Abstract
A taxonomic and annotated functional description of microbial life was deduced from 53 Mb of metagenomic sequence retrieved from a planktonic fraction of the Neotropical high Andean (3,973 meters above sea level) acidic hot spring El Coquito (EC). A classification of unassembled metagenomic reads using different databases showed a high proportion of Gammaproteobacteria and Alphaproteobacteria (in total read affiliation), and through taxonomic affiliation of 16S rRNA gene fragments we observed the presence of Proteobacteria, micro-algae chloroplast and Firmicutes. Reads mapped against the genomes Acidiphilium cryptum JF-5, Legionella pneumophila str. Corby and Acidithiobacillus caldus revealed the presence of transposase-like sequences, potentially involved in horizontal gene transfer. Functional annotation and hierarchical comparison with different datasets obtained by pyrosequencing in different ecosystems showed that the microbial community also contained extensive DNA repair systems, possibly to cope with ultraviolet radiation at such high altitudes. Analysis of genes involved in the nitrogen cycle indicated the presence of dissimilatory nitrate reduction to N2 (narGHI, nirS, norBCDQ and nosZ), associated with Proteobacteria-like sequences. Genes involved in the sulfur cycle (cysDN, cysNC and aprA) indicated adenylsulfate and sulfite production that were affiliated to several bacterial species. In summary, metagenomic sequence data provided insight regarding the structure and possible functions of this hot spring microbial community, describing some groups potentially involved in the nitrogen and sulfur cycling in this environment.
Introduction
The Colombian Andean region is characterized by high volcanic activity, comprising part of the region called the “Ring of Fire”, and is considered a hotspot for biodiversity [1]. This region has several hot springs that represent unique and extreme ecosystems due to their high elevation and exposure to ultraviolet (UV) light. El Coquito (EC) spring is located within the National Natural Park Los Nevados, it has a low pH (2.7) and water temperature of approximately 29°C, which is considerably higher than ambient temperature (∼9°C) and allows us to classify it as a hot spring [2]. A previous analysis of the microbial community at EC hot spring showed that it is dominated by Bacteria rather than Archaea, with predominance of Proteobacteria, Firmicutes and Planctomycetes. The planktonic community contained putative chemotrophic bacteria potentially involved in cycling of ferrousiron and sulfur-containing minerals and phototrophic organisms (mostly eukaryotic micro-algae) [3].
Microbial diversity in hot springs is dictated by environmental physicochemical characteristics (pH, redox potential, temperature and concentration of trace elements) [4]–[6]. In acidic hot springs the most representative genera described are Acidithiobacillus, Acidimicrobium, Sulfobacillus, Thiomonas, Leptospirillum and Hydrogenobaculum [7], [8]. These chemolithotrophic acidophiles are often the predominant primary producers and may also contribute to iron and sulfur cycling via oxidization of reduced inorganic sulfur and ferrous iron compounds [9]. Other acidic hot springs with mesophilic temperature (30–35°C) are dominated by Acidiphilium mesophilic heterotrophs and Acidithiobacillus autotrophic thermotolerant sulfur oxidizers [10]. In extremely acidic and UV light-irradiated environments, primary production may also be mediated by mesophilic phototrophic acidophiles (mainly eukaryotic micro-algae) [11]. Many of these studies have assessed microbial diversity by 16S rRNA gene analysis [12]–[14], which is useful but does not provide information on ecologically relevant genes involved in various biogeochemical cycles.
Metagenomic analyses using high throughput sequencing or library construction have been extremely valuable for describing microbial structure and functionality in extreme ecosystems [15]–[17] and for identifying novel genes [18]–[20]. Comparative metagenomic studies have also characterized microbial communities and shown differences in functionality in several ecosystems [21], [22]. The current and most frequently used tools for taxonomic and functional classification of metagenomic reads are based on local alignments (BLAST) against different databases and associating best hits to taxa, specific genes, functional identifiers or metabolic pathways. However, a more comprehensive picture of the genes and functions in a metagenomic dataset can be obtained using different algorithms, parameters and databases for read assignment [23].
In this work we analyzed the sequences obtained from a metagenome of the EC high mountain (Paramo ecosystem) acidic spring to obtain a deeper view of the genes present and the functional-based structure of the microbial community in the planktonic fraction. The taxonomic and functional profile obtained from metagenomic unassembled reads differed depending on the database used. The microbial community was dominated by Proteobacteria (Gammaproteobacteria and Alphaproteobacteria), with some sequences mapping within the genomes of the acidophiles Acidiphilium cryptum and Acidithiobacillus caldus, a finding that broadens the repertoire of natural environments where these organism are found. A more in depth analysis of the nitrogen and sulfur cycles using KEGG pathways to associate best hits to taxa and specific genes showed some of the processes involved in denitrification, nitrogen fixation, and sulfide oxidation, the latter likely linked to the acidity of the environment.
Materials and Methods
Ethics Statement
The studied locations are in a state owned National Park. The study did not involve endangered or protected species. All necessary permits were obtained for the described field studies from the corresponding national authorities, MAVDT (contract number 15, 2008) for access to genetic resources and UAESPNN (research permit code DTNO-N-20/2007).
Sample collection and processing
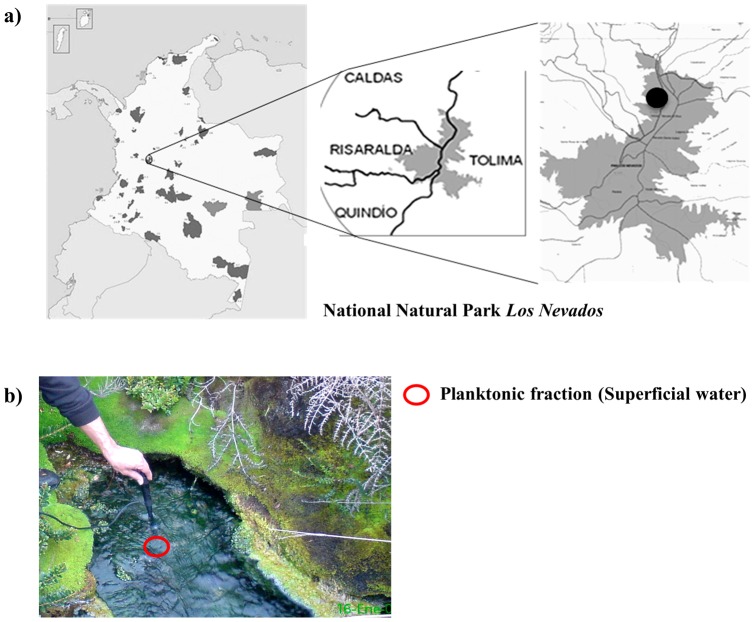
Surface water (15 L) was collected in separate sterile plastic containers for biological and physicochemical analyses in April 2008 (rainy season) at EC hot spring, located at 3,973 meters above sea level (masl) (04°52′27′′ N; 75°15′51.4′′ W) in the National Natural Park Los Nevados (Figure 1a). Due to the difficulty in accessing this location, the samples were kept and transported at 4°C to the laboratory and processed within 18 h for further physicochemical analysis (SO4 2−, Ca2+, Mg2+, Na+, K+, Fe2+, Fe3+ CaCO3+, NO3+, Chloride, PO4+ and total dissolved solids) and DNA isolation [3]. Temperature and pH were recorded in situ using a Hach pH-meter equipped with a pH and temperature probe.
Figure 1. Location of EC hot spring in the National Natural Park Los Nevados.
a) Geographical location. b) Photographs of the acidic hot spring El Coquito (EC), circle indicates the planktonic fraction.
DNA extraction and pyrosequencing
Water (10 L) was filtered through 5.0 µm cellulose filters (Fisherbrand Q5), to remove particles and large cells, and then through 0.22 µm polycarbonate filters (GTTP, Millipore). Planktonic cells on filters were processed to obtain the DNA, as previously described [3]. A total of 20 ng of metagenomic DNA was amplified with pHi29 polymerase using the isothermal multiple displacement amplification (MDA) system (REPLI-g, Qiagen) by incubating at 30°C for 1.5 hrs. This step was monitored for contamination using a negative control tube without DNA. The reaction was stopped by heating at 65°C for 3 min, and the final product was purified using UltraClean GelSpin DNA Extraction Kit (MoBio Laboratories Inc., Carlsbad, CA, USA), resulting in 46.2 µg of DNA (∼1,850 ng/µl). A total of 12.2 µg of metagenomic DNA was used for library preparation using emulsion PCR and pyrosequencing using 454 GS FLX Titanium technology on ½ plate (Engencore, University of South Carolina, Columbia, SC, USA). The total reads obtained (292,559) from a SSF (standard flowgram file) were filtered and trimmed based on length and quality using an in-house python script (www.corpogen.org/tools/clean454.zip). Sequences with a minimum length of 30 bp were evaluated using a sliding window of 20 bp and only those sequences with a recommended quality score of ≥20, were retained [24], [25]. All sequences are available in the metagenomic RAST (MG-RAST) server under project ID 4449206.3.
Taxonomic assignment of metagenomic sequences
The taxonomic assignment of the unassembled metagenomic dataset was performed using BLASTX [26] on MG-RAST v3.0 [27] against the GenBank (NCBI-nr), RefSeq and SEED databases using a cut-off E-value of 1e-10 and minimum alignment of 50 bp. WebCARMA v1.0 online system [28] was also used with previously proposed parameters [23], [24]. SSU rRNA (16S rRNA) reads were extracted from the dataset using a HMMER search against a hidden Markov model built based on multiple sequence alignments [29], aligned using the NAST align tool (batch size for NAST: 5; minimum percentage identity: 75) (http://greengenes.lbl.gov/cgi-bin/nph-NAST_align.cgi), and taxonomically classified using the classification tool for aligned SSU rRNA sequences (http://greengenes.lbl.gov/cgi-bin/nph-classify.cgi), which has been shown to have the highest accuracy for assigning taxonomy to short pyrosequencing reads when compared with other methods [30].
Recruitment plots to draft genomes from acidophilic bacteria
A fragment recruitment plot of the EC hot spring metagenome was performed against the draft microbial genomes of A. cryptum JF-5 (349163.4), Legionella pneumophila str. Corby (400673.6) and A. caldus (33059.1), using BLASTX in the MG-RAST v2.0 platform. The criteria for counting a hit were: cut-off E-value of 1e-5, minimum alignment length of 50 bp and minimum identity of 40%.
Functional analysis using COG and KEGG identifiers
A functional-based classification of EC metagenome was conducted using BLASTX and RPS-BLAST (cut-off E-value of 1e-10) against the COG database [31] downloaded from the NCBI ftp site and the NCBI (nr/nt) local database. Annotation results for BLASTX against the NCBI-nr database were loaded into the MEtaGenome ANalyzer software (MEGAN v4.0) and classified using KEGG identifiers [32], according to suggested parameters for the Lowest Common Ancestor algorithm (LCA) (maximum number of match per read: 5; min support: 5; min score: 35; and top percent: 10) [33].
Metabolic mapping of energy metabolism
The allocation of reads into metabolic maps was obtained using BLASTX (cut-off E-value of 1e-10) against the NCBI-nr database and analyzed with the MEGAN v4.0 software. The metabolic pathways involved in nitrogen and sulfur transformations were recognized using KEGG identifiers. The number of reads in each pathway (oxidative phosphorylation, methane metabolism, nitrogen metabolism, carbon fixation pathways in prokaryotes, and carbon fixation in photosynthetic organism, sulfur metabolism and photosynthesis) were recorded. Extracted sequences were given taxonomic assignments by performing BLASTX against the NCBI-nr and RefSeq databases, and analyzed with the MEGAN v4.0 software and the MG-RAST v3.0 server.
Comparison with other metagenomes
We performed a taxonomic and functional comparison of the EC hot spring metagenome using BLASTX (cut-off E-value of 1e-10) against the RefSeq, COG, KEGG and SEED databases. Comparisons were carried out with different datasets obtained by pyrosequencing in different ecosystems: acid and non-acid mines, coral reefs, marine waters, tropical forest soil, Andean forest acid soil and pristine mangrove sediment (Table 1). A double hierarchical dendrogram was created using the Bray-Curtis distance metric and normalized values in the MG-RAST v3.0 server.
Table 1. Characteristics of datasets used for comparative metagenomic analysis.
| RSM (4440281.3) | BSM (4440282.3) | POCR (4440039.3) | SW (4443702.3) | PO (4443713.3) | TFS (4446153.3) | HAFS (4445417.3) | PMS (4451036.3) | AHEC (4449206.3) | |
| Number of reads | 334,386 | 388,627 | 351,205 | 209,073 | 221,744 | 782,404 | 619,288 | 217,605 | 280,753 |
| Average size read (bp) | 105±17 | 99±16 | 105±17 | 226±60 | 239±55 | 411±103 | 310±118 | 222±107 | 190±95 |
| Total Mbp | 35.5 | 38.5 | 37.0 | 47.2 | 53.0 | 322.2 | 192.3 | 48.5 | 53.5 |
| Mean GC content (%) | 49±12 | 44±10 | 46±10 | 40±9 | 39±9 | 59±6 | 62±7 | 54±10 | 52±10 |
| Taxonomy classified reads (%) a | 0.0068 | 0.038 | 0.021 | 47.5 | 55.3 | 59.4 | 58.7 | 33.5 | 8,7 |
| % KEGG matches b | 0.0095 | 0.055 | 0.037 | 19.8 | 23.1 | 16.9 | 19.5 | 12,0 | 1,8 |
| % SEED matches b | 0.0185 | 0.108 | 0.100 | 47.6 | 55.7 | 37.6 | 43.7 | 29.0 | 4.2 |
| Reference | [39] | [39] | [80] | [81] | Unpublished | [82] | Unpublished | [24] | This study |
RSM: Red Soudan Mine (acidic); BSM: Black Soudan Mine; POCR: Pacific Ocean (coral reefs); SW: Sea Water; PO: Pacific Ocean; TFS: Tropical Forest Soil; HAFS: High Andean Forest Soil; PMS: Pristine Mangrove Sediments; AHEC: Acidic Hot Spring EC. a Using RefSeq database (Bacteria, Archaea, Eukarya and Virus) (cut-off E-value 1e-10); b cut-off E-value 1 e-10.
Results and Discussion
In this study, we carried out a metagenomic analysis (taxonomic and functional-basis assignment of metagenomic unassembled reads) of the planktonic microbial community present in EC hot spring located in the Colombian Andes (Figure 1). The target hot spring is surrounded by endemic vegetation and characterized by an acidic pH (2.7), high solar radiation (approximately 9–11 mW/cm2 nm UV-B) [34], high mineral content (1,003 mg SO4 2− L−1, 320 mg Ca2 L−1, 56.6 mg Cl− L−1, 8.27 mg of total iron L−1 and 45.2 mg Na+ L−1) and high total dissolved solids (2,280 mg L−1). As a complement to a previous 16S rRNA gene analysis [3], and in order to assess the microbial community structure using direct sequencing, the metabolic pathways were evaluated and the major players involved in nitrogen and sulfur transformations were identified. Total metagenomic DNA eluted from the filter was equivalent to 116 ng of bacterial DNA per liter, which could be indicative of a low cell density [35], and is consistent with the low planktonic biomass detected previously [3]. Due to the low amount of recovered metagenomic DNA (580 ng), amplification was performed using phi29 polymerase prior to 454 pyrosequencing, using the appropriate controls (negative) recommended by the manufacturers to diminish the risk of contamination from the starting material [36]. So far this is the most advanced and reliable method available to achieve these descriptions from very limited initial metagenomic DNA [37]. From a total of 292,559 sequences obtained, 280,753 metagenomic reads with an average size of 190±95 bp (equivalent to 53 Mb of DNA) were retained after quality assessment and trimming using an in-house python script. The amount of data falls within what has been reported for some other high throughput sequencing datasets: hot spring (12 Mb) [38], acid mine (35 Mb) [39], canine fecal sample (53 Mb) [21], mangrove sediments (215 Mb) [24] and Antarctic permafrost (388 Mb) [40]. In our dataset most sequences (>90%) had a GC content between 45–60% (mean of 52±10%). An additional 85,409 sequences failed to pass the Quality Control (QC) in MG-RAST pipeline. Of the sequences that passed QC, 28,304 sequences (10.1%) contained predicted proteins with known functions and 131,697 sequences (46.9%) contained predicted proteins of unknown function.
Taxonomic assignment of metagenomic sequences
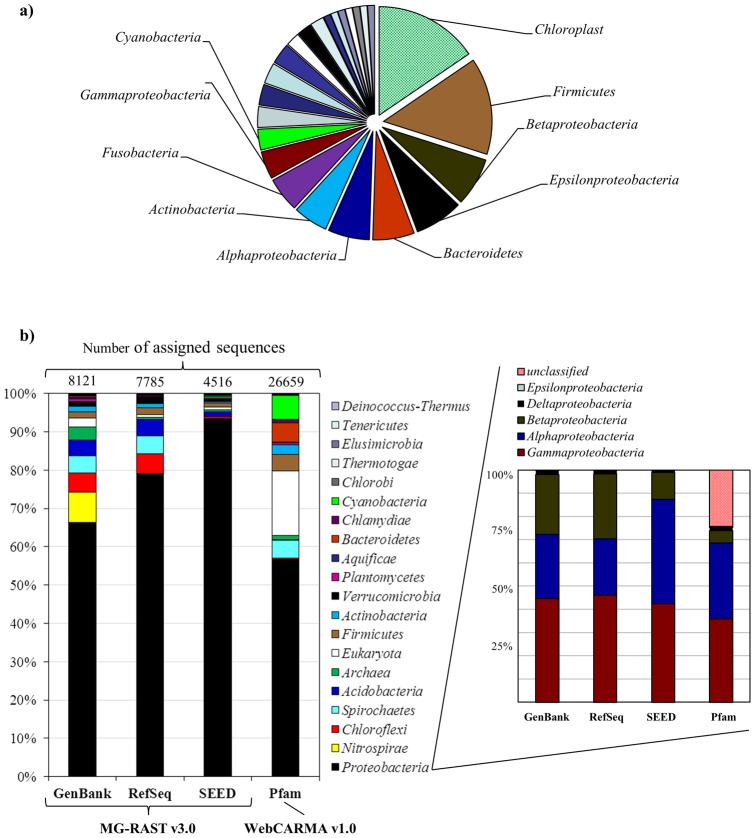
To provide a framework for the metagenomic analyses, we first compared the taxonomic assignments obtained by retrieving SSU rRNA reads and by using BLASTX to classify total reads. Using the hidden Markov model approach, 97 SSU rRNA sequences were identified within the metagenomic dataset. These SSU rRNA sequences, which are free from PCR and cloning biases, showed a prevalence of Proteobacteria (Gammaproteobacteria > Alphaproteobacteria > Betaproteobacteria) (∼25%), followed by micro-algae chloroplast ribosomal DNA (∼15%), Firmicutes (∼14%) and Bacteroidetes (∼6%) (Figure 2a; Supplementary Table S1). These results are consistent with the previous characterization of this site based on clone library and pyrotag sequence data [3]. The low number of SSU sequences recovered (0.03% in our metagenomic dataset) is consistent with other reports [22], [24]. This could be due to drawbacks with sequence assignment using short sequence reads (<100 bp) [41], to secondary structure conformations [42], problems due to low coverage using the 454 platform, absence of assembly or possible MDA biases [43]. These results also highlight the need to develop more refined bioinformatics tools to precisely and efficiently extract sequence reads belonging to the gene family of interest [44], [45].
Figure 2. Classification of metagenomic reads from EC hot spring.
a) Taxonomic affiliation based on 97 SSU rRNA extracted sequences using the Greengenes database; b) Comparison of the taxonomic assignment of metagenomic sequences, based on predicted proteins using the MG-RAST v3.0 server and WebCARMA v1.
In addition to SSU rRNA gene analysis, a taxonomic affiliation was also carried out in the MG-RAST server using all the metagenomic reads to obtain a broad idea of the groups present in this ecosystem. This analysis showed differences in the number of sequences classified depending on the database used: 2.9% (8,121 reads) for the GenBank database, followed by RefSeq with 2.7% (7,785 reads) and SEED with 1.6% (4,516 reads). With WebCARMA (Pfam database) we assigned 26,659 reads (9.4%) (Figure 2b). These low values contrast with the Amazon River metagenome reads classified using BLASTX against NCBI-nr in MG-RAST, 49% of 1.1 million pyrosequencing reads [46]. These low values indicate that the EC hot spring could contain a great amount of newly described sequences, as suggested by the high number of unclassified 16S rRNA pyrotags reported previously [3], or could be explained by a large amount of noncoding DNA present in the genomes (especially in micro-Eukaryotes). A major bias using this approach is that the current databases contain mostly sequences from cultivable microorganisms. In addition, the accuracy also depends on the representation of the different taxonomic groups in a database [15]. Thus, up to 90% of the sequences in a metagenomic dataset may remain unidentified due to the lack of reference sequences [47]. Hopefully, sequence analysis and taxonomic affiliation will improve as more genome sequencing projects become available and bioinformatics software tools are developed [48]–[50].
The predominant phylum in all databases, detected by total read assignment using BLASTX, was Proteobacteria (∼76%). This was followed by Nitrospirae (10%), which was assigned only in the GenBank database, Chloroflexi, Spirochaetes and Acidobacteria (Figure 2b). The highest percentage of sequences assigned to Eukarya (13%) was obtained using WebCARMA, which also assigned 7% of the sequences to the phylum Cyanobacteria (Figure 2b). These latter sequences most probably correspond to micro-algae, as it was observed previously for this ecosystem [3], or could be indicating that this group of oxygenic phototrophs could be present both in planktonic and in mat communities [51]. The fact that few phototrophic bacteria (Chlorobi, Cyanobacteria, and Chloroflexi) were detected in EC hot spring, both in this and the previous study based on 16S rRNA analysis [3], is consistent with the notion that Cyanobacteria are sensitive to metals and solutes found in acidic waters [52]. The eukaryotic micro-algae detected might play important roles in primary production by using solar energy at the surface, similar to what occurs in surface acid streamers and other acidic extreme environments [53]. A Pearson analysis showed limited correlation (0.73) between taxonomic affiliation based on SSU rRNA (Greengenes database) and total metagenomic read assignment (RefSeq database), which could be partially explained by the fact that most of phylotypes identified by SSU rRNA sequences belonged to linages for which genome sequences are not yet available [44], [54], or by variation in SSU rRNA copies and/or genome size [55].
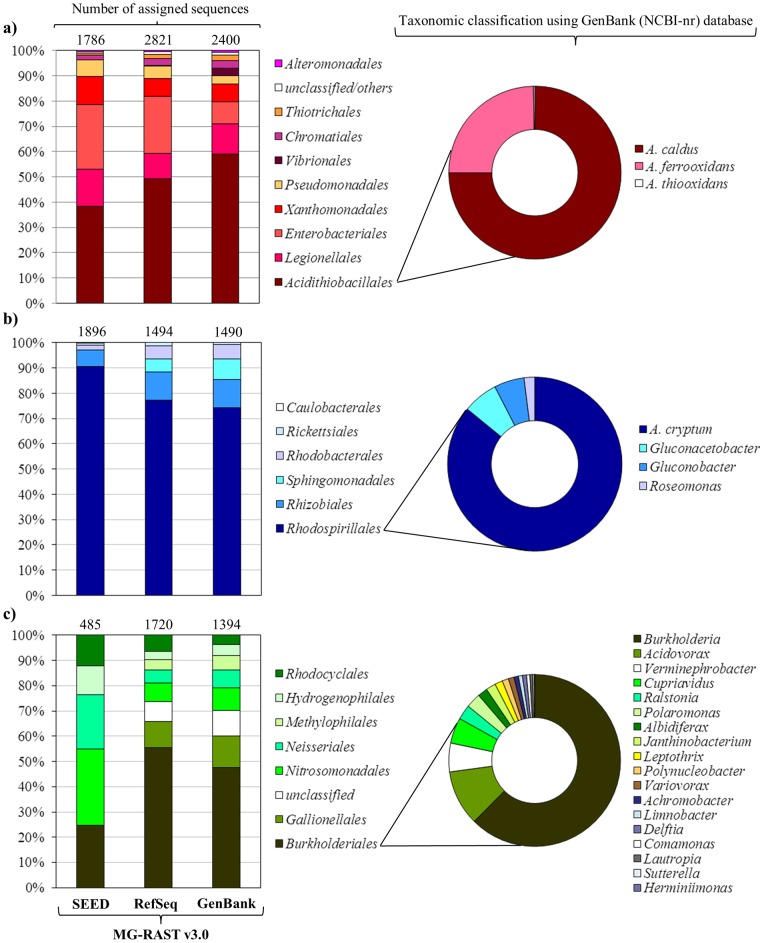
Gammaproteobacteria was the dominant annotated class (∼40%) in all databases (based on predicted proteins). Using the RefSeq and GenBank databases we observed a high number of sequences related to Acidithiobacillales (∼54%) (represented by sequences related to Acidithiobacillus caldus, Acidithiobacillus ferroxidans and Acidithiobacillus thiooxidans), followed by Legionellales (Figure 3a). Acidithiobacillus, which have been found in extremely acidophilic sulfur-oxidizing biofilms “snottites”, can grow on reduced inorganic sulfur and iron compounds as the energy sources [56] and oxidize sulfur via sulfide–quinone reductase and the sox pathway [17]. In other environments, these microorganisms play an important role in sulfur mobilization by catalyzing metal sulfide oxidation at low pH and by releasing sulfate into solution [57]. Several species of infectious and non-infectious Legionella have been found using molecular techniques in hot springs, acid mines and rivers [58]–[61]. Alphaproteobacteria was the second most dominant class (∼30%), with sequences related to the order Rhodospirillales (∼80%) (Figure 3b) that included A. cryptum (1,681 and 1,017 assigned reads using SEED and GenBank, respectively). Acidiphilium, one of the most abundant and versatile genera found in acid mine drainage [60], represents autotrophic and heterotrophic, sulfur oxidizing mesophilic bacteria able to grow at pH between 2.5–6.0 [62]. Acidiphilium species can also carry out photosynthesis using Zn-BChl (photopigments) [63], which may be advantageous for growth and survival in oligotrophic environments. Sequences assigned to Betaproteobacteria (∼20%) included the orders Burkholderiales (between 25–55%), reported in warm and hot acid-sulfate springs [64], [65], followed by Gallionellales, Nitrosomonadales, Rhodocyclales and Hydrogenophilales (Figure 3c). Despite the short length of reads and the possibility of compositional bias associated with sample handling, these results provide a general view of the taxa involved and, interestingly, are consistent with the previous study of the diversity in this ecosystem based on SSU rRNA analysis, which showed the predominance of the orders Burkholderiales, Rhodocyclales, Legionellales, Rhodospirillales, Clostridiales, Plantomycetales and Nitrospirales [3].
Figure 3. Taxonomic comparison within Proteobacteria of total metagenomic sequences based on predicted proteins using BLASTX against different databases in MG-RAST v3.0 server.
a) Gammaproteobacteria b) Alphaproteobacteria c) Betaproteobacteria.
Comparison with draft genomes from acidophilic bacteria
The reads for predicted proteins in our dataset were next compared to genomes belonging to taxa that contained a large number of affiliated reads, such as A. cryptum, A. caldus and Legionella sp. (Figure 3). Of the total metagenomic reads compared to the A. cryptum JF-5 genome, 1,063 reads were mapped to 312 of 3,612 features (putative genes) (Supplementary Table S2). Some of the genes that mapped in highest proportion and with lowest E-value (less than 1e-50) encoded enzymes involved in transposition and integration of mobile genetic elements (transposases). Genes that encoded phage portal proteins were identified, indicating the presence of A. cryptum phages or prophages, as has been found in the Amazon River metagenome [46] and in acidophilic microbial communities [66]. The presence of genes potentially involved in lateral gene transfer might indicate that horizontal exchange of genetic material could be a common occurrence in EC hot spring, as has been suggested for hydrothermal chimneys [16], [67]. Other sequences that mapped to A. cryptum genes were involved in energy production, amino sugar metabolism, fatty acid biosynthesis, RNA synthesis, as well as ATP carrier proteins, ABC transporters, hydrolytic enzymes and hypothetical proteins. The metagenomic reads were also compared with the L. pneumophila and the A. caldus genomes. Of the total metagenomic reads, 624 mapped to 447 out of 3,423 features from the L. pneumophila str. Corby genome, and 365 reads mapped into the A. caldus genome (Supplementary Table S2). In the L. pneumophila genome sequences mapped to genes associated with various processes such as posttranslational modification and RNA synthesis, among others, and to a sequence that encodes a helicase responsible for regulating RNA synthesis at low temperatures [68]. Finally, in the A. caldus genome, we observed sequences associated with transposases (as occurred for A. cryptum) and histone-like mobilization proteins.
Functional analysis using COG, KEGG and SEED identifiers
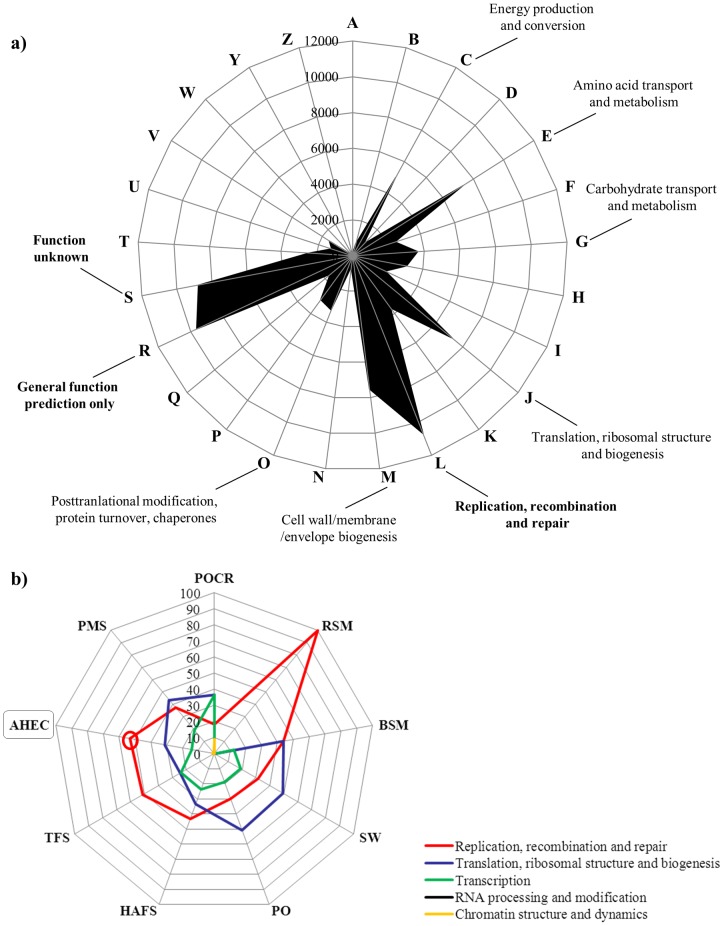
The proportion of matches to the COG database exceeded the KEGG and SEED matches, similar to previous reports [22], [24]. A total of 87,023 reads (30.9%) were assigned to 25 COG categories and most of the sequences were related to replication, recombination and repair (L) (10,712 reads, ∼12%) (Figure 4a). Some of the other COG categories identified included general function prediction only (R) (9,637 reads), function unknown (S) (8,777 reads) and energy production and conversion (C) (5,001 reads) (Figure 4a). Using BLASTX against the NCBI-nr database and the MEGAN software, 19,876 sequences (∼7.0%) were associated with KEGG pathways, specifically to metabolism of carbohydrates (2,623), amino acids (2,584), energy (1,920) and nucleotides (1,431) (Supplementary Figure S1). Some functions were found using both COG and KEGG identifiers, such as DNA repair, translation, transcription, replication, homologous recombination and carbohydrate and energy metabolism while others, such as photosynthesis, regulation and cell signaling, metabolism (protein, sulfur, RNA), carbohydrates and respiration were identified using the SEED database (Supplementary Figure S2b). Even though there might be biases associated with the sampling strategy and with MDA, suggesting that these functions may be important in this ecosystem. The presence of recombination, replication and repair systems, that were evident in the COG analysis and by comparison with other metagenomes (Figure 4b), could also be important in this ecosystem where high UV radiation (at ∼4000 masl), acidic pH and high water temperature may cause significant damage to DNA. Reports of enrichment for genes involved in mismatch DNA repair and homologous recombination in deep sea hydrothermal vent chimneys and hot springs suggest that the microbial communities have evolved extensive DNA repair systems to cope with extreme conditions that have potential deleterious effects on their genomes [51], [16]. Finally, in this study we also identified sequences associated with quorum sensing and cellular communication in biofilms, which could form on the surface of the EC hot spring (Figure 1b).
Figure 4. Functional assignment using BLASTX against the COG database.
a) Analysis of metagenomic sequences obtained from EC hot spring, the numbers indicate the amount of sequences affiliated to the predominant COG identifiers. b) Comparison of various metagenomes (RSM: Red Soudan Mine; BSM: Black Soudan Mine; POCR: Pacific Ocean (coral reefs); SW: Sea Water; PO: Pacific Ocean; TFS: Tropical Forest Soil; HAFS: High Andean Forest Soil; PMS: Pristine Mangrove Sediments; AHEC: Acidic Hot Spring EC) using the percentage of annotated reads in the specified COG categories.
Energy metabolism and mapping of nitrogen and sulfur transformations
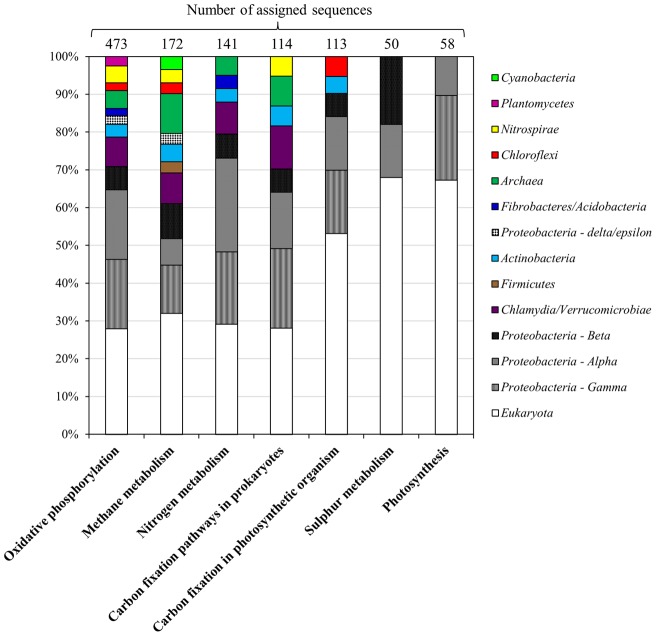
To gain more insight regarding possible functions in this community, we looked at reads associated with different metabolic pathways. A total of 1,920 reads were mapped to energy metabolism using BLASTX against the NCBI-nr database, and corresponded to both Eukarya and Bacteria. Most of the bacterial reads were assigned to Alphaproteobacteria, Gammaproteobacteria and Betaproteobacteria. Sequences involved in various pathways, such as oxidative phosphorylation, methane, nitrogen, and carbon fixation, were associated with Chlamydia/Verrucomicrobia, Actinobacteria, Nitrospirae and Archaea. Also, reads associated with Planctomycetes, Cyanobacteria and Chloroflexi were observed in oxidative phosphorylation, methane metabolism and carbon fixation in photosynthetic organisms, respectively (Figure 5).
Figure 5. Taxonomic assignment of metagenomic reads obtained from EC hot spring related to energy metabolism (KEGG identifiers).
The taxonomic affiliation was performed by BLASTX against NCBI-nr and analyzed using the MEGAN v 4.0 (LCA algorithm), which provides phylogenetic classification at different levels, depending on the sequence read. Classification is shown at the kingdom level for Archaea and Eukaryota; the phyla shown are from Bacteria and only for Proteobacteria were reads classified to the class level: Alpha (Alphaproteobacteria), Beta (Betaproteobacteria), Gamma (Gammaproteobacteria), delta/epsilon (Deltaproteobacteria/Epsilonproteobacteria).
We also looked more closely at pathways involved in nitrogen and sulfur metabolism, since these may be important in habitats where terminal electron acceptors other than O2 may be important, such as nitrate, ferric iron, arsenate, thiosulfate, elemental S, sulfate or CO2. A total of 268 and 127 sequences were associated with nitrogen and sulfur metabolism, respectively. Approximately 80% of the genes associated with the nitrogen cycle were related to Alphaproteobacteria, Gammaproteobacteria, Betaproteobacteria and Eukarya. A detailed analysis using the MEGAN software showed the presence of narGHI genes (14 reads) (dissimilatory reduction of nitrate) (Table 2; Supplementary Figure S3a). Based on current models of dissimilatory nitrate reduction in bacteria, a nitrite reductase (nirK or nirS) would be required to produce NO, which serves as a substrate for nitric oxide reductase (norB) to produce N2O [38]. Several sequences from EC spring were associated with nirS and norBDQ, with nosZ (associated with magnetotactic bacteria), which is important for the conversion of N2O to N2, and 13 sequences were associated with ferredoxin-nitrite reductase (nirA). The balance among these pathways is influenced greatly by environmental conditions, such as temperature, pH, oxygen, nitrate availability, and organic matter content. Finally, we also identified nifK (associated with sulfate reducing Thermodesulfovibrio and sulfur reducing bacteria Desulfitobacterium), a gene involved in the synthesis of molybdenum dependent (Mo-dependent) nitrogenase, suggesting that in addition to denitrification, nitrogen fixation could also be taking place in EC hot spring [69], [70] (Table 2; Supplementary Figure S3a). Enzymes such as ammonium monooxygenase subunit A and glutamine synthetase were not detected in our dataset, due either to the low depth of sequencing achieved or to the fact that they may not be present in EC hot spring, similar to what has been reported previously in Yellowstone hot springs [38]. Based on the taxonomic affiliation of the identified reads, we propose that dissimilatory nitrate reduction is most likely carried out by Proteobacteria-like organisms and assimilatory reduction of nitrate is carried out mostly by acidophilic micro-algae, Acidobacteria, Spartobacteria and Alphaproteobacteria (Table 2; Supplementary Figure S3a).
Table 2. Sequences associated with specific functions and taxa within the nitrogen cycle using KEGG pathways.
| Number of reads assigned | E.C Number | Orthology KEGG | Enzyme name | Gene | Organism | Class | E-value c |
| 14 | 1.7.99.4 | K00370 | nitrate reductase 1, alpha subunit | narG | Pseudomonas stutzeri | Gammaproteobacteria | 1 e–25 |
| Thiomonas itermedia | Betaproteobacteria | 4 e–41 | |||||
| Desulfococcus oleovorans | Deltaproteobacteria | 8 e–27 | |||||
| K00371 | nitrate reductase 1, beta subunit | narH | Thiomonas sp. | Betaproteobacteria | 6 e–35 | ||
| Oligotropha carboxidovorans | Alphaproteobacteria | 1 e–68 | |||||
| 1 e–26 | |||||||
| K00374 | nitrate reductase 1, gamma subunit | narI | Pseudomonas fluorescens | Gammaproteobacteria | 1 e–12 | ||
| 4 | 1.3.12.16 | K00459 | nitronate monooxygenase | Candidatus Koribacter versatilis | Acidobacteria | 7 e–16 | |
| Legionella pneumophila | Gammaproteobacteria | 6 e–18 | |||||
| 10 | 1.7.99.7 | K04561 | nitric-oxide reductase, cytochrome b-containing subunit I | norB | Legionella longbeachae | Gammaproteobacteria | 1 e–16 |
| Legionella pneumophila | 2 e–20 | ||||||
| 2 e–14 | |||||||
| K02164 | nitric-oxide reductase NorE protein | norE | Agrobacterium tumefaciens | Alphaproteobacteria | 8 e–46 | ||
| K02305 | nitric-oxide reductase, cytochrome c-containing subunit II | norC | Bradyrhizobium sp. | Alphaproteobacteria | 1 e–14 | ||
| K04748 | nitric-oxide reductase NorQ protein | norQ | Thiobacillus denitrificans | Betaproteobacteria | 1 e–54 | ||
| 2 | 1.7.99.6 | K00376 | nitrous-oxide reductase | nosZ | Magnetospirillum magneticum | Alphaproteobacteria | 6 e–28 |
| Cardiobacterium hominis | Gammaproteobacteria | 3 e–15 | |||||
| 7 | 1.18.6.1 | K02586 | nitrogenase molybdenum- iron protein alpha chain | nifD | Bradyrhizobium sp. | Alphaproteobacteria | 7 e–24 |
| K02591 | nitrogenase molybdenum- iron protein beta chain | nifK | Desulfitobacterium hafniense | Firmicutes | 8 e–31 | ||
| Thermodesulfovibrio yellowstonii | Nitrospira | 3 e–18 | |||||
| 4 | 1.7.1.4 | K00362 | nitrite reductase (NAD(P)H) large subunit | nirB * | Sphigomonas sp. | Alphaproteobacteria | 1 e–18 |
| 7 | 1.7.7.1 | K00366 | ferredoxin-nitrite reductase | nirA * | Candidatus Koribacter versatilis | Acidobacteria | 1 e–18 |
| Acidobacterium capsulatum | Acidobacteria | 7 e–26 | |||||
| Pseudochlorella sp. a | Trebouxiophyceae | 3 e–15 | |||||
| Thalassiosira pseudonana b | Coscinodiscophyceae | 3 e–15 | |||||
| Chthoniobacter flavus | Spartobacteria | 2 e–18 | |||||
| 2 | 1.7.2.1 | K00368 | nitrite reductase | nirS | Nitrosococcus halophilus | Gammaproteobacteria | 5 e–36 |
assimilatory nitrate reduction; a micro-algae; b diatom: c (cut-off E-value 1e -10).
Most of the genes involved in the sulfur cycle were related to the conversion of sulfate into adenylylsulfate and to the further generation of sulfite and H2S (Supplementary Figure S3b), although, we also observed sequences related to serine O-acetyltransferase production. Genes were identified for cysteine synthase AB (cysKM) and for formation of adenylysulfate (Table 3; Supplementary Figure S3b). We also detected a bi-functional enzyme (sulfate adenylytransferase and adenylysulfate kinase) (cysNC). In dissimilatory sulfate reduction and sulfur oxidation, adenosine-5′-phosphosulfate (APS) reductase (Apr) is considered a key enzyme. In the sulfate-reducing pathway, sulfate has to be activated to APS by ATP-sulfurylase at the expense of ATP, Apr converts the APS to sulfite, and then, sulfite is reduced to sulfide by dissimilatory sulfite reductase (Dsr) [71]. The alpha subunits of Apr enzymes are found in all known sulfate reducing and most of sulfur oxidizing prokaryotes. We detected genes involved in conversion of adenylylsulfate to sulfite (aprAB; cysH), in sulfite reduction and H2S formation (cysI), and in the oxidation of sulfite to sulfate (sulfite oxidase enzyme) (Table 3; Supplementary Figure S3b).
Table 3. Sequences associated with specific functions and taxa within the sulfur cycle using KEGG pathways.
| Number of reads assigned | E.C Number | Orthology KEGG | Enzyme name | Gene | Organism | Class | E-value e |
| 8 | 2.3.1.30 | K00640 | serine O-acetyltransferase | cysE | Gemmata obscuriglobus | Planctomycetacia | 6 e–36 |
| Bryantella formatexigens | Clostridia | 1 e–32 | |||||
| 37 | 2.7.7.4 | K00956 | sulfate adenylyltransferase subunit 1 | cysN | Laribacter hongkongensis | Betaproteobacteria | 2 e–15 |
| Stigmatella aurantiaca | Deltaproteobacteria | 6 e–11 | |||||
| Bacterium Ellin514 | Verrucomicrobiae | 3 e–11 | |||||
| K00957 | sulfate adenylyltransferase subunit 2 | cysD | Clostridium cellulovorans | Clostridia | 2 e–24 | ||
| Uncultured Archeon | Archaea | 2 e–18 | |||||
| Rhodomicrobium vannielii | Alphaproteobacteria | 4 e–10 | |||||
| Opitutaceae bacterium | Opitutae | 6 e–20 | |||||
| Marinobacter aquaeolei | Gammaproteobacteria | 2 e–19 | |||||
| Clostridium cellulovorans | Clostridia | 7 e–16 | |||||
| K00958 | sulfate adenylyltransferase | cysDN | Leptospirillum ferrodiazotrophum | Nitrosospira | 1 e–75 | ||
| Micromonas pusilla a | Prasinophyceae | 2 e–21 | |||||
| Planctomyces maris | Planctomycetacia | 4 e–29 | |||||
| Thialkalivibrio sp. | Gammaproteobacteria | 5 e–26 | |||||
| Rhodothermus marinus | Sphingobacteria | 5 e–23 | |||||
| Silicibacter lacuscaerulensis | Alphaproteobacteria | 2 e–17 | |||||
| Syntrophobacter fumaroxidans | Deltaproteobacteria | 2 e–18 | |||||
| Thiobacilllus denitrificans | Betaproteobacteria | 6 e–15 | |||||
| K00955 | bifunctional enzyme CysN/CysC | cysNC | Acidiphilium cryptum | Alphaproteobacteria | 7 e–72 | ||
| Xenorhabdus nematophila | Gammaproteobacteria | 6 e–41 | |||||
| 8 | 1.8.3.1 | K00387 | sulfite oxidase * | X | Phaeodactylum tricornutum b | Bacillariophyceae | 1 e–14 |
| Nematostella vectensis c | Anthozoa | 8 e–15 | |||||
| 9 | 1.8.99.2 | K00395 | adenylylsulfate reductase, subunit B | aprB | Thiobacillus aquaesulis | Betaproteobacteria | 3 e–32 |
| Thermodesulfovibrio yellowstonii | Nitrosospira | 5 e–45 | |||||
| K00394 | adenylylsulfate reductase, subunit A | aprA | Thiobacilllus denitrificans | Betaproteobacteria | 9 e–24 | ||
| Desulfirivibrio alkaliphilus | Deltaproteobacteria | 3 e–14 | |||||
| Thiobacilllus denitrificans | Betaproteobacteria | 2 e–22 | |||||
| 13 | 2.7.1.25 | K00955 | bifunctional enzyme CysN/CysC | cysNC | Acidiphilium cryptum | Alphaproteobacteria | 7 e–72 |
| Xenorhabdus nematophila | Gammaproteobacteria | 6 e–41 | |||||
| K00860 | adenylylsulfate kinase | cysC | Geobacter sp. | Deltaproteobacteria | 7 e–15 | ||
| Burkholderia sp. | Betaproteobacteria | 3 e–15 | |||||
| Chloroflexus sp. | Chloroflexi | 8 e–22 | |||||
| Chlamydomonas reinhardtii a | Chlorophyceae | 1 e–24 | |||||
| 4 e–33 | |||||||
| 1 e–32 | |||||||
| 23 | 2.5.1.47 | K12339 | cysteine synthase B | cysM | Thioalkalivibrio sp. | Gammaproteobacteria | 5 e–16 |
| Streptomyces clavuligerus | Actinobacteria | 4 e–24 | |||||
| K01738 | cysteine synthase A | cysK | Desulfirivibrio alkaliphilus | Deltaproteobacteria | 3 e–12 | ||
| Chlamydomonas reinhardtii a | Chlorophyceae | 2 e–13 | |||||
| Thalassiosira pseudonana b | Coscinodiscophyceae | 5 e–16 | |||||
| Gemmata obscuriglobus | Planctomycetacia | 3 e–12 | |||||
| Chlamydomonas reinhardtii a | Cholorophyceae | 5 e–22 | |||||
| Polytomella parva a | Cholorophyceae | 1 e–15 | |||||
| Symbiobacterium thermophilum | Clostridia | 2 e–15 | |||||
| 2 e–15 | |||||||
| 4 | 1.8.4.8 | K00390 | phosphoadenosine phosphosulfate reductase | cysH | Acidobacterium capsulatum | Acidobacteria | 4 e–22 |
| 10 | 1.8.1.2 | K00381 | sulfite reductase (NADPH) hemoprotein beta-component | cysI | Haliangium achraceum | Deltaproteobacteria | 2 e–24 |
| Azoarcus sp. | Betaproteobacteria | 5 e–30 | |||||
| Citomicrobium bathyomarinum | Alphaproteobacteria | 1 e–32 | |||||
| K00380 | sulfite reductase (NADPH) flavoprotein alpha-component | cysJ | Chlamydomonas reinhardtii a | Chlorophyceae | 1 e–17 | ||
| 3 | 1.8.7.1 | K00392 | sulfite reductase (ferredoxin) | sir | Physcomitrella patens d | Embryophyta | 1 e–13 |
| Micromonas pusilla a | Prasinophyceae | 2 e–12 |
sulfur oxidation; a micro-algae; b diatom; c sea anemone; d moss; e (cut-off E-value 1e -10).
Comparison with other metagenomes
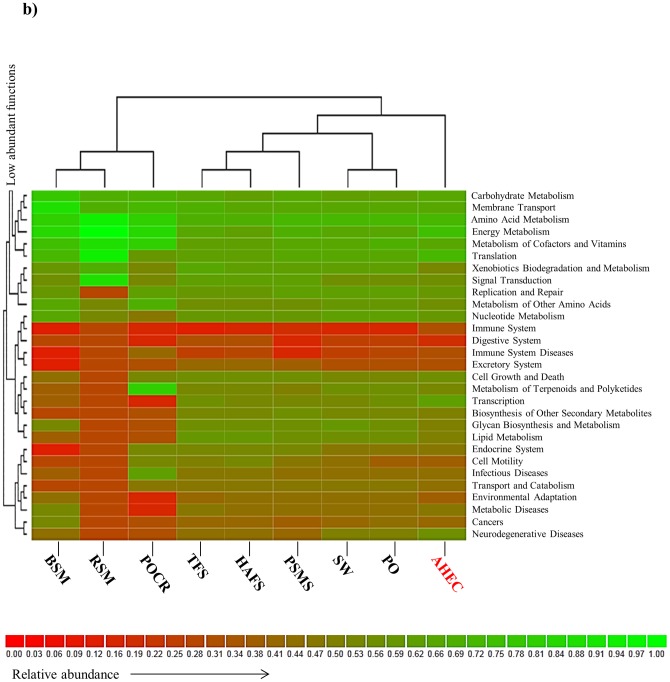
In order to uncover unique features and highlight relevant taxa or possible microbial functions of the EC metagenome, we compared the dataset from EC hot spring against a collection of selected metagenomes obtained by pyrosequencing (to minimize potential biases because of differences in sequence length) (Table 1). The taxonomic composition of the nine metagenomes done using BLASTX against the RefSeq database, showed differences at the phylum level and a closer association of EC hot spring with ocean metagenomes (except coral reefs) (Supplementary Figure S2a). This result was consistent with a hierarchical functional comparison based on KEGG and SEED identifiers (Figure 6; Supplementary Figure S2b). In soil samples and in mangrove sediments, Acidobacteria, Verrucomicrobia and Planctomycetes were found in a high relative proportion. Interestingly, EC hot spring had a higher proportion of sequences related with Streptophyta, Bacillariophyta, unclassified sequences derived from Eukarya, Cyanobacteria, Chloroflexi, Spirochaetes, Acidobacteria, Euryarchaeota and Nitrospirae, when compared with other metagenomes (Figure 6a). In particular, the metagenome from EC differed in processes related to energy metabolism, translation, photosynthesis, cell signaling and replication-repair (Figure 6b; Figure 4b).
Figure 6. Functional clustering based on total reads of various metagenomes (RSM: Red Soudan Mine; BSM: Black Soudan Mine; POCR: Pacific Ocean (coral reefs); SW: Sea Water; PO: Pacific Ocean; TFS: Tropical Forest Soil; HAFS: High Andean Forest Soil; PMS: Pristine Mangrove Sediments; AHEC: Acidic Hot Spring EC).
The data was compared to KEGG databases. Dendrogram linkages are based on relative abundance of the metabolic identifiers (KEGG database) within the samples.
Concluding remarks and future perspectives
In this metagenomic study, the taxonomic and functional features (metabolic pathways) of the microbial community present in a Colombian acidic hot spring EC were analyzed. Unexplored and extreme ecosystems such as this have been previously shown to be of interest as a source of new biotechnological products [72], [73], species and ecological features (as homologous recombination in speciation processes) [74]–[76]. Only a small proportion of the sequences in EC hot spring had matches against the available databases, suggesting that there is a high proportion of novel DNA sequences, consistent with analysis of extreme environments using pyrosequencing data and possibly due to a high proportion of novel taxa, as suggested by previous analysis based on 16S rRNA pyrotags [3], or to the limited length of reads and sequence depth achieved. The taxa identified both by classification based on SSU read affiliation and total read assignment (annotated portion of the microbial community), such as Acidiphilium, Burkholderia, Acidovorax, Acidithiobacillus, Nitrosospira, Legionella, Thiobacillus, Desulfitobacterium and acidophilic micro-algae were comparable to those identified based on PCR amplification of 16S rRNA genes [3]. The annotated portion of the microbial community indicated the presence of DNA repair systems that may be involved in homologous recombination and adaptation processes to extreme environments. The identification of genes coding for nitrogen and sulfur cycling indicate a population involved in dissimilatory-assimilatory reduction of nitrate, and conversion of sulfate into adenylylsulfate and sulfite. Future studies will target the comparison between metatrascriptomic and metagenomic analysis [77]–[79], structure dynamics of microbial communities (especially micro-eukaryotes), and analysis of other pathways or ecological processes, like the carbon cycle, photosynthesis and the oxidation or reduction of iron, which can lead to further understanding of these communities. Overall, this sequence-based exploration of the metagenomic content in an Andean hot spring goes beyond the identification of taxa using 16S rRNA gene analysis and provides insight into both taxonomical composition and metabolic potential. However a greater depth of sequencing will be required to more fully assess the functional diversity present in this ecosystem, the association of metabolic routes with particular taxa and their relevance to community dynamics.
Supporting Information
Functional assignment of metagenomic sequences using BLASTX against NCBI-nr. The data was analyzed using MEGAN v4.0 software and KEGG identifiers.
(PPT)
Taxonomic and functional clustering based on total reads of various metagenomes (RSM: Red Soudan Mine; BSM: Black Soudan Mine; POCR: Pacific Ocean (coral reefs); SW: Sea Water; PO: Pacific Ocean; TFS: Tropical Forest Soil; HAFS: High Andean Forest Soil; PMS: Pristine Mangrove Sediments; AHEC: Acidic Hot Spring EC). The data were compared to a) RefSeq and b) SEED databases. Dendrogram linkages are based on relative abundance of the phylum level (RefSeq) and relative abundance of the metabolic identifiers (Subsystems, SEED database) within the samples.
(PPT)
Partial a) nitrogen and b) sulfur pathways identified by KEGG affiliation of the sequences from EC hot spring. Boxes indicate the KEGG characteristic identified and numbers in gray circles indicate the amount of sequence reads affiliated to the KEGG function.
(PPT)
Phylogenetic affiliation of SSU rRNA sequences. The aligned reads were classified taxonomically using “classify a batch of sequences against multiple taxonomies tool” from Greengenes.
(XLS)
Metagenomic reads obtained from EC hot spring that mapped to the A. cryptum JF-5, L. pneumophila str. Corby and A. caldus draft genomes.
(XLS)
Acknowledgments
We thank G. Lopez, L. Bohorquez and L. Delgado for help with sampling and J.R. Bustos for technical assistance. This research was done under permits from MAVDT (contract number 15, 2008) for access to genetic resources) and UAESPNN (research permit code DTNO-N-20/2007).
Funding Statement
This research was supported by COLCIENCIAS – SENA (project number 6570-392-19990). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403: 853–858. [DOI] [PubMed] [Google Scholar]
- 2. Rzonca B, Schulze-Makuch D (2003) Correlation between microbiological and chemical parameters of some hydrothermal springs in New Mexico, USA. J Hidrol 280: 272–284. [Google Scholar]
- 3. Bohórquez LC, Delgado-Serrano L, López G, Osorio-Forero C, Klepac-Ceraj V, et al. (2012) In-depth characterization via complementing culture-independent approaches of the microbial community in an acidic hot spring of the Colombian Andes. Microb Ecol 63: 103–115. [DOI] [PubMed] [Google Scholar]
- 4. Siering PL, Clarke JM, Wilson MS (2006) Geochemical and biological diversity of acidic, hot springs in Lassen volcanic National Park. Geomicrobiol J 23 (2): 129–141. [Google Scholar]
- 5. Mathur J, Bizzoco RW, Ellis DG, Lipson DA, Poole AW, et al. (2007) Effects of abiotic factors on the phylogenetic diversity of bacterial communities in acidic thermal springs. Appl Environ Microbiol 73 (8): 2612–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lau MCY, Aitchison JC, Pointing SB (2009) Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles 13: 139–149. [DOI] [PubMed] [Google Scholar]
- 7.Norris PR (2001) Acidophiles. Wiley J and Sons (eds). In: Encyclopedia of life sciences. http://els.net, 1–6. Accessed 11 November 2011. doi: 10.1038/npg.els.0000336.
- 8. Stout LM, Blake RE, Greenwood JP, Martini AM, Rose EC (2009) Microbial diversity of boron-rich volcanic hot springs of St. Lucia, Lesser Antilles. FEMS Microbiol Ecol 70 (3): 402–412. [DOI] [PubMed] [Google Scholar]
- 9. Hamamura N, Olson SH, Ward DM, Inskeep WP (2005) Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at rainbow springs, Yellowstone National Park. Appl Environ Microbiol 71 (10): 5943–5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burton NP, Norris PR (2000) Microbiology of acidic, geothermal springs of Montserrat: environmental rDNA analysis. Extremophiles 4 (5): 315–320. [DOI] [PubMed] [Google Scholar]
- 11. Aguilera A, Souza-Egipsy V, González-Toril E, Rendueles O, Amils R (2010) Eukaryotic microbial diversity of phototrophic microbial mats in two Icelandic geothermal hot springs. Int Microbiol 13 (1): 21–32. [DOI] [PubMed] [Google Scholar]
- 12. Tomova I, Dimitrina-Lyutskanova M, Pascual J, Petrov P, Kambourova M (2010) Phylogenetic analysis of the bacterial community in a geothermal spring, Rupi Basin, Bulgaria. World J Microbiol Biotechnol 26 (11): 2019–2028. [Google Scholar]
- 13. Mirete S, De Figueras CG, González-Pastor JE (2011) Diversity of Archaea in Icelandic hot springs based on 16S rRNA and chaperonin genes. FEMS Microbiol Ecol 77 (1): 165–175. [DOI] [PubMed] [Google Scholar]
- 14. Kato S, Itoh T, Yamagishi A (2011) Archaeal diversity in a terrestrial acidic spring field revealed by a novel PCR primer targeting archaeal 16S rRNA genes. FEMS Microbiol Lett 319 (1): 34–43. [DOI] [PubMed] [Google Scholar]
- 15. Simon C, Wiezer A, Strittmatter AW, Daniel R (2009) Phylogenetic diversity and metabolic potential revealed in a glacier ice metagenome. Appl Environ Microbiol 75 (23): 7519–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xie W, Wang F, Guo L, Chen Z, Sievert SM, et al. (2011) Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J 5: 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones DS, Albrecht HL, Dawson KS, Schaperdoth I, Freeman KH, et al. (2012) Community genomic analysis of an extremely acidophilic sulfur-oxidizing biofilm. ISME J 6: 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tirawongsaroj P, Sriprang R, Harnpicharnchai P, Thongaram T, Champreda V, et al. (2008) Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol 133: 42–49. [DOI] [PubMed] [Google Scholar]
- 19. Steele HE, Jaeger JE, Daniel R, Streit WR (2009) Advances in recovery of novel biocatalysts from metagenomes. J Mol Microbiol Biotechnol 16 (1–2): 25–37. [DOI] [PubMed] [Google Scholar]
- 20. Jiménez DJ, Montaña JS, Alvarez D, Baena S (2012) A novel cold active esterase derived from high Andean forest soil metagenome. World J Microbiol Biotechnol 28: 361–370. [DOI] [PubMed] [Google Scholar]
- 21. Swanson KS, Dowd SE, Suchodolski JS, Middelbos IS, Vester BM, et al. (2011) Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J 5: 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quaiser A, Zivanovic Y, Moreira D, Lopez-Garcia P (2011) Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J 5 (2): 285–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Montaña JS, Jiménez DJ, Hernández M, Ángel T, Baena S (2012) Taxonomic and functional assignment of cloned sequences from high Andean forest soil metagenome. A van Leeuw J Microb 101: 205–215. [DOI] [PubMed] [Google Scholar]
- 24. Andreote FD, Jiménez DJ, Chaves D, Dias ACF, Luvizotto DM, et al. (2012) The Microbiome of Brazilian Mangrove Sediments as Revealed by Metagenomics. PLoS One 7 (6): e38600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 (17): 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, et al. (2008) The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9 (386): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gerlach W, Jünemann S, Tille F, Goesmann A, Stoye J (2009) WebCARMA: A web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics 10: 430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang Y, Li W, Finn PW, Perkins DL (2009) Ribosomal RNA identification in metagenomic and metatranscriptomic datasets. In: de Bruijn, FL (ed). Handbook of Molecular Microbial Ecology I: Metagenomics and Complementary Approaches, 784.
- 30. Liu Z, DeSantis TZ, Andersen GL, Knight R (2008) Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 36 (18): e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tatusov RL, Galperin MY, Natale DA, Koonin EV (2001) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucl Acids Res 28 (1): 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: D277–D280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17: 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.IDEAM (2005) Atlas de radiación solar en Colombia. Mapas de radiación ultravioleta banda 305nm. https://documentacion.ideam.gov.co/openbiblio/Bvirtual/019649/4-RadiacionUltravioleta.pdf. Accessed 16 April 2012.
- 35. Lawrence ME, Possingham JV (1986) Direct measurement of femtogram amounts of DNA in cells and chloroplasts by quantitative microspectrofluorometry. J Histochem Cytochem 34: 761. [DOI] [PubMed] [Google Scholar]
- 36. Silander K, Saarela J (2008) Whole genome amplification with Phi29 DNA polymerase to enable genetic or genomic analysis of samples of low DNA yield. Methods Mol Biol 439: 1–18. [DOI] [PubMed] [Google Scholar]
- 37. Hosono S, Faruqi AF, Dean FB, Du Y, Sun Z, Wu X, Du J, Kingsmore SF, Egholm M, Lasken RS (2003) Unbiased whole-genome amplification directly from clinical samples. Genome Re 13 (5): 954–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inskeep WP, Rusch DB, Jay ZJ, Herrgard MJ, Kozubal MA, et al. (2010) Metagenomes from high-temperature chemotrophic systems reveal geochemical controls on microbial community structure and function. PLoS One 5 (3): e9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, et al. (2006) Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yergeau E, Hogues H, Whyte LG, Greer CW (2010) The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J 4: 1206–1214. [DOI] [PubMed] [Google Scholar]
- 41. Wommack KE, Bhavsar J, Ravel J (2008) Metagenomics: Read length matters. Appl Environ Microbiol 74 (5): 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Snyder TM, Tse BN, Liu DR (2008) Effects of template sequence and secondary structure on DNA-templated reactivity. J Am Chem Soc 130: 1392–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yilmaz S, Allgaier M, Hugenholtz P (2010) Multiple displacement amplification compromises quantitative analysis of metagenomes. Nature Methods 7: 943–944. [DOI] [PubMed] [Google Scholar]
- 44. Fan L, McElroy K, Thomas T (2012) Reconstruction of Ribosomal RNA Genes from Metagenomic Data. PLoS One7 (6): e39948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mitra S, Stärk M, Huson DH (2011) Analysis of 16S rRNA environmental sequences using MEGAN. BMC Genomics 12 (Suppl 3)S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ghai R, Rodriguez-Valera F, McMahon KD, Toyama D, Rinke R, et al. (2011) Metagenomics of the water column in the pristine upper course of the Amazon River. PLoS One 6 (8): e23785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huson DH, Richter DC, Mitra S, Auch AF, Schuster SC (2009) Methods for comparative metagenomics. BMC Bioinformatics 10 Suppl 1S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bohnebeck U, Lombardot T, Kottmann R, Glöckner FO (2008) MetaMine – A tool to detect and analyses gene patterns in their environmental context. BMC Bioinformatics 9: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weingart U, Persi E, Gophna U, Horn D (2010) Deriving enzymatic and taxonomic signatures of metagenomes from short read data. BMC Bioinformatics 11: 390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gori F, Folino G, Jetten MSM, Marchiori E (2011) MTR: taxonomic annotation of short metagenomic reads using clustering at multiple taxonomic ranks. Bioinformatics 27 (2): 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Klatt CG, Wood JM, Rusch DB, Bateson MM, Hamamura N, et al. (2011) Community ecology of hot spring cyanobacterial mats: predominant populations and their functional potential. ISME J 5: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Elshahed MS, Senko JM, Najar FZ, Kenton SM, Roe BA, et al. (2003) Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl Environ Microbiol 69: 5609–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rowe OF, Sanchez-España J, Hallberg KB, Johnson DB (2007) Microbial communities and geochemical dynamics in an extremely acidic, metal-rich stream at an abandoned sulfide mine (Huelva, Spain) underpinned by two functional primary production systems. Environ Microbiol 9: 1761–1771. [DOI] [PubMed] [Google Scholar]
- 54.Dini-Andreote F, Andreote FD, Araújo WL, Trevors JT, van Elsas JD (2012) Bacterial genomes: Habitat specificity and uncharted organisms. Microb Ecol: In press. [DOI] [PMC free article] [PubMed]
- 55. Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M (2011) Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics 12 Suppl 2S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rzhepishevska OI, Valdés J, Marcinkeviciene L, Gallardo CA, Meskys R, et al. (2007) Regulation of a novel Acidithiobacillus caldus gene cluster involved in metabolism of reduced inorganic sulfur compounds. Appl Environ Microbiol 73 (22): 7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schrenk MO, Edwards KJ, Goodman RM, Hamers RJ, Banfield JF (1998) Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxidans: Implications for generation of acid mine drainage. Science 279 (5356): 1519–1522. [DOI] [PubMed] [Google Scholar]
- 58. Kurosawa H, Fujita M, Kobatake S, Kimura H, Ohshima M, et al. (2010) A case of Legionella pneumonia linked to a hot spring facility in Gunma Prefecture, Japan. Jpn J Infect Dis 63 (1): 78–79. [PubMed] [Google Scholar]
- 59. Furuhata K, Ogihara K, Ishizaki N, Oonaka K, Yoshida Y, et al. (2010) Identification of Legionella londiniensis isolated from hot spring water samples in Shizuoka, Japan, and cytotoxicity of isolates. J Infect Chemother 16 (5): 367–371. [DOI] [PubMed] [Google Scholar]
- 60. Hao C, Wang L, Gao Y, Zhang L, Dong H (2010) Microbial diversity in acid mine drainage of Xiang Mountain sulfide mine, Anhui Province, China. Extremophiles. 14 (5): 465–74. [DOI] [PubMed] [Google Scholar]
- 61. Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, et al. (2011) Microbial community structure across the tree of life in the extreme Río Tinto. ISME J 5 (1): 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Johnson DB, Hallberg KB (2009) Carbon, iron and sulfur metabolism in acidophilic micro-organisms. Adv Microb Physiol 54: 201–255. [DOI] [PubMed] [Google Scholar]
- 63.Hiraishi A, Imhoff JF (2005) Genus Acidiphilium. In: Boone, DR, Castenholz RW, Garrity GM (ed). Bergey's manual of systematic bacteriology, vol 2, 2nd edn. Springer, New York, 54–61.
- 64. Tekere M, Lötter A, Olivier J, Jonker N, Venter S (2011) Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo, South Africa. Afr. J. Biotechnol. 10 (78): 18005–18012. [Google Scholar]
- 65. Kozubal MA, Macur RE, Jay ZJ, Beam JP, Malfatti SA, et al. (2012) Microbial iron cycling in acidic geothermal springs of Yellowstone National Park: integrating molecular surveys, geochemical processes, and isolation of novel Fe-active microorganisms. Front. Microbio. 3: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Belnap CP, Pan C, Denef VJ, Samatova NF, Hettich RL, et al. (2011) Quantitative proteomic analyses of the response of acidophilic microbial communities to different pH conditions. ISME J 5: 1152–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brazelton WJ, Baross JA (2009) Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J 3 (12): 1420–1424. [DOI] [PubMed] [Google Scholar]
- 68. Rocak S, Linder P (2004) DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5 (3): 232–241. [DOI] [PubMed] [Google Scholar]
- 69. Dos Santos PC, Fang Z, Mason SW, Setubal JC, Dixon R (2012) Distribution of nitrogen fixation and nitrogenase-like sequences amongst microbial genomes. BMC Genomics 13: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ju X, Zhao L, Sun B (2008) Nitrogen fixation by reductively dechlorinating bacteria. Environ Microbiol 9 (4): 1078–1083. [DOI] [PubMed] [Google Scholar]
- 71. Meyer B, Kuever J (2008) Homology modeling of dissimilatory APS reductases (AprBA) of sulfur-33 oxidizing and sulfate-reducing prokaryotes. PLoS One 3 (1): e1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gomes J, Steiner W (2004) The biocatalytic potential of extremophiles and extremozymes. Food Technol Biotechnol 42 (4): 223–235. [Google Scholar]
- 73. Kumar L, Awasthi G, Singh B (2011) Extremophiles: a novel source of industrially important enzymes. Biotech 10 (2): 121–135. [Google Scholar]
- 74. McCready S, Müller JA, Boubriak I, Berquist BR, Ng WL, et al. (2005) UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Systems 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18 (1): 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vos M (2009) Why do bacteria engage in homologous recombination? Trends Microbiol 17 (6): 226–232. [DOI] [PubMed] [Google Scholar]
- 77. Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, et al. (2009) Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol 11 (6): 1358–1375. [DOI] [PubMed] [Google Scholar]
- 78. Shi Y, Tyson GW, Eppley JM, DeLong EF (2011) Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Liu Z, Klatt CG, Wood JM, Rusch DB, Ludwig M, et al. (2011) Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J 5: 1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, et al. (2008) Microbial Ecology of Four Coral Atolls in the Northern Line Islands. PLoS One 3 (2): e1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gilbert JA, Field D, Huang Y, Edwards R, Li W, et al. (2008) Detection of Large Numbers of Novel Sequences in the Metatranscriptomes of Complex Marine Microbial Communities. PLoS One 3 (8): e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. De Angelis KM, Gladden JM, Allgaier M, Dhaeseleer P, Fortney JL, et al. (2010) Strategies for enhancing the effectiveness of metagenomic based enzyme discovery in lignocellulolytic microbial communities. Bioenerg Res 3 (2): 146–158. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional assignment of metagenomic sequences using BLASTX against NCBI-nr. The data was analyzed using MEGAN v4.0 software and KEGG identifiers.
(PPT)
Taxonomic and functional clustering based on total reads of various metagenomes (RSM: Red Soudan Mine; BSM: Black Soudan Mine; POCR: Pacific Ocean (coral reefs); SW: Sea Water; PO: Pacific Ocean; TFS: Tropical Forest Soil; HAFS: High Andean Forest Soil; PMS: Pristine Mangrove Sediments; AHEC: Acidic Hot Spring EC). The data were compared to a) RefSeq and b) SEED databases. Dendrogram linkages are based on relative abundance of the phylum level (RefSeq) and relative abundance of the metabolic identifiers (Subsystems, SEED database) within the samples.
(PPT)
Partial a) nitrogen and b) sulfur pathways identified by KEGG affiliation of the sequences from EC hot spring. Boxes indicate the KEGG characteristic identified and numbers in gray circles indicate the amount of sequence reads affiliated to the KEGG function.
(PPT)
Phylogenetic affiliation of SSU rRNA sequences. The aligned reads were classified taxonomically using “classify a batch of sequences against multiple taxonomies tool” from Greengenes.
(XLS)
Metagenomic reads obtained from EC hot spring that mapped to the A. cryptum JF-5, L. pneumophila str. Corby and A. caldus draft genomes.
(XLS)