Abstract
Objectives
To identify the genes responsible for tetracycline resistance in a strain of Streptococcus australis isolated from pooled saliva from healthy volunteers in France. S. australis is a viridans Streptococcus, originally isolated from the oral cavity of children in Australia, and subsequently reported in the lungs of cystic fibrosis patients and as a cause of invasive disease in an elderly patient.
Methods
Agar containing 2 mg/L tetracycline was used for the isolation of tetracycline-resistant organisms. A genomic library in Escherichia coli was used to isolate the tetracycline resistance determinant. In-frame deletions and chromosomal repair were used to confirm function. Antibiotic susceptibility was determined by agar dilution and disc diffusion assay.
Results
The tetracycline resistance determinant from S. australis FRStet12 was isolated from a genomic library in E. coli and DNA sequencing showed two open reading frames predicted to encode proteins with similarity to multidrug resistance-type ABC transporters. Both genes were required for tetracycline resistance (to both the naturally occurring and semi-synthetic tetracyclines) and they were designated tetAB(46).
Conclusions
This is the first report of a predicted ABC transporter conferring tetracycline resistance in a member of the oral microbiota.
Keywords: antibiotic resistance, oral microflora, oral streptococci
Introduction
Tetracyclines are broad-spectrum antibiotics that are used extensively to combat bacterial infections in humans and animals, and have been used as growth promoters in animals, agriculture and aquaculture.1–3 Bacterial resistance to tetracycline is primarily mediated through acquired genes encoding one of three main mechanisms: active efflux, ribosomal protection proteins (RPPs), or enzyme-mediated drug inactivation.4 Within the oral cavity, ribosomal protection [e.g. tet(M), tet(Q) and tet(O)] is the most commonly observed mechanism, whereas tetracycline-inactivating enzymes and efflux mechanisms occur less frequently.5,6
Tetracycline efflux systems have been reported in both Gram-negative and Gram-positive bacteria.7,8 The best-studied tetracycline efflux genes [e.g. tet(A) and tet(B)] encode membrane-associated, energy-dependent proteins belonging to the major facilitator superfamily (MFS) in which proton-motive force is used to drive efflux.4 However, other tetracycline efflux proteins not belonging to the MFS group have been reported: (i) Tet(35) of Vibrio harveyi9 is a member of the H+ antiporter (NhaC) family and confers resistance to tetracycline, oxytetracycline and minocycline; and (ii) OtrC of Streptomyces rimosus is predicted to encode, on distinct polypeptides, the nucleotide-binding domain (NBD) and membrane-spanning domain (MSD) typical of members of the ABC transporter family. No functional information is available for OtrC.4
In this study, we characterize a novel tetracycline resistance determinant in Streptococcus australis FRStet12, isolated from pooled saliva from healthy French subjects as part of a study investigating antibiotic resistance in bacteria colonizing adult humans.10 We show that two proteins, each encoding predicted ABC transporter subunits, are both required for tetracycline resistance.
Materials and methods
Sample collection and culture
Saliva samples (∼5 mL) were collected from 20 healthy adult volunteers who had not received antibiotic therapy in the previous 3months, from two centres in France (Faculté de Parmacie, Université Paris Sud and INRA-UEPSD, Domain de Vilvert), as previously described.10 The samples were pooled in a sterile 200 mL Duran bottle and processed within 48 h of collection. A 10-fold dilution series was prepared from 1 mL of the sample in Luria–Bertani (LB) broth and spread onto Iso-Sensitest agar (Oxoid) supplemented with 5% defibrinated horse blood (E&O Laboratories, Bonnybridge, UK) and 2 mg/L tetracycline. The plates were incubated in air enriched with 5% CO2 for up to 72 h.10 Growth at concentrations >2 mg/L is defined as resistant by the BSAC.11
Identification of the resistance genes
Genomic DNA from S. australis FRStet12 was hybridized to a macroarray containing 23 known tetracycline resistance genes [9 RPP genes—M, O, B(P), Q, S, T, W, 32 and 36; 12 efflux genes—A, B, C, D, E, G, H, J, A(P), Y, Z and 30; and 2 enzymatic inactivation genes—tet(X) and tet(34)], as previously described.12 The genomic DNA was also tested for the presence of RPP-encoding genes by PCR using a set of universal RPP primers.13 To clone the resistance genes, S. australis FRStet12 genomic DNA was partially digested with HindIII, ligated into HindIII-digested, dephosphorylated pUC19 and transformed into Escherichia coli JM109 competent cells, according to the supplier's instructions (Promega). Bacteria were spread on to LB agar supplemented with 100 mg/L ampicillin and 5 mg/L tetracycline and incubated at 37°C in air for up to 36 h. A tetracycline-resistant clone, designated P9, was isolated and the insert sequenced.
Species identification
Amplification and sequencing of a manganese-dependent superoxide dismutase (sodA) gene fragment was carried out according to Poyart et al.,14 using the primers listed in Table S1 (available as Supplementary data at JAC Online).
DNA sequencing
To sequence the HindIII genomic DNA fragment of S. australis in pUC19 from clone P9 (pP9), a walking strategy was employed using the primers listed in Table S1 (available as Supplementary data at JAC Online).
Mutagenesis
In-frame deletions in tetA(46) and tetB(46) were created using ‘splicing by overlapping extension’ (SOEing) PCR.15 The hybrid fragments were ligated into pGEM-T-Easy (Promega) and verified by DNA sequencing. The ΔtetA(46) fragment was created using two sets of primer pairs: ABC1-1F/ABC1-2R and ABC1-3F/ABC1-4R; primers ABC1-2R and ABC1-3F shared 24 nucleotides of complementary sequence to facilitate the ligation of the two amplicons (Table S1 available as Supplementary data at JAC Online). The ligated mutant fragment, when recombined into the genome, resulted in a 51 bp in-frame deletion (bp 1073–1123 inclusive) in tetA(46). The ΔtetB(46) fragment was created in the same way, using two sets of primer pairs: ABC2-1F/ABC2-2R and ABC2-3F/ABC2-4R; similarly, recombination of this mutagenic fragment resulted in a 51 bp in-frame deletion (bp 1090–1140 inclusive) in tetB(46). The ΔtetA(46) or ΔtetB(46) mutagenic fragments were co-transformed with pVA83816 into S. australis FRStet12 to provide selection for successful transformation (ErmR).
Transformation
Genetic competence was induced in S. australis FRStet12 using a modified version of the method reported by Hudson and Curtiss.17 A single colony was inoculated into 10 mL of Todd–Hewitt broth (THB) containing 10% horse serum and incubated at 37°C, in air + 5% CO2, for 18 h. A 1/40 dilution of the overnight culture was grown in THB plus 10% horse serum under the same conditions until the optical density at 600 nm was between 0.1 and 0.2. For co-transformation experiments, 1 μg of either the ΔtetA(46) or ΔtetB(46) mutant fragments plus ∼250 ng of pVA83816 were added to 1 mL of culture, mixed and incubated for 3 h in the CO2-enriched atmosphere. pVA838 is a streptococcal shuttle plasmid that carries an erythromycin resistance gene as a selectable marker.16 Aliquots of 100 μL were then spread on to brain heart infusion (BHI) agar supplemented with 5% defibrinated horse blood plus 10 mg/L erythromycin and incubated at 37°C, in air + 5% CO2, for 24 h. Colonies were then transferred to BHI agar plates supplemented with 5% defibrinated horse blood plus 10 mg/L erythromycin and replica plated on to BHI agar plates supplemented with 5% defibrinated horse blood plus 10 mg/L erythromycin and 5 mg/L tetracycline. All plates were incubated at 37°C, in air + 5% CO2, for 24–48 h.
Construction of plasmids for chromosome repair
The wild-type tetA(46) or tetB(46) genes were PCR amplified, ligated into pVA838 and transformed into the mutant strains, as described above. The tetA(46) gene was amplified using primers ABC1-8F and ABC1-9R (Table S1 available as Supplementary data at JAC Online), each containing XbaI restriction sites. The tetB(46) gene was amplified using primers ABC2-7F and ABC2-8R, each containing SphI restriction sites (Table S1 available as Supplementary data at JAC Online). The amplified products were digested with their respective restriction endonucleases and ligated into either the XbaI or SphI sites of pVA838, creating the recombinant plasmids pABC1 and pABC2, respectively. The plasmids were transformed into E. coli α-select bronze competent cells (Bioline) and the presence of the wild-type gene confirmed by DNA sequencing. Both E. coli strains were grown overnight in LB broth supplemented with 80 mg/L chloramphenicol, at 37°C, aerobically with shaking at 200 rpm. Plasmid DNA was extracted using a HiSpeed Plasmid Midi Kit (Qiagen).
Antibiotic susceptibility testing
The MICs of tetracycline, oxytetracycline, doxycycline, chlortetracycline and tigecycline were determined according to BSAC guidelines.18 The recommended medium and inoculum (104 cfu/spot) was also used for determination of the acriflavine and ethidium bromide MICs. Triplicate individual colonies of each strain were inoculated into 10 mL BHI broth and incubated at 37°C, in air + 5% CO2, for 18 h. The cells were diluted and then spotted, using a multipoint inoculator, on to Iso-Sensitest agar supplemented with 5% defibrinated horse blood or Iso-Sensitest agar supplemented with 5% defibrinated horse blood plus the appropriate antibiotic at concentrations of 0.25–32 mg/L. The plates were incubated at 37°C, in air + 5% CO2, for up to 48 h.
Agar diffusion assays were carried out according to BSAC guidelines.11 Discs containing the following amounts of antibiotics were laid on to agar: tetracycline (10 μg), ciprofloxacin (1 μg), metronidazole (5 μg), azithromycin (15 μg), ampicillin (2 μg), methicillin (5 μg), oxacillin (1 μg), penicillin (1 μg) and gentamicin (10 μg). Plates were incubated at 37°C, in air + 5% CO2, for 20 h and the zones of inhibition measured.
Results
S. australis FRStet12 was isolated on agar containing 2 mg/L tetracycline.19 Sequencing of a sodA gene fragment showed that the closest relative was the S. australis type strain CIP 107167 (DQ132987), with 95.2% identity. A phylogenetic tree showing the relationship between the sodA gene fragments of S. australis FRStet12 and other streptococcal species is shown in the supplementary data (Figure S1, available as Supplementary data at JAC Online). To determine whether tetracycline resistance was conferred by a previously described mechanism, genomic DNA from S. australis FRStet12 was hybridized to a macroarray containing known tetracycline resistance genes and the genomic DNA was also tested for the presence of RPP-encoding genes by PCR.13 The genomic DNA failed to hybridize to the macroarray and was negative in the RPP PCR, suggesting the presence of a rare or novel tetracycline resistance determinant. In our study, of a total of 69 Gram-positive, facultatively anaerobic, tetracycline-resistant isolates, S. australis FRStet12 was one of only two isolates that failed to hybridize with the array.19 The other isolate that failed to hybridize has not been investigated.
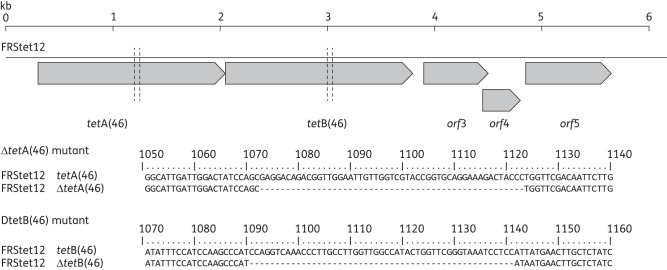
To identify the gene(s) conferring resistance in S. australis FRStet12, a library of HindIII-digested genomic DNA was created in pUC19, selecting transformants on agar containing 100 mg/L ampicillin and 5 mg/L tetracycline. One tetracycline-resistant transformant was selected for further study and designated E. coli P9. The insert from the plasmid in P9 (pP9) was completely sequenced (accession number HQ652506) and found to contain five putative open reading frames (orfs), two of which encoded predicted proteins with similarity to multidrug resistance (MDR)-type ABC transporters (Figure 1). These genes, which were both subsequently shown to encode resistance to tetracycline (see below), have ≤79% amino acid identity to previously characterized tet genes and were assigned tetA(46) and tetB(46) under the current naming standards.20 They are predicted to encode non-identical polypeptides of 574 and 578 amino acids, respectively, each containing an NBD and an MSD characteristic of the ABC transporter superfamily. TetA(46) and TetB(46) are most closely related to YheI (36% amino acid identity) and YheH (35% identity) of Bacillus subtilis, respectively, which, when overexpressed in E. coli, form a heterodimeric MDR-type ABC transporter. TetA(46) and TetB(46) show sequence similarity with other heterodimeric ABC transporters with experimentally proven function, including LmrCD of Lactococcus lactis and EfrAB of Enterococcus faecalis, as well as related systems in the Gram-negative bacteria, Serratia marcescens and E. coli (Table 1).
Figure 1.
Diagram of the cloned genomic DNA fragment from S. australis FRStet12, indicated by the black line. Five putative orfs are indicated in grey [tetA(46); tetB(46); orf3, encodes a putative metalloprotease; orf4, encodes a putative diacylglycerol kinase; and orf5, encodes a putative GTP-binding protein—accession number HQ652506]. The vertical broken black lines indicate the point of the in-frame deletions in tetA(46) and tetB(46) and the sequence of the deletion is given beneath.
Table 1.
Comparison between S. australis TetA(46)/TetB(46) and ABC transporters with experimentally proven function
| Percentage identical (similar) residues |
|||
|---|---|---|---|
| Organism (reference) | ABC transporter subunit | TetA(46) | TetB(46) |
| S. australis (this study) | TetA(46) | — | 23.3 (52.7) |
| TetB(46) | 23.3 (52.7) | — | |
| B. subtilis21 | YheI | 36.4 (65.3) | 26.7 (51.1) |
| YheH | 21.4 (50.1) | 34.9 (57.5) | |
| S. marcescens22 | SmdA | 30.3 (58.6) | 25.5 (49.6) |
| SmdB | 23.3 (51.0) | 29.1 (54.0) | |
| E. coli23 | MdlA | 30.3 (57.8) | 25.0 (50.2) |
| MdlB | 23.0 (51.6) | 28.4 (52.6) | |
| L. lactis24 | LmrC | 26.2 (55.1) | 23.5 (51.5) |
| LmrD | 21.4 (47.2) | 26.1 (48.5) | |
| E. faecalis25 | EfrA | 23.6 (55.0) | 25.3 (51.5) |
| EfrB | 25.3 (53.4) | 27.7 (56.6) | |
GenBank proteins numbers: YheI, NP_388852; YheH, NP_388853; SmdA, BAF79679; SmdB, BAF79680; MdlA, P77265; MdlB, P0AAG5; LmrC, Q9CIP6; LmrD, Q9CIP5. For the other sequences, Swiss-Prot entries (in parentheses) were used: EfrA (Q82ZX7_ENTFA); and EfrB (Q82ZX8_ENTFA). Percentage identity and similarity were obtained by sequence alignment using Clustal W at http://npsa-pbil.ibcp.fr/.
To determine the function of each gene, 51 bp in-frame deletions were created within the sequences predicted to encode the Walker A box of the ATP-binding site of each gene product using SOEing PCR.15 The mutant gene fragments were co-transformed26 into S. australis FRStet12 along with plasmid pVA838,16 making use of the erythromycin resistance gene on the plasmid for selection. This plasmid was chosen for co-transformation since our experience showed it to be rapidly lost from other Streptococcus species in the absence of selection. Of 90 erythromycin-resistant clones examined following transformation with the tetA(46) mutant fragment and pVA838, five were unable to grow on 5 mg/L tetracycline and sequence analysis confirmed the presence of the ΔtetA(46) allele in all five (Figure 1). The tetracycline-resistant transformants contained wild-type tetA(46) and were erythromycin resistant, indicating they had only taken up pVA838. Despite repeated subculturing without erythromycin (14 passages in total), pVA838 remained in the mutant strains. Therefore, to allow comparison between wild-type and mutant strains, an isogenic strain was created by transforming the wild-type with pVA838. Of 72 erythromycin-resistant clones examined following transformation of the wild-type with the tetB(46) mutant fragment and pVA838, 7 were unable to grow on 5 mg/L tetracycline and sequencing confirmed the presence of the ΔtetB(46) allele (Figure 1), while those that remained resistant to tetracycline contained wild-type tetB(46).
To determine whether tetracycline resistance could be restored in the FRStet12ΔtetA(46) and FRStet12ΔtetB(46) mutants, a chromosome repair was carried out. The wild-type tetA(46) and tetB(46) genes were cloned into pVA838 to create the recombinant plasmids pABC1 and pABC2, respectively. In three independent experiments, transformation of pABC1 into FRStet12ΔtetA(46) resulted in colonies on agar containing 5 mg/L tetracycline, whereas transformation with pVA838 alone did not. Sequence analysis of two tetracycline-resistant transformants from each of the three independent experiments, using primers flanking tetA(46) in the chromosome, revealed that in each case the wild-type tetA(46) had replaced the mutant allele within the chromosome, demonstrating that wild-type tetA(46) is required for tetracycline resistance. The transformants contained empty pVA838 and the mutant ΔtetA(46) allele was not detectable by PCR. Transformation of pABC2 into FRStet12ΔtetB(46) restored the ability of the mutant strain to grow on 5 mg/L tetracycline, whereas transformation with pVA838 did not. Sequence analysis of these transformants confirmed that wild-type tetB(46) had replaced the mutant allele within the chromosome, demonstrating that wild-type tetB(46) is also essential for tetracycline resistance.
The MIC of tetracycline was determined for S. australis FRStet12, S. australis FRStet12 containing pVA838, the ΔtetA(46) and ΔtetB(46) isogenic mutants and their corresponding complemented strains. The MIC for the wild-type and the complemented strains was 8 mg/L, compared with <0.25 mg/L for the ΔtetA(46) and ΔtetB(46) mutants. MIC determination of the other tetracyclines (Table 2) showed that the mutants were 2- to 8-fold more sensitive to oxytetracycline, doxycycline, chlortetracycline and tigecycline, indicating that TetAB(46) is also able to export these molecules. As MDR-type transporters often are capable of extrusion of other toxic compounds,21,22,24 we determined the MIC of acriflavine and ethidium bromide but found no difference between the wild-type and mutant strains. In addition, we found no difference in the zone size in disc diffusion assays with ciprofloxacin, metronidazole, azithromycin, ampicillin, methicillin, oxacillin, penicillin or gentamicin, indicating that TetAB(46) is specific for the transport of tetracyclines.
Table 2.
MICs of tetracyclines for S. australis FRStet12 and the tetAB(46) mutant strains
| Strain | Oxytetracycline | Doxycycline | Chlortetracycline | Tigecycline |
|---|---|---|---|---|
| FRStet12 | 8 | 0.5 | 4 | 1 |
| FRStet12 ΔABC1 | 1 | <0.25 | 0.5 | <0.25 |
| FRStet12 ΔABC2 | 2 | <0.25 | 0.5 | <0.25 |
To determine whether similar genes were present in other bacteria, BLAST searches were performed. This analysis revealed orthologues of TetAB(46) in the one draft genome of S. australis present in the database (NCTC 13166) and in all six of the Streptococcus parasanguinis draft genomes present in the database (ATCC 903, ATCC 15912, F0405, F0449, FW213 and SK236). The predicted proteins within these streptococcal genomes share ≥95% amino acid identity with TetA(46) and TetB(46). Since both S. australis NCTC 13166 and S. parasanguinis NCTC 55898 (ATCC 15912) are resistant to tetracycline (2 mg/L), it is possible that these tetAB(46) orthologues confer this phenotype.
Discussion
Most tetracycline resistance genes reported in oral bacteria encode RPPs,6,27,28 whereas efflux genes are rarely detected. Here, we report the discovery of a predicted tetracycline efflux determinant, tetAB(46) from S. australis. This species, first isolated from the oral cavities of children in Australia,29 is an opportunist pathogen reported in sputum samples of adult cystic fibrosis patients30 and in a case of invasive infection, a community-acquired meningitis in an elderly patient.31
Most tetracycline efflux proteins belong to the MFS family of transporters, which are membrane located and exchange a proton for a tetracycline–cation complex against a concentration gradient. These have been described in both Gram-positive and Gram-negative bacteria.7 ABC transporters conferring resistance to tetracyclines have also been reported. There is one example currently in the tet nomenclature database: OtrC from S. rimosus,4 which consists of an NBD and an MSD encoded by separate genes. While other ABC transporters capable of exporting tetracycline have been reported, e.g. SmdAB in S. marcescens,22 these are capable of exporting a number of other compounds and have not therefore been given a tetracycline resistance gene designation.
Analysis of S. australis FRStet12 revealed two orfs responsible for tetracycline resistance, which are most closely related to YheI and YheH of B. subtilis. YheI and YheH are non-identical ABC transporter subunits, each containing an NBD and an MSD, which were shown to interact to form a heterodimeric multidrug ABC transporter capable of transporting several structurally dissimilar drugs, such as fluorescently labelled ethidium bromide and daunomycin.21 Another related MDR transporter is LmrCD of L. lactis, also shown to be capable of extrusion of a range of structurally unrelated drugs.24 Expression of E. faecalis EfrA and EfrB together in E. coli confers resistance to a range of drugs, including acriflavine, norfloxacin and doxycycline. Further, energy-dependent efflux of acriflavine in E. coli harbouring efrAB was also demonstrated.25 MDR-type ABC transporters from S. marcescens and E. coli are also related to TetAB(46): although there are no functional data for the E. coli system,23 SmdAB of S. marcescens has been shown to confer multidrug, including tetracycline, resistance on E. coli and to be inhibited by ATPase inhibitors.22
Of the tetracycline (tet) and oxytetracycline (otr) resistance genes currently listed in the tetracycline gene nomenclature database (http://faculty.washington.edu/marilynr/), 28 code for active efflux, 12 for ribosomal protection, 3 for enzymatic drug inactivation and 1 has an unknown mechanism. The two genes required for tetracycline resistance in S. australis FRStet12 were designated tetA(46) and tetB(46) under the current naming standards.20
In-frame deletions in either tetA(46) or tetB(46) demonstrated that both were required for tetracycline resistance in S. australis. The fact that the genes encode non-identical proteins, each containing a predicted MSD and NBD, suggests that they may function as a heterodimeric ABC transporter, although confirmation of this requires demonstration of a physical interaction between the two proteins, experiments that are beyond the scope of the present study.
During the course of this work, we made two observations on the molecular biology of S. australis that are worthy of discussion. Firstly, we report the maintenance of pVA838 after multiple passages in the absence of selection, which is unusual in our experience with this plasmid in other streptococci. The stability of ‘empty’ pVA838 in the mutant strains may have contributed to the second unexpected result obtained in this work: in experiments designed to introduce the wild-type alleles in trans, transformation of both mutants with the wild-type alleles cloned in pVA838 resulted in chromosome repair, i.e. double crossover recombination occurred, resulting in replacement of the mutant allele with the wild-type allele in the chromosome, and this restored the tetracycline resistance phenotype. However, instead of detecting the mutant allele in the plasmid as expected following double crossover, we detected only pVA838 without an insert in these strains and did not detect the presence of the mutant alleles. This result was reproduced in triplicate for both mutants. One explanation is that following allelic exchange, the plasmid carrying the mutant allele was lost from the population because of the fitness disadvantage associated with its replication, compared with replication of the plasmid without the insert.
Despite homology to proven multidrug transporters, TetA(46) and TetB(46) were found to confer resistance only to tetracyclines. The MFS efflux systems transport only the naturally occurring tetracyclines, with the exception of TetA(B), which also transports the semi-synthetic analogue, minocycline.7 The reduced MICs in the tetA(46) and tetB(46) mutants indicate that TetAB(46) is able to transport both tetracycline and its semi-synthetic derivatives. We have previously shown that S. australis FRStet12 is sensitive to minocycline (MIC of 0.25 mg/L).32 Since MDR-type ABC transporters have been shown to extrude toxic molecules other than antibiotics,24,25 we determined the MICs of acriflavine and ethidium bromide, but found no difference between the wild-type and mutant strains. Thus, our data suggest that TetAB(46) is specific for tetracyclines.
In conclusion, we have identified a novel tetracycline resistance determinant, tetAB(46), in an oral viridans Streptococcus species. TetAB(46) is related to known MDR-type ABC transporters in both Gram-positive and Gram-negative bacteria, but confers resistance only to tetracyclines. Genes highly related to tetAB(46) are present in the genomes of other tetracycline-resistant oral streptococci, including several strains of S. parasanguinis and another strain of S. australis.
Funding
This study has been carried out with financial support from the Wellcome Trust and the Commission of the European Communities, specifically the Infectious Diseases research domain of the Health theme of the 7th Framework Programme, contract 241446, ‘The effects of antibiotic administration on the emergence and persistence of antibiotic-resistant bacteria in humans and on the composition of the indigenous microbiotas at various body sites’ and the 5th Framework Research and Technological Development (RTD) Program ‘Quality of Life and Management of Living Resources’, QLK2-CT-2002-00843, ‘Antimicrobial resistance transfer from and between Gram-positive bacteria of the digestive tract and consequences for virulence’.
Transparency declarations
None to declare.
Supplementary data
Acknowledgements
We wish to thank Dr Arthur Hosie and Dr Alex Webb for their advice on ABC transporter structure.
References
- 1.McManus PS, Stockwell VO, Sundin GW, et al. Antibiotic use in plant agriculture. Annu Rev Phytopathol. 2002;40:443–65. doi: 10.1146/annurev.phyto.40.120301.093927. [DOI] [PubMed] [Google Scholar]
- 2.Miranda CD, Kehrenberg C, Ulep C, et al. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrob Agents Chemother. 2003;47:883–8. doi: 10.1128/AAC.47.3.883-888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goossens H, Ferech M, Vander Stichele R, et al. ESAC Project Group. Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet. 2005;365:579–87. doi: 10.1016/S0140-6736(05)17907-0. [DOI] [PubMed] [Google Scholar]
- 4.Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Torres ML, McNab R. Novel tetracycline resistance determinant from the oral metagenome. Antimicrob Agents Chemother. 2003;47:1430–2. doi: 10.1128/AAC.47.4.1430-1432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villedieu A, Diaz-Torres ML, Hunt N, et al. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob Agents Chemother. 2003;47:878–82. doi: 10.1128/AAC.47.3.878-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chopra I, Roberts M. Tetracycline antibiotics: mode of action, application, molecular biology and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;65:232–60. doi: 10.1128/MMBR.65.2.232-260.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellido HLM, Guirao GY, Zufiaurre NGM, et al. Efflux-mediated antibiotic resistance in Gram-positive bacteria. Rev Med Microbiol. 2002;13:1–13. [Google Scholar]
- 9.Teo JW, Tan TM, Poh CL. Genetic determinants of tetracycline resistance in Vibrio harveyi. Antimicrob Agents Chemother. 2002;46:1038–45. doi: 10.1128/AAC.46.4.1038-1045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seville L, Patterson AJ, Scott KP, et al. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15:159–66. doi: 10.1089/mdr.2009.0916. [DOI] [PubMed] [Google Scholar]
- 11.Andrews JM. BSAC standardized disc susceptibility testing method. J Antimicrob Chemother. 2001;48(Suppl 1):S43–57. doi: 10.1093/jac/48.suppl_1.43. [DOI] [PubMed] [Google Scholar]
- 12.Patterson AJ, Colangeli R, Spigaglia P, et al. Distribution of specific tetracycline and erythromycin resistance genes in environmental samples assessed by macroarray detection. Environ Microbiol. 2007;9:703–15. doi: 10.1111/j.1462-2920.2006.01190.x. [DOI] [PubMed] [Google Scholar]
- 13.Warburton P, Roberts AP, Allan E, et al. Characterization of tet(32) genes from the oral metagenome. Antimicrob Agents Chemother. 2009;53:273–6. doi: 10.1128/AAC.00788-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poyart C, Quesne G, Coulon S, et al. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–7. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton RM, Cai ZL, Ho SN, et al. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–35. [PubMed] [Google Scholar]
- 16.Macrina FL, Tobian JA, Jones KR, et al. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982;19:345–53. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 17.Hudson MC, Curtiss R., III Regulation of expression of Streptococcus mutans genes important to virulence. Infect Immun. 1990;58:464–70. doi: 10.1128/iai.58.2.464-470.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48(Suppl 1):S5–16. doi: 10.1093/jac/48.suppl_1.5. [DOI] [PubMed] [Google Scholar]
- 19.Seville L. University College London; 2007. Searching for novel antibiotic resistance genes and their genetic supports in the oral and faecal microbiota of six European countries. PhD Thesis. [Google Scholar]
- 20.Levy SB, McMurry LM, Barbosa TM, et al. Nomenclature for new tetracycline resistance determinants. Antimicrob Agents Chemother. 1999;43:1523–4. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres C, Galián C, Freiberg C, et al. The YheI/YheH heterodimer from Bacillus subtilis is a multidrug ABC transporter. Biochim Biophys Acta. 2009;1788:615–22. doi: 10.1016/j.bbamem.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo T, Chen J, Minato Y, et al. SmdAB, a heterodimeric ABC-type multidrug efflux pump, in Serratia marcescens. J Bacteriol. 2008;190:648–54. doi: 10.1128/JB.01513-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allikmets R, Gerrard B, Court D, et al. Cloning and organization of the abc and mdl genes of Escherichia coli: relationship to eukaryotic multidrug resistance. Gene. 1993;136:231–6. doi: 10.1016/0378-1119(93)90470-n. [DOI] [PubMed] [Google Scholar]
- 24.Lubelski J, de Jong A, van Merkerk R, et al. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol. 2006;61:771–81. doi: 10.1111/j.1365-2958.2006.05267.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee E-W, Huda MN, Kuroda T, et al. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob Agents Chemother. 2003;47:3733–8. doi: 10.1128/AAC.47.12.3733-3738.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas I, Drake L, Johnson S, et al. Unmarked gene modification in Streptococcus mutans by a cotransformation strategy with a thermosensitive plasmid. Biotechniques. 2007;42:487–90. doi: 10.2144/000112414. [DOI] [PubMed] [Google Scholar]
- 27.Lancaster H, Ready D, Mullany P, et al. Prevalence and identification of tetracycline-resistant oral bacteria in children not receiving antibiotic therapy. FEMS Microbiol Lett. 2003;228:99–104. doi: 10.1016/S0378-1097(03)00740-7. [DOI] [PubMed] [Google Scholar]
- 28.Lancaster H, Bedi R, Wilson M, et al. The maintenance in the oral cavity of children of tetracycline-resistant bacteria and the genes encoding such resistance. J Antimicrob Chemother. 2005;56:524–31. doi: 10.1093/jac/dki259. [DOI] [PubMed] [Google Scholar]
- 29.Willcox MD, Zhu H, Knox KW. Streptococcus australis sp. nov., a novel oral Streptococcus. Int J Syst Evol Microbiol. 2001;51:1277–81. doi: 10.1099/00207713-51-4-1277. [DOI] [PubMed] [Google Scholar]
- 30.Tazumi A, Maeda Y, Goldsmith CE, et al. Molecular characterization of macrolide resistance determinants [erm(B) and mef(A)] in Streptococcus pneumoniae and viridans group streptococci (VGS) isolated from adult patients with cystic fibrosis (CF) J Antimicrob Chemother. 2009;64:501–6. doi: 10.1093/jac/dkp213. [DOI] [PubMed] [Google Scholar]
- 31.Héry-Arnaud G, Rouzic N, Doloy A, et al. Streptococcus australis meningitis. J Med Microbiol. 2011;60:1701–4. doi: 10.1099/jmm.0.030114-0. [DOI] [PubMed] [Google Scholar]
- 32.Ciric L, Mullany P, Roberts AP. Antibiotic and antiseptic resistance genes are linked on a novel mobile genetic element: Tn6087. J Antimicrob Chemother. 2011;66:2235–9. doi: 10.1093/jac/dkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.