Abstract
Thaxtomin phytotoxins produced by plant-pathogenic Streptomyces species contain a nitro group that is essential for phytotoxicity. The N,N’-dimethyldiketopiperazine core of thaxtomins is assembled from L-phenylalanine and L-4-nitrotryptophan by a nonribosomal peptide synthetase and nitric oxide synthase-generated NO is incorporated into the nitro group, but the biosynthesis of the non-proteinogenic amino acid L-4-nitrotryptophan is unclear. Here we report that TxtE, a unique cytochrome P450, catalyzes L-tryptophan nitration using NO and O2.
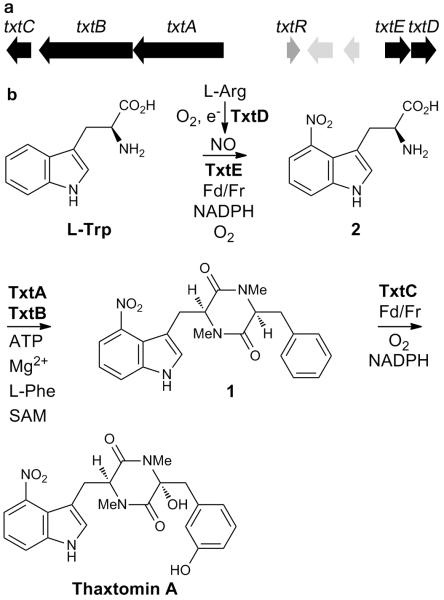
Thaxtomin A exerts its phytotoxicity by inhibiting cellulose biosynthesis in plants1. Conserved pathogenicity islands on the chromosomes of Streptomyces turgidiscabies, Streptomyces scabies and Streptomyces acidiscabies contain the thaxtomin biosynthetic gene cluster (Fig. 1a)2. The txtD gene within this gene cluster encodes a nitric oxide synthase (NOS) that generates NO from L-arginine (Fig. 1b)3. Deletion of txtD in S. turgidiscabies severely attenuates thaxtomin A production3. A 15N label from the guanidino group of L-arginine is specifically incorporated into the nitro group of thaxtomin A and NO donors boost thaxtomin production in the txtD mutant, indicating that part of the nitro group is derived from NO3,4. The txtA and txtB genes encode nonribosomal peptide synthetases that are implicated in the assembly of the thaxtomin N,N’-dimethyldiketopiperazine core 1 from the unique non-proteinogenic amino acid L-4-nitrotryptophan 2, L-phenylalanine and S-adenosyl-L-methionine, using ATP and Mg2+ as cofactors5. However, the mechanism for biosynthesis of L-4-nitrotryptophan remains to be elucidated. Nitro groups in natural products usually result from oxidation of an amino group6. While biomimetic syntheses and feeding experiments with labeled precursors implicate direct nitration in the biosynthesis of two metabolites6, a natural product biosynthetic enzyme that is capable of direct nitration has never been reported. Peroxidases and globins can catalyze nitration of L-tryptophan in the presence of NO −2/H2O2 and NO/O2, respectively, but the reactions are unselective, resulting in several nitrotryptophan regioisomers7. Similarly, in Deinococcus radiodurans a NOS in partnership with an adjacently-encoded t-RNA synthetase can catalyze regioselective nitration of L-tryptophan. However, the catalytic efficiency of this transformation is very low8. In both cases, the biological significance of the reactions is unknown; it is unclear whether nitration is anything more than an adventitious side reaction. Here we report that txtE encodes a unique cytochrome P450 (CYP) that catalyzes direct regiospecific 4-nitration of L-tryptophan, the first committed step in thaxtomin A biosynthesis, using NO and O2 as substrates and spinach ferredoxin (Fd) and ferredoxin reductase (Fr) as surrogate electron donors.
Figure 1.
Thaxtomin biosynthesis. a: Organization of the thaxtomin biosynthetic gene cluster in S. turgidiscabies. Biosynthetic genes are in black; txtR, which encodes a cellobiose-responsive pathway specific activator, is in dark gray; two genes encoding putative transposases are in light gray. The txtAB genes encode nonribosomal peptide synthetases. The txtC and txtE genes encode CYPs. The txtD gene encodes a NOS that produces NO from L-arginine. b: Proposed pathway for thaxtomin A biosynthesis.
CYPs are ubiquitous heme-dependent enzymes that activate molecular oxygen to catalyse a vast array of oxidative transformations including epoxidation, C- and N-hydroxylation and oxidative dealkylation9,10. Coordination of NO to the heme iron atom causes reversible inhibition of CYPs. Subsequent reaction with O2 forms reactive nitrogen species that nitrate amino acid residues such as tyrosine, irreversibly inhibiting enzyme activity7. The inhibition of CYP monooxygenases by NO significantly affects signal transduction, cellular inflammation, neurodegeneration, and drug metabolism11,12. P450nor (CYP55A1) from Fusarium oxysporum is an interesting exception, because it reduces bound NO to N2O using electrons from NADH 13.
We hypothesized that TxtE and TxtD are together responsible for 4-nitration of L-tryptophan in thaxtomin A biosynthesis. Thus, we expected the genes encoding these enzymes to be co-transcribed. S1 nuclease protection analysis confirmed co-transcription of txtE and txtD (Supplementary Methods; Supplementary Fig. 12). No additional protected RNA transcripts were observed, suggesting that txtD transcription is solely dependent on the txtE promoter. The transcriptional start site of txtE is located 232 bp upstream of the translational start site. The predicted DNA binding regions for the transcription factor are both shifted upstream of the −10 and −35 positions. A comparison of the txtE promoter region with other previously-characterized Streptomyces promoters revealed a potential core promoter motif CANNAT and a putative −35 core promoter motif, GNTTNC14. A 9 bp inverted repeat in the vicinity of the −10 and −35 regions in S. turgidiscabies (Supplementary Fig. 12) may indicate an additional level of transcriptional regulation of txtE.
To determine if TxtE is required for thaxtomin biosynthesis, txtE was deleted from S. turgidiscabies Car8 using marker-exchange mutagenesis. Thaxtomin A production was abolished in the ΔtxtE strain and partially restored by complementation with txtE expressed under the control of the thiostrepton-inducible tipA promoter in plasmid pIJ8600txtE (Supplementary Results; Supplementary Table 1). Partial restoration of thaxtomin A production in the ΔtxtEpIJ8600txtE strain and improved complementation with addition of NO donors is consistent with a polar effect on txtD (Table 1). Reverse-transcriptase PCR was used to confirm the absence of txtD expression in S. turgidiscabies ΔtxtE. The low level of thaxtomin A production in the strain lacking txtD is likely due to the production of NO by nitrate/nitrite reductases in S. turgidiscabies. In order to assess the individual contribution of TxtE to thaxtomin A biosynthesis, txtD was introduced into S. turgidiscabies ΔtxtE using pIJ8600txtD3. Ectopic expression of txtD did not restore thaxtomin A production nor did addition of NO donors, indicating that deletion of txtE alone is responsible for the loss of thaxtomin A production in this strain (Table 1 and Supplementary Table 1).
Table 1.
Thaxtomin A production by S. turgidiscabies strains Car8 (wild type), ΔtxtD, ΔtxtE and the complemented txtE mutant, ΔtxtEpIJ8600txtE, at 7 days after addition of DEANO (1 mM) or 15 μg of L-4-nitrotryptophan (4-NO2Trp), at 3 days after inoculation.
| Thaxtomin A (μg culture−1) a | ||||
|---|---|---|---|---|
| Strain | - DEANO | + DEANO | − 4-NO2Trp | + 4-NO2Trpc |
| No inoculum | ND | ND | - | - |
| Car8 | 33.0 ± 3.5 | 18.9 ± 0.8 | 24.9 ± 0.5 | 34.0 ± 0.6 (34.2) |
| Δ txtD | 0.15 ± 0.01 | 14.3 ± 1.0 | - | - |
| Δ txtE | ND | ND | ND | 8.0 ± 0.2 (30.1) |
| ΔtxtEpIJ8600txtEb | 0.4 ± 0.1 | 2.4 ± 0.4 | - | - |
mean ± standard deviation, n=6. ND = not detected
complementation construct induced by addition of 10 μg/ml thiostrepton
numbers in parentheses are the percentage of added 4-NO2Trp converted to thaxtomin A
To further analyze the respective roles of TxtD and TxtE in thaxtomin A production, thiostrepton-inducible txtE and txtD were introduced into S. turgidiscabies Car8 and the ability of these constructs to modulate thaxtomin A production was examined. Overexpression of txtE in the wild type strain resulted in a 34% increase in thaxtomin A production (Supplementary Table 1), suggesting that TxtE is a rate-limiting enzyme in its biosynthesis. In contrast, overexpression of txtD in wild type resulted in a 23% decrease in thaxtomin A production, consistent with NO cytotoxicity.
Since cultures of the ΔtxtE strain did not accumulate non-nitrated thaxtomin derivatives, we hypothesized that TxtE-catalyzed 4-nitration of l-tryptophan is the first committed step in thaxtomin A biosynthesis. Addition of L-4-nitrotryptophan to cultures of the ΔtxtE strain resulted in the restoration of thaxtomin A production (Table 1). This result is consistent with the hypothesis that L-4-nitrotryptophan is a biosynthetic intermediate utilized by the nonribosomal peptide synthetase TxtB for thaxtomin A biosynthesis (Fig. 1b)5.
To directly examine the role of TxtE in L-tryptophan nitration we cloned and overexpressed txtE in E. coli. Attempted overproduction of TxtE from S. turgidiscabies was not successful. Thus, we chose to work with the ortholog from S. scabies 87-22 (94% similarity, 87% identity). Recombinant TxtE was produced as an N-terminal His6-tagged fusion protein, which was purified to homogeneity by nickel affinity and gel filtration chromatography (Supplementary Fig. 3). The latter indicated that the protein exists as a monomer in solution. Purified TxtE was analysed by UV-Vis spectroscopy (Supplementary Fig. 4). The Soret band at 447 nm in the ferrous minus ferrous-CO difference spectrum of TxtE confirmed that it is a CYP (Supplementary Fig. 4)15.
The substrate specificity of TxtE was examined by investigating changes in the UV-Vis spectrum upon titration with different amino acids. Characteristic type I binding spectra were observed (Supplementary Fig. 5) in the presence of l-tryptophan, arising from conversion of the heme iron atom from low to high spin upon substrate binding and resulting in a λmax shift from 417 nm to 390 nm16. We estimated that the dissociation constant for the L-tryptophan-TxtE complex is 60 ±6 μM from a plot of the difference in absorbance at 390 nm and 420 nm with increasing L-tryptophan concentrations (Supplementary Fig. 5). No shift in λmax was observed with d-tryptophan suggesting that TxtE is stereospecific.
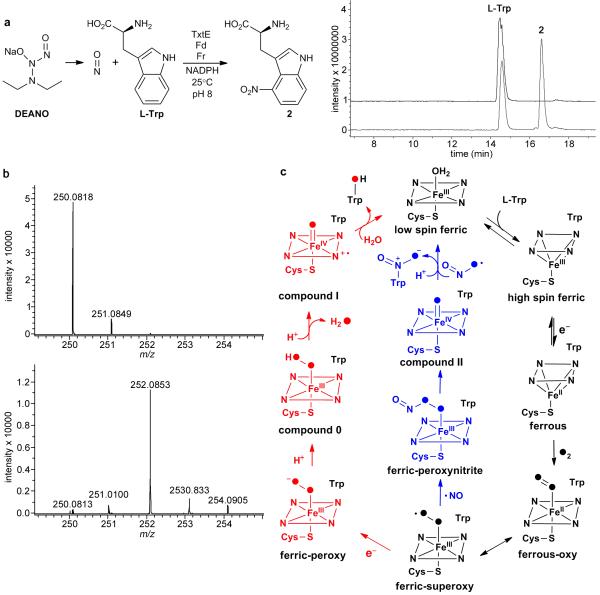
Assuming that, like many other bacterial CYPs, TxtE requires the electron transfer enzymes Fd and Fr for activity, we incubated TxtE with spinach Fd and Fr, NADPH, the NO donor 2-(N,N-diethylamino)-diazenolate 2-oxide (DEANO), and L-tryptophan. The reaction was carried out in air at room temperature for 90 minutes, over which time the solution became yellow. LC-MS analyses identified a product, absent from control reactions, with m/z 250 and a λmax at 400 nm, consistent with L-4-nitrotryptophan (Fig. 2a and Supplementary Fig. 6)8. The reaction was scaled up and the product was purified from the reaction by HPLC. The retention time, the ESI-MS/MS spectrum and the aromatic region of the 1H NMR spectrum of this compound were identical to those for synthetic DL-4-nitrotryptophan (Supplementary Methods; Supplementary Figs. 7 and 8). No products with m/z corresponding to hydroxylated tryptophan derivatives were observed in either the enzymatic reaction or the control reaction lacking DEANO, suggesting that TxtE is not capable of hydroxylating L-tryptophan. Furthermore, compounds with m/z corresponding to 4-nitrosotryptophan, a potential intermediate in the nitration of L-tryptophan, were not detected in the enzymatic reaction.
Figure 2.
Characterization of purified recombinant TxtE. a: Extracted ion chromatograms at m/z = 204 and 250 (right) from LC-MS analyses of the enzymatic nitration reaction (left). The bottom chromatogram is from the enzymatic reaction and the top chromatogram is from a negative control containing boiled TxtE. b: High resolution mass spectra of L-4-nitrotryptophan obtained from incubation of L-tryptophan with TxtE, DEANO, ferredoxin, ferredoxin reductase and NADPH in air (top spectrum), and in an 18O2 atmosphere (bottom spectrum). The m/z = 250.0818 ion corresponds to [M+H]+ for L-4-nitrotryptophan. An ion corresponding to [M+H]+ for L-4-nitrotryptophan containing a single 18O label (m/z calculated for C11H12N3O +4: 252.0865; found: 252.0853) predominates (>90%) in the bottom spectrum c: Comparison of the proposed mechanism of TxtE-catalyzed nitration and the mechanism of CYP-catalyzed hydroxylation. Intermediates that differ in CYP-catalyzed hydroxylation and nitration are highlighted in red and blue, respectively. Common intermediates are black. The atoms of O2 are filled to illustrate their fate in the reactions.
CYPs usually utilize O2 as a co-substrate. O2 binds to the iron atom of the ferrous heme resulting from reduction of the ferric enzyme-substrate complex by Fd/Fr (using electrons from NADPH). The resulting ferrous-O2/ferric-superoxide complex undergoes further reduction and dehydration to form a ferryl complex (Fe(IV)=O porphyrin cation radical; often referred to as “compound I”) that oxidizes the substrate10. To examine the involvement of O2 in the TxtE-catalyzed nitration reaction, we incubated the enzyme with L-tryptophan, NADPH, DEANO and spinach Fd/Fr under an 18O2 atmosphere. High resolution MS analysis of the resulting L-4-nitrotryptophan showed that it was >90% labeled with a single 18O atom (Fig. 2b). No labeling of L-4-nitrotryptophan was observed in a control reaction in which 18O2 was replaced with air (Fig. 2b), or H218O (Supplementary Fig. 9). Moreover, no turnover of L-tryptophan to L-4-nitrotryptophan was observed when air was excluded from the reaction (Supplementary Fig. 10). These data are consistent with the utilization of O2 as a co-substrate by TxtE.
Taken together, our results prompt us to propose a catalytic mechanism for the formation of potential nitrating species, based on the well-established catalytic mechanism of CYP monooxygenases (Figure 2c)9,10. L-tryptophan binds to the active site of TxtE, triggering loss of the water ligand from the ferric heme, resulting in the observed spin state change. This promotes reduction of the heme to its ferrous form by Fr/Fd and O2 binds to the heme iron atom, forming a ferric-superoxide complex. Reaction of NO with the ferric-superoxide complex affords a ferric-peroxynitrite complex, which can undergo homolytic cleavage to yield NO2 and an Fe(IV)=O species (often referred to as “compound II”). Nitration of the bound L-tryptophan could then occur via NO2 addition and compound II-mediated hydrogen atom abstraction, resulting in formation of an Fe(III)-OH species (or vice versa). An alternative mechanism for nitration involving protonation-triggered heterolytic cleavage of the ferric-peroxynitrite complex to yield NO2+ and an Fe(III)-OH species, followed by classical electrophilic aromatic substitution, is also possible. Irrespective of the nitration mechanism, protonation of the Fe(III)-OH species formed is required to regenerate the Fe(III)-OH2 resting state of the enzyme. The observed incorporation of predominantly a single oxygen atom from 18O2 into L-4-nitrotryptophan is consistent with all of the mechanistic scenarios outlined above and further experiments will be required to discriminate between them.
In conclusion, TxtE is a unique new member of the CYP superfamily that catalyzes regiospecific 4-nitration of L-tryptophan using NO and O2. Sequence alignments with several structurally-characterized CYPs indicate differences in several functionally-important regions of TxtE (Supplementary Fig. 2), providing clues to the origin of its dramatically different catalytic activity. TxtE may prove to be a useful nitration biocatalyst, given that in organic synthesis direct nitration of indoles is often carried out under harsh conditions and lacks regioselectivity17-19.
Supplementary Material
Acknowledgements
Prakash Patel is thanked for assistance with fitting the L-tryptophan binding data to the one site binding model. The UK BBSRC (Grant Ref. BB/H006281/1 to G.L.C.) and the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (Grant No. 2008-35319-19202 to R. Loria) are thanked for funding this research. The Bruker MaXis mass spectrometer used in this research was obtained through Birmingham Science City: Innovative Uses for Advanced Materials in the Modern World with support from Advantage West Midlands (AWM) and part funded by the European Regional Development Fund (ERDF).
Footnotes
Competing financial interests
The authors declare no competing financial interests
Author Information
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to G.L.C. (G.L.Challis@warwick.ac.uk) or R.L. (rl21@cornell.edu)
References
- 1.Scheible WR, et al. Plant Cell. 2003;15:1781–1794. doi: 10.1105/tpc.013342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kers JA, et al. Mol. Microbiol. 2005;55:1025–1033. doi: 10.1111/j.1365-2958.2004.04461.x. [DOI] [PubMed] [Google Scholar]
- 3.Kers JA, et al. Nature. 2004;429:79–82. doi: 10.1038/nature02504. [DOI] [PubMed] [Google Scholar]
- 4.Wach MJ, Kers JA, Krasnoff SB, Loria R, Gibson DM. Nitric Oxide. 2005;12:46–53. doi: 10.1016/j.niox.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Johnson EG, et al. Mol. Microbiol. 2009;73:409–418. doi: 10.1111/j.1365-2958.2009.06780.x. [DOI] [PubMed] [Google Scholar]
- 6.Winkler R, Hertweck C. ChemBioChem. 2007;8:973–977. doi: 10.1002/cbic.200700042. [DOI] [PubMed] [Google Scholar]
- 7.Roncone R, Barbieri M, Monzani E, Casella L. Coord. Chem. Rev. 2006;250:1286–1293. [Google Scholar]
- 8.Buddha MR, et al. J. Biol. Chem. 2004;279:49567–49570. doi: 10.1074/jbc.C400418200. [DOI] [PubMed] [Google Scholar]
- 9.Sono M, Roach MP, Coulter ED, Dawson JH. Chem. Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 10.Denisov IG, Makris TM, Sligar SG, Schlichting I. Chem. Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 11.Quaroni LG, et al. Biochemistry. 2004;43:16416–16431. doi: 10.1021/bi049163g. [DOI] [PubMed] [Google Scholar]
- 12.Franke A, et al. J. Am. Chem. Soc. 2004;126:4181–4191. doi: 10.1021/ja038774d. [DOI] [PubMed] [Google Scholar]
- 13.Nakahara K, et al. J. Biol. Chem. 1993;268:8350–8355. [PubMed] [Google Scholar]
- 14.Bourn WR, Babb B. Nucleic Acids Res. 1995;23:3696–3703. doi: 10.1093/nar/23.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omura T, Sato R. J. Biol. Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 16.Schenkman JB, Remmer H, Estabrook RW. Mol. Pharmacol. 1967;3:113–123. [PubMed] [Google Scholar]
- 17.Ishii Y, et al. J. Pharm. Biomed. Anal. 2007;44:150–159. doi: 10.1016/j.jpba.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 18.Ottoni O, Cruz R, Krammer NH. Tetrahedron Lett. 1999;40:1117–1120. [Google Scholar]
- 19.Osborne AS, Som P, Metcalf JL, Phillips RS. Bioorg. Med. Chem. Lett. 2008;18:5750–5752. doi: 10.1016/j.bmcl.2008.09.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.