ann Arbor, MI), ephrin type-A2 (Novus Biologicals, Littleton, CO), and huntingtin (Novus Biologicals). The proteins separated by SDS-PAGE were electrophoretically transferred to a nitrocellulose membrane (Bio-Rad, Hercules, CA). Following a blocking step, membranes were probed with patient sera (1:500) or IgG elutes (15 μg), and the secondary antibody incubation used antihuman IgG conjugated with horse-radish peroxidase (1:2000; Sigma-Aldrich). Immunoreactive bands were visualized by enhanced chemiluminescence using commercial reagents (GE Healthcare, Pittsburgh, PA). For densitometry of immunoblots, images were acquired using a scanning device with a linear dynamic range (Typhoon 9400; GE Healthcare). Image analysis used a specific software (ImageQuant; GE Healthcare), background correction was applied to each lane by baseline subtraction, and integral values were selected as measurement parameters.
Results
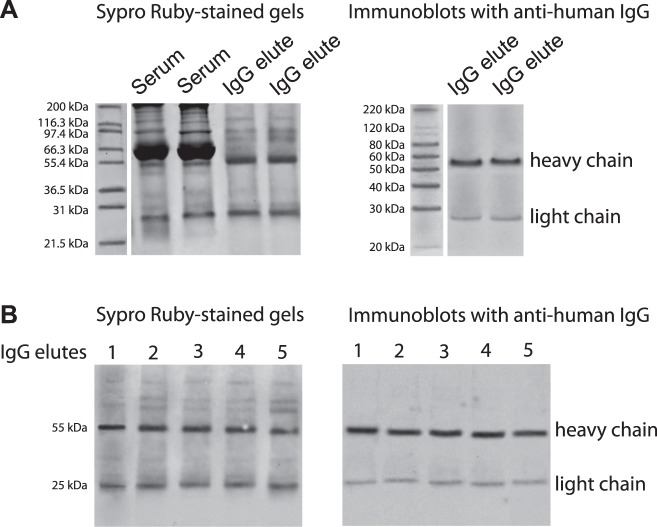
Prior to their mass spectrometric analysis, we validated isolated serum IgG elutes by SDS-PAGE and immunoblotting (Fig. 1). Based on SDS-PAGE, eluted factions exhibited a prominent removal of most serum proteins except for the remaining few bands. Western blot analysis using antihuman IgG antibody verified IgG heavy and light chains in these elutes. The other bands detectable by the SDS-PAGE of isolated IgG elutes were thought to correspond to serum proteins co-eluted with IgG. When the serum IgG elutes were subjected to LC-MS/MS analysis, we identified a list of proteins co-eluted with IgG (Table). Our additional data (presented later in Fig. 4) indicated that the glaucomatous serum samples and isolated IgG elutes were indeed reactive to co-eluted proteins. These findings collectively support that the serum proteins co-eluted with IgG and subsequently identified by LC-MS/MS are most likely to reflect antigenic targets of IgG, not artifacts of incomplete purification. Since two alternative methods for serum IgG isolation using Melon gel spin columns or protein G-Dynabeads indicated consistent results, the following data are those from spin column purification.
Figure 1. .
Serum IgG-containing protein mixtures. To identify antigenic targets of serum antibodies for high-throughput characterization, serum proteins co-eluted with IgG were analyzed by LC-MS/MS. (A) SDS-PAGE was performed to verify serum IgG elutes prior to further analysis. Shown Sypro Ruby-stained gels include samples of the elutes whole serum and the IgG elutes isolated using two alternative methods: Melon gel spin column purification and protein G-Dynabeads-based purification, respectively. Immunoblots of these elutes with anti-human IgG antibody detected IgG heavy (approximately 55 kDa) and light (approximately 25 kDa) chains. Since two alternative methods for serum IgG isolation using Melon gel spin columns or protein G-Dynabeads indicated consistent results, the following analysis used the samples eluted by spin column purification. (B) Sypro Ruby-stained gels of IgG elutes and corresponding immunoblots with anti-human IgG antibody represent randomly selected five glaucomatous sera that were subjected to following LC-MS/MS analysis.
Table. .
Serum Proteins Co-Eluted with IgG
|
Protein Name |
Accession Number |
| Integrin alpha-9 precursor | gi|52485941 |
| Disintegrin and metalloproteinase domain-containing protein 9 precursor* | gi|4501915 |
| Neural cell adhesion molecule 2 precursor | gi|33519481 |
| SH3 and multiple ankyrin repeat domains protein 3* | gi|122937241 |
| Extended synaptotagmin-1 isoform 2* | gi|14149680 |
| SLIT and NTRK-like protein 3 precursor | gi|40217820 |
| Microtubule-actin cross-linking factor 1 isoform b | gi|33188443 |
| Calmodulin-regulated spectrin-associated protein 2 | gi|44955929 |
| Vesicle-associated membrane protein 7 isoform 3* | gi|297747292 |
| Synphilin-1 | gi|76563940 |
| Huntingtin | gi|90903231 |
| Cytoplasmic dynein 1 heavy chain 1 | gi|33350932 |
| Dynein heavy chain 2, axonemal | gi|75677365 |
| Dynein heavy chain 5, axonemal | gi|19115954 |
| Dynein heavy chain 6, axonemal | gi|194353966 |
| Dynein heavy chain 7, axonemal | gi|151301127 |
| Dynein heavy chain 10, axonemal | gi|198442844 |
| Ephrin type-A receptor 5 isoform a precursor | gi|221625401 |
| Ephrin type-A receptor 8 isoform 1 precursor | gi|10140845 |
| Ephrin type-A receptor 10 isofom 3 | gi|150456460 |
| V-type proton ATPase subunit B, brain isoform | gi|19913428 |
| Kv channel-interacting protein 2 isoform 2 | gi|27886670 |
| Tax1-binding protein 1 isoform b* | gi|119943086 |
| Serine/threonine-protein phosphatase 2A regulatory subunit A alpha isoform | gi|21361399 |
| Adenylate cyclase type 8 | gi|4557257 |
| rho guanine nucleotide exchange factor 11 isoform 2 | gi|38026934 |
| Activated CDC42 kinase 1 isoform 1 | gi|56549666 |
| Pleckstrin homology domain-containing family A member 4 isoform 1* | gi|238859651 |
| Nuclear factor related to kappa-B-binding protein isoform 1 | gi|219802034 |
| Paired box protein Pax-2 isoform a* | gi|34878699 |
| Zinc finger protein 496 | gi|14249386 |
| Zinc finger protein 561* | gi|206725458 |
| Zinc finger protein 611 isoform a | gi|239787080 |
| CREB-binding protein isoform b | gi|119943102 |
| Protein argonaute-2 isoform 2* | gi|257467482 |
| Histone-lysine N-methyltransferase NSD3 isoform long* | gi|13699811 |
| E3 ubiquitin-protein ligase PDZRN3* | gi|57529737 |
| E3 ubiquitin-protein ligase RBBP6 isoform 2 | gi|33620716 |
| 26S proteasome non-ATPase regulatory subunit 1 isoform 1 | gi|25777600 |
| Apoptosis-inducing factor 1, mitochondrial isoform 4* | gi|195927004 |
| Baculoviral IAP repeat-containing protein 3 | gi|33946285 |
| Baculoviral IAP repeat-containing protein 6 | gi|153792694 |
| Protein S100-A8 | gi|21614544 |
| Annexin A1 | gi|4502101 |
| NACHT, LRR and PYD domains-containing protein 2 isoform 1* | gi|291463275 |
| Complement C3 precursor | gi|115298678 |
| Complement C5 preproprotein | gi|38016947 |
| Complement component C8 alpha chain precursor | gi|4557389 |
| Complement component C8 beta chain preproprotein | gi|4557391 |
| Complement factor H-related protein 1 precursor* | gi|118442839 |
To identify co-eluted proteins, the serum IgG elutes isolated from five randomly selected glaucomatous and nonglaucomatous sera were individually analyzed by quantitative LC-MS/MS. Listed are 50 serum proteins detected in the glaucomatous serum IgG elutes and also known to be present in the glaucomatous human retina. All listed proteins were identified with high confidence (< 99.0% probability assigned by the Protein Prophet algorithm) based on at least two identified peptides.
Methionine oxidation increased or detectable only in glaucomatous samples.
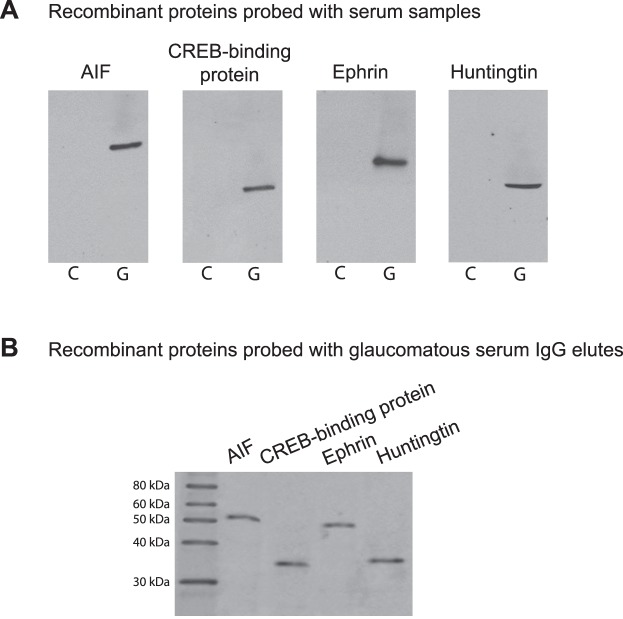
Figure 4. .
Serum immunoreactivity to candidate biomarkers. (A) To test serum immunoreactivity by Western blot analysis, recombinant proteins were probed with glaucomatous (G) and nonglaucomatous control (C) sera (1:500), IgG elutes of which were analyzed by LC-MS/MS. Western blots indicated prominent immunoreactivity of the glaucomatous serum to studied proteins, including AIF (approximately 57 kDa), CREB-binding protein (approximately 37 kDa), ephrin (approximately 46 kDa), and huntingtin (approximately 38 kDa). (B) When similarly tested against recombinant proteins, isolated IgG elutes of the glaucomatous sera (15 μg) also exhibited reactivity to these proteins. Presented are the most representative immunoreactivities. The secondary antibody incubation used anti-human IgG conjugated with horse-radish peroxidase (1:2000).
Our LC-MS/MS analysis of IgG-containing protein mixtures identified 231 proteins by two peptides or more at the 0.4% peptide and 0.1% protein false discovery rates. A complex panel of proteins co-eluted with serum IgG included those with potential relevance to glaucoma as well as seemingly irrelevant proteins localized elsewhere. Among these proteins, 106 exhibited higher amounts in samples isolated from glaucomatous sera compared to those obtained from nonglaucomatous controls, while 63 proteins were detected only in glaucomatous samples. Unlike proteins bound by normal serum IgG, proteins bound to glaucomatous serum IgG included a diverse group known to be expressed by neurons. To attempt to reduce the list of candidate autoantigens based on their potential relevance to glaucoma, we determined whether the MS/MS data obtained from the glaucomatous serum IgG elutes overlapped with our high-throughput proteomic data obtained from the glaucomatous human retina.20–23 Ninety-six proteins overlapped between the serum and retinal proteomic data. Figure 2 summarizes the quantities of overlapping proteins. The Table lists 50 newly identified antigens in glaucomatous serum IgG elutes, which are also known to be present in the glaucomatous human retina.
Figure 2. .
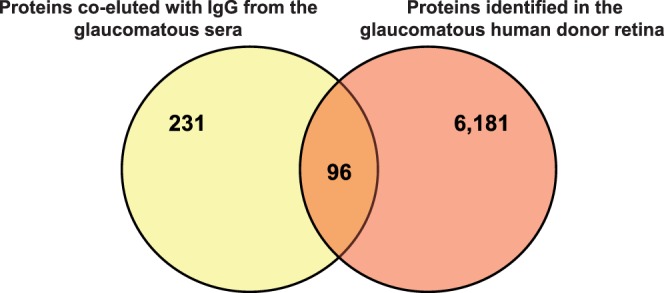
Candidate biomarkers of glaucoma. As a narrowing-down strategy, we determined whether the MS/MS data obtained from the glaucomatous serum IgG elutes overlapped with our previous high-throughput proteomic data obtained from the glaucomatous human retina. Ninety-six proteins overlapped between the serum and retinal proteomic data.
When we compared methionine oxidation of the identified serum proteins, we found greater abundances in samples isolated from the glaucomatous sera compared with age-matched controls. Methionine oxidation counts were two or more in 23% and 11% of proteins identified in glaucomatous and control samples, respectively, while for 62 proteins (27%), methionine oxidation was detectable only in glaucomatous samples. The Table also indicates the oxidation status of listed proteins.
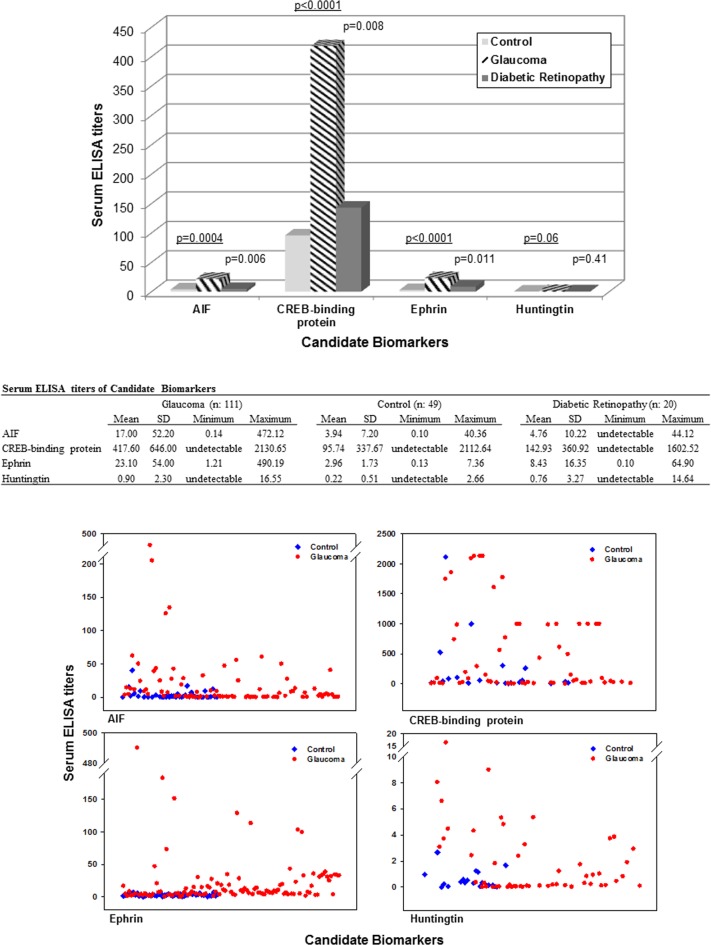
We selected 10 of the listed serum proteins as candidate biomarkers for initial validation studies and measured their serum titers in all of our glaucomatous and control samples by specific ELISAs. In addition, to test the specificity of these biomarker candidates for glaucoma, we have also included an additional group of 20 patients with diabetic retinopathy. Proteins selected based on their known relevance to glaucoma included AIF, annexin A1, IAP 6, CREB-binding protein, dynein heavy chain 1, ephrin type-A, huntingtin protein, neural cell adhesion molecule, NLRP2, and NF-κB-binding protein. As shown in Figure 3, three of these candidate biomarkers, including AIF, CREB-binding protein, and ephrin type-A receptor, exhibited significantly higher ELISA titers in the glaucomatous sera compared to serum samples from healthy controls or patients with diabetic retinopathy (Mann-Whitney rank sum test, P < 0.01). In addition, many samples from patients with glaucoma (and some samples from patients with diabetic retinopathy) exhibited higher ELISA titers for huntingtin protein than healthy controls, but the overall difference was not statistically significant (Mann-Whitney rank sum test, P > 0.05). ELISA titers of other selected candidates, including annexin A1, IAP 6, dynein heavy chain 1, neural cell adhesion molecule, NLRP2, and NF-κB-binding protein, were not significantly different between the glaucomatous and control sera studied (Mann-Whitney rank sum test, P > 0.05).
Figure 3. .
Serum ELISA titers of candidate biomarkers. Serum levels of 10 candidate biomarkers were measured in glaucomatous and nonglaucomatous samples by specific ELISAs. In addition to healthy controls, nonglaucomatous samples also included those collected from an additional group of patients with diabetic retinopathy. As shown in the bar graph of mean values and the Table summarizing the serum ELISA data, glaucomatous samples exhibited higher ELISA titers (ng/mL) for four biomarker candidates. The P values underlined indicate the statistical difference between glaucomatous samples versus samples from healthy controls; other P values indicate the statistical difference between glaucomatous samples versus samples from patients with diabetic retinopathy (Mann-Whitney rank sum test). The scatter graphs of serum ELISA titers in glaucomatous and control samples present individual variability.
After identification of biomarker candidates, we aimed to confirm whether these proteins identified by LC-MS/MS analysis of serum IgG elutes were antigenic targets of IgG or contaminant of purification. To determine whether glaucomatous serum was immunoreactive to proteins identified in serum IgG elutes, we probed recombinant proteins with glaucomatous and nonglaucomatous sera. As shown in Figure 4, glaucomatous sera exhibited prominent immunoreactivity to AIF, CREB-binding protein, ephrin type-A, and huntingtin. As also presented in Figure 4, isolated IgG elutes were similarly reactive to these recombinant proteins. These observations further support that the serum proteins co-eluted with IgG and identified by LC-MS/MS were antigenic targets of IgG in the glaucomatous sera.
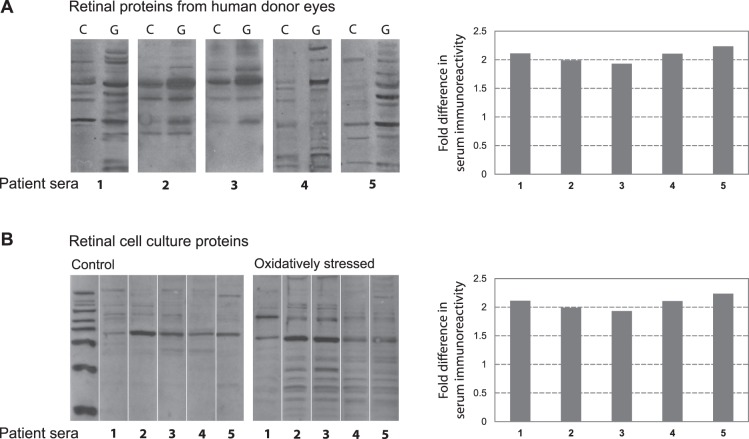
The prominent overlap detected between serum IgG-bound proteins in the glaucomatous sera and upregulated proteins in the glaucomatous human retina, along with a relative increase in oxidative modification of proteins in glaucomatous samples, motivated us to further determine whether glaucoma-related factors affect serum immunoreactivity to retinal proteins. Therefore, we tested the immunoreactivity of patient sera against retinal proteins isolated from glaucomatous versus nonglaucomatous human retinas (Fig. 5). When tested by immunoblotting, glaucomatous serum samples exhibited a complex pattern of immunoreactivity to human retinal proteins. However, serum immunoreactivity to retinal proteins obtained from glaucomatous human donors was greater than the immunoreactivity to human retinal proteins obtained from nonglaucomatous donors. In order to test whether oxidative stress affects serum immunoreactivity, we also conducted in vitro experiments with primary cultures of rat retinal cells incubated in the presence or absence of H2O2-induced oxidative stress. Findings of these experiments provided additional evidence that the immunoreactivity of glaucomatous sera against oxidatively stressed retinal cell culture proteins was more than their immunoreactivity to control proteins. Band intensities of serum IgG immunoreactivity to glaucomatous human retinal proteins, or oxidatively stressed cell culture proteins, were over 2-fold greater than the band intensities detected with retinal proteins obtained from nonglaucomatous control donors or control cell cultures.
Figure 5. .
Serum immunoreactivity to retinal proteins. To test serum immunoreactivity by Western blot analysis, retinal proteins were separated by SDS-PAGE, and membranes were cut into strips to probe with different serum samples. (A) Glaucomatous serum samples (1–5) exhibited a greater immunoreactivity to retinal proteins obtained from glaucomatous human donors (G) relative to retinal proteins obtained from non-glaucomatous controls (C). (B) We also tested serum immunoreactivity to retinal protein samples obtained from cell cultures. Immunoreactivity of the glaucomatous patient sera (1–5) against oxidatively stressed retinal cell culture proteins was greater than their immunoreactivity to controls. Bar graphs show fold increase in serum immunoreactivity to glaucomatous human retinal proteins (relative to nonglaucomatous retinal proteins), or fold increase in serum immunoreactivity to oxidatively stressed retinal proteins (relative to control proteins) as calculated based on band intensities. Serum dilution was 1:500, and the secondary antibody incubation used anti-human IgG conjugated with horse-radish peroxidase (1:2000).
Discussion
This immunoproteomic study delineated a variety of new antigens targeted by IgG in the glaucomatous sera. The serum proteins co-eluted with IgG closely mirrored the up-regulated proteins previously identified in the glaucomatous retina. In addition, we took advantage of our MS/MS data to determine the oxidation status of the identified serum proteins.29–31 In particular, we determined methionine oxidation, because methionine residues are an important target for protein oxidation by almost all forms of reactive oxygen species32,33 and have been useful for examining the extent of oxidative stress.34 We detected greater abundance of methionine oxidation in serum proteins isolated from the glaucomatous sera compared with age-matched healthy controls. We then initiated validation studies for selected 10 candidate molecules, four of which, including AIF, CREB-binding protein, ephrin type-A receptor, and huntingtin protein, exhibited higher serum ELISA titers in our glaucomatous patient cohort.
Complementary experiments aimed to determine whether glaucoma-related protein modifications affect serum immunoreactivity. We tested the immunoreactivity of patient sera against human retinal proteins isolated from glaucomatous versus nonglaucomatous human donors and against rat retinal proteins obtained from cell cultures incubated in the absence or presence of oxidative stress. We detected greater immunoreactivity of the patient sera to glaucomatous retinal proteins or oxidatively stressed cell culture proteins.
The importance of our findings is 2-fold. First, the identified serum proteins may serve as candidate biomarkers to be validated for future clinical applications. Second, glaucoma-related protein modifications, such as increased expression or oxidation, may be immunostimulatory.
Candidate Serum Biomarkers of Glaucoma
Protein biomarkers are under intensive investigation to facilitate early disease detection, prognosis prediction, and follow-up of treatment responses. A number of techniques with their own advantages and drawbacks, including serologic identification of antigens by recombinant expression cloning, serologic proteome analysis, protein microarrays, and open reading frame phage display, have been used to identify disease-associated antigens and their cognate autoantibodies in cancer, autoimmune diseases,35,36 and uveitis.37 In glaucoma, antibody-based approaches have included immunoblotting-based analysis or protein arrays,3–5 and direct analysis of potential proteomic biomarkers has only been applied to profile protein patterns in aqueous humor or tear samples.38,39 An important factor interfering with serum biomarker detection is the much lower abundance of most proteomic biomarkers than some disease-irrelevant serum proteins, such as albumin or transferrin. Isolation of serum IgG-containing fractions for mass spectrometric analysis should have minimized this limitation in our study.
Although IgG isolation methods we employed are commonly and effectively utilized, they may have technical limitations if not used properly. These methods may also have limitations in complete purification due to co-elution of IgG-bound antigenic proteins. Although co-eluted proteins may limit IgG purification studies, it was an advantage for this study that particularly aimed to identify antigenic targets of serum IgG. Furthermore, our SDS-PAGE and immunoblotting data presented in Figure 1, along with the immunoreactivity data presented in Figure 4, validated the efficiency of IgG isolation and supported that the serum proteins co-eluted with IgG reflect antigenic targets of IgG, not artifacts of incomplete purification. However, it is also worth noting that even if some of the biomarker candidates were serum proteins contaminating the IgG purification, they would still serve for the primary purpose of this study for biomarker discovery, and any possibility of imperfect purification would not diminish the value of the presented data.
Similar to our study, the commonly used approach to identify protein biomarkers is to initially select biomarker candidates and then pursue validation studies in larger patient populations to verify their value. Immunoassays, multiplex assays,40,41 and mass spectrometry-based multiple reaction monitoring assays42 provide attractive options to quantitate candidate molecules in large numbers of samples. We initiated a similar approach to test selected biomarker candidates by specific ELISAs or serum samples.
Many of the selected candidates, including AIF,23,43 IAP,44 dynein heavy chain,45 ephrin receptor,46 neural cell adhesion molecule,47 NLRP,23 and NF-κB,23,48 have been associated with glaucoma. Another selected candidate was annexin A1, which is a calcium-binding protein with anti-inflammatory properties.49,50 We have previously detected over 2-fold increased abundance of this protein (RefSeq: NP_000691) in the glaucomatous human retinal proteome. Serum antibodies to annexin A1 have also been suggested to be a diagnostic marker for inflammatory diseases.51 Two other candidate biomarkers, CREB-binding protein and huntingtin protein, were similarly selected based on their consistent detection in the retinal proteome of glaucomatous human donor eyes (RefSeq: NP_004371 and NP_002102, respectively). Among the selected 10 candidates, four proteins, including AIF, CREB-binding protein, ephrin type-A receptor, and huntingtin protein, exhibited higher serum titers in our glaucomatous samples relative to controls, although higher ELISA titers for huntingtin protein were not found statistically significant.
The AIF involved in the mitochondrial apoptosis pathway has been linked to RGC death in in vitro43,52 and in vivo models53 of glaucoma and found to be significantly up-regulated in the glaucomatous human retina.23
The CREB-binding protein was first isolated as a nuclear protein binding to CREB that has been linked to astrocyte responses to elevated pressure,54 while cAMP signaling is critical for RGC survival.55 The CREB-binding protein with intrinsic histone acetyltransferase activity is now known to be involved in the transcriptional coactivation of many different transcription factors. A recent immunohistochemical localization study has demonstrated that this protein is expressed in retinal neurons and glial cells,56 and its reduced phosphorylation has been linked to retinal degeneration in an animal model of excitotoxicity.57
Ephrin receptors expressed in the retina, mainly including RGCs, regulate developmental axon guidance and synapse formation and determine the outcome of injury in the adult brain.58,59 Our previous studies have detected different subtypes of ephrin type-A receptor in the human retinal proteome (RefSeq: NP_001396, NP_005224, NP_004429, NP-004431, NP_065387). Based on proteomic analysis of serum samples, ephrin receptor A2 has recently been indicated as a candidate biomarker for acute retinal injury in a primate model.60
Regarding the huntingtin protein expressed in neurons, a mutant form is implicated in Huntington's disease. Failure in clearance of the proteolytic fragments of huntingtin (which is mainly through autophagy) eventually results in altered intracellular Ca(2+) homeostasis, organelle dysfunction, disrupted intracellular trafficking, and impaired gene transcription.61,62 Interestingly, animal models of Hungtington's disease exhibit progressive retinal degeneration and dysfunction in which mutant huntingtin aggregates in the nucleus of all neurons, including RGCs.63 Although we detected huntingtin and a number of associated/interacting proteins (including optineurin)12 in the retinal proteome of glaucomatous human donor eyes, any connection of this protein to pathogenic mechanisms of glaucomatous neurodegeneration remains unknown. In this study, many glaucomatous serum samples as well as some samples from patients with diabetic retinopathy exhibited higher ELISA titers for huntingtin protein; however, the overall difference between these and control samples was not statistically significant. These findings suggest that aging-related or other mechanisms leading to dysfunction of the ubiquitin-proteasome and autophagy-lysosome pathways may provide links to similar mechanisms of neuropathology in Huntington's disease and other neuropathies, such as glaucoma or diabetic retinopathy.
The list of biomarker candidates obtained through an antibody-based approach in this study demands further studies for validation. It should be recognized that the assessment of the clinical value of identified molecules is a major standing challenge common for biomarker discovery through different approaches. Although the discovery phase normally takes place using samples from a small well-characterized cohort, when proceeding to the validation phase, larger heterogeneous cohorts are needed. This is critical to eliminate false positivity and calculate the sensitivity and specificity of candidate molecules for clinical prediction. In the case of glaucoma, validation studies of biomarker candidates should include patients with different types and stages of glaucoma with different demographics and comorbidities, and also nonglaucomatous patients with other ocular and neurodegenerative diseases and disease-free controls. Particularly respecting the individual variability in serum ELISA titers, once their clinical correlations are established, a set of biomarkers in an array format, rather than a single molecule, may potentially be more useful as a diagnostic and/or prognostic clinical tool in glaucoma.
Glaucoma-Related Protein Modifications and Increased Serum Immunoreactivity
We detected a greater immunoreactivity of the patient serum to glaucomatous human retinal proteins relative to retinal proteins obtained from nonglaucomatous control donors. This observation suggests that glaucomatous alterations in protein expression affect the antigenic features of proteins, increase the exposure or presentation of retinal antigens to the immune system, and stimulate immune response by autoantibody production. Previous evidence consistent with the increased recognition of ocular antigens by immune system cells in human glaucoma includes the activation of glial immunoregulatory functions and antigen presenting abilities through increased expression and activation of various immune mediators/regulators, such as MHC class II molecules,64 toll-like receptors,22 pro-inflammatory cytokines,12,54 NF-κB,12 and inflammasome components.12 Oxidative stress in glaucoma21,65 appears to constitute an important component of the immunostimulatory signals arising from glaucomatous tissues.66 As also supported by present findings, oxidative protein modifications67 may particularly be important to increase the antigenicity of retinal proteins during glaucomatous neurodegeneration. It is not unexpected that the immune system reacts to post-translationally modified epitopes in glaucoma. However, this aspect, consistent with the self-tolerance of the immune system, encourages further research to better determine whether modified serum proteins, as opposed to unmodified native proteins, are particularly useful as disease-specific biomarkers of glaucoma.
Conclusions
Despite increasing interest in the immune system involvement in glaucoma6 and ongoing efforts to validate its causative importance,9,10,15 immunogenic mechanisms of glaucomatous neurodegeneration and the pathogenic importance of serum antibodies remain unclear. It is also unclear whether the IgG-bound serum proteins identified in this study reflect disease-causing antigens. However, findings of this study support that analysis of serum antibodies may lead us to biomarker discovery for future clinical applications. Our initial findings delineating a list of potential biomarker candidates await further validation studies to determine the value of these molecules as a clinical tool to determine diagnosis, prognosis, and/or treatment responses in glaucoma.
Acknowledgments
The authors thank Tongalp H. Tezel, University of Louisville, for kindly providing the serum samples collected from patients with diabetic retinopathy.
Footnotes
Supported by Grants from the National Eye Institute, Bethesda, Maryland (R01 EY013813 and R01 EY017131); from Research to Prevent Blindness, Inc., New York, New York; and the John and Dorothy Roberts Research Fund of the New York Glaucoma Research Institute, New York, New York.
Disclosure: G. Tezel, None; I.L. Thornton, None; M.G. Tong, None; C. Luo, None; X. Yang, None; J. Cai, None; D.W. Powell, None; J.B. Soltau, None; J.M. Liebmann, None; R. Ritch, None
References
- 1.Tezel G. Immune regulation towards immunomodulation for neuroprotection in glaucoma. Curr Opin Pharmacol. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama I, Ohguro H, Ikeda Y. Retinal ganglion cells recognized by serum autoantibody against gamma-enolase found in glaucoma patients. Invest Ophthalmol Vis Sci. 2000;41:1657–1665 [PubMed] [Google Scholar]
- 3.Tezel G, Wax MB. Glaucoma. Chem Immunol Allergy. 2007;92:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reichelt J, Joachim SC, Pfeiffer N, Grus FH. Analysis of autoantibodies against human retinal antigens in sera of patients with glaucoma and ocular hypertension. Curr Eye Res. 2008;33:253–261 [DOI] [PubMed] [Google Scholar]
- 5.Dervan EW, Chen H, Ho SL, et al. Protein macroarray profiling of serum autoantibodies in pseudoexfoliation glaucoma. Invest Ophthalmol Vis Sci. 2010;51:2968–2975 [DOI] [PubMed] [Google Scholar]
- 6.Tezel G. The role of glia, mitochondria, and the immune system in glaucoma. Invest Ophthalmol Vis Sci. 2009;50:1001–1012 [DOI] [PubMed] [Google Scholar]
- 7.Joachim SC, Grus FH, Kraft D, et al. Complex antibody profile changes in an experimental autoimmune glaucoma animal model. Invest Ophthalmol Vis Sci. 2009;50:4734–4742 [DOI] [PubMed] [Google Scholar]
- 8.Wax MB, Tezel G, Edward PD. Clinical and ocular histopathological findings in a patient with normal-pressure glaucoma. Arch Ophthalmol. 1998;116:993–1001 [DOI] [PubMed] [Google Scholar]
- 9.Tezel G, Wax MB. The mechanisms of hsp27 antibody-mediated apoptosis in retinal neuronal cells. J Neurosci. 2000;20:3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laspas P, Gramlich OW, Muller HD, et al. Autoreactive antibodies and loss of retinal ganglion cells in rats induced by immunization with ocular antigens. Invest Ophthalmol Vis Sci. 2011;52:8835–8848 [DOI] [PubMed] [Google Scholar]
- 11.Joachim SC, Gramlich OW, Laspas P, et al. Retinal ganglion cell loss is accompanied by antibody depositions and increased levels of microglia after immunization with retinal antigens. PLoS One. 2012;7:e40616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding QJ, Cook AC, Dumitrescu AV, Kuehn MH. Lack of immunoglobulins does not prevent C1q binding to RGC and does not alter the progression of experimental glaucoma. Invest Ophthalmol Vis Sci. 2012;53:6370–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coutinho A, Kazatchkine MD, Avrameas S. Natural autoantibodies. Curr Opin Immunol. 1995;7:812–818 [DOI] [PubMed] [Google Scholar]
- 14.Shoenfeld Y, Toubi E. Protective autoantibodies: role in homeostasis, clinical importance, and therapeutic potential. Arthritis Rheum. 2005;52:2599–2606 [DOI] [PubMed] [Google Scholar]
- 15.Wax MB, Tezel G, Yang J, et al. Induced autoimmunity to heat shock proteins elicits glaucomatous loss of retinal ganglion cell neurons via activated T-cell-derived fas-ligand. J Neurosci. 2008;28:12085–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joachim SC, Wax MB, Seidel P, Pfeiffer N, Grus FH. Enhanced characterization of serum autoantibody reactivity following HSP 60 immunization in a rat model of experimental autoimmune glaucoma. Curr Eye Res. 2010;35:900–908 [DOI] [PubMed] [Google Scholar]
- 17.Boehm N, Wolters D, Thiel U, et al. New insights into autoantibody profiles from immune privileged sites in the eye: a glaucoma study. Brain Behav Immun. 2011;26:96–102 [DOI] [PubMed] [Google Scholar]
- 18.Wax MB, Tezel G, Kawase K, Kitazawa Y. Serum autoantibodies to heat shock proteins in glaucoma patients from Japan and the United States. Ophthalmology. 2001;108:296–302 [DOI] [PubMed] [Google Scholar]
- 19.Grus FH, Joachim SC, Bruns K, Lackner KJ, Pfeiffer N, Wax MB. Serum autoantibodies to alpha-fodrin are present in glaucoma patients from Germany and the United States. Invest Ophthalmol Vis Sci. 2006;47:968–976 [DOI] [PubMed] [Google Scholar]
- 20.Tezel G, Yang X, Luo C, et al. Hemoglobin expression and regulation in glaucoma: insights into retinal ganglion cell oxygenation. Invest Ophthalmol Vis Sci. 2010;51:907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tezel G, Yang X, Luo C, et al. Oxidative stress and the regulation of complement activation in human glaucoma. Invest Ophthalmol Vis Sci. 2010;51:5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo C, Yang X, Kain AD, Powell DW, Kuehn MH, Tezel G. Glaucomatous tissue stress and the regulation of immune response through glial toll-like receptor signaling. Invest Ophthalmol Vis Sci. 2010;51:5697–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Luo C, Cai J, et al. Neurodegenerative and inflammatory pathway components linked to TNF-alpha/TNFR1 signaling in the glaucomatous human retina. Invest Ophthalmol Vis Sci. 2011;52:8442–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392 [DOI] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658 [DOI] [PubMed] [Google Scholar]
- 26.Powell DW, Weaver CM, Jennings JL, et al. Cluster analysis of mass spectrometry data reveals a novel component of SAGA. Mol Cell Biol. 2004;24:7249–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tezel G, Wax MB. Increased production of tumor necrosis factor-alpha by glial cells exposed to simulated ischemia or elevated hydrostatic pressure induces apoptosis in cocultured retinal ganglion cells. J Neurosci. 2000;20:8693–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tezel G, Yang X, Luo C, Peng Y, Sun SL, Sun D. Mechanisms of immune system activation in glaucoma: oxidative stress-stimulated antigen presentation by the retina and optic nerve head glia. Invest Ophthalmol Vis Sci. 2007;48:705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies MJ, Fu S, Wang H, Dean RT. Stable markers of oxidant damage to proteins and their application in the study of human disease. Free Radic Biol Med. 1999;27:1151–1163 [DOI] [PubMed] [Google Scholar]
- 30.Houde D, Kauppinen P, Mhatre R, Lyubarskaya Y. Determination of protein oxidation by mass spectrometry and method transfer to quality control. J Chromatogr A. 2006;1123:189–198 [DOI] [PubMed] [Google Scholar]
- 31.Fedorova M, Kuleva N, Hoffmann R. Identification of cysteine, methionine and tryptophan residues of actin oxidized in vivo during oxidative stress. J Proteome Res. 2010;9:1598–1609 [DOI] [PubMed] [Google Scholar]
- 32.Stadtman ER, Moskovitz J, Levine RL. Oxidation of methionine residues of proteins: biological consequences. Antioxid Redox Signal. 2003;5:577–582 [DOI] [PubMed] [Google Scholar]
- 33.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703:93–109 [DOI] [PubMed] [Google Scholar]
- 34.Long LH, Wu PF, Guan XL, et al. Determination of protein-bound methionine oxidation in the hippocampus of adult and old rats by LC-ESI-ITMS method after microwave-assisted proteolysis. Anal Bioanal Chem. 2011;399:2267–2274 [DOI] [PubMed] [Google Scholar]
- 35.Desmetz C, Maudelonde T, Mange A, Solassol J. Identifying autoantibody signatures in cancer: a promising challenge. Expert Rev Proteomics. 2009;6:377–386 [DOI] [PubMed] [Google Scholar]
- 36.Georgieva Y, Konthur Z. Design and screening of M13 phage display cDNA libraries. Molecules. 2011;16:1667–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y, Caberoy NB, Alvarado G, Davis JL, Feuer WJ, Li W. Identification of Hnrph3 as an autoantigen for acute anterior uveitis. Clin Immunol. 2011;138:60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan X, Xue P, Wang N, Dong Z, Lu Q, Yang F. Proteomic analysis of aqueous humor from patients with primary open angle glaucoma. Mol Vis. 2010;16:2839–2846 [PMC free article] [PubMed] [Google Scholar]
- 39.Bouhenni RA, Al Shahwan S, Morales J, et al. Identification of differentially expressed proteins in the aqueous humor of primary congenital glaucoma. Exp Eye Res. 2011;92:67–75 [DOI] [PubMed] [Google Scholar]
- 40.Matt P, Fu Z, Fu Q, Van Eyk JE. Biomarker discovery: proteome fractionation and separation in biological samples. Physiol Genomics. 2008;33:12–17 [DOI] [PubMed] [Google Scholar]
- 41.Hanash SM, Pitteri SJ, Faca VM. Mining the plasma proteome for cancer biomarkers. Nature. 2008;452:571–579 [DOI] [PubMed] [Google Scholar]
- 42.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588 [DOI] [PubMed] [Google Scholar]
- 43.Tezel G, Yang X. Caspase-independent component of retinal ganglion cell death, in vitro. Invest Ophthalmol Vis Sci. 2004;45:4049–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKinnon SJ, Lehman DM, Tahzib NG, et al. Baculoviral IAP repeat-containing-4 protects optic nerve axons in a rat glaucoma model. Mol Ther. 2002;5:780–787 [DOI] [PubMed] [Google Scholar]
- 45.Martin KR, Quigley HA, Valenta D, Kielczewski J, Pease ME. Optic nerve dynein motor protein distribution changes with intraocular pressure elevation in a rat model of glaucoma. Exp Eye Res. 2006;83:255–262 [DOI] [PubMed] [Google Scholar]
- 46.Schmidt JF, Agapova OA, Yang P, Kaufman PL, Hernandez MR. Expression of ephrinB1 and its receptor in glaucomatous optic neuropathy. Br J Ophthalmol. 2007;91:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ricard CS, Pena JD, Hernandez MR. Differential expression of neural cell adhesion molecule isoforms in normal and glaucomatous human optic nerve heads. Brain Res Mol Brain Res. 1999;74:69–82 [DOI] [PubMed] [Google Scholar]
- 48.Agapova OA, Kaufman PL, Hernandez MR. Androgen receptor and NFkB expression in human normal and glaucomatous optic nerve head astrocytes in vitro and in experimental glaucoma. Exp Eye Res. 2006;82:1053–1059 [DOI] [PubMed] [Google Scholar]
- 49.Pupjalis D, Goetsch J, Kottas DJ, Gerke V, Rescher U. Annexin A1 released from apoptotic cells acts through formyl peptide receptors to dampen inflammatory monocyte activation via JAK/STAT/SOCS signalling. EMBO Mol Med. 2011;3:102–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Z, Huang L, Zhao W, Rigas B. Annexin 1 induced by anti-inflammatory drugs binds to NF-kappaB and inhibits its activation: anticancer effects in vitro and in vivo. Cancer Res. 2010;70:2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kretz CC, Norpo M, Abeler-Dorner L, et al. Anti-annexin 1 antibodies: a new diagnostic marker in the serum of patients with discoid lupus erythematosus. Exp Dermatol. 2010;19:919–921 [DOI] [PubMed] [Google Scholar]
- 52.Li GY, Osborne NN. Oxidative-induced apoptosis to an immortalized ganglion cell line is caspase independent but involves the activation of poly(ADP-ribose)polymerase and apoptosis-inducing factor. Brain Res. 2008;1188:35–43 [DOI] [PubMed] [Google Scholar]
- 53.Munemasa Y, Kitaoka Y, Kuribayashi J, Ueno S. Modulation of mitochondria in the axon and soma of retinal ganglion cells in a rat glaucoma model. J Neurochem. 2010;115:1508–1519 [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Lukas TJ, Hernandez MR. Hydrostatic pressure-dependent changes in cyclic AMP signaling in optic nerve head astrocytes from Caucasian and African American donors. Mol Vis. 2009;15:1664–1672 [PMC free article] [PubMed] [Google Scholar]
- 55.Meyer-Franke A, Kaplan MR, Pfrieger FW, Barres BA. Characterization of the signaling interactions that promote the survival and growth of developing retinal ganglion cells in culture. Neuron. 1995;15:805–819 [DOI] [PubMed] [Google Scholar]
- 56.Oh H, Kim H, Ahn M, et al. Immunohistochemical localization of cyclic AMP-responsive element binding protein (CREB)-binding protein in the pig retina during postnatal development. Anat Cell Biol. 2011;44:143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mali RS, Zhang XM, Chintala SK. A decrease in phosphorylation of cAMP-response element-binding protein (CREBP) promotes retinal degeneration. Exp Eye Res. 2011;92:528–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldshmit Y, McLenachan S, Turnley A. Roles of Eph receptors and ephrins in the normal and damaged adult CNS. Brain Res Rev. 2006;52:327–345 [DOI] [PubMed] [Google Scholar]
- 59.Bevins N, Lemke G, Reber M. Genetic dissection of EphA receptor signaling dynamics during retinotopic mapping. J Neurosci. 2011;31:10302–10310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunmire JJ, Bouhenni R, Hart ML, et al. Novel serum proteomic signatures in a non-human primate model of retinal injury. Mol Vis. 2011;17:779–791 [PMC free article] [PubMed] [Google Scholar]
- 61.Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat Rev Neurosci. 2005;6:919–930 [DOI] [PubMed] [Google Scholar]
- 62.Li XJ, Li S. Proteasomal dysfunction in aging and Huntington disease. Neurobiol Dis. 2011;43:4–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Helmlinger D, Yvert G, Picaud S, et al. Progressive retinal degeneration and dysfunction in R6 Huntington's disease mice. Hum Mol Genet. 2002;11:3351–3359 [DOI] [PubMed] [Google Scholar]
- 64.Yang J, Yang P, Tezel G, Patil RV, Hernandez MR, Wax MB. Induction of HLA-DR expression in human lamina cribrosa astrocytes by cytokines and simulated ischemia. Invest Ophthalmol Vis Sci. 2001;42:365–371 [PubMed] [Google Scholar]
- 65.Tezel G, Luo C, Yang X. Accelerated aging in glaucoma: immunohistochemical assessment of advanced glycation end products in the human retina and optic nerve head. Invest Ophthalmol Vis Sci. 2007;48:1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tezel G. The immune response in glaucoma: a perspective on the roles of oxidative stress. Exp Eye Res. 2011;93:178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tezel G, Yang X, Cai J. Proteomic identification of oxidatively modified retinal proteins in a chronic pressure-induced rat model of glaucoma. Invest Ophthalmol Vis Sci. 2005;46:3177–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]